Abstract
Few studies have investigated the interactions between HDL-C-related SNPs identified by genome-wide association (GWA) study and physical activity (PA) on HDL-C. First, we conducted a sex-stratified GWA study in a discovery sample (2,231 men and 2,431 women) and replication sample (2,599 men and 3,109 women) to identify SNPs influencing log-transformed HDL-C in Japanese participants in the baseline survey of the Japan Multi-Institutional Collaborative Cohort Study. We also replicated previously reported HDL-C-related SNPs in a combined (discovery plus replication) sample (4,830 men and 5,540 women). We then analyzed the interactions of the HDL-C-related SNPs with PA on HDL-C. The sex-stratified GWA analyses identified 11 and 10 HDL-C-related SNPs in men and women as targets for an interaction analysis. Among these, only one interaction of ABCA1 rs1883025 with PA was statistically significant in men, after Bonferroni correction [P-interaction = 0.001 (α = 0.05/21 = 0.002)]. The per-major-allele (C allele) increase in log-transformed HDL-C was lost in men with low PA (β = 0.008) compared with those with medium (β = 0.032) or high PA (β = 0.034). These findings suggest that the benefit of carrying a C allele of ABCA1 rs1883025 on enhancing HDL-C may be attenuated in inactive men.
Keywords: epidemiology, exercise, genetics, high density lipoprotein-cholesterol, polymorphisms, cholesterol efflux, adenosine 5′-triphosphate binding cassette transporter A1
Higher levels of circulating HDL-C have been consistently associated with a reduced risk of cardiovascular disease (1). HDL, the lipoprotein carrying HDL-C, plays a major role in reverse cholesterol transport (RCT), wherein the cholesterol accumulated in macrophages in the artery vessel walls is extracted and transported to the liver and subsequently to the intestine for excretion from body (2, 3). Thus, increased HDL-C concentrations and enhanced RCT are generally considered to be important for preventing atherosclerosis and cardiovascular diseases.
The serum HDL-C levels are known to be strongly influenced by genetic factors (4). Previous genome-wide association (GWA) studies have identified at least 79 SNPs that are associated with circulating HDL-C levels (4–6). However, the levels of serum HDL-C are also influenced by other modifiable factors, such as physical activity (PA) (7), alcohol consumption (8), and cigarette habits (9). Habitual PA in particular is a low-cost nonpharmacological measure that can increase the serum HDL-C concentration (7).
While there have been a few reports on the GWA meta-analyses of HDL-C in populations predominantly of European ancestry (5) or East-Asian ancestry, in which Japanese individuals were partially included as a part of the total sample (6), there are few GWA studies of HDL-C in entirely Japanese populations. Ahmad et al. (10) showed that the effects of GWA analysis-derived HDL-C-related SNPs in genes involved in lipid metabolism [rs10096633 in LPL, rs1800588 in hepatic lipase (LIPC), and rs1532624 in cholesteryl ester transfer protein (CETP)] on HDL-C were significantly modified by habitual PA in women of European ancestry. However, gene-PA interactions are known to be influenced by subject characteristics, including sex and ethnicity (11). To date, there has been no GWA study that has investigated in both men and women the effects of interaction between HDL-C-related SNPs (identified by GWA analysis) and PA on the HDL-C concentration in an understudied Japanese population.
Therefore, in the present study, we investigated the potential interactions of the HDL-C-related SNPs derived from a GWA analysis and habitual PA on serum HDL-C levels in Japanese men and women.
MATERIALS AND METHODS
Subjects
The study participants were middle-aged Japanese people 35–69 years of age who voluntarily participated in the baseline survey of the Japan Multi-Institutional Collaborative Cohort (J-MICC) study on the gene-environment interactions of lifestyle-related diseases (12, 13). They were recruited from 12 different investigation areas (Chiba, Okazaki, Shizuoka-Daiko, Takashima, Kyoto, Sakuragaoka, Aichi, Saga, Kagoshima, Tokushima, Fukuoka, and Kyushu-KOPS) throughout Japan from 2004 to 2013 (the dataset version of January 11, 2018 was used in the present study). Information on the variables of interest, including the subjects’ medical history, medications, and lifestyle factors (habitual PA, alcohol drinking, and cigarette smoking) were obtained via questionnaire, and blood samples were collected at the baseline survey.
A total of 14,555 subjects of J-MICC study were genotyped (by a method described below). Among these 14,555 subjects, 26 samples with inconsistent sex information between the questionnaire and an estimate from genotyping were excluded. The identity-by-descent method implemented in the PLINK 1.9 software program (14) detected 388 close relationship pairs [pi-hat (an index of the degree of genetic similarity between two individuals) > 0.1875], and one sample of each pair was excluded. A principal component analysis (15, 16) with a 1000 Genomes reference panel (phase 3) (17) detected 34 subjects whose estimated ancestries were outside of the Japanese population (18), and these 34 subjects were also excluded. SNPs with a genotype call rate of <0.98 and/or a Hardy-Weinberg equilibrium exact test P-value < 1 × 10−6 were removed, leaving 14,091 individuals and 873,254 SNPs.
Of these 14,091 individuals who passed genotype quality control filtering, 2,435 were excluded because their HDL-C data (as outcome) were not available. In addition, another 1,100 subjects were excluded because they had a history of taking cholesterol-lowering drugs. Consequently, 10,556 subjects remained.
To detect SNPs that are associated with serum HDL-C levels in the current GWA analysis, we restricted our sample to the 10,370 individuals (4,830 men and 5,540 women) for whom we had data on all potential confounding factors, including the BMI, alcohol consumption, and cigarette habit (numbers of subjects missing data for the three covariates are described in the Statistical analyses section). We used the data of 4,662 subjects (men, n = 2,231; women, n = 2,431) from the five investigation areas of Okazaki, Shizuoka-Daiko, Takashima, Kyoto, and Sakuragaoka for the discovery stage, while the data of 5,708 subjects (men, n = 2,599; women, n = 3,109) from the other five areas of Fukuoka, Saga, Kagoshima, Tokushima, and Kyushu-KOPS were used for the replication stage.
Written informed consent was obtained from all participants, and the study protocol was approved by the Ethics Committees of the Nagoya University Graduate School of Medicine and other institutions participating in the J-MICC study. The study was conducted in accordance with the Declaration of Helsinki principles.
