Abstract
The bile acid (BA) composition in mice is substantially different from that in humans. Chenodeoxycholic acid (CDCA) is an end product in the human liver; however, mouse Cyp2c70 metabolizes CDCA to hydrophilic muricholic acids (MCAs). Moreover, in humans, the gut microbiota converts the primary BAs, cholic acid and CDCA, into deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. In contrast, the mouse Cyp2a12 reverts this action and converts these secondary BAs to primary BAs. Here, we generated Cyp2a12 KO, Cyp2c70 KO, and Cyp2a12/Cyp2c70 double KO (DKO) mice using the CRISPR-Cas9 system to study the regulation of BA metabolism under hydrophobic BA composition. Cyp2a12 KO mice showed the accumulation of DCAs, whereas Cyp2c70 KO mice lacked MCAs and exhibited markedly increased hepatobiliary proportions of CDCA. In DKO mice, not only DCAs or CDCAs but also DCAs, CDCAs, and LCAs were all elevated. In Cyp2c70 KO and DKO mice, chronic liver inflammation was observed depending on the hepatic unconjugated CDCA concentrations. The BA pool was markedly reduced in Cyp2c70 KO and DKO mice, but the FXR was not activated. It was suggested that the cytokine/c-Jun N-terminal kinase signaling pathway and the pregnane X receptor-mediated pathway are the predominant mechanisms, preferred over the FXR/small heterodimer partner and FXR/fibroblast growth factor 15 pathways, for controlling BA synthesis under hydrophobic BA composition. From our results, we hypothesize that these KO mice can be novel and useful models for investigating the roles of hydrophobic BAs in various human diseases.
Keywords: CRISPR-Cas9, cytochrome P450, CYP2A12, CYP2C70, cholesterol 7α-hydroxylase, 7α-hydroxy-4-cholesten-3-one 12α-hydroxylase, cytokines, farnesoid X receptor, knockout mouse, pregnane X receptor
A mouse is the most commonly used laboratory animal to extrapolate investigations regarding human metabolism. However, numerous differences have been reported between mice and humans. Bile acids (BAs), the end products of cholesterol catabolism, take part in the intestinal digestion and absorption and are recycled via the enterohepatic circulation. The BA composition is a primary indicator of the metabolic difference between mice and humans (1). Certain BAs are ligands of nuclear and transmembrane G protein-coupled receptors and regulate lipid and carbohydrate metabolism, inflammation, fibrosis, and carcinogenesis (2). Therefore, the BA composition can be a crucial factor for creating relevant mouse models of human diseases (3, 4).
Although there are several differences between humans and mice with respect to BA metabolism (5), the following two reactions determine the characteristic phenotype associated with the BA composition in mice. First, most of chenodeoxycholic acid (CDCA), an end product in human liver, is further metabolized to muricholic acids (MCAs) by CDCA 6β-hydroxylase in the mouse and rat liver (Fig. 1A). CDCA is a cytotoxic BA (6, 7) and is the most potent physiological agonist of the FXR (NR1H4) (8, 9). In contrast, MCAs are cytoprotective (7) and have antagonistic effects on FXR (10).
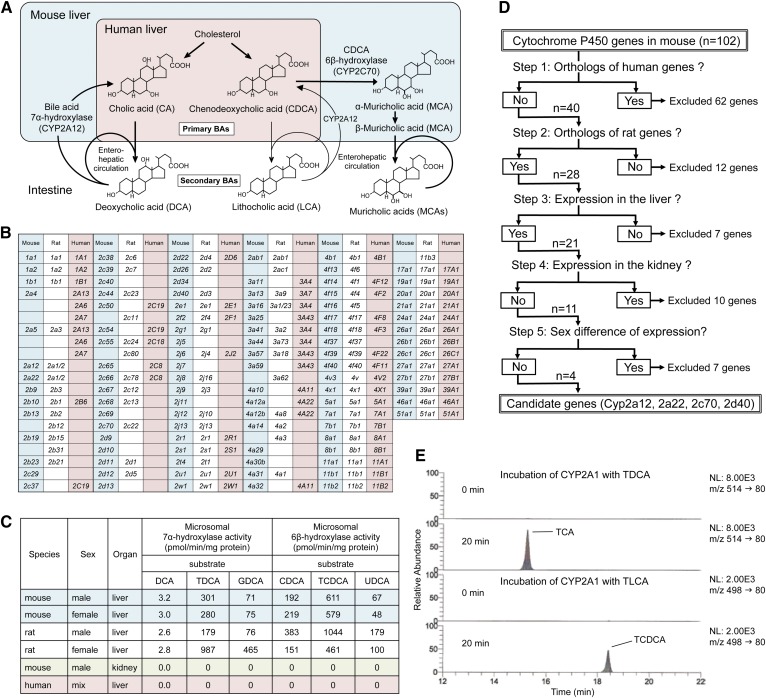
Fig. 1.
