Abstract
Homoharringtonine, a plant alkaloid, has been reported to suppress protein synthesis and has been approved by the US Food and Drug Administration for the treatment of chronic myeloid leukemia. Here we show that in acute myeloid leukemia (AML), homoharringtonine potently inhibits cell growth/viability and induces cell cycle arrest and apoptosis, significantly inhibits disease progression in vivo, and substantially prolongs survival of mice bearing murine or human AML. Strikingly, homoharringtonine treatment dramatically decreases global DNA 5-hydroxymethylcytosine abundance through targeting the SP1/TET1 axis, and TET1 depletion mimics homoharringtonine’s therapeutic effects in AML. Our further 5hmC-seq and RNA-seq analyses, followed by a series of validation and functional studies, suggest that FLT3 is a critical down-stream target of homoharringtonine/SP1/TET1/5hmC signaling, and suppression of FLT3 and its downstream targets (e.g. MYC) contributes to the high sensitivity of FLT3-mutated AML cells to homoharringtonine. Collectively, our studies uncover a previously unappreciated DNA epigenome-related mechanism underlying the potent antileukemic effect of homoharringtonine, which involves suppression of the SP1/TET1/5hmC/FLT3/MYC signaling pathways in AML. Our work also highlights the particular promise of clinical application of homoharringtonine to treat human AML with FLT3 mutations, which accounts for more than 30% of total cases of AML.
Introduction
Homoharringtonine (HHT, also known as omacetaxine mepesuccinate) is a cytotoxic alkaloid originally isolated from the cephalotaxus hainanensis.1 It has been approved by the US Food and Drug Administration (FDA) for treatment of chronic myeloid leukemia (CML) with resistance and/or intolerance to imatinib or other tyrosine kinase inhibitors.2 However, it is still awaiting approval for use in the treatment of acute myeloid leukemia (AML). In China, HHT has been used in the treatment of leukemia for more than 30 years due to its low price and high efficiency.3 In a pilot clinical trial we launched in 2006 in Zhejiang Province, China, we used an HHT-based induction regimen, namely HAA (HHT 4 mg/m2/day, days 1-3; cytarabine 150 mg/m2/day, days 1-7; aclarubicin 12 mg/day, days 1-7) to treat de novo AML and achieved a complete remission rate of 83%.4 Afterwards, a multi-center, open-label, randomized, controlled phase III trial was carried out in China and confirmed modified HAA regimen (HHT 2 mg/m2/day, days 1-7; cytarabine 100 mg/m2/day, days 1-7; aclarubicin 20 mg/day, days 1-7) as an alternative therapeutic strategy in treating de novo AML, especially for those patients with favorable and intermediate prognosis.5 A potential mechanism by which HHT exerts its biological function is through its binding to the A site of the ribosome, resulting in the inhibition of protein synthesis.6 However, it is unclear whether there is any other mechanism(s) underlying antileukemic effect of HHT, in particular in AML.
Acute myeloid leukemia is one of the most common and fatal forms of hematopoietic malignancies, characterized by blockage of myeloid differentiation and malignant proliferation of immature myeloid blasts.7 With contemporary therapies, the vast majority (over 70%) of patients with AML cannot survive over five years. Despite the common myeloid background, molecular and cytogenetic alterations contribute to the heterogeneity of the disease and the variable responses to treatment. For instance, mutations in FLT3, including internal-tandem duplications (ITD) and tyrosine kinase domain (TKD) point mutations, occur in over 30% of AML cases and are often associated with poor prognosis.7–9 Meanwhile, overexpression of FLT3 has also been reported in more than 60% of AML with a variety of AML subtypes, such as AML carrying FLT3-ITD or t(11q23) [i.e. chromosome rearrangements involving the mixed lineage leukemia (MLL) gene].9 FLT3-ITD activates multiple signaling pathways, leading to the aberrant overexpression of a set of oncogenes including MYC.10 Despite extensive efforts in developing and testing various FLT3 tyrosine kinase inhibitors (TKI) in clinical trials, the long-term therapeutic effects are still not promising,11,12 indicating that the development of more effective therapeutics to treat FLT3-mutated AML remains an unmet need.
The Ten-eleven translocation (TET) proteins (including TET1/2/3) are a family of methylcytosine dioxygenases that convert 5-methylcytosine (5mC) to 5-hydrox-ymethylcytosine (5hmC), leading to active or passive DNA demethylation.13 TET1, the founding member of the TET family, was first identified as a fusion partner of the MLL gene associated with t(10;11)(q22;q23) in AML.14,15 In contrast to the frequent loss-of-function mutations and tumor-suppressor role of TET2 observed in hematopoietic malignancies,16 we reported recently that TET1 plays a critical oncogenic role in the pathogenesis of various subtypes of AML and represents a promising therapeutic target for AML treatment.17–19 The oncogenic role of Tet1 in the development of myeloid malignancies was also observed by others.20
In the present study, we show that HHT exhibits potent anti-AML effects both in vitro and in vivo, and affects DNA epigenome by directly targeting SP1 and inhibiting SP1- mediated transcriptional regulation of TET1 expression, thereby reducing global 5hmC levels. Moreover, we demonstrate that FLT3 is a direct target of the HHT┤SP1/TET1/5hmC axis, and therefore HHT treatment substantially inhibits the FLT3/MYC pathways. Consistently, human primary FLT3-ITD AML cell samples display particularly high sensitivity to HHT treatment. Taken together, our studies reveal a previously unrecognized mechanism involving HHT-induced 5hmC reduction in treating AML, and suggest that HHT-based regimens hold great therapeutic potential for the treatment of AML, especially that carrying FLT3 mutations.
Methods
Cell lines and cell culture
MA9.3ITD (MLL-AF9 fusion gene plus FLT3-ITD mutation; MLL, also known as KMT2A) and MA9.3RAS (MLL-AF9 fusion gene plus NRASG12D mutation) were established by Dr. James Mulloy and cultured in IMDM supplemented with 20% FBS.21 MONOMAC 6, MV4-11, MOLM13, Kasumi-1, THP-1, SKNO-1 and ML-2 are obtained and maintained as previously described.22 Homoharringtonine purchased from Sigma-Aldrich was used in this study.
Mouse model
B6.SJL-Ptprc (CD45.1) mice and NOD/LtSz-scid IL2RG-SGM3 (NSGS) mice were obtained from the Jackson Lab (Bar Harbor, ME, USA), and bone marrow transplantation (BMT) or xeno-transplantation assays were carried out as previously described.17,19,22,23 All mice were maintained in the animal core facility of the University of Cincinnati. All experiments were conducted according to our research protocol which was approved by the Institutional Animal Care and Use Committee. In all mouse models, HHT (1 mg/kg body weight) or phosphate-buffered saline (PBS) was intraperitoneally injected daily for ten consecutive days from day 11. The mice were euthanized by CO2 inhalation once typical leukemic symptoms, i.e. paralysis, hunched posture, labored breathing and decreased activity, had been observed. The peripheral blood (PB), bone marrow (BM), spleen and liver samples were harvested for further analysis.
Patients’ samples
Bone marrow aspirates and PB samples were obtained from AML patients with their written informed consent. Mononuclear cells (MNC) were isolated and used for subsequent experiments. The genetic mutations were tested by the First Affiliated Hospital, Zhejiang University College of Medicine. The study was approved by the Ethics Committee of the First Affiliated Hospital, Zhejiang University College of Medicine.
DNA 5hmC sequencing
DNA samples were collected and sent for 5hmC sequencing. 5hmC library construction and sequencing were performed by Dr. Chunxiao Song’s lab as previously reported.24 The identification of 5hmC peaks in each sample was performed using MACS2,25 gene expression was analyzed by RSEM,26 5hmC and expression target analysis was performed by BETA.27 5hmC sequencing data have been deposited in the Gene Expression Omnibus (GEO) repository with the accession number GSE103144.