Genotyping of polymorphisms and genotype imputation
The buffy coat fraction was collected from the blood samples (EDTA-2Na), and the extracted DNA was stored at −80°C. The genomic DNA was extracted from the buffy coat using a BioRobot M48 Workstation (QIAGEN Group, Tokyo, Japan) at the central J-MICC study office, except for the blood samples obtained from two investigation areas (Fukuoka and Kyushu-KOPS). Regarding the blood samples obtained from those two areas, DNA was extracted locally (at the respective investigation sites) from whole blood using an automatic nucleic acid isolation system (NA-3000; Kurabo Co., Ltd., Osaka, Japan). Genotyping was performed at RIKEN Center for Integrative Medical Science using a HumanOmniExpressExome BeadChip array (Illumina Inc., San Diego, CA), as previously described (19–21). Genotype imputation was conducted using the SHAPEIT (22) and Minimac3 (23) software programs based on the 1000 Genomes reference panel (phase 3) (17). After the genotype imputation, strict quality control filters were applied as previously described (24); namely, variants with an imputation R2 < 0.8 and a minor allele frequency < 0.01 were excluded, resulting in 7,094,228 variants.
Heritability estimation
The narrow-sense heritability of the natural log-transformed HDL-C level was estimated based on the genotyping data using a mixed linear model (25) with adjustment for age, sex, and investigation area. In brief, the model assumes that a genetic random effect for each individual is drawn from a multivariate normal distribution with mean 0 and a variance-covariance matrix calculated from genotype data [i.e., genetic relationship matrix (GRM)]. Similarly, a nongenetic random effect is assumed to be normally distributed with mean 0 and an identity variance-covariance matrix. Adjustment variables were modeled as fixed-effect variables. The relative contribution of genetic and nongenetic effects was estimated using the restricted maximum likelihood method. To calculate the GRM, additional quality control filtering was applied according to a previous study (Hardy-Weinberg exact test P-value ≥ 0.05, and minor allele frequency ≥ 0.01) (7, 26), and the remaining 482,567 directly genotyped SNPs on autosomal chromosomes were used. Calculation of the GRM and heritability estimation were performed using the GCTA software program, version 1.24.2 (27).
Assessments of habitual PA, alcohol consumption, and cigarette habit by a questionnaire, and anthropometric measurements
Data on habitual PA, alcohol consumption, cigarette habit, current medications, and disease history were collected using a self-administered questionnaire. The total amount of habitual PA, which corresponds to an activity intensity of ≥3 metabolic equivalents (METs), was assessed as previously described (28). Habitual PA was calculated as the sum of PA in daily life and leisure time. Each of the self-reported PA types was assigned to a specific MET value according to Compendium of Physical Activities (29), in which, for instance, walking (assumed to be 4.86 km/h) is assigned as 3.3 METs. The usual PA in daily life was estimated by multiplying the daily time spent for walking and engaged in physically heavy work by their assigned MET intensities (walking, 3.3 METs; physically heavy work, 4.5 METs), while the leisure-time PA was estimated by multiplying the time spent per day by the assigned MET intensities for light activity (3.3 METs), moderate activity (4.0 METs), and vigorous activity (8.0 METs). One MET is defined as oxygen consumption of 3.5 ml/kg weight/min, which represents the average energy expenditure at rest.
Alcohol consumption was categorized as never, former, and current 0.1–22.9 g, 23.0–45.9 g, or ≥46 g ethanol/day [based on the fact that one glass of Japanese sake (180 ml) contains approximately 23 g ethanol]. The ethanol consumption per day was calculated using data obtained via a validated short food frequency questionnaire (30–32).
Regarding the smoking habit, subjects were first asked about their smoking status from the past to the present. Current smokers were then requested to report their usual cigarette consumption (cigarettes/day). The smoking status was categorized as never, former, current 1–19, or current ≥20 cigarettes/day (based on the fact that one pack of cigarettes commonly contains 20 cigarettes in Japan).
Anthropometric measurements and blood sampling were conducted as part of the health checkup or for research purposes at the institutions participating in the J-MICC study (12). Height and weight were measured to the nearest 0.1 cm and 0.1 kg, respectively. The BMI was determined by dividing the body weight in kilograms by the square of height in meters.
Measurement of serum HDL-C levels
Venous blood samples were drawn from each participant, and serum was separated by centrifugation and stored at −80°C until further use. The participants were not required to be in a fasting state. Nonfasting blood measures should not affect HDL-C levels (33). Serum HDL-C levels were measured via a polyanion-polymer/detergent method. The HDL-C levels were examined as a part of the health checkup or for research purposes at the institutions affiliated with the J-MICC study (34).
Statistical analyses
From the 10,556 subjects whose data remained (as mentioned above in the Subjects section), we further excluded those with any of the following conditions from the current GWA analysis stratified by sex: missing age data (n = 1), missing BMI data (n = 1), missing alcohol drinking data (n = 172), and missing cigarette smoking data (n = 12). Self-reported BMI data (calculated by self-reported weight and height) were used in place of objectively measured BMI data (calculated by measured weight and height) for subjects with missing objective BMI data (n = 232). Consequently, 10,370 individuals (men: n = 4,830, women: n = 5,540) were ultimately included in the sex-stratified GWA analysis.
The association between genetic variants and the natural log-transformed HDL-C level was examined separately by sex using a mixed linear model association method (25) with adjustment for age (continuous), investigation site (categorical), BMI (continuous), alcohol drinking (categorical), and cigarette smoking (categorical). Alcohol drinking status was coded as follows: never (0), former (1), current [2 (<23 g ethanol/day)], current [3 (23.0–45.9 g ethanol/day)], or current [4 (≥46 g ethanol/day)]. The cigarette smoking status was coded as follows: never (0), former (1), current [2 (1–19 cigarettes/day)], or current [3 (≥20 cigarettes/day)]. The same GRM used for heritability estimation was applied. Adjustment covariates and dosage data for each variant were modeled as fixed-effect variables, and genetic and nongenetic effects were modeled as random effect variables. The genome-wide significance level was set at a P-value of <5 × 10−8. Gene variants that achieved genome-wide significance in the discovery stage were further investigated in the replication stage.
In addition, we conducted a replication in the Japanese population using data of large-scale meta-analyses in populations predominantly of European ancestry (5) and in East Asian population including Japanese individuals (6). The former meta-analysis reported 157 loci associated with lipid levels at P < 5 × 10−8, 71 of which were associated with HDL-C levels. The latter meta-analysis identified three variants in East Asian individuals and five variants in trans-ancestry analyses with European individuals (eight SNPs in total). Among these 79 variants, in the current J-MICC samples, 10 variants showed low minor allele frequencies (<0.01), and 2 showed a low imputation quality (<0.8). We therefore examined the association between the remaining 67 variants and the log-transformed HDL-C levels in the current J-MICC samples (discovery ± replication samples). Bonferroni’s corrected α for the replication was set at 0.00075 (0.05/67). SNPs identified in the replication of previous studies were excluded from the target for the testing of interaction with PA, if they were in linkage disequilibrium (R2 > 0.9) (35) with the SNPs identified by the current GWA analysis.
As the main part of the current statistical analysis, analyses of interactions between the HDL-C-related SNPs (identified by the current GWA study or identified by the replication of previous meta-analysis studies) and habitual PA on the serum HDL-C levels were performed separately by sex using the SAS software program (version 9.4 for Windows; SAS Institute, Cary, NC).
