Abstract
Recent advances in synthetic biology have led to the development of nucleic acid polymers with backbone structures distinct from those found in nature, termed xeno-nucleic acids (XNAs). Several unique properties of XNAs make them attractive as nucleic acid therapeutics, most notably their high resistance to serum nucleases and ability to form Watson–Crick base pairing with DNA and RNA. The ability of XNAs to induce immune responses has not been investigated. Threose nucleic acid (TNA), a type of XNA, is recalcitrant to nuclease digestion and capable of undergoing Darwinian evolution to produce high affinity aptamers; thus, TNA is an attractive candidate for diverse applications, including nucleic acid therapeutics. In this study, we evaluated a TNA oligonucleotide derived from a cytosine-phosphate-guanine oligonucleotide sequence known to activate toll-like receptor 9-dependent immune signaling in B cell lines. We observed a slight induction of relevant mRNA signals, robust B cell line activation, and negligible effects on cellular proliferation.
Keywords: threose nucleic acid, toll-like receptor 9, immunogenicity, nucleic acid therapeutic, chemical modification, B cell
Introduction
Chemical modifications have been utilized extensively to improve the binding energy, stability, and tolerability of potential therapeutics, including antisense oligonucleotides, siRNAs, and aptamers, among others. Phosphorothioate modifications in particular have been effective due to their ability to enhance cellular uptake and evade nuclease-mediated degradation in serum. More recently, advances in synthetic biology have led to the development of an expanded set of nucleic acid polymers with backbone structures distinct from those found in nature, termed xeno-nucleic acids (XNAs) [1,2].
XNAs are particularly attractive in the field of nucleic acid therapeutics due to several unique properties of several XNAs, mostly notably their high resistance to serum nucleases [2] and their ability to form Watson–Crick base pairing with DNA and RNA [1]. Based on these properties, XNA modifications are under exploration for a variety of applications, such as selection of aptamers targeted to specific proteins, development of nanostructures for therapeutic delivery of payloads, and diagnostics [2–7]. Thus, XNA-based nucleic acid-based tools hold much promise in the field.
However, XNAs have not been extensively evaluated for their ability to induce innate immune responses, and such evaluations will be critical for further therapeutic development of XNAs. Notably, some chemical modifications have been shown to abolish or decrease immune signaling (e.g., locked nucleic acid and 2’-Fluoro substitutions) [8], while others robustly induce immune signaling (e.g., phosphorothioate cytosine-phosphate-guanine [CpG] oligonucleotides [ODNs]). Both phenotypic classes are highly relevant to therapeutics, as some conditions may benefit from stimulation of the immune system (e.g., vaccine adjuvants and anticancer applications), while others may benefit from inhibition of immune signaling (e.g., autoimmune disease) or avoidance of immune signaling (e.g., antisense, XNA-mediated payload delivery) [3,4,9–11].
Threose nucleic acids (TNAs) are XNAs in which the natural ribose sugar found in RNA has been replaced by a four-carbon α-L-threofuranose sugar [12]. TNA adopts an A-like helical geometry, facilitating stable, anti-parallel Watson–Crick duplexes with complementary strands of DNA and RNA [13,14]. Due to a high level of resistance to serum nucleases [15] and recent advances in the use of engineered polymerases that can copy genetic information back and forth between TNA and DNA [1,16], TNA has become an attractive tool for a variety of applications. In fact, in vitro selection of TNA aptamers targeting human thrombin [16], HIV reverse transcriptase [5], and ochratoxin A [6] has recently been reported, as well as an antisense TNA targeted to green fluorescent protein (GFP) that suppresses GFP expression in live cells [17].
Despite recent progress in the utilization of XNAs such as TNA for therapeutic applications, the ability of various XNAs to either avoid or initiate immune responses remains largely unexplored. In this study, we set out to evaluate the ability of TNAs to induce innate immune responses in B cell lines by utilizing TNA sequences derived from ligands recognized by human toll-like receptor 9 (TLR9).
TLR9 is a pattern recognition receptor that specifically recognizes DNA. Chemical modifications, specifically phosphorothioate modifications, have long been incorporated into ligands for TLR9, and these ligands have been investigated extensively for a variety of therapeutic applications (e.g., adjuvants and anticancer drugs) due to their ability to induce innate immune responses and prime the adaptive immune response. Initiation of innate immune responses through TLR9 by self-DNA [18], unmethylated CpG DNA [19–22], RNA-DNA duplexes [23], and synthetic oligonucleotide sequences containing phosphorothioate-modified CpG ODN motifs [19–22] has been documented.
Notably, most of the work to date evaluating TLR9-specific responses has utilized phosphorothioate-modified CpG ODNs [24], demonstrating that TLR9 ligands are amenable to chemical modification while retaining functionality. However, the nature, magnitude, and cell specificity of innate immune induction can differ significantly between the chemically modified phosphorothioate and the natural phosphodiester backbones [25–27], suggesting that there is more to learn about interactions of various natural and unnatural nucleic acids with nucleic acid-sensing receptors.
Currently, there are three classes of CpG ODNs. Class A CpG ODNs are characterized by a phosphodiester palindromic motif containing a central CpG motif and a phosphorothioate-modified poly-G string on the 3′ end that has been shown to increase cellular uptake and provide resistance to serum nucleases [20,21,28,29]. Class A ODNs induce high levels of interferon (IFN) production in plasmacytoid dendritic cells, but only weakly stimulate signaling through TLR9-dependent nuclear factor kappa B (NFκB) activation [21,29]. Class B CpG ODNs contain a full phosphorothioate backbone with one or more CpG dinucleotides. Unlike Class A ODNs, Class B ODNs strongly stimulate NFκB signaling, but only weakly stimulate IFN signaling [21,30,31]. Class C CpG ODNs are a combination of classes A and B, with a complete phosphorothioate backbone and a CpG-containing palindromic motif, with the capability to induce IFN secretion and activate NFκB signaling [21,22].