RNA sequencing and RNA-sequencing analysis
RNA sequencing was performed with total RNA samples isolated from MA9.3RAS and MA9.3ITD AML cells with or without HHT treatment (5 or 10 ng/mL for 48 hours) by The Genomics, Epigenomics and Sequencing Core of the University of Cincinnati. The Gene Set Enrichment Analysis (GSEA)28 was used to analyze the enriched signaling pathway in PBS or HHT treated cell samples. The RNA sequencing data have been deposited in the GEO repository with the accession number GSE103143.
Statistical analysis
Data were analyzed with GraphPad Prism 6 and were presented as mean±Standard Deviation as indicated. Two-tailed Student t-test was used to compare means between groups as indicated. P<0.05 was considered significant. The Kaplan-Meier survival curves were produced with GraphPad Prism 6 and P-values were calculated using the log rank test. The densitometric analysis of the bands from Western blot or dots from dot blot were performed with Gel-Pro analyzer and normalized to the loading controls.
A detailed description of all materials and methods is available in the Online Supplementary Appendix.
Results
Homoharringtonine potently inhibits cell growth and viability and promotes cell cycle arrest, apoptosis and differentiation in human acute myeloid leukemia
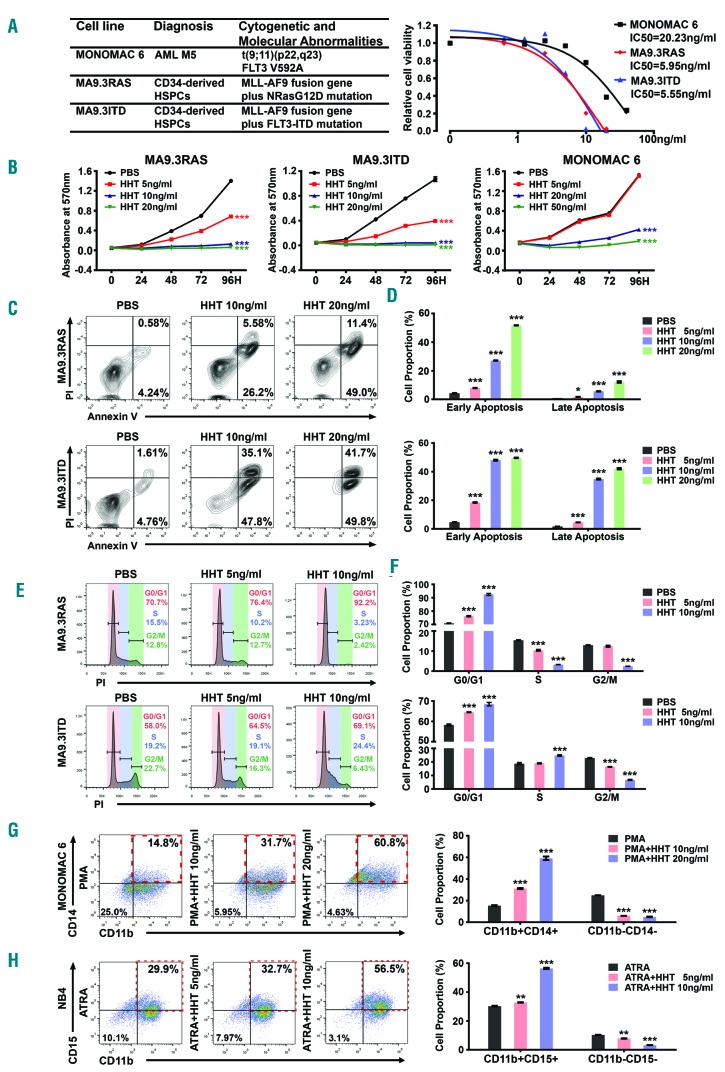
To systematically investigate the therapeutic potential of HHT in AML, we first analyzed the responses of human AML cells to HHT in vitro. Three AML cell lines with various backgrounds, including MONOMAC 6, MA9.3ITD and MA9.3RAS, were included for the analyses (Figure 1A). Remarkably, we found that all three AML cell lines were highly sensitive to HHT treatment, with very low IC50 values (5~20 ng/mL; or 9.2~36.7 nM) (Figure 1A), and HHT significantly inhibited their growth and viability in a dose- and time-dependent manner (Figure 1B and Online Supplementary Figure S1A). HHT dramatically induced apoptosis (Figure 1C and D, and Online Supplementary Figure S1B and C) and cell cycle arrest in G0/G1 phase (Figure 1E and F, Online Supplementary Figure S1D and E) in AML cells. Furthermore, we also assessed the potential effect of HHT on myeloid differentiation of MONOMAC 6 and NB4 (carrying t(15;17)/PML-RARA) AML cells. Notably, HHT also significantly promoted myeloid differentiation of AML cell as detected by both flow cytometry and qualitative polymerase chain reaction (qPCR) (Figure 1G and H, Online Supplementary Figure S1F and G), including PMA-induced monocytic differentiation and ATRA-induced granulocytic differentiation. Thus, HHT exhibits a broad-spectrum antileukemic activity involving the inhibition of AML cell growth/viability and the promotion of apoptosis, cell cycle arrest, and myeloid differentiation.
Figure 1.
Acute myeloid leukemia (AML) cells display high sensitivity to homoharringtonine (HHT) treatment in vitro. (A) (Left) Genetic information of MA9.3RAS, MA9.3ITD and MONOMAC 6 and (right) the inhibitory concentration of 50% (IC50) values with HHT treatment at 48 hours (h) for these three AML cell lines. (B) Effects of HHT treatment on cell growth/proliferation in MA9.3RAS, MA9.3ITD and MONOMAC 6 at different time points (0, 24, 48, 72 and 96 h). The colors represent different HHT concentrations (0, 5, 10, 20 ng/mL; or, 0, 9.2, 18.3, 36.7 nM). (C) Effect of HHT on apoptosis in AML cells. All the cells were treated with HHT for 48 h and representative flow cytometric plots and percentages of cell apoptosis are shown. (D) Statistical apoptosis analysis from three independent experiments determined by flow cytometry. (E) Function of HHT on cell cycle arrest in AML cells. All the cells were treated with HHT for 48 h and representative flow cytometric percentages of cell cycle phases are shown. (F) Statistical analysis of cell cycle assays from three independent experiments determined by flow cytometry. (G) Staining of CD11b and CD14 in MONOMAC 6 cells upon HHT treatment for 96 h during PMA-induced monocytic differentiation (left panel), along with statistical analysis of cell proportions of CD11b+CD14+ and CD11b−CD14− in MONOMAC 6 (right panel). (H) Staining of CD11b and CD15 in NB4 cells [carrying t(15;17)/PML-RARA; AML-M3] upon treatment with HHT for 96 h during ATRA-induced granulocytic differentiation (left panel), along with statistical analysis of cell proportions of CD11b+CD15+ and CD11b−CD15− in NB4 (right panel). Red boxes in (G) and (H) represent the differentiated cell population with double positive markers. *P<0.05; **P<0.01; ***P<0.001; t-test. Error bar, mean±Standard Deviation.
