Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disability that demonstrates impaired social interactions, communication deficits, and restrictive and repetitive behaviors. ASD has a strong genetic basis and many ASD-associated genes have been discovered thus far. Our previous work has shown that loss of expression of the X-linked gene NEXMIF/KIDLIA is implicated in patients with autistic features and intellectual disability (ID).
Keywords: autism spectrum disorder, ASD, NEXMIF, brain development, synapse
Abstract
Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disability that demonstrates impaired social interactions, communication deficits, and restrictive and repetitive behaviors. ASD has a strong genetic basis and many ASD-associated genes have been discovered thus far. Our previous work has shown that loss of expression of the X-linked gene NEXMIF/KIDLIA is implicated in patients with autistic features and intellectual disability (ID). To further determine the causal role of the gene in the disorder, and to understand the cellular and molecular mechanisms underlying the pathology, we have generated a NEXMIF knock-out (KO) mouse. We find that male NEXMIF KO mice demonstrate reduced sociability and communication, elevated repetitive grooming behavior, and deficits in learning and memory. Loss of NEXMIF/KIDLIA expression results in a significant decrease in synapse density and synaptic protein expression. Consistently, male KO animals show aberrant synaptic function as measured by excitatory miniatures and postsynaptic currents in the hippocampus. These findings indicate that NEXMIF KO mice recapitulate the phenotypes of the human disorder. The NEXMIF KO mouse model will be a valuable tool for studying the complex mechanisms involved in ASD and for the development of novel therapeutics for this disorder.
SIGNIFICANCE STATEMENT Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disorder characterized by behavioral phenotypes. Based on our previous work, which indicated the loss of NEXMIF/KIDLIA was associated with ASD, we generated NEXMIF knock-out (KO) mice. The NEXMIF KO mice demonstrate autism-like behaviors including deficits in social interaction, increased repetitive self-grooming, and impairments in communication and in learning and memory. The KO neurons show reduced synapse density and a suppression in synaptic transmission, indicating a role for NEXMIF in regulating synapse development and function. The NEXMIF KO mouse faithfully recapitulates the human disorder, and thus serves as an animal model for future investigation of the NEXMIF-dependent neurodevelopmental disorders.
Introduction
Autism spectrum disorders (ASD), estimated to affect 1 in 59 individuals in the United States (Baio et al., 2018), is characterized by reduced social interactions, impaired language and communication, and stereotyped, repetitive behaviors (Lord et al., 1989). ASD often presents with other comorbid psychiatric and medical conditions such as intellectual disability (ID), epilepsy, gross and fine motor deficits, anxiety, gastrointestinal issues and attention-deficit/ hyperactivity disorder (ADHD) (Towbin et al., 2005; Zafeiriou et al., 2007; Reiersen and Todd, 2008; Storch et al., 2012; Tye et al., 2018).
ASD is among the most heritable neuropsychiatric disorders, and the genetic influences implicated in ASD are highly heterogeneous (Persico and Bourgeron, 2006; Pinto et al., 2010; Geschwind, 2011; Jeste and Geschwind, 2014; Krumm et al., 2015; Ziats and Rennert, 2016; Grove et al., 2019). Analysis of the Simons Foundation Autism Research Initiative (SFARI) gene database identifies a vast diversity of hundreds of ASD associated genes (Banerjee-Basu and Packer, 2010; Abrahams et al., 2013). Among these genes, a large number play roles in the development and function of synapses, such as neuroligin 3 (NLGN3), neuroligin 4X (NLGN4X), neurexin 1 (NRXN1), and SH3 and multiple ankyrin repeat domains 3 (SHANK3) (Jamain et al., 2003; Feng et al., 2006; Durand et al., 2007; Tabuchi et al., 2007). These and other reports show that synaptic dysregulation and imbalances in neuronal activity are among the most common cellular alterations in ASD (Zoghbi, 2003; Berkel et al., 2010; Pfeiffer et al., 2010; Bourgeron, 2015; Schwede et al., 2018; Bagni and Zukin, 2019; Romero-Garcia et al., 2019).
The NEXMIF gene (also known as KIDLIA, KIAA2022, or Xpn) is a novel gene, localized on the X chromosome at Xq13.2, with little known biological function. Previous studies have demonstrated that NEXMIF mRNA expression starts as early as E10.5, increases throughout development with a peak at P3, and continues at a lower level into adulthood (Cantagrel et al., 2009; Ishikawa et al., 2012). NEXMIF mRNA shows notably strong expression in the cortex, hippocampus, cerebellum, and olfactory bulb (Allen Institute for Brain Science, 2004; Cantagrel et al., 2009). At the protein level, NEXMIF is specifically distributed in the nucleus of postmitotic neurons but not in glia. Strong protein expression can be detected from E17 through adulthood in mice (Gilbert and Man, 2016), indicating a role for NEXMIF in brain development.
The NEXMIF gene was first implicated in ASD as one of the candidate genes in two male members of a family who had autistic phenotypes and ID (Cantagrel et al., 2004). Our earlier work in collaboration with colleagues investigated the pathogenic mutations of NEXMIF in patients with similar inherited phenotypes (Van Maldergem et al., 2013). These patients showed repetitive behaviors, impaired language, seizures, and ID, with several cases of microcephaly (Cantagrel et al., 2004, 2009; Van Maldergem et al., 2013). Multiple additional reports have confirmed the loss of NEXMIF by gene mutation or deletion in ASD patients (Charzewska et al., 2015; Kuroda et al., 2015; Farach and Northrup, 2016; Webster et al., 2017; de Lange et al., 2016; Lambert et al., 2018; Lorenzo et al., 2018; Alarcon-Martinez et al., 2019). NEXMIF has been listed as an ASD gene in the SFARI database. We have recently shown that NEXMIF knock-down in rat hippocampal neurons impairs neurite outgrowth via a disruption of N-cadherin/δ-catenin signaling and actin dynamics, and loss of NEXMIF caused defects in neuron migration in vivo (Van Maldergem et al., 2013; Gilbert and Man, 2016). However, to date, little is known regarding the function of NEXMIF gene, as well as the cellular and molecular mechanisms underlying the disorder caused by the loss of NEXMIF.
Here, we report the creation of the NEXMIF knock-out (KO) mouse. The NEXMIF KO mice showed autism-like behaviors including deficits in social behaviors, increased repetitive self-grooming, impaired communication, and deficiency in learning and memory. The KO phenotype recapitulated the symptoms reported in loss-of-NEXMIF human patients. Cultured hippocampal neurons with NEXMIF knock-down, as well as neurons from NEXMIF KO brains, showed a significant decrease in spine density with an increase in immature spines. Also, loss of NEXMIF resulted in a significant decrease in synaptic proteins such as AMPAR, PSD-95, and gephyrin. Consistently, electrophysiological recordings demonstrated suppressed synaptic transmission in both cultured neurons and the KO mouse brain. This study establishes the NEXMIF KO mouse as a new ASD model in which we provide the first evidence for synaptopathy as an underlying pathology for NEXMIF-dependent ASD.
Materials and Methods
Generation of NEXMIF KO mouse
C77370tm1 (KOMP) Wtsi (KIAA2022) KO mice were created by blastocyst injection of targeted embryonic stem (ES) cell clone EPD0411_7_H02 from the NIH-funded Knock-out Mouse Project (KOMP) (International Mouse Knockout Consortium et al., 2007; Pettitt et al., 2009). The ES cell clone was the result of deletion of exon 4 of the NEXMIF gene. A chimeric animal was generated through the Boston University Transgenic Core and mouse colonies were maintained in a C57BL/6J genetic background. Female mice heterozygous for NEXMIF were crossed with wild-type male mice and KO male mice were used in experiments. All WT (+/+) mice were randomized littermate controls of the KO mice. Transgenic mice were backcrossed to C57BL/6J mice >5 times before use.
Animal care and use
All the procedures involving animal use were in compliance with the policies of the Institutional Animal Care and Use Committee (IACUC) at Boston University. Mouse colonies were maintained in the Laboratory Animal Care Facility (LACF) at Boston University, Charles River Campus. Mice aged 8–12 weeks were used in all behavioral studies to avoid variability due to changes during adolescent periods. Male mice were used for all experiments because NEXMIF is located on the X chromosome and loss of NEXMIF function was identified as an X-linked disorder in male patients.
Genotyping
A single tail snip was collected from mice at the time of weaning (postnatal day 21. [P21]) and placed into an Eppendorf tube. Then, 75 μl of a base solution (25 mm NaOH, 0.2 m EDTA) was added to each sample and the tubes were incubated at 95°C for 30 min to extract genomic DNA. The tubes were allowed to cool to room temperature and 75 μl of a neutralization solution (40 mm Tris-HCl, pH 5) was added and the mixture was vortexed. Touchdown PCR was performed using 2 μl of the DNA extract with a 2720 Thermo Cycler (Applied Biosystems) with the following conditions: initial hold at 94°C for 5 min, followed by 40 extension cycles of a 15 s denaturing step at 94°C, with 10 cycles with an annealing step for 60 s annealing from 65−55°C (1°C stepdown/ cycle) followed by 30 cycles with a 60 s annealing step at 55°C, and an extension step of 40 s at 72°C, followed by a final extension step at 72°C for 5 min. PCR fragments were run on a 1% agarose gel with ethidium bromide to label DNA and visualized under ultraviolet light.
Two sets of primers were used to genotype NEXMIF KO animals. One set was targeted against the deleted exon 4 region using the primers: 5′-AGGACTTGCTTAGGTTGCTTCATGGAA-3′ and 5′-CTTAAATTGCTCTACCTCAAGACCACCA-3′ with an expected PCR fragment of 949 bp. The other set was targeted against the KO cassette using the primers: 5′-ACACCTCCCCCTGAACCTGAAA-3′ and 5′-TGGGCAAACAGTGCATAAACTACTCG-3′ with an expected PCR fragment of 1154 bp.
Three-chamber social test
A three-chambered box was constructed from 0.75 in thick white plastic board measuring 65 × 28 × 28 cm. The walls to the center chamber had 4 × 4 in cut-out doors allowing movement between chambers. The two side chambers contained small wire cages to later house social mice. Before the test, mice were habituated to the apparatus with empty cages in both side chambers, over a 3 d period for 5 m each session, and allowed to move freely between all three chambers. On the testing day, mice were singly placed into the center chamber, with the doors blocked with white plastic boards and a social mouse (Mouse 1) was placed into either of the side chambers under the wire cage. The doors were unblocked and the mouse was allowed to move freely within the apparatus for 5 min. The test mouse was then returned to the center chamber, the doors were blocked again and a second mouse (Novel Mouse) was placed into the other side chamber. The center doors were unblocked and the test mouse was allowed to move freely within the apparatus for another 5 min. The entire apparatus was wiped with 70% ethanol between mice to eliminate odor cues between animals. Video recordings were captured with a Logitech c920 webcam during the test and the time spent interacting with each mouse or empty cage (nose ≤ 2 cm), time spent in each chamber, and locomotion tracks were scored using the MTrack plugin in ImageJ, blinded to the genotype.
Marble-burying test
Marble burying was conducted in the animal's home cage with a 3 in thick layer of fresh pine chip bedding. Sixteen shiny glass marbles (0.25 in diameter) were arranged in a 4 × 4 grid on top of the bedding. Mice were singly placed back into the cage and allowed to move freely and bury marbles. Video recordings were captured with a Logitech c920 webcam during the test and the number of marbles buried after 5, 10, 15 and 25 min was quantified.
Open-field test
The open-field test was conducted in a chamber constructed from 0.75-inch-thick white plastic board measuring 28 × 28 × 28 cm. The box was placed on a large white plastic board with a 9 × 9 grid drawn on the floor for later analysis. Lights in the testing room were off except for a small desk lamp in the corner allowing the experimenter to see. Mice were habituated to the testing room over 3 d and handled for 5 min each session. On the test day, each mouse was singly placed into the center of the box and allowed to freely explore the chamber. Video recordings were captured with a Logitech c920 webcam during the test and the time spent in the center square of the grid, within 3 cm of the walls of the chamber, track lengths and grooming behavior were quantified blinded to the genotype of the animals.