For the purposes of this study, we utilized ODN sequence 2395 (5′-tcgtcgttttcggcgc:gcgccg-3′), one of several commonly studied Class C TLR9 ligands, which contains nuclease-resistant phosphorothioate nucleotides and is shown with the palindrome underlined. This ODN has been demonstrated to induce both IFN and pro-inflammatory responses [21,32,33] and to activate B cells [34]. Our experiments were performed with sequence 2395 synthesized using a fully phosphorothioate backbone (CpG 2395), a fully phosphodiester backbone (DNA 2395), and a fully TNA backbone (TNA 2395) to enable comparison of the induction of innate immune responses among the three backbones in B cell lines.
We then measured induction of various innate mRNAs by quantitative real time-polymerase chain reaction (qRT-PCR), B cell line activation by upregulation of surface-expressed CD86, and proliferative responses. Our data indicate that while TNA 2395 induces only low-level upregulation of IFN and pro-inflammatory genes at the dosages utilized in this study, TNA 2395 is capable of robust B cell line activation, similar to both CpG 2395 and DNA 2395. While the specific receptor involved in TNA 2395-mediated signaling remains unknown, these data clearly establish that TNA 2395 is capable of inducing innate immune responses in B cell lines.
Materials and Methods
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Poly I:C was purchased from Invivogen (San Diego, CA). The synthetic TLR9 ligand, CpG ODN 2395, a type C ODN, was purchased from Invivogen. DNA 2395 was purchased from Integrated DNA Technologies (Coralville, Iowa). TNA molecules were synthesized on an Applied Biosystems 3400 DNA synthesizer using standard β-cyanoethyl phosphoramidite chemistry with chemically synthesized TNA phosphoramidites [35]. QUANTI-Blue detection reagent for use with the HEK-Blue hTLR9 secreted alkaline phosphatase (SEAP) reporter cell line was purchased from Invivogen (San Diego, CA).
Cell lines
The human B cell lines, Ramos (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Ramos Cells from Drs. Li Wu and Vineet KewalRamani) [36] and Raji (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Raji from Drs. Li Wu and Vineet KewalRamani) [36], were maintained in Roswell Park Memorial Institute (RPMI) medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and 2 mM l-glutamine (Gibco; Life Technologies, Grand Island, NY). The human cell line expressing TLR9, HEK-Blue hTLR9 (Invivogen), was maintained according to the manufacturer's instructions. All cell lines were maintained at 37°C in 5% carbon dioxide.
Cell stimulation
For stimulation experiments, Ramos and Raji cells were plated in 1 mL serum-free medium or serum-containing medium at a density of 2 × 105 cells per well in 12-well plates. HEK-Blue hTLR9 cells were plated at 5 × 104 cells per well in 96-well plates. Following plating (Ramos and Raji) or overnight incubation to allow adherence to the plates (HEK-Blue hTLR9), cells were stimulated with the indicated ligands (untreated/vehicle only, poly I:C, CpG ODN 2395, DNA 2395, or TNA 2395) in nuclease-free water at the indicated concentrations.
RNA isolation and quantitation
For isolation of total cellular RNA, Ramos cells treated with or without ligands for specified TLRs were collected by centrifugation 24 h poststimulation and washed twice with 1 × phosphate buffered saline (PBS), followed by RNA isolation using TRIzol reagent (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions. RNA was precipitated twice and washed with 70% ethanol to remove any contaminants resulting from TRIzol RNA isolation procedures. Isolated RNA was subjected to DNase treatment using TURBO DNase (Ambion; Life Technologies, Grand Island, NY) and quantified on a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Real time quantitative PCR
Total cellular was used to synthesize cDNA using random hexamer primers with the ImProm II Reverse Transcription System (Promega, Madison, WI) as per the manufacturer's instructions. The cDNA synthesis was performed using 500 ng RNA. “No RT” and “non-template” (NT) controls were included. Before performing real time quantitative PCR (RT-qPCR), cellular and viral cDNA, including “no RT” and “NT” controls, were validated by end point PCR (40 cycles) using primers specific for 18S rRNA to ensure that “no RT” and “NT” controls were free of contaminating signal.
RT-qPCR was performed on each set of cDNA using iTaq Universal SYBR Green Master Mix (Bio-Rad, Hercules, CA) with primers specific to 18S rRNA (forward 5′-CAGCCACCCGAGATTGAGCA-3′ and reverse 5′-TAGTAGCGACGGGCGGTGTG-3′), interleukin 8 (IL-8; forward 5′-CTTCTAGGACAAGAGCCAGGAAGAAACC-3′ and reverse 5′-GTCCAGACAGAGCTGTCTTCCATCAGAA-3′), tumor necrosis factor alpha (TNFα) (TNFα; forward 5′-GAGTGACAAGCCTGTAGCCCATGTTGTA-3′ and reverse 5′-GCAATGATCCCAAAGTAGACCTGCCCAG-3′), IFN beta (IFNβ; forward 5′-TTGTGCTTCTCCACGACAGC-3′ and reverse 5′-GCACAGTGACTGTCCTCCT-3′), IFN-induced protein with tetratricopeptide repeats 1 (IFIT-1; forward 5′-GTAACTGAAAATCCACAAGACAGAATAG-3′ and reverse 5′-GCAAATGTTCTCCACCTTGTCCA-3′), and 2′-5′-oligoadenylate synthetase 1 (OAS; forward 5′-TGTTTCCGCATGCAAATCAACCATGC-3′ and reverse 5′-GCCAGTCAACTGACCCAGGGCA-3′), according to the manufacturer's instructions.