Cyp2a12 and Cyp2c70 are the genes responsible for hepatic BA 7α-hydroxylation and CDCA 6β-hydroxylation. A: Comparison of BA metabolism between human and mouse. B: Orthology among mouse, rat, and human CYP genes. C: Species difference, sexual dimorphism, and tissue distribution of microsomal BA 7α-hydroxylase and CDCA 6β-hydroxylase activities. GDCA, glycodeoxycholic acid. Data are represented as means of duplicate assay. D: Methods for narrowing down candidate mouse genes encoding BA 7α-hydroxylase and CDCA 6β-hydroxylase using orthology, tissue distribution, and sexual dimorphism data of Cyp. E: LC-selected reaction monitoring chromatograms (24) of taurocholic acid (TCA) and TCDCA in extracts obtained from the incubation mixture of recombinant rat CYP2A1, NADPH generating system, and TDCA or TLCA. Q10
Second, the primary BAs, cholic acid (CA) and CDCA, are 7α-dehydroxylated by intestinal bacteria and transformed into the secondary BAs, deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. In mice and rats, these secondary BAs are converted to primary BAs by the hepatic BA 7α-hydroxylase (Fig. 1A). Compared with primary BAs, DCA and LCA are more effective in activating the Takeda G protein-coupled receptor 5 (TGR5) (11). They are also more cytotoxic (6) and promote carcinogenesis (12, 13). However, the biliary proportion of these secondary BAs in mice is markedly low (less than 3%) (14) compared with that in humans (∼20–30%) (15).
The specific rodent genes responsible for the CDCA 6β-hydroxylation and BA 7α-rehydroxylation were not determined for a long time. However, Takahashi et al. (16) recently found that MCAs were not detected in liver samples of Cyp2c-cluster-null mice (17). They, therefore, concluded that Cyp2c70 was necessary for the 6β-hydroxylation of CDCA in mice. However, the specific rodent genes responsible for hepatic BA 7α-rehydroxylation were still not determined until recently. An early study on the purification and characterization of rat liver taurodeoxycholic acid (TDCA) 7α-hydroxylase showed that BA 7α-rehydroxylation was catalyzed by a cytochrome P450 (CYP) enzyme (18). Another study using liver-specific CYP oxidoreductase KO mice showed that biliary TDCA levels were markedly elevated after feeding CA (19), suggesting that 7α-rehydroxylation of BAs is catalyzed by CYP enzyme(s).
In this study, we first identified the mouse and rat genes encoding BA 7α-hydroxylase and CDCA 6β-hydroxylase through a new approach using orthology, tissue distribution, and sexual dimorphism data of CYP (20, 21). Then, we generated double KO (DKO) mice to examine BA metabolism. These mice showed BA composition just as we had expected, and the BA pool was markedly reduced. However, much to our surprise, FXR was not activated.
MATERIALS AND METHODS
Materials
αMCA, βMCA, ωMCA, tauro-αMCA, tauro-βMCA, and tauro-ωMCA were purchased from Steraloids (Newport, RI). Taurohyodeoxycholic acid (THDCA), taurolithocholic acid 3-sulfate (TLCA-3S), and LCA 3-sulfate (LCA-3S) were obtained from Cayman Chemical (Ann Arbor, MI). Pooled male mouse liver microsomes (CD1), pooled female mouse liver microsomes (CD1), pooled male rat liver microsomes (Sprague-Dawley), pooled female rat liver microsomes (Sprague-Dawley), and pooled human liver microsomes were purchased from BD Biosciences (Franklin Lakes, NJ), and male CD1 mouse kidney microsomes were purchased from Sekisui XenoTech (Kansas City, KS).
Animals
Cyp2a12−/−Cyp2c70−/− DKO mice were generated using the CRISPR-Cas9 system by the Laboratory Animal Resource Center, University of Tsukuba (Ibaraki, Japan) and Charles River Laboratories Japan, Inc. (Kanagawa, Japan). Mice were kept under pathogen-free conditions and a regular 12 h light-dark cycle (light period: 0600–1800), with free access to standard chow and water. This project was approved by the Animal Experiment Committees of the University of Tsukuba, Charles River Laboratories Japan, and Tokyo Medical University.
Generation of Cyp2a12−/−, Cyp2c70−/−, and Cyp2a12−/−Cyp2c70−/− mice
The oligos, Cyp2a12 intron 2 CRISPR F (5′-caccATAGTTAGGGGAAGCGACAT-3′) and Cyp2a12 intron 2 CRISPR R (5′-aaacATGTCGCTTCCCCTAACTAT-3′), Cyp2a12 intron 4 CRISPR F (5′-caccGTCTTACAATCCAGGCGAGG-3′) and Cyp2a12 intron 4 CRISPR R (5′-aaacCCTCGCCTGGATTGTAAGAC-3′), Cyp2c70 intron 1 CRISPR F (5′-caccAGATGATTATTAGTGTACAG-3′) and Cyp2c70 intron 1 CRISPR R (5′-aaacCTGTACACTAATAATCATCT-3′), and Cyp2c70 intron 2 CRISPR F (5′-caccTGGAACAGTGACAAGAGCGA-3′) and Cyp2c70 intron 2 CRISPR R (5′-aaacTCGCTCTTGTCACTGTTCCA-3′) were annealed and inserted into the BbsI restriction site of the pX330 vector (Addgene plasmid 42230). Constructed plasmids (circular) were designated pX330-Cyp2a12 intron 2, pX330-Cyp2a12 intron 4, pX330-Cyp2c70 intron 1, and pX330-Cyp2c70 intron 2.
