Abstract
The brain is highly enriched in the long-chain omega-3 (n-3) PUFA DHA. Due to the limited capacity for local DHA synthesis in the brain, it relies on a continual supply from the circulation to replenish metabolized DHA. Previous studies investigating brain DHA turnover and metabolism have relied on isotope tracers to determine brain fatty acid kinetics; however, this approach is cumbersome and costly. We applied natural abundance carbon isotope ratio analysis via high-precision gas chromatography combustion isotope ratio mass spectrometry, without the use of labeled tracers, to determine the half-life of brain DHA in mice following a dietary switch experiment. Mice fed diets containing either α-linolenic acid (ALA) or DHA as the sole dietary n-3 PUFA were switched onto diets containing ALA, DHA, or ALA + DHA at 6 weeks of age, while control mice were maintained on their respective background diet. We measured brain DHA carbon isotope ratios (reported as δ13CDHA signatures) over a 168-day time course. Brain δ13CDHA signatures of control mice maintained on background diets over the time course were stable (P > 0.05). Brain δ13CDHA signatures of mice switched to the DHA or ALA + DHA diet from the ALA diet changed over time, yielding brain incorporation half-lives of 40 and 34 days, respectively. These half-lives determined by natural abundance carbon isotope ratio analysis were consistent with estimates from kinetic isotope tracer studies. Our results demonstrate the feasibility of natural abundance carbon isotope ratio analysis in the study of fatty acid metabolism without the use of isotopically labeled fatty acid tracers.
Keywords: lipids, omega-3, metabolism, diet, docosahexaenoic acid, mass spectrometry, fatty acids
The composition of fatty acids in the brain is unique compared with most other tissues in that the brain is highly enriched in the long-chain omega-3 (n-3) PUFA DHA (22:6 n-3) (1, 2). Mounting evidence indicates a role of DHA and its bioactive products in the support and regulation of optimal neuronal and synaptic function (3). Furthermore, consistent with the hypothesis that brain DHA homeostasis is critical in supporting optimal brain function, postmortem analyses suggest that brain DHA levels may be altered, either as cause or consequence, in many neurological and neuropsychiatric disorders (1). Although it appears to be vital in supporting brain function, DHA is not efficiently synthesized locally within the brain, and therefore DHA must be incorporated preformed from circulating plasma pools to replenish that which has been metabolized (4, 5).
Rigorous kinetic tracer studies have been conducted to better understand the incorporation and metabolism of DHA in the brain, as well as the factors that may influence its metabolism. Modeling the loss of radioactivity following intracerebroventricular injections of radiolabeled DHA, the brain DHA half-life in rats has been determined to range between 30 and 65 days and is extended to 90 days during n-3 PUFA dietary deprivation (6–8). Acute intravenous infusions of radiolabeled DHA have been used to investigate the effects of mood-stabilizing agents on brain DHA uptake and metabolism, including turnover (9–12), as well as to elucidate the preferred plasma lipid pool supplying the brain (8, 13–15). In addition to the use of radioactive isotopes, stably labeled DHA tracers have also been applied to probe metabolic questions regarding brain DHA and its origin (16–18). Although these techniques allow for the modeling of brain DHA kinetics, they are often constrained to acute study designs and a single time-point analysis due to the significant expense associated with the custom synthesis of high-purity tracer compounds.
Natural abundance isotopic analysis may present an alternative to the use of synthetic isotopic tracers when investigating fatty acid incorporation and metabolism. Because the carbon isotopic makeup of a molecule is conserved following incorporation from the diet, highly precise natural abundance carbon isotope ratio measurements (13C/12C; expressed as δ13C in units of mUr) by gas chromatography isotope ratio mass spectrometry can provide insight into the origin of a compound of interest. Importantly, innate differences in the carbon fixation pathways of C3 and C4 photosynthesis result in characteristic average δ13C values of −27 and −13 mUr for C3 and C4 plants, respectively (19, 20). Through the use of well-controlled dietary switch experiments (Fig. 1), ecologists have used the natural variation in carbon isotope ratios between C3 and C4 plants to measure tissue turnover rates to better construct models of resource utilization and food-web analysis (21, 22). Similarly, previous dietary intervention studies in infants have used variations in natural abundance carbon isotope ratios to investigate the contribution of endogenous synthesis from 18-carbon fatty acids to the long-chain PUFA arachidonic acid (20:4n-6) and DHA in infants (23, 24).
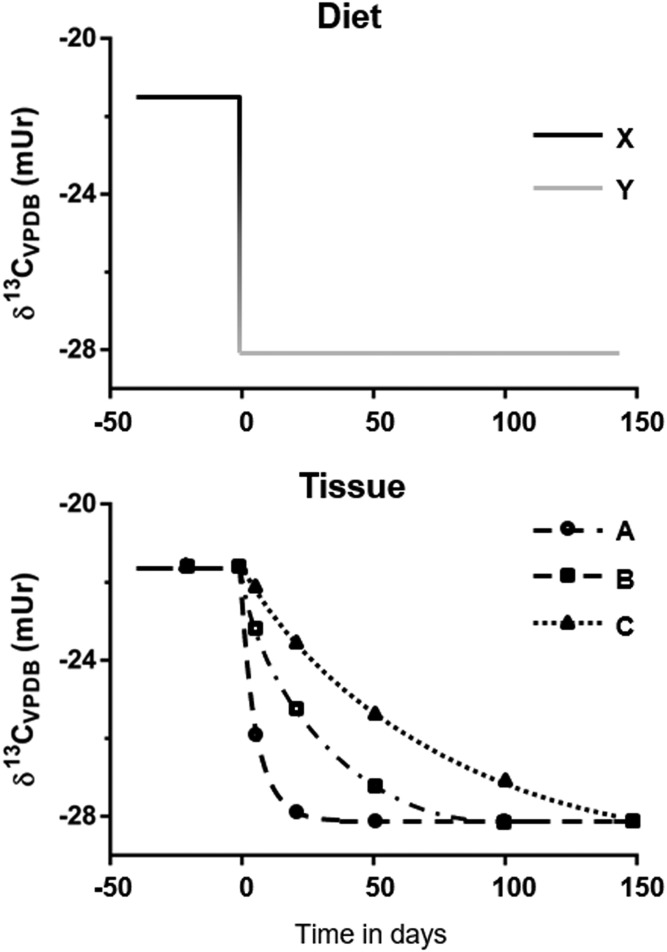
Fig. 1.
Graphical representation of a dietary switch experiment. The top panel is an example of the isotopic dietary parameters of a dietary switch experiment. Animals are equilibrated to study diet X with an isotope ratio of −21.5 mUr and then switched to diet Y with an isotope ratio of −28 mUr at time 0. The bottom panel depicts the resulting changes in tissue carbon isotope ratios in response to the dietary switch. As the compound of interest is metabolized it is replaced with that from the diet, which has a different δ13C value. By measuring and plotting changes in tissue δ13C signatures over time, tissue-specific turnover can be modeled.
Our laboratory has published on the use of natural abundance carbon isotope ratios measured by compound-specific isotope analysis (CSIA) to determine the contribution of retroconversion to plasma DHA levels in rats (25) and in humans (26) and to identify the dietary origin of brain DHA (27, 28). By using the naturally occurring variation in carbon isotope ratios between terrestrial plant-sourced and marine-sourced n-3 PUFA, we previously demonstrated that brain DHA, established by feeding either a diet containing α-linolenic acid (ALA) or DHA, differed by ∼6 mUr (27). In the current study, we applied a dietary switch experiment in which dietary n-3 PUFA sources and their respective carbon isotope ratios were altered, and corresponding changes in brain, liver, and adipose DHA carbon isotope ratios (referred to as δ13CDHA) were measured over a 168-day time course. The resulting changes in δ13CDHA signatures were modeled by a one-phase decay function to determine experimental incorporation half-lives. Our results indicate that brain incorporation half-lives generated from the dietary switch experiment were consistent with those previously determined from radiolabeled kinetic modeling. Additionally, we found that preformed DHA is the preferred source of DHA for the brain when ALA is provided at equals levels in the diet. These results highlight the utility of natural abundance CSIA to study fatty acid metabolism kinetics in a chronic feeding model without the administration of labeled tracer compounds.
MATERIALS AND METHODS
Materials
All solvents used were American Chemical Society or HPLC grade and were purchased from either Sigma-Aldrich (Mississauga, Canada) or Thermo Fischer Scientific (Ottawa, Canada). Fatty acid standards and reference fatty acid methyl ester (FAME) mixtures were purchased from Nu-Chek Prep, Inc. (Elysian, MN). Isotope reference materials USGS70, USGS71, and USGS72 were obtained from the Reston Stable Isotope Laboratory (Reston, VA).