Each sample was assayed in triplicate to determine technical variability. Relative quantities were determined using the relative quantity (2−ΔΔCT) method as previously described [37]. Samples were normalized to the specified endogenous control (18S rRNA) to determine expression level relative to total cellular RNA, and these values were plotted relative to the corresponding value for the negative controls (untreated, set to 1). Samples were assayed on the ABI 7500 (Applied Biosystems, Foster City, CA) and analyzed using ABI 7500 Software Version 2.3. Experiments were repeated thrice.
Flow cytometry
Activation of Ramos and Raji B cell lines was determined using flow cytometry to detect surface-expressed CD86 after 24 or 72 h poststimulation. A phycoerythrin (PE)-labeled antibody for CD86 (B7–2) and a corresponding isotype control, mouse IgG2b kappa, were purchased from eBiosciences (now Thermo Fisher Scientific, Grand Island, NY). Following stimulation of Ramos and Raji cells for 72 h as described in “cell stimulation,” cells were collected by centrifugation and resuspended in 1 × PBS. Cells were incubated with αCD86-PE or isotype control for 30 min at 4°C followed by washing twice in 1 × PBS. Cells were then analyzed by flow cytometry using a BD Accuri C6 Flow Cytometer (BD Biosciences, San Jose, CA).
Proliferation assays
Proliferation of Ramos cells in response to cell stimulation was assessed by performing cell counts 24 or 72 h poststimulation. Cell counts were performed by collecting cells, staining with Trypan blue to exclude nonviable cells, and counting using hemacytometer. Cell counts were confirmed by counting using a BD Accuri C6 Flow Cytometer (BD Biosciences).
SEAP reporter assays
HEK-Blue hTLR9 cells were seeded at a density of 5 × 104 cells per well in 96-well plates. Cells were stimulated in triplicate with the indicated ligands at the indicated concentrations for either 24 or 72 h. Following stimulation, QUANTI-Blue reagent was added to a separate 96-well plate according to the manufacturer's instructions. To assess SEAP activity, 20 μL of supernatant from each well was added to the QUANTI-Blue reagent (180 μL). Reactions were incubated for 2 h at 37°C, and absorbance was detected using a PerkinElmer Plate Reader at an optical density of 655 nm.
Results and Discussion
Differential mRNA upregulation by low and high doses of TLR9 ligands in Ramos cells
To explore the impact of phosphorothioate, phosphodiester, and TNA backbones on the ability of sequence 2395 to induce innate immune responses, we first sought to determine the responsiveness of the Ramos cells to low and high doses of the previously characterized, commercially available TLR9 ligand, CpG 2395 (phosphorothioate backbone), and the phosphodiester form of CpG 2395, DNA 2395. Cells were also stimulated with the TLR3 ligand, Poly I:C, which is also known to induce innate immune responses in Ramos cells [38], as a positive control.
Ramos cells were plated in serum-free medium and stimulated with either 1 or 10 μg/mL ligand for 24 h. Stimulated samples were then subjected to analysis by qRT-PCR to determine RNA levels of a combination of innate immune genes, including IFIT-1, IFNβ, IL-8, OAS, and TNFα. These genes were chosen to represent both the IFN response initiated through IFN regulatory factor 7 (IFIT-1, IFNβ, OAS) and the pro-inflammatory response initiated through NFκB (IL-8 and TNFα) that can be elicited by Class C ODNs, such as CpG 2395.
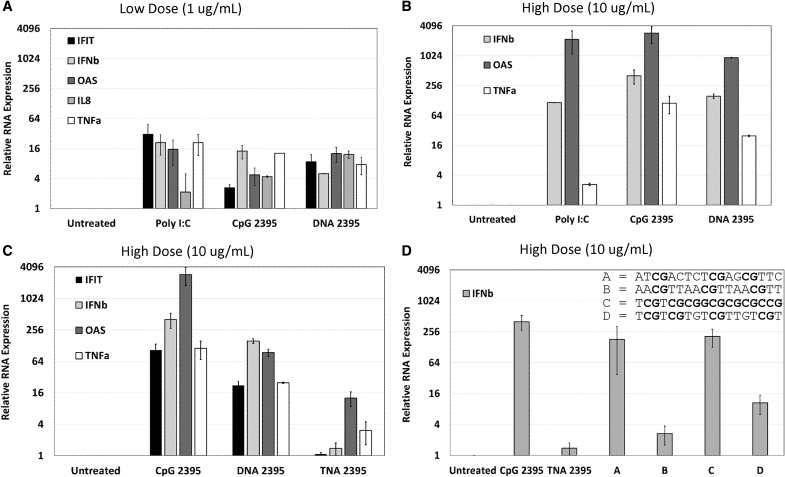
As shown in Fig. 1A and B, we observed minimal RNA upregulation with the low dose ligand, while we observed high levels of RNA upregulation with high dose ligands. Notably, the observed response profiles were very similar for CpG 2395 and DNA 2395. These results demonstrate that both the phosphorothioate and phosphodiester forms of sequence 2395 are capable of upregulating IFN and pro-inflammatory genes upon stimulation in Ramos cells and that the magnitude of the response is dose dependent.
FIG. 1.
