Abstract
Alterations of the tumor suppressor gene TP53 are found in different cancers, in particular in carcinomas of adults. In pediatric acute lymphoblastic leukemia (ALL), TP53 mutations are infrequent but enriched at relapse. As in most cancers, mainly DNA-binding domain missense mutations are found, resulting in accumulation of mutant p53, poor therapy response, and inferior outcome. Different strategies to target mutant p53 have been developed including reactivation of p53’s wildtype function by the small molecule APR-246. We investigated TP53 mutations in cell lines and 62 B-cell precursor ALL samples and evaluated the activity of APR-246 in TP53-mutated or wildtype ALL. We identified cases with TP53 missense mutations, high (mutant) p53 expression and insensitivity to the DNA-damaging agent doxorubicin. In TP53-mutated ALL, APR-246 induced apoptosis showing strong anti-leukemia activity. APR-246 restored mutant p53 to its wildtype conformation, leading to pathway activation with induction of transcriptional targets and re-sensitization to genotoxic therapy in vitro and in vivo. In addition, induction of oxidative stress contributed to APR-246-mediated cell death. In a preclinical model of patient-derived TP53-mutant ALL, APR-246 reduced leukemia burden and synergized strongly with the genotoxic agent doxorubicin, leading to superior leukemia-free survival in vivo. Thus, targeting mutant p53 by APR-246, restoring its tumor suppressive function, seems to be an effective therapeutic strategy for this high-risk group of TP53-mutant ALL.
Introduction
Although most pediatric patients diagnosed with acute lymphoblastic leukemia (ALL) have a favorable prognosis, achievement of long-term survival remains a major clinical challenge, particularly at relapse.1 Alterations of cell death programs cause treatment failure and resistance in many cancers including leukemia. The nuclear phosphoprotein p53 is a transcription factor that controls cellular responses to stress, including DNA damage. Originally identified more than three decades ago,2,3 p53 was characterized as a tumor suppressor negatively regulating cell cycle and growth, inhibiting the cancer cell’s oncogenic potential.4,5 The gene coding for p53 (TP53) is localized on the short arm of chromosome 17 (17p13) and it is the most frequently mutated gene across different cancers.6,7 Both deletions and point mutations have been described and mutations often co-occur with loss of the corresponding wildtype allele.8,9 The majority are TP53 missense mutations found within the DNA-binding domain coding region (codons 100-300, exons 5-8) and affect the structural integrity and DNA-binding ability of p53, leading to accumulation of dysfunctional p53 protein and increased oncogenic potential.10–13
TP53 mutations are found frequently, in up to 95% of carcinomas, typically in older patients.7,8 In ALL, recent studies identified alterations of TP53 in subsets of up to 16%, with higher rates in T-ALL, at relapse, and in elderly patients.14–18 Moreover, more than 90% of ALL cases with a low hypodiploid karyotype (including loss of chromosome 17) carry somatic TP53 alterations19,20 and TP53 germline mutations confer a high risk for hypodiploid ALL.21 In pediatric ALL, TP53 alterations are associated with poor response to chemotherapy and an inferior outcome, particularly at relapse, identifying TP53-mutant B-cell precursor (BCP)-ALL patients as a high-risk subgroup with a particular need for alternative therapies.14,16–18,22
Different strategies to interfere with the p53 pathway have been evaluated. Inhibition of the interaction of p53 and its negative regulator, mouse double minute 2 (MDM2), leads to sustained p53 transcriptional activity, but requires the presence of wildtype p53.23 Therefore, direct targeting of mutant p53 has been investigated, identifying small molecules that reactivate p53 function.24 In line, anti-tumor activity has been observed in murine lymphoma and liver cancer models upon genetic restoration of p53, supporting the principle of p53 reactivation as a therapeutic strategy.25,26 APR-246 (PRIMA-1Met), the structural analog of PRIMA-1 (p53 reactivation and induction of massive apoptosis) is a small molecule, identified in a screen for mutant p53-dependent growth suppression in sarcoma cells, showing activity on both structural and DNA-binding mutants.27 APR-246 is a prodrug that is converted into methylene quinuclidinone, which binds covalently to the core domain of mutant p53 interacting with thiol groups of cysteines, restoring p53 wildtype conformation and function.28,29 In addition, induction of oxidative stress has been reported as a second activity of APR-246, deriving from glutathione depletion, thioredoxin reductase inhibition and other effects.30–33
APR-246 demonstrated preclinical antitumor activity and synergism with DNA-damaging drugs in different cancers32,34–39 and showed very moderate side effect profiles in a first-in-human phase I/IIa clinical trial in patients with refractory prostate cancer, acute myeloid leukemia, chronic lymphocytic leukemia, multiple myeloma and lymphoma.40 Accordingly, APR-246 is currently being investigated in ovarian and esophageal cancer, myeloid neoplasms and melanoma in phase II clinical trials (ClinicalTrials.gov).41 However, mutant p53 has so far not been addressed as a target for therapeutic intervention in ALL.
In this study, we investigated a large cohort of patient-derived pediatric BCP-ALL primograft samples identifying TP53-mutated cases and analyzed the effects of APR-246 in TP53-mutated (TP53mut) and TP53-wildtype (TP53wt) BCP-ALL. We identified strong and selective antileukemia activity of APR-246 in TP53mut ALL providing the basis to develop personalized therapy regimens for this high-risk subgroup of ALL.
Methods
Additional detailed information is provided in the Online Supplementary Data.