Animals
Experimental animal protocols were approved by the University of Toronto Animal Ethics Committee and conducted in accordance with the policy and guidelines of the Canadian Council on Animal Care and the Regulations of Animals Research Act of Ontario (protocol # 20011661). All animals were housed in a temperature-controlled environment under a 14 h/10 h light/dark cycle with ad libitum access to food and water.
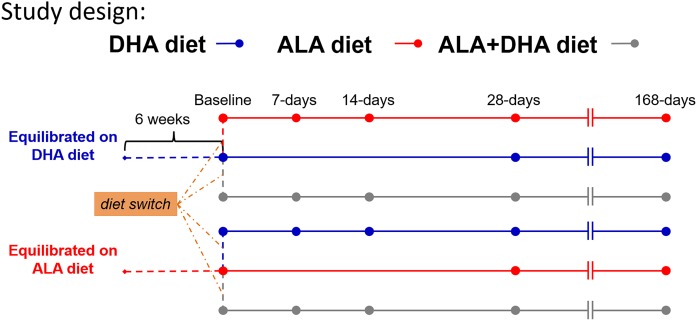
Two colonies of BALB/c mice were maintained on a diet containing either ALA or DHA as the only n-3 PUFA for two generations. Our laboratory has demonstrated previously that brain δ13CDHA signatures are equilibrated with the δ13C signature of dietary n-3 PUFA following two generations of breeding. At 6 weeks of age equilibrated male mice were used for a dietary switch experiment (Fig. 2). Mice on the ALA diet were randomly switched onto either the DHA diet or ALA + DHA diet. Conversely, those on the DHA diet were randomly placed onto either the ALA diet or ALA + DHA diet. In addition to the mice undergoing the dietary switch, a small subset of mice were maintained on their respective background diet, either the ALA diet or DHA diet, to evaluate the stability of tissue δ13CDHA signatures over the course of the experiment. Mice were euthanized over a time course of 0, 7, 14, 28, 56, 112, and 168 days.
Fig. 2.
Study design schematic for the dietary switch experiment. Prior to the dietary switch male BALB/c mice were equilibrated to either the ALA diet or DHA diet by breeding over two generations while maintained on the respective study diet. From the DHA-equilibrated group, mice were randomly placed onto either the ALA diet or the ALA + DHA diet at 6 weeks of age. Similarly, mice from the ALA diet were switched onto either the DHA diet or the ALA + DHA diet. Control arms from both equilibrated groups remained on their respective diets over the study time course. Mice were euthanized at 7, 14, 28, 56, 112, and 168 days after the dietary switch. Control mice were euthanized at 28 and 112 days. A group of mice from both equilibrated diet groups was euthanized killed at 6 weeks of age for baseline measurements. ALA, α-linolenic acid.
All mice were euthanized by cardiac puncture under terminal anesthesia with 0.25 mg/g body weight of 2,2,2-tribromoethanol (Sigma-Aldrich). Brain (left hemisphere), liver (left lateral lobe), and adipose (epididymal) tissues were collected, flash-frozen in liquid nitrogen, and stored at −80°C until further processing.
Diets
For detailed information on the composition of the experimental diet, see Lacombe et al. (27). Briefly, study diets were based on a modified low AIN-93 custom low n-3 PUFA rodent diet (Dyets, Inc., Bethlehem, PA). Background oils were derived from safflower and fully hydrogenated coconut oils and added fatty acid ethyl esters (32.8%, 63.2%, and 4.0% by weight, respectively). For the ALA, DHA, and ALA + DHA diets added oils were 2% ALA ethyl ester (BASF Pharma Callanish Ltd., Isle of Lewis, United Kingdom) and 2% DHA ethyl ester (BASF Pharma Callanish Ltd.). Oleate ethyl ester (Nu-Chek Prep, Inc.) was added to both ALA and DHA diets at 2% of the fatty acids by weight to maintain equivalent fatty acid content. All added fatty acid ethyl esters were determined to be >98% pure by gas chromatography flame ionization detection (GC/FID). The carbon isotope ratios of ALA and DHA from the study diets were determined to be −28.2 ± 0.3 and −21.8 ± 0.2 mUr, respectively (27).
Lipid extraction and transmethylation
Total lipid extracts were isolated from harvested tissues by methodologies adapted from Folch, Lees, and Sloane Stanley (29). Brain and adipose tissue (∼200 and 50 mg, respectively) were homogenized in 6 ml of a chloroform-methanol solution (2:1; v/v) containing known quantities of docosatrienoic ethyl ester (22:3n-3; Nu-Chek Prep, Inc.) and either heptadecanoic acid (17:0; Nu-Chek Prep, Inc.) or triheptadecanoin (Nu-Chek Prep, Inc.) for fatty acid quantification. Liver tissue was cryopulverized by mortar and pestle in the presence of liquid nitrogen prior to lipid extraction. For lipid extraction, ∼50 mg of pulverized liver was vortexed in 6 ml of a 2:1 chloroform-methanol solution (v/v) containing docosatrienoic acid ethyl ester. All brain, adipose, and liver homogenates were held at 4°C overnight, following which 1.75 ml of 0.88% aqueous potassium chloride was added. Samples were then vortexed vigorously, and phase separation was achieved through centrifugation at 500 g for 10 min. The lower lipid containing the chloroform phase was collected into clean glass test tubes and stored at −80°C until methylation. Transesterification of extracted lipids by 14% boron trifluoride was carried out following previously published methods (27). The transesterification of lipids in the presence of excess alcohol proceeds quantitatively; therefore, no isotopic fractionation effect is expected to be imparted by this process (30).
FAME quantification
FAMEs were quantified by GC/FID on a Varian 430 gas chromatograph (Bruker, Billerica, MA) equipped with a Supelco SP-2560 capillary column (100 m × 0.25 mm i.d. × 0.20 µm df; Sigma-Aldrich). For GC parameters see Lacombe et al. (27). Samples were concentrated to ∼1 mg of total FAME per milliliter of hexane before GC/FID analysis. FAMEs were identified by comparing the retention time to the GLC-569 external reference standard (Nu-Chek Prep, Inc.) and quantified by comparing the peak area to that of the docosatrienoic ethyl ester internal standard.
Adipose FAME fractionation by silver ion chromatography
Due to the low relative abundance of DHA in adipose fatty acids, DHA and other PUFAs were fractionated and concentrated prior to carbon isotope ratio analysis. Silver ion chromatography allows for the separation of FAMEs by number, configuration, and position of double bonds, thereby permitting the isolation of low abundant PUFA species from adipose total lipids (31). Separation was carried out using Discovery Ag-Ion solid-phase extraction cartridges (750 mg bed wt.; 6 ml volume; Sigma-Aldrich) affixed in an Agilent Vac Elut SPS 24 Manifold (Agilent Technologies, Santa Clara, CA). The solvent system for the elution of FAMEs was adapted from the methods of Kramer et al. (32). Prior to sample loading, cartridges were washed with 4 ml acetonitrile followed by 4 ml hexane, allowing the solvent to flow under gravity. Approximately 1 mg FAME from adipose TLE was dissolved in 1 ml hexane and loaded onto the cartridges and run through. Cartridges were then washed sequentially with 6 ml each of hexane-acetone (99:1; v/v), hexane-acetone (90:10; v/v), and acetone (100%), allowing each solvent to flow under gravity with 3 s of vacuum applied between each solvent step. FAMEs containing three or more double bonds were eluted with 6 ml acetone-acetonitrile (60:40; v/v). Samples were then evaporated under a gentile stream of nitrogen and reconstituted in 100 µl heptane for isotopic analysis. A representative chromatogram of the adipose sample before and after silver ion fractionation can be found in supplemental Fig. 1.
Carbon isotope ratio analysis
CSIA of FAMEs by gas chromatography combustion isotope ratio mass spectrometry (GC/C/IRMS) was conducted similar to previously published methods (27) and as described briefly below. FAMEs (1 μl) were injected onto a 100 m SP-2560 capillary column (Sigma-Aldrich) in a Trace 1310 gas chromatograph (Thermo Fisher Scientific) using a TriPlus RSH autosampler (Thermo Fisher Scientific). Analyte peaks were separated using the same thermal and pneumatic program described previously (27), yielding baseline resolutions of analyte peaks of interest. The GC effluent was swept by helium carrier gas to a GC Iso Link II combustion interface (Thermo Fisher Scientific) held at 1,000°C featuring nickel and copper catalysts and interfaced to a Delta V Plus IRMS (Thermo Fisher Scientific) via a ConFlo IV (Thermo Fisher Scientific) continuous-flow interface. CO2 gas produced through the quantitative combustion of isolated analytes was dried via flowing gas through a Nafion dryer prior to entering the IRMS ion source (Dupont, Wilmington, DE).