Homoharringtonine substantially inhibits murine and human acute myeloid leukemia progression in vivo
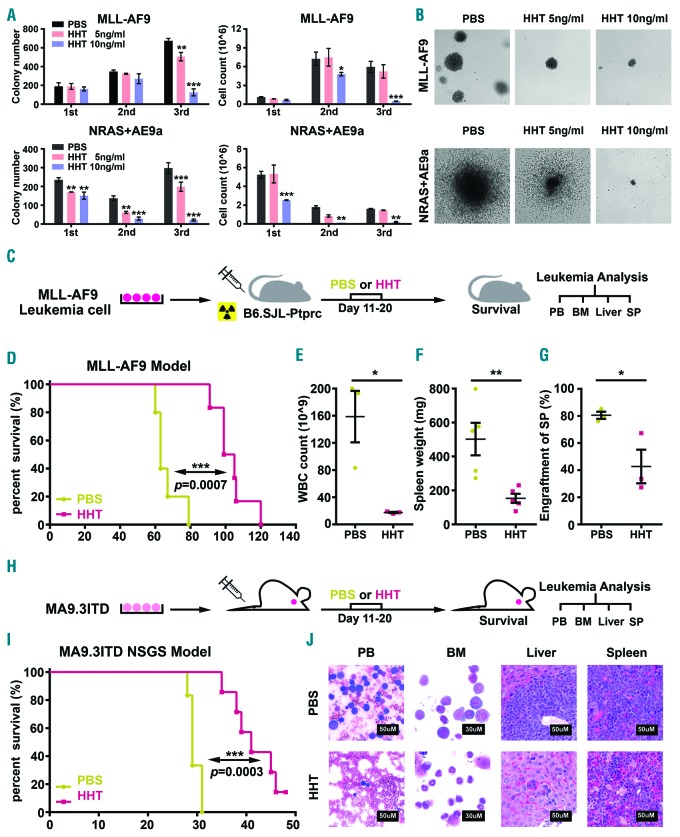
We next examined the effect of HHT on survival and proliferation of primary mouse AML cells via colony forming assays. Leukemic BM blast cells collected from primary BMT recipient mice carrying MLL-AF9- or NRAS+AE9a (AML1-ETO9a fusion gene29 plus NRASG12D)-induced full-blown AML were seeded into semi-solid medium containing PBS or HHT (5 ng/mL or 10 ng/mL) for serial plating. After three rounds of plating, HHT significantly suppressed colony-forming activity and decreased cell proliferation of primary AML cells in a dose-dependent manner (Figure 2A). HHT treatment also markedly reduced the colony size (Figure 2B).
Figure 2.
Homoharringtonine (HHT) inhibits the progression of acute myeloid leukemia (AML) in vivo. (A) Effects of HHT on colony forming activity of mouse hematopoietic stem/progenitor cells (HSPC) transformed by MLL-AF9 or NRAS plus AML-ETO9a (AE9a). Colony numbers (left panel) and cell counts (right panel) from colony forming assay (CFA) were displayed. (B) Representative images of the 3rd generation of colonies under treatment with different HHT concentrations (0, 5 and 10 ng/mL) (5× microscope). (C) Schematic illustration of secondary MLL-AF9 AML transplantation mouse model coupled with HHT or phosphate-buffered saline (PBS) treatment. (D) Kaplan-Meier curves of PBS- and HHT-treated mice that were transplanted with mouse MLL-AF9 AML cells. (E-G) White blood cell (WBC) count (E), spleen (SP) weight (F), and the engraftment ratio of leukemic cells into SP (G) at the end point of the PBS- or HHT-treated MLL-AF9 AML mice. (H) Schematic illustration of the MA9.3ITD AML xenograft NOD/LtSz-scid IL2RG-SGM3 (NSGS) model coupled with HHT or PBS treatment. (I) Kaplan-Meier curves of PBS- and HHT-treated NSGS mice that were xenotransplanted with human MA9.3ITD AML cells. (J) Wright-Giemsa staining of mouse peripheral blood (PB) and bone marrow (BM), and Hematoxylin and Eosin (H&E) staining of liver and spleen (SP) from PBS- or HHT-treated MA9.3ITD leukemic mice. Bars represent 50 mM for PB, SP and liver; 30 mM for BM. *P<0.05; **P<0.01; ***P<0.001; t-test. Error bar, mean±Standard Deviation. For Kaplan-Meier curve, P-values were calculated by log-rank test.
We then utilized leukemic mouse BMT model to assess the effect of HHT on AML progression in vivo. Briefly, primary mouse MLL-AF9 AML cells (CD45.2) were injected via tail vein (i.v.) into semi-lethally irradiated recipient mice (CD45.1). Ten days post transplantation, the recipients were treated with either HHT (1 mg/kg body weight) or PBS once daily for ten consecutive days (Figure 2C). As expected, HHT treatment significantly inhibited AML progression and substantially prolonged survival in the AML mice (102 days vs. 63 days; P=0.0007) (Figure 2D). Compared to the PBS-treated control group, HHT treatment dramatically reduced leukemic burden in PB, BM, spleen and liver in mice (Figure 2E-G and Online Supplementary Figure S2A and B).
We also employed “human-in-mouse” xenograft models to further evaluate the effect of HHT on human AML progression in vivo. Human MA9.3ITD AML cells were i.v. injected into NSGS mice and ten days post xenotransplantation, the mice were treated with PBS or HHT (1 mg/kg body weight) for ten days (Figure 2H). HHT substantially delayed leukemia progression and prolonged survival of treated mice, associated with significantly inhibited engraftment of human AML cells and remarkably reduced leukemia burden in recipient mice (Figure 2I and Online Supplementary Figure S2C-F). Pathological morphologies also identified a significant decrease of leukemia blasts in PB, BM, liver, and spleen tissues in HHT-treated group compared with the control group (Figure 2J). Similar effects of HHT treatment were also observed in NSGS mice xeno-transplanted with human MONOMAC 6 AML cells (Online Supplementary Figure S2G-I). Collectively, HHT treatment can substantially inhibit leukemia progression and prolong survival of mice carrying human or murine AML, demonstrating the potent therapeutic efficacy of HHT in treating AML.
Homoharringtonine down-regulates global 5hmC level by targeting SP1/TET1 in acute myeloid leukemia cells
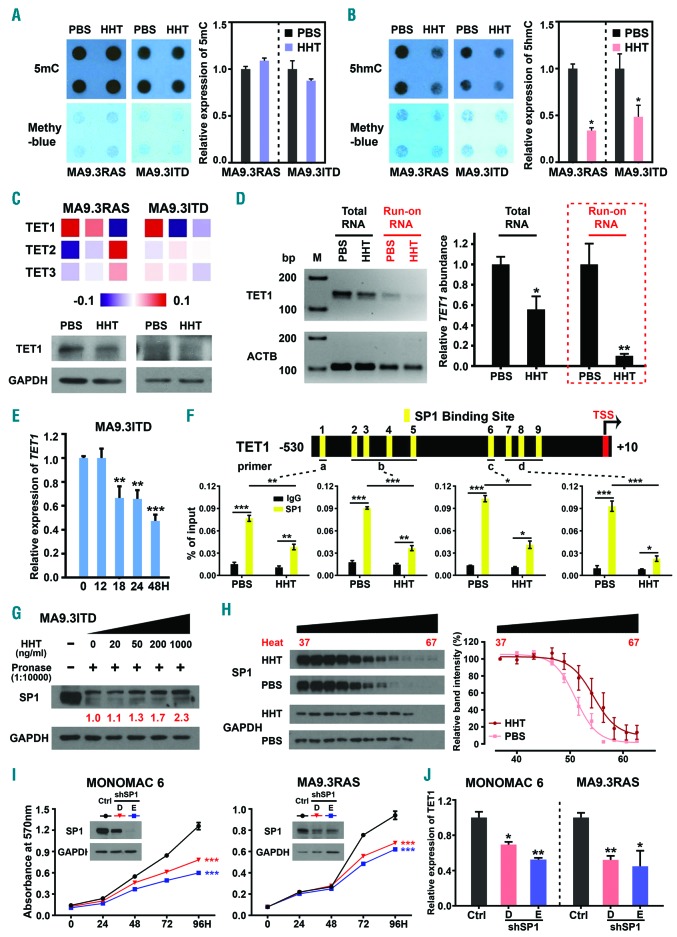
Altered epigenetic modification at DNA levels is a well-known feature of AML and displays critical effects during AML initiation, progression, and prognosis.30,31 Strikingly, we found that HHT treatment significantly reduced global 5hmC level, but not 5mC level, both in MA9.3RAS and MA9.3ITD AML cells (Figure 3A and B). Notably, amongst the genes encoding methylcytosine dioxygenase TET proteins (including TET1, TET2, and TET3) that convert 5mC to 5hmC,13 we found that only TET1, but not TET2 or TET3, was significantly down-regulated upon HHT treatment (in a dose-dependent manner) in AML cells as detected by qPCR (Online Supplementary Figure S3A). Furthermore, we also confirmed HHT-induced suppression of TET1 expression through our RNA-seq data analysis and Western blotting assay (Figure 3C). Notably, our qPCR results indicated that the significant downregulation of TET1 started as early as at 18 hours and continued afterwards in MA9.3ITD upon HHT treatment (Figure 3E). Thus, HHT-induced decrease of 5hmC level is owing to the downregulation of TET1. To further determine whether HHT-mediated TET1 inhibition is due to transcriptional inhibition, we employed nuclear run-on assay,32 with biotin-labeled uridine 5′-triphosphate (UTP) (Online Supplementary Figure S3B) and showed that HHT treatment significantly decreased the transcriptional rate of TET1 (Figure 3D), suggesting transcriptional inhibition largely contributes to HHT-induced downregulation of TET1. To elucidate the molecular mechanism by which HHT inhibits the transcription of TET1, we examined the potential binding of transcription factors (TF) on the promoter region of TET1 (−530 to +10 bp) and identified mul tiple putative binding sites of SP1 (Figure 3F, top panel, and Online Supplementary Figure S3C). SP1 is an important TF in AML and mediates drug resistance to chemotherapy.33,34 Our ChIP-qPCR confirmed the direct binding of SP1 on TET1 promoter region and such binding was remarkably decreased upon HHT treatment in AML cells (Figure 3F, bottom panel). Furthermore, we conducted drug affinity responsive target stability (DARTS) assay and cellular thermal shift assay (CETSA) to clarify whether SP1 is a potential direct binding target of HHT.35,36 Indeed, the DARTS result suggests that HHT directly binds with SP1 protein and confers drug-induced pronase-resistance for SP1 (Figure 3G). Moreover, the CETSA result confirms the direct association of HHT to SP1 and leads to the shift thermal stability of SP1 protein (Figure 3H). Finally, we showed that depletion of SP1 expression significantly inhibited AML cell growth and suppressed TET1 expression (Figure 3I and J), recapitulating the effects of HHT treatment (Figures 1B and 3C, and Online Supplementary Figure S3A). Thus, by inhibiting the binding of SP1 on TET1 promoter, HHT negatively regulates transcription of TET1. Collectively, our data reveal a previously unrecognized signal pathway involving HHT, SP1 and TET1, and identify a novel mechanism by which HHT inhibits TET1 expression and thereby reduces global 5hmC modification in AML cells.