Sixty-two patient-derived xenograft samples established by transplantation of patients’ ALL cells onto NOD.CB17-Prkdcscid/J mice42 and six BCP-ALL cell lines were studied. Leukemia samples were obtained from pediatric BCP-ALL patients at diagnosis or relapse upon informed consent from the children and/or their legal guardians in accordance with the institution’s ethical review boards. All animal experiments were approved by the appropriate authority (Regierungspräsidium Tübingen) and carried out following the national animal welfare guidelines. TP53 mutations were analyzed by denaturing high-performance liquid chromatography and confirmed by Sanger sequencing, 17p deletions were assessed by fluorescence in situ hybridization. Mutation information was matched to the IARC-TP53 database.43 The sensitivity of leukemia samples to doxorubicin, APR-246 (kindly provided by Aprea Therapeutics, Stockholm, Sweden) or the combination was assessed after incubation of ALL cells with increasing drug concentrations, analyzing cell death by flow-cytometry according to forward- and side-scatter criteria. Data from three independent experiments performed in triplicate (cell lines) or of one experiment performed in triplicate (primografts) were analyzed by t-test, and differences of half maximal inhibitory concentrations (IC50) titrations by F-test. P values ≤0.05 were considered statistically significant. Synergies of drug combinations were assessed calculating combination indices (CI), indicating strong synergism (CI 0.1-0.3), synergism (CI <1), an additive effect (CI=1) or antagonism (CI>1). Apoptosis was analyzed assessing annexin-V-FLUOS positivity and caspase-3 activity. Proteins (p53, PUMA, p21, NOXA, GAPDH) were detected by western blot analysis using the respective antibodies. The wildtype conformation of p53 was detected by immunoprecipitation using a conformation-specific anti-p53 wildtype antibody (PAb1620) followed by western blot analysis with an anti-p53 (total) antibody (DO-7). An immunoglobulin light chain-specific peroxidase conjugated binding protein was used for western blot analyses carried out following immuno-precipitation. Depletion of p53 was achieved by lentiviral shRNA-mediated knockdown or siRNA-mediated downregulation in TP53mut or TP53wt ALL cells. For in vivo treatment, transplanted recipients showing >5% human ALL cells in peripheral blood were randomized and treated (for 3 weeks) with solvent, APR-246 (days 1-5), doxorubicin (day 1), or the combination (APR-246 days 1-5, doxorubicin day 5) and sacrificed at the end of treatment for analysis of leukemia loads. For survival analyses, recipients were followed up after treatment until onset of leukemia-related morbidity and sacrificed. High loads of human ALL cells were detected in bone marrow and spleen in all cases, confirming reoccurrence of manifest leukemia.
Results
Identification of TP53 mutations in B-cell precursor acute lymphoblastic leukemia
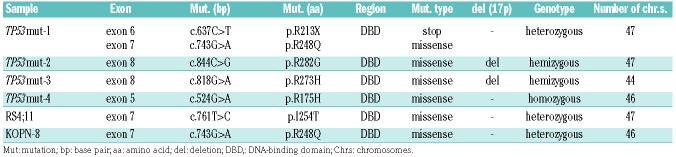
We investigated 62 patient-derived pediatric BCP-ALL samples, which were established in our NOD/SCID/huALL xenograft model from patients at diagnosis (n=53) or relapse (n=9). TP53mut cases were identified by denaturing high-performance liquid chromatography and confirmed by Sanger sequencing (exons 4-10). Four TP53mut cases were found, one derived from a patient at second relapse (TP53mut-1) and three at diagnosis (TP53mut-2, -3, -4) (Online Supplementary Table S1). In parallel, we characterized six BCP-ALL cell lines and identified two TP53mut (RS4;11, KOPN-8) and four TP53wt (MUTZ-5, EU-3, UoCB-6 and NALM-6) lines. All samples carried missense mutations previously described (p53.iarc.fr),15,43 localized within the region encoding the DNA-binding domain, suggesting loss of p53’s tumor suppressive function (Figure 1A, Table 1). In the TP53mut samples, the second allele carried a nonsense mutation (TP53mut-1), was absent (loss of 17p, TP53mut-2, -3), or carried the same missense mutation (TP53mut-4) (Table 1). Somatic and germline TP53mut are associated with (low) hypodiploid ALL.15,19–21 One primo-graft sample (TP53mut-3) showed a hypodiploid karyotype with 44 chromosomes (Table 1). In line with disrupted degradation and accumulation of mutant p53 protein, TP53mut cases showed higher p53 protein levels compared to TP53wt leukemias (Figure 1B).
Figure 1.
TP53-mutated acute lymphoblastic leukemias are DNA-damage resistant but sensitive to APR-246. (A) All TP53mut B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) primograft and cell lines harbor missense mutations (filled circles) localized in the DNA-binding domain of TP53. The primograft sample TP53mut-1 carries an additional stop mutation (R213X, open circle). See also Table 1. (B) Increased p53 protein expression in TP53mut compared to TP53wt ALL in primograft (left) and cell line (right) leukemia samples. Western blot, anti-p53 antibody (total, clone DO-7) with GAPDH as a loading control. (C-F) Significantly higher half maximal inhibitory concentrations (IC50) for doxorubicin in TP53mut (red curves) primograft (C) and cell line (D) BCP-ALL, and significantly lower IC50 values for APR-246 in TP53mut primograft (E) and cell line (F) samples, indicating insensitivity to the DNA-damaging agent doxorubicin but sensitivity to APR-246 in TP53mut BCP-ALL. Dose-response curves reflect cell death induction in response to increasing concentrations summarizing one (primografts, 24 h; C, E) or three (cell lines, 48 h; D, F) independent experiments, each performed in triplicate. Comparison of sensitivities of TP53wt and TP53mut leukemias, F-test, ***P<0.001. See also Online Supplementary Table S2.
Table 1.
TP53 mutations in acute lymphoblastic leukemia cell lines and primograft samples.
