Abstract
Persistent dysregulation of IL-6 production and signaling have been implicated in the pathology of various cancers. In systemic mastocytosis, increased serum levels of IL-6 associate with disease severity and progression, although the mechanisms involved are not well understood. Since systemic mastocytosis often associates with the presence in hematopoietic cells of a somatic gain-of-function variant in KIT, D816V-KIT, we examined its potential role in IL-6 upregulation. Bone marrow mononuclear cultures from patients with greater D816V allelic burden released increased amounts of IL-6 which correlated with the percentage of mast cells in the cultures. Intracellular IL-6 staining by flow cytometry and immunofluorescence was primarily associated with mast cells and suggested a higher percentage of IL-6 positive mast cells in patients with higher D816V allelic burden. Furthermore, mast cell lines expressing D816V-KIT, but not those expressing normal KIT or other KIT variants, produced constitutively high IL-6 amounts at the message and protein levels. We further demonstrate that aberrant KIT activity and signaling are critical for the induction of IL-6 and involve STAT5 and PI3K pathways but not STAT3 or STAT4. Activation of STAT5A and STAT5B downstream of D816V-KIT was mediated by JAK2 but also by MEK/ERK1/2, which not only promoted STAT5 phosphorylation but also its long-term transcription. Our study thus supports a role for mast cells and D816V-KIT activity in IL-6 dysregulation in mastocytosis and provides insights into the intracellular mechanisms. The findings contribute to a better understanding of the physiopathology of mastocytosis and suggest the importance of therapeutic targeting of these pathways.
Introduction
Mastocytosis defines a group of heterogeneous disorders characterized by the accumulation of neoplastic/clonal mast cells in the skin, bone marrow (BM) and other organs.1 Mastocytosis is clinically subdivided into systemic (SM) and cutaneous (CM) mastocytosis, both of which are comprised of several variants defined in accordance with histological and clinical parameters and organ involvement.1 Somatic variants in the receptor for stem cell factor (SCF), KIT, that render it constitutively active often associate with SM, particularly p.(D816V), a missense in the tyrosine kinase domain of KIT. D816V-KIT may be accompanied by variants in other genes that further contribute to the oncogenic expansion of mast cells.2–4
Interleukin-6 (IL-6) is a pleiotropic cytokine produced by several cell types including stromal, hematopoietic and tumor cells. In addition to its involvement in normal inflammatory processes and host immune defense mechanisms, IL-6 may contribute to malignancy in a range of cancers including multiple myeloma, B-cell and non-B-cell leukemias and lymphomas,5,6 by modulating cellular development, growth, apoptosis, metastasis and/or cellular resistance to chemotherapy.6 As elevated IL-6 levels in the serum of patients with such malignancies have been associated with poor clinical outcomes, blocking IL-6 or its synthesis in these patients is viewed as a potential therapeutic avenue.7,8
In SM, the levels of serum IL-6 are higher in patients with aggressive versus indolent variants of SM and have been associated with adverse clinical features of mastocytosis such as accumulation of mast cells in the BM, organomegaly, elevated tryptase levels,9,10 osteoporosis and/or bone pain.11 Although progression into more aggressive disease within patients with indolent SM (ISM) occurs only in a subset of patients, IL-6 plasma levels significantly correlate with disease progression and lower progression-free survival, suggesting that blockade of IL-6 synthesis or function may be beneficial in cases with aberrant IL-6 pathways.10 Other studies have shown that IL-6 promotes the differentiation, growth and degranulation of normal mast cells,12 and induces the production of reactive oxygen species by malignant mast cells and their accumulation in tissues in a model of mastocytosis.13 Despite the potential implications for disease pathology, the cell types and the mechanisms that may contribute to the constitutively elevated IL-6 levels in mastocytosis are not known.
In this study, we test the hypothesis that cells expressing gain of function variants of KIT, particularly D816V-KIT, confer the ability to constitutively produce IL-6. As will be shown, ex vivo BM mast cells from patients with SM release IL-6 in correlation with the allelic frequency of D816V-KIT. We further demonstrate that expression of D816V-KIT causes persistent IL-6 induction by mechanisms independent of autocrine feed-forward loops involving IL-6 and signal transducer and activator of transcription 3 (STAT3) described in other malignant cells, but dependent on oncogenic KIT-derived signals. These signals include phosphatidylinositide 3-kinase (PI3K) pathways and oncogenic STAT5 activation by both janus kinase 2 (JAK2) and, unexpectedly, by the mitogen-activated protein kinase MEK/ERK1/2 pathways. These data expand our understanding of the potential mechanisms initiating enhanced IL-6 production in mastocytosis and emphasize targets for therapeutic intervention in cases of high IL-6 profiles and suspected disease progression.
Methods
A detailed description of the methods used in this study can be found in Online Supplementary Appendix.
Study patients
Bone marrow samples were obtained when clinically indicated from patients with SM classified according to the World Health Organization (WHO) guidelines1,3,14 (Online Supplementary Table S1). All human samples were obtained after informed consent, on clinical protocols approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases (02-I-0277 and 08-I-0184) in agreement with the Declaration of Helsinki. D816V-KIT mutation analysis and its allele burden in the BM (D816V-KIT frequency) were determined by allele-specific polymerase chain reaction (PCR) from patient blood and BM genomic DNA.15
Cell lysates
Cell lines were cultured as described in the Online Supplementary Appendix. To obtain lysates for western blots, 3x106 cells were plated in 6-well plates and incubated with or without the indicated inhibitors for 2 hours (h) in serum-free media. Cells were lysed as previously described.16
IL-6 measurements
Cells (3x106) were plated in 6-well plates for 2 h to overnight in 6 mL of serum-free media to exclude the possibility that any extrinsic stimulant present in the serum would influence the results. IL-6 released into the media was measured by ELISA (R&D Systems). Human colorectal carcinoma HCT116 cells were stimulated with 20 ng/mL PMA plus 1 mM ionomycin overnight and the supernatants then collected for IL-6 measurements.
Mononuclear cells in BM aspirates from patients were cultured in StemPro-34 medium with human recombinant SCF (100 ng/mL) for 2-4 days. IL-6 released into the media was determined by ELISA. Alternatively, IL-6 expression in single cells was determined by flow cytometry using a LSRII flow cytometer. BM cells were incubated with Brefeldin A for 4 h and stained with an antibody cocktail containing anti-CD3-QDOT605, anti-CD34-APC, anti-KIT-BV605 and anti-FcεRI-FITC, for 30 minutes (min). Cells were fixed, permeabilized and stained with anti-IL-6-PE for 30 min. Expression of IL-6 in mast cells (CD3−/CD34−/KIT+/FcεRI+) was analyzed using FlowJo software.
Quantitative real-time polymerase chain reaction
HMC-1.2 cells (3x106) were plated in 6-well plates in 6 mL and incubated for 2 h in serum-free media. Cellular RNA was extracted and reverse-transcribed into cDNA. cDNA was then amplified using TaqMan® Gene Expression Master Mix and Taqman® Gene Expression Assays for IL-6, STAT3, STAT4, STAT5A, STAT5B or GAPDH as described in the Online Supplementary Appendix.