Upregulation of mRNA in response to CpG 2395, DNA 2395, TNA 2395, and a TNA panel of variable sequence composition in Ramos cells. For all panels, Ramos cells were plated at a density of 2 × 105 cells in 12-well plates with serum-free medium and stimulated with the indicated ligands (negative control, untreated/vehicle only; positive control, Poly I:C for stimulation of TLR3; test ligands, CpG 2395, DNA 2395, TNA 2395, and Sequences A, B, C, and D) at the indicated concentrations (A, 1 μg/mL; B–D, 10 μg/mL). After 24 h, cells were collected, pelleted, and washed in 1 × PBS. Total cellular RNA was then harvested from each sample. Following DNase treatment of the total cellular RNA, cDNA was synthesized, and quantitative real time-polymerase chain reaction was performed to examine expression of the indicated genes (IFIT, IFNβ, IL-8, OAS, and/or TNFα) and the 18S rRNA housekeeping gene for normalization. Relative quantities were determined using the relative quantity (2−ΔΔCT) method as described in Materials and Methods section. Average quantities are shown for technical replicates measured in triplicate. Error bars represent the standard deviation for three technical replicates within each experiment. A representative experiment is shown in each panel. Experiments were repeated thrice. TNA, threose nucleic acid; CpG, cytosine-phosphate-guanine; PBS, phosphate buffered saline.
TNA 2395 displays lower levels of mRNA upregulation in Ramos cells compared to CpG 2395 and DNA 2395
We next wanted to determine whether the 2395 sequence synthesized with a TNA backbone was able to upregulate innate immune genes in the Ramos cells. As shown in Fig. 1C, while TNA 2395 was capable of upregulating OAS and TNFα, very little upregulation was observed for IFIT and IFNβ. The low level of IFNβ upregulation observed in Fig. 1C was surprising, given the higher level upregulation of OAS, as OAS is an IFN-stimulated gene. It is possible that TNA 2395 induces another type I IFN, such as a subtype of interferon alpha (IFNα), which could serve to upregulate OAS. However, as a variety of different IFNα subtypes could be responsible for the subsequent upregulation of OAS, determination of the specific type I IFN responsible was not investigated in this study.
Collectively, the results in Fig. 1C demonstrate that while TNA 2395 was capable of eliciting an innate immune response at the level of mRNA through upregulation of IFN (OAS) and pro-inflammatory (TNFα) genes in the Ramos cells, the magnitude of the response was significantly lower than the levels observed for CpG 2395 and DNA 2395.
TNA-induced upregulation of mRNA varies based on sequence
To examine whether mRNA upregulation of innate immune genes was a generalizable phenomenon of TNA or whether the responses may be sequence specific, we evaluated a small set of TNA molecules containing different sequences and variable numbers and spacing of CpG motifs for their abilities to upregulate IFNβ, as TNA 2395 demonstrated little ability to upregulate IFNβ. Notably, variation in the ability of CpG-containing sequences to upregulate interferons (IFNs) through TLR9 has been documented for the different classes of commercially available TLR9 ligands [21,32,33]. The set of sequences tested here, with CpG motifs in bold, included the following: Sequence A (ATCGACTCTCGAGCGTTC), Sequence B (AACGTTAACGTTAACGTT), Sequence C (TCGTCGCGGCGCGCGCCG), and Sequence D (TCGTCGTGTCGTTGTCGT).
As shown in Fig. 1D, we observed variable IFNβ upregulation for the different TNA sequences, suggesting that some TNAs containing CpG motifs are able to significantly upregulate IFNβ. Interestingly, the sequence most similar to TNA 2395, Sequence C (lacking the internal tttt motif present in TNA 2395), was able to robustly induce IFNβ mRNA. Sequence B, similarly to TNA 2395, was not a strong IFNβ inducer. A commonality between the two sequences is the four nucleotide spacing between CpG motifs (all CpGs for Sequence B are separated by four nucleotides, while the second CpG and palindrome sequence of TNA 2395 is separated by four nucleotides).
Notably, the three stimulatory TNAs, Sequence A, C, and (to a lesser extent) D, contain CpG motifs in closer proximity and lack a palindromic sequence. Thus, it is possible that spacing of CpG motifs may play a role in sequence specificity for the TNA sequences and may enable tuning of TNA sequences for either innate immune stimulation or evasion of stimulation. Future work beyond the scope of this study should examine sequence specificity constraints for TNA-mediated signaling, as well as determine whether TLR9 is responsible for TNA-induced signaling outcomes.
Ramos and Raji cells upregulate CD86 in response to CpG 2395, DNA 2395, and TNA 2395
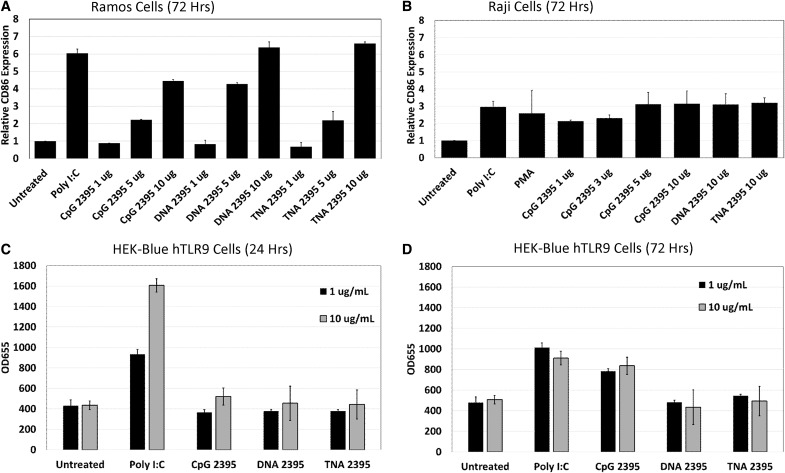
CpG ODNs are known to trigger both activation and proliferation of primary B cells and various cell lines derived from B cell malignancies [34,39–42]. Therefore, we next set out to determine whether stimulation of Ramos and Raji B cell lines by CpG 2395, DNA 2395, and TNA 2395 led to upregulation of the well-known B cell activation marker, CD86 [28,42,43]. Ramos cells were stimulated with ligands ranging from 1 to 10 μg/mL. After 72 h, cells were collected and analyzed for CD86 expression by flow cytometry. As shown in Fig. 2A, Ramos cells were robustly activated by all three 2395 sequences. The highest ligand dose utilized yielded 4.5-fold induction by CpG 2395 and a 6.5-fold induction for both DNA 2395 and TNA 2395.