TP53-mutated leukemias are sensitive to APR-246 but not to genotoxic therapy
In response to genotoxic agents and stress, wildtype p53 suppresses cellular viability and proliferation. However, dysfunctional, mutant p53 fails to mediate tumor-suppressive functions such as induction of cell death. Therefore, we analyzed cell death in TP53mut and TP53wt ALL primografts (TP53mut n=4, TP53wt n=4) and cell lines (TP53mut n=2, TP53wt n=4) in response to increasing concentrations of the DNA-damaging agent doxorubicin, a standard genotoxic drug regularly used in ALL treatment protocols, and to APR-246. All TP53mut primografts and cell lines showed, as expected, insensitivity to doxorubicin indicated by significantly higher IC50 values, in contrast to doxorubicin-sensitive TP53wt leukemias (Figure 1C, D; Online Supplementary Table S2A, B). An opposite effect was observed upon exposure to APR-246 with high sensitivity and cell death induction in all TP53mut leukemias, but low APR-246 sensitivity in TP53wt ALL (Figure 1E, F; Online Supplementary Table S2C, D). Interestingly, diagnosis- (TP53mut-2, -3, -4) or relapse-derived (TP53mut-1) primograft samples did not show differences in APR-246 or doxorubicin sensitivity.
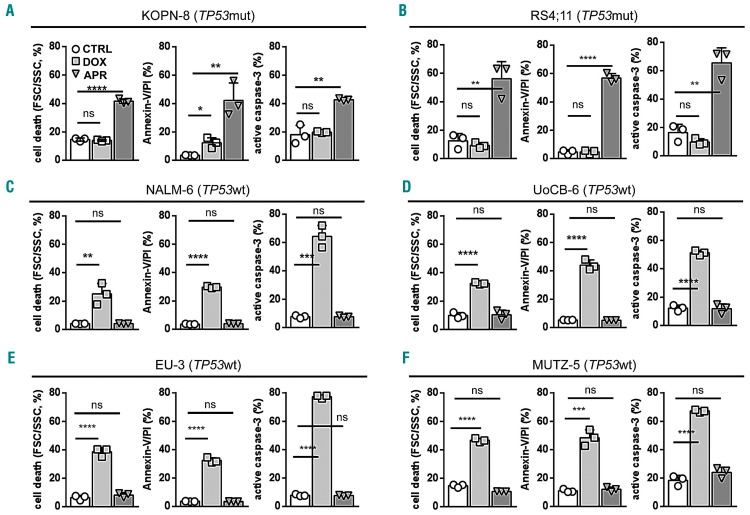
Activation of the p53 pathway results in apoptosis induction. Along with cell death, APR-246 led to annexin-V/propidium iodide positivity and caspase-3 activation indicating apoptosis induction in TP53mut ALL. In contrast, apoptosis was induced by doxorubicin but not APR-246 in TP53wt cells (Figure 2 and Online Supplementary Figure S1).
Figure 2.
APR-246 induces apoptosis in TP53-mutated acute lymphoblastic leukemia. (A-F) Induction of cell death (left diagrams, forward/side scatter criteria, flow cytometry), annexin-V/propidium iodide (PI) positivity (middle diagrams) and caspase-3 activation (right diagrams) by APR-246 in TP53mut cell lines KOPN-8 (A) and RS4;11 (B), in contrast to cell death and apoptosis induction by doxorubicin in TP53wt lines NALM-6 (C), UoCB-6 (D), EU-3 (E), and MUTZ-5 (F). Proportions of cells after 48 h exposure to solvent (CTRL), APR-246 (APR, 5 mM), or doxorubicin (DOX, 15 ng/mL). Mean values ± standard deviation of three independent experiments, each performed in triplicate. Student t-test, ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; n.s., not significant.
Thus, all identified TP53mut leukemias carried missense mutations in the DNA-binding domain, showed accumulation of p53 indicative of dysfunctional mutant p53, resistance to the genotoxic agent doxorubicin, and were highly sensitive to APR-246-induced apoptosis.
APR-246 restores p53’s wildtype conformation reactivating tumor suppressive functions
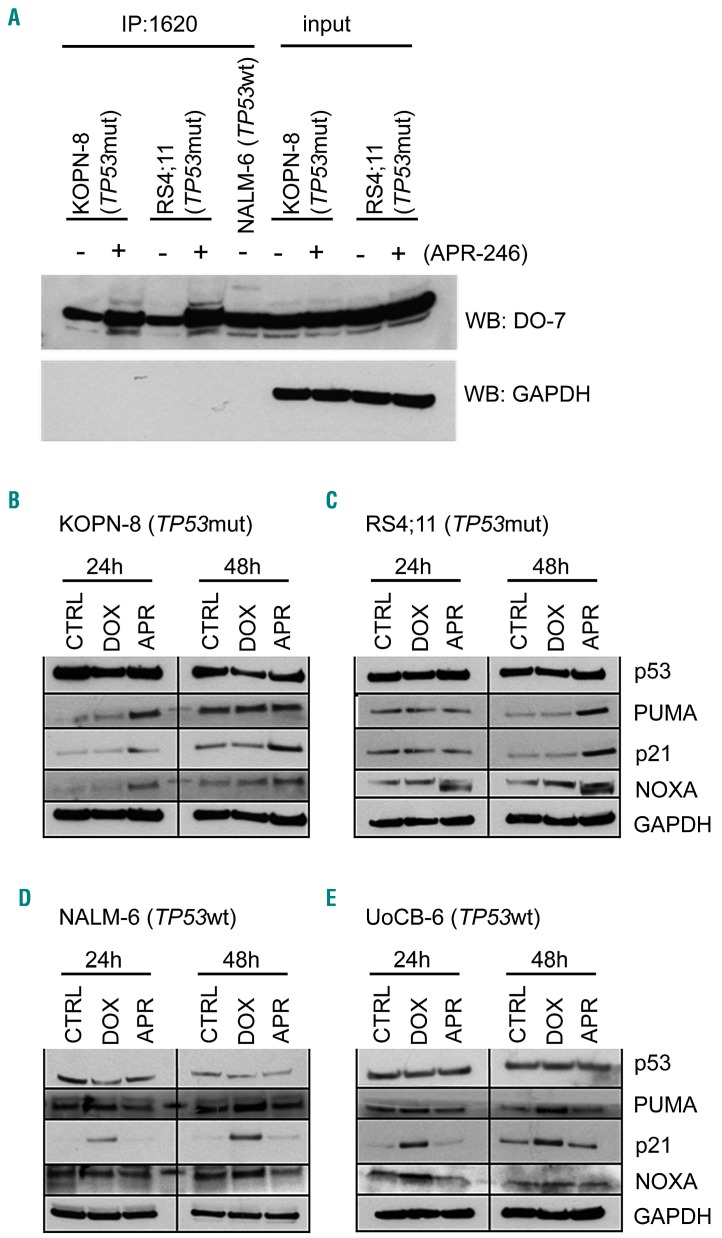
We further addressed the mode of action of APR-246 in TP53mut ALL and examined the conformation of p53 and activation of the pathway in response to APR-246. Using a p53 wildtype conformation-specific antibody (PAb1620), larger amounts of p53 with wildtype conformation were immunoprecipitated from lysates of TP53mut ALL cells exposed to APR-246, indicating reconstitution of p53 wildtype conformation in TP53mut ALL by APR-246 (Figure 3A). However, this effect was not observed in TP53wt leukemia cells (Online Supplementary Figure S2). Next, we assessed expression of the p53 transcriptional targets PUMA (P53-Upregulated Modulator of Apoptosis), p21 (Cyclin Dependent Kinase Inhibitor 1A, CDKN1), and NOXA upon APR-246 or doxorubicin treatment in TP53mut (KOPN-8, RS4;11) and TP53wt (NALM-6, UoCB-6) ALL lines. In TP53mut ALL, APR-246 led to induction of all p53 targets (Figure 3B, C and Online Supplementary Figure S3). In contrast, an opposite picture of activation of p53 transcriptional targets in TP53wt but not in TP53mut leukemias was observed upon incubation with doxorubicin (Figure 3D, E and Online Supplementary Figure S3). Thus, APR-246 induces restoration of mutant p53 to wildtype conformation, transcriptional target expression, and apoptosis in TP53mut ALL.