Barnes maze spatial memory test
The Barnes maze was constructed from a circular, 0.75-inch-thick, white plastic board with a 48 inch diameter. Twenty holes with a diameter of 2 inches were evenly spaced around the perimeter of the maze with a distance of 1 inch to the edge. The maze was mounted on a pedestal that was 30 inches above the ground and could rotate at its center. The escape cage was made from a black plastic box with a ramp connected beneath the escape hole for easy access. Four bright ceiling lamps and an alarm were used as aversive stimuli during the test. The maze and escape cage were thoroughly cleaned with 70% ethanol between testing sessions to avoid any olfactory cues and the maze was rotated randomly after every mouse to avoid intramaze odor or visual cues.
During the test, the mice were habituated to the maze on day 1. For each session, one mouse was placed in the center of the maze, covered with an opaque cardboard chamber for 15 s then slowly guided to the escape hole with a 3 L glass beaker. Each mouse was given 3 min to enter the escape hole on its own if not, the mouse was nudged gently into the hole or placed directly into the hole. Afterward, the mouse was allowed to stay in the escape hole for 2 min. The ceiling lights and white noise remained on while the mouse was exploring the maze and turned off immediately after they entered the escape hole. This procedure was repeated for all mice. On days 2–5, the mice were trained to ensure a strong memory for the escape location (4 times/mouse/d, 16 times in total) and to learn to enter the escape hole by themselves. During these trials, the mice were allowed to stay in the escape cage for 1 min during each training trial. Memory of the escape hole location was probed 1 and 5 d later. During the probe sessions, the escape hole was covered and spatial memory for the subject mice was tested. Video recordings were captured with a Logitech c920 webcam. The primary latency, total latency, correct/error nose pokes, and traces of locomotion were scored in ImageJ using the plugin MTrack blinded to the animal's identity.
Ultrasonic vocalization (USV) recording
To record USVs, pups were separated from the dam and littermates and isolated in a recording chamber. Mouse vocalizations were recorded every 2 d (P5, P7, P9) in a random order for each litter using a CM16/CMPA microphone (Avisoft Bioacoustics) placed 15 cm above the pups. The recording chamber was cleaned with 70% ethanol and dried between each pup. Audio recordings were done at a sampling rate of 300 kHz in 16-bit format. The microphone was connected to a preamplifer UltraSoundGate 116Hb (Avisoft Bioacoustics) and digitized sonograms were stored in a computer. Recordings were analyzed using SASLab Pro 5.2.12 (Avisoft Bioacoustics). Spectrograms were generated using fast Fourier transform (256 FFT length, 100% frame, FlatTop window, and 50% window overlap) and a high-pass filter was applied to eliminate background noise <30 kHz. USVs were detected automatically (threshold: −47 ± 10 dB, hold time: 7 ms) followed by manual inspection to ensure all detected calls were legitimate. Several acoustic parameters of calls were measured including number of calls, mean call duration, total time spent calling, peak frequency, and peak amplitude. Calls were classified based on their acoustic features into the following groups; simple: short, flat, upward, downward, chevron, U shape, and complex: modulated, frequency jump, multiple jumps, harmonic.
Fear-conditioning test
The fear-conditioning tests consisted of two parts: contextual- and cue-dependent fear memory. Before the test, mice were habituated in a separate cage with a similar context as the testing cage (bars on the floor, white and smooth walls, pinewood bedding with an 8% acetic acid odor, lights on) for three continuous days (5 min/d). On experimental day 1, for each trial, one mouse was placed into the testing cage and allowed to explore the cage freely for 3 min, followed by a 30 s tone (80 db) and an electrical foot shock (2 s, 0.6 mV) at the end of the tone. The tone and foot shock were repeated three times in each trial with three 1 min intervals. The procedure was repeated for every mouse in the experiment. The whole cage was thoroughly cleaned with 70% ethanol and 8% acetic acid after every mouse to avoid potential olfactory cues from other mice. On experimental day 2, the mice were placed back into the same cage with exactly the same environment and acetic acid odor to test their contextual fear memory without any foot shock. The mice were allowed to explore the cage for 5 min. On experimental day 3, the context of the cage was completely changed (smooth white floor, back wall decorated with dots, corn bedding with 20% vanilla odor, lights off) and the mice were allowed to explore the cage freely for 3 min followed by another 2 min with the same tone from day 1 played. The motion index and freezing percentages of the mice were recorded and analyzed by Video Freeze software (Med Associates).
Immunocytochemistry of cultured neurons
Hippocampal neurons were washed twice in ice-cold ACSF and fixed for 10 min in a 4% paraformaldehyde/4% sucrose solution at room temperature. Cell membranes were permeabilized for 10 min in 0.3% Triton X-100 (Fisher Biotec) in PBS, rinsed three times in PBS, then subjected to a blocking procedure (5% goat serum in PBS) for 1 h. After blocking, cells were incubated with primary antibodies (in 5% goat serum PBS) for 2 h at room temperature, washed and incubated with Alexa Fluor-conjugated fluorescent secondary antibodies (1:700, Life Technologies) for an additional hour. Cells were then mounted to microscopy glass slides with Prolong Gold anti-fade mounting reagent (Life Technologies) for subsequent visualization.
Immunohistochemistry of brain slices
Mice older than P0 underwent transcardial perfusion with ice-cold PBS before removing the brain from the skull. For P0 animals, brains were left in the skull for fixation and slicing.
Brains were fixed in 4% paraformaldehyde for 4–6 h and cryoprotected in 30% sucrose/PBS. The tissue was then frozen in OCT (Tissue-Tek) and cut into 20 μm sections using a CM1850 cyrostat (LEICA Biosystems). Sections were mounted onto SuperFrost microscope slides (Fisher Scientific) and stored at −80°C for future use. Before immunostaining, sections were rinsed with PBS for a minimum of 2 h and antigen retrieval was performed by microwaving brain sections in sodium citrate buffer (10 mm, pH 6) at 800 W for 1 min followed by 80 W for 10 min. Sections were then blocked in 5% goat serum, 0.3% Triton X-100/PBS for 1 h followed by incubation with primary antibodies overnight at 4°C. The following day, sections were washed with PBS and incubated with the appropriate Alexa Fluor-conjugated secondary antibodies. Brain slices were then washed three times with PBS, with the first wash containing Hoescht (1:3000, Thermo Fisher Scientific). Sections were then mounted with ProLong-Gold mounting medium (Invitrogen), allowed to dry at room temperature overnight, and stored at −20°C.
Golgi impregnation
Golgi impregnation of whole brains from C57BL/6J wild-type and NEXMIF KO mice was performed with the FD Rapid GolgiStainTM Kit (FD NeuroTechnologies), following the manufacturer's protocol. Mice were killed at the indicated time in a 4% CO2 chamber for 5 min and the brains were immediately removed and rinsed in MilliQ water. Brains were then immersed in a Golgi–Cox solution containing potassium dichromate, mercuric chloride, and potassium chromate. The mixture of solutions was replaced once after 16 h of initial immersion, then stored at room temperature in the dark for 2 weeks. After the immersion period in the Golgi-Cox solution, the embedded brains were transferred to a cryoprotectant solution and stored at 4°C for at least 1 week in the dark before cutting. The brain slices were sectioned in the coronal plane at 100–200 μm thickness on a microtome. Brain slices were transferred onto gelatin-coated slides and were air dried at room temperature in the dark overnight before further processing. After drying, sections were rinsed with distilled water and stained in a developing solution and subsequently dehydrated with 50%, 75%, 95%, and 100% ethanol. Finally, the sections were defatted in xylene substitute and mounted onto coverslips with Permount mounting liquid (Fisher Scientific). Images were acquired with a Carl Zeiss Axiovert 200M microscope and AxioVision software (Release 4.5). Each neuron was scanned under high magnification (63×, oil-immersion, numerical aperture 1.4) by varying the depth of the Z plane, to ensure that all parts of the cell (especially dendrites) were intact. The number and size of apical and basal spines and the length of dendrites on hippocampal CA1 pyramidal neurons was measured blinded to the genotype with ImageJ software.
Primary neuronal culture
Hippocampal and cortical brain tissue were dissected out of E18 rat embryo brains of either sex and used for primary culture. Tissues were digested in papain (0.5 mg/ml in Hanks balanced salt solution, Sigma-Aldrich; catalog #4762) for 15 min at 37°C, then triturated in a trituration buffer [0.1% DNase (catalog #PA5–22017 RRID:AB_11153259), 1% ovomucoid (Sigma-Aldrich; catalog #T2011)/1% bovine serum albumin (Sigma-Aldrich; catalog #05470) in DMEM] to fully dissociate neurons. Dissociated neurons were counted and plated on 18 mm circular coverslips (Carolina Biological Supply; catalog #633013, no. 0) in 60 mm Petri dishes (five coverslips/dish) that had been coated in poly-l-lysine (Sigma-Aldrich; catalog #P2636; 100 μg/ml in borate buffer) overnight at 37°C then washed three times with sterile deionized water and left in plating medium [minimal essential medium (500 ml) containing 10% fetal bovine serum (Atlanta Biologicals; catalog #S11550), 5% horse serum (Atlanta Biologicals; catalog #S12150), 31 mg of l-cysteine, 1% penicillin/streptomycin (Corning; catalog #30–002-Cl), and l-glutamine (Corning; catalog #25–005-Cl) before cell plating. The day after plating, plating medium was replaced by feeding medium (neurobasal medium supplemented with 1% horse serum, 2% B-27, and 1% penicillin/streptomycin and l-glutamine) which was supplemented with 5′-fluoro-2′-deoxyuridine (10 μm; Sigma-Aldrich; catalog #F0503) after 5 d in vitro to suppress glial growth.
Viral infection
Two recombinant lentiviruses for NEXMIF shRNA and a scrambled shRNA sequence were designed using the small interfering RNA (siRNA) Wizard v3.1 (Invivogen) and cloned into the pLKO.1-TRC cloning vector (Addgene; catalog #10878, RRID:SCR_005907) using AgeI and EcoRI sites. They were produced by transfecting HEK293T cells with plasmids for the shRNA constructs with viral packaging and envelope proteins (pRSV/REV, pMDLg/RRE, and pVSVG) using polyethylenimine reagent (Polysciences; catalog #23966). Conditioned medium containing lentivirus was harvested after 48 h, centrifuged at 1000× g for 10 min, filtered through a 0.45 μm filter, and stored at −80°C. Neurons were infected with lentivirus on the day of plating, and medium was changed 1 d later to feeding medium.
Microscopy
For in vitro analysis of neuronal morphology, an inverted fluorescent microscope (Carl Zeiss Axiovert) was used to collect images with a 40× oil-immersion objective (numerical aperture, 1.3) and collected with AxioVision Release 4.5 software. Neuron images were quantified in using ImageJ software (see below).
Brain sections were imaged using a Zeiss LSM 700 laser scanning confocal microscope using the Zen software package.
Image analysis
Mounted coverslips were kept overnight in the dark at room temperature before imaging with a 63× oil-immersion objective. Images were collected with Axiovision 4.5 software and exposure time for the fluorescence signal was adjusted manually so the signals were within a full dynamic range. Either the glow scale look-up table or the histogram was used to monitor the saturation level. Once the parameters were set, they were fixed and used throughout the experiment. For accurate quantification, all images were collected in 8-bit grayscale and saved for raw data analysis with ImageJ software. Synaptic puncta size was calculated as the product of size and fluorescence intensity of each puncta. Colocalization of presynaptic and postsynaptic fluorescent puncta was assessed using the “puncta analyzer” plugin.
Cell count analysis
For cell counts in cortex and hippocampus, brain slice images of nuclei stained with Hoescht were uploaded to ImageJ software and converted to 8-bit greyscale. Each image was then manually thresholded to highlight all nuclei and the images were converted to a binary black and white file. To separate nuclei that merged together during the thresholding process, the watershed tool was used followed by manual inspection to discriminate individual nuclei. Cells were then counted using the “analyze particles” function with parameters set to include particles between 5 and 20 μm in size.