Figure 3.
Homoharringtonine (HHT) substantially reduces global 5hmC abundance via targeting SP1/TET1 in acute myeloid leukemia (AML). (A and B) Effects of HHT on global 5mC (A) and 5hmC (B) abundance in MA9.3RAS and MA9.3ITD AML cells upon treatment with 5 ng/mL HHT for 48 hours (h). (C) Relative expression of TET1 in HHT-treated MA9.3ITD at different time points, including 0, 12, 18, 24 and 48 h. (D) Heat map showing gene expression of the individual TET family members in MA9.3RAS and MA9.3ITD AML cells treated with phosphate-buffered saline (PBS) or HHT (5 or 10 ng/mL) for 48 h as detected by RNA-sequencing (RNA-seq) (top panel), along with western blotting result of TET1 in MA9.3RAS and MA9.3ITD AML cells treated with PBS or HHT (5 ng/mL) for 48 h (bottom panel). (E) RNA levels of TET1 and ACTB in Total RNA (in black) and Run-on RNA (in red) were determined by reverse transcription polymerase chain reaction (RT-PCR) (left panel, M, Marker). Qualitative PCR (qPCR) analysis of relative TET1 abundance in Total RNA and Run-on RNA isolated from PBS- or HHT-treated MA9.3ITD cells (right panel). (F) Schematic presentation of SP1 binding sites within the promoter region of TET1 (top panel). Chromatin immune-precipitation (ChIP)-qPCR assay was used to determine the binding of SP1 to the TET1 promoter in MA9.3ITD treated with PBS or 5 ng/mL HHT for 48 h. IgG was used as a negative control. (G) Identification of direct binding between HHT and SP1 via DARTS assay in MA9.3ITD cells. Western blot analysis of the DARTS samples. (H) Identification of direct binding between HHT and SP1 via CETSA assay in MA9.3ITD cells. Western blot analysis of the CETSA samples. (I) Western blotting analysis of SP1 knockdown efficacy and effects of SP1 knockdown on the growth/proliferation of MA9.3RAS and MONOMAC 6 AML cells. (J) Relative expression of TET1 in MA9.3RAS and MONOMAC 6 AML cells with or without SP1 knockdown. *P<0.05; **P<0.01; ***P<0.001; t-test. Error bar, mean±Standard Deviation.
TET1 is a functionally important target of homoharringtonine that mediates homoharringtonine effects in acute myeloid leukemia
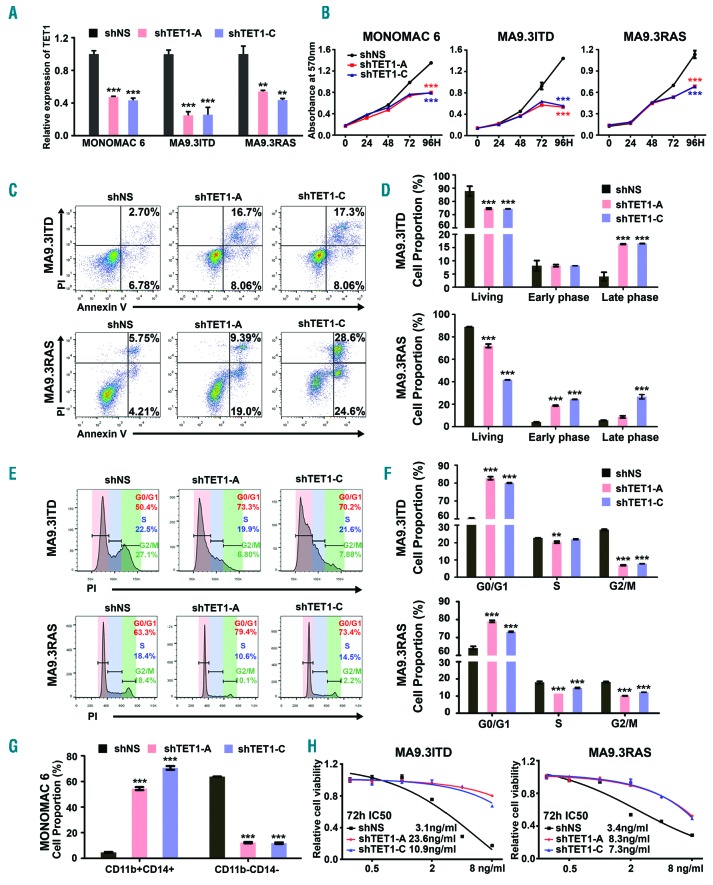
We next investigated whether knockdown of TET1 can mimic the effects of HHT (TET1 inhibition) and whether the treatment efficacy of HHT is dependent on its inhibition on TET1 expression in AML. As expected, depletion of TET1 expression by shRNA (Figure 4A) could largely mimic the effects of HHT treatment in AML cells, including inhibiting cell growth, inducing apoptosis, blocking cell cycle, and promoting myeloid differentiation (Figure 4B-G, and Online Supplementary Figure S4A-C). Furthermore, knockdown of TET1 dramatically reduced the sensitivity of AML cells to HHT treatment, with IC50 values increased to more than 2-fold (Figure 4H), suggesting that TET1 is a critical target of HHT that mediates the effects of HHT treatment in AML cells. Together, our data indicate that HHT induced cell growth inhibition, cell apoptosis, cell cycle arrest, and cell differentiation largely owing to its inhibition on TET1 expression/function in AML.
Figure 4.