Figure 3.
Conformational and functional restoration of mutant p53 by APR-246. (A) Increased levels of p53 with wildtype conformation in TP53mut ALL (KOPN-8, mutation R248Q; RS4;11, mutation I254T) upon exposure to APR-246 (5 mM, 24 h). Immunoprecipitation (IP, anti-wt p53 specific antibody PAb1620) and western blot analysis (WB, anti-p53 antibody DO-7, light chain-specific goat anti-mouse peroxidase conjugated binding protein), GAPDH expression in input lysates and absence in precipitates, NALM-6 serves as a wildtype p53 positive control. (B-E) Expression of p53 transcriptional targets PUMA, p21 and NOXA in (B, C) TP53mut ALL upon APR-246 treatment and in (D, E) TP53wt ALL upon doxorubicin treatment. Western blot, exposure to solvent (CTRL), doxorubicin (DOX, 15 ng/mL), or APR-246 (APR, 5 mM) for the indicated times, with GAPDH as a loading control. The results of one representative out of two independent experiments are shown. See also Online Supplementary Figures S2 and S3.
Induction of oxidative stress contributes to APR-246-mediated cellular death
Induction of oxidative stress has been described as a second activity of APR-246 in different cancers.30–32 APR-246 was reported to interfere with different regulators of the cellular redox system, such as thioredoxin reductase, thioredoxin and glutathione, and with the transcription factor NRF2, leading to induction of reactive oxygen species (ROS).28,30–33,44–47 Given the activity of APR-246 in TP53mut ALL, we addressed whether TP53mut and TP53wt ALL display distinct sensitivities in response to ROS generation. Upon treatment with 3-morpholinosyd-nonimine, a spontaneous generator of reactive oxygen and nitrogen species, and the oxidant tert-butyl hydroxyperoxide, increased ROS levels were observed in both TP53mut and TP53wt ALL cells, leading to similar cell death rates (Online Supplementary Figure S4).
Next, we investigated whether induction of oxidative stress is involved in APR-246-mediated cell death in TP53mut ALL. Importantly, methyl quinuclidinone, the active drug spontaneously formed from APR-246, binds covalently to cysteine residues in the core domain of p53, but also to cysteines in the widely used antioxidant and ROS inhibitor N-acetylcysteine (NAC).28,44 Thus, NAC directly blocks APR-246 activity and cannot be used to investigate the role of ROS in APR-246-mediated cell death. Therefore, the synthetic antioxidant compound and ROS inhibitor superoxide dismutase mimetic Mn (III) tetrakis (5, 10, 15, 20-benzoic acid) porphyrin (MnTBAP) was used. Cell death was analyzed together with ROS levels in TP53mut (KOPN-8 and RS4;11) and TP53wt (NALM-6 and UoCB6) leukemia cells exposed to APR-246 with or without NAC or MnTBAP. Similar ROS levels were observed upon APR-246 treatment in both TP53mut and TP53wt ALL (Online Supplementary Figure S5A, C, E, G), however induction of cell death was only seen in TP53mut cells (Online Supplementary Figure S5F, H) but not in TP53wt cells (Online Supplementary Figure S5B, D). Interestingly, ROS inhibition by MnTBAP partially inhibited APR-246-induced cell death in TP53mut ALL, indicating that ROS contribute to APR-246- induced cell death. It was also interesting that, even in the presence of MnTBAP, i.e. in the absence of ROS, APR-246 retained a statistically significant cytotoxic effect (Online Supplementary Figure S5F, H). In line with previous reports,44 the activity of APR-246 was completely blocked by NAC.