Elemental analysis (EA) was carried out on heptadecanoic acid and methyl heptadecanoate that was derivatized with each batch of samples. This was carried out to determine the carbon isotopic ratio of methyl groups esterified to generate FAMEs from TLE. Aliquots (∼150 µg of total material) were transferred to clean, smooth, thin-walled tin capsules (Elemental Microanalysis, Okehampton, UK), and the solvent was evaporated overnight at room temperature. A Zero Blank autosampler (Costech Analytical Technologies Inc., Valencia, CA) was used to introduce capsules into the Flash 2000 Elemental Analyzer (Thermo Fisher Scientific). Temperatures were held at 1,020°C and 650°C for the oxidation and reduction reactors, respectively. Similar to the GC/C/IRMS setup, CO2 gas was introduced to the Delta V Plus IRMS via a ConFlo IV continuous-flow interface (Thermo Fisher Scientific). Isodat Workplace 3.0 (Thermo Fisher Scientific) was used to process all isotopic data.
Carbon isotope ratio normalization and methyl corrections
All carbon isotope ratios were normalized using consensus-validated 20-carbon FAME reference material USGS70, USGS71, and USGS72 (Reston Stable Isotope Laboratory) using multipoint linear normalization similar to methods previously described (27, 33). Consensus-derived carbon isotope ratio values of the reference materials are −30.53 ± 0.04‰, −10.50 ± 0.03‰, and −1.54 ± 0.03‰ for USGS70, USGS71, and USGS72, respectively, and are expressed relative to Vienna Peedee Belemnite on a scale normalized to primary reference materials NBS 19 and LSVEC (34). Reference materials were injected twice throughout each batch of samples analyzed by GC/C/IRMS and EA/IRMS under identical analytical conditions to the samples being analyzed. Normalizing equations were generated for each batch of samples from linear regression lines fit through the measured and accepted δ13C values of reference materials. R2 values for all equations were ≥0.9998.
IRMS measures carbon isotope ratios from CO2 gas that is generated through the quantitative combustion of sample material or specific organic analytes and therefore does not distinguish from the derivatized carbon units added to fatty acids during transmethylation. To account for the contribution of derivatized carbon units to measured carbon isotope ratios and allow the reporting of true values, methyl corrections were carried using mass balance equations:
| (Eq. 1) |
where n refers to the number of moles of carbon in the corresponding FAMEs and fatty acids and the subscripts FAME, ME, and FA refer to the measured carbon isotope ratios of FAMEs, the methyl group, and fatty acids, respectively. For methyl corrections of measured DHA carbon isotope ratios and equal 23 and 22, respectively. Carbon isotope ratios of the methyl group was determined for each batch of samples by EA of methylated and free heptadecanoic acid standards (Nu-Chek Prep Inc.) as previously described (27). Methyl group carbon isotope signatures ranged from −40.5 to −39.1 mUr.
Statistics and calculations
All data reported herein are expressed as means ± SDs unless otherwise stated. The sample size for the time course ranged from three to five mice per time point. Brain, liver, and adipose fatty acid concentrations of mice after the dietary switch were compared using two-way ANOVA for the interaction of time and dietary group. As the a priori design of this experiment was to assess changes in brain carbon isotope ratios over a 168-day time course for the determination of DHA half-lives, multiple comparisons analysis was not carried out because sample sizes for individual time points were underpowered.
To test for the stability of tissue δ13CDHA signatures over time, carbon isotope ratio values of tissue from the dietary control arms were plotted, and slopes of linear regressions were tested for deviation from zero. Statistical significance was determined at P < 0.05. Tissue δ13CDHA signatures from mice following the dietary switch were plotted over time and modeled with a one-phase decay function to generate a curve of best fit:
| (Eq. 2) |
where y0 describes the best-fit curve at time zero, plateau describes the value of the best-fit curve at an infinite time, and k defines the decay constant. An exception was made for DHA-background mice that were switched onto the ALA + DHA combined diet due to the relatively unchanged δ13CDHA signatures over the time course. Instead, carbon isotope ratios generated from this arm of the dietary switch were modeled by linear regression, and slopes were tested for deviation from zero. Incorporation half-lives for tissue δ13CDHA signatures were determined from the best-fit curves using the following formula:
| (Eq. 3) |
where k is the decay constant from the best-fit curve modeled by a one-phase decay function. When applicable, the rate of loss of DHA from the brain DHA pool, Jout (µmol/g brain/day), was calculated using the following formula (6, 7, 35):
| (Eq. 4) |
where CFA is the baseline DHA concentration from the given tissue lipid pool and t1/2 is the corresponding experimentally derived half-life. The Jout may be used as an estimate of the net incorporation rate of DHA given that the net rate of loss and the net incorporation rate are assumed to be equal when the brain DHA pool is at steady state.
RESULTS
Brain DHA concentrations are less influenced by diet than liver and adipose
Brain, liver, and adipose DHA concentrations of the ALA and DHA diet control arms are presented in supplemental Fig. 2. With respect to brain DHA concentrations, in the absence of an interaction there were significant main effects observed for diet (P < 0.0001) and time (P = 0.006), indicating slightly greater (6% to 8%) brain DHA concentrations in mice maintained on the DHA diet compared with those on the ALA diet. Liver DHA concentrations were ∼2-fold higher in mice on the DHA diet compared with mice on the ALA diet across all time points (main effect of diet: P < 0.0001). A significant time diet interaction (P = 0.0002) was observed when comparing adipose DHA concentrations between control mice maintained on either the ALA diet or DHA diet. DHA concentrations in adipose total lipids ranged from 3- to 5-fold higher in mice on the DHA diet compared with those on the ALA diet.
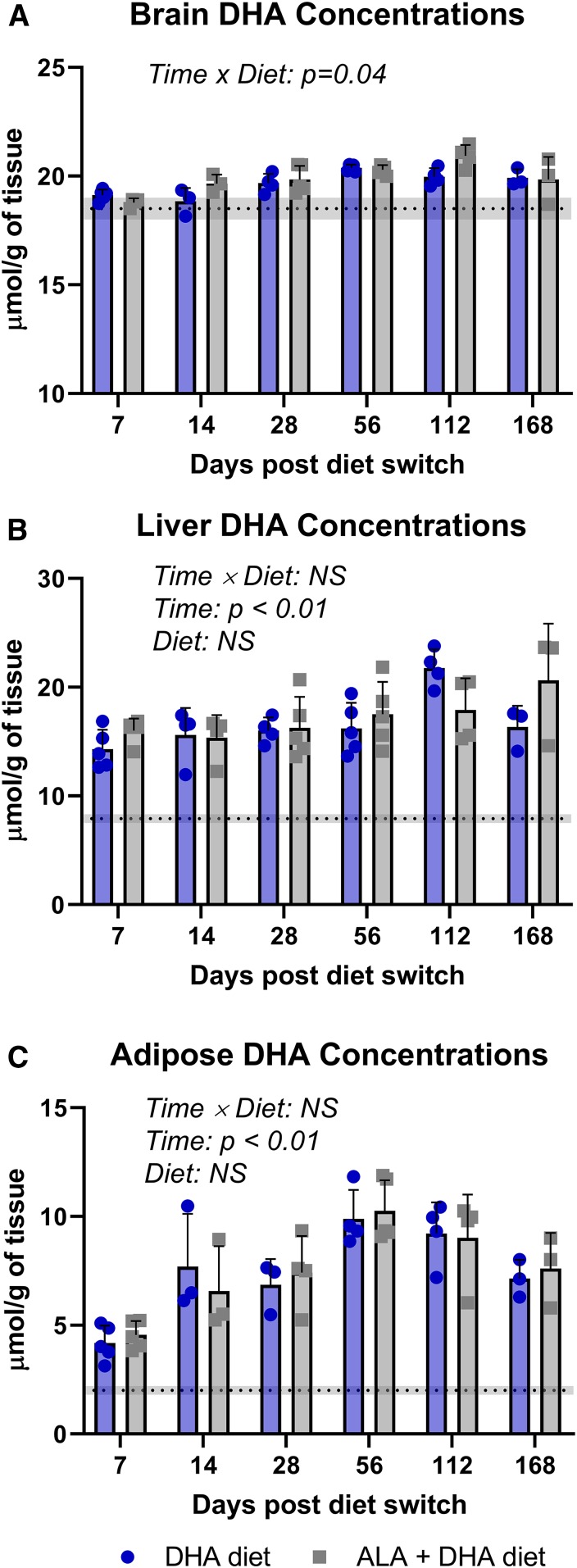
Data on DHA concentrations following the dietary switch are presented in Figs. 3 and 4 for mice equilibrated to the ALA diet and DHA diet, respectively. With respect to the brain, following the dietary switch there was a significant time × diet interaction effect observed in both ALA- and DHA-background groups (P = 0.04 and P = 0.02, respectively). Brain DHA concentrations of mice from the ALA-background diet switched onto the DHA and ALA + DHA diets increased over time, albeit modestly (mean difference over baseline at 168 days after the dietary switch: +1.4 and +1.3 µmol, respectively; Fig. 3A). In mice equilibrated to the DHA background dietary brain DHA concentrations remained relatively stable over the time course in both ALA and ALA + DHA diet arms.