Knockdown of STAT transcription factors
Knockdown of STAT3 and STAT4 was performed by lentiviral-mediated transduction of small hairpin RNA (sh-RNA) (Sigma-Aldrich, St. Louis, MO, USA) as previously described.17 STAT5 mRNA was silenced by a small interference-RNA (si-RNA) “ON-TARGET” pool from Dharmacon (Lafayette, CO, USA), introduced into cells by electroporation.
Statistical analysis
Data were expressed as mean±Standard Error of Mean (SEM). Values were from at least three independent experiments, each performed at least in duplicate. Statistically significant differences were calculated by using the Student t-test (unpaired). Statistical significance was indicated as follows: *P<0.01; **P<0.01; ***P<0.001; ****P<0.0001.
Results
Release of IL-6 from patient’s bone marrow cells and its association with D816V-KIT and mast cell frequencies
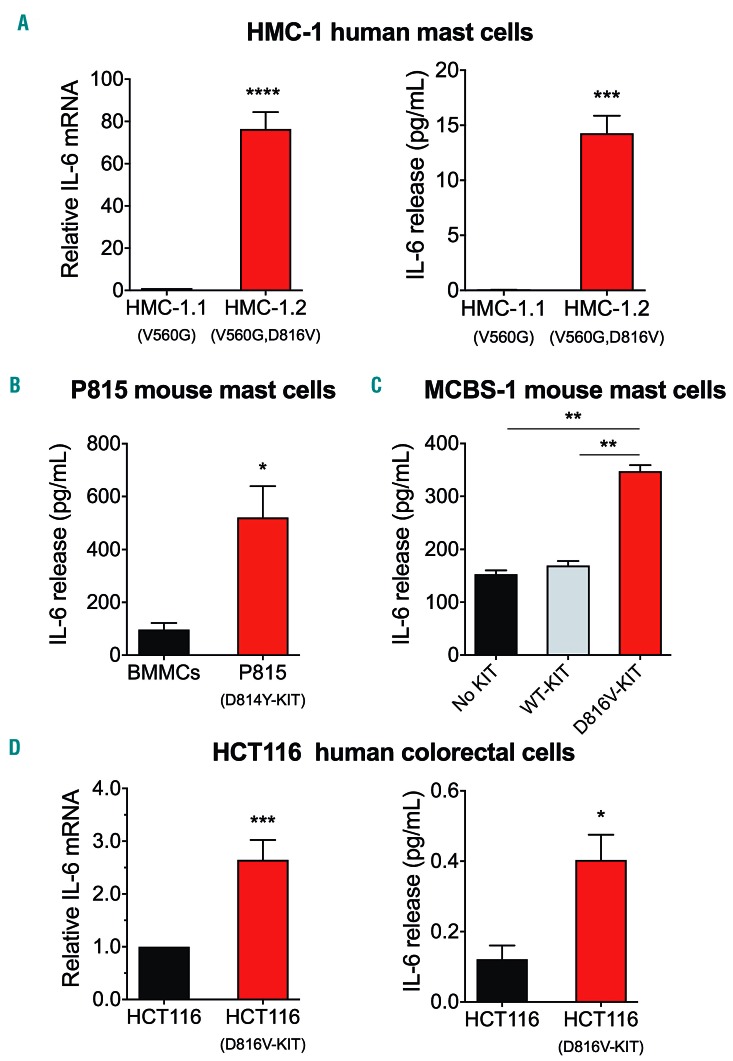
The levels of IL-6 in serum9 as well as the allelic frequency of D816V-KIT18 correlate with the levels of tryptase, a surrogate marker of mast cell burden. As mast cells often accumulate in the BM in SM, we tested the ability of BM cells to produce IL-6 in short-term cultures. BM mononuclear cells isolated from patients with SM showed varied ability to release IL-6 into the culture media after 2-4 days, with more release observed in cells from patients with a higher BM D816V-KIT allelic frequency (Figure 1A). Although cells other than mature mast cells may express D816V-KIT,18 the release of IL-6 into the media correlated with the percentage of mast cells within BM live cells (Figure 1B), suggesting a contributory role for mast cells in IL-6 production. In addition, we analyzed intracellular IL-6 staining in single BM mast cells by flow cytometry from three separate patients (see Online Supplementary Table S1 for patients’ characteristics). Patient 1 had idiopathic anaphylaxis and did not meet criteria for SM and thus was used as a control. This patient had no detectable D816V-KIT, 0.098% of BM cells were CD3−/CD34−/KIT+/FcεRI+ (mast cells) and a minor percentage of these were IL-6 positive (0.063%) (Figure 1C, left panel). However, CD3−/CD34−/KIT+/FcεRI+ cells from Patients 2 and 3, with BM D816V frequencies of 2.7% and 5.5%, were 77% and 99% positive for IL-6, respectively (Figure 1C, middle and right panels, respectively). In this ex vivo experiment, where BM cells were cultured up to 4 days (d) in the presence of SCF, cell lineages other than mast cells (KIT+/FcεRI−, KIT−/FcεRI+ and KIT−/FcεRI−) also showed positive intracellular IL-6 staining (Online Supplementary Table S2). However, the highest IL-6 mean fluorescence intensity (MFI) was associated with KIT+ cells. As SCF in the media may induce IL-6 directly or through autocrine/paracrine signals in clonal and non-clonal cells in these cultures, we examined the expression of IL-6 in BM biopsies by immunofluorescence (IF). IF images of BM of patients with SM indicated that IL-6 intracellular content was mostly associated with mast cells (Online Supplementary Figure S1). Thus, the combination of ex vivo and in situ experiments suggests mast cells as the predominant producers of IL-6, with variable participation of other cell lineages. Data are also consistent with an association between increased IL-6 expression by BM mast cells and D816V-KIT frequency.
Figure 1.
Production of IL-6 by bone marrow cells and mast cells is enhanced in patients with systemic mastocytosis (SM) in association with the D816V-KIT allelic burden. (A) IL-6 release from bone marrow mononuclear cells isolated from five patients with SM with D816V-KIT bone marrow allelic frequencies of <5 (Patients 4 and 5 in Online Supplementary Table S1) or >5 (Patients 6, 7, and 10). BM aspirates were cultured for 2-4 days and IL-6 released into the culture media was measured by ELISA. Data are the mean±Standard Error of Mean (SEM). (B) Correlation between IL-6 released into the media by BM cells and the percentage of mast cells in those cultures which was determined by flow cytometry. (C) Flow cytometry histograms showing intracellular IL-6 staining in BM mast cells. Mast cells were gated as CD3−/CD34−/KIT3/FcεRI+ within the BM cells of three patients (Patients 1-3 in Online Supplementary Table S1), with D816V-KIT allelic frequencies in the bone marrow also indicated in the figure. Patient 1 had idiopathic anaphylaxis and did not meet criteria for SM but was used as a control. The percentage of IL-6 positive cells within the mast cell population is indicated in the histograms. ISM: indolent SM.