Taken together, these data show that induction of oxidative stress might contribute to APR-246-mediated cell death in ALL, in line with previously reported data of a dual mode of action of APR-246 in other malignancies.28,30–33,44–46
APR-246 activity depends on mutant p53
Activity of APR-246 was observed in TP53mut but not TP53wt ALL. Therefore, we analyzed the effect of APR-246 in TP53mut and TP53wt cell lines upon lentiviral shRNA-mediated knockdown of p53 (Figure 4A-C). In both TP53mut lines (KOPN-8 and RS4;11) depletion of p53 led to APR-246 insensitivity and cell death resistance, in contrast to dose-dependent cell death induction in control-transduced cells (Figure 4D, E). However, TP53wt cells with p53 depletion were unresponsive to APR-246, like the corresponding control transduced cells (Figure 4F). A similar result was observed upon siRNA-mediated p53 downregulation with clearly lower cell death induction in TP53mut ALL but no effect in TP53wt cells (Figure 4G-J and Online Supplementary Figure S6). Together with our observations on APR-246 insensitivity in TP53wt ALL (Figure 1E, F) and the absence of p53 transcriptional target expression upon APR-246 treatment in TP53wt ALL (Figure 3D, E), these findings indicate that the activity of APR-246 is associated with the presence of mutant p53. Accordingly, distinct sensitivities to APR-246 were found in four primary ALL samples obtained from patients with therapy-resistant disease or relapse carrying different TP53 mutations (Figure 4L, Table 2). Robust dose-dependent cell death induction was observed in leukemia cells from patients 2, 3, and 4 carrying missense mutations resulting in expression of mutant p53 (Figure 4L, N-P), whereas APR-246 did not induce cell death in ALL cells of patient 1 carrying a hemizygous splice site mutation without detectable expression of p53 protein (Figure 4L, M).
Figure 4.
APR-246 activity depends on mutant p53. (A-C) Stable lentiviral shRNA-mediated p53 knockdown in TP53mut (RS4;11 and KOPN-8) and TP53wt (NALM-6) cell lines. Western blot, anti-p53 antibody DO-7, with GAPDH as a loading control, non-transduced cells (ctrl), cells transduced with scrambled control (scr-ctrl) and TP53-specific shRNA (sh-p53). (D-F) Increasing cell death (forward/side scatter criteria, flow cytometry) in control transduced cells and abrogated cell death induction upon p53 knockdown at increasing concentrations of APR-246 (APR, 48 h) in TP53mut but not TP53wt cells. Mean values ± standard deviation (SD) of three independent experiments, each performed in triplicate. Student t-test, *P<0.05; **P<0.01. (G-J) si-RNA-mediated p53 downregulation in TP53mut (KOPN-8), (G) and TP53wt (NALM-6), (I) cells leading to clearly lower cell death induction upon p53 downregulation as compared to higher cell death in control cells (H), while p53 downregulation in TP53wt cells did not affect cell death induction upon APR-246 treatment (J). Mean values ± SD of three independent experiments. Student t-test, *P<0.05; **P<0.01. (K) TP53 mutations identified in primary samples from patients with acute lymphoblastic leukemia (ALL): the mutations were localized in the DNA-binding domain with one splice site mutation (open circle, Patient-1) and three missense mutations (filled circles, Patients -2, -3, -4). (L) No detectable p53 protein in ALL cells from Patient-1 (western blot, anti-p53 antibody DO-7, GAPDH as a loading control), and (M) no APR-246 activity in these cells (Patient-1), in contrast to cell death induction in cases carrying missense hot spot TP53 mutations (N, O, P; Patients-2, -3, -4). Mean values ± SD, measurements performed in triplicate. Student t-test, ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; n.s., not significant.
Table 2.
TP53 mutations in primary samples from patients with acute lymphoblastic leukemia.
APR-246 re-sensitizes TP53-mutated acute lymphoblastic leukemia to doxorubicin
TP53mut cancer cells show resistance to DNA damage. Therefore, we analyzed whether reactivation of mutant p53 re-sensitizes TP53mut ALL to the DNA-damaging agent doxorubicin, which is also used in treatment of pediatric ALL. TP53mut and TP53wt ALL cell lines and primograft samples were exposed to APR-246, doxorubicin, or to combinations of both at increasing concentrations. Strongly increased cell death rates were observed in all four TP53mut primografts and two cell lines upon com bination treatment with APR-246, as compared to APR-246 or doxorubicin alone, indicating synergistic activity for APR-246 and doxorubicin in TP53mut ALL (Figure 5A-F, Online Supplementary Table S3A). In TP53wt leukemias however, only doxorubicin showed cell death-inducing activity, which was not increased by adding APR-246 (Figure 5G-L, Online Supplementary Table S3B).
Figure 5.
APR-246 synergizes with doxorubicin. Synergistic activity of APR-246 in combination with the DNA-damage-inducing agent doxorubicin in TP53mut B-cell precursor (BCP)-acute lymphoblastic leukemia (ALL) cell lines (A, B) and TP53mut primograft leukemias (C-F) leading to doxorubicin re-sensitization, in contrast to no increased activity compared to treatment with doxorubicin alone in TP53wt cell lines (G, H) and TP53wt primograft ALL (I-L). Cell death (forward/side scatter criteria, flow cytometry) after exposure (primografts 24 h, cell lines 48 h) at indicated concentrations of APR-246 (APR), doxorubicin (DOX) or the combination (COMBI, 3 h APR-246 pre-incubation). Mean values ± standard deviation (SD) of three independent experiments, each performed in triplicate (cell lines: A, B, G, H). Mean values ± SD, three measurements (primografts: C-F, I-L). Combination indices (CI) indicating a strong synergistic (CI 0.1-0.3), a synergistic (CI <1), an additive (CI=1) or an antagonistic effect (CI>1) upon combination.
We also addressed whether induction of oxidative stress would increase the antileukemia activity of APR-246. In contrast to clearly increased cell death upon treatment with APR-246 together with the DNA-damaging agent doxorubicin, combining APR-246 with the ROS inducers 3-morpholinosydnonimine and tert-butyl hydroxyperoxide did not lead to clearly increased cell death (Online Supplementary Figure S7). Thus, APR-246 effectively synergizes with doxorubicin and re-sensitizes TP53mut ALL to DNA-damage-induced cell death, while additional ROS induction did not increase APR-246-mediated leukemia cell death.