Pregnant mare serum gonadotropin and human chorionic gonadotropin were injected into female C57BL/6J mice at 48 h intervals, and the mice were mated with male C57BL/6J mice. Fertilized ova were collected from the oviducts, and 5 ng/μl each of pX330-Cyp2a12 intron 2 and pX330-Cyp2a12 intron 4 or pX330-Cyp2c70 intron 1 and pX330-Cyp2c70 intron 2 were injected into the pronuclei according to standard protocols (22). The injected one-cell embryos were transferred into pseudopregnant CD1 mice.
Using genomic DNA obtained from tail clippings, founder (F0) mice were selected by PCR followed by direct sequencing as described by Hoshino et al (23). The sequences of the oligonucleotide primer pairs used were forward (F): 5′-GAGAGGCAAATGGGAACAAA-3′ and reverse (R): 5′-AACAGGCAGAAGCAGGGATA-3′ for WT Cyp2a12, F: 5′-GAGAGGCAAATGGGAACAAA-3′ and R: 5′-AGGACCTCGGGATGAGAAGT-3′ for mutant Cyp2a12, F: 5′-TCTTCTTGCCTTCAACAGCA-3′ and R: 5′-AACCATTGCACAGAGCACAG-3′ for WT Cyp2c70, F: 5′-TCTTCTTGCCTTCAACAGCA-3′ and R: 5′-GAAAGCCCATGAGAGAGGAA-3′ for mutant Cyp2c70, and F: 5′-AGTTCATCAAGCCCATCCTG-3′ and R: 5′-GAAGTTTCTGTTGGCGAAGC-3′ for Cas9 detection.
F0 mice for Cyp2a12-null and Cyp2c70-null were bred with WT C57BL/6J mice to determine their germline competency. A male F1 Cyp2a12+/− mouse was crossed with female C57BL/6J mice by in vitro fertilization using CARD HyperOva (Kyudo Co., Saga, Japan), and the resulting female Cyp2a12+/− mice were crossed with a male F1 Cyp2c70+/− mouse by in vitro fertilization using CARD HyperOva. Then, these double heterozygous Cyp2a12+/−Cyp2c70+/− animals were crossed to obtain Cyp2a12−/−Cyp2c70+/+ (2a12KO), Cyp2a12+/+Cyp2c70−/− (2c70KO), and Cyp2a12−/−Cyp2c70−/− (DKO) mice. All experiments reported here were performed with subsequent generations of these animals.
Sample collection from mice
Mice that were 10–12 weeks old [11.1 ± 0.8 (mean ± SD), n = 44] were used. After making them fast for 4 h with free access to water, they were euthanized between 1100 and 1600 under combination anesthesia with medetomidine, midazolam, and butorphanol. Their gallbladder, blood (serum), liver, small intestine, cecal contents, and feces were collected immediately and frozen at −80°C.
Determination of liver function tests
Serum activities of alanine transaminase (ALT) and alkaline phosphatase (ALP) were determined by colorimetric assays using Transaminase CII-Test Wako and LabAssay ALP (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan).
Determination of BA concentrations
BA concentrations were determined as described by Murakami et al. (24) with minor modifications. Liver, small intestine, cecal contents, and feces were solubilized in 5% KOH/water at 80°C for 20 min; this heating step was omitted for serum and bile samples. After the addition of internal standards and 0.5 M potassium phosphate buffer (pH 7.4), BAs were extracted with Bond Elut C18 cartridges and quantified by LC-MS/MS. Chromatographic separation was performed using a Hypersil GOLD column (200 × 2.1 mm, 1.9 μm; Thermo Fisher Scientific) at 40°C. The mobile phase consisted of (A) 20 mM ammonium acetate buffer (pH 7.5)-acetonitrile-methanol (70:15:15, v/v/v) and (B) 20 mM ammonium acetate buffer (pH 7.5)-acetonitrile-methanol (30:35:35, v/v/v). The following gradient program was used at a flow rate of 150 μl/min: 0–50% B for 20 min, 50–100% B for 10 min, hold 100% B for 15 min, and re-equilibrate to 100% A for 10 min. Detailed LC-MS/MS conditions are presented in supplemental Table S1.
Determination of lipid concentrations
Serum and hepatic concentrations of total cholesterol and triglycerides were measured by colorimetric assays using Cholesterol E-Test Wako and Triglyceride E-Test Wako (FUJIFILM Wako Pure Chemical Corporation), respectively. Biliary cholesterol and phospholipid concentrations were determined by Cholesterol E-Test Wako and Phospholipid C-Test Wako, respectively. Sterol and oxysterol concentrations in the liver and serum were quantified using our previously described LC-MS/MS method (25). Briefly, 5 μl of serum or 5 mg of liver tissue were incubated with internal standards in 1 N ethanolic KOH at 37°C for 1 h. Sterols were extracted with n-hexane, derivatized to picolinyl esters, and analyzed by LC-MS/MS.