FIG. 2.
CpG 2395, DNA 2395, and TNA 2395 activate Ramos and Raji cells, while only CpG 2395 induces SEAP reporter activity in HEK-Blue hTLR9 cells. (A, B) Ramos and Raji cells were plated at a density of 2 × 105 cells in 12-well plates and stimulated with the indicated ligands at the indicated concentrations. After 72 h, cells were harvested for flow cytometric analysis of surface-expressed CD86 expressing using a PE-labeled antibody specific for CD86. A PE-labeled isotype antibody was included as a control. Data are shown relative to the untreated control for Ramos (A) and Raji (B) cells. Error bars represent the standard deviation between two replicates within a single experiment. A representative experiment is shown in each panel. Experiments were performed 3 times. (C, D) HEK-Blue hTLR9 cells were plated at a density of 5 × 104 in 96-well plates and allowed to adhere overnight. Cells were then stimulated with the indicated ligands at the indicated concentrations. After 24 (A) or 72 (B) hours, supernatants were harvested and tested for SEAP activity using QUANTI-Blue reagent. Samples were assayed in triplicate. Error bars represent standard deviations for three replicates within a single experiment. A representative experiment is shown in each panel. Experiments were performed 3 times. PE, phycoerythrin; SEAP, secreted alkaline phosphatase.
Interestingly, although TNA 2395 induced only low-level upregulation of innate immune-associated mRNAs explored in the context of this study, activation of Ramos cells through upregulation of CD86 expression by TNA 2395 was similar to that of both CpG 2395 and DNA 2395, indicating that TNA 2395 is able to robustly activate Ramos cells.
In addition to activation of the Ramos cells, the 2395 sequences were analyzed for their ability to activate Raji cells. Notably, Raji cells display a higher level of CD86 expression in their unstimulated state, presumably due to the chronic Epstein–Barr Virus infection of the cell line [44]. Similarly to the Ramos cells, CpG 2395, DNA 2395, and TNA 2395 were all able to upregulate CD86 expression in the Raji cells above the background level after 72 h stimulation, although to a lesser extent (threefold) (Fig. 2B).
Together, these results confirm prior studies demonstrating that CpG ODNs containing phosphorothioate and phosphodiester backbones are able to activate B cell lines. In addition, these data also show that a TNA backbone is able to robustly activate B cell lines, despite only low-level upregulation of innate immune-associated genes examined in this study compared to CpG 2395 and DNA 2395.
HEK-Blue hTLR9 cells respond to CpG 2395, but fail to respond to DNA 2395 and TNA 2395
HEK-Blue hTLR9 cells are a commercially available cell line specifically designed to analyze the immunological properties of TLR9-specific ligands. These cells stably express SEAP under control of the NFκB promoter. Originally, we planned to utilize a lentiviral vector expressing a TLR9-specific guide RNA and CRISPR/Cas9 to knockout TLR9 in the HEK-Blue hTLR9 cells to determine whether TNA 2395 was specifically signaling through TLR9, as B cell lines are notoriously difficult to transduce. However, as shown in Fig. 2C and D, while the HEK-Blue hTLR9 cells respond to Poly I:C after 24 and 72 h of stimulation and to CpG 2395 after 72 h of stimulation, they fail to respond to either DNA 2395 or TNA 2395. Thus, we were unable to examine the ability of TNA 2395 to specifically stimulate TLR9 within the scope of this study.
Interestingly, a recent report demonstrated that expression of human macrophage scavenger receptor 1 gene (hMSR1), which is not expressed in the HEK-Blue hTLR9 cells, was necessary for induction of SEAP by CpG ODN containing a phosphodiester backbone in the HEK-Blue hTLR9 cells [45]. When hMSR1 was transfected into the HEK-Blue hTLR9 cells, uptake and SEAP activity of CpG DNA was shown to be equivalent to that of CpG ODN with a phosphorothioate backbone [45]. Thus, it is possible that addition of hMSR1, or some other receptor, to our HEK-Blue hTLR9 cells could allow uptake of DNA 2395 and TNA 2395, conveying the ability of the cells to respond to these ligands. However, the investigation of whether hMSR1 or some other cell surface receptor facilitates uptake of DNA 2395 or TNA 2395 was beyond the scope of this study.