Preclinical antileukemia activity of APR-246 and in vivo synergy with genotoxic therapy
Based on our findings, we investigated the antileukemia activity of APR-246 in a preclinical setting in vivo. Mice were transplanted with the TP53mut primograft (TP53mut-4; R175H). Upon manifestation of leukemia, as indicated by 5% or more human CD19+ ALL cells in the recipients’ peripheral blood, mice were treated with APR-246 (25, 50 or 100 mg/kg) or vehicle until control-treated animals showed signs of leukemia-related morbidity (3 weeks, days 1-5) (Figure 6A). Upon APR-246 treatment, a clear dose-dependent reduction of leukemia loads was observed in all three organ compartments: spleen, bone marrow and central nervous system (Figure 6B-D). Moreover, in leukemia cells isolated from these APR-246-treated animals, dose-dependent increases in mutant p53 with wildtype conformation and expression of PUMA and p21 were detected (Figure 6E, F), indicating restoration of wildtype p53 conformation and function in vivo.
Figure 6.
Anti-leukemia activity of APR-246 and synergy with genotoxic therapy in TP53-mutated acute lymphoblastic leukemia in vivo. (A) Schematic representation of the experimental procedure: endpoint analysis assessing leukemia loads in differently treated recipients. (B-D) Dose-dependent reduction of leukemia load in bone marrow (BM) (B), spleen (S) (C) and central nervous system (CNS) (D) upon treatment of mice bearing TP53mut-4 (mutation R175H) acute lymphoblastic leukemia (ALL) with solvent or increasing doses of APR-246 for 3 weeks as indicated (n=3 recipients per group, except n=2 for BM 100 mg/kg). Student t-test, *P<0.05; n.s., not significant. (E) Restoration of p53 wildtype conformation upon in vivo APR-246 therapy, immunoprecipitation (IP: anti-wt p53 specific antibody PAb1620, western blot: anti-p53 antibody DO-7, light chain-specific goat anti-mouse peroxidase conjugated binding protein, GAPDH as a loading control), and (F) dose-dependent induction of p53 transcriptional targets PUMA and p21 (western blot, GAPDH as a loading control). (G-I) Significant reduction of leukemia load in bone marrow (BM) (G), spleen (S) (H) and central nervous system (CNS) (I) upon treatment of TP53mut-3 (mutations R248Q, R213X) ALL-bearing mice with APR-246 (APR, 100 mg/kg) or solvent (CTRL) for 3 weeks, n=6 mice per group, Student t-test, *P<0.05. (J) Schematic representation of the experimental procedure: survival analysis. (K) Superior survival of animals treated with APR-246 (APR, 50 mg/kg, 3 weeks, days 1-5; n=6) as compared to doxorubicin (DOX, 2 mg/kg, 3 weeks, day 1; n=7) or vehicle (CTRL, 3 weeks, days 1-5; n=7) (P<0.0001); and synergy of the combination of APR-246 and doxorubicin (COMBI, APR-246, 50 mg/kg, days 1-5, and doxorubicin, 2 mg/kg, day 5; n=7) leading to increased survival as compared to APR-246 treatment alone (P=0.0005). Kaplan-Meier analysis, log-rank test.
Furthermore, APR-246 demonstrated strong in vivo antileukemia activity in another TP53mut ALL primograft sample (TP53mut-1; R248Q/R213X) leading to significantly reduced leukemia loads in the spleen, bone marrow and central nervous system upon therapy of leukemia-bearing recipients (Figure 6G-I).
We also addressed the effects of APR-246 in combination with doxorubicin in vivo. Recipients with manifest ALL (TP53mut-1; R248Q/R213X; 5% or more human ALL cells in the peripheral blood) were treated with APR-246, doxorubicin, or the combination of both for 3 weeks. After treatment, the animals were followed up and the time until onset of ALL-related morbidity was analyzed for each animal (Figure 6J). Upon sacrifice, high loads of human ALL were detected in the spleen and bone marrow of all recipients, confirming recurrence of manifest leukemia at clinical onset. Importantly, in addition to clear antileukemia activity as a single agent, leading to increased post-treatment survival (P<0.0001), APR-246 synergized strongly with doxorubicin and re-sensitized TP53mut ALL to genotoxic therapy in vivo, resulting in significantly prolonged survival as compared to APR-246 alone (P=0.0005) (Figure 6K). In all treatment experiments, application of APR-246 was well tolerated and no side effects were observed in the recipients.
Taken together, our findings in ALL carrying TP53 missense mutations in the DNA-binding domain, which lead to accumulation of dysfunctional p53, indicate that targeting mutant p53 with APR-246 results in refolding of mutant p53 into its native wildtype conformation, induction of p53 transcriptional targets, involvement of oxidative stress, induction of apoptosis, sensitization to DNA damage and, most importantly, preclinical antileukemia activity with significant reduction of leukemia loads, re-sensitization to genotoxic therapy and clearly prolonged survival in vivo. Thus, application of APR-246 can provide an effective strategy for directed therapeutic intervention in the high-risk subtype of TP53mut BCP-ALL.
Discussion
Investigating a large cohort of 62 patient-derived BCP-ALL samples, all identified TP53mut cases showed missense mutations leading to alterations in the DNA-binding domain of p53, high levels of p53 protein and insensitivity to doxorubicin. Interestingly, APR-246 demonstrated robust antileukemia activity in these cases, including induction of apoptosis, effective reduction of leukemia loads, and sensitization to doxorubicin in an in vivo model of TP53mut ALL. Both in vitro and in vivo experiments showed that treatment with APR-246 led to restored conformation and activation of mutant p53, and induction of transcriptional targets.