Enzyme assays
Microsomes and mitochondria were prepared from livers by differential ultracentrifugation (26). Microsomal activities of BA 6β-hydroxylase and BA 7α-hydroxylase were measured as follows: microsomes (100 μg of protein) were incubated for 20 min at 37°C with 200 μM of each BA (dissolved in 10 μl of 50% acetone in water), NADPH (1.2 mM), glucose-6-phosphate (3.6 mM), 1 unit of glucose-6-phosphate dehydrogenase, and 100 mM of potassium phosphate buffer (pH 7.4) containing 0.1 mM of EDTA in a total volume of 250 μl. The incubation was stopped by the addition of 10 μl of 8.9 M aqueous KOH solution. After the addition of internal standards and 0.5 M potassium phosphate buffer (pH 7.4), BAs were extracted with Bond Elut C18 cartridges and quantified by LC-MS/MS as described above. Instead of using microsomes, recombinant rat CYP2A1 (Supersome) prepared from insect cells (Corning, NY) was used to determine BA 7α-hydroxylation.
Microsomal HMG-CoA reductase activity was measured by LC-MS/MS method, as described previously (27). The activities of microsomal cholesterol 7α-hydroxylase (CYP7A1) (28), mitochondrial cholesterol 27-hydroxylase (CYP27A1) (28), and microsomal oxysterol 7α-hydroxylase (CYP7B1) (29) were measured according to our stable-isotope dilution MS method except that LC-MS/MS was used instead of GC-MS to quantify 7α-hydroxycholesterol, 27-hydroxycholesterol, 7α,27-dihydroxycholesterol, and their isotopic variants (25). Microsomal 7α-hydroxy-4-cholesten-3-one 12α-hydroxylase (CYP8B1) activity was determined as described previously (30) except that [2H7]7α,27-dihydroxycholesterol was used as an internal standard and LC-MS/MS was employed instead of HPLC to quantify 7α,12α-dihydroxy-4-cholesten-3-one (25).
mRNA measurements
An aliquot of the liver and terminal ileum specimen were collected in RNAlater (Thermo Fisher Scientific) and stored at −80°C until RNA isolation. Total RNA was extracted using an RNeasy Plus Mini Kit (QIAGEN). Reverse transcription was performed on 4 μg of total RNA using a Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany). Real-time quantitative PCR was performed on cDNA aliquots with the FastStart DNA MasterPLUS SYBR Green I and a LightCycler (Roche). The sequences of the oligonucleotide primer pairs used to amplify mRNAs are shown in supplemental Table S2. PCR amplification began with a 10 min preincubation step at 95°C, followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 62°C for 10 s, and elongation at 72°C for 16 s. The relative concentration of the PCR product derived from the target gene was calculated by the comparative Ct method, and results were standardized to the expression of Gapdh. The specificity of each PCR product was assessed by melting curve analysis.
Determination of serum FGF15 concentrations
Serum concentrations of fibroblast growth factor 15 (FGF15) were measured using mouse FGF15 ELISA kit (catalog #MBS2700661; MyBiosource, Inc., San Diego, CA), according to the manufacturer’s instruction.
Determination of serum LPS and TNFα concentrations
Serum concentrations of lipopolysaccharides (LPSs; endotoxin) and TNFα were quantified using ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit (catalog #L00350; GenScript USA Inc., Piscataway, NJ) and LBIS Mouse TNF-α ELISA Kit (catalog #AKMTNFA-011; FUJIFILM Wako Shibayagi, Gunma, Japan), respectively, according to the manufacturers’ instructions.
Histopathological examination
An aliquot of the liver was fixed in 10% neutral buffered formalin and embedded in a paraffin block. Each paraffin block was sectioned at 3 μm and the paraffin sections were stained using hematoxylin/eosin.
Statistics
Data are expressed as the mean ± SEM. The statistical significance of differences between the results in the different groups was evaluated using the Tukey-Kramer test or the Dunnett’s test. For all analyses, significance was accepted at the level of P < 0.05. Correlations were tested by calculating parametric Pearson’s correlation coefficient, r, and nonparametric Spearman’s correlation coefficient, rs. All statistical analyses were conducted using JMP (version 10.0) software (SAS Institute, Cary, NC).
RESULTS
Cyp2a12 is responsible for BA 7α-rehydroxylation
We first explored the mouse and rat genes encoding BA 7α-hydroxylase. To confirm the usefulness of our new method, genes encoding CDCA 6β-hydroxylase [Cyp2c70 is strongly suggested as a responsible gene (16)] were also examined along with those encoding BA 7α-hydroxylase. Mouse and rat genes encoding BA 7α-hydroxylase and CDCA 6β-hydroxylase must fall under any of 102 mouse and 87 rat Cyp genes (20, 21) (Fig. 1B). Species difference, sexual dimorphism, and tissue distribution with respect to both enzymes were determined using commercially available microsomal fractions. Neither of the two enzyme activities was detected in the human liver or mouse kidney. Furthermore, sexual dimorphism of both enzyme activities was not observed in the mouse liver but was apparent in the rat liver (Fig. 1C). From the 102 mouse Cyp genes, we narrowed down to four candidate genes, Cyp2a12, Cyp2a22, Cyp2c70, and Cyp2d40 (Fig. 1D, supplemental Table S3), using previously reported orthology, tissue distribution, and sex difference data (20, 21) as well as our experimental observations (Fig. 1C).
