Abstract
The gut microbiota is a unique ecosystem of microorganisms interacting with the host through several biochemical mechanisms. The endocannabinoidome (eCBome), a complex signaling system including the endocannabinoid system, approximately 50 receptors and metabolic enzymes, and more than 20 lipid mediators with important physiopathologic functions, modulates gastrointestinal tract function and may mediate host cell-microbe communications there. Germ-free (GF) mice, which lack an intestinal microbiome and so differ drastically from conventionally raised (CR) mice, offer a unique opportunity to explore the eCBome in a microbe-free model and in the presence of a reintroduced functional gut microbiome through fecal microbiota transplant (FMT). We aimed to gain direct evidence for a link between the microbiome and eCBome systems by investigating eCBome alterations in the gut in GF mice before and after FMT. Basal eCBome gene expression and lipid profiles were measured in various segments of the intestine of GF and CR mice at juvenile and adult ages using targeted quantitative PCR transcriptomics and LC-MS/MS lipidomics. GF mice exhibited age-dependent modifications in intestinal eCBome gene expression and lipid mediator levels. FMT from CR donor mice to age-matched GF male mice reversed several of these alterations, particularly in the ileum and jejunum, after only 1 week, demonstrating that the gut microbiome directly impacts the host eCBome and providing a cause-effect relationship between the presence or absence of intestinal microbes and eCBome signaling. These results open the way to new studies investigating the mechanisms through which intestinal microorganisms exploit eCBome signaling to exert some of their physiopathologic functions.
Keywords: endocannabinoids, gut microbiome, germ-free phenotype, intestine, fecal microbiota transplant, gene expression, lipidomics.
The gut microbiome represents an array of different microorganisms that unevenly colonize the surfaces of the gastrointestinal tract. Considering its active biomass and metabolic function, it can be considered as a true organ in the body (1), exerting a profound impact on adult and early human health and playing a potential role in specific diseases (2). Intestinal bacteria have specific functions in host nutrient, xenobiotic, and drug metabolism, and the maintenance of the structural integrity and immune-competence of the gut mucosal barrier (3). They influence several other organ systems, including the central nervous system, adipose tissue, and the liver (4–6). Individual populations of microbiota exist within characteristically distinct regions along the length of the gastrointestinal tract, with the colon having the greatest proportion (7, 8). These individual populations can be affected by host and host-related characteristics, such as gender, age, and genetics, as well as the diet and other environmental factors (9–11).
Germ-free (GF) mice provide a useful experimental tool to understand microbiota-host interactions and dissect their molecular mechanism. These mice have a characteristic phenotype, showing several developmental and physiological differences with respect to conventionally raised (CR) mice (12). At the gastrointestinal level, they display alterations in gut morphology, motility, absorption, secretion, and immune function. The cecum of GF mice is enlarged by 4- to 8-fold because of mucus and undigested fiber accumulation, and the small intestine is less developed (12) with reduced epithelial cell turnover and irregular microvilli (13) along with decreased gut motility, which is partly due to decreased innervation with sensory neurons (14). Despite the absence of microbially produced short chain fatty acids, GF mice were also demonstrated to have increased levels of colon-derived and circulating glucagon-like peptide-1 (GLP-1) that did not improve the incretin response but instead slowed small intestinal transit, potentially as an adaptive response, promoting nutrient absorption in the face of insufficient energy availability (15). GF mice also exhibit decreased or absent expression of several Toll-like receptors, essential for the recognition of pathogen-associated molecular patterns (16), poorly formed Peyer’s patches, and alterations of the composition of CD4+ T cells and IgA-producing B cells in the lamina propria, with impaired development and maturation of isolated lymphoid follicles (17).
Some perturbations of the gut microbiota, generally defined as “dysbiosis,” may cause a decrease of intestinal barrier integrity, and the subsequent entry into the bloodstream of bacterial constituents such as lipopolysaccharides from gram-negative bacteria, which trigger the onset of low-grade inflammation contributing to, among others, insulin resistance (18). It has been suggested that this pathological condition occurs via the overactivation of the cannabinoid receptor type-1 (CB1) (19), thus highlighting the potential relationship between microbiota dysbiosis and the endocannabinoid (eCB) system. This is a pleiotropic signaling system composed of CB1 and cannabinoid receptor type-2 (CB2), their two lipid ligands, the eCBs N-arachidonoylethanolamine (AEA, anandamide) and 2-arachidonoylglycerol (2-AG), and eCB anabolic and catabolic enzymes. These enzymes include, among others, N-acylphosphatidylethanolamine-hydrolyzing phospholipase D (NAPE-PLD) and fatty acid amide hydrolase (FAAH), respectively, for AEA, and diacylglycerol lipase (DAGL)α, DAGLβ, and monoacylglycerol lipase, respectively, for 2-AG (20). It is well-established that AEA and 2-AG are often accompanied in tissues by their congeners, the N-acylethanolamines (NAEs) and 2-acylglycerols, as well as by other amides of long chain fatty acids with amino acids or neurotransmitters (21). Both eCB congeners, which share the same anabolic and catabolic enzymes with the eCBs, and other eCB-related amides, which occasionally can be inactivated by FAAH, interact with receptors other than CB1 and CB2, including some transient receptor potential channels, PPARs, and some orphan G protein-coupled receptors, which are also activated by other non-endocannabinoidome (eCBome) ligands. Oxidation of AEA and 2-AG and some of their arachidonic acid (AA)-containing congeners by AA-metabolizing enzymes also leads to yet another series of emerging bioactive lipids (21). This wide range of lipid mediators, receptors, and enzymes, including and extending the eCB system, has been defined as the eCBome (22, 23).
The eCBome modulates several physiological functions, including gastrointestinal tract function, energy homeostasis, and metabolism (24). Tissue and circulating levels of eCBome molecules are altered by dietary factors (particularly, in view of their chemical nature, by the fatty acid composition of the diet) as well as by gut microbiota. For example, a high fat diet alters gut microbiota, which control intestinal eCBome tone by altering Napepld, Cnr1, and Faah expression and AEA levels resulting in increased intestinal permeability and decreased adipogenesis in obese mice (19). A recent study showed that chronic treatment with antibiotics, leading to dysbiosis without erasing the gut microbiota population, can alter the levels of NAEs and N-acyl-serotonins in the small intestine (25). On the other hand, Napepld deletion in white adipocytes or intestinal epithelial cells induces dysbiosis and related dysmetabolism, possibly also due to the reduction of the local tissue concentrations of some NAEs and subsequently impaired signaling of non-cannabinoid receptors (26, 27).
So far, however, no study has been conducted to define how the eCBome is altered in the increasingly used GF mouse model, which lacks an intestinal microbiome and offers the unique possibility, through fecal microbiota transplant (FMT), to look at the effects on animal physiological pathways of reintroducing, in part or completely, a functional gut microbiome. In the present study, we therefore assessed the basal eCBome gene profiles within GF and CR male mice at juvenile and adult ages, and we correlated these results with eCBome lipid mediator levels. We also assessed the effect of a FMT on the potentially altered eCBome. We focused on the small and large intestine where the largest microbial populations reside in mammals, and the most profound alterations due to their absence are likely to occur.
MATERIALS AND METHODS
Animals and housing
CR and GF C57BL/6NTac mice were purchased from Taconic (Taconic Bioscience) and maintained in the animal facility of the Institut Universitaire de Cardiologie et Pneumologie de Québec (IUCPQ; Québec, Canada). All animals were grouped with three to four mice per cage under a 12:12 h light-dark cycle with ad libitum access to NIH-31 Open Formula Autoclavable Diet (Zeigler) and water. GF mice were housed in axenic status and fecal samples were tested weekly for microbes and parasites by the facility’s staff to ensure that the GF unit was indeed sterile. Both GF and CR mice were acclimatized for at least 1 week prior to the start of the procedures.
In order to confirm gene expression data, we also used mice housed within a different animal facility.
C57BL/6J 8-week-old male mice were purchased from Charles River Laboratories (France) and were housed in a specific and opportunistic pathogen-free animal facility with controlled temperature and humidity and a 12 h light-dark cycle (6:00 AM light/6:00 PM dark). The mice had free access to water and food (A04, Villemoisson sur Orge, France). Swiss Webster CR mice or GF male mice (7 weeks old) from Taconic were also used in the transplantation study.
Animal experiments and FMT
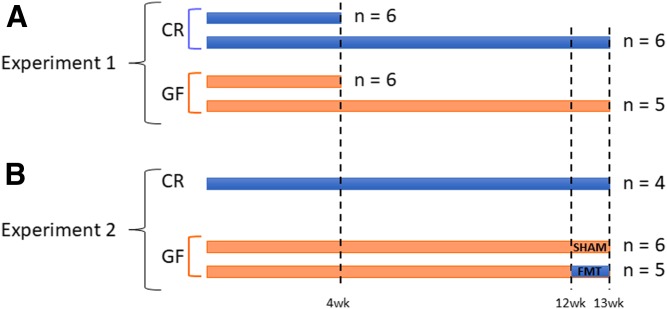
Six male CR and GF mice at 4 and 13 weeks of age were intraperitoneally anesthetized with a cocktail of ketamine/xylazine/acepromazine at a dose of 50/10/1.7 mg/kg body weight and euthanized by intra-cardiac puncture. Whole blood was collected in K3-EDTA tubes. The abdominal cavity was opened and the whole digestive tract was carefully aligned from the stomach up to the colon. Once the stomach was removed, the small intestine (duodenum, jejunum, ileum) and large intestine (cecum and colon) were carefully excised and separated, and the intestinal contents were harvested by flushing with 1 ml of sterile PBS without Ca/Mg (Thermo Fisher Scientific) and snap-frozen. Sections of small and large intestine were both stored either in RNALater (Thermo Fisher Scientific) for RNA stabilization or immediately snap-frozen and stored at −80°C for further analysis. This common procedure was concluded for each mouse within a maximum of 15 min, a time frame that allowed for the preservation of mRNA and lipid for further analysis. All the experimental protocols were validated and approved by Laval University animal ethics committee (CPAUL 2018010-1).
For the FMT experiments, GF mice were randomly divided into two groups at the age of 12 weeks; those gavaged with sterile PBS (SHAM; five mice) and those gavaged with fecal microbiota (FMT; six mice). Material gavaged for FMT consisted of the intestinal contents and stools of a single mouse and four CR donor mice, respectively. Briefly, intestinal contents of the duodenum, jejunum, ileum, colon, and cecum were collected from one 12-week-old CR donor mouse and were mixed with stool pellets from all the CR mice to be used as controls. The mixture was well homogenized, weighed, suspended 1:10 in sterile PBS, and centrifuged at 805 g for 10 min at room temperature. The supernatant was used to gavage the mice (200 μl of homogenate per mouse). The FMT mice were then housed (three per cage) for 1 week in conventional conditions in cages contaminated with used litter coming from the cages of donor mice. CR mice were euthanized the day of the gavages, while SHAM and FMT mice were euthanized 1 week after the gavage; plasma, tissues, and intestinal contents were collected from all the animals as previously described (for an outline of the experimental protocol, see Fig. 1).
Fig. 1.