Expression of D816V-KIT causes IL-6 upregulation and secretion
Given these associations and that mast cells and their progenitors often carry somatic D816V-KIT variants in SM, we investigated the hypothesis that D816V-KIT expression intrinsically promotes IL-6 production. Thus, we analyzed the production of IL-6 at the protein and message levels in various cell lines expressing or not D816V-KIT. In agreement with our previous observations, the HMC-1.2 mastocytosis mast cell line which harbors KIT with D816V plus another missense variant in the juxta membrane domain of KIT (V560G), showed markedly higher IL-6 mRNA synthesis13 (Figure 2A, left panel) and IL-6 release than HMC-1.1 cells, which express only the monoallelic V560G-KIT variant (Figure 2A, right panel).
Figure 2.
Cells with D816V-KIT constitu-tively express and release IL-6. (A) IL-6 mRNA expression (left) and IL-6 released into the media (right) by the mastocytosis cell lines HMC-1.1 (with V560G) and HMC-1.2 (with V560G and D816V) after 2 hours (h) in serum-free media. (B and C) Comparison of IL-6 released into the media by the mouse P815 mastocytoma mast cell line (with D814Y-KIT) compared to normal murine bone marrow mononuclear cells (BMMC) (B), and by the murine mast cell line MCBS-1 (which lacks c-Kit) transfected with human KIT or D816V-KIT compared to MCBS-1 transfected with vector alone (C). (D) Comparison of IL-6 mRNA expression (left) and IL-6 released into the media (right) by the human colorectal carcinoma cell line HCT116 expressing or not D816V-KIT. HCT116 cells were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin overnight. IL-6 mRNA expression was determined by quantitative-real-time polymerase chain reaction (q-RT-PCR) and relative expression was calculated in relationship to the expression of GAPDH using the DCt method and expressed as fold change compared to HMC-1.1 (A, left), or the HCT116 parental cell line (D). All data (A-D) are the results of three independent experiments done in triplicates. *P=0.01; **P<0.01; ***P<0.001 and ****P<0.0001.
The mastocytoma mouse mast cell line P815 carrying the homolog to the D816V-KIT variant (D814Y-KIT), also released significantly more IL-6 than primary mouse BMMC (Figure 2B). Furthermore, expression of human D816V-KIT in an immortalized mouse mast cell line that lacks KIT,13,19 unlike cells expressing normal KIT or vector alone, released significant amounts of IL-6 into the media (Figure 2C). In aggregate, the results using the various mast cell lines demonstrate an association between expression of D816V-KIT in mast cells and persistent transcription and release of IL-6. This may not be restricted to mast cells, since introduction of D816V-KIT by CRISPR in the colorectal carcinoma cell line HCT116 also promoted IL-6 transcription and release (Figure 2D, left and right panels, respectively) when cells were stimulated with PMA and ionomycin, indicating that in these cells D816V-KIT primes HCT116 cells for more robust IL-6 production.
KIT tyrosine kinase activity is required for IL-6 production in mast cells
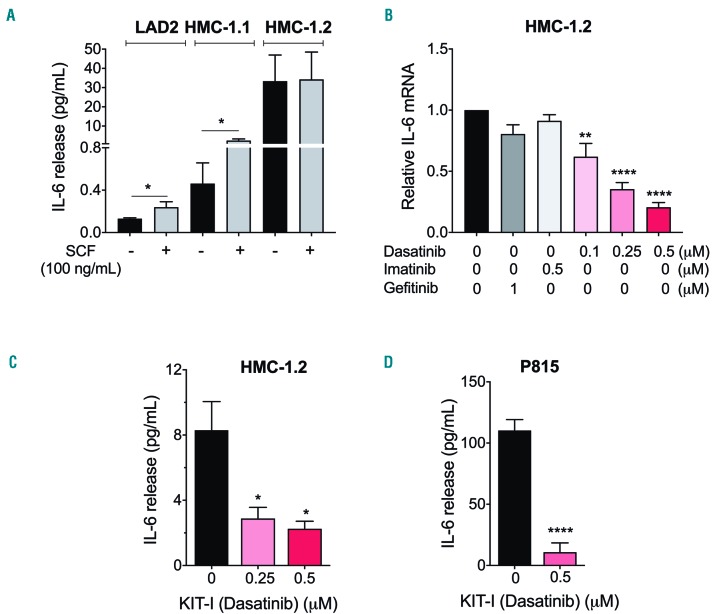
Ligand-activated KIT signaling induces IL-6 production in mast cells and enhances IgE-receptor-driven IL-6 production.20 Indeed, stimulation of KIT by SCF in the LAD2 cell line, which expresses normal KIT, induced IL-6 release, albeit the amounts were quantitatively limited (Figure 3A). HMC-1.1 showed higher production of IL-6 than LAD2 cells, particularly when stimulated with SCF (Figure 3A). However, not only was IL-6 production approximately 100-fold higher in unstimulated HMC-1.2 than in LAD2 or HMC-1.1 cells, but ligand-induced stimulation of KIT did not cause any further IL-6 secretion by HMC-1.2 cells (Figure 3A). These data are consistent with the conclusion that ligand-independent signals induced by oncogenic KIT activity, particularly those from D816V-KIT, are more effective in inducing IL-6 secretion than ligand-activated KIT signals.
Figure 3.
D816V-KIT oncogenic activity drives ligand-independent IL-6 induction. (A) IL-6 released to the extracellular media was measured in LAD2, HMC-1.1 and HMC-1.2 cells incubated for 48 hours (h) (37C, 5%CO2) in serum-free medium in the presence or absence of 100 ng/mL stem cell factor (SCF). (B) IL-6 mRNA levels were measured in HMC-1.2 after 2 h incubation in serum-free medium in the presence or absence of the KIT inhibitors, dasatinib and imatinib, or the EGFR inhibitor gefitinib at the indicated concentrations. Relative expression of IL-6 mRNA was obtained by normalizing to the expression of GAPDH using the DCt method and the results are expressed as fold change compared to untreated cells. The effect of the KIT inhibitor dasatinib (0.5 mM) on the secretion of IL-6 by HMC-1.2 (C) or by P815 mast cells (D) was determined after 6 h incubation in serum free medium. All data are the mean±Standard Error of Mean of three independent experiments done in triplicates. SCF: stem cell factor. *P=0.01, **P<0.01 and ****P<0.0001.