If Cyp2c70 encodes for CDCA 6β-hydroxylase (16), the gene encoding BA 7α-hydroxylase would be either Cyp2a12, Cyp2a22, or Cyp2d40. Because mouse CYP2A12, rat CYP2A1, and hamster CYP2A9 have homologous amino acid sequences and similar testosterone 7α-hydroxylase activity (31), we incubated recombinant rat CYP2A1 with TDCA and TLCA. As a result, CYP2A1 catalyzed the 7α-rehydroxylation of TDCA and TLCA (Fig. 1E), suggesting that Cyp2a12 is responsible for BA 7α-rehydroxylation in mice. It is important to note that Cyp2a22 may also encode for BA 7α-hydroxylase in mice because it is highly homologous to Cyp2a12 (96.2% mRNA identity). However, the hepatic expression level of Cyp2a22 is extremely low (less than 3%) compared with that of Cyp2a12 (21). Therefore, we finally concluded that Cyp2a12 was primarily responsible for BA 7α-rehydroxylation in mice.
Generation of Cyp2a12−/−, Cyp2c70−/−, and Cyp2a12−/−Cyp2c70−/− mice
We generated Cyp2a12/Cyp2c70 DKO mice using the CRISPR-Cas9 system (Fig. 2A, B). Founder (F0) mice were selected by PCR genotyping (Fig. 2C, D) followed by the detection of Cas9 (supplemental Fig. S1) and direct sequencing (supplemental Table S4). Finally, #4, #5, #44, and #47 mice for Cyp2a12-null and #17, #19, and #32 mice for Cyp2c70-null were selected as F0 mice. The F0 mice were bred with WT C57BL/6J mice, and PCR genotyping followed by sequencing assay demonstrated that mice from #4, #5, and #47 lines and those from #19 line had DNA sequences at CRISPR target sites, as predicted (supplemental Table S5). Finally, a male F1 Cyp2a12+/− mouse from the #4 line and a male F1 Cyp2c70+/− mouse from the #19 line were used for subsequent studies. Amino acid sequences of mutated CYP2A12 and CYP2C70 were compared with those of the WTs (supplemental Table S6). After Cyp2a12+/−Cyp2c70+/− mice were produced, these double heterozygous animals (28 males and 28 females) were crossed, and a total of 129 mice including Cyp2a12−/−Cyp2c70+/+ (2a12KO), Cyp2a12+/+Cyp2c70−/− (2c70KO), and Cyp2a12−/−Cyp2c70−/− (DKO) mice were obtained (Fig. 2E, supplemental Fig. S2).
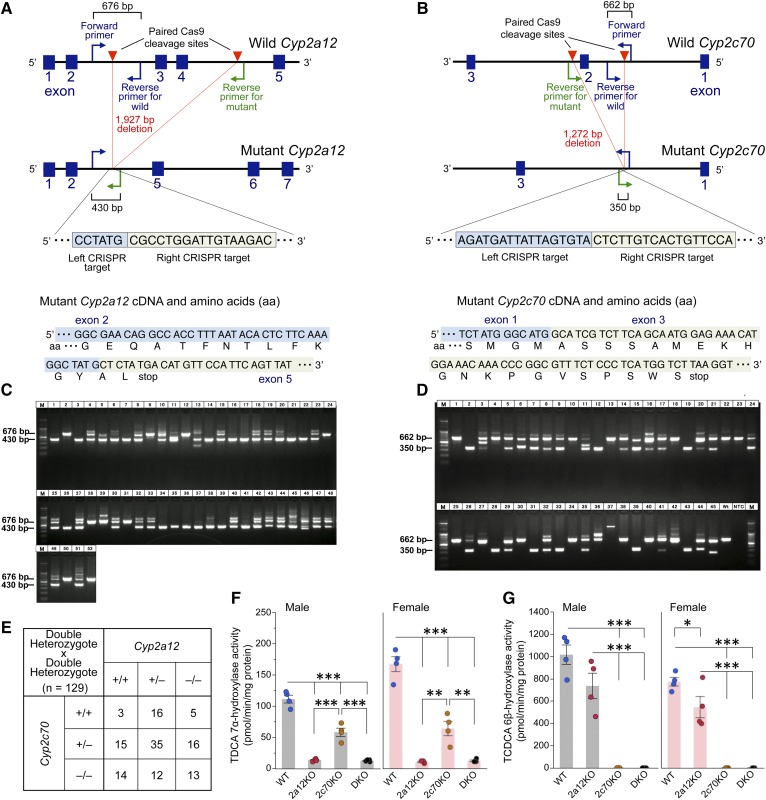
Fig. 2.