Alterations in TP53 have been described in diverse cancers at high frequencies of up to 95%.7 In our cohort, TP53 mutations were identified in four out of 62 cases (6.5%), in line with reported rates in ALL of 6-16%.14–16,18 All mutations identified in the primograft and cell line samples were missense mutations localized in the DNA-binding domain, with additional loss of the second allele in some of the cases, consistent with mutational patterns reported throughout different cancer types.10,43 One TP53mut sample showed hypodiploidy, in line with reported associations of hypodiploidy with germline and somatic TP53 mutations.19–21
Mutated dysfunctional p53 results in resistance to therapy-induced DNA damage48 and poor patient outcome.14,16–18,22 Correspondingly, increased numbers of TP53 alterations are seen at ALL relapse16 and all TP53mut leukemias were insensitive to the DNA-damaging agent doxorubicin. Importantly, these TP53mut leukemias were sensitive to APR-246, likely by reactivation of high levels of dysfunctional p53 accumulated in the cells. Most importantly, in line with reports in ovarian cancer,32,39 APR-246 clearly synergized with doxorubicin in vitro, ex vivo and in vivo, re-sensitizing initially resistant TP53mut ALL to DNA damage. Therefore, combining functional p53 restoration with genotoxic therapies triggering the p53-mediated DNA-damage response would be the rationale to apply APR-246 together with doxorubicin, a classical DNA-damage-inducing agent used in ALL treatment regimens. Importantly, a favorable pharmacological profile and anti-tumor effects were observed upon first clinical use of APR-246 in patients with refractory cancers40 and APR-246 is being tested in combination with anticancer agents, including doxorubicin, in ongoing phase II trials (ClinicalTrials.gov).41
We addressed the molecular mechanism of action of APR-246 and demonstrated restoration of p53 wildtype conformation, p53 pathway activation with induction of downstream transcriptional targets, and a contribution of oxidative stress leading to apoptosis of TP53mut BCP-ALL cells. Importantly, this antileukemia effect was also observed in vivo in TP53mut ALL, but not in TP53wt ALL, upon p53 knockdown or in a patient’s sample with a splice site mutation and loss of p53 protein expression. High levels of misfolded mutant p53 were described to be associated with APR-246 sensitivity in cancer cell lines,38,49,50 consistent with our observation of high APR-246 activity in TP53mut ALL with high p53 expression. However, evaluation of larger cohorts of patients together with outcome data would be required to explore the value of the level of expression of p53 as an indicator of APR-246 responsiveness in TP53mut ALL.
APR-246 activity has been reported to be mediated independently of p53 by induction of oxidative stress in other types of cancer, including acute myeloid leukemia and multiple myeloma.30–33 However, we only observed cell death in TP53mut ALL, although TP53mut and TP53wt ALL showed no differences in induction of and sensitivity to oxidative stress. Interestingly, APR-246 activity in TP53mut ALL was partially inhibited by ROS neutralization. This suggests that induction of oxidative stress contributes to APR-246-mediated cell death in ALL, in line with reports on a dual mode of action of APR-246,30–32 which might vary between tumor types and cellular context.
The presence of p53 in a mutated, dysfunctional form, as typically is the case for missense mutations in the DNA-binding core domain, enables binding of the active moiety of APR-246, leading to activity in BCP-ALL.11,28 This is of clinical relevance, since the majority of TP53 mutations in BCP-ALL are missense hot spot mutations in the DNA-binding domain15,43 resulting in accumulation of misfolded p53 protein, which is targeted by APR-246. However, the precise mechanism of activity on DNA contact mutations is not yet known. Importantly, we and others 27 have demonstrated antitumor activity on structural and contact mutants including clear preclinical antileukemia activity on TP53mut ALL carrying either a structure (R175H) or contact (R248Q) mutation.
Taken together, our study shows that the small molecule APR-246 exhibits profound antileukemia activity in TP53mut BCP-ALL, targeting non-functional mutant p53 resulting from missense mutations in the DNA-binding domain of TP53, the most frequent mutation type reported throughout different malignancies. Mechanistically, we showed that APR-246 led to restoration of p53’s wildtype conformation, pathway activation with expression of transcriptional targets and induction of apoptosis in TP53mut ALL. Moreover, we found a clear synergism between APR-246 and doxorubicin treatment, strongly suggesting that the combination of p53 reactivation and DNA-damage induction could be an effective antileukemia strategy for BCP-ALL patients with TP53 missense mutations. Hence, targeting mutant p53 appears to be a promising, directed treatment for this high-risk subgroup of TP53mut ALL.
Acknowledgments
The authors would like to thank Aprea Therapeutics (Stockholm, Sweden) for kindly providing APR-246 for the study, S. Volk and S. Essig for excellent technical assistance, the Ulm University Sorting and Animal Facilities and Pharmacy of the Ulm University Medical Center, and the INFORM study group. The authors would also like to thank the International Graduate School in Molecular Medicine Ulm (SD, EB), Madeleine-Schickedanz-Stiftung and “Förderverein für Krebskranke Kinder Tübingen” (ME, RH), Swedish Research Council and Swedish Childhood Cancer Society (GS), EU COST Action CA16223 (GtK), the German Research Foundation, SFB 1074 B6 (LHM, KMD) and B1 (SS) for supporting the work.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/1/170
References
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. [DOI] [PubMed] [Google Scholar]
- 2.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. [DOI] [PubMed] [Google Scholar]
- 3.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. [DOI] [PubMed] [Google Scholar]
- 4.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57(7):1083–1093. [DOI] [PubMed] [Google Scholar]
- 5.Chen PL, Chen YM, Bookstein R, Lee WH. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250(4987):1576–1580. [DOI] [PubMed] [Google Scholar]
- 6.Isobe M, Emanuel BS, Givol D, Oren M, Croce CM. Localization of gene for human p53 tumour antigen to band 17p13. Nature. 1986;320(6057):84–85. [DOI] [PubMed] [Google Scholar]
- 7.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342(6250):705–708. [DOI] [PubMed] [Google Scholar]
- 9.Baker SJ, Fearon ER, Nigro JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244(4901):217–221. [DOI] [PubMed] [Google Scholar]