We next blocked D816V-KIT activity in HMC-1.2 cells with dasatinib, a tyrosine kinase inhibitor that effectively suppresses the activity of the active, open conformation of D816V-KIT. Dasatinib markedly reduced IL-6 mRNA levels (Figure 3B) and IL-6 secretion (Figure 3C), while concentrations of the tyrosine kinase inhibitor imatinib that are ineffective in blocking D816V-KIT, did not alter IL-6 mRNA levels (Figure 3B), further suggesting an involvement of D816V-KIT signaling. Inhibition by gefitinib of the epidermal growth factor receptor (EGFR), a tyrosine kinase receptor that can drive IL-6 production in transformed cells,21 had no significant effects on IL-6 mRNA levels in HMC-1.2 cells (Figure 3B). Similar to HMC-1.2 cells, dasatinib effectively inhibited IL-6 release in P815 murine mastocytoma cells (Figure 3D). In contrast, persistent IL-6 production in HMC-1.2 cells did not appear to be mediated via feed-forward loops of activation by other receptors as those reported for the IL-6 receptor (IL-6R),22 sphingosine-1-phosphate receptors23 or the TGFβ receptor24 in other neoplastic cells, since it was not altered by specific blockage of these receptors (Online Supplementary Figure S2A-C). Overall, these data suggest that signals from D816V-KIT can promote ligand-independent IL-6 production without involving some of the most common autocrine feed-forward loops described in malignant cells.
The constitutive release of IL-6 by unstimulated HMC-1.2 was enhanced by stimuli such as complement component 5a (C5a), IL-1b, 10% FBS and PMA/ionomycin (Online Supplementary Figure S3A, left panel) suggesting that the production and secretion of IL-6 due to D816V-KIT can synergize with other complementary signals or environmental cues. Of interest, the intracellular content of IL-6 protein was only approximately 10% of the total IL-6 released (Online Supplementary Figure S3A), and although dasatinib did not affect this percentage, it also reduced the total intracellular content. Thus, dasatinib inhibited the transcription, intracellular content and release of IL-6 (Figure 3B and C, and Online Supplementary Figure S3A, right panel), consistent with the conclusion that D816V-KIT signals cause constitutive de novo production and release of IL-6 without regulating storage. In addition, we demonstrate that IL-6 protein released by HMC-1.2 cells is biologically active since conditioned media of these cells caused IL-6R-mediated STAT3 phosphorylation in LAD2 cells which express and respond to IL-6R activation12 (Online Supplementary Figure S3B).
D816V-KIT-induced IL-6 production is dependent on JAK2, ERK and PI3K pathways
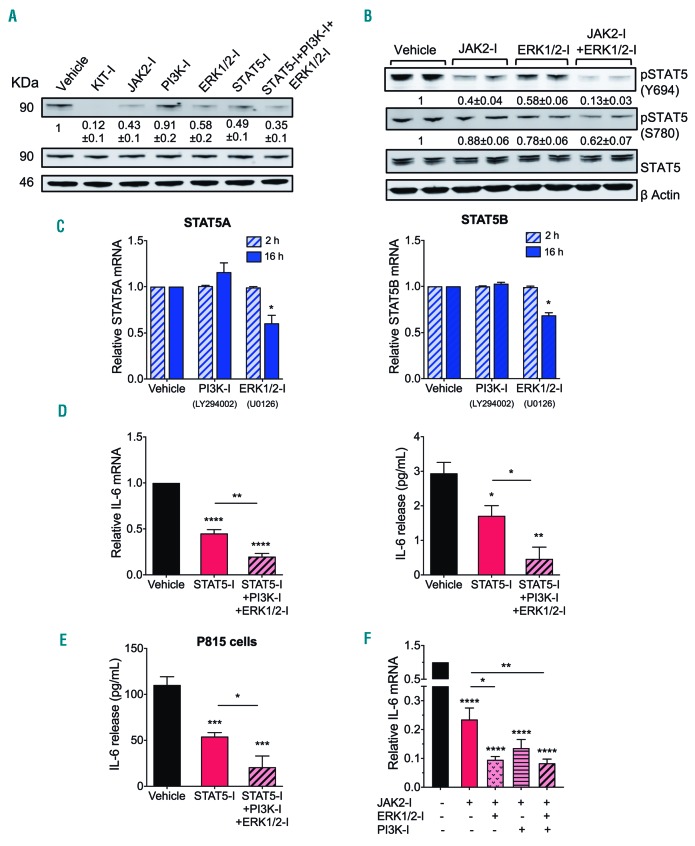
Mitogen-activated protein kinase (ERK1/2 and p38) and PI3K pathways are part of the oncogenic signals derived from D816V-KIT activity.2,25 Since these pathways may affect IL-6 expression,26–28 we investigated their potential roles in D816V-KIT-induced IL-6 production. While inhibition of p38 with SB203580 had minor effects on IL-6 synthesis, inhibition of the ERK1/2 pathway using a MEK1/2 inhibitor (U0126) (Figure 4A) or inhibition of the PI3K pathway by the PI3Kα/δ/β inhibitor LY294002 (Figure 4B), caused a 50-60% reduction, respectively, in IL-6 production at the message (Figure 4B, left panel) and protein levels (Figure 4B, right panel).
Figure 4.
MEK/ERK-, PI3K- and JAK2-mediated pathways are independently activated by D816V-KIT and contribute to ligand-independent IL-6 induction. (A and B) Effect of inhibition of MEK/ERK1/2 (U0126), p38 (SB203580) (A) or PI3K pathways (LY294002) (B) on the expression of IL-6 mRNA (left) after treatment for 2 hours (h) and the release of IL-6 into the media (right) by HMC-1.2 cells for 16 h. (C) Effect of the JAK2 inhibitor fedratinib at the indicated concentrations on the expression of IL-6 mRNA (left) and the release of IL-6 into the media (right) by HMC-1.2 cells. (D) Effect of various concentrations of inhibitors for JAK1/2 (ruxolitinib) and JAK3 (tofacitinib) on the expression of IL-6 mRNA by HMC-1.2 cells. (E) Effect of inhibition of MEK/ERK1/2, PI3K or JAK2 on the production of IL-6 by mouse P815 cells after 6 h of incubation. (F) Effect of inhibitors for KIT (KIT-I: dasatinib; 0.5 mM), JAK2 (JAK2-I: fedratinib; 1 mM), PI3K (PI3K-I: LY294002; 10 mM) and MEK/ERK1/2 (ERK-I: U0126; 10 mM) on the phosphorylation of KIT, JAK2, AKT and ERK1/2 in the indicated amino acid residues. Inhibitors were incubated for 2 h in serum-free medium. Numbers under each phosphorylated band are Standard Error of Mean (SEM) of at least three experiments and represent fold changes in the relative band fluorescence (normalized to the corresponding total expression) compared to untreated cells. Relative expression of IL-6 mRNA was obtained by comparing to the expression of GAPDH using the DCt method and the results were expressed as fold change compared to untreated cells. Data represent the mean±SEM and are from three independent experiments. **P<0.01, ***P<0.001 and ****P<0.0001.
The JAK/STAT axis is also known to be prominently up-regulated by D816V-KIT activity.29,30 JAK2 is activated by SCF31 leading to STAT phosphorylation and translocation into the nucleus, where it exerts its transcriptional activity.32 Inhibition of JAK2 by the JAK2 selective inhibitor fedratinib (TG101348) markedly blocked IL-6 constitutive transcription and cytokine release (Figure 4C, left and right panels, respectively). Ruxolitinib which inhibits JAK1 in addition to JAK2, similarly inhibited IL-6 expression, although at higher concentrations than fedratinib (Figure 4D). However, tofacitinib, a pan-JAK inhibitor preferential for JAK3 and to a lesser extent JAK1, was less effective (Figure 4D). Similar to HMC-1.2 cells, in mouse P815 cells inhibition of MEK/ERK1/2, PI3K or JAK2 pathways markedly reduced IL-6 release in mouse P815 cells (Figure 4E). The data thus implicate JAK2 in D816V-KIT induced IL-6 production.