Knockdown of TET1 expression recapitulates effects of homoharringtonine (HHT) treatment in acute myeloid leukemia (AML) cells. (A) Qualitative polymerase chain reaction (qPCR) analysis of TET1 knockdown efficacy in MONOMAC 6, MA9.3ITD and MA9.3RAS cells. (B) Effects of TET1 knockdown on cell growth/proliferation of these three AML cell lines at different time points [0, 24, 48, 72 and 96 hours (h)]. (C) Effects of TET1 knockdown on apoptosis in MA9.3ITD and MA9.3RAS AML cells. (D) Statistical analysis of apoptosis assay in AML cells from three independent experiments determined by flow cytometry. (E) Effects of TET1 knockdown on cell cycle arrest in AML cells. (F) Statistical analysis of cell cycle assays from three independent experiments determined by flow cytometry. (G) Statistical analysis of cell proportions of CD11b+CD14+ and CD11b−CD14− in TET1 knockdown or control MONOMAC 6 cells. (H) HHT IC50 in MA9.3ITD and MA9.3RAS cells with or without TET1 knockdown. These cells were exposed to HHT for 72 h. *P<0.05; **P<0.01; ***P<0.001; t-test. Error bar, mean±Standard Deviation.
FLT3 is a direct target of TET1 and is suppressed by the homoharringtonine⊣TET1/5hmC/axis
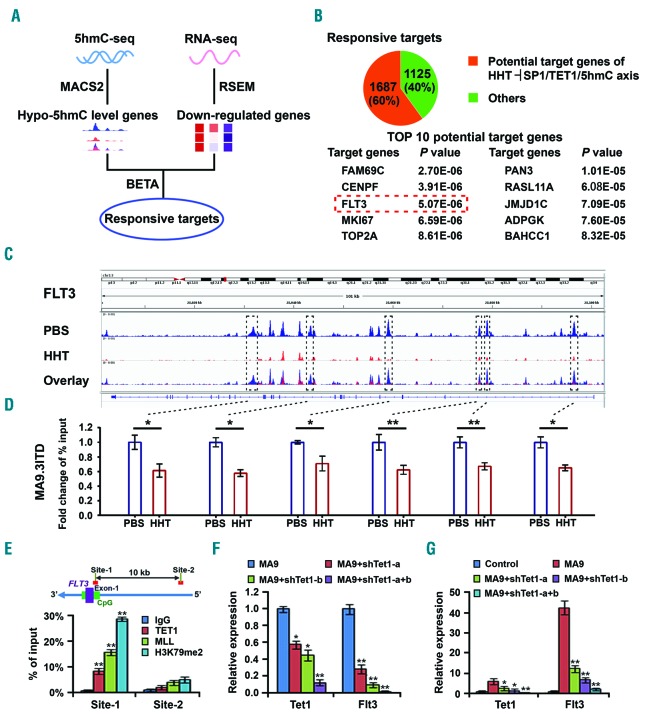
We next performed 5hmC-seq and RNA-seq of AML cells with or without HHT treatment to identify downstream targets that are regulated by the HHT⊣TET1/5hmC axis. As TET1 has been reported to positively regulate expression of many target genes through a 5hmC-dependent mechanism,17,37 we sought to identify responsive targets that exhibit reduced 5hmC level and downregulation in expression upon HHT treatment (Figure 5A). By overlapping such responsive targets with the list of TET1 potential direct targets as detected by ChIP-on-chip or ChIP-seq in the mammalian genome,37–39 we identified 1,687 potential TET1 targets that showed significant decreases in 5hmC level and significant downregulation in expression (Figure 5B, top panel, and Online Supplementary Table S3). Across the genomic locus of these potential targets, the top ten targeted genes with the most significant decreases in 5hmC levels were listed (Figure 5B, bottom panel). Notably, FLT3, a well-recognized oncogene related to leukemogenesis,7,9 is in the top list, associated with substantially decreased 5hmC abundance and significant downregulation in expression after HHT treatment (Figure 5C and Online Supplementary Figure S5A). Our ChIP-qPCR further confirmed the decreased 5hmC abundance on FLT3 gene locus upon HHT treatment in AML cells (Figure 5D).
Figure 5.
FLT3 is a critical target of the homoharringtonine (HHT) SP1/TET1/5hmC axis. (A) Scheme of identification of response targets of the HHT⊣SP1/TET1/5hmC axis by 5hmC-sequencing (5hmC-seq) and RNA-seq of MA9.3 RAS and MA9.3ITD acute myeloid leukemia (AML) cells treated with phosphate-buffered saline (PBS) or HHT (10 ng/mL for 5hmC-seq samples) for 48 hours (h). Responsive targets refer to genes with downregulation in both 5hmC abundance and RNA level upon HHT treatment. (B) Potential HHT⊣SP1/TET1/5hmC targets found by overlap analysis of the responsive targets and putative TET1 targets (top panel). Top ten target genes were shown (bottom panel). (C) The view of 5hmC abundance across FLT3 genomic locus in MA9.3ITD cells with PBS or HHT (10 ng/mL) treatment. (D) The verification of decreased 5hmC abundance on FLT3 via Chromatin immune-precipitation (ChIP)-qPCR analysis with different primers covering corresponding 5hmC peaks shown in boxes in (C). (E) ChIP-qPCR analysis of the binding of TET1, as well as MLL-fusion proteins, to the loci of FLT3 in MONOMAC 6 cells. Green bar represents the CpG island and purple bar represents exons of FLT3. Both MLL and H3K79me2 were used as positive controls. (F and G) The effects of knockdown of Tet1 on expression of Flt3 in MLL-AF9 transformed colony cells (F) and in leukemic bone marrow (BM) blast cells of bone marrow transplantation (BMT) recipient mice that developed MLL-AF9-induced AML (G). *P<0.05; **P<0.01; ***P<0.001; t-test. Error bar, mean±Standard Deviation.
The ChIP-seq data reported previously38 showed that Flt3 is a direct target gene of Tet1 in mouse embryonic stem cells (Online Supplementary Figure S5B). To validate whether FLT3 is also a direct target of TET1 in human AML cells, we performed ChIP-qPCR in MONOMAC 6 cells and showed that TET1 was especially enriched at the CpG area (Site 1) rather than the distal upstream area (Site 2) of FLT3 (in Figure 5E MLL and H3K79me2 are included as positive controls). Moreover, we showed that knockdown of Tet1 resulted in decreased expression of Flt3 in MLL-AF9 transformed colony cells (Figure 5F) and in BM cells of MLL-AF9 leukemia mice (Figure 5G). Conversely, forced expression of wild-type Tet1 (but not catalytically inactive mutant Tet1) in non-MLL rearranged human AML cells, such as Kasumi-1 cells (carrying t(8;21)/AML1-ETO), results in a significantly elevated expression of FLT3 (Online Supplementary Figure S5C). Similarly, in human CD34+ hematopoietic stem/progenitor cells (HSPC), we observed a strong positive correlation between FLT3 and TET1 in expression during both granulocytic and monocytic differentiation models (Online Supplementary Figure S5D). Furthermore, both HHT treatment and TET1 knockdown suppressed FLT3 expression in human AML cells (Online Supplementary Figure S5E). Taken together, our results suggest that FLT3 is a direct target of TET1 and HHT treatment-induced TET1 inhibition or knockdown of TET1 suppresses FLT3 expression through a 5hmC-related mechanism.
We previously reported that HOXA9 and MEIS1 were directly targeted by TET1 in AML cells.17 Consistently, here we showed that HHT treatment could also decrease expression of HOXA9 and MEIS1 in human AML cells (Online Supplementary Figure S5F). Interestingly, we and others have also reported previously that HOXA9/MEIS1 and FLT3 may each positively regulate the expression of the other.23,40,41 Indeed, here we showed that forced expression of either wild-type FLT3 or FLT3-ITD could significantly up-regulate expression of HOXA9 and MEIS1 in human 293T cells (Online Supplementary Figure S5G) in a manner similar to FLT3-ITD-mediated upregulation of HOXA9 and MEIS1 in human MONOMAC 6 AML cells, as we had reported previously.23 Conversely, we also showed that forced expression of HOXA9 and MEIS1 could substantially increase FLT3 level in mouse bone marrow progenitor cells (Online Supplementary Figure S6A). Thus, there may be a reciprocal positive regulatory loop between TET1 targets, such as FLT3 and HOXA9/MEIS1.