Experimental protocols. A: Six male CR and GF mice were euthanized at 4 and 13 weeks of age. B: For FMT, 12-week-old GF mice were gavaged with either intestinal contents and stools coming from one and four donor mice, respectively (FMT group; n = 6), or sterile PBS (SHAM group; n = 5). For both experiments, sections of duodenum, jejunum, ileum, cecum, and colon were isolated, and the intestinal contents were harvested.
The mice utilized for Fig. 2E and F were anesthetized using intraperitoneal ketamine and xylazine at concentrations of 100 and 10 mg/kg, respectively, after a 5 h fasting period. Tissues were harvested for further analysis. Mice were euthanized by cervical dislocation. The intestinal segments (jejunum, ileum, and colon) were precisely dissected, immersed in liquid nitrogen, and stored at −80°C for further analysis.
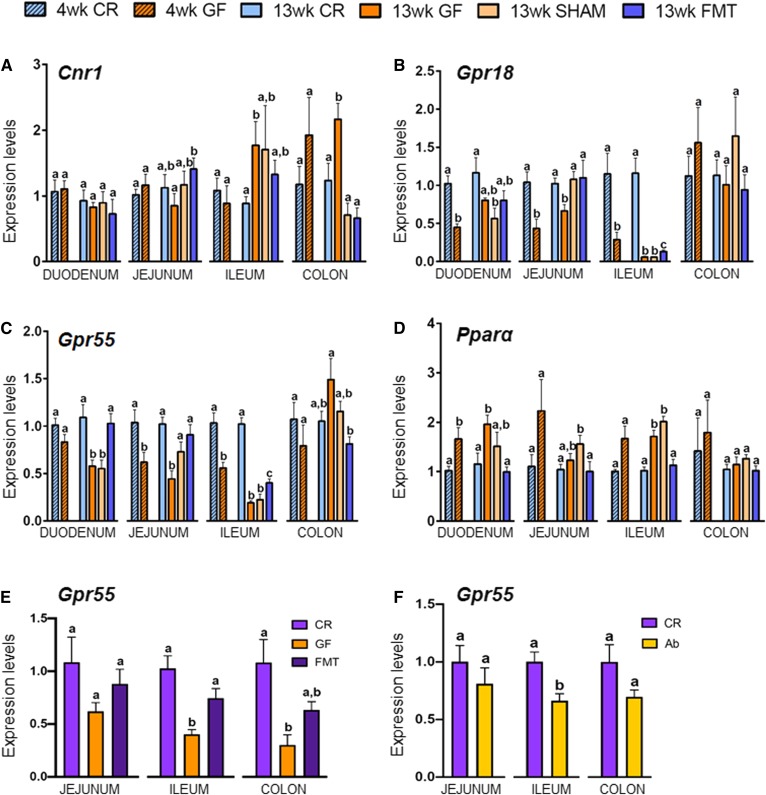
Fig. 2.
Intestinal expression levels of the eCBome receptors Cnr1 (A), Gpr18 (B), Gpr55 (C), and Pparα (D) in 4- and 13-week-old CR and GF mice and in 13-week-old male GF mice gavaged with sterile PBS (SHAM) or fecal microbiota (FMT). The mRNA expression levels were measured by qPCR array in duodenum, jejunum, ileum, and colon for all experimental groups and are expressed relative to CR mice for each age within each tissue. Values are mean ± SEM of n = 5–10 mice. Bars marked with different letters were significantly different at the P ≤ 0.05 level. E, F: Gpr55 gene expression in 7-week-old CR, GF, and FMT Swiss Webster mice (E) and control (CR) or antibiotic-treated (Ab) C57BL/6J mice (F).
Swiss Webster GF mice were conventionalized with the gut microbes (i.e., cecal contents) from male Swiss Webster CR mice and maintained in individualized ventilated cages (IVC AERO GM500, Tecnilab-BMI).
Antibiotic treatment
C57BL/6J mice were treated with ampicillin (1 g/l; Sigma) and neomycin (0.5 g/l; Sigma) in their drinking water for 2 weeks (19).
RNA isolation, reverse transcription, and qPCR-based TaqMan open array
RNA was extracted from the duodenum, jejunum, ileum, and colon samples with the RNeasy Plus mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions and eluted in 50 μl of UltraPure distilled water (Invitrogen). The concentration and purity of RNA was determined by measuring the absorbance of the RNA in a Biodrop at 260 nm and 280 nm, and RNA integrity was assessed by an Agilent 2100 bioanalyzer, using the Agilent RNA 6000 Nano kit (Agilent Technologies). One microgram of total RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) in a reaction volume of 20 μl. Sixty nanograms of starting RNA were used to evaluate the expression of the 52 eCBome-related genes and four housekeeping genes (supplemental Table S1) using a custom-designed quantitative (q)PCR-based TaqMan open array on a QuantStudio 12K Flex real-time PCR system (Thermo Fisher Scientific) following the manufacturer’s instructions. The mRNA expression levels of 14 eCBome genes were validated using the same cDNA and specific forward and reverse primer pairs (IDT) (supplemental Table S2) on a CFX384 Touch qPCR system (Bio-Rad) using PowerUp SYBR Green qPCR master mix (Thermo Fisher Scientific) in duplicate reactions. Hprt1 and Tbp were used as reference genes for all qPCR assays. Based on the fact that the threshold cycles (Ct) of the reference genes were the same in all of the 13-week-old control samples (first and FMT experiments), we merged the two control sets into one 13 week control group. Gene expression levels were evaluated by the 2−ΔΔCt method and represented as fold change with respect to baseline within each tissue section for each age. Target genes were, however, differentially expressed in the different intestinal segments, as shown by differences in Ct (summarized in supplemental Table S3).
The intestinal gene expression levels of Gpr55 were also assessed in another institute as follows: total RNA was prepared from tissues using TriPure reagent (Roche); cDNA was synthesized from 1 μg of total RNA using a reverse transcription kit (Promega Corp.); and qPCR was performed by using a StepOnePlus instrument and software (Applied Biosystems) according to the manufacturer’s instructions. Rpl19 RNA was chosen as the housekeeping gene, and data were analyzed according to the 2−ΔΔCt method. The identity and purity of the amplified product were assessed by melting curve analysis at the end of amplification.
Lipid extraction and HPLC-MS/MS for the analysis of eCBome mediators
Lipids were extracted from tissue samples according to the Bligh and Dyer method (28). Briefly, about 10 mg of each tissue were homogenized in 1 ml of Tris-HCl 50 mM (pH 7) and methanol (1/1) using a tissue homogenizer; 200 μl of the mixture were added to 800 μl (1:5 dilution) of the same Tris-HCl/methanol solution containing 0.1 M acetic acid and 5 ng of deuterated standards. One milliliter of chloroform was then added to each sample, vortexed for 30 s, and centrifuged at 3,000 g for 5 min. This was repeated twice for a total addition of 3 ml of chloroform. The organic phases were collected and evaporated under a stream of nitrogen and then suspended in 50 μl of mobile phase containing 50% of solvent A (water + 1 mM ammonium acetate + 0.05% acetic acid) and 50% of solvent B [acetonitrile/water (95/5) + 1 mM ammonium acetate + 0.05% acetic acid]. Forty microliters of each sample were finally injected onto an HPLC column (Kinetex C8, 150 × 2.1 mm, 2.6 μm; Phenomenex) and eluted at a flow rate of 400 μl/min using a discontinuous gradient of solvent A and solvent B (27). Quantification of eCBome-related mediators (supplemental Table S4) was carried out by HPLC system interfaced with the electrospray source of a Shimadzu 8050 triple quadrupole mass spectrometer and using multiple reaction monitoring in positive ion mode for the compounds and their deuterated homologs.
The 13-week-old control results were assembled into a single control group. In the case of unsaturated monoacylglycerols, the data are presented as 2-monoacylglycercols but represent the combined signals from the 2- and 1(3)-isomers because the latter are most likely generated from the former via acyl migration from the 2- to the sn-1 or sn-3 position.
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from intestinal contents using the QIAmp PowerFecal DNA kit (Qiagen) according to the manufacturer’s instructions. The DNA concentrations of the extracts were measured fluorometrically with the Quant-iT PicoGreen dsDNA kit (Thermo Fisher Scientific), and the DNAs were stored at −20°C until 16S rDNA library preparation. Briefly, 1 ng of DNA was used as template, and the V3-V4 region of the 16S rRNA gene was amplified by PCR using the QIAseq 16S Region Panel protocol in conjunction with the QIAseq 16S/ITS 384-Index I (Sets A, B, C, D) kit (Qiagen). The 16S metagenomic libraries were eluted in 30 μl of nuclease-free water and 1 μl was qualified with a Bioanalyser DNA 1000 Chip (Agilent) to verify the amplicon size (expected size ∼600 bp) and quantified with a Qubit (Thermo Fisher Scientific). Libraries were then normalized and pooled to 2 nM, denatured, and diluted to a final concentration of 6 pM and supplemented with 5% PhiX control (Illumina). Sequencing (2 × 300 bp paired-end) was performed using the MiSeq Reagent Kit V3 (600 cycles) on an Illumina MiSeq system. Sequencing reads were generated in less than 65 h. Image analysis and base calling were carried out directly on the MiSeq. Data were processed using the DADA2 pipeline (29) and microbiota composition assessed by calculating α- and β-diversity indexes and intra- and inter-individual variations in microbial composition using PERMANOVA (vegan R package) (30). Reads were rarefied to 8,000 reads to account for depth bias for total FMT pool, colon CR, colon FMT, cecum CR, and cecum FMT. Total read counts were used for ileum CR (8,088), ileum FMT (4,072), jejunum CR (5,164), and jejunum FMT (716). Statistical analysis was only performed on colon and cecum, as read counts for ileum and jejunum were not sufficiently high to carry out this analysis.
Statistical analysis
Data are expressed as the mean ± SEM, as specified in the individual tables and figures.
For gene expression analysis, the differences between the CR and GF of 4- and 13-week-old mice were assessed separately and each intestinal tract was analyzed independently. Particularly, the differences between the CR and GF of 4-week-old male mice and the differences between CR and Ab mice were assessed using an unpaired Mann-Whitney t-test on ddCt, while for the 13-week-old mice and the Swiss Webster mice, one-way ANOVA followed by Fisher’s least significant difference test was used. With regard to eCBome mediator levels (picomoles per milligram), superscript letters represent the differences, in each intestinal tract, between CR and GF 4-week-old mice as determined by an unpaired Mann-Whitney t-test and between CR, GF, SHAM, and FMT 13-week-old mice as determined by a one-way ANOVA followed by Fisher’s least significant difference test. The changes of the CR mediator levels between the different intestinal segments at both 4 weeks and 13 weeks were also assessed using unpaired Mann-Whitney t-test. For the 4- and 13-week-old analysis, CR and Ab mouse experiments, and Swiss Webster mouse experiments, the differences between experimental groups were considered statistically significant for P ≤ 0.05 and groups with different superscript letters indicate significant differences. Data were analyzed using GraphPad Prism 6.01 software (La Jolla, CA).
Principal component analysis (PCA) was performed using the FactoMineR R package (31).