Since JAK2 has also been reported in certain cells to activate PI3K or ERK,32,33 we further investigated the potential inter-relationships between JAK2 and the ERK and PI3K pathways. While, as expected, inhibition of KIT by dasatinib blocked the phosphorylation of JAK2, AKT and ERK1/2 signaling pathways downstream of the receptor by 40-80% (Figure 4F), inhibitors of JAK2, PI3K/AKT or MEK1/2/ERK1/2 reduced phosphorylation of their respective targets but did not show significant effects on any of the others, suggesting that these signals are activated independently from each other.
D816V-KIT-induced IL-6 production is dependent on STAT5
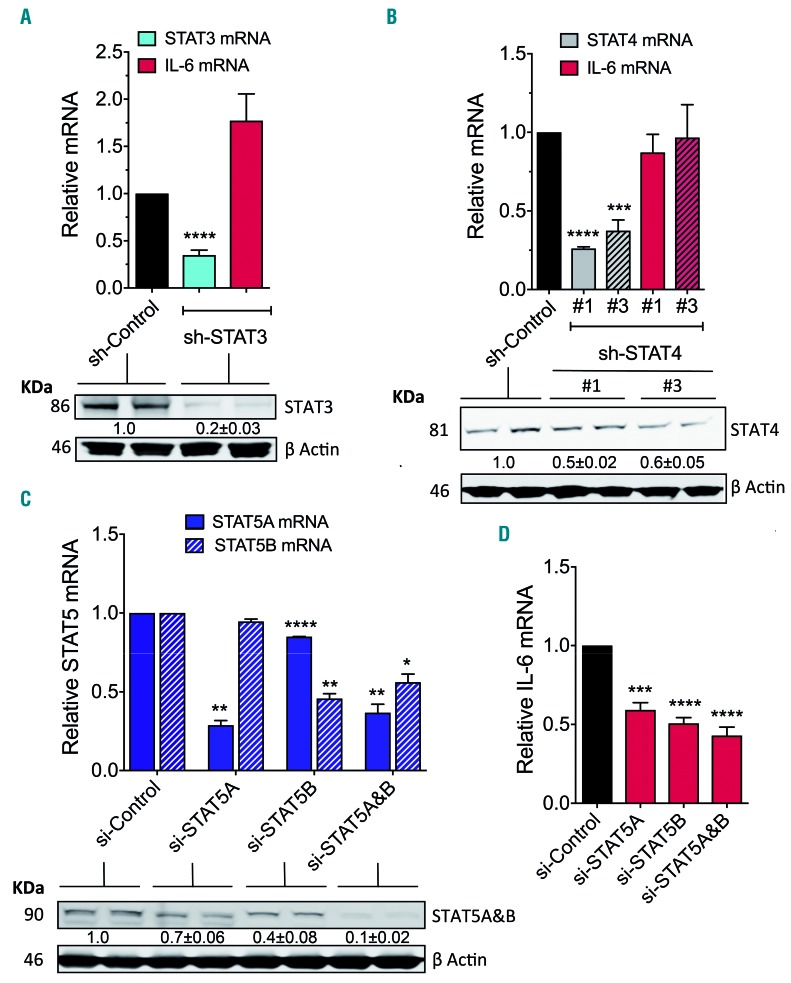
JAK phosphorylates STAT, and STAT family members such as STAT3,34,35 STAT436 and STAT537 have been implicated in the regulation of IL-6 transcription in various cells and conditions. As the levels of expression or phosphorylation of STAT3, STAT4 and STAT5 (Online Supplementary Figure S4A-C) are increased in HMC-1.2 and other cells with D816V-KIT30 and STAT4 and STAT5 are up-regulated in BM mast cells from patients with SM,29,38,39 we investigated their possible involvement in the induction of IL-6 transcription by silencing STAT3-5 expression using sh-RNA or si-RNA.
Sh-RNA-mediated STAT3 silencing resulted in >75% reduction in STAT3 at the messenger and protein levels (Figure 5A) but did not affect IL-6 production by HMC-1.2 (Figure 5A, red bar). Similarly, a selective small inhibitor molecule for STAT3, C188-9, at concentrations that caused >80% reduction in STAT3 phosphorylation (Online Supplementary Figure S5A, top panel) did not alter constitutive IL-6 production by these cells (Online Supplementary Figure S5A, bottom panel). Neutralizing antibodies for the IL-6R, which signals through STAT3, were also ineffective on IL-6 production (Online Supplementary Figure S2A). Reduction of STAT4 message and protein by >50% using sh-RNA knockdown was also inconsequential for IL-6 persistent production by these cells (Figure 5B).
Figure 5.
STAT5A and B are key in mediating D816V-KIT-induced persistent IL-6 production. Effect of STAT3 (A) and STAT4 (B) knockdown by lentiviral sh-RNA on their respective targets and on the expression of IL-6 mRNA by HMC-1.2 cells. For sh-STAT4, the effect of two different constructs are shown (#1 and #3). Western blot gels underneath the bar graphs represent the effect of sh-STAT3 (A) or sh-STAT4 (B) constructs on the protein levels of STAT3 and STAT4, respectively. Lysates from duplicate samples are shown. The numbers under each pair are mean±Standard Error of Mean (SEM) of at least three separate experiments and represent fold change in the relative band fluorescence compared to the sh-RNA non-target (control). Actin content was used as loading control. (C and D) Effect of silencing STAT5A, STAT5B or the combination of both STAT5A&B by si-RNA on the expression of STAT5A and STAT5B mRNA (C) and of IL-6 mRNA (D) in HMC-1.2. The blots under the bar graph in (C) show the effect of STAT5 silencing in the protein levels of STAT5A/B and the numbers under the blot, the fold change as in (A) and (B). Of note, the antibody used recognizes both STAT5A and B. *P=0.01, **P<0.01, ***P<0.001 and ****P<0.0001.
However, specific reduction in the mRNA for STAT5A or STAT5B messages by >50% and in protein expression (Figure 5C) using STAT5-specific si-RNA pools, resulted in concomitant reductions in IL-6 transcription (Figure 5D). Simultaneous knockdown of STAT5A and B did not accomplish significantly greater effects on IL-6 expression than silencing each individually (Figure 5C and D), suggesting a redundant function for the two isoforms. The selective STAT5 inhibitor (CAS 285986-31-4), at a concentration that inhibited STAT5 phosphorylation by 50% (50 mM) (Online Supplementary Figure S5B, top panel) also significantly reduced IL-6 transcription and IL-6 protein synthesis by 50% (Online Supplementary Figure S5B, bottom panel, and S5C), a result that was also confirmed in P815 cells (Online Supplementary Figure S5D). As STAT5 is over-expressed and hyperactivated in cells carrying D816V (Online Supplementary Figure S4C)29,38,39 and STAT5 mRNA was not depleted even when both STAT5A and B were silenced (Figure 5C), we treated STAT5-knockdown cells with the STAT5 inhibitor, which showed a reduction in IL-6 mRNA expression by 80% (Online Supplementary Figure S5E) consistent with the reduction observed by inhibition of JAK2. The results point towards JAK2/STAT5 as a major pathway leading to the constitutive expression of IL-6 in D816V-KIT expressing cells.