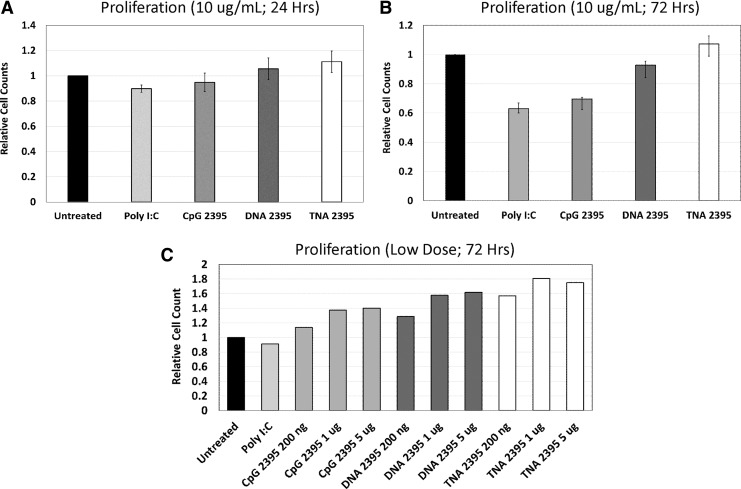
CpG 2395 and DNA 2395 inhibit Ramos cell proliferation at high doses, but stimulate proliferation at low doses
In addition to activation of B cells, CpG ODNs have been shown to induce proliferation of primary B cells and some cell lines derived from B cell malignancies [28,34,39,41,43]. However, stimulation with CpG ODN has also been demonstrated to induce apoptosis in other B cell lines [11,41]. Therefore, we next sought to determine whether stimulation of Ramos cells by CpG 2395, DNA 2395, and TNA 2395 leads to a proliferative response. We examined proliferative responses by cell counting at both 24 and 72 h at a ligand dose of 10 μg/mL. As shown in Figure 3, at 24 h poststimulation, we did not observe a difference in cell counts. This was expected, as the cells would not have undergone many rounds of cell division. At 72 h, we observed a reduction in cell number compared to the untreated control for Poly I:C and CpG 2395.
FIG. 3.
CpG 2395 decreases B cell proliferation, while DNA 2395 and TNA 2395 do not alter B cell proliferation. Ramos cells were plated at a density of 2 × 105 cells in 12-well plates and stimulated with the indicated ligands at the indicated concentrations. After 24 (A) or 72 (B, C) hours, cells were collected and counted using a hemacytometer. Cell counts were confirmed using a flow cytometer to ensure counting accuracy. Cell counts relative to the untreated control are shown. Experiments were repeated 3 times.
Poly I:C is a well-known inducer of apoptosis through TLR3-mediated signaling, so the observed decrease was expected. CpG ODNs have specifically been shown to induce apoptosis in Burkitt's Lymphoma-derived cell lines, such as the Ramos cells [46]. DNA 2395 demonstrated a slight reduction in cell counts at 72 h, but not to the magnitude of Poly I:C nor CpG 2395. TNA 2395 cell numbers remained equivalent to the untreated control, suggesting that TNA 2395 does not alter proliferative responses despite robust activation of B cell lines, similarly to responses observed for ligation of TLR7 in B cells [40].
In conclusion, the data presented here demonstrate that similar to CpG 2395 and DNA 2395, the 2395 sequence composed entirely of TNA is able to stimulate some innate immune responses. Notably, the ability of TNAs to induce immune responses has not previously been examined. Similar to sequences composed of phosphorothioate backbones, the TNA equivalents are highly resistant to serum nucleases [15], making them an attractive tool for the future development of nucleic acid-based therapeutics.
Recent advances in TNA synthesis using engineered polymerases have allowed for the selection of TNAs that target human thrombin [16] and HIV reverse transcriptase, opening the door for selections against a variety of other protein targets. In addition, a recent study demonstrated that antisense TNA oligonucelotides are able to effectively suppress GFP expression in living cells [17].
While a potential benefit of chemically modified nucleic acids is the ability to evade the immune response (e.g., antisense applications), chemically modified nucleic acid therapeutics such as TNAs could also be highly useful as stimulators of immune responses (e.g., adjuvants) or inhibitors of immune responses (e.g., anti-inflammatory therapies). Thus, further exploration of the utility of TNAs and other chemical modifications is needed, including their ability to stimulate or inhibit immune responses and the specificity of these responses.
Whether TNA 2395, or any of the other TNA sequences tested here, signals specifically through TLR9, as with CpG 2395 and DNA 2395, remains to be determined and is beyond the scope of this study. TNA 2395 demonstrated a lower level of mRNA upregulation compared to CpG 2395 and DNA 2395, but robustly activated B cells through CD86 upregulation. Other TNA sequences tested here, however, displayed a robust ability to regulate IFNβ mRNA, suggesting that sequence composition plays a role in the ability of TNA sequences to induce immune responses.
TLR9 has been shown to respond differentially to various CpG-containing sequences, as well as non-CpG sequences, and could be responsible for TNA-induced immune signaling [19,21,22,32,33,47,48]. However, TNA is capable of antiparallel, Watson–Crick base pairing with complementary DNA, RNA, and TNA oligonucleotides, and the self-paired TNA/TNA duplex occurs through an A-like helical geometry [7,12,49,50]. Thus, it is possible that the TNAs tested in this study could also signal through endosomal TLRs that recognize RNA, such as TLRs 3, 7, or 8, or even a variety of other nucleic acid-sensing receptors should the TNA escape the endosome. Further experiments beyond the scope of this study will be required to determine the specificity of TNA-induced signaling, specifically, the sequence motif requirements (CpG versus non-CpG) and the receptor responsible for TNA-induced signaling outcomes.
Author Disclosure Statement
The authors, Drs. M.J.L., D.H.B., and J.C.C., report no competing financial interests for the work detailed in this article.