RESULTS
GF mice exhibit important changes in eCBome signaling that are partially reversed by FMT
To investigate how the absence of the gut microbiota can affect the eCBome, we measured eCBome gene expression and mediator levels within juvenile (4 weeks) and adult (13 weeks) GF and CR male mice in the duodenum, jejunum, ileum, and proximal colon. Then, to assess the potential for gut microbiota to subsequently reverse the observed changes in the eCBome, we conventionalized the mice through FMT in 12-week-old GF mice and reassessed the eCBome after 1 week. Although qPCR analyses from different ages and intestinal segments cannot be quantitatively compared as they were run in separate experiments, a comparison of the Cts for each gene (see supplemental Table S3) indicated that the mRNA expression of none of the genes investigated was strongly altered when passing from 4 to 13 weeks of age.
While several alterations were found in GF mice, we shall discuss hereafter only those that were reversed at some level by the FMT, suggesting that the observed changes in GF mice are directly due to a lack of microbiota and not developmental consequences resulting from their absence or adaptive mechanisms.
Genes encoding eCBome receptors.
We observed changes in the expression of Cnr1, Gpr18, Gpr55, and Pparα within the intestines of GF mice. In general, these changes showed similar trends in young and adult GF mice with respect to CR controls in the duodenum, jejunum, ileum, and colon; and in some instances, such as for Gpr18 and Gpr55 in the ileum, the differences in gene expression became markedly more pronounced with age (Fig. 2B, C).
With regard to Cnr1 (encoding CB1), while there were no changes in expression at 4 weeks in any intestinal section, there was a trend to increased expression in the colon, which became statistically significant by 13 weeks (Fig. 2A). Similarly, ileal Cnr1 expression increased by ∼2-fold in 13-week-old GF mice. Notably, even though not statistically significant, the FMT partially reversed this change (Fig. 2A). In contrast, the FMT did reverse increased Cnr1 expression within the colon; however, puzzlingly, the SHAM controls showed the exact same effect, despite having been confirmed to be GF at the point of tissue harvesting.
As for the two orphan G protein-coupled receptors, Gpr18 and Gpr55 (Fig. 2B, C), their expression in GF mice was decreased in all tracts of the small intestine at 4 and 13 weeks, except in the duodenum for Gpr55 at 4 weeks and Gpr18 at 13 weeks. As with Cnr1 in the colon, FMT reversed the observed changes for both targets in the jejunum, but SHAMs showed similar effects. However, in the ileum, FMT partially but significantly rescued the expression of both genes, while no effects were observed in SHAMs. Instead, Gpr55 expression in the duodenum was fully and specifically rescued only by FMT (Fig. 2B, C).
In order to confirm the effects observed for Gpr55 in the ileum, and in view of the increasing importance of the protein encoded by this gene as a target for both eCBs and plant cannabinoids, particularly in immune and inflammatory response, we assayed its expression in 7-week-old GF Swiss Webster mice housed within a different animal facility. Consistent with results obtained in C57BL/6NTac mice, Gpr55 expression was significantly reduced in the ileum of these mice, and conventionalization resulted in a significant reversion of this decrease (Fig. 2E). The regulation of Gpr55 expression by gut microbes in the ileum was further confirmed through the use of antibiotics, which significantly decreased Gpr55 expression (Fig. 2F). In GF Swiss Webster mice or in CR C57BL/6J mice treated with antibiotics, Gpr55 expression was also reduced in the colon, where again this effect showed a strong tendency toward reversion by conventionalization (Fig. 2E). This supports the identification of a trend observed in the colon of 4-week-old C57BL/6NTac mice (Fig. 2C), and suggests a potential age-dependent effect of the lack of gut microbiota on this gene. Finally, in the jejunum, Swiss Webster and C57BL/6J mice also showed a trend toward reduction of Gpr55 expression following GF raising or treatment with antibiotics, respectively (Fig. 2E, F), in partial agreement with the results obtained in both 4- and 13-week-old C57BL/6NTac mice.
Pparα expression was increased in all the tracts of the small intestine of GF mice at 4 weeks, but remained increased only in the duodenum and ileum at 13 weeks (Fig. 2D). These latter decreases were totally and specifically reversed by the FMT (Fig. 2D).
eCBome mediators.
The tone of a given receptor can be altered either by its expression levels and coupling to downstream signaling or by changes in the concentration of its ligand or both. We therefore next measured a panel of eCBome ligands (supplemental Table S4) in all tracts of the small intestine and colon to determine whether they were altered in 13-week-old GF mice and whether their levels could subsequently be affected by FMT. Not all mediator levels were within the detection limits of the method. Moreover, we present data only for those mediators that, apart from being detectable and quantifiable, underwent alterations in GF, SHAM, or FMT mice.
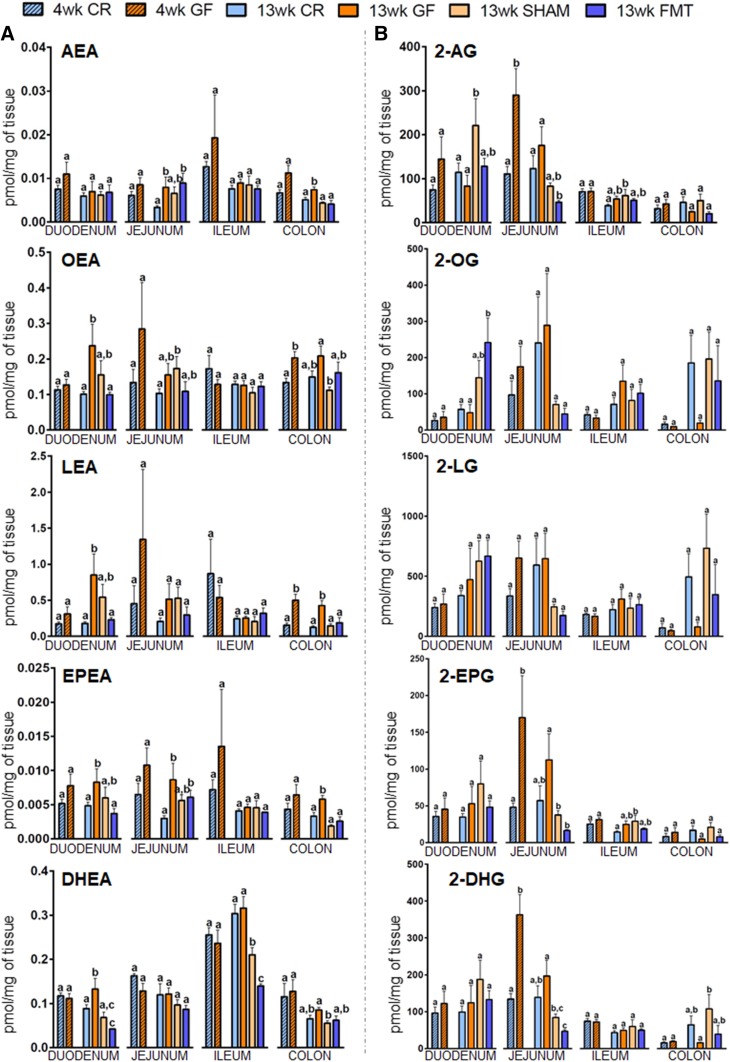
First, we checked to determine whether the tissue levels of the mediators varied between 4- and 13-week-old CR mice, and found that in most of the cases when the differences were statistically significant, the concentrations were higher at 4 weeks, as in the case of AEA and N-eicosapentaenoyl-ethanolamide (EPEA) in the jejunum (P = 0.011 and P = 0.016, respectively) and ileum (P = 0.04 and P = 0.02, respectively), 2-AG in the ileum (P = 0.001), N-docosahexaenoyl-ethanolamide (DHEA) in jejunum, ileum, and colon (P = 0.022, P = 0.016, and P = 0.024, respectively), and N-linoleoyl-ethanolamide (LEA), 2-EPG, and 2-DHG in the ileum (P = 0.007, P = 0.02, and P = 0.004, respectively). The only mediator found to increase was 2-oleoyl-glycerol (2-OG), which was significantly higher at 13 weeks, although only in the colon (P = 0.022). We also observed several statistically significant differences between the levels of the mediators in different intestinal segments at both 4 and 13 weeks (supplemental Table S5A, B). In general, 2-acylglycerols were more abundant in proximal intestinal regions than distal ones [the most frequent exception to this rule being 2-OG and 2-linoleoyl-glycerol (2-LG)]. By contrast, NAEs tended to show an opposite trend, especially at 4 weeks along the small intestine, with AEA, LEA, and DHEA being highest in the ileum.
Next, we analyzed differences due to the GF condition. AEA levels did not show any significant alterations in young GF mice; however, they were significantly increased in the jejunum and colon of GF mice at 13 weeks. While in the jejunum there was no FMT-reversible change observed, the colon displayed a statistical reversion by FMT, though the SHAM showed similar results (Fig. 3A). On the other hand, 2-AG levels were only transiently increased in the jejunum at 4 weeks, returning to CR levels at 13 weeks, and not significantly increased in the ileum at 13 weeks. Interestingly however, FMT resulted in a significant decrease in jejunal 2-AG compared with GF mice, to levels that were even lower than those in CR mice. This suggests that the production or metabolism of 2-AG within the jejunum, despite no longer being significantly increased in 13-week-old CR mice, becomes more sensitive to the introduction of bacteria. These results for 2-AG are partly consistent with alterations in the expression of the 2-AG catabolic enzymes, monoacylglycerol lipase (supplemental Table S6) and Ppt1 at 4 weeks, and for Ppt1 at 13 weeks (see below and supplemental Table S6).
Fig. 3.
Concentrations in intestinal tissues of eCBs and select congeners in 4- and 13-week-old CR or GF mice and in 13-week-old male GF mice gavaged with sterile PBS (SHAM) or fecal microbiota (FMT). Levels of NAEs (A) and 2-acylglycerols (B) are expressed as picomoles per milligram of tissue and were determined in duodenum, jejunum, ileum, and colon. Values are the mean ± SEM of n = 5–10. Bars marked with different letters were significantly different at the P ≤ 0.05 level. 2-AG, the three enantiomers of mono-arachidonoylglycerol (added together because presumably coming from the isomerization of 2-arachidonoylglycerol); 2-EPG, the three enantiomers of mono-eicosapentaenoyl-glycerol (added together because presumably coming from the isomerization of 2-eicosapentaenoyl-glycerol); 2-DHG, the three enantiomers of mono-docosahexaenoyl-glycerol (added together because presumably coming from the isomerization of 2-docosahexaenoyl -glycerol).
With regard to NAEs (Fig. 3A), the most robust changes were observed for N-oleoylethanolamide (OEA), LEA, EPEA, and DHEA. Indeed, the significant increases of OEA, LEA, and EPEA in the duodenum of GF adult mice were fully reversed by FMT but not SHAM treatments, while for DHEA, the SHAM also significantly reversed the observed increase in GF mice, though FMT resulted in a stronger effect to levels below those observed in CR mice. The jejunum showed the same trend for OEA, LEA, and EPEA, though the increase was only significant for the latter, and in all cases, FMT only resulted in statistically insignificant trends toward decreased levels. In the colon, both OEA and LEA were significantly increased in 4-week-old GF mice, though this was only maintained in the case of LEA at 13 weeks, and while FMT completely reversed this effect, so did the SHAM. This same pattern in 13-week-old mice was observed for EPEA at 13 weeks.