ERK contributes to STAT5 phosphorylation and expression while the effect of the PI3K pathway on IL-6 is STAT5-independent
As MAP kinases have been implicated in the phosphorylation and the activation of STAT family members,40 we sought to determine whether ERK1/2 may contribute to the induction of IL-6 by enhancing STAT5 phosphorylation. While inhibition of ERK1/2 did not affect JAK2 phosphorylation (Figure 4F), it substantially inhibited STAT5 tyrosine phosphorylation (Figure 6A) and reduced STAT5 serine (Ser780) phosphorylation (Figure 6B), with the effects on STAT5 tyrosine phosphorylation being less pronounced than those of the JAK2 and STAT5 inhibitors (Figure 6A). Simultaneous inhibition of both JAK2 and ERK1/2 markedly blunted STAT5 phosphorylation (Figure 6B). In addition, inhibition of MEK/ERK also reduced STAT5A and STAT5B mRNA expression by 16 h, an effect that was not seen by 2 h (Figure 6C). The results implicate MEK/ERK1/2 as a dual regulator of STAT5 activity (via JAK2-independent phosphorylation) and STAT5 transcription. In contrast, inhibition of PI3K had no effect on STAT5 phosphorylation or transcription in HMC-1.2 cells (Figure 6A and C), indicating the PI3K pathway regulates IL-6 independently of STAT5.
Figure 6.
MEK/ERK signaling pathway co-operates with JAK2 in the regulation of STAT5 to induce IL-6 while PI3K regulates IL-6 independently of STAT5. (A) Effect of inhibitors for KIT (KIT-I: dasatinib; 0.5 mM), JAK2 (JAK2-I: fedratinib; 1 mM), PI3K (PI3K-I: LY294002; 10 mM), MEK/ERK1/2 (ERK-I: U0126; 10 mM), STAT5 (STAT5-I: CAS285986-31-4; 50 mM), or their combination on the protein and phosphorylation levels of STAT5 in HMC-1.2 cells. (B) Effect of inhibitors of JAK2 and ERK1/2 alone or in combination on STAT5 phosphorylation in HMC-1.2 cells. (A and B) Lysates were obtained after 2 hours (h) incubation with the indicated inhibitors in serum-free medium. Data under the blots are the mean±Standard Error of Mean (SEM) of three independent experiments carried out in duplicates, and represent fold change in the relative band fluorescence compared to the untreated. STAT5 content was used to normalize the data. (C) Inhibition of MEK/ERK1/2 reduces STAT5A and STAT5B expression ((left and right, respectively) after 16 h incubation. Relative expression of STAT5 mRNA was obtained by comparison with the expression of GAPDH using the DCt method and the results are expressed as fold change compared to untreated cells at each time (2 h or 16 h). (D and E) Inhibitors for STAT5, PI3K and MEK/ERK1/2 additively prevent IL-6 mRNA induction (D, left) and IL-6 release in HMC-1.2 cells (D, right) and P815 cells (E). The release of IL-6 into the media was determined after 6 h. (F) Inhibitors for JAK2, PI3K and MEK/ERK1/2 additively prevent IL-6 mRNA induction in HMC-1.2 cells. All data represent mean±Standard Error of Mean of at least three independent experiments. *P=0.01, **P<0.01, ***P<0.001 and ****P<0.0001.
Combination treatment of inhibitors for STAT5, ERK1/2 and PI3K markedly suppressed constitutive IL-6 expression (Figure 6D, left panel) and release (Figure 6D, right panel) in HMC-1.2 cells and in P815 cells (Figure 6E). Similarly, the JAK2 inhibitor fedratinib, currently in clinical trials, nearly ablated IL-6 production in HMC-1.2 cells when in combination with either (or both) PI3K and ERK1/2 inhibitors (Figure 6F), indicating the separate contributions of these pathways to persistent IL-6 induction.
Discussion
IL-6 plays important roles in host defense, but if produced in an uncontrolled or persistent manner it may be detrimental and contribute to the development of inflammatory diseases (e.g. rheumatoid arthritis and inflammatory bowel disease) and malignancies.7,8,41 Although little is known about the exact role of IL-6 in mastocytosis and how it is dysregulated, the levels of IL-6 in circulation correlate with mast cell burden and tissue involvement, osteoporosis,9,11 and risk of progression.10 Our data suggest a contributory role for mast cells in IL-6 production in mastocytosis and implicate aberrant signaling of D816V-KIT as an initial event promoting persistent IL-6 transcription and consequent protein secretion. Among these aberrant signals, we identified increased JAK2 activity and MEK/ERK- and PI3K-derived signals as drivers of IL-6 transcription, the former two by regulating the activation/expression of STAT5. The study provides the first clues into mechanisms leading to persistent IL-6 production in mastocytosis and potential target molecules for therapeutic intervention.
Increased expression of IL-6 and its signaling through STAT3 in many malignancies can be driven by overexpression of IL-6R, co-receptors and regulators (such as JAK and STAT3), polymorphisms in the IL-6 promoter and/or oncogenic signaling from tyrosine kinase receptors such as EGFR/HER, and may occur when negative regulation is not fully effective.34 Often, in malignant cells, production of IL-6 and constitutive STAT3 activation drive their own expression in feed-forward regulatory loops that are key for tumorigenesis.22,23,34,41 Here, we show that upregulation of IL-6 in mast cells with the D816V-KIT missense variant does not involve the IL-6R and co-receptor gp130 (Online Supplementary Figure S2A) or feedback loops involving STAT3 (Figure 5). Moreover, STAT4, shown to enhance IL-6 transcription in human fibroblasts,36 had no role in IL-6 upregulation in HMC-1.2 mast cells. Instead, we demonstrate that persistent IL-6 production was dependent on oncogenic D816V-KIT activity and aberrant STAT5 activation, as it was suppressed by tyrosine kinase inhibitors that effectively block D816V-KIT tyrosine kinase activity and by STAT5 silencing or inhibition. In addition, BM mast cells from patients with mastocytosis produced and released IL-6 ex vivo in correlation with D816V-KIT allelic burden (Figure 1). As STAT5 expression and phosphorylation are also up-regulated in BM mast cells of patients with SM,29,38,39 our data suggest that oncogenic STAT5 activation may be a priming event contributing to the elevated serum IL-6 levels in mastocytosis. This does not exclude the involvement of other mechanisms such as IL-6-mediated feed-forward loops on other cell types in the surrounding tissue, which may drive IL-6 production further. In addition, it is important to note that, although most of the intracellular IL-6 staining was associated with mast cells in patient’s BM biopsies (Online Supplementary Figure S1), enhanced production of IL-6 in SM may not be restricted to mast cells as the presence of D816V-KIT may also induce or promote IL-6 production in other clonal cells (Figure 2D and Online Supplementary Table S2). Furthermore, additional signals in the local environment could crosstalk with oncogenic KIT signals in mast cells or other hematopoietic clonal cells in SM with associated multilineage involvement, further contributing to IL-6 dysregulation.