Disruption of Cyp2a12 and Cyp2c70 genes markedly reduces hepatic TDCA 7α-hydroxylase and TCDCA 6β-hydroxylase activities. A: CRISPR target site in the Cyp2a12 gene and predicted cDNA and amino acid sequences of mutant Cyp2a12. Primers were designed for genotyping of mice to detect the 1,927 bp deletion of Cyp2a12 induced by Cas9. The forward primer and reverse primer for WT amplify the WT allele (676 bp), and the forward primer and reverse primer for mutant amplify the mutant allele (430 bp). B: CRISPR target site in the Cyp2c70 gene and predicted cDNA and amino acid sequences of mutant Cyp2c70. Primers were designed for genotyping of mice to detect the 1,272 bp deletion of Cyp2c70 induced by Cas9. The forward primer and reverse primer for WT amplify the WT allele (662 bp), and the forward primer and reverse primer for mutant amplify the mutant allele (350 bp). C: PCR genotyping of Cyp2a12−/− (430 bp), Cyp2a12+/− (430 bp and 676 bp), and Cyp2a12+/+ (676 bp) in pups (n = 52) for the screening of F0 mice. D: PCR genotyping of Cyp2c70−/− (350 bp), Cyp2c70+/− (350 bp and 662 bp), and Cyp2c70+/+ (662 bp) in pups (n = 45) for the screening of F0 mice. E: Genotypes of mice (n = 129) obtained by crossing double heterozygous Cyp2a12+/−Cyp2c70+/− animals. F0 mice were bred with WT C57BL/6J mice to obtain F1 heterozygous Cyp2a12+/− and Cyp2c70+/− mice, and these were crossed to produce Cyp2a12+/−Cyp2c70+/− mice. Then, these double heterozygous mice were crossed to obtain Cyp2a12−/−Cyp2c70+/+ (2a12KO), Cyp2a12+/+Cyp2c70−/− (2c70KO), and Cyp2a12−/−Cyp2c70−/− (DKO) mice. F, G: Hepatic TDCA 7α-hydroxylase (F) and TCDCA 6β-hydroxylase (G) activities in WT, 2a12KO, 2c70KO, and DKO mice (n = 4). Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.005, and ***P < 0.0001 were considered significantly different by the Tukey-Kramer test.
To confirm that Cyp2a12 and Cyp2c70 are indeed responsible for BA 7α-rehydroxylation and CDCA 6β-hydroxylation, respectively, enzyme activities were determined in the liver of WT, 2a12KO, 2c70KO, and DKO mice. TDCA 7α-hydroxylase activities were reduced to ∼10% in 2a12KO and DKO mice compared with those in the WT mice (Fig. 2F), whereas taurochenodeoxycholic acid (TCDCA) 6β-hydroxylase activities were almost absent in 2c70KO and DKO mice (Fig. 2G).
BA composition in Cyp2a12−/−, Cyp2c70−/−, and Cyp2a12−/−Cyp2c70−/− mice
Analysis of BA composition in the liver, gallbladder, small intestine, and serum of 2a12KO, 2c70KO, and DKO mice revealed that 2a12KO induced the accumulation of DCAs, whereas 2c70KO caused a complete replacement of MCAs by CDCAs in both sexes (Fig. 3A–C; supplemental Figs. S3B, S4A, B, D). Notably, not only DCAs or CDCAs but also DCAs, CDCAs, and LCAs were all increased in the tissues of DKO mice. In feces (Fig. 3D, supplemental Fig. S4C) and cecum (supplemental Fig. S3A), almost all BAs were deconjugated, and most primary BAs were transformed into secondary BAs (ωMCA, DCA, and LCA) regardless of the genotypes.
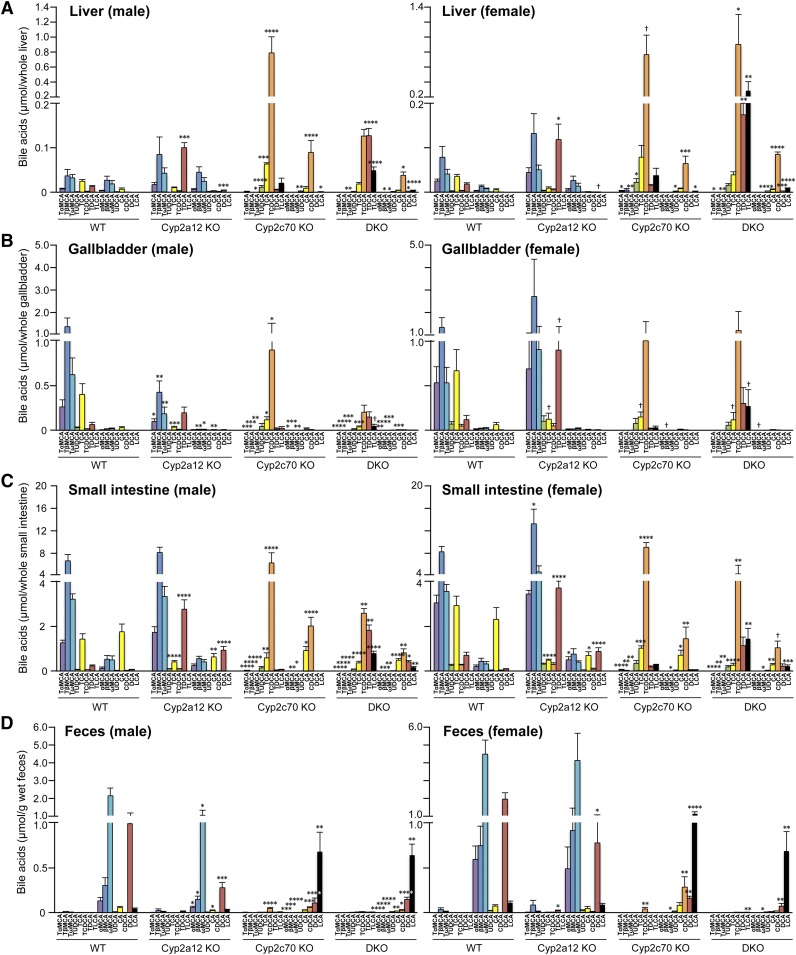
Fig. 3.