Regarding monoacylglycerols, while there were no significant changes in 2-OG and 2-LG along the entire intestine at both ages, 2-EPG and 2-DHG levels were significantly increased in the jejunum of 4-week-old GF mice (Fig. 3B). The same trend was present in the adult mice. FMT reduced their levels from those in GF mice and, in the case of 2-DHG, to levels lower than those in CR mice, as observed for 2-AG. However, once again, SHAMs showed similar effects. In the ileum, 2-EPG levels were not significantly increased at 13 weeks in GF and SHAM mice, and decreased by FMT.
AA, a lipid mediator itself and the ultimate precursor (and metabolite) for both AEA and 2-AG, was significantly increased in the jejunum of young GF mice and the same trend was present in adult mice (Fig. 4A), while in the other parts of the small intestine only nonsignificant trends were observed. However, interestingly, in the ileum, FMT significantly increased AA to levels higher than observed in 13-week-old CR mice. This, however, did not correlate with alterations in AEA or 2-AG levels (Fig. 3A, B). In the colon, AA levels were also significantly increased in GF mice and reduced by FMT, although the SHAM treatment also produced a similar reduction.
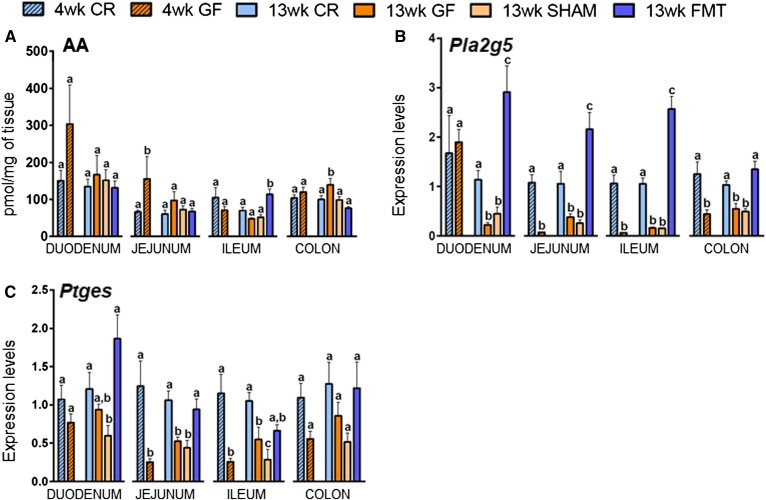
Fig. 4.
Concentrations of AA expressed as picomoles per milligram of tissue (A) and mRNA expression levels of Pla2g5 (B) and Ptges (C) in 4- and 13-week-old male CR or GF mice and in 13-week-old male GF mice gavaged with sterile PBS (SHAM) or fecal microbiota (FMT). Values are the mean ± SEM of n = 5–10. Bars marked with different letters were significantly different at the P ≤ 0.05 level.
Genes encoding eCBome metabolic enzymes.
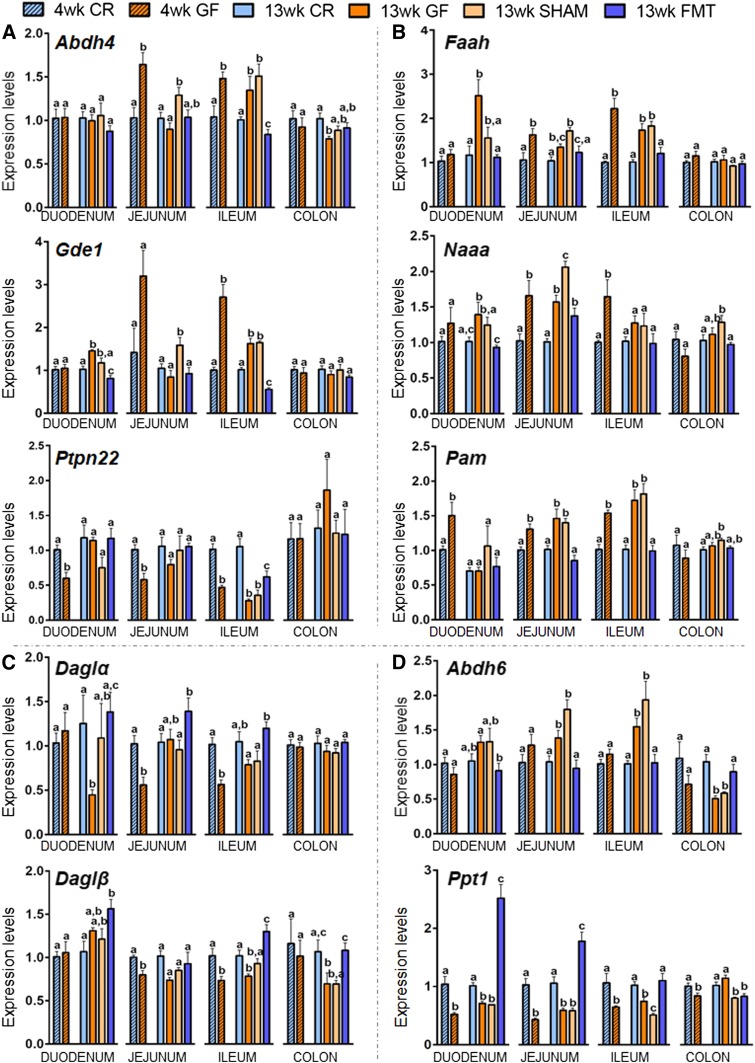
As for the eCBome anabolic enzymes for AEA and other NAEs, the most significant results were observed for Abhd4, Gde1, and Ptpn22. Abhdd4 and Gde1 encode enzymes that in combination provide a NAPE-PLD-independent pathway for NAE synthesis. Both genes were significantly increased in the ileum of 4-week-old GF mice, which was maintained only in the ileum at 13 weeks and completely reversed to levels below those of CR mice specifically by FMT (Fig. 5A). Gde1 expression was similarly affected in the duodenum at 13 weeks, although less markedly. Here too, FMT reduced gene expression levels to those below CR mice, while SHAMs had a nonsignificant decrease (Fig. 5A). Most interestingly, the protein tyrosine phosphatase, Ptpn22, encoding a multi-function phosphatase suggested to be involved in AEA biosynthesis (32), had the mirror opposite expression in response to GF status to those described above. Ptpn22 expression was, in fact, decreased in all sections of the small intestine at 4 weeks in GF mice, an effect that was only maintained in the ileum at 13 weeks. Here again, the FMT was specifically able to reverse this effect, but not completely as with Abhd4 and Gde1 (Fig. 5A). These data suggest that two alternate pathways for NAE biosynthesis respond differently to the GF status and subsequent conventionalization.
Fig. 5.
Expression levels of select intestinal eCBome-related anabolic (A, C) and catabolic (B, D) enzymes for N-aminoacylethanolamines and 2-acylglycerols in 4- and 13-week-old male CR and GF mice and in 13-week-old male GF mice gavaged with sterile PBS (SHAM) or fecal microbiota (FMT). The mRNA expression levels of all the genes represented were measured by qPCR array in the duodenum, jejunum, ileum, and colon for all experimental groups. Values are the mean ± SEM of n = 5–10. Bars marked with different letters were significantly different at the P ≤ 0.05 level.
Concerning the expression levels of eCBome catabolic enzymes for AEA and other NAEs, the expression levels of Faah and Naaa responded almost identically to the GF status and FMT, and although results were not always statistically significant, trends were maintained (Fig. 5B). The expression of both genes was increased in the duodenums of 13-week-old GF mice, which was completely reversed by FMT, while SHAMs only showed nonsignificant decreases. Faah and Naaa expression also increased in the jejunums and ileums of 4- and 13-week-old GF mice, though this effect was only maintained in a statistically significant manner for Faah in the ileums of older mice. Within the jejunums, FMT decreased the expression of both Faah and Naah from those observed in SHAMs, but not to levels below those observed in GF mice. However, for Faah in the ileum, the increase was completely reversed to CR levels by FMT.
Regarding the gene encoding the enzyme, peptidylglycine α-amidating monooxygenase (Pam), responsible for the biosynthesis of primary fatty acid amides from the oxidative catabolism of the corresponding N-acyl-glycines, its expression was increased in all regions of the small intestine of 4-week-old GF mice. At 13 weeks, Pam expression behaved exactly like Faah and Naaa in the jejunum and ileum of GF mice. Furthermore, in both tissues, FMT always induced a significant reversion to levels observed in CR mice, while SHAMs showed no effect (Fig. 5B).
As for the metabolic enzymes for 2-AG and other 2-acyl-glycerols, the most relevant results were found for Daglα and Daglβ, encoding for enzymes that biosynthesize these compounds by catalyzing the hydrolysis of the corresponding diacylglycerols (33, 34), and for the serine hydrolases Abhd6 and Ppt1, which encode for enzymes that have been described to play a role in 2-acyl-glycerol inactivation under certain conditions (35–37) (Fig. 5C, D). In 4-week-old GF mice, Daglα displayed a significant decrease in the jejunum and ileum and only in the latter did FMT reverse the trend toward decreased expression at 13 weeks. Daglβ expression was similarly affected in the jejunum and ileum at both ages, though here the decrease and complete reversion by FMT in ileums was statistically significant and specific (i.e., not observed with SHAMs) (Fig. 5C). The expression levels of Daglβ were also decreased in the colons of 13-week-old GF mice and then totally reverted by the FMT, while SHAM treatment produced no changes (Fig. 5C). For Abhd6 and Ppt1, only the latter showed changes of gene expression in the small intestine at 4 weeks, where it was consistently downregulated in all regions (Fig. 5D). Interestingly, at 13 weeks, Abhd6 and Ppt1 often showed mirror opposite changes in gene expression within individual sections of the small intestine; Abhd6 is increased, while Ppt1 is decreased in all regions. FMT resulted in specific and full rescue of these alterations in all cases. Indeed, in the duodenum and jejunum, FMT increased Ppt1 expression to levels significantly higher than those observed in CR mice at 13 weeks.
Among the other genes encoding for lipid mediator metabolic enzymes, surprising results came from the analysis of Pla2g5 (Fig. 4B). This gene encodes a calcium-dependent phospholipase A2 involved in AA mobilization (and eicosanoid biosynthesis), phospholipid remodeling, and subsequent formation of AEA biosynthetic precursors. The expression of this gene was drastically reduced in a manner significantly and specifically reversed by FMT in all the tracts of the small intestine and colon, with the only exception in the duodenum of 4-week-old GF mice. Even though the results were not always statistically significant, Ptges, encoding the biosynthesizing enzyme for prostaglandins and prostamide E2, a bioactive metabolite of AEA, also showed a significant reduction (although less marked) in the jejunum and ileum in both 4- and 13-week-old GF mice, with a similar trend observed in the duodenum. These decreases were totally reversed by FMT but not in SHAMs (Fig. 4C).