Western blot
Cortical neurons cultured in either 60 mm (3 × 106 cells/dish) or 6-well plates (106 cells/well) were treated with virus at the time of plating and any other drugs as stated. Neurons were lysed in Laemmli 2X sample buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 m Tris HCl) and boiled for 10 min at 95°C for SDS-PAGE.
Brains were dissected on ice immediately after sacrificing animals at the appropriate time points. For a ∼5 mg piece of tissue, ∼300 μl of ice-cold lysis buffer (50 mm Tris-Cl, 150 mm NaCl, 1% NP40, 1% sodium deoxycholate (SDOC), 1% SDS) supplemented with a protease inhibitor mixture complete tablets (Roche) was added and samples were homogeneneized mechanically with a pestle, followed by sonication (10× for 10 s each). Samples were then head-to-toe rotated for 2 h at 4°C. Brain lysates were then centrifuged for 20 min at 13,000 rpm at 4°C in a microcentrifuge. The tubes were placed on ice and the supernatant was carefully aspirated and placed into a fresh tube kept on ice. Samples were subjected to a BCA assay according to the manufacturer's protocol (Pierce) to determine protein concentrations. Protein levels were normalized with RIPA lysis buffer cells, an equal volume of 2× Laemmli sample buffer and the lysates were boiled for 10 min at 95°C to prepare for SDS-PAGE.
SDS-PAGE was performed to separate proteins of interest using standard procedures. Proteins were transferred to PVDF membranes and blocked for 1 h in 5% milk prepared in PBS. Following block, membranes were probed with the appropriate primary antibody overnight at 4°C in Tris buffered saline supplemented with 0.05% Tween (TBST). Membranes were washed 3X in TBST and then incubated with the appropriate secondary antibody for 1 h. After secondary incubation, membranes were washed three times with TBST, followed by one time with PBS to remove excess Tween. Blots visualized using a chemiluminescence detection system (GE Healthcare) and exposed to Fuji medical X-ray films (Fisher Scientific), scanned, and analyzed using ImageJ.
Patch-clamp recordings in cultured neurons
For in vitro mEPSC recordings, rat hippocampal neurons cultured on coverslips were recorded at 14–16 d in vitro (DIV). The coverslip was transferred to a recording chamber with an extracellular solution containing the following (in mm): 140 NaCl, 3 KCl, 1.5 MgCl2, 2.5 CaCl2, 11 glucose, and 10 HEPES (305 mOsm, pH 7.4,), which was supplemented with TTX (1 μm) to block action potentials, APV (50 μm) to block NMDARs, and bicuculline (20 μm) to block GABAA receptor-mediated currents. Whole-cell voltage-clamp recordings were made with patch pipettes filled with intracellular solution containing the following (in mm): 110 Cs-methanesulfonate, 10 CsCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.3 Na2-GTP, and 10 sodium phosphocreatine (295 mOsm, pH 7.4), with the membrane potential clamped at −70 mV. Recordings started 5 min after establishing whole-cell configuration to ensure equilibration between the pipette solution and the cytosol. mEPSCs were recorded with an Axopatch 200B amplifier and displayed and recorded digitally on a computer for subsequent off-line analysis with Clampfit (Molecular Devices). For cumulative probabilities, the relationship between control and experimental amplitude and interevent interval distributions were determined by ranking an equal number of randomly selected values from all cells recorded from each condition in ascending order. These amplitude distributions were linearly interpolated to plot each condition.
Extracellular field potential recordings on hippocampal brain slices
Hippocampal slices (300 μm) were prepared from 15- to 17-week-old animals. The experimenter was blinded to mouse genotype until all data were collected and analysis began. Mice were anesthetized with isoflurane and decapitated after disappearance of the righting reflex. The brain was rapidly removed and submerged for 1 min in ice-cold dissection buffer containing the following (in mm): 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 7 MgCl2·6H2O, 1 CaCl2·2H2O, 119 choline chloride, 30 glucose, 3 sodium pyruvate, 1 kynurenic acid, and 1.3 ascorbic acid. The brain was quickly dissected and hippocampi were removed. Hippocampus slices were made using a vibratome (Microtome HM 650 V or Leica VT1200S) in ice-cold dissection buffer. Slices were transferred to a reservoir filled with ACSF containing the following (in mm): 124 NaCl, 26 NaHCO3, 3.3 KCl, 1.3 NaH2PO4, 4 MgCl2·6H2O, 4 CaCl2·2H2O, and 11 glucose. Slices were allowed to recover for 1 h at 35°C and then remained at room temperature. The ACSF and dissection buffer were equilibrated with 95% O2 and 5% CO2.
For recording, slices were transferred to a submerged recording chamber, maintained at 29°C, and perfused continuously with ACSF containing picrotoxin (50 μm) at a rate of 2 ml/min. Field potentials were recorded with extracellular recording electrodes filled with ACSF and placed in stratum radiatum of area CA1. Field potentials were evoked by monophasic stimulation (duration, 100 μs) of Schaffer-collateral/commissural afferents with an extracellular stimulating pipette fabricated from borosilicate theta glass (World Precision Instruments) and filled with ACSF. To generate input/output curves, five test stimuli of different intensities were chosen to elicit field EPSPs (fEPSPs) and five sweeps at each stimulus intensity were used to calculate the average fEPSP slope and fiber volley amplitude for that stimulus intensity. For paired-pulse ratio experiments, test stimuli that evoked fEPSPs at 40–50% of the maximal fEPSP slope were used to test paired-pulse facilitation using interstimulus intervals of 25, 50, 100, 200, and 400 ms. Five sweeps at each interstimulus interval were used to calculate an average. Data were collected using a MultiClamp 700B amplifier (Molecular Devices) digitized at 10 kHz with the analog-to-digital converter ITC-18 computer interface (Heka Instruments). Data were acquired and analyzed on-line using custom routines written (by Richard Gerkin, PhD) with Igor Pro software (Wavemetrics). Using custom written algorithms, the fEPSP slope was measured by assessing 10–90% of the total fEPSP slope, and the fEPSP amplitude and fiber volley were measured by assessing the deflection from the baseline following the test stimulus.
Statistical analysis
Data from multiple trials were averaged to obtain the mean for each experiment. Multiple means of the same conditions were averaged to obtain the SEM, as indicated by the error bars in histograms. Statistical analysis for parametric data was performed using the two-population student's t test and ANOVA with Tukey's post test. Kolmogorov–Smirnov test was used to analyze cumulative probabilities of mEPSC frequency and amplitude. All statistical analyses were performed using GraphPad Prism 6 software. p-values ≤ 0.05 were considered statistically significant. All post hoc power values are shown in Fig. 1-1.
Results
Generation of a NEXMIF KO mouse
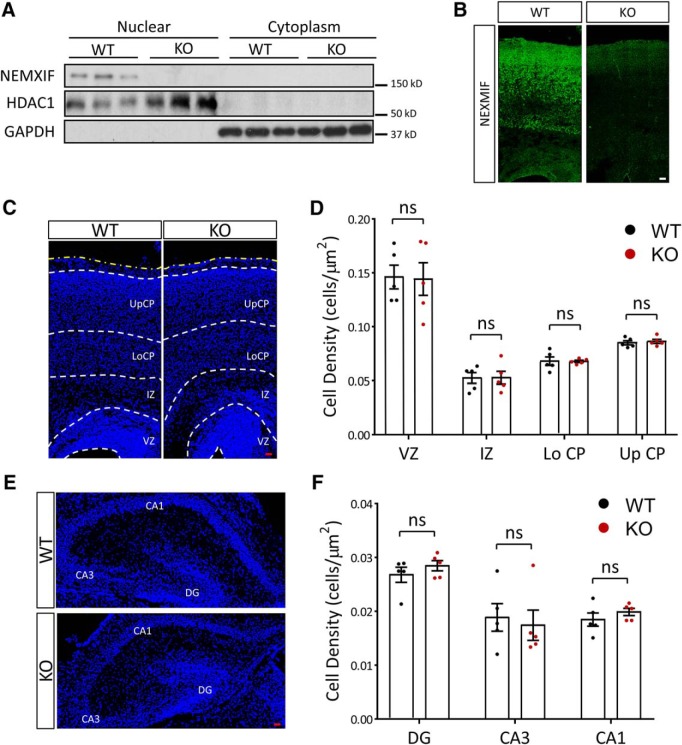
Recently we have shown impaired dendritic outgrowth and actin dynamics after shRNA-mediated knock-down of NEXMIF in rat neuronal cultures (Gilbert and Man, 2016). To extend our analyses on the function of NEXMIF in neurodevelopment and brain function, we generated a transgenic mouse with complete KO of NEXMIF (NEXMIF KO). Mice were generated with blastocyst injection of KIAA2022 KO embryonic stem (ES) cells purchased from the KO mouse project (KOMP, clone EPD0411_7_H02) and maintained in a C57BL/6J genetic background. Complete KO of the NEXMIF gene was not embryonically lethal. Because the NEXMIF gene is on the X-chromosome, heterozygous female breeders (X−X+) were mated to wild-type (WT) males (X+Y) to produce KO male mice (X−Y) and randomized WT littermates, which were used as controls. Loss of the NEXMIF protein product was confirmed in Western blot analyses with expression only detected in the nuclear fraction of WT mouse cortical lysates but not in cytoplasmic or KO animal lysates (Fig. 1A) and in immunohistochemistry (IHC) (Fig. 1B), with only minimal background staining observed in brain slices from KO animals. As we've reported previously, NEXMIF expression was only observed in the nuclear fraction of cell lysates from WT animals and was found to colocalize with the nuclear marker Hoechst in IHC (Gilbert and Man, 2016).
Figure 1.
Molecular characterization of the NEXMIF KO mouse. A, NEXMIF expression was observed only in the nuclear fraction of wild-type mouse cortical lysates but not in cytoplasmic or KO animal lysates. B, Immunohistochemistry in P0 mouse cortical brain slices shows NEXMIF signal in the cortical plate from WT animals, with only background signal in brain slices from KO animals. Scale bar, 20 μm. C–F, Cell density in cortex (C,D) or in hippocampus (E,F) was similar in KO and WT animals at P0. UpCP, Upper cortical plate; LoCp, lower cortical plate; IZ, intermediate zone; VZ, ventricular zone; CA1, cornu ammonis area 1; CA3, cornu ammonis area 3; DG, dentate gyrus. The yellow dashed line indicates the pia. Scale bars, 20 μm. Data are represented as average ± SEM. ns, Not significant. For post hoc power analyses, see Figure 1-1.
Post hoc power analysis. Two-sample, continuous endpoint post hoc power analyses were done with the assumption of equal variance and using a Type I error (alpha) of 0.05. All values are reported as % power. Download Figure 1-1, DOCX file (15.8KB, docx)
NEXMIF KO animals displayed good general health and normal food consumption, however animals began to exhibit seizures around 3 months of age that were sometimes fatal at older ages. There was no difference in total body weight between WT and NEXMIF KO animals at P0 (data not shown, WT: 1.71 ± 0.06 g, n = 11; KO: 1.68 ± 0.04 g, n = 12, p = 0.68). We did not detect major alterations in overall brain structure, cortical layer distribution, and cell density in neocortex or hippocampus at P0 (VZ: t(8) = 0.10, p = 0.92; IZ: t(8) = 0.03, p = 0.98; Lo CP: t(8) = 0.06, p = 0.95; Up CP: t(8) = 0.48, p = 0.65; DG: t(8) = 0.99, p = 0.35; CA3: t(8) = 0.38, p = 0.71; CA1: t(8) = 1.01, p = 0.34) (Fig. 1C–F, Fig. 1-1).