Fig. 3.
Brain (A), liver (B), and adipose (C) DHA concentrations following the dietary switch from mice equilibrated to the ALA diet. Data are shown as means ± SDs and were compared by two-way ANOVA for the interaction of time and diet (n = 3–5 per time point). P < 0.05 was considered statistically significant. Baseline DHA concentrations are represented by the dashed line with the gray band indicating ± SD. ALA, α-linolenic acid; NS, not significant.
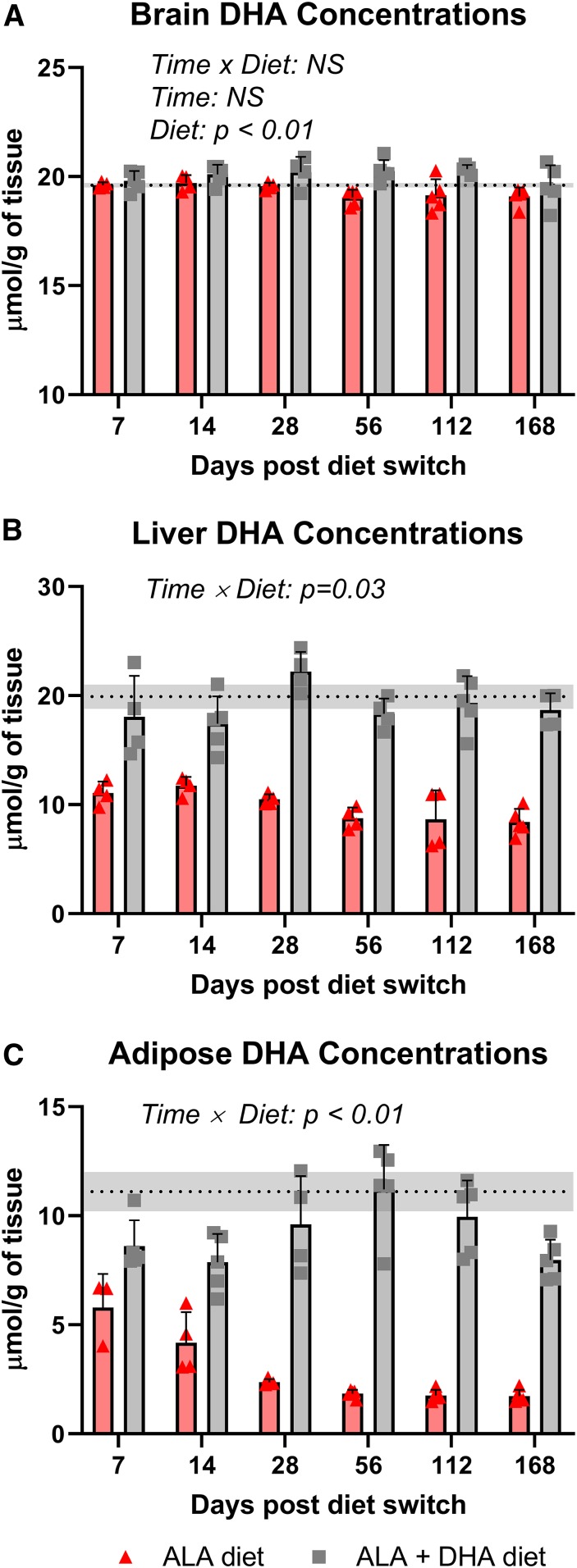
Fig. 4.
Brain (A), liver (B), and adipose (C) DHA concentrations following the dietary switch from mice equilibrated to the DHA diet. Data are shown as means ± SDs and were compared by two-way ANOVA for the interaction of time and diet (n = 3–5 per time point). P < 0.05 was considered statistically significant. Baseline DHA concentrations are represented by the dashed line with the gray band indicating ± SD. ALA, α-linolenic acid; NS, not significant.
Liver and adipose DHA pools responded rapidly to changes in n-3 PUFA intakes. In mice equilibrated to the ALA diet there was a significant main effect of time in both the liver and adipose DHA pools (P < 0.003 and P < 0.0001, respectively), indicating a trend toward increasing DHA concentrations when switched onto the DHA and ALA + DHA diets (Fig. 3B, C). In the liver total lipid pool, DHA concentrations doubled over baseline levels after 1 week of feeding both the DHA and ALA + DHA diets. Adipose DHA concentrations reached a maximal 4-fold increase over baseline concentrations at 56 days and remained ∼2.5-fold higher at 168 days in mice on the DHA and ALA + DHA diets.
Mice equilibrated to the DHA diet experienced a large drop in DHA concentration in both liver and adipose total lipid pools when switched to the ALA diet, which was not observed in the ALA + DHA diet arm. In the liver, DHA concentrations decreased by ∼50% after 7 days of being on the ALA diet and stayed ∼50% lower than mice on the ALA + DHA diet across all time points (Fig. 4B). In the adipose pool, DHA concentrations were 50% lower than baseline levels after 7 days on the ALA diet and plateaued at ∼2 µmol/g at 28 days, ∼80% lower than baseline concentrations (Fig. 4C). Adipose DHA concentrations remained relatively stable in mice on the ALA + DHA diet over the 168-day time course.
Tissue δ13C signatures model the replacement of DHA in tissue total fatty acid pool
In control mice equilibrated and maintained on their respective ALA and DHA diets over the course of the study, brain and adipose δ13CDHA signatures remained stable. Brain δ13CDHA signatures over the time course averaged −26.9 ± 0.2 and −21.8 ± 0.7 mUr for ALA and DHA control groups, respectively. Adipose δ13CDHA signatures averaged −29.6 ± 1.1 and −23.5 ± 0.4 mUr over the time course for ALA and DHA control mice, respectively. Slopes of linear regression lines of δ13CDHA signatures in brain and adipose total lipids did not significantly differ from zero in control mice maintained on either the ALA or DHA diet (Figs. 5, 6). Liver δ13CDHA signatures were stable in control mice maintained on the DHA diet (P > 0.05; 23.7 ± 0.5 mUr); however, δ13CDHA signatures from ALA became more enriched over the course of the study based on linear regression analysis (P = 0.004).
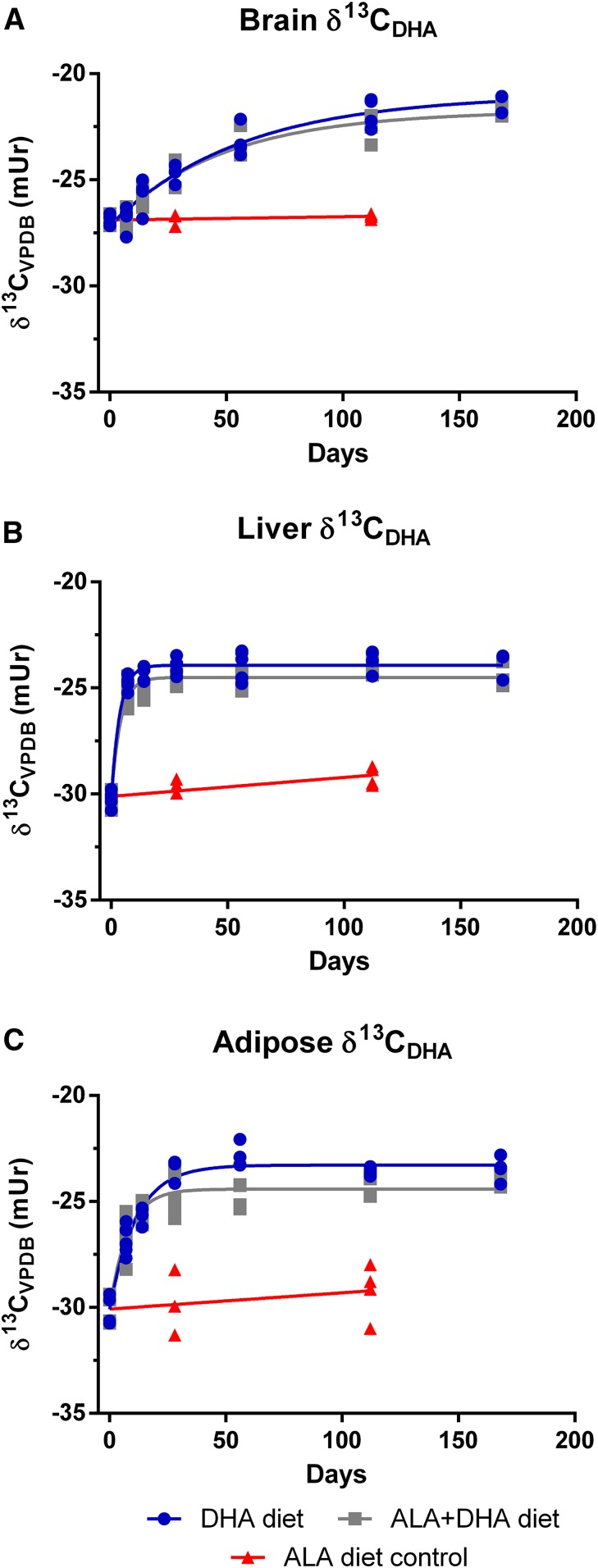
Fig. 5.