Global changes in eCBome gene expression in response to GF status
We found that GF mice are characterized by significant changes in gene expression of several eCBome receptors and metabolic enzymes, both at 4 and 13 weeks of age and in the various parts of the gastrointestinal tract. Most importantly, the successful colonization by gut microbiota partially reversed the changes in expression of several eCBome genes in GF mice, particularly in the various tracts of the small intestine. Accordingly, PCA of the qPCR array data for all genes showed that for all small intestine segments, CR and FMT mice clustered closely, while GF and SHAM mice were found in the same quadrant, clustered separately but closer to each other than to CR and FMT mice (an overview of all relative gene expression changes is shown in supplemental Table S5, and the corresponding PCA is shown in supplemental Fig. S1). To validate the qPCR array results, we examined the expression of selected genes by single RT-qPCR and, as shown in supplemental Fig. S2, the results confirmed the qPCR array expression data for all the examined genes.
16S metagenomics analyses show that a gut microbiome was reintroduced in GF mice following FMT
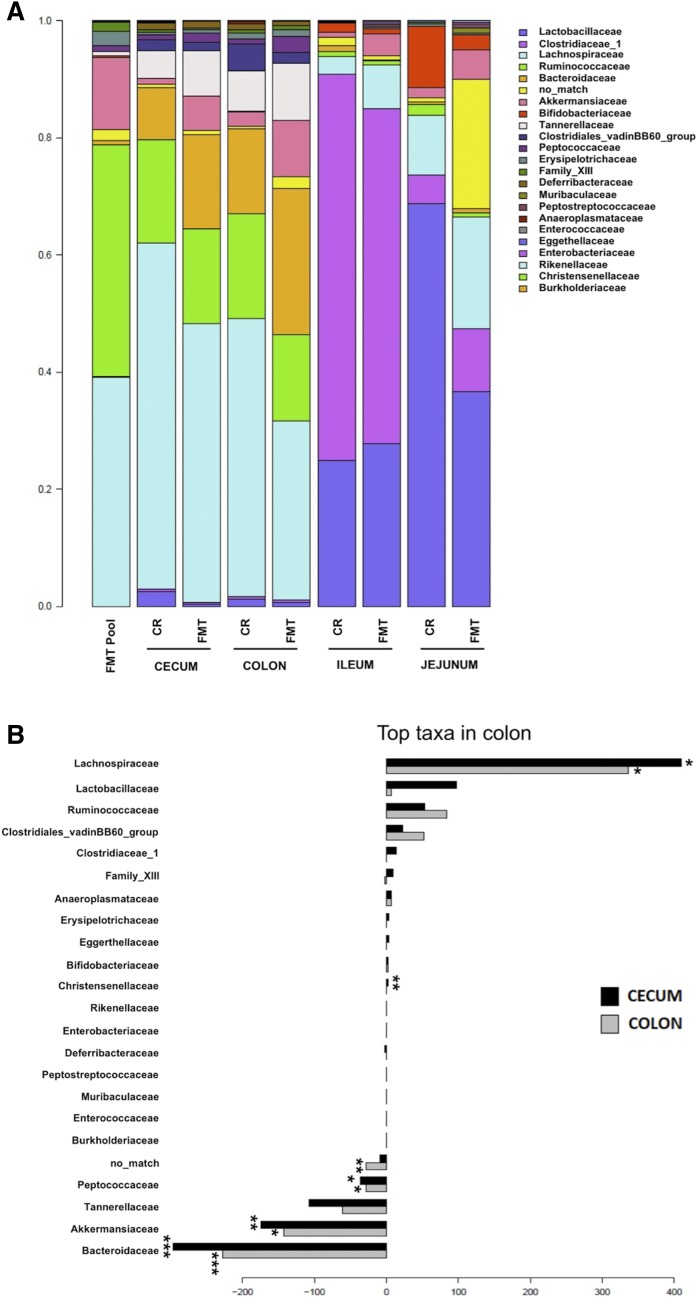
Metataxonomic profiling of GF mice subjected to FMT using microbiota samples from CR mice showed an overall microbiome composition very similar to that of CR mice for colon, cecum, ileum, and jejunum (Fig. 6A). However, specific taxa were associated to compositional differences, as R-squared of the PERMANOVA was 45.7% for colon and 30.2% for cecum. Statistical comparison of FMT mice to CR mice for cecum and colon showed that some taxa were differentially abundant between the two groups; i.e., Bacteroidaceae, Akkermansiaceae, and Peptococcaceae were significantly increased in the FMT group (Fig. 6B). In opposition, Lachnospiraceae were reduced in the FMT group for colon and cecum, and Christensenellaceae were significantly reduced in the FMT group in the cecum only.
Fig. 6.
Composition of the intestinal microbiome of 13-week-old male GF mice after FMT and in CR mice. A: Average bacterial family composition of different intestinal segments (jejunum, ileum, colon, and cecum) in CR and FMT mice and in the intestinal contents and feces pool (FMT pool) used for microbiota transfer. Family names (right) are presented from most (top) to least (bottom) abundant. B: Comparison of colon and cecum microbiota composition of FMT mice and CR mice. Barplots show the coefficient associated bacterial families after PERMANOVA. Positive values indicate a higher prevalence in CR mice and negative indicate a higher prevalence in FMT mice. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005.
DISCUSSION
The present study demonstrates that GF mice exhibit significant modifications in eCBome gene expression and mediator levels in the large and, particularly, small intestine of young and adult male mice. Many of these modifications seem to be directly due to the lack of microbiota because the successful transplantation of intestinal microbiota from donor to age-matched GF mice resulted, after only 1 week, in the partial or complete reversal of many of these changes. These results are unique and important because they provide a cause-effect relationship between the presence or absence of intestinal microbes, especially those that were successfully reintroduced in GF mice by FMT, and eCBome signaling. Based on the known intestinal functions of eCBome receptors and their endogenous ligands that were altered in GF mice (Table 1), our findings represent the basis for future studies aimed at investigating whether such alterations may explain part of the peculiar intestinal phenotype of these mice.
TABLE 1.
Principal features of investigated eCBome molecular targets and lipid ligands
| Most Studied eCBome Targets | eCBome Mediators that Activate the Target | Role of the Target in the Small or Large Intestine | Role in Inflammation of the Target | Anabolic or Catabolica Enzymes Reported to Regulate the Levels of the eCBome Mediators, and Found to be Altered in this Study (22) |
| CB1 | AEA (Ki = 61 nM), 2-AG (Ki = 472 nM)>, DHEA (Ki = 324 nM) (63–65) | ↓ Motility (66) | Pro-inflammatory (70) | GDE1, ABHD4, PTPN22 (for NAEs) |
| ↓ Satiety in response to fat intake (67) | DAGLα, DAGLβ (for 2-acylglycerols) | |||
| ↑ Paracellular permeability (68, 69) | FAAH (for NAEs) | |||
| ABHD6, PPT1 (for 2-acylglycerols) | ||||
| CB2 | 2-AG (Ki = 1400nM)> AEA (Ki = 1930nM) ≥ DHEA (63, 65, 71, 72) | ↓ Motility in gut inflammatory states (73) | Anti-inflammatory (74) | GDE1, ABHD4, PTPN22 (for NAEs) |
| DAGLα, DAGLβ (for 2-acylglycerols) | ||||
| FAAH (for NAEs) | ||||
| ABHD6, PPT1 (for 2-acylglycerols) | ||||
| TRPV1 | Unsaturated and polyunsaturated NAEs and 2-MAGs, including AEA and 2-AG (54, 55) | ↓ Paracellular permeability (75) | Anti-inflammatory (due to its rapid desensitization) (77) | GDE1, ABHD4, PTPN22 (for NAEs) |
| ↑ GLP-1 release (76) | DAGLα, DAGLβ (for 2-acylglycerols) | |||
| FAAH (for NAEs) | ||||
| ABHD6, PPT1 (for 2-acylglycerols) | ||||
| PPARα | OEA > PEA > DHEA (78) | ↑ Satiety in response to fat intake (51) | Anti-inflammatory (79, 80) | GDE1, ABHD4, PTPN22 |
| ↓ Paracellular permeability (75) | FAAH, NAAA | |||
| PPARγ | AEA, DHEA (81, 82) | ↑ Lipid metabolism (83) | Anti-inflammatory (84) | GDE1, ABHD4, PTPN22, FAAH |
| GPR55 | PEA (85) | ↓ Motility (86) | Anti-inflammatory (87) | GDE1, ABHD4, PTPN22, NAAA |
| GPR18 | N-arachidonoyl-glycine | ↑ Correct immune response of the intestine (43) | Possibly anti-inflammatory (44) | FAAH, PAM |
| GPR119 | OEA, LEA, 2-LG, 2-OG (52) | ↑ GLP-1 release (88, 89) | Unknown | GDE1, ABHD4, PTPN22 (for NAEs) |
| DAGLα, DAGLβ (for 2-acylglycerols) | ||||
| FAAH (for NAEs) | ||||
| ABHD6 (for 2-acylglycerols) |
Reference citations appear in parentheses.
Catabolic enzymes are indicated by italic type.
The significant increase in the expression levels of Cnr1 in 13-week-old GF mice that we observed in the ileum and colon along with the trends toward increases in 2-AG levels in the former tissue and AEA levels in the latter tissue, together with previous studies showing that activation of the receptor encoded by this gene inhibits intestinal motility and secretion (38–40), may explain in part why these functions are inhibited in GF mice (14). Our results in GF C57BL/6NTac mice recapitulate those obtained by Muccioli et al. (19) in GF Swiss Webster mice in which Cnr1 expression was similarly increased in the colon, while a nonsignificant decrease was observed in the jejunum. However, the effect of FMT on these alterations was not investigated. Now, we report that FMT partially reversed the increased Cnr1 expression in the ileum and fully reversed it, but not selectively over SHAM treatment, in the colon. These data support the notion that microbiota directly regulate Cnr1 expression in the intestines of mice.
Contrary to Cnr1, the expression of the gene encoding for the recently proposed eCB and plant cannabinoid target, GPR55, was significantly decreased in GF mice in all three small intestinal segments analyzed and, except for the jejunum, in both young and adult mice. This effect was reversed by FMT fully and selectively over SHAM treatment in the duodenum, fully but not completely selectively in the jejunum, and partly and selectively in the ileum. Because a similarity between GF C57BL/6NTac and 7-week-old Swiss Webster mice was observed (and discussed above) for Cnr1 (19), we sought to confirm the Gpr55 expression results in this latter strain, where indeed the GF status resulted again in decreased expression in the ileum and, not significantly, in the jejunum (the duodenum was not analyzed). GF Swiss Webster mice also presented reduced Gpr55 expression in the colon, similar to the non-statistically significant trend observed in GF C57BL/6NTac mice at 4 weeks but not 13 weeks. The reason for this difference may lie in the genetic and age differences between the mice used to perform the experiments. Importantly, however, also in GF Swiss Webster mice gut microbiome, conventionalization resulted in rescued Gpr55 expression. Furthermore, antibiotic treatment of C57BL/6J mice also reduced Gpr55 mRNA expression in the ileum and, as a non-statistically significant trend, the jejunum and colon. These results, taken together, clearly indicate that the gut microbiome as a whole positively controls the expression of Gpr55, particularly in the ileum. The involvement of this receptor in the physiology and pathophysiology of the rodent gut have been described (41), although the description of tissues and cell types expressing this receptor is still lacking. The higher expression of Gpr55 in rats treated with lipopolysaccharides suggested a potential pro-inflammatory role of this receptor in the gut. Thus, decreased GPR55 signaling in the small intestine could be one of the mechanisms underlying decreased gut inflammation in GF mice (42). Future experiments, which are beyond the scope of this study, are needed to address the role of reduced GPR55 signaling in the GF mouse small intestine.