The significance of the univariate correlations between PA and the basic characteristics of the study subjects was assessed according to Spearman’s rank correlation coefficient (ρ). To examine the potential interaction of the HDL-C-related SNPs (identified by the GWA analysis) and habitual PA, an additional interaction term (each SNP × PA) (both SNP and PA were treated as continuous variables) was included in a multiple regression model with adjustment for the age (continuous), investigation site (categorical), BMI (continuous), alcohol drinking (categorical), and cigarette smoking (categorical). To avoid the false positive interactions due to multiple testing, Bonferroni’s corrected α was used for the interaction analysis (0.05/21 = 0.002) [number of target SNPs in men (n = 11) + women (n = 10) = 21]. An ANCOVA was used to calculate the adjusted means of log-transformed serum HDL-C (and 95% confidence interval) and to compare the adjusted geometric means of HDL-C among the three genotype groups in the three different PA groups divided into tertiles (low, medium, or high).
Replication of the interaction between HDL-C-related SNPs derived from the GWA analysis and habitual PA on HDL-C levels in another sample
Replication of the interaction was performed in subjects from the Saga region in the J-MICC study (4,013 men and 5,722 women). Study participants from the Saga region who were included in the original interaction analysis were completely excluded from the analysis for the replication of interaction. Similar exclusion criteria to the original analysis were applied to the replication. In these subjects, ABCA1 SNP rs1883025 was analyzed by a TaqMan® SNP genotyping assay using a StepOne Plus real-time PCR system (Applied Biosystems), and habitual PA was objectively measured by single-axis accelerometers (Life-corder; Suzuken Co. Ltd., Nagoya, Japan). PA level (PAL) was used as an index of habitual PA, as previously described in detail (36). Serum HDL-C levels and the four covariates (age, BMI, alcohol drinking, and cigarette smoking) were assessed in a manner similar to the original interaction analysis. In the statistical model for the replication of interaction, the serum HDL-C levels were log-transformed, and the accelerometer wear time (minutes per day) was further added as a covariate in addition to the four above-mentioned covariates.
RESULTS
The sex-stratified GWA analysis of serum HDL-C levels in the Japanese population
The baseline characteristics of the current subjects in the discovery and replication samples are shown in supplemental Table S1. The narrow-sense heritability of the natural log-transformed HDL-C was estimated to be 30.3% (SE 3.2%). We identified five and four SNPs influencing the HDL-C levels in men and women, respectively, at the genome-wide level of statistical significance (supplemental Table S2).
Replication of previous GWA studies
As a result of replication of previous GWA studies, seven variants in men and seven variants in women were significantly associated with the serum HDL-C levels, after performing Bonferroni’s correction [Bonferroni-corrected α = 0.00075 (0.05/67 previously reported SNPs)] (supplemental Table S3). Among these identified SNPs, an SNP (CETP rs3764261) was excluded from the target SNPs for the testing of interaction with PA because the CETP rs3764261 was in strong linkage disequilibrium with CETP rs56156922 (R2 = 0.92), which was identified in men and women by the sex-stratified GWA analysis.
Characteristics of the study subjects according to PA tertiles and correlations between PA and variables of characteristics
Characteristics of the study subjects analyzed for the interaction between the HDL-C-related SNPs selected based on the GWA analysis and habitual PA are shown by three different PA levels (low, medium, and high) in Table 1. Higher PA was correlated with a higher age (in men and women), lower BMI (in men), higher alcohol drinking (in men), and higher HDL-C (in men and women). In contrast, the PA was not correlated with the total cholesterol in either men or women.
TABLE 1.
Characteristics of the study subjects analyzed for the interaction between the SNPs selected based on the GWA analysis and habitual PA, according to PA tertiles
| Characteristics | Men (n = 4,830) | Women (n = 5,540) | ||||||||
| Low PA (n = 1,581) | Medium PA (n = 1,642) | High PA (n = 1,607) | ρa | P | Low PA (n = 1,815) | Medium PA (n = 1,870) | High PA (n = 1,855) | ρa | P | |
| Age (years) | 54.1 (9.0) | 54.9 (9.5) | 55.0 (9.7) | 0.047 | 0.001 | 53.2 (9.5) | 53.9 (9.5) | 54.4 (9.0) | 0.049 | 0.0003 |
| Height (cm) | 168.2 (6.3) | 167.6 (6.2) | 166.6 (6.5) | −0.105 | <0.0001 | 155.1 (5.8) | 155.0 (5.7) | 154.3 (5.9) | −0.063 | <0.0001 |
| Weight (kg) | 67.9 (10.0) | 67.1 (10.5) | 65.8 (9.9) | −0.098 | <0.0001 | 54.2 (8.6) | 53.0 (7.6) | 53.6 (8.5) | −0.031 | 0.019 |
| BMI (kg/m2) | 24.0 (3.1) | 23.9 (3.3) | 23.7 (3.1) | −0.051 | 0.0004 | 22.6 (3.5) | 22.1 (3.2) | 22.5 (3.4) | 0.002 | 0.867 |
| Smoking status [n (%)] | −0.004 | 0.785 | 0.016 | 0.222 | ||||||
| Never | 417 (26.38) | 464 (28.3) | 439 (27.3) | — | — | 1556 (85.7) | 1635 (87.4) | 1561 (84.2) | — | — |
| Former | 596 (37.7) | 659 (40.1) | 597 (37.2) | — | — | 111 (6.1) | 115 (6.2) | 146 (7.9) | — | — |
| Current | ||||||||||
| 1‒19 (cigarettes/day) | 167 (10.56) | 155 (9.4) | 152 (9.5) | — | — | 90 (5.0) | 89 (4.8) | 102 (5.5) | — | — |
| 20+ (cigarettes/day) | 401 (25.36) | 364 (22.2) | 419 (26.1) | — | — | 58 (3.2) | 31 (1.7) | 46 (2.5) | — | — |
| Alcohol consumption [n (%)] | 0.060 | <0.0001 | 0.020 | 0.145 | ||||||
| Never | 356 (22.52) | 322 (19.6) | 305 (19.0) | — | — | 1164 (64.1) | 1153 (61.7) | 1144 (61.7) | — | — |
| Former | 48 (3.0) | 44 (2.7) | 41 (2.6) | — | — | 33 (1.8) | 31 (1.7) | 29 (1.6) | — | — |
| Current | ||||||||||
| 0.1‒22.9 (g/day) | 554 (35.0) | 617 (37.6) | 518 (32.2) | — | — | 526 (29.0) | 596 (31.9) | 592 (31.9) | — | — |
| 23.0‒45.9 (g/day) | 294 (18.6) | 323 (19.7) | 327 (20.4) | — | — | 63 (3.5) | 71 (3.8) | 63 (3.4) | — | — |
| 46.0+ (g/day) | 329 (20.81) | 336 (20.5) | 416 (25.9) | — | — | 29 (1.6) | 19 (1.0) | 27 (1.5) | — | — |
| PA (MET·h/day) | 3.16 (1.54) | 10.17 (2.58) | 30.92 (13.37) | 0.943 | <0.0001 | 3.51 (1.97) | 11.06 (2.72) | 28.61 (12.34) | 0.943 | <0.0001 |
| HDL-C (mg/dl)b | 54.0 (1.3) | 54.9 (1.3) | 57.4 (1.3) | 0.098 | <0.0001 | 64.9 (1.3) | 65.8 (1.3) | 65.9 (1.3) | 0.022 | 0.105 |
| Total cholesterol (mg/dl)c | 206.3 (32.8) | 205.2 (32.4) | 205.4 (33.3) | −0.011 | 0.473 | 216.2 (37.3) | 216.0 (35.2) | 215.8 (35.0) | −0.001 | 0.943 |
Values are mean (SD) for continuous variables and number (percentage) for categorical variables.