The NEXMIF KO mouse shows typical autistic features in social behavior
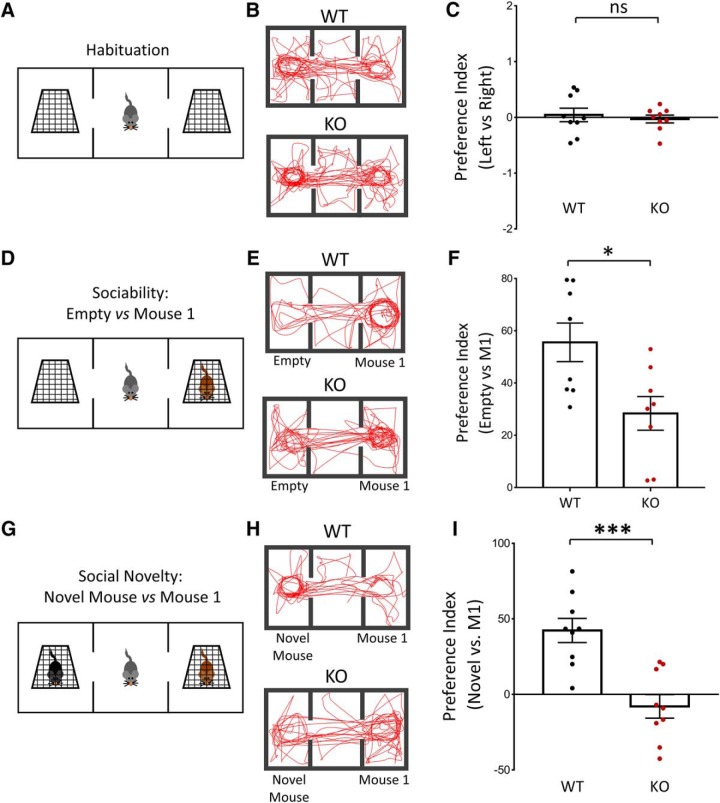
Loss of NEXMIF expression in humans produces behavioral deficits typical of ASD, including repetitive behavior, decreased social behavior and ID (Cantagrel et al., 2004, 2009; Van Maldergem et al., 2013; Kuroda et al., 2015; Farach and Northrup, 2016; Webster et al., 2017; de Lange et al., 2016). We therefore first sought to characterize the social behavior of the NEXMIF KO mouse in the three-chamber social task, a test broadly used for assessing mouse sociability and interest in social novelty (Crawley, 2007; Moy et al., 2009). Animals were habituated to the apparatus by allowing the animals to explore all three chambers, including the empty cages, before testing (Left vs Right: WT: t(32) = 0.61, p = 0.55; KO: t(32) = 0.29, p = 0.77) (Fig. 2A–C). To examine sociability, first a stranger mouse (Mouse 1) was placed into one of the chambers and the test mouse was allowed to freely navigate within the three-chamber apparatus (Fig. 2D). Representative track lengths during the test show that NEXMIF KO mice spent significantly less time interacting with Mouse 1 compared with the empty cage (Fig. 2E). A significant decrease in the preference index for Mouse 1 was observed, indicating an impairment in sociability (WT = 55.57 ± 7.37, n = 8; KO = 28.38 ± 6.46, n = 8; t(6) = 2.71, p = 0.035) (Fig. 2F). To examine the NEXMIF KO animal's interest in social novelty, a second mouse (Novel Mouse) was placed into the empty cage after the test mouse had been allowed to interact with Mouse 1 (Fig. 2G). Strikingly, NEXMIF KO animals showed no preference for the Novel Mouse as shown in the representative track lengths (Fig. 2H) with a preference index significantly smaller compared with the WT animals, indicating a lack of interest in social novelty (WT = 42.30 ± 8.00, n = 9; KO = −7.93 ± 7.81, n = 9; t(8) = 3.84, p = 0.005) (Fig. 2I).
Figure 2.
NEXMIF KO mice show impaired social behavior in the three-chamber social test. A, Habituation to the three-chamber apparatus. Mice were released from the center chamber, with empty cages in the adjacent chambers, and allowed to move freely within the apparatus for 5 min before beginning the sociability and social novelty tests. B, Traces of animal track paths in the habituation phase for WT and KO mice. C, Neither WT or KO animals showed any preference for either side chamber during habituation. D, Paradigm for the sociability test. An unfamiliar mouse (Mouse 1) was placed into either of the side chambers and the test mouse was allowed to move freely within the apparatus. E, Traces of animal track paths in the sociability test for WT and KO mice. F, Quantification of the preference index (see methods) showed a decrease in preference for Mouse 1 in NEXMIF KO animals compared with WT controls. G, Paradigm for the social novelty test. A second mouse (Novel Mouse) was placed into the remaining empty chamber opposite to Mouse 1, and the test mouse was allowed to interact with both mice. H, Traces of animal track paths in the social novelty test for WT and KO mice. I, In the social novelty test, KO animals showed no preference for the novel animal. Data are represented as average ± SEM. *p < 0.05; ***p < 0.001. ns, Not significant.
NEXMIF KO animals show increased repetitive behavior and hyperactivity
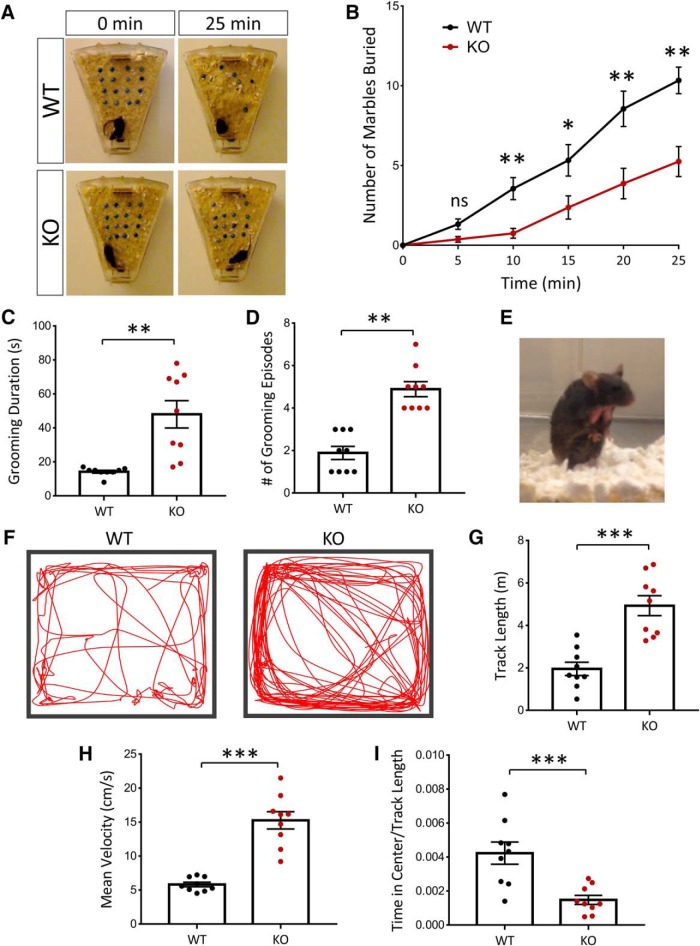
To further examine behavioral phenotypes in the NEXMIF KO animals, we used the marble burying task. WT mice placed in a cage with fresh bedding and marbles will spontaneously bury the marbles and perform digging behaviors. We found that adult NEXMIF KO mice buried significantly fewer marbles than WT mice across all time points observed during a 25 min observation period (Fig. 3A,B). Previous studies have indicated that marble burying depends on interest in the external environment and burying behavior does not change with increased experience to the marbles or habituation to the marble-burying test (Thomas et al., 2009; Greco et al., 2013). These findings therefore suggest that loss of NEXMIF expression dramatically decreases the animal's interest in the external environment, a phenotype consistent with ASD.
Figure 3.
NEXMIF KO mice show decreased interest in the external environment and increased repetitive grooming behavior and hyperactivity. A, B, In the marble-burying test, KO animals buried significantly fewer marbles, indicating a decreased interest in the environment. C, D, NEXMIF KO mice showed increased time spent grooming and an increase in grooming episodes. E, Example of the resulting fur loss due to excessive grooming behavior in the KO animals. F, Track paths of WT and KO animals in the open-field test. G–I, KO animals showed increased track lengths, increased mean velocities and decreased relative time in center in the open-field test. Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. ns, Not significant.
To assess grooming behavior, mice were observed in their home cages and the amount of time and number of grooming episodes were quantified. In contrast to a decrease in marble-burying behavior, NEXMIF KO animals showed ∼3× increase in the time spent self-grooming (WT = 14.2 ± 0.85 s, n = 9; KO = 48.0 ± 8.03 s, n = 9; t(16) = 4.18, p < 0.0001) (Fig. 3C) and the number of grooming episodes (WT = 1.88 ± 0.31, n = 9; KO = 4.89 ± 0.35, n = 9; t(16) = 6.41, p < 0.0001) (Fig. 3D). Indeed, due to over self-grooming, we observed that NEXMIF KO animals developed patches of fur loss due to excessive repetitive grooming behaviors in the same body areas (Fig. 3E). Mice were group housed with WT control littermates and only NEXMIF KO animals displayed fur loss on the chest, suggesting it was not the result of barbering.
We next assessed WT and NEXMIF KO animals in the open-field test. The KO animals showed no sign of locomotion deficits, but had a significant increase in their amount of movement during the test as shown in the representative tracks (Fig. 3F). Quantification of the total track length showed that NEXMIF KO animals traveled a larger distance during the test (WT = 1.96 ± 0.31 m, n = 9; KO = 4.93 ± 0.47 m, n = 9; t(16) = 5.27, p < 0.0001) (Fig. 3G) and KO animals showed a significant increase in average velocity (WT = 5.83 ± 0.34 cm/s, n = 9; KO = 15.26 ± 1.27, n = 9 cm/s; t(16) = 7.2, p < 0.0001) (Fig. 3H). Furthermore, quantification of the time spent in the center of the open field normalized to the total distance traveled showed that NEXMIF KO animals spent significantly less time in the center (WT = 1.00 ± 0.15, n = 9; KO = 0.35 ± 0.06, n = 9; t(16) = 3.91, p = 0.0012) (Fig. 3I). Together, these results indicate that NEXMIF KO animals were much more mobile with tendencies of center avoidance in the open field, indicative of a phenotype of increased anxiety and/or hyperactivity.