Even though canonical binding sites for STAT3, but not STAT5, are recognized in the IL-6 promoter, stimulation of STAT5 by the IgE receptor in mast cells was reported to mediate IL-6 production37 and constitutively active STAT5 mutants to induce IL-6 expression.42 Whether active STAT5 in D816V-KIT mast cells binds directly to the IL-6 promoter driving transcription, or does so indirectly by binding other transcription factors42 or by causing chromatin remodeling,43 needs further evaluation. Regardless of this, as STAT5 hyperactivation is critical for neoplastic D816V-KIT mast cell growth and survival39,44 and for normal mast cell development,45 our description of a novel regulatory role for STAT5 on constitutive IL-6 expression supports targeting STAT5 for treatment of patients with mastocytosis and high IL-6 levels. This could also be a potential treatment for patients with other hematologic malignancies such as chronic myelogenous leukemia where serum IL-6 levels represent a predictor of outcome46,47 and pathogenesis is in part driven by constitutive STAT5 activation.48
The activation of STAT5 by D816V-KIT was complex and involved an interplay of JAK2- and MEK/ERK1/2-mediated pathways. JAK2-selective inhibitors such as fedratinib markedly reduced constitutive IL-6 production, while the effectiveness of other panjakinib correlated with their relative selectivity for JAK2, suggesting that JAK1 and 3 have no significant role. The specific involvement of JAK2 agrees with the notion that STAT5 is a major target for JAK2 in hematopoietic cells29 and that activation of KIT by SCF causes JAK2 phosphorylation.31 Unlike the known effect on JAK2 inhibition, the effect of MEK/ERK1/2 inhibition on STAT5 activity and transcript abundance was unexpected. Phosphorylation in serine/threonine residues of STAT1/3/4 by MAPK was reported to affect STAT1/3/4 activity.33,40 Although serine phosphorylation at the C-terminal tail of STAT5 proteins is essential for leukemogenesis49 and for growth hormone-induced gene expression,50 the serine/threonine kinases that mediate this type of phosphorylation or the exact functional implications are not well understood. Surprisingly, in cells with D816V-KIT, inhibition of MEK/ERK1/2 caused a more pronounced effect on STAT5 phosphorylation at tyrosine 694 (a target for JAK2) than at serine 780 (Figure 6), even though JAK2 activity was not affected by the ERK inhibitor (Figure 4). A similar JAK-independent role for ERK in STAT3 tyrosine phosphorylation was reported in plasma cells for IL-6,27 but the mechanism for such regulation, and what potential kinases or phosphatase may be involved, need further evaluation.
We also found a role for PI3K/AKT-dependent pathways on IL-6 induction in D816V-KIT mast cells, but in a STAT5-independent manner, indicating that persistent IL-6 expression likely involves multiple transcription factors. Similar to STAT5,39,44 oncogenic growth and survival of mast cells with D816V-KIT is also dependent on constitutive activation of AKT.2,30 Thus, combined targeting of STAT5 and PI3K/AKT pathways may be desirable to treat patients with mastocytosis and elevated IL-6, as was proposed for drug-resistant chronic myelogenous leukemia where both pathways are critical for disease evolution.44 Alternatively, since small molecule inhibitors for JAK2 such as fedratinib and the dual JAK1/2 inhibitor ruxolitinib inhibit STAT5 activity and IL-6 upregulation in neoplastic mast cells (Figures 4 and 6), and both drugs are approved by the US Federal Drug Administration for various blood disorders, they are also feasible drug candidates alone or in combination with other therapies for increased therapeutic index.
The work presented here links expression of the D816V-KIT variant with IL-6 persistent activation in mast cells and sheds light into the molecular mechanisms driving dysregulated IL-6. Our data underscore a role for constitutive STAT5 activation, achieved by both JAK2 and ERK- mediated activities, and PI3K/AKT signals on IL-6 dysregulation. Together, our findings establish the groundwork for exploring new potential therapeutic combinations targeting the mentioned kinases in the treatment of patients with mastocytosis.
Acknowledgments
We thank Robin Eisch in her role as protocol study coordinator, Linda Scott, the nurse practitioner on the study protocols, Pahul Hanjra and Irina Maric for providing information on patients and Daly Cantave for arranging for patient bone marrow samples.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/1/124
Funding
This work was supported by the Division of Intramural Research within the National Institute of Allergy and Infectious Diseases (NIAID), at the National Institutes of Health.
References
- 1.Valent P, Akin C, Hartmann K, et al. Advances in the Classification and Treatment of Mastocytosis: Current Status and Outlook toward the Future. Cancer Res. 2017;77(6):1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruse G, Metcalfe DD, Olivera A. Functional deregulation of KIT: link to mast cell proliferative diseases and other neoplasms. Immunol Allergy Clin North Am. 2014;34(2):219–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valent P, Akin C, Metcalfe DD. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood. 2017;129(11):1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metcalfe DD, Mekori YA. Pathogenesis and Pathology of Mastocytosis. Annu Rev Pathol. 2017;12:487–514. [DOI] [PubMed] [Google Scholar]
- 5.Burger R. Impact of Interleukin-6 in Hematological Malignancies. Transfus Med Hemoth. 2013;40(5):336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26(1):54–74. [DOI] [PubMed] [Google Scholar]
- 7.Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for translational therapeutics. Cancer. 2007;110(9):1911–1928. [DOI] [PubMed] [Google Scholar]
- 8.Rossi JF, Lu ZY, Jourdan M, Klein B. Interleukin-6 as a therapeutic target. Clin Cancer Res. 2015;21(6):1248–1257. [DOI] [PubMed] [Google Scholar]
- 9.Brockow K, Akin C, Huber M, Metcalfe DD. IL-6 levels predict disease variant and extent of organ involvement in patients with mastocytosis. Clin Immunol. 2005;115(2):216–223. [DOI] [PubMed] [Google Scholar]
- 10.Mayado A, Teodosio C, Garcia-Montero AC, et al. Increased IL6 plasma levels in indolent systemic mastocytosis patients are associated with high risk of disease progression. Leukemia. 2016;30(1):124–130. [DOI] [PubMed] [Google Scholar]
- 11.Theoharides TC, Boucher W, Spear K. Serum interleukin-6 reflects disease severity and osteoporosis in mastocytosis patients. Int Arch Allergy Immunol. 2002;128(4):344–350. [DOI] [PubMed] [Google Scholar]
- 12.Desai A, Jung MY, Olivera A, et al. IL-6 promotes an increase in human mast cell numbers and reactivity through suppression of suppressor of cytokine signaling 3. J Allergy Clin Immunol. 2016;137(6):1863–1871 e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DK, Beaven MA, Kulinski JM, et al. Regulation of Reactive Oxygen Species and the Antioxidant Protein DJ-1 in Mastocytosis. PLoS One. 2016;11(9): e0162831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horny HP, Metcalfe DD, Akin C, et al. Mastocytosis. In: Swerdlow SH, Campo E, Harris NL, Jaffee ES, Pileri SA, Stein H, et al., eds. WHO classification of tumours of haematopoietic and lymphoid tissues. 2017 ed. Lyon, France: IARC Press, 2017:62–69. [Google Scholar]