Spearman’s rank correlation coefficient.
Geometric mean (geometric SD).
Based on 4,429 males and 5,091 females.
Interactions of the GWA analysis-derived SNPs and habitual PA on the serum HDL-C levels
As shown in Table 2, among the 11 HDL-C-related SNPs identified in men, only one significant interaction of ABCA1 SNP rs1883025 with habitual PA was detected, after Bonferroni’s correction [P-interaction = 0.0010 (Bonferroni corrected α = 0.05/21 = 0.002)]. No such significant interaction of ABCA1 rs1883025 with PA was observed in women (Table 3). The other interactions examined in men and women did not reach statistical significance.
TABLE 2.
P for interactions between the 11 HDL-C-related SNPs identified by the GWA analysis and habitual PA on the serum HDL-C levels in men
| SNP | Gene | P-Interaction |
| Five SNPs identified by the current GWA analysis | ||
| rs651821 | APOA5 | 0.318 |
| rs1077834 | LIPC | 0.169 |
| rs1800588a | LIPC | 0.184 |
| rs56156922 | CETP | 0.996 |
| rs2303790a | CETP | 0.691 |
| Six SNPs identified by the replication analysis of previously-reported SNPs | ||
| rs12678919a | LPL | 0.721 |
| rs2293889a | TRPS1 | 0.370 |
| rs1883025a | ABCA1 | 0.0010 |
| rs964184a | APOA1 | 0.470 |
| rs1532085a | LIPC | 0.520 |
| rs737337a | ANGPTL8 | 0.735 |
ANGPTL8, angiopoietin-like 8; LIPG, endothelial lipase; TRPS1, transcriptional repressor GATA binding 1.
Directly genotyped.
TABLE 3.
P for interactions between the 10 HDL-C related SNPs identified by the GWA analysis and habitual PA on the serum HDL-C levels in women
| SNP | Gene | P-Interaction |
| Four SNPs identified by the current GWA analysis | ||
| rs662799 | APOA5 | 0.613 |
| rs1800588a | LIPC | 0.348 |
| rs56156922 | CETP | 0.311 |
| rs2303790a | CETP | 0.956 |
| Six SNPs identified by the replication analysis of previously-reported SNPs | ||
| rs12678919a | LPL | 0.348 |
| rs1883025a | ABCA1 | 0.796 |
| rs581080a | TTC39B | 0.672 |
| rs964184a | APOA1 | 0.874 |
| rs838880a | SCARB1 | 0.948 |
| rs1532085a | LIPC | 0.948 |
SCARB1, scavenger receptor class B member 1; TTC39B, tetratricopeptide repeat domain 39B.
Directly genotyped.
The genotype distribution of the ABCA1 rs1883025 in the Japanese population and correlations between the ABCA1 SNP rs1883025 and characteristic variables of the subjects
The genotype distribution of the ABCA1 SNP rs1883025 in the current subjects [men, TT: 8.0% (n = 385), CT: 40.3% (n = 1,948), CC: 51.7% (n = 2,497); women, TT: 8.5% (n = 469), CT: 40.7% (n = 2,254), CC: 50.8% (n = 2,817) was confirmed to be very similar to the data for the Japanese general population (n = 172) reported in the public database for dbSNP (HapMap-JPT, TT: 8.1%, CT: 40.7%, CC: 51.2%). Correlations between the ABCA1 SNP rs1883025 and characteristic variables are shown in Table 4. While the ABCA1 rs1883025 was not correlated with PA, it was significantly correlated with the serum HDL-C level in both men and women. It was confirmed that a higher number of C alleles (effect allele or major allele) of ABCA1 rs1883025 was associated with higher serum HDL-C levels.
TABLE 4.
Correlations between the characteristic variables and the ABCA1 SNP rs1883025, according to sex
| Characteristics | Men (n = 4,830) | Women (n = 5,540) | ||
| ρa | P | ρa | P | |
| Age (years) | −0.019 | 0.178 | −0.039 | 0.003 |
| Height (cm) | 0.006 | 0.676 | −0.004 | 0.791 |
| Weight (kg) | −0.0005 | 0.973 | −0.005 | 0.733 |
| BMI (kg/m2) | −0.005 | 0.720 | −0.002 | 0.866 |
| Smoking status [n (%)]b | −0.015 | 0.300 | −0.025 | 0.064 |
| Alcohol drinking [n (%)]c | −0.007 | 0.616 | 0.036 | 0.008 |
| PA (MET·h/day) | −0.008 | 0.590 | 0.018 | 0.183 |
| HDL-C (mg/dl) | 0.056 | <0.0001 | 0.096 | <0.0001 |
The ABCA1 SNP rs1883025 was coded as 0 (TT), 1 (CT), or 2 (CC).
Spearman’s rank correlation coefficient.
Never, former, and current smoker of 1–19 or 20+ cigarettes per day.
Never or former, and current drinker of 0.1–22.9, 23.0‒45.9, or 46.0+ g ethanol per day.