Notably, MA9.3ITD cell line was established from human cord blood CD34+ cells virally transduced with MLL-AF9 and FLT3-ITD.21 Thus, one may expect that ectopic expression of FLT3-ITD in this cell line would not be suppressed by HHT/TET1 and thereby should show resistance to HHT, which is somewhat opposite to what we observed (e.g. Figure 1A and B). We presumed that such a discrepancy might be due to the possibility that virally transduced FLT3-ITD in MA9.3ITD cells was by chance integrated to a locus that is also under control of TET1. Actually, there are a total of 11,632 genes that are associated with Tet1 enrichment in their promoter regions [−2kb to +2kb relative to annotated transcription start sites (TSS)] as detected by at least 2 out of 3 genome-wide ChIP-on-chip or ChIP-seq analyses in mouse embryonic stem cells (mESC).37–39 Therefore, although many of such putative targets identified from mESC might not be genuine targets of TET1 in human AML cells, there is still a good chance that the virally transduced FLT3-ITD in MA9.3ITD cell line was integrated, by chance, to a locus that is also under control of TET1. Indeed, HHT treatment could dramatically decrease the overall FLT3 (including FLT3-ITD) protein level, suggesting that is highly likely that FLT3-ITD expression in MA9.3ITD cell line is also under control of HHT/TET1 (Figure 6E). To determine whether non-TET1-controlled ectopic expression of FLT3 or FLT3-ITD can cause resistance to HHT in transduced AML cells, we virally transduced human Kasumi-1 AML cells with a high titer of FLT3 or FLT3-ITD viruses. In this way, each transduced cell had multiple copies of FLT3 or FLT3-ITD, and thus there would be a good chance that at least one copy was integrated into a locus not controlled by TET1. We sorted transduction-positive cells (i.e. RFP+ cells) 48 hours post transduction and then treated the cells with HHT or PBS for 24 hours. Forced expression of FLT3 or FLT3-ITD conferred at least partial resistance in transduced Kasumi-1 cells to HHT, while TET1 expression was still suppressed by HHT (Online Supplementary Figure 5H-J). Taken together, our data suggest that FLT3 is a downstream target of HHT/TET1 and mediates the sensitivity of AML cells to HHT.
Figure 6.
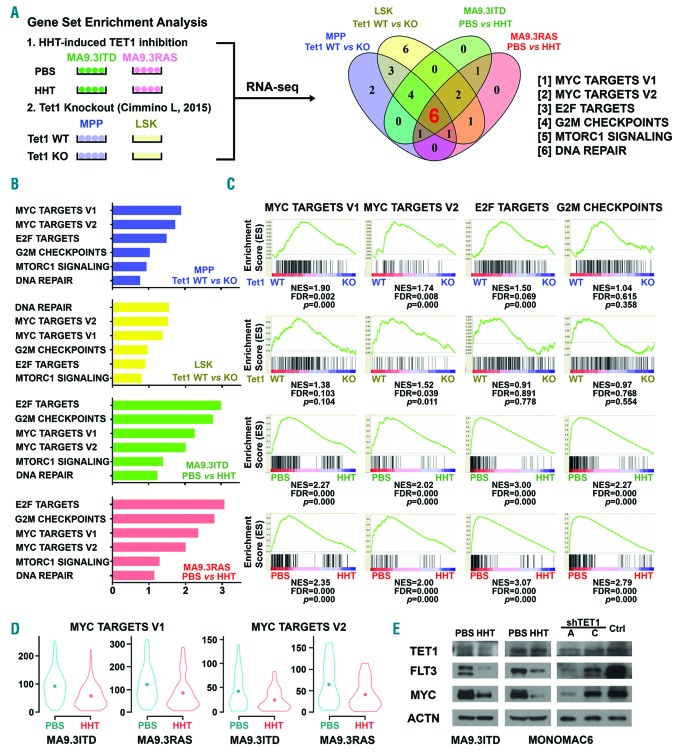
Pathways affected by the homoharringtonine (HHT)-SP1/TET1/5hmC axis. (A) Integrative analysis of our HHT-treatment RNA-sequencing (RNA-seq) data with published Tet1 knockout RNA-seq data42 to identify pathways or gene sets that were commonly affected by both HHT treatment and Tet1 knockout. RNA-seq data from MA9.3RAS and MA9.3ITD AML cells treated with phosphate-buffered saline (PBS) or HHT (10 ng/mL) for 48 hours (h), along with RNA-seq data from mouse BM Lin−/c-Kit+/Sca1+ (LSK) and multipotent progenitor (MPP) cells with or without Tet1 knockout,42 were used in the analysis. Six gene sets were identified to be affected by both HHT treatment and Tet1 knockout in all four conditions. (B) Normalized enrichment score (NES) of the six gene sets. (C) Among the six signaling pathways, MYC targets V1, MYC targets V2, E2F targets, and G2M checkpoints gene sets were significantly suppressed upon both HHT treatment and Tet1 knockout. (D) Decreased relative expression levels of genes of the MYC targets V1/V2 gene sets in MA9.3ITD and MA9.3RAS upon HHT treatment. The dot inside represents the median expression levels of the gene sets. (E) Western blot analysis of TET1, FLT3, and MYC in PBS- or HHT-treated MA9.3ITD and MONOMAC-6 cells and in MONOMAC-6 cells with or without TET1 knockdown. ACTIN was used as an endogenous control.
MYC signaling is a major downstream pathway affected by the homoharringtonine⊣SP1/TET1/5hmC axis
To further identify downstream pathways affected by the HHT⊣TET1/5hmC axis, we conducted an integrative analysis of our RNA-seq data of HHT-induced TET1 inhibition in AML cells and RNA-seq data of Tet1 knockout in mouse HSPCs [Lin−/c-Kit+/Sca1+ (LSK) and multipotent progenitor (MPP) cells] reported by Cimmino et al.42 Through gene set enrichment analysis (GSEA), we identified six gene sets strongly enriched in both HHT-induced TET1 inhibition and Tet1 knockout, including MYC targets V1, MYC targets V2, E2F targets, G2M checkpoints, MTORC1 signaling, and DNA repair (Figure 6A). The normalized enrichment score (NES) of the six co-enriched signaling pathways in all four pairs of samples are shown in Figure 6B. Among the six gene sets, MYC targets V1, MYC targets V2, E2F targets, and G2M checkpoints were significantly suppressed upon HHT treatment and Tet1 knockout (Figure 6C). The violin plots showed the down-regulated expression of the clustering genes in MYC targets V1 and MYC targets V2 after HHT treatment in AML cell lines (Figure 6D). Among these suppressed signal path ways, MYC functions as universal transcriptional amplifier and directly and indirectly regulates expression of multiple core enriched genes.43 More interestingly, MYC was reported as a downstream target of FLT3, and was significantly enriched in FLT3 constitutively activated cells and largely suppressed with the treatment of FLT3 inhibitor.10,12 Indeed, we showed that forced expression of either FLT3 or FLT3-ITD can substantially increase expression of MYC (Online Supplementary Figure S6B). In addition, consistent with previous studies showing that HOXA9/MEIS1 can up-regulate expression of MYC,44 we found that forced expression of HOXA9 and MEIS1 could also substantially increase MYC level (Online Supplementary Figure S6A). Our Western blot results also confirmed the decreased expression of TET1, FLT3 and MYC in HHT-treated or TET1-knockdown human MLL-rearranged or non-MLL-rearranged AML cells (Figure 6E and Online Supplementary Figure S6C). These findings suggest that MYC is an essential downstream target of the HHT⊣SP1/TET1/5hmC/FLT3-HOXA9-MEIS1 axis and MYC signaling is a major pathway inhibited by HHT treatment in AML.