Similar to Gpr55, we observed a robust and general decrease in the expression levels of Gpr18 in the small intestine in GF mice of both ages, an effect that FMT could reverse specifically over SHAM treatment in almost all analyzed parts of this organ. This, in the absence of any significant change in the levels of the proposed ligand for this receptor, i.e., N-arachidonoyl-glycine (which was below detection levels of 5 fmol), suggests that the tonic activity of GPR18 may be correspondingly decreased in GF mice. This receptor is required cell-intrinsically for the reconstitution of small intestine intraepithelial lymphocyte subsets (43), whose functionality in inflammatory bowel disease is altered in conjunction with dysbiosis (44). Thus, decreased GPR18 tone in GF mice may influence small intestine inflammatory responses by altering intraepithelial lymphocyte activity.
GF mice exhibit a lean phenotype due to increased energy expenditure, and hence consume more food than controls to maintain the same body weight (45) and are resistant to diet-induced obesity and insulin resistance, showing decreased adiposity and hepatic triglyceride levels (46, 47). Although the underlying mechanisms are still to be elucidated, it is known that GF mice have increased fatty acid oxidation and decreased lipogenesis (48, 49). In our study, we found an increase in the expression levels of the gene encoding the nuclear receptor PPARα in all the tracts of the small intestine and particularly in the duodenum and ileum of both young and adult GF mice; these changes were totally reversed 1 week after FMT, again specifically over SHAM treatment. PPARα is well-known to play an important role in several metabolic pathways and in lipid metabolism in particular, promoting fatty acid oxidation, lipid transport, and also ketogenesis and gluconeogenesis (50). Although few reports exist about the role of PPARα in the intestine, the activation of this receptor by OEA in the jejunum was shown to inhibit the intake of dietary fats (51). In accordance with these data, a recent study demonstrated that mice lacking intestinal epithelial Napepld, the enzyme mainly responsible of the synthesis of NAEs, display lower levels of OEA and an increase of lipid absorption and fat mass gain leading to a high fat diet-induced fatty liver, thus highlighting the importance of the intestinal NAEs in the regulation of energy homeostasis and to the disruption of signaling leading to the development of obesity and hepatic steatosis (27). We hypothesize that the FMT-reversible increased expression of Pparα in GF mice could, on the one hand, support the “positive” metabolic phenotype of the GF mice and, on the other hand, represent an adaptive response counteracting the increased food intake displayed by these mice.
Indeed, increased Pparα expression was sustained by, and possibly due to, the simultaneous FMT-reversible increases in the levels of the potent endogenous PPARα agonist, OEA, at least in the duodenum of 13-week-old GF mice. Other mediators were also found to be increased in some sections of the small intestine and then decreased back to control levels by FMT. These included the OEA congener, LEA, which, like OEA, acts as a GPR119 agonist (52). GPR119 is a major intestinal stimulator of GLP-1 release, which upregulates insulin and downregulates glucagon levels and thus controls glucose homeostasis, including in response to OEA (53). Interestingly, Wichmann et al. (15) showed that GF mice are characterized by a 3-fold increase in GLP-1 production, contributing to a better glucose tolerance, while FMT completely abolished the increased GLP-1 production and colonic L-cell number. Hence, it is tempting to speculate from our data that increased OEA and LEA may underlie increased production of endogenous GLP-1 in GF mice and contribute to their resistance to high fat diet-induced glucose intolerance. Finally, long chain NAEs, such as OEA and LEA, also activate TRPV1 channels (54, 55), which were shown to inhibit food-intake, stimulate insulin sensitivity, and release and exert both pro- and anti-inflammatory effects (the latter especially following ligand-induced desensitization) (56). Importantly, neither GPR119 nor TRPV1 expression was found here to be affected by lack of the intestinal microbiota (supplemental Table S5).
Two major genes encoding for enzymes for NAE hydrolysis, i.e., Faah and Naaa, showed an FMT-reversible increase along all the small intestinal tracts. However, NAE concentrations were not significantly reduced in the intestinal tissues of GF mice. This could be due to the fact that genes encoding NAE anabolic enzymes, i.e., Abhd4 and, especially, Gde1, also exhibited increased expression in GF mice, thus possibly counteracting the effects of increased degradation and even underlying the increase of select NAEs such as OEA and LEA. On the other hand, there was a decrease in the expression of the gene encoding for a potential AEA biosynthetic enzyme, Ptpn22 (32). However, only scant evidence exists for the involvement in the biosynthesis of NAEs other than AEA for this enzyme, which, as such or through some of its polymorphisms, is instead implicated in intestinal inflammation in a manner dependent on the gut microbiota (57).
Interestingly, two eCBome genes that are more marginally involved in NAE biosynthesis or degradation were instead strongly affected by the lack of microbiota in GF mice. Ptges, encodes prostaglandin E synthase, an enzyme involved in the biosynthesis of PGE2 and also in the transformation of AEA or 2-AG into prostamide E2 and prostaglandin glycerol ester E2 (58, 59) (which, with our method, were below the detection limit of 5 and 25 fmol, respectively). Pla2g5, encodes a calcium-dependent secretory PLA2 catalyzing the release of AA from membrane phospholipids (59). Both these genes were significantly downregulated in all tracts of the small and large intestine, in both young and adult mice and in a manner significantly reversed by FMT. For some of these “multi-task” enzymes, we hypothesize their involvement in the altered immune response of GF mice, which may be related to decreased pro-inflammatory eicosanoid production.
Regarding the anabolic enzymes for 2-acyl-glycerols, both Daglα and Daglβ were reduced, in a manner reversed by FMT, in young and adult GF mice in the ileum, and, in the case of the latter enzyme, also in jejunum and colon, despite the observed elevation of 2-AG and 2-EPG levels in this tissue. Intriguingly, the expression of the gene encoding what is considered the second most important enzyme for 2-AG hydrolysis, Abhd6, was instead increased in the ileum (as well as in the duodenum and jejunum, and decreased in the colon) in an FMT-reversible manner, again in apparent disagreement with the increases observed with some 2-acylglycerols. Among the observed alterations in anabolic or catabolic genes, the latter effect could be due to the reduction in the expression of Ppt1, encoding an enzyme that, however, was shown so far to be involved in 2-AG degradation in only one study (35). In summary, the alterations in monoacylglycerol metabolic enzymes found here in GF mice would have predicted a decrease in the levels of 2-acylglycerols in these mice and, therefore, do not seem to account for the increases observed for some members of this lipid family in some intestinal segments.
The fact that the modifications of the eCBome anabolic and catabolic enzyme expression do not necessarily correlate with the changes observed in lipid mediators (when these latter could be/were measured) may be justified by the redundancy of the metabolic pathways and enzymes able to synthesize and degrade these molecules. This redundancy does not always allow for the prediction of changes in metabolic product concentrations based on changes in enzyme expression (60). A further complication derives from the possibility that the concentrations of these lipid mediators are also regulated by the availability of their ultimate phospholipid precursors (34). Thus, the expression of genes encoding for enzymes may also represent adaptive responses to feedback on mediator changes. Finally, the observed alterations in enzyme expression may have impacted upon the concentrations of mediators that we did not measure, such as, for example, the fatty acid primary amides. These bioactive mediators are produced from the action of the enzyme PAM, whose gene we found here to be significantly upregulated in the jejunum and ileum of both young and adult GF mice and SHAM controls.
Regardless of their role, or lack thereof, in contributing to the gut phenotype or intestinal eCBome mediator levels, the observed mRNA expression levels of eCBome receptors or metabolic enzymes in GF and SHAM mice, on the one hand, and CR and FMT mice, on the other hand, overall clustered together or very close in PCAs, at least in the small intestine where most of the FMT-induced reversals were observed. This observation, together with our metagenomics data showing that the FMT did indeed restore a gut bacterial population very similar to that found in CR mice in GF mice, reinforces the hypothesis of a direct link between the gut microbiota and the eCBome, at least in this part of the gut. This hypothesis is also supported by previous findings by Everard and colleagues showing that: 1) mice lacking, specifically in intestinal epithelial cells, the key toll-like receptor adaptor protein MyD88, which mediates some of the systemic inflammatory effects of gut microbiota-derived lipopolysaccharides, and 2) wild-type mice under a high fat diet treated with the metabolically beneficial commensal bacterium, Akkermansia muciniphila, both exhibit higher intestinal levels of eCBome mediators with anti-inflammatory and GLP-1-stimulating effects (61, 62).
In conclusion, we showed here that GF mice, starting at 4 weeks of age and until at least 13 weeks of age, undergo important changes in eCBome signaling that a partial introduction of the intestinal microbiota is able to reverse after only 1 week. These findings provide unprecedented direct evidence of how the eCBome in the small intestine is subject to regulation by commensal microorganisms. Further studies are now needed to determine whether the changes observed here are responsible for some of the intestinal phenotypes of GF mice and to assess whether the manipulation of gut microorganisms by different diets and pre-, pro-, and symbiotics or, more selectively, with antibiotics and bacteriophages, may impact on the eCBome and, hence, the physiopathological conditions in which this complex signaling system is involved.
Supplementary Material
Acknowledgments
The authors thank Dr. Francesco Tinto for the synthesis of the standard lipid mediator N-oleoyl-l-serine, Cynthia Faubert for being responsible for the GF facility at IUCPQ, Dr. Dominique Boudreau for his helpfulness in the quality control of the samples for the 16S analysis, and all the colleagues, particularly Mélissa Shen of Prof. Cristoforo Silvestri’s team, for their help. Computing was performed on Compute Canada infrastructure (RRG2734).