Change in brain (A), liver (B), and adipose (C) δ13CDHA signatures following the dietary switch from mice equilibrated to the ALA diet. Changes in carbon isotope ratios were fit with a one-phase decay function on untransformed data (n = 3–5 per time point). ALA, α-linolenic acid; VPDB, Vienna Peedee Belemnite.
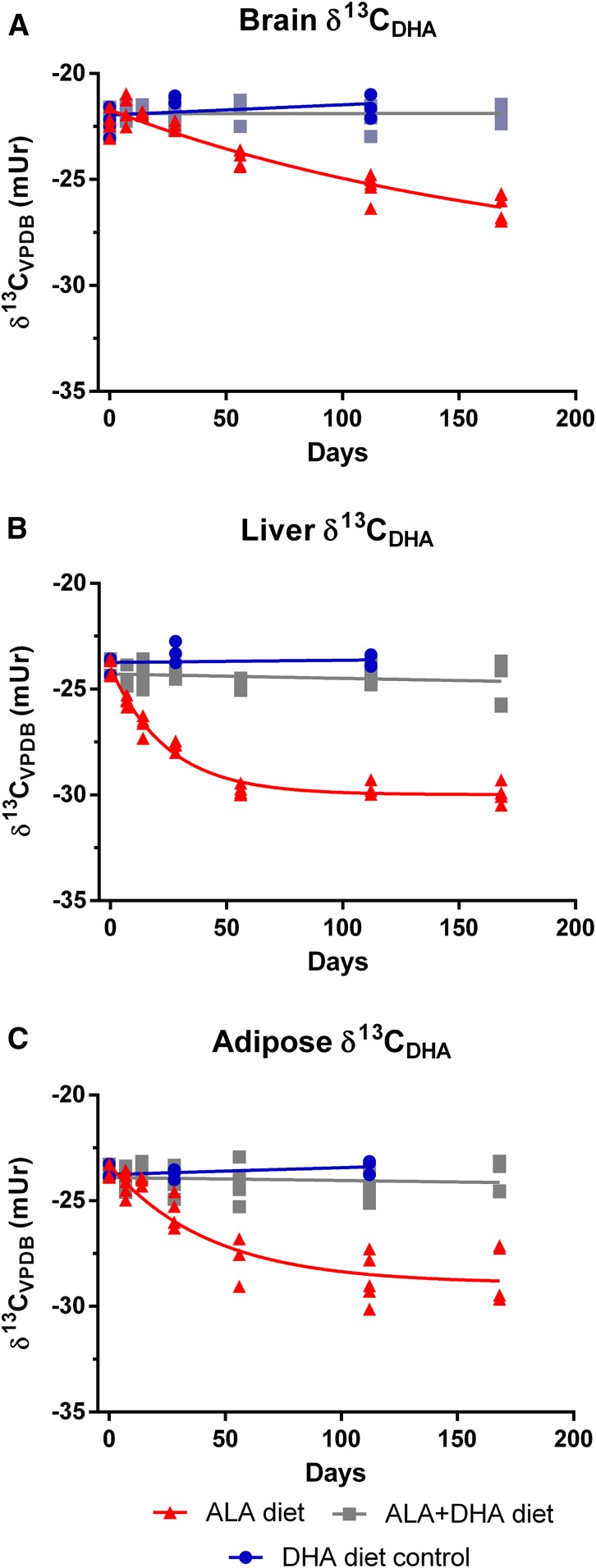
Fig. 6.
Change in brain (A), liver (B), and adipose (C) δ13CDHA signatures following the dietary switch from mice equilibrated to the DHA diet. Changes in carbon isotope ratios for the ALA diet group were fit with a one-phase decay function on untransformed data (n = 3–5 per time point). Carbon isotope ratios from the ALA + DHA group were not adequately modeled by a one-phase decay function and were instead fit by linear regression; slopes from linear regression analyses from all tissues did not deviate from zero (P > 0.05). ALA, α-linolenic acid; VPDB, Vienna Peedee Belemnite.
To model DHA turnover, brain, liver, and adipose δ13CDHA signatures measured over the 168-day time course were fit with a one-phase decay function (Figs. 5 and 6 for ALA and DHA equilibrated mice, respectively). δ13CDHA signatures in the brain, liver, and adipose of mice equilibrated on the DHA diet and switched to the ALA + DHA diet did not change over the time course, therefore carbon isotope ratios from this group were fit with linear regression. Slopes of linear regression lines were determined to not significantly deviate from zero (P > 0.05; Fig. 6); therefore, δ13CDHA signatures were determined to be stable.
Incorporation half-lives were determined for each tissue and diet arm using equation 3 (see Materials and Methods) from experimentally generated curves and are reported, along with parameters of best-fit curves, for mice equilibrated on the ALA and DHA diet in Table 1. Half-lives and curve parameters could not be determined for DHA-equilibrated mice on the ALA + DHA diet because δ13CDHA signatures from this group were not adequately modeled by a one-phase decay function. Liver DHA incorporation half-lives were determined to be between 1- and 2-fold shorter than adipose and between 12- and 15-fold shorter than brain DHA half-lives. Interestingly, brain, liver, and adipose DHA half-lives were longest in mice equilibrated on the DHA diet and switched to the ALA diet. Notably, in ALA-equilibrated mice, DHA incorporation half-lives did not appreciably differ between mice on either the DHA or ALA + DHA diet.
TABLE 1.
Kinetic parameters from best-fit curves of δ13CDHA signature response to dietary switch
| Tissue | Diet | Y0, mUr | Plateau, mUr | k,a days−1 × 10−3 | t1/2,b days (95% CI) | Jout,c µmol/g brain/day |
| Equilibrated to ALA diet | ||||||
| Brain | DHA | −27.2 ± 0.2 | −21.0 ± 0.4 | 177.8 ± 3.4 | 39.0 (27.9, 65.0) | 0.32 |
| ALA + DHA | −27.1 ± 0.2 | −21.8 ± 0.3 | 205.5 ± 3.5 | 33.7 (24.9, 52.2) | 0.38 | |
| Liver | DHA | −30.2 ± 0.2 | −23.9 ± 0.1 | 277.1 ± 37.8 | 2.5 (2.0, 3.5) | ND |
| ALA + DHA | −30.2 ± 0.2 | −24.5 ± 0.1 | 252.5 ± 35.0 | 2.7 (2.1, 3.8) | ND | |
| Adipose | DHA | −30.1 ± 0.3 | −23.3 ± 0.2 | 128.3 ± 21.8 | 7.9 (6.5, 10.0) | ND |
| ALA + DHA | −30.1 ± 0.4 | −24.4 ± 0.2 | 87.6 ± 9.0 | 5.4 (4.0, 8.3) | ND | |
| Equilibrated to DHA diet | ||||||
| Brain | ALA | −21.7 ± 0.2 | −29.3 ± 2.8 | 5.6 ± 3.2 | 124.4 (57.1, ∞) | 0.11 |
| ALA + DHA | ND | ND | ND | ND | ND | |
| Liver | ALA | −24.0 ± 0.2 | −30.0 ± 0.1 | 41.0 ± 3.7 | 16.9 (14.2, 20.8) | ND |
| ALA + DHA | ND | ND | ND | ND | ND | |
| Adipose | ALA | −23.3 ± 0.4 | −28.9 ± 0.5 | 23.0 ± 6.6 | 30.1 (19.0, 72.5) | ND |
| ALA + DHA | ND | ND | ND | ND | ND | |
Values are SEMs unless otherwise indicated. Kinetic parameters were generated from best-fit curves plotted through δ13CDHA signatures using a one-phase decay function. ALA, α-linolenic acid; ND, nondeterminable.
Decay constant generated from best-fit curves.
Generated from best-fit curves using equation 1.
Generated from best-fit curves using equation 2 and baseline DHA concentrations.