Homoharringtonine treatment represents a promising therapeutic strategy for the treatment of acute myeloid leukemia with FLT3 mutations
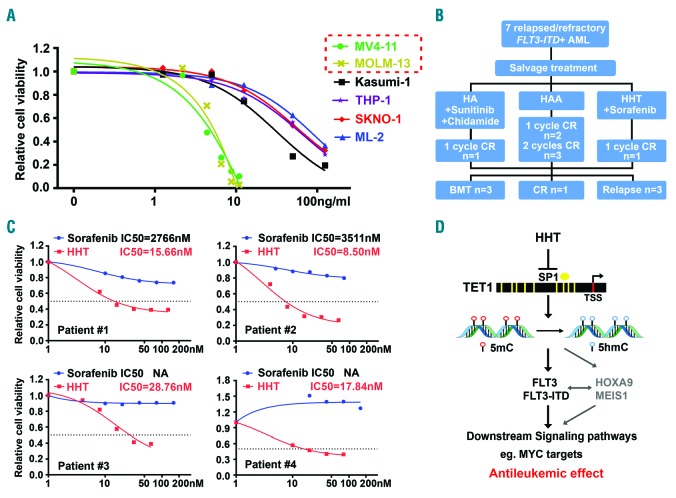
In line with the above discoveries, we found that human AML cell lines with FLT3 mutations are indeed much more sensitive to HHT than those without (Figure 7A). Next, we collected four primary AML samples from de novo or relapse/refractory patients with FLT3-ITD mutation (Online Supplementary Table S4). Notably, all the primary AML samples are highly sensitive to HHT treatment, with IC50 values < 30 nM; in contrast, these AML samples are relatively resistant to sorafenib, a tyrosine kinase inhibitor that was usually recommended to patients with FLT3-ITD mutation in clinic,45 with IC50 values >2.7 μM (Figure 7C). The superior effect of HHT, relative to sorafenib, might be owing to the fact that HHT can suppress expression of not only FLT3 but also other critical oncogenic targets of TET1 (e.g. HOXA9 and MEIS1), as mentioned above. Furthermore, we have successfully applied the HHT-based salvage chemotherapy in treating relapse/refractory patients in Zhejiang, China, and some of them were successfully bridged to BMT (Figure 7B and Jie et al., unpublished data). Together with our mechanistic studies described above, our data suggest that the high sensitivity of HHT in AML with FLT3 mutations is largely attributed to the HHT-induced inhibition of FLT3/HOXA9/MEIS1 expression/function through the HHT⊣SP1/TET1/5hmC axis (Figure 7D).
Figure 7.
Acute myeloid leukemia (AML) with FLT3 mutations are highly sensitive to homoharringtonine (HHT) treatment. (A) The sensitivity of AML cells with and without FLT3 mutations to HHT treatment. The AML cells were treated with a series of concentrations of HHT for 48 hours. (B) The HHT-based treatment regimen used for seven relapsed/refractory FLT3-ITD AML patients in clinic. HA: HHT plus cytarabine; HAA: HHT plus cytarabine and aclarubicin. (C) The IC50 values of HHT and sorafenib in primary FLT3-ITD AML patients’ samples.(C) The HHT-based treatment regimen used for seven relapsed/refractory FLT3-ITD AML patients in clinic. HA: HHT plus cytarabine; HAA: HHT plus cytarabine and aclarubicin. (D) Schematic illustration of the molecular mechanism underlying the anti-tumor effects of HHT mainly through suppression of the SP1/TET1/5hmC/FLT3-HOXA9-MEIS1/MYC axis.
Discussion
Previous studies have reported that HHT-based chemotherapy exhibited a high efficiency in treating de novo AML patients,4,5 but the underlying mechanism has not been well elucidated. In the present study, we showed that HHT treatment alone caused potent inhibition of AML cell growth/survival in vitro and substantial suppression of AML progression in vivo, and such inhibitory effects are likely attributed to HHT-induced cell cycle blockage and apoptosis, as well as enhanced myeloid differentiation. Mechanistically, we showed that, by targeting SP1/TET1, HHT treatment causes a substantial decrease in global 5hmC abundance and thereby markedly changes DNA epigenome and reprograms the downstream pathways. We demonstrated that SP1 is a direct drug target of HHT and a positive transcriptional regulator of TET1, and HHT competitively inhibits the binding of SP1 to the promoter region of TET1 and thereby suppresses SP1-mediated TET1 transcription; knockdown of either SP1 or TET1 can largely recapitulate the effects of HHT in AML. In addition, we have previously showed that depletion or suppression of TET1 expression dramatically inhibited AML progression and substantially prolonged survival in AML mice,17–19 recapitulating the potent in vivo anti-AML effect of HHT. Moreover, depletion of TET1 expression could make AML cells much less sensitive to HHT, further suggesting that the anti-AML activity of HHT relies on the suppression of the SP1/TET1/5hmC axis. However, further systematical studies are warranted to determine which particular sites/domains of SP1 are bound by HHT; such information would help us better understand how HHT disrupts the transcription-factor activity of SP1. Interestingly, SP1 has also been reported to positively regulate expression of BCR-ABL in chronic myeloid leukemia (CML) cells.46 Thus, the antileukemic effect of HHT in CML might not be solely due to its binding to ribosome,6 but likely also through targeting SP1 directly and thereby suppressing SP1-mediated activation of the BCR-ABL and TET1 signaling pathways.
Furthermore, our 5hmC-seq and RNA-seq analyses identified FLT3 as a critical target of the HHT⊣SP1/TET1/5hmC axis; HHT treatment or TET1 knockdown markedly reduced 5hmC abundance on FLT3 locus and decreased FLT3 expression in AML cells, and our ChIP-qPCR assay confirmed that FLT3 is a direct target of TET1. Interestingly, consistent with previous reports,23,40,41 here we showed that FLT3 exhibits a positive reciprocal regulation relationship with HOXA9/MEIS1, two known targets of TET1.17 Thus, our data suggest that, by suppression of TET1 expression, HHT simultaneously inhibits expression of multiple target genes of TET1 (which may form a reciprocal positive regulatory loop) in AML cells and thereby displays a potent antileukemic effect.
FLT3 encodes a class III receptor tyrosine kinase that regulates hematopoiesis and the mutation of FLT3 is the most common driven mutation found in more than 30% of AML patients.47 Both ITD and tyrosine kinase domain mutation of FLT3 result in its constitutive activation and thus lead to leukemogenesis by promoting expression of a number of critical oncogenic downstream targets such as MYC.10–12,48 Despite the extensive efforts in developing and testing FLT3 inhibitors in the clinic, AML patients with high allelic ratio FLT3-ITD are still classified as adverse risk category in 2017 European LeukemiaNet recommendation due to the high relapse rate and poor overall survival.7,11,12 Thus, the development of improved therapeutics for treating FLT3-ITD AML is still an unmet need.
Here we also showed that primary AML patients with FLT3 mutations, including both newly diagnosed and relapsed patients, exhibit a high sensitivity to HHT treatment (with IC50 <30 nM). Consistent with our findings, another group also reported recently that HHT exhibited preferential antileukemic effect against AML carrying FLT3-ITD as detected by an in vitro drug screening on patients’ samples.49 In addition, they conducted a phase II clinical trial in relapsed/refractory FLT3-ITD AML patients, in which 20 out of 24 patients achieved complete remission with sorafenib and HHT combination treatment (median leukemia-free survival and overall survival: 12 and 33 weeks, respectively).49 While they showed sorafenib alone reduced the amount of pFLT3 protein, and HHT alone reduced the amount of both FLT3 and pFLT3 protein in FLT3-ITD AML cell lines, no further mechanistic studies were carried out.49 Our studies elucidated the molecular mechanism underlying the high sensitivity of FLT3-mutated AML to HHT treatment. This mechanism involves HHT-induced reprogramming of DNA epigenome by targeting the SP1/TET1/5hmC axis and thereby inhibition of transcription of a set of critical oncogenic targets, especially FLT3, which in turn leads to the suppression of FLT3 downstream pathways, such as MYC signaling. Notably, it is well known that FLT3-ITD mutation patients under therapy often develop secondary FLT3 mutations which result in drug resistance. Interestingly, we found that MONOMAC 6, which carries the FLT3 V592A mutation,50 was also sensitive to HHT. Moreover, the relapsed/refractory FLT3-ITD AML patient also showed sensitivity to HHT treatment and could be bridged to transplantation in subsequent treatment49 (Figure 7B). Therefore, HHT-based therapeutics (i.e. HHT plus other therapeutic agents such as FLT3 inhibitors and/or standard chemotherapy) represent effective novel treatments for de novo or relapsed/refractory FLT3-mutated AML patients.