Stratified analyses according to different PA levels
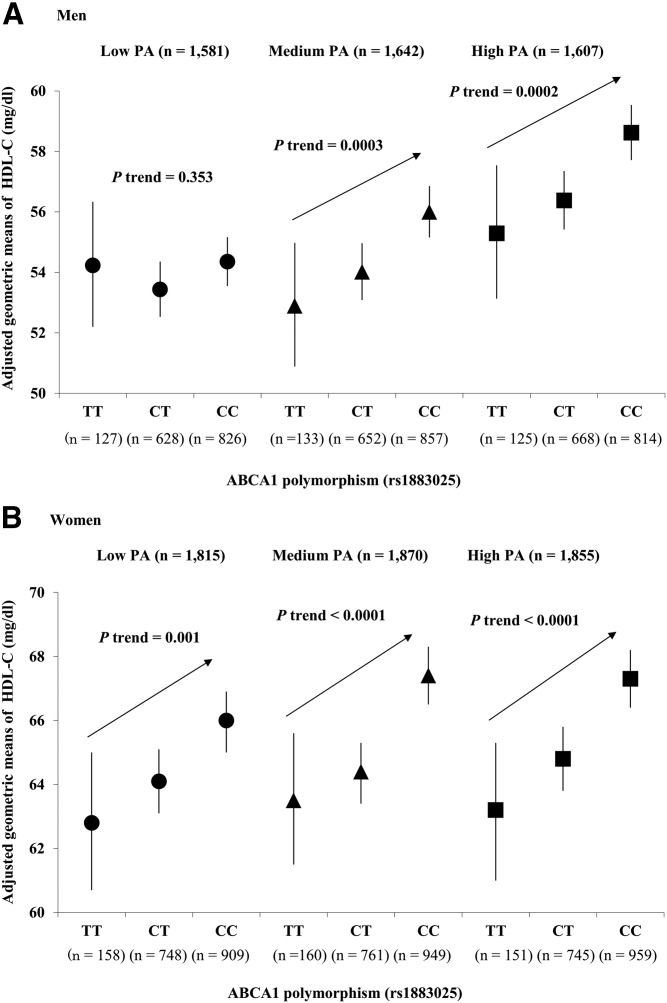
As for the ABCA1 rs1883025, the adjusted geometric means (milligrams per deciliter) of HDL-C by three genotypes (TT, CT, and CC) and regression coefficients (β) were analyzed according to the three different PA levels in men and women. The stratified analyses showed that the regression coefficients of the associations between the ABCA1 rs1883025 and natural log-transformed HDL-C were greater in men with medium PA [β = 0.032 (SE 0.009)] or high PA [β = 0.034 (0.009)] than in men with low PA [β = 0.008 (0.009)] (Fig. 1A). In contrast, the regression coefficients of these associations of the ABCA1 SNP and the log-transformed HDL-C were similar among the three different PA levels in women [low PA, β = 0.026 (SE 0.008); medium PA, β = 0.036 (0.008); high PA, β = 0.034 (0.008)] (Fig. 1B).
Fig. 1.
The associations between the ABCA1 SNP rs1883025 and serum HDL-C levels by the three different PA levels (low, medium, and high) in men (A) and women (B). Plots represent the adjusted means of the serum HDL-C levels and their 95% confidence intervals.
Replication of the interaction between ABCA1 SNP rs1883025 and PA on the serum HDL-C levels
Although the P value for the replication of interaction was marginal (P-interaction = 0.110), the stratified analyses (by three different PA levels) showed a similar pattern to the results obtained by the original interaction analysis in men, as the association between ABCA1 rs1883025 and log-transformed HDL-C levels was most attenuated in physically inactive men (β = 0.021) compared with men with medium (β = 0.026) or high PA (β = 0.043) (Table 5). In contrast, the positive associations between rs1883025 SNP and HDL-C were similar among female subjects with different PA levels (P-interaction = 0.638), which was also similar to the results obtained by the original interaction analysis in women (Table 5).
TABLE 5.
Replication of the interaction between ABCA1 polymorphism (rs1883025) and objectively measured PA on the serum HDL-C levels in subjects from the Saga region in the J-MICC study
| PAL Tertiles | βa | SE | P-Trend | P-Interaction |
| Men (n = 4,013) | 0.110 | |||
| Low PAL (n = 1,336) | 0.021 | 0.010 | 0.030 | — |
| Medium PAL (n = 1,334) | 0.026 | 0.009 | 0.005 | — |
| High PAL (n = 1,343 | 0.043 | 0.009 | <0.0001 | — |
| Women (n = 5,722) | 0.638 | |||
| Low PAL (n = 1,902) | 0.028 | 0.008 | 0.000 | — |
| Medium PAL (n = 1,906) | 0.024 | 0.007 | 0.001 | — |
| High PAL (n = 1,914) | 0.033 | 0.007 | <0.0001 | — |
β indicates regression coefficients of associations between the ABCA1 rs1883025 [coded as 0 (TT), 1 (CT), 2 (CC)] and natural log-transformed HDL-C.
DISCUSSION
In the current study of a Japanese middle-aged population, we found that the association of ABCA1 SNP rs1883025 with the circulating HDL-C levels was modified by habitual PA levels in men, in which the beneficial association of carrying a C allele of ABCA1 SNP rs1883025 with elevated serum HDL-C levels was masked in physically inactive men. The beneficial association of carrying a C allele with elevated HDL-C levels may be mediated by daily PA in middle-aged men. No such interaction of ABCA1 rs1883025 with PA (observed in men) was observed in women. In addition, replication of the interaction in another sample showed a similar result pattern in men, as the association between ABCA1 rs1883025 and objective habitual PA was most strongly attenuated in physically inactive men compared with men with medium or high PA, while no such interaction was observed in women.
Several HDL-C-related SNPs identified in a large-scale meta-analysis in 188,577 individuals (5) or in 22,939 women of European ancestry (10) were not observed in the current GWA analyses. Additionally, among three HDL-C-related SNPs showing significant interaction with PA in women of European ancestry (10), one SNP (rs1800588 in LIPC) was also identified by the current sex-stratified GWA analyses, but its interaction with PA did not reach statistical significance [P-interaction = 0.18 and 0.35 in men and women, respectively (Tables 2, 3)]. These differences from the previous largest-scale GWA studies may be partly explained by less statistical power due to the relatively small sample size of the current study, although our Japanese population was sufficiently large as a single ethnicity cohort. On the other hand, we found a novel interaction between ABCA1 rs1883025 and PA that was not detected in the previous GWA study whose participants were only women (10). The previous GWA study of women might have missed the interaction, likely because it was a male-specific interaction.
The ABCA1 SNP rs1883025 is a well-known genetic factor that can modulate circulating HDL-C levels (4). ABCA1 is an integral membrane protein that plays an important role in RCT (2). For instance, ABCA1 deficiency causes a rare disorder of Tangier disease that is associated with severely impaired RCT and a markedly low level of serum HDL-C (37). Therefore, ABCA1 is considered to be a central molecule for the biosynthesis of HDL-C (2, 37).
Previous studies have suggested the importance of ABCA1 gene induction in the mechanisms of the PA-induced increase in the serum HDL-C concentration. For instance, higher habitual PA was associated with a higher leukocyte ABCA1 mRNA expression, as well as a higher concentration of APOA-I and preβ1-HDL [the first (or immature) particle in the HDL-C formation in the RCT] in Caucasian men (38). Consistently, rodent studies showed that endurance exercise enhanced the ABCA1 gene expression in the liver and small intestine, which was accompanied by increased concentrations of APOA-I, preβ1-HDL, and HDL-C (39–41). In addition, while it is known that pro-inflammatory interferon-γ suppresses the expression of the ABCA1 gene (42), our recent study suggested that exercise-induced increases in HDL-C may be due in part to the reduction in the circulating interferon-γ concentration after exercise (43).