- 10.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. [DOI] [PubMed] [Google Scholar]
- 11.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265(5170):346–355. [DOI] [PubMed] [Google Scholar]
- 13.Kim MP, Zhang Y, Lozano G. Mutant p53: multiple mechanisms define biologic activity in cancer. Front Oncol. 2015;5:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiaretti S, Brugnoletti F, Tavolaro S, et al. TP53 mutations are frequent in adult acute lymphoblastic leukemia cases negative for recurrent fusion genes and correlate with poor response to induction therapy. Haematologica. 2013;98(5):e59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stengel A, Schnittger S, Weissmann S, et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood. 2014;124(2):251–258. [DOI] [PubMed] [Google Scholar]
- 16.Hof J, Krentz S, van Schewick C, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol. 2011;29(23):3185–3193. [DOI] [PubMed] [Google Scholar]
- 17.Richter-Pechanska P, Kunz JB, Hof J, et al. Identification of a genetically defined ultrahigh-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J. 2017;7(2):e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3):705–711. [DOI] [PubMed] [Google Scholar]
- 19.Muhlbacher V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer. 2014;53(6): 524–536. [DOI] [PubMed] [Google Scholar]
- 20.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian M, Cao X, Devidas M, et al. TP53 germline variations influence the predisposition and prognosis of B-cell acute lymphoblastic leukemia in children. J Clin Oncol. 2018;36(6):591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krentz S, Hof J, Mendioroz A, et al. Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 2013;27(2): 295–304. [DOI] [PubMed] [Google Scholar]
- 23.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22(4):730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bykov VJ, Wiman KG. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 2014;588(16):2622–2627. [DOI] [PubMed] [Google Scholar]
- 25.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127(7): 1323–1334. [DOI] [PubMed] [Google Scholar]
- 26.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–288. [DOI] [PubMed] [Google Scholar]
- 28.Lambert JM, Gorzov P, Veprintsev DB, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–388. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Bykov VJN, Wiman KG, Zawacka-Pankau J. APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 2018;9(5):439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tessoulin B, Descamps G, Moreau P, et al. PRIMA-1Met induces myeloma cell death independent of p53 by impairing the GSH/ROS balance. Blood. 2014;124(10): 1626–1636. [DOI] [PubMed] [Google Scholar]
- 31.Ali D, Mohammad DK, Mujahed H, et al. Anti-leukaemic effects induced by APR-246 are dependent on induction of oxidative stress and the NFE2L2/HMOX1 axis that can be targeted by PI3K and mTOR inhibitors in acute myeloid leukaemia cells. Br J Haematol. 2016;174(1):117–126. [DOI] [PubMed] [Google Scholar]
- 32.Mohell N, Alfredsson J, Fransson A, et al. APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 2015;6:e1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng X, Zhang MQ, Conserva F, et al. APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 2013;4:e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zandi R, Selivanova G, Christensen CL, Gerds TA, Willumsen BM, Poulsen HS. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res. 2011;17(9):2830–2841. [DOI] [PubMed] [Google Scholar]
- 35.Nahi H, Merup M, Lehmann S, et al. PRIMA-1 induces apoptosis in acute myeloid leukaemia cells with p53 gene deletion. Br J Haematol. 2006;132(2):230–236. [DOI] [PubMed] [Google Scholar]
- 36.Bykov VJ, Zache N, Stridh H, et al. PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24(21):3484–3491. [DOI] [PubMed] [Google Scholar]
- 37.Izetti P, Hautefeuille A, Abujamra AL, et al. PRIMA-1, a mutant p53 reactivator, induces apoptosis and enhances chemotherapeutic cytotoxicity in pancreatic cancer cell lines. Invest New Drugs. 2014;32(5):783–794. [DOI] [PubMed] [Google Scholar]
- 38.Liu DS, Read M, Cullinane C, et al. APR-246 potently inhibits tumour growth and overcomes chemoresistance in preclinical models of oesophageal adenocarcinoma. Gut. 2015;64(10):1506–1516. [DOI] [PubMed] [Google Scholar]
- 39.Fransson A, Glaessgen D, Alfredsson J, Wiman KG, Bajalica-Lagercrantz S, Mohell N. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant high-grade serous ovarian cancer. J Ovarian Res. 2016;9(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehmann S, Bykov VJ, Ali D, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30(29):3633–3639. [DOI] [PubMed] [Google Scholar]
- 41.Zarin DA, Tse T, Williams RJ, Rajakannan T. Update on trial registration 11 years after the ICMJE policy was established. N Engl J Med. 2017;376(4):383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer LH, Eckhoff SM, Queudeville M, et al. Early relapse in ALL is identified by time to leukemia in NOD/SCID mice and is characterized by a gene signature involving survival pathways. Cancer Cell. 2011;19(2): 206–217. [DOI] [PubMed] [Google Scholar]
- 43.Bouaoun L, Sonkin D, Ardin M, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Human Mutat. 2016;37(9):865–876. [DOI] [PubMed] [Google Scholar]
- 44.Bykov VJ, Zhang Q, Zhang M, Ceder S, Abrahmsen L, Wiman KG. Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient cancer therapy. Front Oncol. 2016;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisek K, Walerych D, Del Sal G. Mutant p53-Nrf2 axis regulates the proteasome machinery in cancer. Mol Cell Oncol. 2017;4(1): e1217967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu DS, Duong CP, Haupt S, et al. Inhibiting the system xC(−)/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun. 2017;8:14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haffo L, Lu J, Bykov VJN, et al. Inhibition of the glutaredoxin and thioredoxin systems and ribonucleotide reductase by mutant p53-targeting compound APR-246. Sci Rep. 2018;8(1):12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hientz K, Mohr A, Bhakta-Guha D, Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2017;8(5):8921–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bykov VJ, Issaeva N, Selivanova G, Wiman KG. Mutant p53-dependent growth suppression distinguishes PRIMA-1 from known anticancer drugs: a statistical analysis of information in the National Cancer Institute database. Carcinogenesis. 2002;23(12):2011–2018. [DOI] [PubMed] [Google Scholar]
- 50.Synnott NC, Murray A, McGowan PM, et al. Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer. 2017;140(1):234–246. [DOI] [PubMed] [Google Scholar]