References
- 1. Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew SY, et al. (2012). Synthetic genetic polymers capable of heredity and evolution. Science 336:341–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joyce GF. (2012). Evolution. Toward an alternative biology. Science 336:307–308 [DOI] [PubMed] [Google Scholar]
- 3. Hunter P. (2013). XNA marks the spot. What can we learn about the origins of life and the treatment of disease through artificial nucleic acids? EMBO Rep 14:410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pinheiro VB. and Holliger P. (2014). Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol 32:321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mei H, Liao JY, Jimenez RM, Wang Y, Bala S, McCloskey C, Switzer C. and Chaput JC. (2018). Synthesis and evolution of a threose nucleic acid aptamer bearing 7-Deaza-7-substituted guanosine residues. J Am Chem Soc 140:5706–5713 [DOI] [PubMed] [Google Scholar]
- 6. Rangel AE, Chen Z, Ayele TM. and Heemstra JM. (2018). In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res 46:8057–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu H, Zhang S. and Chaput JC. (2012). Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem 4:183–187 [DOI] [PubMed] [Google Scholar]
- 8. Vollmer J, Jepsen JS, Uhlmann E, Schetter C, Jurk M, Wader T, Wüllner M. and Krieg AM. (2004). Modulation of CpG oligodeoxynucleotide-mediated immune stimulation by locked nucleic acid (LNA). Oligonucleotides 14:23–31 [DOI] [PubMed] [Google Scholar]
- 9. Porciani D, Cardwell LN, Tawiah KD, Alam KK, Lange MJ, Daniels MA. and Burke DH. (2018). Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat Commun 9:2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scheiermann J. and Klinman DM. (2014). Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 32:6377–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang X, Moseman EA, Farrar MA, Bachanova V, Weisdorf DJ, Blazar BR. and Chen W. (2010). Toll-like receptor 9 signaling by CpG-B oligodeoxynucleotides induces an apoptotic pathway in human chronic lymphocytic leukemia B cells. Blood 115:5041–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schöning K, Scholz P, Guntha S, Wu X, Krishnamurthy R. and Eschenmoser A. (2000). Chemical etiology of nucleic acid structure: the alpha-threofuranosyl-(3′–>2') oligonucleotide system. Science 290:1347–1351 [DOI] [PubMed] [Google Scholar]
- 13. Anosova I, Kowal EA, Sisco NJ, Sau S, Liao JY, Bala S, Rozners E, Egli M, Chaput JC. and Van Horn WD. (2016). Structural insights into conformation differences between DNA/TNA and RNA/TNA chimeric duplexes. Chembiochem 17:1705–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ebert MO, Mang C, Krishnamurthy R, Eschenmoser A. and Jaun B. (2008). The structure of a TNA-TNA complex in solution: NMR study of the octamer duplex derived from alpha-(L)-threofuranosyl-(3'−2′)-CGAATTCG. J Am Chem Soc 130:15105–15115 [DOI] [PubMed] [Google Scholar]
- 15. Culbertson MC, Temburnikar KW, Sau SP, Liao JY, Bala S. and Chaput JC. (2016). Evaluating TNA stability under simulated physiological conditions. Bioorg Med Chem Lett 26:2418–2421 [DOI] [PubMed] [Google Scholar]
- 16. Dunn MR. and Chaput JC. (2016). Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. Chembiochem 17:1804–1808 [DOI] [PubMed] [Google Scholar]
- 17. Liu LS, Leung HM, Tam DY, Lo TW, Wong SW. and Lo PK. (2018). Alpha-l-threose nucleic acids as biocompatible antisense oligonucleotides for suppressing gene expression in living cells. ACS Appl Mater Interfaces 10:9736–9743 [DOI] [PubMed] [Google Scholar]
- 18. Yasuda K, Yu P, Kirschning CJ, Schlatter B, Schmitz F, Heit A, Bauer S, Hochrein H. and Wagner H. (2005). Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol 174:6129–6136 [DOI] [PubMed] [Google Scholar]
- 19. Roberts TL, Sweet MJ, Hume DA. and Stacey KJ. (2005). Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J Immunol 174:605–608 [DOI] [PubMed] [Google Scholar]
- 20. Vollmer J, Jurk M, Samulowitz U, Lipford G, Forsbach A, Wüllner M, Tluk S, Hartmann H, Kritzler A, et al. (2004). CpG oligodeoxynucleotides stimulate IFN-gamma-inducible protein-10 production in human B cells. J Endotoxin Res 10:431–438 [DOI] [PubMed] [Google Scholar]
- 21. Vollmer J, Weeratna R, Payette P, Jurk M, Schetter C, Laucht M, Wader T, Tluk S, Liu M, Davis HL. and Krieg AM. (2004). Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol 34:251–262 [DOI] [PubMed] [Google Scholar]
- 22. Hartmann G, Battiany J, Poeck H, Wagner M, Kerkmann M, Lubenow N, Rothenfusser S. and Endres S. (2003). Rational design of new CpG oligonucleotides that combine B cell activation with high IFN-alpha induction in plasmacytoid dendritic cells. Eur J Immunol 33:1633–1641 [DOI] [PubMed] [Google Scholar]
- 23. Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MA, Lundie RJ, Revuelta A, Davidson DJ, et al. (2014). RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J 33:542–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller T, Hamm S. and Bauer S. (2008). TLR9-mediated recognition of DNA. Handb Exp Pharmacol 183:51–70 [DOI] [PubMed] [Google Scholar]
- 25. Pohar J, Lainšček D, Kunšek A, Cajnko MM, Jerala R. and Benčina M. (2017). Phosphodiester backbone of the CpG motif within immunostimulatory oligodeoxynucleotides augments activation of Toll-like receptor 9. Sci Rep 7:14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts TL, Dunn JA, Sweet MJ, Hume DA. and Stacey KJ. (2011). The immunostimulatory activity of phosphorothioate CpG oligonucleotides is affected by distal sequence changes. Mol Immunol 48:1027–1034 [DOI] [PubMed] [Google Scholar]