The magnitude of the association of HDL-C with PA differs among individuals. For example, in Japanese subjects, a low-intensity exercise regimen has been shown to be highly effective to increase serum HDL-C levels in older Japanese subjects (44), while a large inter-individual variability in the response of serum HDL-C to the same duration (per week) of low-intensity training was seen in this previous study. The current cross-sectional observational study indicates that the coincidence of two favorable factors (i.e., carrying the C allele of ABCA1 rs1883025 and active engagement with PA) may lead to higher serum HDL-C levels. Further exercise-intervention studies are needed to compare the extent of improvement in the serum HDL-C after longitudinal exercise training among Japanese men carrying different genotypes (TT, CT, CC) of the ABCA1 rs1883025.
To our knowledge, the functional molecular mechanisms underlying how ABCA1 SNP rs1883025 (located in intron 2) modulates the serum HDL-C concentration are unknown at present, despite this SNP being well-known as an HDL-C-related SNP. The biological mechanisms underlying the interaction between the ABCA1 SNP rs1883025 and habitual PA on HDL-C (that was observed only in men) is also currently unclear. Although the physiological and molecular genetic mechanisms by which the variants in the genes exert genotype-dependent differential effects on the improvement of lifestyle-related diseases through engagement with PA remain to be clarified (11), it is conceivable that there are two patterns of results regarding the gene-PA interaction studies. One is that PA attenuates or counteracts the associations of the allele with health outcomes (45, 46), while another is that PA amplifies or clarifies the associations of the allele with the outcomes (11). The current gene-PA interaction found in male subjects exhibits the second pattern, in which carrying the C allele of ABCA1 rs1883025 had only a negligible influence on the HDL-C level in inactive or sedentary men, whereas the beneficial association of carrying the C allele with an elevated HDL-C level is manifested when carrying the C allele is accompanied by habitual engagement with PA.
The current findings prompted us to generate a hypothesis regarding the molecular mechanism underlying the interaction of ABCA1 rs1883025 (located in intron 2) with PA. The expression of ABCA1 has been shown to be strongly regulated by nuclear LXR, which binds to the AGGTCA motif separated by four nucleotides in the ABCA1 promoter region (47, 48), while the motif (AGGTCA) separated by zero to six nucleotides may potentially work as the nuclear LXR element (49). The importance of intragenic intron sequences for the regulation of ABCA1 expression has also been suggested (50). We then realized that the nucleotide sequence around the ABCA1 SNP rs1883025 showed high homology [AGGTGT (CTA) AG G/A TCA] to the LXR biding site (G/A located at third nucleotide in the latter AGGTCA motif is rs1883025). Habitual PA (or exercise training) may upregulate the expression of LXR in white blood cells (51) and liver (52), as well as increase the oxysterol (natural ligand of LXR) levels in the arterial wall (53). We thus hypothesized that LXR protein activated by PA would bind more easily to the G allele [shown as C allele (complementary strand) in the current study] compared with the A allele (shown as T allele in the present study). Consequently, men carrying the G allele might have increased ABCA1 gene expression as well as increased serum HDL-C levels. This hypothesis is more plausible than our initial one, considering that estrogen increases the expression of nuclear LXR (54), because the association of the C allele with increased HDL-C levels was clearly seen in inactive women, who are presumed to have higher estrogen levels than inactive men in the lowest PA tertile.
The strengths of the current study are the large sample size and careful adjustment for potential confounding factors, including the BMI, alcohol consumption, and smoking habit in the interaction analysis. However, the present study also has several limitations. First, because this was a cross-sectional study, we were unable to make any causal inferences regarding the observed associations. Second, habitual PA was simply assessed by a self-administered questionnaire (not objective measurement) in the original interaction analysis. Third, although the concept that HDL function, rather than HDL-C concentration, is important for the anti-atherogenic qualities of HDL has been proposed (55–57), we did not measure the functional aspect of HDL and merely focused on the serum HDL-C levels in the current study. Finally, our subjects were limited to people of Japanese ancestry, and the current results may not be generalizable to other ethnic groups.
In conclusion, the current results suggest that the association of ABCA1 SNP rs1883025 with serum HDL-C was significantly modified by habitual PA in Japanese middle-aged men, with the beneficial association of carrying the C allele of ABCA1 rs1883025 being attenuated in inactive men compared with those with medium or high PA. It appears that middle-aged men carrying the C allele of ABCA1 rs1883025 may enjoy benefits with regard to improved serum HDL-C levels only when they are engaged in habitual PA. In contrast, the association of carrying the C allele of the ABCA1 SNP with elevated serum HDL-C levels was similar or not markedly different among different PA levels in women. The current evidence will contribute to the development of individualized PA programs to optimize the prevention and treatment of dyslipidemia or low HDL-C levels based on the individual genotypes of HDL-C-related SNPs.