Impairments in communication by ultrasonic vocalization in NEXMIF KO mice
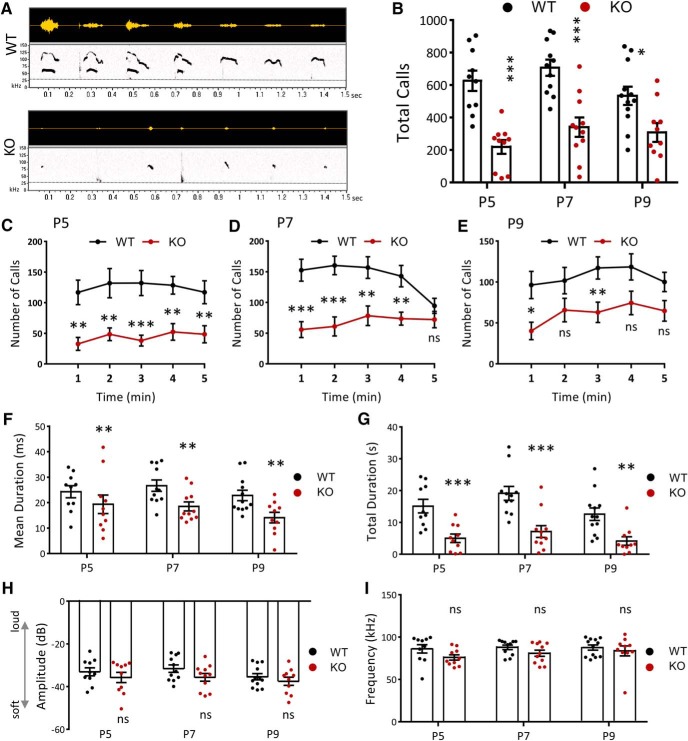
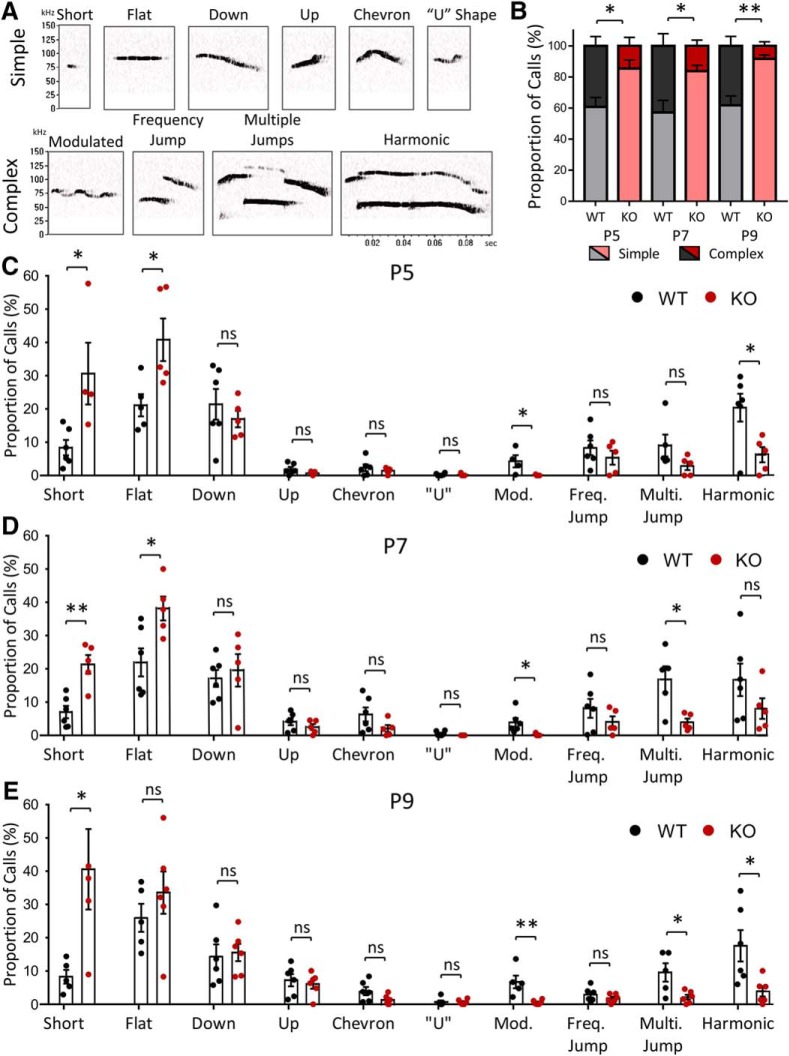
Impairments in communication are a characteristic ASD behavioral deficit. To examine the communication behavior of NEXMIF KO animals, we recorded and analyzed their USVs. Pups isolated from the mother and littermates at P5, P7, and P9 were placed in a recording chamber for 5 min in which a microphone recorded any vocalizations (Fig. 4A). Quantification of the number of calls showed that NEXMIF KO animals made significantly fewer calls overall (P5: t(18) = 5.37, n = 10, p < 0.0001; P7: t(20) = 4.71, n = 11, p = 0.00013; P9: t(20) = 2.78, WT n = 12, KO n = 10, p = 0.012) (Fig. 4B) and fewer calls within each minute of the 5 min recording period, at all time points measured (Fig. 4C–E). Calls made by the NEXMIF KO animals were also of shorter length (P5: t(16) = 3.06, p = 0.0075; P7: t(20) = 2.86, p = 0.0097; P9: t(20) = 2.98, p = 0.0074) (Fig. 4F) resulting in a reduced total time spent calling (P5: t(18) = 4.02, p = 0.0008; P7: t(20) = 4.18, p = 0.00047; P9: t(20) = 3.43, p = 0.0027) (Fig. 4G). There was no change in average peak amplitude or peak frequency of vocalizations between the WT and KO animals (Fig. 4H,I). Further analysis of the call syllable types (Fig. 5A) revealed that NEXMIF KO animals generally made simpler USV calls than WT animals (P7: WT = 57.12 ± 7.86%, n = 6; KO = 83.72 ± 3.69%, n = 5; F(1,9) = 19.27, p = 0.002, ANOVA) (Fig. 5B), making significantly more short and flat calls and fewer modulated and multiple jump calls (P7: Short: t(9) = 4.9, p = 0.0018; Flat: t(9) = 2.86, p = 0.019; Mod.: t(9) = 2.60, p = 0.029; Multi. Jump: t(8) = 3.14, p = 0.014) (Fig. 5C–E). These findings suggest that loss of NEXMIF results in impairment in normal communication, further establishing this as an ASD model.
Figure 4.
NEXMIF KO mice show impairments in communication. A, Representative vocalizations from P7 recordings for WT and KO mice. Top, Raw USV signals. Bottom, Associated spectrograms; all are 1.5 s in length. Dashed line depicts the 30 kHz frequency threshold for eliminating noise. B, Quantification of the call rate shows a decrease in the number of calls made by NEXMIF KO animals compared with WT controls at each developmental time point recorded. C–E, Mean number of calls emitted across the 5 min recording period at each developmental time point P5 (C: WT, n = 10; KO, n = 10), P7 (D: WT, n = 11; KO, n = 11), and P9 (E: WT, n = 12; KO, n = 11). F, G, NEXMIF KO mice showed a decreased mean call syllable duration (F) and a decrease in total time spent calling (G) at each time point. H, I, Quantification of the peak amplitude (H) and peak frequency (I) of calls shows no change between NEXMIF WT and KO animals at any time point. Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. ns, Not significant.
Figure 5.
Syllable classification of USV calls in NEXMIF mice. A, Representative calls of each type used in syllable characterization. B, KO animals showed an increase in Simple type calls and a decrease in Complex type calls compared with WT animals at each time point. C, Call syllable classification at P5 shows an increase in short and flat calls and a decrease in modulated and harmonic calls made by KO animals compared with WT animals. D, KO mice made significantly more short and flat calls compared with WT mice and significantly less modulated and multiple jump calls at P7. E, Call syllable classification at P9 shows an increase in short calls and decrease in modulated, multiple jump, and harmonic calls made by KO animals compared with WT animals. (WT, n = 6; KO, n = 5). Data are represented as average ± SEM. *p < 0.05; **p < 0.01. ns, Not significant.
Impaired spatial memory in NEXMIF KO animals
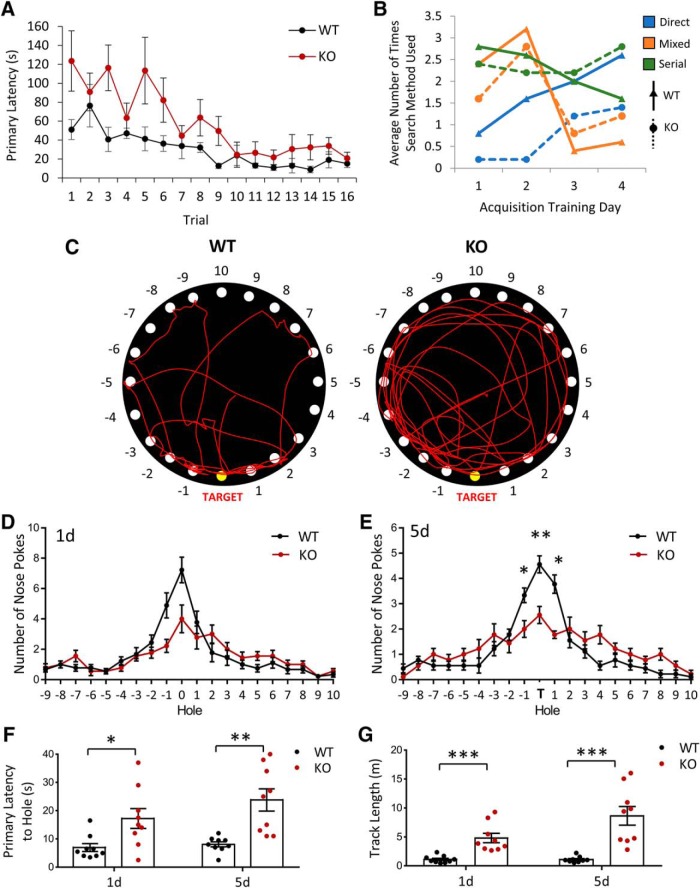
Humans with loss of function mutations or genetic deletions of NEXMIF show autistic phenotypes accompanied with severe ID. We therefore sought to investigate whether our NEXMIF KO mouse showed learning and memory deficits to recapitulate the human condition. The first test we used was the Barnes maze for spatial memory. In this task, mice were trained over a period of 4 d to learn the location of a target escape hole, using spatial cues placed within the testing room, on a board containing 20 holes around its circumference. NEXMIF KO animals showed the ability to learn the target escape hole location during acquisition training periods (Fig. 6A). Additionally, we quantified the search strategy animals used to find the target hole. A “direct” search was counted if the initial search was pointed toward the target hole or either hole directly adjacent to the target. A “mixed” search was classified as an unordered or random search of the maze and a “serial” search strategy was classified as finding the target hole after visiting at least two adjacent holes in a serial manner. Analysis of search strategies used by the mice showed that KO animals were slower to use direct searches compared with WT animals, but did make progress in making more direct searches over the training sessions (Fig. 6B). It is interesting to note that although NEXMIF KO animals relied on direct searches increasingly during training, they still used the serial strategy significantly more than the WT animals, perhaps due to hyperactivity, more anxiety and/or deficits in learning and memory.
Figure 6.
NEXMIF KO mice show impairments in Barnes maze spatial learning and memory. A, Quantification of the primary latency (in seconds) to find the target hole across all training trials (4 trials per day over 4 d) for WT and KO animals. B, Quantification of the types of search strategies used for locating the target hole over the 4 training days. C, Traces of track paths in the Barnes spatial memory maze during the 5 d memory probe for WT and KO animals. D, Distribution of nose pokes on the board showed no difference 24 h after training. E, Quantification of the distribution of nose pokes across different holes on the board showed a significant decrease for the target and adjacent holes in the 5 d memory probe. F, G, Primary latency to the target hole (F) and total track length (G) were increased in the KO animals probing both 1 and 5 d after training Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. ns, Not significant.
After spatial acquisition training, the target hole was covered and the number of nose pokes made at each hole location was quantified 24 h and 5 d later (Fig. 6C). In the 24 h probe for spatial memory, NEXMIF KO animals did not show significant differences in the number of nose pokes made at the target hole (Fig. 6D). Probing spatial memory 5 d after training showed that NEXMIF KO animals had impairments in memory of the target hole location, with significantly less nose pokes at the target and adjacent holes and a broader search of the board (Fig. 6E). Quantification of the primary latency to find the target hole showed that NEXMIF KO animals took much longer to find the target hole (24 h: WT = 6.94 ± 1.41 s, n = 9; KO = 17.22 ± 3.5 s, n = 9; t(32) = 2.64, p = 0.013; 5 d: WT = 8.06 ± 0.90 s, n = 9; KO = 23.78 ± 3.91 s, n = 9; t(32) = 4.04, p = 0.00032) and traveled much longer before making an initial nose poke at the target hole (24 h: WT = 0.11 ± 0.021 m, n = 9; KO = 0.48 ± 0.0.81 m, n = 9; t(16) = 4.41, p = 0.00044; 5 d: WT = 0.11 ± 0.017 m, n = 9; KO = 0.86 ± 0.16 m, n = 9; t(16) = 4.65, p = 0.00027) in both the 24 h and the 5 d tests (Fig. 6F,G). These findings suggest that loss of NEXMIF impairs spatial memory, which may be a contributing factor to the ID observed in humans with loss of NEXMIF and ASD with ID.