DKO mice have human-like hydrophobic BA composition. Individual organs and feces were collected from 44 mice (n = 6, 7, 4, 10, 5, 4, 4, and 4 for male WT, Cyp2a12KO, Cyp2c70KO, DKO, female WT, Cyp2a12KO, Cyp2c70KO, and DKO, respectively) aged 11.1 ± 0.8 (mean ± SD) weeks. Specific BA concentrations in the liver (A), gallbladder (B), small intestine (C), and feces (D) were analyzed. Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significantly different from BA concentrations in WT by the Dunnett’s test.
The amounts of minor BA components in the enterohepatic circulation were calculated by adding the amounts of BA in the liver, gallbladder, and small intestine. The previous report suggests that hyodeoxycholic acid (HDCA) and murideoxycholic acid (MDCA) are synthesized from LCA, and hyocholic acid (HCA) from HDCA (32). However, HDCA and MDCA pools in DKO mice were small compared with those in 2a12KO mice, although the former had a much larger LCA pool than the latter (supplemental Fig. S5). Therefore, bacterial 7α-dehydroxylation and 6-epimerization of αMCA appear to be alternate pathways for the production of MDCA and HDCA (supplemental Fig. S6). It may be possible that HCA is also produced by bacterial 6-epimerization of αMCA. However, because 2c70KO mice had a larger HCA pool than WT mice, HCA is likely to be synthesized by 6α-hydroxylation of CDCA. Sulfation of the 3α-OH position of LCA is catalyzed by sulfotransferase 2A1 expressed predominantly in female mice (32). Indeed, our results showed that TLCA-3S and LCA-3S were detected in female mice in a 2a12KO-dominant manner (supplemental Fig. S5).
Effects of BA composition on BA pool size and cholesterol metabolism
By combining the amounts of all BAs in the liver, gallbladder, and small intestine, we calculated the total BA pool in each group (Fig. 4A). The total pool size was larger in females than in males and not significantly altered in 2a12KO mice. In contrast, 2c70KO and DKO mice had a markedly reduced pool size (by ∼50%) in both sexes. BA content by organ is also shown, and hepatic BA content was significantly elevated in male 2c70KO and female DKO mice in spite of reduced total BA pool size. As a consequence of the reduced pool size, fecal BA concentrations were also decreased in 2c70KO and DKO mice (Fig. 4B). In addition, biliary proportions of cholesterol and phospholipid were significantly elevated with a decrease in the BA pool in male DKO mice (Fig. 4C).
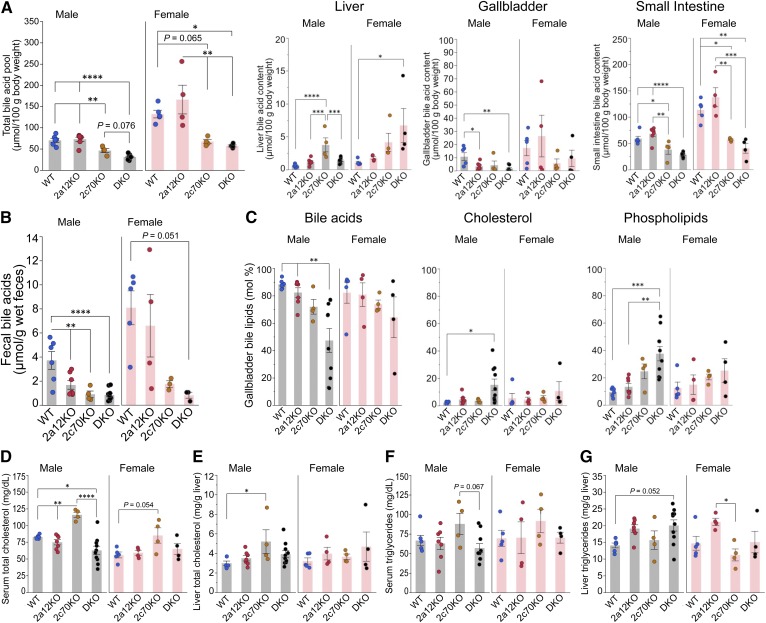
Fig. 4.