Under the assumption that the DHA pool size is stable, estimates of the daily rate of loss (Jout) can be determined using equation 4 based on the notion that the rate of incorporation of DHA into a tissue is approximately the rate out (see Materials and Methods). Given the large and rapid changes in DHA concentrations observed in liver and adipose total lipids resulting from the dietary switch, Jout values were only calculated for brain total lipids (Table 1). The rate of loss of brain DHA in ALA diet equilibrated mice on the DHA or ALA + DHA diets was found to be ∼3-fold faster than DHA-equilibrated mice on the ALA diet.
DISCUSSION
By using a dietary switch study, we demonstrated that changes in tissue δ13C signatures in response to the altered dietary δ13C signature can be used to adequately estimate brain DHA half-lives. For decades, carbon isotope ratio analysis and dietary switch studies have been applied to estimate tissue bulk carbon turnover by ecologists (21, 36, 37); however, to our knowledge this is the first time a dietary switch study was used in conjunction with natural abundance CSIA for fatty acid-specific turnover. By equilibrating the brain δ13CDHA signature with that of the dietary n-3 PUFA (either ALA or DHA) and switching mice to a diet providing n-3 PUFA with a different carbon isotopic signature, changes in brain δ13CDHA signatures were modeled as a function of time, allowing for the determination of brain DHA half-lives and turnover.
In mice equilibrated to the ALA diet and switched to the DHA diet or ALA + DHA diet, we determined brain DHA half-lives to be ∼30 to 40 days (Table 1). These values are consistent with previously published studies that used radioactive tracers administered via intracerebroventricular injection to measure the loss of brain radioactivity over time in rats; the half-lives in these studies were estimated to between 30 and 65 days (6–8). Previous studies investigating the effects of dietary PUFA on brain DHA turnover used diets with low or nondetectable levels of DHA (6, 7). In the current study diets contained DHA at 2% of the total dietary fatty acids, equivalent to 0.2% of the mass consumed. Interestingly, brain DHA turnover values from mice on the ALA background switched to DHA or ALA + DHA diet were similar with the values of rodents not consuming appreciable levels of long chain n-3 PUFAs. This suggests that the addition of DHA to a diet with adequate levels of ALA may not affect brain DHA turnover.
Although both current and previous methods of determining DHA half-lives yield similar estimates, they operate on different underlying assumptions. The radioactive loss method estimates half-lives based on the metabolic loss of the radiotracer from the brain. In contrast, the current method estimates half-lives based on the incorporation of preformed dietary DHA or newly synthesized DHA into the brain lipid pool, replacing that which has been metabolized. By making the assumption that the brain DHA pool is at steady state, then the net rate of DHA metabolic loss must be equal to the net rate of DHA incorporation (38). Although brain DHA concentrations slightly increased over the time course in mice from the ALA-equilibrated group when switched to either of the diets containing DHA, we anticipate that any influence from the small change in pool size (∼6% to 8%) on the determination of brain DHA half-lives would be minor. Furthermore, calculated Jout values in the current study (0.11–0.38 µmol/g brain/day) are broadly similar to the Jin of plasma unesterified DHA determined from steady-state pulse-labeling studies (0.04–1.2 µmol/g brain/day) (8–12, 39). Collectively, the agreement between half-lives and Jout values determined by natural abundance carbon isotope ratio analysis with those previously determined increases the confidence of our estimates and demonstrates that this method can adequately model fatty acid turnover. In comparison with mice equilibrated on the ALA diet, the brain DHA half-life determined from the DHA-equilibrated mice switched onto the ALA diet was nearly three times longer than for either ALA-equilibrated diet arm. Peripheral stores of DHA have been shown to be a significant contributor to brain DHA accretion when feeding diets devoid of preformed DHA during brain development (16). Therefore, it is possible that adipose stores could also be a major contributor of DHA for the adult brain in the absence of preformed DHA in the diet (5). The depletion of DHA from the liver and adipose in mice equilibrated to the DHA diet and switched to the ALA diet may have contributed to the observed longer brain DHA half-life in this group. DHA released from these tissues, with an enriched δ13CDHA signature resembling the dietary DHA, would contribute to maintaining a more isotopically enriched circulating DHA pool available for uptake into the brain, thereby decreasing the measured rate of change in brain δ13CDHA signatures and effectively lengthening the estimated half-life.
The primary focus of the current study was to evaluate the ability of natural abundance CSIA to estimate brain DHA half-lives using a dietary switch study. As a secondary analysis we also examined and modeled the changes in liver and adipose δ13CDHA signatures following each dietary switch and estimated DHA half-lives in these tissues. Using this method, DHA half-lives were found to be between 2.5 and 17 days in the liver and between 5.4 and 30.1 days in the adipose depending on the background-equilibrated diet and dietary switch groups. These results were broadly similar to those reported in mice estimated from the depletion of deuterium in labeled liver and adipose lipid pools (40). However, estimates from concentration depletion and accretion studies in pigs and humans suggest adipose half-lives to be much longer (300–700 days) (41–43). Discrepancies in estimated half-lives may be attributed to species differences as well as considerable differences in the methodological approach. Additionally, it is important to note that the large accretion of DHA in liver and adipose pools in the ALA-equilibrated mice over the course of the study may have contributed to an underestimation of true half-lives of DHA in these tissues.
While few studies have investigated the primary source dietary n-3 PUFA for brain DHA incorporation, there is evidence from well-designed isotopic tracer studies to suggest preformed dietary DHA is the major source of DHA for the brain when provided in combination with shorter-chain n-3 PUFA precursors. Previously, in hand-reared rat pups fed a diet providing isotopically labeled ALA and unlabeled DHA, Lefkowitz et al. (16) showed that ∼90% of brain DHA accumulated over 20 days was from preformed DHA, either from the diet or that which had accumulated in peripheral stores from hepatic synthesis prior to feeding labeled ALA. Furthermore, preformed DHA was found to be 7-fold more effective than ALA for brain DHA accretion in newly born baboons (17). In the current study, brain δ13CDHA signatures in DHA-equilibrated mice switched to the ALA + DHA diet were not altered with the addition of ALA to the diet. Furthermore, curves of brain δ13CDHA signatures from mice equilibrated to the ALA diet and switched to the DHA diet or the ALA + DHA diet were nearly indistinguishable and shared similar DHA incorporation half-lives. Therefore, the contribution of DHA synthesized from dietary ALA and incorporated into the brain over the course of the study is minor (∼4%) compared with the contribution of preformed DHA, similar to the results of Lefkowitz et al. (16).
Because the carbon isotope ratio of fatty acids is more or less conserved following absorption and uptake into tissues, we can use pool-mixing models to estimate the contribution of synthesized DHA from ALA in collected tissues. δ13CDHA signatures of brain, liver, and adipose tissues of mice equilibrated to the ALA diet switched to the ALA + DHA diet were 0.8, 0.6, and 1.1 mUr more deplete, respectively, than mice switched to the DHA diet when δ13CDHA signatures plateaued. The plateaued values generated from best-fit curves can be considered to represent a steady-state equilibrium between diet and tissue. Based on these values, the contribution of DHA synthesized from ALA to the liver and adipose total DHA pool of mice on the ALA + DHA diet can be estimated using the following mass balance equation (44):
| (Eq. 5) |
where δ13Ctissue refers to the measured δ13CDHA signature from the tissue, x refers to the proportional contribution of synthesized DHA to the tissue DHA pool, and and refer to the δ13CDHA signatures measured in the final time point of the ALA diet and DHA diet control arms, respectively. The purpose of using values measured from the control groups as opposed to the carbon isotope ratios of dietary ALA and DHA is to account for any kinetic isotope effect during the synthesis and assimilation of DHA with the tissue fatty acid pool. By applying equation 5 the contribution of ALA to brain, liver, and adipose DHA is estimated to be 4%, 15%, and 18%, respectively. Broadly similar estimates were reported by Lefkowitz et al. when feeding rats pups a diet providing labeled ALA and unlabeled DHA (∼9%, ∼10%, and 8% for the brain, the liver, and white adipose, respectively) (16, 45). The small discrepancies between our estimates and those previously obtained through tracer studies could be attributable to differences in life stage between rodent models or the overall length of dietary exposure.