- 27. Sester DP, Naik S, Beasley SJ, Hume DA. and Stacey KJ. (2000). Phosphorothioate backbone modification modulates macrophage activation by CpG DNA. J Immunol 165:4165–4173 [DOI] [PubMed] [Google Scholar]
- 28. Hartmann G. and Krieg AM. (2000). Mechanism and function of a newly identified CpG DNA motif in human primary B cells. J Immunol 164:944–953 [DOI] [PubMed] [Google Scholar]
- 29. Guiducci C, Ott G, Chan JH, Damon E, Calacsan C, Matray T, Lee KD, Coffman RL. and Barrat FJ. (2006). Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J Exp Med 203:1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klinman DM, Yi AK, Beaucage SL, Conover J. and Krieg AM. (1996). CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci U S A 93:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA. and Klinman DM. (1995). CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546–549 [DOI] [PubMed] [Google Scholar]
- 32. Abel K, Wang Y, Fritts L, Sanchez E, Chung E, Fitzgerald-Bocarsly P, Krieg AM. and Miller CJ. (2005). Deoxycytidyl-deoxyguanosine oligonucleotide classes A, B, and C induce distinct cytokine gene expression patterns in rhesus monkey peripheral blood mononuclear cells and distinct alpha interferon responses in TLR9-expressing rhesus monkey plasmacytoid dendritic cells. Clin Diagn Lab Immunol 12:606–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jurk M, Schulte B, Kritzler A, Noll B, Uhlmann E, Wader T, Schetter C, Krieg AM. and Vollmer J. (2004). C-Class CpG ODN: sequence requirements and characterization of immunostimulatory activities on mRNA level. Immunobiology 209:141–154 [DOI] [PubMed] [Google Scholar]
- 34. Jiang W, Lederman MM, Harding CV, Rodriguez B, Mohner RJ. and Sieg SF. (2007). TLR9 stimulation drives naive B cells to proliferate and to attain enhanced antigen presenting function. Eur J Immunol 37:2205–2213 [DOI] [PubMed] [Google Scholar]
- 35. Sau SP, Fahmi NE, Liao JY, Bala S. and Chaput JC. (2016). A scalable synthesis of alpha-L-threose nucleic acid monomers. J Org Chem 81:2302–2307 [DOI] [PubMed] [Google Scholar]
- 36. Wu L, Martin TD, Carrington M. and KewalRamani VN. (2004). Raji B cells, misidentified as THP-1 cells, stimulate DC-SIGN-mediated HIV transmission. Virology 318:17–23 [DOI] [PubMed] [Google Scholar]
- 37. Lange MJ, Sharma TK, Whatley AS, Landon LA, Tempesta MA, Johnson MC. and Burke DH. (2012). Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol Ther 20:2304–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pohar J, Pirher N, Benčina M, Manček-Keber M. and Jerala R. (2013). The role of UNC93B1 protein in surface localization of TLR3 receptor and in cell priming to nucleic acid agonists. J Biol Chem 288:442–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettengill MA, van Haren SD, Li N, Dowling DJ, Bergelson I, Jans J, Ferwerda G. and Levy O. (2016). Distinct TLR-mediated cytokine production and immunoglobulin secretion in human newborn naive B cells. Innate Immun 22:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hanten JA, Vasilakos JP, Riter CL, Neys L, Lipson KE, Alkan SS. and Birmachu W. (2008). Comparison of human B cell activation by TLR7 and TLR9 agonists. BMC Immunol 9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jahrsdorfer B, Mühlenhoff L, Blackwell SE, Wagner M, Poeck H, Hartmann E, Jox R, Giese T, Emmerich B, et al. (2005). B-cell lymphomas differ in their responsiveness to CpG oligodeoxynucleotides. Clin Cancer Res 11:1490–1499 [DOI] [PubMed] [Google Scholar]
- 42. Jahrsdörfer B, Hartmann G, Racila E, Jackson W, Mühlenhoff L, Meinhardt G, Endres S, Link BK, Krieg AM. and Weiner GJ. (2001). CpG DNA increases primary malignant B cell expression of costimulatory molecules and target antigens. J Leukoc Biol 69:81–88 [PubMed] [Google Scholar]
- 43. Decker T, Schneller F, Sparwasser T, Tretter T, Lipford GB, Wagner H. and Peschel C. (2000). Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood 95:999–1006 [PubMed] [Google Scholar]
- 44. Iskra S, Kalla M, Delecluse HJ, Hammerschmidt W. and Moosmann A. (2010). Toll-like receptor agonists synergistically increase proliferation and activation of B cells by epstein–barr virus. J Virol 84:3612–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohtsuki S, Takahashi Y, Inoue T, Takakura Y. and Nishikawa M. (2017). Reconstruction of Toll-like receptor 9-mediated responses in HEK-Blue hTLR9 cells by transfection of human macrophage scavenger receptor 1 gene. Sci Rep 7:13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bai L, Chen W, Chen J, Li W, Zhou L, Niu C, Han W. and Cui J. (2017). Heterogeneity of Toll-like receptor 9 signaling in B cell malignancies and its potential therapeutic application. J Transl Med 15:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yasuda K, Rutz M, Schlatter B, Metzger J, Luppa PB, Schmitz F, Haas T, Heit A, Bauer S. and Wagner H. (2006). CpG motif-independent activation of TLR9 upon endosomal translocation of “natural” phosphodiester DNA. Eur J Immunol 36:431–436 [DOI] [PubMed] [Google Scholar]
- 48. Bauer S, Kirschning CJ, Häcker H, Redecke V, Hausmann S, Akira S, Wagner H. and Lipford GB. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 98:9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chim N, Shi C, Sau SP, Nikoomanzar A. and Chaput JC. (2017). Structural basis for TNA synthesis by an engineered TNA polymerase. Nat Commun 8:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chaput JC, Ichida JK. and Szostak JW. (2003). DNA polymerase-mediated DNA synthesis on a TNA template. J Am Chem Soc 125:856–857 [DOI] [PubMed] [Google Scholar]