Supplementary Material
Footnotes
Abbreviations:
- CETP
- cholesteryl ester transfer protein
- GRM
- genetic relationship matrix
- GWA
- genome-wide association
- J-MICC
- Japan Multi-Institutional Collaborative Cohort
- LIPC
- hepatic lipase
- MET
- metabolic equivalent
- PA
- physical activity
- PAL
- physical activity level
- RCT
- reverse cholesterol transport
This work was supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer (17015018) and Innovative Areas (221S0001) and by Japan Society for the Promotion of Science KAKENHI Grants JP15H02524 and JP16H06277 from the Japanese Ministry of Education, Culture, Sports, Science and Technology. This study was also supported in part by funding for the BioBank Japan Project from the Japan Agency for Medical Research and Development since April 2015 and the Ministry of Education, Culture, Sports, Science and Technology from April 2003 to March 2015.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bruckert E., and Hansel B.. 2007. HDL-c is a powerful lipid predictor of cardiovascular diseases. Int. J. Clin. Pract. 61: 1905–1913. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi R., Mu H., Wang X., Yao Q., and Chen C.. 2005. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. QJM. 98: 845–856. [DOI] [PubMed] [Google Scholar]
- 3.Cuchel M., and Rader D. J.. 2006. Macrophage reverse cholesterol transport: key to the regression of atherosclerosis? Circulation. 113: 2548–2555. [DOI] [PubMed] [Google Scholar]
- 4.Weissglas-Volkov D., and Pajukanta P.. 2010. Genetic causes of high and low serum HDL-cholesterol. J. Lipid Res. 51: 2032–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spracklen C. N., Chen P., Kim Y. J., Wang X., Cai H., Li S., Long J., Wu Y., Wang Y. X., Takeuchi F., et al. 2017. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 26: 1770–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monda K. L., Ballantyne C. M., and North K. E.. 2009. Longitudinal impact of physical activity on lipid profiles in middle-aged adults: the Atherosclerosis Risk in Communities Study. J. Lipid Res. 50: 1685–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katcher H. I., Hill A. M., Lanford J. L., Yoo J. S., and Kris-Etherton P. M.. 2009. Lifestyle approaches and dietary strategies to lower LDL-cholesterol and triglycerides and raise HDL-cholesterol. Endocrinol. Metab. Clin. North Am. 38: 45–78. [DOI] [PubMed] [Google Scholar]
- 9.Chelland Campbell S., Moffatt R. J., and Stamford B. A.. 2008. Smoking and smoking cessation–the relationship between cardiovascular disease and lipoprotein metabolism: a review. Atherosclerosis. 201: 225–235. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad T., Chasman D. I., Buring J. E., Lee I. M., Ridker P. M., and Everett B. M.. 2011. Physical activity modifies the effect of LPL, LIPC, and CETP polymorphisms on HDL-C levels and the risk of myocardial infarction in women of European ancestry. Circ Cardiovasc Genet. 4: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori M., Higuchi K., Sakurai A., Tabara Y., Miki T., and Nose H.. 2009. Genetic basis of inter-individual variability in the effects of exercise on the alleviation of lifestyle-related diseases. J. Physiol. 587: 5577–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamajima N., and Group J. M. S.. 2007. The Japan Multi-Institutional Collaborative Cohort study (J-MICC study) to detect gene-environment interactions for cancer. Asian Pac. J. Cancer Prev. 8: 317–323. [PubMed] [Google Scholar]
- 13.Wakai K., Hamajima N., Okada R., Naito M., Morita E., Hishida A., Kawai S., Nishio K., Yin G., Asai Y., et al. 2011. Profile of participants and genotype distributions of 108 polymorphisms in a cross-sectional study of associations of genotypes with lifestyle and clinical factors: a project in the Japan Multi-Institutional Collaborative Cohort (J-MICC) study. J. Epidemiol. 21: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., and Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 16.Patterson N., Price A. L., and Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.1000 Genomes Project Consortium, Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., and McVean G. A.. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi-Kabata Y., Nakazono K., Takahashi A., Saito S., Hosono N., Kubo M., Nakamura Y., and Kamatani N.. 2008. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am. J. Hum. Genet. 83: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa-Senda H., Hachiya T., Shimizu A., Hosono S., Oze I., Watanabe M., Matsuo K., Ito H., Hara M., Nishida Y., et al. 2018. A genome-wide association study in the Japanese population identifies the 12q24 locus for habitual coffee consumption: the J-MICC Study. Sci. Rep. 8: 1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimanoe C., Hachiya T., Hara M., Nishida Y., Tanaka K., Sutoh Y., Shimizu A., Hishida A., Kawai S., Okada R., et al. 2019. A genome-wide association study of coping behaviors suggests FBXO45 is associated with emotional expression. Genes Brain Behav. 18: e12481. [DOI] [PubMed] [Google Scholar]
- 21.Hara M., Hachiya T., Sutoh Y., Matsuo K., Nishida Y., Shimanoe C., Tanaka K., Shimizu A., Ohnaka K., Kawaguchi T., et al. 2018. Genome-wide association study of leisure-time exercise behavior in Japanese adults. Med. Sci. Sports Exerc. 50: 2433–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaneau O., Marchini J., and Zagury J. F.. 2011. A linear complexity phasing method for thousands of genomes. Nat. Methods. 9: 179–181. [DOI] [PubMed] [Google Scholar]
- 23.Fuchsberger C., Abecasis G. R., and Hinds D. A.. 2015. minimac2: faster genotype imputation. Bioinformatics. 31: 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hachiya T., Komaki S., Hasegawa Y., Ohmomo H., Tanno K., Hozawa A., Tamiya G., Yamamoto M., Ogasawara K., Nakamura M., et al. 2017. Genome-wide meta-analysis in Japanese populations identifies novel variants at the TMC6-TMC8 and SIX3-SIX2 loci associated with HbA1c. Sci. Rep. 7: 16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., Nyholt D. R., Madden P. A., Heath A. C., Martin N. G., Montgomery G. W., et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanoni P., Khetarpal S. A., Larach D. B., Hancock-Cerutti W. F., Millar J. S., Cuchel M., DerOhannessian S., Kontush A., Surendran P., Saleheen D., et al. 2016. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 351: 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., Lee S. H., Goddard M. E., and Visscher P. M.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higashibata T., Hamajima N., Naito M., Kawai S., Yin G., Suzuki S., Kita Y., Niimura H., Imaizumi T., Ohnaka K., et al. 2012. eNOS genotype modifies the effect of leisure-time physical activity on serum triglyceride levels in a Japanese population. Lipids Health Dis. 11: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ainsworth B. E., Haskell W. L., Whitt M. C., Irwin M. L., Swartz A. M., Strath S. J., O’Brien W. L., Bassett D. R. Jr., Schmitz K. H., Emplaincourt P. O., et al. 2000. Compendium of physical activities: an update of activity codes and MET intensities. Med. Sci. Sports Exerc. 32: S498–S504. [DOI] [PubMed] [Google Scholar]
- 30.Tokudome S., Goto C., Imaeda N., Tokudome Y., Ikeda M., and Maki S.. 2004. Development of a data-based short food frequency questionnaire for assessing nutrient intake by middle-aged Japanese. Asian Pac. J. Cancer Prev. 5: 40–43. [PubMed] [Google Scholar]