The novel approach to measuring brain DHA half-lives in the current work presents some benefits over previous methods (6–8), namely the lack of expensive and potentially hazardous radiolabeled tracers, but it is not without limitations. By avoiding the use of expensive tracers, studies can be carried out more cost-effectively; however, natural abundance CSIA necessitates the use of high-precision GC/C/IRMS instruments that pose a sizable upfront expense. Furthermore, as experienced in the current work and briefly discussed above, switching diets may contribute to altering the concentration of the analyte of interest within a specific tissue pool, which complicates the interpretation of calculated half-lives because the tissue pool may not be at steady state. This can be addressed by taking measures to only manipulate the carbon isotope ratio of the compound of interest in the diet while maintaining the same quantity in the diet. In the case of DHA, this could be carried out by switching between a marine-sourced product and a more isotopically enriched algal product such as DHASCO (24). Additionally, to ensure the quality of CSIA measurements effort must be taken to ensure sample processing does not introduce a kinetic isotope effect that would erode the validity of measured carbon isotope ratios. However, if these concerns are appropriately addressed during the initiation of a study, the application of natural abundance CSIA can be applied as a useful tool for studying complex areas of metabolism.
The natural 13C enrichment of marine-sourced fatty acids, the predominant source of long-chain n-3 PUFAs, compared with relatively 13C-deplete dietary sources of ALA (e.g., canola, soy, flax) presents an opportunity to study the nuances of long-chain PUFA metabolism in humans without tracer compounds. Metherel et al. (26) used natural abundance carbon isotope signatures of plasma long-chain n-3 PUFA following 12 weeks of supplementation with fish oil concentrates to fingerprint the flux of PUFA through the endogenous biosynthetic pathway. In doing so, we were able to demonstrate that increases in plasma EPA pool size following DHA supplementation are not the result of apparent retroconversion, confirming earlier observations made in rodents similarly using natural abundance isotope ratio measurements (25, 26). Additionally, commercially available algal oils with unique 13C enrichment in long-chain PUFAs, even greater than those of marine sources, are favorable candidates to study PUFA metabolism in clinical applications. For example, Carnielli et al. (24) used ARASCO- and DHASCO-fortified infant formula coupled with natural abundance carbon isotope ratio analysis to generate estimates of arachidonic acid and DHA synthesis in infants. Due to the variation in carbon isotope ratios of n-3 PUFAs in the environment, natural abundance carbon isotope ratio analysis is well suited for the investigation of n-3 PUFA metabolism in humans and offers benefits over tracer studies, namely the wide availability of supplemental sources and lower cost.
To conclude, with the use of natural abundance CSIA we determined brain DHA half-lives to be ∼30 to 135 days in mice depending on the diet group. Estimates from ALA-equilibrated mice switched to a diet containing DHA matched those previously determined through the use of radiolabeled decay models and calculated via pulse labeling with unesterified DHA (6–8). The agreement of these findings provides confidence to the estimate that brain DHA in rodents has a half-life of ∼30 days. Furthermore, the ability of peripheral stores to supply the brain with DHA when it is not present in the diet was indicated by the longer half-life of DHA-equilibrated mice switched to the ALA diet. These results aptly highlight the utility of natural abundance carbon isotope ratio analysis in a dietary switch experiment to study the metabolism and turnover of specific fatty acids and present the basis for the greater application of natural abundance isotope measurements for the investigation of fatty acid and lipid metabolism in future studies.
Supplementary Material
Acknowledgments
The authors thank Ashley St. Pierre for assistance with the IRMS analysis.
Footnotes
Abbreviations:
- ALA
- α-linolenic acid
- CSIA
- compound-specific isotope analysis
- EA
- elemental analysis
- FAME
- fatty acid methyl ester
- GC/C/IRMS
- gas chromatography combustion isotope ratio mass spectrometry
- GC/FID
- gas chromatography flame ionization detection
This study was supported by Natural Sciences and Engineering Research Council of Canada Grant 482597. R.P.B. is supported by grant funding through the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada and holds a Canada Research Chair in Brain Lipid Metabolism. R.J.S.L. holds an Ontario Graduate Scholarship and receives funding support through the Peterborough K. M. Hunter Charitable Foundation. R.P.B. has received industrial grants, including those matched by the Canadian government, and/or travel support related to work on brain fatty acid uptake from Arctic Nutrition, Bunge Ltd., DSM, Fonterra, Mead Johnson, and Nestec Inc. Moreover, R.P.B. is on the executive committee of the International Society for the Study of Fatty Acids and Lipids and held a meeting on behalf of fatty acids and cell signaling, both of which rely on corporate sponsorship. R.P.B. has given expert testimony in relation to supplements and the brain. R.J.S.L. and C-C.L. declare that they have no conflicts of interest with the contents of the article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Lacombe R. J. S., Chouinard-Watkins R., and Bazinet R. P.. 2018. Brain docosahexaenoic acid uptake and metabolism. Mol. Aspects Med. 64: 109–134. [DOI] [PubMed] [Google Scholar]
- 2.Fliesler S. J., and Anderson R. E.. 1983. Chemistry and metabolism of lipids in the vertebrate retina. Prog. Lipid Res. 22: 79–131. [DOI] [PubMed] [Google Scholar]
- 3.Bazinet R. P., and Layé S.. 2014. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15: 771–785. [DOI] [PubMed] [Google Scholar]
- 4.Demar J. C., Ma K., Chang L., Bell J. M., and Rapoport S. I.. 2005. Alpha-linolenic acid does not contribute appreciably to docosahexaenoic acid within brain phospholipids of adult rats fed a diet enriched in docosahexaenoic acid. J. Neurochem. 94: 1063–1076. [DOI] [PubMed] [Google Scholar]
- 5.Domenichiello A. F., Kitson A. P., and Bazinet R. P.. 2015. Is docosahexaenoic acid synthesis from alpha-linolenic acid sufficient to supply the adult brain? Prog. Lipid Res. 59: 54–66. [DOI] [PubMed] [Google Scholar]
- 6.DeMar J. C. Jr., Ma K., Bell J. M., and Rapoport S. I.. 2004. Half-lives of docosahexaenoic acid in rat brain phospholipids are prolonged by 15 weeks of nutritional deprivation of n-3 polyunsaturated fatty acids. J. Neurochem. 91: 1125–1137. [DOI] [PubMed] [Google Scholar]
- 7.Lin L. E., Chen C. T., Hildebrand K. D., Liu Z., Hopperton K. E., and Bazinet R. P.. 2015. Chronic dietary n-6 PUFA deprivation leads to conservation of arachidonic acid and more rapid loss of DHA in rat brain phospholipids. J. Lipid Res. 56: 390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C. T., Kitson A. P., Hopperton K. E., Domenichiello A. F., Trépanier M-O., Lin L. E., Ermini L., Post M., Thies F., and Bazinet R. P.. 2015. Plasma non-esterified docosahexaenoic acid is the major pool supplying the brain. Sci. Rep. 5: 15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazinet R. P., Rao J. S., Chang L., Rapoport S. I., and Lee H. J.. 2006. Chronic carbamazepine decreases the incorporation rate and turnover of arachidonic acid but not docosahexaenoic acid in brain phospholipids of the unanesthetized rat: relevance to bipolar disorder. Biol. Psychiatry. 59: 401–407. [DOI] [PubMed] [Google Scholar]
- 10.Bazinet R. P., Rao J. S., Chang L., Rapoport S. I., and Lee H. J.. 2005. Chronic valproate does not alter the kinetics of docosahexaenoic acid within brain phospholipids of the unanesthetized rat. Psychopharmacology (Berl.). 182: 180–185. [DOI] [PubMed] [Google Scholar]
- 11.Lee H-J., Ghelardoni S., Chang L., Bosetti F., Rapoport S. I., and Bazinet R. P.. 2005. Topiramate does not alter the kinetics of arachidonic or docosahexaenoic acid in brain phospholipids of the unanesthetized rat. Neurochem. Res. 30: 677–683. [DOI] [PubMed] [Google Scholar]
- 12.Chang M. C., Bell J. M., Purdon A. D., Chikhale E. G., and Grange E.. 1999. Dynamics of docosahexaenoic acid metabolism in the central nervous system: lack of effect of chronic lithium treatment. Neurochem. Res. 24: 399–406. [DOI] [PubMed] [Google Scholar]