Loss of NEXMIF affects hippocampal and amygdala-dependent memory in fear-conditioning tests
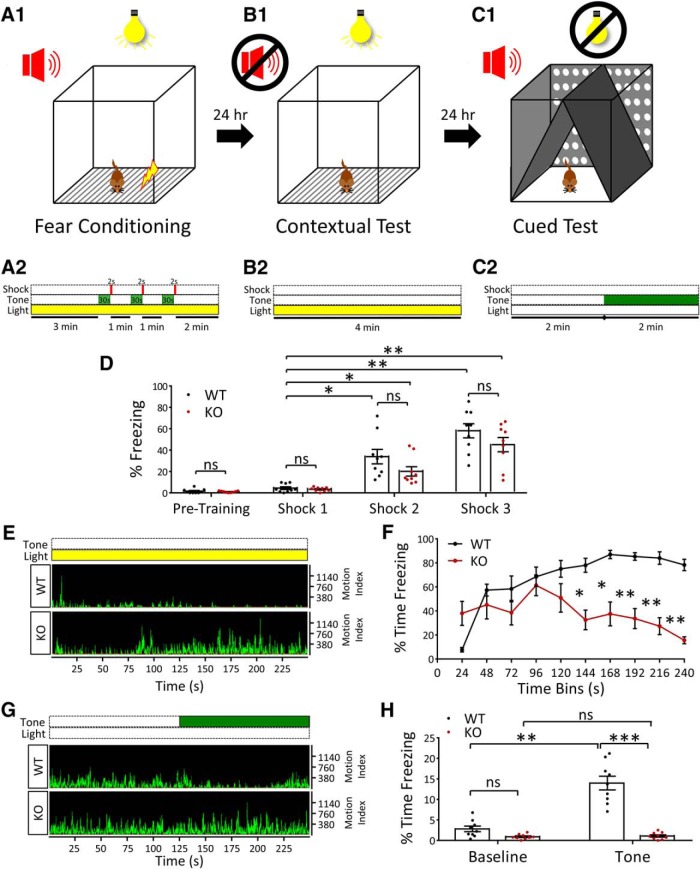
We next performed a fear-conditioning test to probe learning and memory behavior in NEXMIF KO animals. On day 1, mice were placed into the fear conditioning chamber and subjected to a stimulation protocol with a 2 s foot shock followed by a 30 s tone, repeated three times, to condition a fear response. One day later, the animals were placed back into the same box under the same conditions, and the contextual memory was assessed by quantifying the amount of time spent with no motion (freezing). The following day, the entire environment of the box was changed including lights, odorants, wall, floor, and ceiling materials/design. The cued memory was then measured by quantifying the amount of freezing time after the conditioned tone was played (Fig. 7A–C).
Figure 7.
NEXMIF KO mice show impairments in fear conditioning learning and memory. A–C, Paradigm used for fear conditioning, contextual and cued memory tests. On day 1 after habituation, animals were placed into the box (A1) and given three 2 s foot shocks after playing a 30 s tone (A2). On day 2, contextual memory was tested by placing animals into the same box (B1) with no tone played (B2). On day 3, the environment in the box was completely changed (C1) and animals were played the tone used during fear conditioning (C2). D, Quantification of freezing time in WT and KO animals before fear conditioning showed no freezing behavior and during the 30 s tones played during fear conditioning training showed that both WT and KO animals learned to associate the tone with the fear stimulus (foot shock). E, Representative motion indices during the contextual memory test for WT and KO animals. F, Quantification of the time spent freezing (%) throughout the contextual memory test. G, Representative motion indices during the cued memory test for WT and KO animals. H, Quantification of the time spent freezing (%) pretone (Baseline) and after playing the cue (Tone) during the cued memory test. Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. ns, Not significant.
Before fear conditioning, both WT and KO mice did not show significant freezing behavior when placed in the test chamber. Additionally, both mice learned to associate the tone with the foot shock during the conditioning paradigm, indicated with an increase in freezing time with each subsequent tone played during the conditioning paradigm (F(1,16) = 36.81, p = 0.000016, ANOVA) (Fig. 7D). On day 2, NEXMIF KO mice showed a significant impairment in hippocampal-based, contextual fear memory compared with WT animals, with an increase in the motion index and decrease in freezing time across trial blocks (Fig. 7E,F). The most striking difference, however, was the large decrease in amygdala-dependent, cued fear memory in NEXMIF KO animals. Both WT and KO animals showed minimal freezing behavior when placed into the chamber with a completely new setup, indicating they did not have a fear association in the new environment. However, when the tone was played, WT animals showed a robust increase in freezing behavior, while KO animals showed no change in movement compared with their pretone activity (F(3,48) = 67.17, p < 0.0001, ANOVA) (Fig. 7G,H). These results indicate that both contextual and cued fear memory was impaired in the absence of NEXMIF expression, suggesting major deficits in learning and memory.
Neurons of NEXMIF KO mice show deficits in spine density, spine morphology, and synaptic protein composition
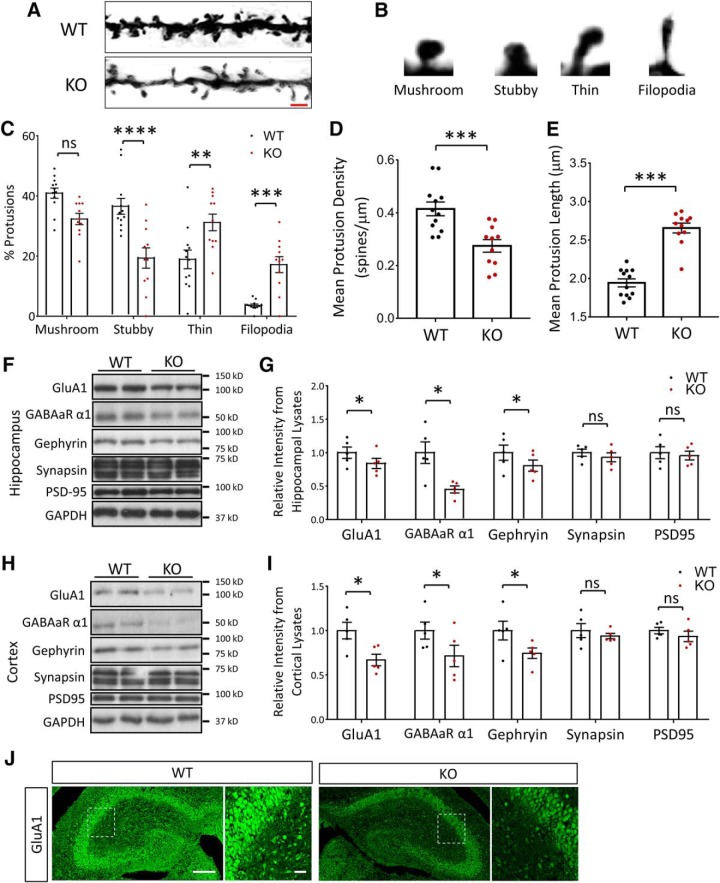
We hypothesized that the autistic phenotypes and severe impairment in learning and memory in NEXMIF KO mice may result from aberrations in spine formation and/or synapse maturation. To investigate this, we first performed Golgi staining on P90 animals, and quantified spine density and morphology in the hippocampus (Fig. 8A). The morphology of dendritic spines can be classified from “thin” (and long) to “stubby” and “mushroom” types, with the more latter representing mature and more stable spines. The more elongated filopodia (>3 μm in length) are immature dendritic protrusions that are highly dynamic and not considered to be functional spines (Fig. 8B). In hippocampus, quantification of the Golgi impregnation showed a significant increase in filopodia and thin protrusions, with less stubby spines in the NEXMIF KO compared with age-matched WT mice (F(3,63) = 29.52, p < 0.0001, ANOVA) (Fig. 8C). Additionally, a 35% reduction in the density of dendritic protrusions (WT = 0.41 ± 0.03 spines/μm, KO = 0.27 ± 0.03 spines/μm, t(21) = 3.95, p = 0.00073, n = 11–12 cells) (Fig. 8D) and a 33% increase in the average length of protrusions (WT = 1.95 ± 0.05 μm, KO = 2.64 ± 0.07 μm, t(21) = 8.69, p < 0.0001, n = 11–12 cells) (Fig. 8E) was observed in the KO animals. Thus, loss of NEXMIF expression resulted in a significant reduction in mature spines in hippocampus, suggesting that abnormal synaptic connectivity may underlie some of the behavioral deficits observed.
Figure 8.
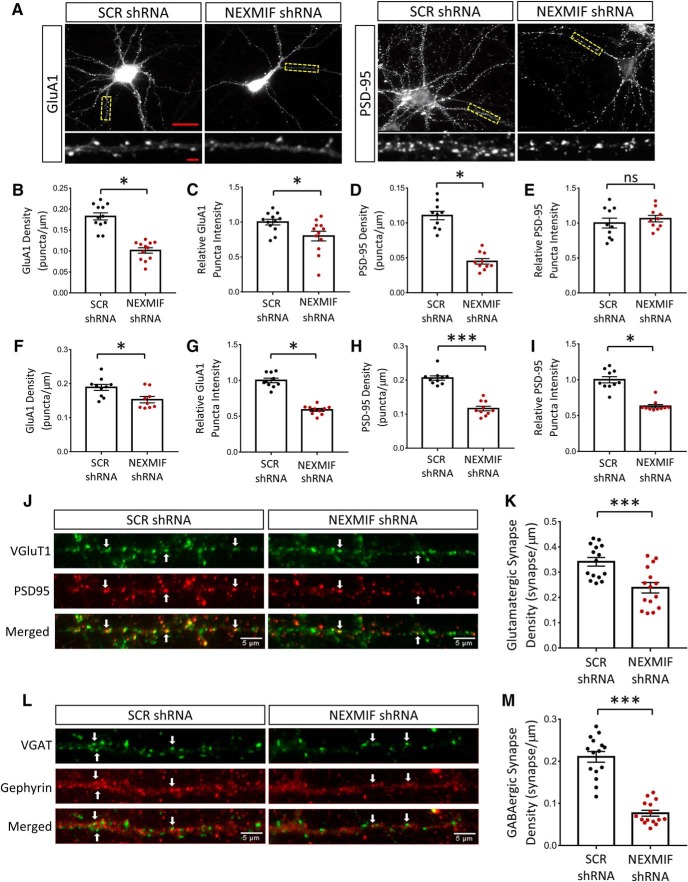
Altered spine morphology and synaptic protein composition in the NEXMIF KO mice. A, Golgi staining of spines from pyramidal neurons in P90 mouse hippocampus. Scale bar, 3 μm. B, Representative morphologies of the different dendritic protrusion types classified. C, Quantification of protrusion morphologies in WT and KO animals showed an increase in immature morphologies. D, E, Spine density (D) was decreased and spine length (E) increased in KO animals. F, Western blot of synaptic-related proteins from WT and NEXMIF KO hippocampus. G, Quantification of Western blot data from hippocampus showed a reduction in GluA1, GABAα1, gephyrin, no change in synapsin and no change in PSD-95 intensity. H, Western blot of synaptic-related proteins from WT and NEXMIF KO cortex. I, Quantification of Western blot data from cortex showed a reduction in GluA1, GABAα1, gephyrin, no change in synapsin and no change in PSD-95 intensity. J, Immunohistochemistry of KO and WT hippocampal brain slices at P0 shows a clear decrease in total GluA1 staining intensity Scale bars, 100 μm (full picture); 50 μm (enlarged area). Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns, Not significant.
To investigate changes in synaptic protein expression, we next performed Western blot of synaptic proteins. We found a decrease in the expression of AMPAR subunit GluA1, the GABAA receptor α1 subunit, and the inhibitory synaptic scaffolding protein gephyrin, while no changes were detected in the amount of the excitatory synaptic scaffolding protein PSD-95, and the presynaptic vesicular protein synapsin, in either hippocampal (Fig. 8F,G) or cortical brain lysates (Fig. 8H,I). The same change in GluA1 was also detected by immunostaining of hippocampal brain slices (Fig. 8J).
Loss of NEXMIF results in deficits in synapse formation
To examine synaptic changes in more detail we used an in vitro model with lentiviral-mediated shRNA knock-down of NEXMIF, and analyzed GluA1 and PSD-95 synaptic puncta via ICC at DIV 14 in primary hippocampal neurons (Fig. 9A). Strikingly, a 45% reduction in GluA1 puncta density (WT = 1.0 ± 0.07, KO = 0.56 ± 0.04, t(22) = 7.59, p < 0.0001, n = 10–12 cells) (Fig. 9B) and 20% reduction in GluA1 puncta intensity were observed after shRNA-mediated knock-down of NEXMIF compared with scrambled shRNA controls (WT = 1.0 ± 0.04, KO = 0.80 ± 0.05, t(22) = 2.50, p = 0.021, n = 10–12 cells) (Fig. 9C). Analysis of the immunostaining of PSD-95 showed a 60% reduction in puncta density with no change in synaptic intensity (Fig. 9D, WT = 1.0 ± 0.05, KO = 0.41 ± 0.04, t(18) = 9.05, p < 0.0001, n = 10–12 cells; Fig. 9E, WT = 1.0 ± 0.07, KO = 1.06 ± 0.05, t(18) = 0.75, p = 0.46, n = 10–12 cells) (Fig. 9D,E). Analysis of GluA1 puncta in primary cortical neurons showed similar reductions in puncta density of 20% and in puncta intensity of 37% (Fig. 9F,G), whereas PSD-95 puncta showed a 45% reduction puncta density and a 40% reduction in puncta intensity (Fig. 9G,I). We then coimmunostained the presynaptic marker VGluT1, as well as the postsynaptic marker PSD-95, to assess changes in glutamatergic synapse formation. After shRNA-mediated knock-down of NEXMIF, glutamatergic synapse density was reduced by 30% (t(28) = 3.8, p = 0.0007, n = 15 cells) (Fig. 9J,K). These findings indicate that loss of NEXMIF expression has a profound effect on glutamatergic synapse formation, with a large decrease in spine and synapse number.