- 31.Tokudome Y., Goto C., Imaeda N., Hasegawa T., Kato R., Hirose K., Tajima K., and Tokudome S.. 2005. Relative validity of a short food frequency questionnaire for assessing nutrient intake versus three-day weighed diet records in middle-aged Japanese. J. Epidemiol. 15: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imaeda N., Goto C., Tokudome Y., Hirose K., Tajima K., and Tokudome S.. 2007. Reproducibility of a short food frequency questionnaire for Japanese general population. J. Epidemiol. 17: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordestgaard B. G., Langsted A., Mora S., Kolovou G., Baum H., Bruckert E., Watts G. F., Sypniewska G., Wiklund O., Boren J., et al. ; European Atherosclerosis Society (EAS) and the European Federation of Clinical Chemistry and Laboratory Medicine (EFLM) Joint Consensus Initiative . 2016. Fasting is not routinely required for determination of a lipid profile: clinical and laboratory implications including flagging at desirable concentration cut-points-a joint consensus statement from the European Atherosclerosis Society and European Federation of Clinical Chemistry and Laboratory Medicine. Eur. Heart J. 37: 1944–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marott S. C., Nordestgaard B. G., Tybjaerg-Hansen A., and Benn M.. 2016. Components of the metabolic syndrome and risk of type 2 diabetes. J. Clin. Endocrinol. Metab. 101: 3212–3221. [DOI] [PubMed] [Google Scholar]
- 35.Machiela M. J., and Chanock S. J.. 2015. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 31: 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida Y., Higaki Y., Taguchi N., Hara M., Nakamura K., Nanri H., Imaizumi T., Sakamoto T., Horita M., Shinchi K., et al. 2014. Objectively measured physical activity and inflammatory cytokine levels in middle-aged Japanese people. Prev. Med. 64: 81–87. [DOI] [PubMed] [Google Scholar]
- 37.Clee S. M., Kastelein J. J., van Dam M., Marcil M., Roomp K., Zwarts K. Y., Collins J. A., Roelants R., Tamasawa N., Stulc T., et al. 2000. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Invest. 106: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoang A., Tefft C., Duffy S. J., Formosa M., Henstridge D. C., Kingwell B. A., and Sviridov D.. 2008. ABCA1 expression in humans is associated with physical activity and alcohol consumption. Atherosclerosis. 197: 197–203. [DOI] [PubMed] [Google Scholar]
- 39.Ghanbari-Niaki A., Ghanbari-Abarghooi S., Rahbarizadeh F., Zare-Kookandeh N., Gholizadeh M., Roudbari F., and Zare-Kookandeh A.. 2013. Heart ABCA1 and PPAR-alpha genes expression responses in male rats: effects of high intensity treadmill running training and aqueous extraction of black Crataegus-pentaegyna. Res. Cardiovasc. Med. 2: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghanbari-Niaki A., Khabazian B. M., Hossaini-Kakhak S. A., Rahbarizadeh F., and Hedayati M.. 2007. Treadmill exercise enhances ABCA1 expression in rat liver. Biochem. Biophys. Res. Commun. 361: 841–846. [DOI] [PubMed] [Google Scholar]
- 41.Khabazian B. M., Ghanbari-Niaki A., Safarzadeh-Golpordesari A., Ebrahimi M., Rahbarizadeh F., and Abednazari H.. 2009. Endurance training enhances ABCA1 expression in rat small intestine. Eur. J. Appl. Physiol. 107: 351–358. [DOI] [PubMed] [Google Scholar]
- 42.Harvey E. J., and Ramji D. P.. 2005. Interferon-gamma and atherosclerosis: pro- or anti-atherogenic? Cardiovasc. Res. 67: 11–20. [DOI] [PubMed] [Google Scholar]
- 43.Nishida Y., Tanaka K., Hara M., Hirao N., Tanaka H., Tobina T., Ikeda M., Yamato H., and Ohta M.. 2015. Effects of home-based bench step exercise on inflammatory cytokines and lipid profiles in elderly Japanese females: A randomized controlled trial. Arch. Gerontol. Geriatr. 61: 443–451. [DOI] [PubMed] [Google Scholar]
- 44.Sunami Y., Motoyama M., Kinoshita F., Mizooka Y., Sueta K., Matsunaga A., Sasaki J., Tanaka H., and Shindo M.. 1999. Effects of low-intensity aerobic training on the high-density lipoprotein cholesterol concentration in healthy elderly subjects. Metabolism. 48: 984–988. [DOI] [PubMed] [Google Scholar]
- 45.Li S., Zhao J. H., Luan J., Ekelund U., Luben R. N., Khaw K. T., Wareham N. J., and Loos R. J.. 2010. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 7: e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boer J. M., Kuivenhoven J. A., Feskens E. J., Schouten E. G., Havekes L. M., Seidell J. C., Kastelein J. J., and Kromhout D.. 1999. Physical activity modulates the effect of a lipoprotein lipase mutation (D9N) on plasma lipids and lipoproteins. Clin. Genet. 56: 158–163. [DOI] [PubMed] [Google Scholar]
- 47.Costet P., Luo Y., Wang N., and Tall A. R.. 2000. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275: 28240–28245. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz K., Lawn R. M., and Wade D. P.. 2000. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem. Biophys. Res. Commun. 274: 794–802. [DOI] [PubMed] [Google Scholar]
- 49.Martin L. J., and Tremblay J. J.. 2010. Nuclear receptors in Leydig cell gene expression and function. Biol. Reprod. 83: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singaraja R. R., Bocher V., James E. R., Clee S. M., Zhang L. H., Leavitt B. R., Tan B., Brooks-Wilson A., Kwok A., Bissada N., et al. 2001. Human ABCA1 BAC transgenic mice show increased high density lipoprotein cholesterol and ApoAI-dependent efflux stimulated by an internal promoter containing liver X receptor response elements in intron 1. J. Biol. Chem. 276: 33969–33979. [DOI] [PubMed] [Google Scholar]
- 51.Butcher L. R., Thomas A., Backx K., Roberts A., Webb R., and Morris K.. 2008. Low-intensity exercise exerts beneficial effects on plasma lipids via PPARgamma. Med. Sci. Sports Exerc. 40: 1263–1270. [DOI] [PubMed] [Google Scholar]
- 52.Rahmati-Ahmadabad S., Shirvani H., Ghanbari-Niaki A., and Rostamkhani F.. 2018. The effects of high-intensity interval training on reverse cholesterol transport elements: A way of cardiovascular protection against atherosclerosis. Life Sci. 209: 377–382. [DOI] [PubMed] [Google Scholar]
- 53.Ferreira G. S., Pinto P. R., Iborra R. T., Del Bianco V., Santana M. F. M., Nakandakare E. R., Nunes V. S., Negrao C. E., Catanozi S., and Passarelli M.. 2017. Aerobic exercise training selectively changes oxysterol levels and metabolism reducing cholesterol accumulation in the aorta of dyslipidemic mice. Front. Physiol. 8: 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H., Liu Y., Zhu L., Wang W., Wan Z., Chen F., Wu Y., Zhou J., and Yuan Z.. 2014. 17β-Estradiol promotes cholesterol efflux from vascular smooth muscle cells through a liver X receptor alpha-dependent pathway. Int. J. Mol. Med. 33: 550–558. [DOI] [PubMed] [Google Scholar]
- 55.Iqbal F., Baker W. S., Khan M. I., Thukuntla S., McKinney K. H., Abate N., and Tuvdendorj D.. 2017. Current and future therapies for addressing the effects of inflammation on HDL cholesterol metabolism. Br. J. Pharmacol. 174: 3986–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asztalos B. F., Tani M., and Schaefer E. J.. 2011. Metabolic and functional relevance of HDL subspecies. Curr. Opin. Lipidol. 22: 176–185. [DOI] [PubMed] [Google Scholar]
- 57.Kraus W. E., Houmard J. A., Duscha B. D., Knetzger K. J., Wharton M. B., McCartney J. S., Bales C. W., Henes S., Samsa G. P., Otvos J. D., et al. 2002. Effects of the amount and intensity of exercise on plasma lipoproteins. N. Engl. J. Med. 347: 1483–1492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.