Footnotes
Abbreviations:
- AA
- arachidonic acid
- AEA
- N-arachidonoylethanolamine (anandamide)
- 2-AG
- 2-arachidonoylglycerol
- CB1
- cannabinoid receptor type-1
- CB2
- cannabinoid receptor type-2
- CR
- conventionally raised
- Ct
- threshold cycle
- DAGL
- diacylglycerol lipase
- DHEA
- N-docosahexaenoyl-ethanolamide
- eCB
- endocannabinoid
- eCBome
- endocannabinoidome
- EPEA
- N-eicosapentaenoyl-ethanolamide
- FAAH
- fatty acid amide hydrolase
- FMT
- fecal microbiota transplant
- GF
- germ-free
- GLP-1
- glucagon-like peptide-1
- LEA
- N-linoleoyl-ethanolamide
- 2-LG
- 2-linoleoyl-glycerol
- NAE
- N-acylethanolamine
- NAPE-PLD
- N-acylphosphatidylethanolamine-hydrolyzing phospholipase D
- OEA
- N-oleoylethanolamide
- 2-OG
- 2-oleoyl-glycerol
- PCA
- principal component analysis
This study was supported by the Canada Research Excellence Chair in the Microbiome-Endocannabinoidome Axis in Metabolic Health (CERC-MEND), which is funded by the Tri-Agency of the Canadian Federal Government (The Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Social Sciences and Humanities Research Council of Canada (SSHRC), to V.D., as well as by the Canadian Foundation of Innovation (to V.D.) and the Sentinelle Nord-Apogée program (to Université Laval). C.M. received a post-doctoral grant from the Joint International Research Unit for Chemical and Biomolecular Research on the Microbiome and its impact on Metabolic Health and Nutrition (UMI-MicroMeNu), which is supported by the Sentinelle Nord program. This work was supported by Fonds Baillet Latour (Grant for Medical Research 2015), Fonds de la Recherche Scientifique - FNRS (FNRS FRFS-WELBIO: WELBIO-CR-2019C-02R), and European Research Council Starting Grant 2013 (336452-ENIGMO). P.D.C. is a senior research associate at FRS-FNRS (Fonds de la Recherche Scientifique), Belgium. All other authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Mazidi M., Rezaie P., Kengne A. P., Mobarhan M. G., and Ferns G. A.. 2016. Gut microbiome and metabolic syndrome. Diabetes Metab. Syndr. 10: S150–S157. [DOI] [PubMed] [Google Scholar]
- 2.Clemente J. C., Ursell L. K., Parfrey L. W., and Knight R.. 2012. The impact of the gut microbiota on human health: an integrative view. Cell. 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jandhyala S. M. 2015. Role of the normal gut microbiota. World J. Gastroenterol. 21: 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Asmakh M., Anuar F., Zadjali F., Rafter J., and Pettersson S.. 2012. Gut microbial communities modulating brain development and function. Gut Microbes. 3: 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., and Pettersson S.. 2012. Host-gut microbiota metabolic interactions. Science. 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 6.Björkholm B., Bok C. M., Lundin A., Rafter J., Hibberd M. L., and Pettersson S.. 2009. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One. 4: e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringel Y., Maharshak N., Ringel-Kulka T., Wolber E. A., Sartor R. B., and Carroll I. M.. 2015. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 6: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pédron T., Mulet C., Dauga C., Frangeul L., Chervaux C., Grompone G., and Sansonetti P. J.. 2012. A crypt-specific core microbiota resides in the mouse colon. MBio. 3: e00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich J. K., Waters J. L., Poole A. C., Sutter J. L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J. T., et al. 2014. Human genetics shape the gut microbiome. Cell. 159: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins M. J., Sharp R., and Macfarlane G. T.. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 48: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez I., Stegen J. C., Maldonado-Gómez M. X., Eren A. M., Siba P. M., Greenhill A. R., and Walter J.. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Reports. 11: 527–538. [DOI] [PubMed] [Google Scholar]
- 12.Al-Asmakh M., and Zadjali F.. 2015. Use of germ-free animal models in microbiota-related research. J. Microbiol. Biotechnol. 25: 1583–1588. [DOI] [PubMed] [Google Scholar]
- 13.Round J. L., and Mazmanian S. K.. 2009. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husebye E., Hellström P. M., Sundler F., Chen J., and Midtvedt T.. 2001. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am. J. Physiol. Gastrointest. Liver Physiol. 280: G368–G380. [DOI] [PubMed] [Google Scholar]
- 15.Wichmann A., Allahyar A., Greiner T. U., Plovier H., Lundén G. Ö., Larsson T., Drucker D. J., Delzenne N. M., Cani P. D., and Bäckhed F.. 2013. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 14: 582–590. [DOI] [PubMed] [Google Scholar]
- 16.Akira S., and Hemmi H.. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 85: 85–95. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson A. J., and Harris N. L.. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4: 478–485. [DOI] [PubMed] [Google Scholar]
- 18.Cani P. D., Amar J., Iglesias M. A., Poggi M., Knauf C., Bastelica D., Neyrinck A. M., Fava F., Tuohy K. M., Chabo C., et al. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 56: 1761–1772. [DOI] [PubMed] [Google Scholar]
- 19.Muccioli G. G., Naslain D., Bäckhed F., Reigstad C. S., Lambert D. M., Delzenne N. M., and Cani P. D.. 2010. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 6: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Marzo V., and De Petrocellis L.. 2012. Why do cannabinoid receptors have more than one endogenous ligand? Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3216–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Marzo, V., and J. Wang, editors. The Endocannabinoidome: The World of Endocannabinoids and Related Mediators. 1st edition. Academic Press, New York.
- 22.Di Marzo V. 2018. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 17: 623–639. [DOI] [PubMed] [Google Scholar]
- 23.Piscitelli F., and Di Marzo V.. 2015. The ever-expanding world of the endocannabinoids: a concise introduction. In Cannabinoids in Neurologic and Mental Disease. Accessed June 26, 2019, at http://www.sciencedirect.com/science/article/pii/B9780124170414000242. [Google Scholar]
- 24.Massa F., and Monory K.. 2006. Endocannabinoids and the gastrointestinal tract. J. Endocrinol. Invest. 29: 47–57. [PubMed] [Google Scholar]
- 25.Guida F., Turco F., Iannotta M., De Gregorio D., Palumbo I., Sarnelli G., Furiano A., Napolitano F., Boccella S., Luongo L., et al. 2018. Antibiotic-induced microbiota perturbation causes gut endocannabinoidome changes, hippocampal neuroglial reorganization and depression in mice. Brain Behav. Immun. 67: 230–245. [DOI] [PubMed] [Google Scholar]
- 26.Geurts, L., A. Everard, M. Van Hul, A. Essaghir, T. Duparc, S. Matamoros, H. Plovier, J. Castel, R. G. Denis, M. Bergiers, et al. 2015. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nat. Commun. 11: 6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everard A., Plovier H., Rastelli M., Van Hul M., de Wouters d’Oplinter A., Geurts L., Druart C., Robine S., Delzenne N. M., Muccioli G. G., et al. 2019. Intestinal epithelial N-acylphosphatidylethanolamine phospholipase D links dietary fat to metabolic adaptations in obesity and steatosis. Nat. Commun. 10: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 29.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson M. J. 2017. Permutational multivariate analysis of variance (PERMANOVA). Accessed June 26, 2019, at http://onlinelibrary.wiley.com/doi/abs/10.1002/9781118445112.stat07841.
- 31.Lê S., Josse J., and Husson F.. 2008. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25: 1–18. [Google Scholar]
- 32.Liu J., Wang L., Harvey-White J., Osei-Hyiaman D., Razdan R., Gong Q., Chan A. C., Zhou Z., Huang B. X., Kim H-Y., et al. 2006. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. USA. 103: 13345–13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bisogno T., Howell F., Williams G., Minassi A., Cascio M. G., Ligresti A., Matias I., Schiano-Moriello A., Paul P., Williams E-J., et al. 2003. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 163: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Marzo V. 2006. Endocannabinoids: synthesis and degradation. In Reviews of Physiology, Biochemistry and Pharmacology. Vol. 160. S. G. Amara, E. Bamberg, B. Fleischmann, et al., editors. pp. 1–24., Springer, Berlin, Heidelberg. 1–24. [DOI] [PubMed] [Google Scholar]
- 35.Wang R., Borazjani A., Matthews A. T., Mangum L. C., Edelmann M. J., and Ross M. K.. 2013. Identification of palmitoyl protein thioesterase 1 in human THP-1 monocytes/macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 52: 7559–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrs W. R., Blankman J. L., Horne E. A., Thomazeau A., Lin Y. H., Coy J., Bodor A. L., Muccioli G. G., Hu S. S-J., Woodruff G., et al. 2010. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 13: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navia-Paldanius D., Savinainen J. R., and Laitinen J. T.. 2012. Biochemical and pharmacological characterization of human α/β-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res. 53: 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto L., Izzo A. A., Cascio M. G., Bisogno T., Hospodar-Scott K., Brown D. R., Mascolo N., Di Marzo V., and Capasso F.. 2002. Endocannabinoids as physiological regulators of colonic propulsion in mice. Gastroenterology. 123: 227–234. [DOI] [PubMed] [Google Scholar]
- 39.Izzo A. A., Capasso F., Costagliola A., Bisogno T., Marsicano G., Ligresti A., Matias I., Capasso R., Pinto L., Borrelli F., et al. 2003. An endogenous cannabinoid tone attenuates cholera toxin-induced fluid accumulation in mice. Gastroenterology. 125: 765–774. [DOI] [PubMed] [Google Scholar]
- 40.Izzo A. A., Fezza F., Capasso R., Bisogno T., Pinto L., Iuvone T., Esposito G., Mascolo N., Di Marzo V., and Capasso F.. 2001. Cannabinoid CB1-receptor mediated regulation of gastrointestinal motility in mice in a model of intestinal inflammation. Br. J. Pharmacol. 134: 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schicho R., and Storr M.. 2012. A potential role for GPR55 in gastrointestinal functions. Curr. Opin. Pharmacol. 12: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernández-Chirlaque C., Aranda C. J., Ocón B., Capitán-Cañadas F., Ortega-González M., Carrero J. J., Suárez M. D., Zarzuelo A., Sánchez de Medina F., and Martínez-Augustin O.. 2016. Germ-free and antibiotic-treated mice are highly susceptible to epithelial injury in DSS colitis. J. Crohns Colitis. 10: 1324–1335. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Sumida H., and Cyster J. G.. 2014. GPR18 is required for a normal CD8αα intestinal intraepithelial lymphocyte compartment. J. Exp. Med. 211: 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regner E. H., Ohri N., Stahly A., Gerich M. E., Fennimore B. P., Ir D., Jubair W. K., Görg C., Siebert J., Robertson C. E., et al. 2018. Functional intraepithelial lymphocyte changes in inflammatory bowel disease and spondyloarthritis have disease specific correlations with intestinal microbiota. Arthritis Res. Ther. 20: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wostmann B. S., Larkin C., Moriarty A., and Bruckner-Kardoss E.. 1983. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab. Anim. Sci. 33: 46–50. [PubMed] [Google Scholar]