Figure 9.
Altered synaptic protein composition in cultured neurons with shRNA-mediated NEXMIF knock-down. A, Representative images of immunohistochemistry for GluA1 and PSD-95 at DIV 15 in rat hippocampal cultures after infection with scrambled (SCR shRNA) or NEXMIF shRNA at DIV0. Scale bars, 15 μm (full picture); 5 μm (enlarged area). B, C, Quantification of GluA1 immunostaining in hippocampal neurons showing a decrease in puncta density and puncta intensity. D, E, Quantification of PSD-95 immunostaining in hippocampal neurons showing a decrease in PSD-95 density with no significant change in PSD-95 intensity. F, G, Quantification of GluA1 immunostaining in cortical neurons showing a decrease in puncta density and intensity from shRNA mediated NEXMIF knock-down. H, I, Quantification of PSD-95 immunostaining in cortical neurons showing a decrease in puncta density and intensity from shRNA mediated NEXMIF knock-down (n = 10 cells). J, Glutamatergic synapses were identified from colocalized VGluT1 and PSD95 puncta florescent signals. K, Quantification showing that shRNA-mediated NEXMIF knock-down resulted in a decrease in glutamatergic synapse density. L, GABAergic synapses were identified from colocalized VGAT and gephyrin puncta florescent signals. M, Quantification showing shRNA mediated NEXMIF knock-down resulted in a decrease in GABAergic synapse density. Examples of colocalization are marked by arrows (n = 15 cells). Data are represented as average ±SEM. *p < 0.05; ***p < 0.001. ns, Not significant.
To examine changes in GABAergic synapses, we used lentiviral-mediated shRNA knock-down of NEXMIF and coimmunostained gephyrin and VGAT to analyze GABAergic synaptic puncta. We found that, compared with scrambled shRNA controls, knock-down of NEXMIF resulted in a 60% reduction in GABAergic synapse density (t(28) = 9.26, p < 0.0001, n = 15 cells) (Fig. 9L,M). These findings demonstrate that loss of NEXMIF also negatively affected the formation of GABAergic synapses, an effect similar to that observed on glutamatergic synapses.
Loss of NEXMIF results in deficits in synaptic transmission
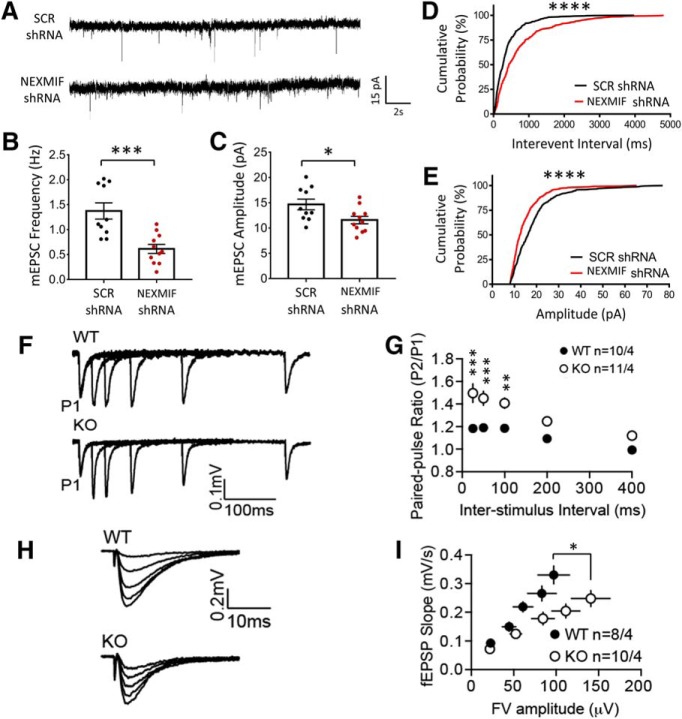
To investigate the functional consequence of NEXMIF knock-down, hippocampal neurons were incubated with lentiviral NEXMIF shRNA at DIV0, and mEPSCs were recorded at DIV 14–16 (Fig. 10A). We found that application of shRNA virus produced a significant reduction in mEPSC frequency (SCR shRNA = 1.37 ± 0.16 Hz; NEXMIF shRNA 0.62 ± 0.09; n = 10–11 cells/group, t(19) = 4.20, p = 0.00048) (Fig. 10B) and amplitude (Con 14.67 ± 1.05; NEXMIF shRNA 11.59 ± 1.17; n = 10–11 cells/group, t(19) = 2.43, p = 0.025) (Fig. 10C). Additionally, plots of the cumulative distribution showed a rightward shift in interevent interval (D(500) = 0.24, p < 0.0001, KS test) (Fig. 10D) and a leftward shift in amplitude (D(500) = 0.23, p < 0.0001, KS test) (Fig. 10E) for the NEXMIF shRNA treated neurons. The decrease in mEPSC amplitude is consistent with reductions the level of AMPAR expression in neurons after NEXMIF knock-down as well as the KO mouse brain. The decrease in mEPSC frequency could be due to either the decrease in synapse number, consistent with the reduction in spine density in neurons after NEXMIF knock-down, or to reduction of basal presynaptic release probability.
Figure 10.
Loss of NEXMIF results in altered synaptic transmission. A, mEPSC recordings from rat hippocampal neurons after infection with lentiviral scrambled (SCR shRNA) or NEXMIF shRNA, together with GFP. B, C, Quantification of mEPSC recordings showing a significant decrease in frequency and amplitude. D, E, Cumulative probability plots of mEPSCs showing a leftward shift in amplitude and a rightward shift in interevent interval for NEXMIF shRNA treated neurons. F, Representative traces from recordings performed in acute hippocampal slices from NEXMIF KO and WT mice to measure the paired-pulse ratio using an interstimulus interval of 25, 50, 100, 200, and 400 ms. G, Quantification of the paired-pulse ratio from recordings shown in I showed a significant increase in KO animals compared with WT controls (n = 10–11). H, Input/output curves from acute hippocampal slices from KO and WT hippocampal brain slices. Five stimulus intensities were chosen to elicit field excitatory postsynaptic responses. I, Quantification of the input/output curves showed that KO mice had a reduction in basal synaptic transmission compared with WT controls (n = 8–10). Data are represented as average ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
We next examined synaptic transmission in NEXMIF KO mice. In hippocampal brain slices, fEPSPs of the CA3–CA1 synapses were recorded at ∼50% maximal stimulation. We found that paired-pulse ratio was markedly higher in KO mice, especially at high-frequency stimulation (n = 10–11) (Fig. 10F,G), indicating abnormalities in presynaptic function, presumably in reduced release probability. This is consistent with the reduced mEPSC frequency seen in neurons after NEXMIF knock-down. We additionally measured the input-output relationship at CA3-CA1 synapses (Fig. 10H). Analysis revealed a weakened input-output relationship in KO mice, compared with the WT controls (n = 8–10) (Fig. 10I), indicating reduced excitatory synaptic strength in KO mice. These results suggest significant impairment in excitatory synaptic function in the KO mouse.
Discussion
In this study, we successfully established a mouse model for NEXMIF-dependent ASD and ID. The NEXMIF KO mouse displays the characteristics observed in human patients with loss of NEXMIF and can therefore be used as an animal model. NEXMIF KO mice display significant impairments in the three-chamber social task, with decreased sociability and decreased interest in social novelty. KO mice spent significantly less time interacting with a novel mouse. Additionally, NEXMIF KO animals showed significant communication impairments shown by USVs, with increased use of simpler and shorter call types as well as fewer calls and less total time spent making calls. We also found significant impairments in the marble-burying test, with NEXMIF KO animals burying significantly fewer marbles during the task, a phenotype that suggests a decreased interest in their surrounding environment. NEXMIF KO mice also displayed significantly increased repetitive grooming and hyperactivity, common phenotypes observed in ASD mouse models (Silverman et al., 2010; Kalueff et al., 2016).
In line with findings of moderate-to-severe ID in human patients with NEXMIF mutations, NEXMIF KO mice also displayed major deficits in both spatial and fear memory (Cantagrel et al., 2004, 2009; Van Maldergem et al., 2013; Kuroda et al., 2015; Lambert et al., 2018; Lorenzo et al., 2018). KO mice were able to acquire the target hole location over the course of the Barnes maze training, suggesting no major impairments in visual system function, but showed impairments when probing their memory 5 d later. Additionally, KO mice were able to associate a tone with a foot shock during the first day of the fear conditioning paradigm, indicating their auditory system was functional. Therefore, the deficits observed in the learning and memory tasks should not be a result of impaired sensory input, but rather impairments in cognitive processing.
The reports of human ASD patients with loss of NEXMIF also indicate seizures as a comorbid phenotype (Cantagrel et al., 2004, 2009; Van Maldergem et al., 2013; Charzewska et al., 2015; de Lange et al., 2016; Farach and Northrup, 2016; Webster et al., 2017; Lambert et al., 2018; Lorenzo et al., 2018). These seizures vary in type but include generalized, tonic-clonic, absence, and myoclonic seizures. The onset of seizures also varied between patients but began as early as < 1 year old and as late as the teens. Some patients do not demonstrate seizures or have only had a single episode, whereas others could have up to hundreds of episodes per day. Our NEXMIF KO mice also demonstrate severe seizures typically in response to animal handling. Onset varied with the earliest observation at 3 months and the majority beginning at ages >4 months. Some seizures in mice aged 6 months or older have resulted in fatalities. This complex phenotype will require further studies for more detailed analysis. These findings provide strong evidence that the NEXMIF KO mouse recapitulates the autistic and ID phenotypes observed in human patients.
Abnormalities in synaptogenesis and synaptic function are key underlying factors that lead to social and cognitive deficits during brain development (Zoghbi and Bear, 2012). Synaptopathy is associated with a number of brain disorders, including Rett Syndrome, Fragile-X mental retardation, Angelman Syndrome, and Phelan-McDermid Syndrome (Belmonte et al., 2004; Shepherd and Katz, 2011; Garner and Wetmore, 2012; Ricciardi et al., 2012). We observed a major dysregulation in synaptic protein expression with a reduction in both glutamatergic and GABAergic synaptic proteins including AMPARs, PSD-95, and gephyrin from neurons of NEXMIF knock-down and the KO brain tissue. Additionally, Golgi analysis from NEXMIF KO mouse brains revealed significantly decreased spine densities with an increase in immature spines. In this study, we chose to assess spine density in the KO animals at P90 to correlate with the age of the mice used for behavioral tests. It will be valuable for future studies to explore changes in spine and synapse formation throughout developmental stages. Given that we've previously shown loss of NEXMIF impairs neurite outgrowth and actin dynamics, future studies with knock-down of NEXMIF after neurite growth and synaptogenesis could clarify whether synaptic and spine deficits result from aberrant structural growth and if NEXMIF plays a role in synapse maintenance and function after synaptogenesis. In addition to spine formation, the observed reduction in synapse density may also result from facilitated synaptic pruning during brain/neuron development.