- 13.Thiés F., Delachambre M. C., Bentejac M., Lagarde M., and Lecerf J.. 1992. Unsaturated fatty acids esterified in 2-acyl-1-lysophosphatidylcholine bound to albumin are more efficiently taken up by the young rat brain than the unesterified form. J. Neurochem. 59: 1110–1116. [DOI] [PubMed] [Google Scholar]
- 14.Thies F., Pillon C., Moliere P., Lagarde M., and Lecerf J.. 1994. Preferential incorporation of sn-2 lysoPC DHA over unesterified DHA in the young rat brain. Am. J. Physiol. 267: R1273–R1279. [DOI] [PubMed] [Google Scholar]
- 15.Chouinard-Watkins R., Chen C. T., Metherel A. H., Lacombe R. J. S., Thies F., Masoodi M., and Bazinet R. P.. 2017. Phospholipid class-specific brain enrichment in response to lysophosphatidylcholine docosahexaenoic acid infusion. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862: 1092–1098. [DOI] [PubMed] [Google Scholar]
- 16.Lefkowitz W., Lim S. Y., Lin Y., and Salem N.. 2005. Where does the developing brain obtain its docosahexaenoic acid? Relative contributions of dietary alpha-linolenic acid, docosahexaenoic acid, and body stores in the developing rat. Pediatr. Res. 57: 157–165. [DOI] [PubMed] [Google Scholar]
- 17.Su H. M., Bernardo L., Mirmiran M., Ma X. H., Corso T. N., Nathanielsz P. W., and Brenna J. T.. 1999. Bioequivalence of dietary α-linolenic and docosahexaenoic acids as sources of docosahexaenoate accretion in brain and associated organs of neonatal baboons. Pediatr. Res. 45: 87–93. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Bartke N., Van Daele H., Lawrence P., Qin X., Park H. G., Kothapalli K., Windust A., Bindels J., Wang Z., et al. 2014. Higher efficacy of dietary DHA provided as a phospholipid than as a triglyceride for brain DHA accretion in neonatal piglets. J. Lipid Res. 55: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutton T. W. 1991. Stable carbon isotope ratios of natural materials. I. Sample preparation and mass spectrometric analysis. In Carbon Isotope Techniques. Coleman D. C. and Fry B., editors. Academic Press, San Diego, CA: pp. 173–187. [Google Scholar]
- 20.O’Leary M. H. 1981. Carbon isotope fractionation in plants. Phytochemistry. 20: 553–567. [Google Scholar]
- 21.Tieszen L. L., Boutton T. W., Tesdahl K. G., and Slade N. A.. 1983. Fractionation and turnover of stable carbon isotopes in animal tissues: implications for δ13C analysis of diet. Oecologia. 57: 32–37. [DOI] [PubMed] [Google Scholar]
- 22.Dalerum F., and Angerbjörn A.. 2005. Resolving temporal variation in vertebrate diets using naturally occurring stable isotopes. Oecologia. 144: 647–658. [DOI] [PubMed] [Google Scholar]
- 23.Demmelmair H., Schenck U. v., Behrendt E., Sauerwald T., and Koletzko B.. 1995. Estimation of arachidonic acid synthesis in full term neonates using natural variation of 13C content. J. Pediatr. Gastroenterol. Nutr. 21: 31–36. [DOI] [PubMed] [Google Scholar]
- 24.Carnielli V. P., Simonato M., Verlato G., Luijendijk I., De Curtis M., Sauer P. J. J., and Cogo P. E.. 2007. Synthesis of long-chain polyunsaturated fatty acids in preterm newborns fed formula with long-chain polyunsaturated fatty acids. Am. J. Clin. Nutr. 86: 1323–1330. [DOI] [PubMed] [Google Scholar]
- 25.Metherel A. H., Chouinard-Watkins R., Trépanier M-O., Lacombe R. J. S. J. S., and Bazinet R. P.. 2017. Retroconversion is a minor contributor to increases in eicosapentaenoic acid following docosahexaenoic acid feeding as determined by compound specific isotope analysis in rat liver. Nutr. Metab. (Lond.). 14: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metherel A. H., Irfan M., Klingel S. L., Mutch D. M., and Bazinet R. P.. 2019. Compound-specific isotope analysis reveals no retroconversion of DHA to EPA but substantial conversion of EPA to DHA following supplementation: a randomized control trial. Am. J. Clin. Nutr. 110: 823–831. [DOI] [PubMed] [Google Scholar]
- 27.Lacombe R. J. S., Giuliano V., Colombo S. M., Arts M. T., and Bazinet R. P.. 2017. Compound-specific isotope analysis resolves the dietary origin of docosahexaenoic acid in the mouse brain. J. Lipid Res. 58: 2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giuliano V., Lacombe R. J. S., Hopperton K. E., and Bazinet R. P.. 2018. Applying stable carbon isotopic analysis at the natural abundance level to determine the origin of docosahexaenoic acid in the brain of the fat-1 mouse. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1863: 1388–1398. [DOI] [PubMed] [Google Scholar]
- 29.Folch J., Lees M., and Sloane Stanley G.. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 30.Rieley G. 1994. Derivatization of organic compounds prior to gas chromatographic-combustion-isotope ratio mass spectrometric analysis: identification of isotope fractionation processes. Analyst (Lond.). 119: 915–919. [Google Scholar]
- 31.Dobson G., Christie W. W., and Nikolova-Damyanova B.. 1995. Silver ion chromatography of lipids and fatty acids. J. Chromatogr. B, Biomed. Appl. 671: 197–222. [DOI] [PubMed] [Google Scholar]
- 32.Kramer J. K. G., Hernandez M., Cruz-Hernandez C., Kraft J., and Dugan M. E. R.. 2008. Combining results of two GC separations partly achieves determination of all cis and trans 16:1, 18:1, 18:2 and 18:3 except CLA isomers of milk fat as demonstrated using ag-ion SPE fractionation. Lipids. 43: 259–273. [DOI] [PubMed] [Google Scholar]
- 33.Paul D., Skrzypek G., and Fórizs I.. 2007. Normalization of measured stable isotopic compositions to isotope reference scales–a review. Rapid Commun. Mass Spectrom. 21: 3006–3014. [DOI] [PubMed] [Google Scholar]
- 34.Schimmelmann A., Qi H., Coplen T. B., Brand W. A., Fong J., Meier-Augenstein W., Kemp H F., Toman B., Ackermann A., Assonov S., et al. 2016. Organic reference materials for hydrogen, carbon, and nitrogen stable isotope-ratio measurements: caffeines, n-alkanes, fatty acid methyl esters, glycines, l-valines, polyethylenes, and oils. Anal. Chem. 88: 4294–4302. [DOI] [PubMed] [Google Scholar]
- 35.Rapoport S. I. 2001. In vivo fatty acid incorporation into brain phospholipids in relation to plasma availability, signal transduction and membrane remodeling. J. Mol. Neurosci. 16: 243–261. [DOI] [PubMed] [Google Scholar]
- 36.O’Brien D., Schrag D., and Martinez del Rio C.. 2000. Allocation to reproduction in a hawkmoth: a quantitative analysis using stable carbon isotopes. Ecology. 81: 2822–2831. [Google Scholar]
- 37.O’Brien D. M., Fogel M. L., and Boggs C. L.. 2002. Renewable and nonrenewable resources: amino acid turnover and allocation to reproduction in Lepidoptera. Proc. Natl. Acad. Sci. USA. 99: 4413–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson P. J., Noronha J., DeGeorge J. J., Freed L. M., Nariai T., and Rapoport S. I.. 1992. A quantitative method for measuring regional in vivo fatty-acid incorporation into and turnover within brain phospholipids: review and critical analysis. Brain Res. Brain Res. Rev. 17: 187–214. [DOI] [PubMed] [Google Scholar]
- 39.Contreras M. A., Greiner R. S., Chang M. C. J., Myers C. S., Salem N. Jr., and Rapoport S. I.. 2000. Nutritional deprivation of alpha-linolenic acid decreases but does not abolish turnover and availability of unacylated docosahexaenoic acid and docosahexaenoyl-CoA in rat brain. J. Neurochem. 75: 2392–2400. [DOI] [PubMed] [Google Scholar]
- 40.Stetten D. J., and Grail G. F.. 1943. The rates of replacement of depot and liver fatty acids in mice. J. Biol. Chem. 148: 509–515. [Google Scholar]
- 41.Anderson D. B., Kauffman R. G., and Benevenga N. J.. 1972. Estimate of fatty acid turnover in porcine adipose tissue. Lipids. 7: 488–489. [DOI] [PubMed] [Google Scholar]
- 42.Hirsch J., Farquhar J. W., Ahrens E. H., Peterson M. L., and Stoffel W.. 1960. Studies of adipose tissue in man. A microtechnic for sampling and analysis. Am. J. Clin. Nutr. 8: 499–511. [DOI] [PubMed] [Google Scholar]
- 43.Dayton S., Hashimoto S., Dixon W., and Pearce M. L.. 1966. Composition of lipids in human serum and adipose tissue during prolonged feeding of a diet high in unsaturated fat. J. Lipid Res. 7: 103–111. [PubMed] [Google Scholar]
- 44.Martínez del Río C., and Wolf B.. 2005. Mass balance models for animal isotopic ecology. In Physiological and Ecological Adaptations to Feeding in Vertebrates. Science Publishers, Enfield, NH. pp. 141–174. [Google Scholar]
- 45.DeMar J. C., DiMartino C., Baca A. W., Lefkowitz W., and Salem N.. 2008. Effect of dietary docosahexaenoic acid on biosynthesis of docosahexaenoic acid from alpha-linolenic acid in young rats. J. Lipid Res. 49: 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.