- 46.Bäckhed F., Manchester J. K., Semenkovich C. F., and Gordon J. I.. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan M. J., Gerasimidis K., Edwards C. A., and Shaikh M. G. Role of gut microbiota in the aetiology of obesity: proposed mechanisms and review of the literature. J. Obes. Epub ahead of print. September 15, 2016; doi:10.1155/2016/7353642. [DOI] [PMC free article] [PubMed]
- 48.Bäckhed M. J., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., and Gordon J. I.. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velagapudi V. R., Hezaveh R., Reigstad C. S., Gopalacharyulu P., Yetukuri L., Islam S., Felin J., Perkins R., Borén J., Orešič M., et al. 2010. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 51: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy J. K., and Hashimoto T.. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz G. J., Fu J., Astarita G., Li X., Gaetani S., Campolongo P., Cuomo V., and Piomelli D.. 2008. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 8: 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syed S. K., Bui H. H., Beavers L. S., Farb T. B., Ficorilli J., Chesterfield A. K., Kuo M-S., Bokvist K., Barrett D. G., and Efanov A. M.. 2012. Regulation of GPR119 receptor activity with endocannabinoid-like lipids. Am. J. Physiol. Endocrinol. Metab. 303: E1469–E1478. [DOI] [PubMed] [Google Scholar]
- 53.Lauffer L. M., Iakoubov R., and Brubaker P. L.. 2009. GPR119 is essential for oleoylethanolamide-induced glucagon-like peptide-1 secretion from the intestinal enteroendocrine L-cell. Diabetes. 58: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raboune S., Stuart J. M., Leishman E., Takacs S. M., Rhodes B., Basnet A., Jameyfield E., McHugh D., Widlanski T., and Bradshaw H. B.. 2014. Novel endogenous N-acyl amides activate TRPV1–4 receptors, BV-2 microglia, and are regulated in brain in an acute model of inflammation. Front. Cell. Neurosci. 8: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Petrocellis L., Nabissi M., Santoni G., and Ligresti A.. 2017. Actions and regulation of ionotropic cannabinoid receptors. Adv. Pharmacol. 80: 249–289. [DOI] [PubMed] [Google Scholar]
- 56.Christie S., Wittert G. A., Li H., and Page A. J.. 2018. Involvement of TRPV1 channels in energy homeostasis. Front. Endocrinol. 9: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pike K. A., and Tremblay M. L.. 2018. Protein tyrosine phosphatases: regulators of CD4 T cells in inflammatory bowel disease. Front. Immunol. 9: 2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matias I., Chen J., Petrocellis L. D., Bisogno T., Ligresti A., Fezza F., Krauss A. H-P., Shi L., Protzman C. E., Li C., et al. 2004. Prostaglandin ethanolamides (prostamides): in vitro pharmacology and metabolism. J. Pharmacol. Exp. Ther. 309: 745–757. [DOI] [PubMed] [Google Scholar]
- 59.Astudillo A. M., Balboa M. A., and Balsinde J.. 2019. Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1864: 772–783. [DOI] [PubMed] [Google Scholar]
- 60.Piscitelli, F., and V. Di Marzo. 2012. “Redundancy” of endocannabinoid inactivation: new challenges and opportunities for pain control. ACS Chem. Neurosci. 3: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Everard A., Geurts L., Caesar R., Van Hul M., Matamoros S., Duparc T., Denis R. G. P., Cochez P., Pierard F., Castel J., et al. 2014. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat. Commun. 5: 5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., Guiot Y., Derrien M., Muccioli G. G., Delzenne N. M., et al. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 110: 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugiura T., Kondo S., Sukagawa A., Nakane S., Shinoda A., Itoh K., Yamashita A., and Waku K.. 1995. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 215: 89–97. [DOI] [PubMed] [Google Scholar]
- 64.Sugiura T., Kodaka T., Nakane S., Miyashita T., Kondo S., Suhara Y., Takayama H., Waku K., Seki C., Baba N., et al. 1999. Evidence that the cannabinoid CB1 receptor is a 2-arachidonoylglycerol receptor. Structure-activity relationship of 2-arachidonoylglycerol, ether-linked analogues, and related compounds. J. Biol. Chem. 274: 2794–2801. [DOI] [PubMed] [Google Scholar]
- 65.Thakur G. A., Nikas S. P., and Makriyannis A.. 2005. CB1 cannabinoid receptor ligands. Mini Rev. Med. Chem. 5: 631–640. [DOI] [PubMed] [Google Scholar]
- 66.Cluny N. L., Keenan C. M., Duncan M., Fox A., Lutz B., and Sharkey K. A.. 2010. Naphthalen-1-yl-(4-pentyloxynaphthalen-1-yl)methanone (SAB378), a peripherally restricted cannabinoid CB1/CB2 receptor agonist, inhibits gastrointestinal motility but has no effect on experimental colitis in mice. J. Pharmacol. Exp. Ther. 334: 973–980. [DOI] [PubMed] [Google Scholar]
- 67.DiPatrizio N. V., Astarita G., Schwartz G., Li X., and Piomelli D.. 2011. Endocannabinoid signal in the gut controls dietary fat intake. Proc. Natl. Acad. Sci. USA. 108: 12904–12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karwad M. A., Couch D. G., Theophilidou E., Sarmad S., Barrett D. A., Larvin M., Wright K. L., Lund J. N., and O’Sullivan S. E.. 2017. The role of CB1 in intestinal permeability and inflammation. FASEB J. 31: 3267–3277. [DOI] [PubMed] [Google Scholar]
- 69.Alhamoruni A., Wright K. L., Larvin M., and O’Sullivan S. E.. 2012. Cannabinoids mediate opposing effects on inflammation-induced intestinal permeability. Br. J. Pharmacol. 165: 2598–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grunewald Z. I., Lee S., Kirkland R., Ross M., and de La Serre C. B.. 2019. Cannabinoid receptor type-1 partially mediates metabolic endotoxemia-induced inflammation and insulin resistance. Physiol. Behav. 199: 282–291. [DOI] [PubMed] [Google Scholar]
- 71.Balvers M. G. J., Verhoeckx K. C. M., Plastina P., Wortelboer H. M., Meijerink J., and Witkamp R. F.. 2010. Docosahexaenoic acid and eicosapentaenoic acid are converted by 3T3-L1 adipocytes to N-acyl ethanolamines with anti-inflammatory properties. Biochim. Biophys. Acta. 1801: 1107–1114. [DOI] [PubMed] [Google Scholar]
- 72.Sugiura T., Kondo S., Kishimoto S., Miyashita T., Nakane S., Kodaka T., Suhara Y., Takayama H., and Waku K.. 2000. Evidence that 2-arachidonoylglycerol but not N-palmitoylethanolamine or anandamide is the physiological ligand for the cannabinoid CB2 receptor. Comparison of the agonistic activities of various cannabinoid receptor ligands in HL-60 cells. J. Biol. Chem. 275: 605–612. [DOI] [PubMed] [Google Scholar]
- 73.Duncan, M., A. Mouihate, K. Mackie, C. M. Keenan, N. E. Buckley, J. S. Davison, K. D. Patel, Q. J. Pittman, and K. A. Sharkley. 2018. Cannabinoid CB2 receptors in the enteric nervous system modulate gastrointestinal contractility in lipopolysaccharide-treated rats. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G78–G87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parlar A., Arslan S. O., Doğan M. F., Çam S. A., Yalçin A., Elibol E., Özer M. K., Üçkardeş F., and Kara H.. 2018. The exogenous administration of CB2 specific agonist, GW405833, inhibits inflammation by reducing cytokine production and oxidative stress. Exp. Ther. Med. 16: 4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karwad M. A., Macpherson T., Wang B., Theophilidou E., Sarmad S., Barrett D. A., Larvin M., Wright K. L., Lund J. N., and O’Sullivan S. E.. 2017. Oleoylethanolamine and palmitoylethanolamine modulate intestinal permeability in vitro via TRPV1 and PPARα. FASEB J. 31: 469–481. [DOI] [PubMed] [Google Scholar]
- 76.Lee E., Jung D. Y., Kim J. H., Patel P. R., Hu X., Lee Y., Azuma Y., Wang H-F., Tsitsilianos N., Shafiq U., et al. 2015. Transient receptor potential vanilloid type-1 channel regulates diet-induced obesity, insulin resistance, and leptin resistance. FASEB J. 29: 3182–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ambrosino P., Soldovieri M. V., Russo C., and Taglialatela M.. 2013. Activation and desensitization of TRPV1 channels in sensory neurons by the PPARα agonist palmitoylethanolamide. Br. J. Pharmacol. 168: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sagar D. R., Kendall D. A., and Chapman V.. 2008. Inhibition of fatty acid amide hydrolase produces PPAR-alpha-mediated analgesia in a rat model of inflammatory pain. Br. J. Pharmacol. 155: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silveira L. S., Pimentel G. D., Souza C. O., Biondo L. A., Teixeira A. A. S., Lima E. A., Batatinha H. A. P., Rosa Neto J. C., and Lira F. S.. 2017. Effect of an acute moderate-exercise session on metabolic and inflammatory profile of PPAR-α knockout mice. Cell Biochem. Funct. 35: 510–517. [DOI] [PubMed] [Google Scholar]
- 80.Pawlak M., Lefebvre P., and Staels B.. 2015. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 62: 720–733. [DOI] [PubMed] [Google Scholar]
- 81.Rovito D., Giordano C., Vizza D., Plastina P., Barone I., Casaburi I., Lanzino M., De Amicis F., Sisci D., Mauro L., et al. 2013. Omega-3 PUFA ethanolamides DHEA and EPEA induce autophagy through PPARγ activation in MCF-7 breast cancer cells. J. Cell. Physiol. 228: 1314–1322. [DOI] [PubMed] [Google Scholar]
- 82.Bouaboula M., Hilairet S., Marchand J., Fajas L., Le G. F., and Casellas P.. 2005. Anandamide induced PPARgamma transcriptional activation and 3T3-L1 preadipocyte differentiation. Eur. J. Pharmacol. 517: 174–181. [DOI] [PubMed] [Google Scholar]
- 83.Duszka K., Oresic M., Le May C., König J., and Wahli W.. 2017. PPARγ modulates long chain fatty acid processing in the intestinal epithelium. Int. J. Mol. Sci. 18: E2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bassaganya-Riera J., Misyak S., Guri A. J., and Hontecillas R.. 2009. PPAR gamma is highly expressed in F4/80(hi) adipose tissue macrophages and dampens adipose-tissue inflammation. Cell. Immunol. 258: 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kramar C., Loureiro M., Renard J., and Laviolette S. R.. 2017. Palmitoylethanolamide modulates GPR55 receptor signaling in the ventral hippocampus to regulate mesolimbic dopamine activity, social interaction, and memory processing. Cannabis Cannabinoid Res. 2: 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin X-H., Yuece B., Li Y-Y., Feng Y-J., Feng J-Y., Yu L-Y., Li K., Li Y-N., and Storr M.. 2011. A novel CB receptor GPR55 and its ligands are involved in regulation of gut movement in rodents. Neurogastroenterol. Motil. 23: 862-e342. [DOI] [PubMed] [Google Scholar]
- 87.Stančić A., Jandl K., Hasenöhrl C., Reichmann F., Marsche G., Schuligoi R., Heinemann A., Storr M., and Schicho R.. 2015. The GPR55 antagonist CID16020046 protects against intestinal inflammation. Neurogastroenterol. Motil. 27: 1432–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lan H., Lin H. V., Wang C. F., Wright M. J., Xu S., Kang L., Juhl K., Hedrick J. A., and Kowalski T. J.. 2012. Agonists at GPR119 mediate secretion of GLP-1 from mouse enteroendocrine cells through glucose-independent pathways. Br. J. Pharmacol. 165: 2799–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen H. S., Rosenkilde M. M., Holst J. J., and Schwartz T. W.. 2012. GPR119 as a fat sensor. Trends Pharmacol. Sci. 33: 374–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.