In summary, here we show that HHT, approved by the FDA for CML treatment, also exhibits potent therapeutic efficacy in AML and decreases global 5hmC abundance by targeting the SP1/TET1/5hmC axis. Although other mechanisms, such as inhibition of protein synthesis,6 may also contribute to the overall antileukemic effects of HHT in AML, which warrant further systematical studies in the future, our work reveals a novel mechanism involving suppression of the SP1/TET1/5hmC/FLT3-HOXA9-MEIS1/MYC signaling through which HHT exhibits potent therapeutic activity in treating AML. Since AML is characterized by cytogenetic and molecular heterogeneity, targeted therapy is a growing trend for selected subtypes of AML, especially for those with adverse prognosis. Here we provide compelling functional and mechanistic data suggesting that an HHT-based therapeutic approach represents effective target therapeutics to treat AML carrying FLT3 mutations and/or with overexpression of endogenous TET1/FLT3/HOXA9/MEIS1, which accounts for more than 40% of total human AML cases.7
Acknowledgments
The authors would also like to thank Dr. James Mulloy for the generous gift of MA9.3ITD and MA9.3RAS cell lines, and Dr. Ravi Bahtia for the kind gift of MV4-11 and MOLM-13 cell lines.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/1/148
Funding
The work was supported in part by the National Institutes of Health (NIH) R01 Grants CA214965 (JC), CA211614 (JC), CA178454 (JC), CA182528 (JC), and CA236399 (JC) and a R56 grant DK120282 (JC), as well as grants from National Natural Science Foundation of China 81820108004 (JJ) and 81900154 (CL). JC is a Leukemia & Lymphoma Society (LLS) Scholar.
References
- 1.Powell RG, Weisleder D, Smith CR., Jr Antitumor alkaloids for Cephalataxus harringtonia: structure and activity. J Pharm Sci. 1972;61(8):1227–1230. [DOI] [PubMed] [Google Scholar]
- 2.Nazha A, Kantarjian H, Cortes J, Quintas-Cardama A. Omacetaxine mepesuccinate (synribo) - newly launched in chronic myeloid leukemia. Expert Opin Pharmacother. 2013;14(14):1977–1986. [DOI] [PubMed] [Google Scholar]
- 3.Cephalotaxine esters in the treatment of acute leukemia. A preliminary clinical assessment. Chin Med J (Engl). 1976; 2(4):263–272. [PubMed] [Google Scholar]
- 4.Jin J, Jiang DZ, Mai WY, et al. Homoharringtonine in combination with cytarabine and aclarubicin resulted in high complete remission rate after the first induction therapy in patients with de novo acute myeloid leukemia. Leukemia. 2006; 20(8):1361–1367. [DOI] [PubMed] [Google Scholar]
- 5.Jin J, Wang JX, Chen FF, et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2013;14(7):599–608. [DOI] [PubMed] [Google Scholar]
- 6.Gurel G, Blaha G, Moore PB, Steitz TA. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and brucean-tin bound to the ribosome. J Mol Biol. 2009;389(1):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Bugno J, Hu C, et al. Eradication of Acute Myeloid Leukemia with FLT3 Ligand-Targeted miR-150 Nanoparticles. Cancer Res. 2016;76(15):4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KT, Baird K, Davis S, et al. Constitutive Fms-like tyrosine kinase 3 activation results in specific changes in gene expression in myeloid leukaemic cells. Br J Haematol. 2007;138(5):603–615. [DOI] [PubMed] [Google Scholar]
- 11.Leung AY, Man CH, Kwong YL. FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia. 2013;27(2):260–268. [DOI] [PubMed] [Google Scholar]
- 12.Konig H, Levis M. Targeting FLT3 to treat leukemia. Expert Opin Ther Targets. 2015;19(1):37–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002; 62(14):4075–4080. [PubMed] [Google Scholar]
- 15.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17(3):637–641. [DOI] [PubMed] [Google Scholar]
- 16.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, Jiang X, Li Z, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110(29):11994–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Hu C, Arnovitz S, et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun. 2016;7:11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Hu C, Ferchen K, et al. Targeted inhibition of STAT/TET1 axis as a therapeutic strategy for acute myeloid leukemia. Nat Commun. 2017;8(1):2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z, Chen L, Dawlaty MM, et al. Combined Loss of Tet1 and Tet2 Promotes B Cell, but Not Myeloid Malignancies, in Mice. Cell Rep. 2015;13(8):1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wunderlich M, Mizukawa B, Chou FS, et al. AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood. 2013;121(12):e90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su R, Dong L, Li C, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172(1-2):90–105 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012; 22(4):524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydrox-ymethylcytosine. Nat Biotechnol. 2011; 29(1):68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Sun H, Ma J, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013; 8(12):2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan M, Kanbe E, Peterson LF, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12(8):945–949. [DOI] [PubMed] [Google Scholar]
- 30.Conway O’Brien E, Prideaux S, Chevassut T. The epigenetic landscape of acute myeloid leukemia. Adv Hematol. 2014; 2014:103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eriksson A, Lennartsson A, Lehmann S. Epigenetic aberrations in acute myeloid leukemia: Early key events during leukemogenesis. Exp Hematol. 2015;43(8):609–624. [DOI] [PubMed] [Google Scholar]
- 32.Smale ST. Nuclear run-on assay. Cold Spring Harb Protoc. 2009; 2009(11):pdb.prot5329. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Wu LC, Pang J, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17(4):333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Chen HX, Zhou SY, et al. Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Mol Cancer. 2015; 14:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomenick B, Jung G, Wohlschlegel JA, Huang J. Target identification using drug affinity responsive target stability (DARTS). Curr Protoc Chem Biol. 2011; 3(4):163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jafari R, Almqvist H, Axelsson H, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Wu F, Tan L, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, D’Alessio AC, Ito S, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011; 473(7347):389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106(1):254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burillo-Sanz S, Morales-Camacho RM, Caballero-Velazquez T, et al. NUP98-HOXA9 bearing therapy-related myeloid neoplasm involves myeloid-committed cell and induces HOXA5, EVI1, FLT3, and MEIS1 expression. Int J Lab Hematol. 2016; 38(1):64–71. [DOI] [PubMed] [Google Scholar]
- 42.Cimmino L, Dawlaty MM, Ndiaye-Lobry D, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessa J, Tavares MJ, Santos J, et al. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multi-potent cells in the early developing zebrafish eye. Development. 2008; 135(5):799–803. [DOI] [PubMed] [Google Scholar]
- 45.Rollig C, Serve H, Huttmann A, et al. Addition of sorafenib versus placebo to standard therapy in patients aged 60 years or younger with newly diagnosed acute myeloid leukaemia (SORAML): a multicen-tre, phase 2, randomised controlled trial. Lancet Oncol. 2015;16(16):1691–1699. [DOI] [PubMed] [Google Scholar]
- 46.Jin B, Wang C, Shen Y, Pan J. Anthelmintic niclosamide suppresses transcription of BCR-ABL fusion oncogene via disabling Sp1 and induces apoptosis in imatinib-resistant CML cells harboring T315I mutant. Cell Death Dis. 2018;9(2):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brondfield S, Umesh S, Corella A, et al. Direct and indirect targeting of MYC to treat acute myeloid leukemia. Cancer Chemother Pharmacol. 2015;76(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam SS, Ho ES, He BL, et al. Homoharringtonine (omacetaxine mepe-succinate) as an adjunct for FLT3-ITD acute myeloid leukemia. Sci Transl Med. 2016; 8(359):359ra129. [DOI] [PubMed] [Google Scholar]
- 50.Spiekermann K, Dirschinger RJ, Schwab R, et al. The protein tyrosine kinase inhibitor SU5614 inhibits FLT3 and induces growth arrest and apoptosis in AML-derived cell lines expressing a constitutively activated FLT3. Blood. 2003;101(4):1494–1504. [DOI] [PubMed] [Google Scholar]