- 15.Kristensen T, Vestergaard H, Moller MB. Improved detection of the KIT D816V mutation in patients with systemic masto-cytosis using a quantitative and highly sensitive real-time qPCR assay. J Mol Diagn. 2011;13(2):180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DK, Beaven MA, Metcalfe DD, Olivera A. Interaction of DJ-1 with Lyn is essential for IgE-mediated stimulation of human mast cells. J Allergy Clin Immunol. 2017;142(1):195–206 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel AM, Stone KD, Cruse G, et al. Diminished allergic disease in patients with STAT3 mutations reveals a role for STAT3 signaling in mast cell degranulation. J Allergy Clin Immunol. 2013;132(6):1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Moller MB, Mastocytosis Centre Odense University H Serum tryptase correlates with the KIT D816V mutation burden in adults with indolent systemic mastocytosis. Eur J Haematol. 2013;91(2):106–111. [DOI] [PubMed] [Google Scholar]
- 19.Smrz D, Bandara G, Zhang S, et al. A novel KIT-deficient mouse mast cell model for the examination of human KIT-mediated activation responses. J Immunol Methods. 2013;390(1-2):52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Smrz D, Jung MY, et al. Stem cell factor programs the mast cell activation phenotype. J Immunol. 2012;188(11):5428–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao SP, Mark KG, Leslie K, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117(12):3846–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13(1):7–9. [DOI] [PubMed] [Google Scholar]
- 23.Lee H, Deng J, Kujawski M, et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16(12):1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao Z, Fenoglio S, Gao DC, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 2010;107(35):15535–15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harir N, Boudot C, Friedbichler K, et al. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112(6):2463–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, Gerl RE, Schrader JW. Defining the involvement of p38alpha MAPK in the production of anti- and proinflammatory cytokines using an SB 203580-resistant form of the kinase. J Biol Chem. 2003;278(25): 22237–22242. [DOI] [PubMed] [Google Scholar]
- 27.Kopantzev Y, Heller M, Swaminathan N, Rudikoff S. IL-6 mediated activation of STAT3 bypasses Janus kinases in terminally differentiated B lineage cells. Oncogene. 2002;21(44):6791–6800. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Lan T, Zhang C, et al. Reciprocal activation between IL-6/STAT3 and NOX4/Akt signalings promotes proliferation and survival of non-small cell lung cancer cells. Oncotarget. 2015;6(2):1031–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimwade LF, Happerfield L, Tristram C, et al. Phospho-STAT5 and phospho-Akt expression in chronic myeloproliferative neoplasms. Br J Haematol. 2009; 147(4):495–506. [DOI] [PubMed] [Google Scholar]
- 30.Lasho T, Tefferi A, Pardanani A. Inhibition of JAK-STAT signaling by TG101348: a novel mechanism for inhibition of KITD816V-dependent growth in mast cell leukemia cells. Leukemia. 2010;24(7):1378–1380. [DOI] [PubMed] [Google Scholar]
- 31.Morales JK, Falanga YT, Depcrynski A, Fernando J, Ryan JJ. Mast cell homeostasis and the JAK-STAT pathway. Genes Immun. 2010;11(8):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rane SG, Reddy EP. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19(49):5662–5679. [DOI] [PubMed] [Google Scholar]
- 33.Jain N, Zhang T, Fong SL, Lim CP, Cao X. Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene. 1998;17(24):3157–3167. [DOI] [PubMed] [Google Scholar]
- 34.Chang Q, Daly L, Bromberg J. The IL-6 feed-forward loop: a driver of tumorigenesis. Semin Immunol. 2014;26(1):48–53. [DOI] [PubMed] [Google Scholar]
- 35.Yoon S, Woo SU, Kang JH, et al. NF-kappaB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene. 2012; 31(29):3467–3481. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen HN, Noss EH, Mizoguchi F, et al. Autocrine Loop Involving IL-6 Family Member LIF, LIF Receptor, and STAT4 Drives Sustained Fibroblast Production of Inflammatory Mediators. Immunity. 2017; 46(2):220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnstein BO, Li G, Wang Z, et al. Stat5 expression is required for IgE-mediated mast cell function. J Immunol. 2006; 177(5):3421–3426. [DOI] [PubMed] [Google Scholar]
- 38.Teodosio C, Garcia-Montero AC, Jara-Acevedo M, et al. Gene expression profile of highly purified bone marrow mast cells in systemic mastocytosis. J Allergy Clin Immunol. 2013;131(4):1213–1224. [DOI] [PubMed] [Google Scholar]
- 39.Baumgartner C, Cerny-Reiterer S, Sonneck K, et al. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: subcellular distribution and role of the transforming oncoprotein KIT D816V. Am J Pathol. 2009;175(6):2416–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000; 19(21):2628–2637. [DOI] [PubMed] [Google Scholar]
- 41.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15(2):79–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawashima T, Murata K, Akira S, et al. STAT5 induces macrophage differentiation of M1 leukemia cells through activation of IL-6 production mediated by NF-kappaB p65. J Immunol. 2001;167(7):3652–3660. [DOI] [PubMed] [Google Scholar]
- 43.Xu R, Nelson CM, Muschler JL, Veiseh M, Vonderhaar BK, Bissell MJ. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J Cell Biol. 2009;184(1): 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bibi S, Arslanhan MD, Langenfeld F, et al. Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: possible new targets of therapy. Haematologica. 2014;99(3):417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shelburne CP, McCoy ME, Piekorz R, et al. Stat5 expression is critical for mast cell development and survival. Blood. 2003; 102(4):1290–1297. [DOI] [PubMed] [Google Scholar]
- 46.Nievergall E, Reynolds J, Kok CH, et al. TGF-alpha and IL-6 plasma levels selectively identify CML patients who fail to achieve an early molecular response or progress in the first year of therapy. Leukemia. 2016;30(6): 1263–1272. [DOI] [PubMed] [Google Scholar]
- 47.Welner RS, Amabile G, Bararia D, et al. Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell. 2015;27(5):671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson EA, Walker SR, Weisberg E, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117(12):3421–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedbichler K, Kerenyi MA, Kovacic B, et al. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood. 2010;116(9):1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pircher TJ, Petersen H, Gustafsson JA, Haldosen LA. Extracellular signal-regulated kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol Endocrinol. 1999;13(4):555–565. [DOI] [PubMed] [Google Scholar]