We also observed functional changes in synaptic transmission both in vitro and in vivo. Knock-down of NEXMIF in cultured hippocampal neurons resulted in reduced mEPSC frequency and amplitude. fEPSP recordings from hippocampal brain slices of NEXMIF KO mice demonstrated an increase in paired-pulse facilitation and a decrease in the input/output response. While the exact mechanisms remain to be elucidated, the reduced basal synaptic transmission strength is consistent with the observed decreases in spine number and AMPAR expression and release probability. These major abnormalities in synapse formation and function may underlie the behavioral deficits of this animal model. The NEXMIF KO mouse should therefore be of great value to the ASD community given the clean genetic background (single gene deletion) and strong autistic behavioral phenotype. Also, because NEXMIF is exclusively expressed in the nucleus of neurons, NEXMIF probably functions as a transcription factor or a component of regulatory complexes in gene expression. The specific roles for NEXMIF and its potential target genes will be elucidated by future studies.
Footnotes
This work was supported by the National Institutes of Health (Grant R01 MH079407). The authors declare no competing financial interests. We thank Howard Gritton and Xue Han for technical assistance and the Man laboratory members for helpful discussion.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A (2013) SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4:36. 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon-Martinez T, Khan A, Myers KA (2019) Torpedo maculopathy associated with NEXMIF mutation. Mol Syndromol 10:229–233. 10.1159/000498835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen Institute for Brain Science (2004) Allen Mouse Brain Alatas. Experiment 69531127, Gene C77370. Available at brain-map.org/api/index.html.
- Bagni C, Zukin RS (2019) A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron 101:1070–1088. 10.1016/j.neuron.2019.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, Zahorodny W, Robinson Rosenberg C, White T, Durkin MS, Imm P, Nikolaou L, Yeargin-Allsopp M, Lee LC, Harrington R, Lopez M, Fitzgerald RT, Hewitt A, Pettygrove S, et al. (2018) Prevalence of autism spectrum disorder among children aged 8 years: autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill Summ 67:1–23. 10.15585/mmwr.ss6706a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee-Basu S, Packer A (2010) SFARI gene: an evolving database for the autism research community. Dis Model Mech 3:133–135. 10.1242/dmm.005439 [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ (2004) Autism and abnormal development of brain connectivity. J Neurosci 24:9228–9231. 10.1523/JNEUROSCI.3340-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, Bonin M, Riess A, Engels H, Sprengel R, Scherer SW, Rappold GA (2010) Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 42:489–491. 10.1038/ng.589 [DOI] [PubMed] [Google Scholar]
- Bourgeron T. (2015) From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci 16:551–563. 10.1038/nrn3992 [DOI] [PubMed] [Google Scholar]
- Cantagrel V, Lossi AM, Boulanger S, Depetris D, Mattei MG, Gecz J, Schwartz CE, Van Maldergem L, Villard L (2004) Disruption of a new X linked gene highly expressed in brain in a family with two mentally retarded males. J Med Genet 41:736–742. 10.1136/jmg.2004.021626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V, Haddad MR, Ciofi P, Andrieu D, Lossi AM, Maldergem Lv, Roux JC, Villard L (2009) Spatiotemporal expression in mouse brain of Kiaa2022, a gene disrupted in two patients with severe mental retardation. Gene Expr Patterns 9:423–429. 10.1016/j.gep.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Charzewska A, Rzońca S, Janeczko M, Nawara M, Smyk M, Bal J, Hoffman-Zacharska D (2015) A duplication of the whole KIAA2022 gene validates the gene role in the pathogenesis of intellectual disability and autism. Clin Genet 88:297–299. 10.1111/cge.12528 [DOI] [PubMed] [Google Scholar]
- Crawley JN. (2007) Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol 17:448–459. 10.1111/j.1750-3639.2007.00096.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange IM, Helbig KL, Weckhuysen S, Møller RS, Velinov M, Dolzhanskaya N, Marsh E, Helbig I, Devinsky O, Tang S, Mefford HC, Myers CT, van Paesschen W, Striano P, van Gassen K, van Kempen M, de Kovel CG, Piard J, Minassian BA, Nezarati MM, et al. (2016) De novo mutations of KIAA2022 in females cause intellectual disability and intractable epilepsy. J Med Genet 53:850–858. 10.1136/jmedgenet-2016-103909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, et al. (2007) Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 39:25–27. 10.1038/ng1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach LS, Northrup H (2016) KIAA2022 nonsense mutation in a symptomatic female. Am J Med Genet A 170:703–706. 10.1002/ajmg.a.37479 [DOI] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH Jr, Skinner C, Schwartz CE, Sommer SS (2006) High frequency of neurexin 1β signal peptide structural variants in patients with autism. Neurosci Lett 409:10–13. 10.1016/j.neulet.2006.08.017 [DOI] [PubMed] [Google Scholar]
- Garner CC, Wetmore DZ (2012) Synaptic pathology of down syndrome. Adv Exp Med Biol 970:451–468. 10.1007/978-3-7091-0932-8_20 [DOI] [PubMed] [Google Scholar]
- Geschwind DH. (2011) Genetics of autism spectrum disorders. Trends Cogn Sci 15:409–416. 10.1016/j.tics.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J, Man HY (2016) The X-linked autism protein KIAA2022/KIDLIA regulates neurite outgrowth via N-cadherin and delta-catenin signaling. eNeuro 3:ENEURO.0238–16.2016. 10.1523/ENEURO.0238-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco B, Managò F, Tucci V, Kao HT, Valtorta F, Benfenati F (2013) Autism-related behavioral abnormalities in synapsin knock-out mice. Behav Brain Res 251:65–74. 10.1016/j.bbr.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, Awashti S, Belliveau R, Bettella F, Buxbaum JD, Bybjerg-Grauholm J, Bækvad-Hansen M, Cerrato F, Chambert K, Christensen JH, Churchhouse C, et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51:431–444. 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Mouse Knockout Consortium, Collins FS, Rossant J, Wurst W (2007) A mouse for all reasons. Cell 128:9–13. 10.1016/j.cell.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Miyata S, Koyama Y, Yoshikawa K, Hattori T, Kumamoto N, Shingaki K, Katayama T, Tohyama M (2012) Transient expression of xpn, an XLMR protein related to neurite extension, during brain development and participation in neurite outgrowth. Neuroscience 214:181–191. 10.1016/j.neuroscience.2012.04.030 [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T; Paris Autism Research International Sibpair Study (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34:27–29. 10.1038/ng1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH (2014) Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol 10:74–81. 10.1038/nrneurol.2013.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, Fentress JC (2016) Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci 17:45–59. 10.1038/nrn.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE (2015) Excess of rare, inherited truncating mutations in autism. Nat Genet 47:582–588. 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Ohashi I, Naruto T, Ida K, Enomoto Y, Saito T, Nagai J, Wada T, Kurosawa K (2015) Delineation of the KIAA2022 mutation phenotype: two patients with X-linked intellectual disability and distinctive features. Am J Med Genet A 167:1349–1353. 10.1002/ajmg.a.37002 [DOI] [PubMed] [Google Scholar]
- Lambert N, Dauve C, Ranza E, Makrythanasis P, Santoni F, Sloan-Béna F, Gimelli S, Blouin JL, Guipponi M, Bottani A, Antonarakis SE, Kosel MM, Fluss J, Paoloni-Giacobino A (2018) Novel NEXMIF pathogenic variant in a boy with severe autistic features, intellectual disability, and epilepsy, and his mildly affected mother. J Hum Genet 63:847–850. 10.1038/s10038-018-0459-2 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E (1989) Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 19:185–212. 10.1007/BF02211841 [DOI] [PubMed] [Google Scholar]
- Lorenzo M, Stolte-Dijkstra I, van Rheenen P, Smith RG, Scheers T, Walia JS (2018) Clinical spectrum of KIAA2022 pathogenic variants in males: case report of two boys with KIAA2022 pathogenic variants and review of the literature. Am J Med Genet Part A 176:1455–1462. 10.1002/ajmg.a.38667 [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Nonneman RJ, Grossman AW, Murphy DL, D'Ercole AJ, Crawley JN, Magnuson TR, Lauder JM (2009) Social approach in genetically engineered mouse lines relevant to autism. Genes Brain Behav 8:129–142. 10.1111/j.1601-183X.2008.00452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico AM, Bourgeron T (2006) Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci 29:349–358. 10.1016/j.tins.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC (2009) Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 6:493–495. 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM (2010) Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron 66:191–197. 10.1016/j.neuron.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, et al. (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466:368–372. 10.1038/nature09146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiersen AM, Todd RD (2008) Co-occurrence of ADHD and autism spectrum disorders: phenomenology and treatment. Expert Rev Neurother 8:657–669. 10.1586/14737175.8.4.657 [DOI] [PubMed] [Google Scholar]
- Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D, Sessa A, Magagnotti C, Bachi A, Giarda E, Verpelli C, Kilstrup-Nielsen C, Sala C, Kalscheuer VM, Broccoli V (2012) CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 14:911–923. 10.1038/ncb2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia R, Warrier V, Bullmore ET, Baron-Cohen S, Bethlehem RAI (2019) Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry 24:1053–1064. 10.1038/s41380-018-0023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede M, Nagpal S, Gandal MJ, Parikshak NN, Mirnics K, Geschwind DH, Morrow EM (2018) Strong correlation of downregulated genes related to synaptic transmission and mitochondria in post-mortem autism cerebral cortex. J Neurodev Disord 10:18. 10.1186/s11689-018-9237-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM, Katz DM (2011) Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and met. Curr Opin Neurobiol 21:827–833. 10.1016/j.conb.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN (2010) Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 11:490–502. 10.1038/nrn2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch EA, Arnold EB, Jones AM, Ale CM, Wood JJ, Ehrenreich-May J, Lewin AB, Mutch PJ, Murphy TK (2012) The role of co-occurring disruptive behavior in the clinical presentation of children and adolescents with anxiety in the context of autism spectrum disorders. Child Psychiatry Hum Dev 43:734–746. 10.1007/s10578-012-0294-1 [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC (2007) A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318:71–76. 10.1126/science.1146221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R (2009) Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl) 204:361–373. 10.1007/s00213-009-1466-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin KE, Pradella A, Gorrindo T, Pine DS, Leibenluft E (2005) Autism spectrum traits in children with mood and anxiety disorders. J Child Adolesc Psychopharmacol 15:452–464. 10.1089/cap.2005.15.452 [DOI] [PubMed] [Google Scholar]
- Tye C, Runicles AK, Whitehouse AJO, Alvares GA (2018) Characterizing the interplay between autism spectrum disorder and comorbid medical conditions: an integrative review. Front Psychiatry 9:751. 10.3389/fpsyt.2018.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maldergem L, Hou Q, Kalscheuer VM, Rio M, Doco-Fenzy M, Medeira A, de Brouwer AP, Cabrol C, Haas SA, Cacciagli P, Moutton S, Landais E, Motte J, Colleaux L, Bonnet C, Villard L, Dupont J, Man HY (2013) Loss of function of KIAA2022 causes mild to severe intellectual disability with an autism spectrum disorder and impairs neurite outgrowth. Hum Mol Genet 22:3306–3314. 10.1093/hmg/ddt187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R, Cho MT, Retterer K, Millan F, Nowak C, Douglas J, Ahmad A, Raymond GV, Johnson MR, Pujol A, Begtrup A, McKnight D, Devinsky O, Chung WK (2017) De novo loss of function mutations in KIAA2022 are associated with epilepsy and neurodevelopmental delay in females. Clin Genet 91:756–763. 10.1111/cge.12854 [DOI] [PubMed] [Google Scholar]
- Zafeiriou DI, Ververi A, Vargiami E (2007) Childhood autism and associated comorbidities. Brain Dev 29:257–272. 10.1016/j.braindev.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM (2016) The evolving diagnostic and genetic landscapes of autism spectrum disorder. Front Genet 7:65. 10.3389/fgene.2016.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. (2003) Postnatal neurodevelopmental disorders: meeting at the synapse? Science 302:826–830. 10.1126/science.1089071 [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF (2012) Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 4:a009886. 10.1101/cshperspect.a009886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Post hoc power analysis. Two-sample, continuous endpoint post hoc power analyses were done with the assumption of equal variance and using a Type I error (alpha) of 0.05. All values are reported as % power. Download Figure 1-1, DOCX file (15.8KB, docx)