BA pool size and cholesterol metabolism are markedly affected in 2c70KO and DKO mice. A total of 44 mice (n = 6, 7, 4, 10, 5, 4, 4, and 4 for male WT, 2a12KO, 2c70KO, DKO, female WT, 2a12KO, 2c70KO, and DKO, respectively) were euthanized at 11.1 ± 0.8 (mean ± SD) weeks of age, and serum, individual organs, and feces were analyzed. A: The total amount of BAs in the enterohepatic circulation was calculated by adding total BAs in the liver, gallbladder, and small intestine. B: BA concentrations in feces. C: Proportion (mole percent) of BAs, cholesterol, and phospholipids in gallbladder bile. D–G: Total cholesterol concentrations in serum (D) and the liver (E) and total triglyceride concentrations in serum (F) and the liver (G). Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significantly different by the Tukey-Kramer test.
Serum total cholesterol concentrations increased by 40% in male 2c70KO mice and decreased by 24% in male DKO mice (Fig. 4D). Liver total cholesterol concentrations also increased by 74% in male 2c70KO mice but were unchanged in male DKO mice (Fig. 4E). In females, the total cholesterol concentrations tended to be increased only in 2c70KO mouse serum; however, this increase was not statistically significant. In contrast, no significant change in serum and liver triglyceride concentrations was observed in either of the KO groups (Fig. 4F, G).
To assess how the BA composition influences cholesterol metabolism, cholesterol-related sterol concentrations were quantified in serum and the liver. Levels of serum lathosterol and desmosterol, surrogate markers for cholesterol biosynthesis, did not change significantly among the groups (supplemental Fig. S7A). However, intestinal cholesterol absorption, as estimated by serum sitosterol and campesterol, was reduced in all the KO groups (supplemental Fig. S7A). Serum 4β-hydroxycholesterol, which is synthesized by CYP3A and a marker for CYP3A activity, was significantly elevated in DKO mice (supplemental Fig. S7A). Other oxysterols and desmosterol in the liver are physiological ligands of the LXRα (NR1H3) (33), and the only desmosterol in male DKO mice was significantly increased (supplemental Fig. S7B).
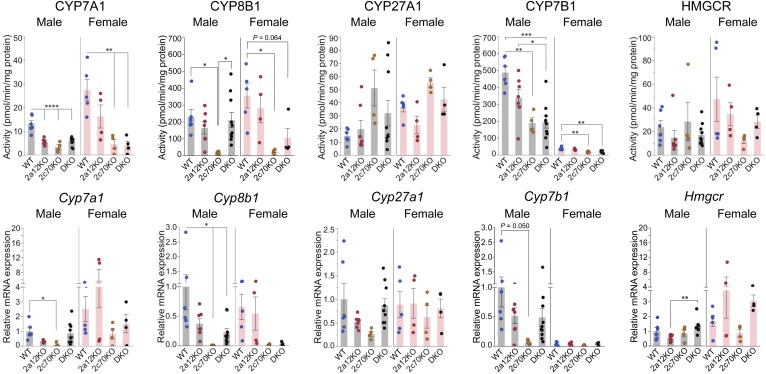
Effects of BA composition on key enzymes in BA synthesis
To clarify the reason for the reduction of BA pool size in 2c70KO and DKO mice, we measured hepatic activities of key enzymes in the BA biosynthetic pathways (Fig. 5). CYP7A1, the rate-limiting enzyme in the classic BA biosynthetic pathway, was female-dominant and decreased by more than 50% in male 2c70KO and DKO mice and by more than 80% in female 2c70KO and DKO mice. CYP8B1, an essential enzyme for CA production, was markedly decreased by ∼93% in both sexes of 2c70KO mice, but not significantly decreased in DKO mice. CYP27A1, the first enzyme in the alternative BA biosynthetic pathway, was not significantly changed among the groups. CYP7B1, an oxysterol 7α-hydroxylase in the alternative BA biosynthetic pathway, was male-dominant and was markedly decreased by ∼60% in male 2c70KO and DKO mice and by ∼50% in female 2c70KO and DKO mice. Although the hepatic mRNA expression levels of these key enzymes were not completely consistent with the enzyme activities, they were enough to indicate the change in enzyme activities. Under most physiological conditions, there is a covariation between BA biosynthesis and cholesterol biosynthesis (34). However, HMG-CoA reductase activities and mRNA expression levels of related genes were not decreased in 2c70KO and DKO mice (Fig. 5), and this result was supported by normal serum lathosterol and desmosterol concentrations (supplemental Fig. S7A). Therefore, it is suggested that cholesterol biosynthesis is preserved under reduced BA synthesis owing to the compensation for reduced cholesterol absorption (supplemental Fig. S7A).
Fig. 5.
BA biosynthesis is downregulated in 2c70KO and DKO mice. Liver samples were collected from 44 mice (n = 6, 7, 4, 10, 5, 4, 4, and 4 for male WT, 2a12KO, 2c70KO, DKO, female WT, 2a12KO, 2c70KO, and DKO, respectively) at 11.1 ± 0.8 (mean ± SD) weeks of age. RNA was extracted and hepatic microsomal and mitochondrial fractions were prepared for enzyme assays. Hepatic activities and mRNA expression levels of key enzymes in the BA and cholesterol biosynthetic pathways were determined. mRNA expression levels were standardized to those of Gapdh, and the mean expression levels in male WT mice were set to 1.0. Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significantly different by the Tukey-Kramer test.
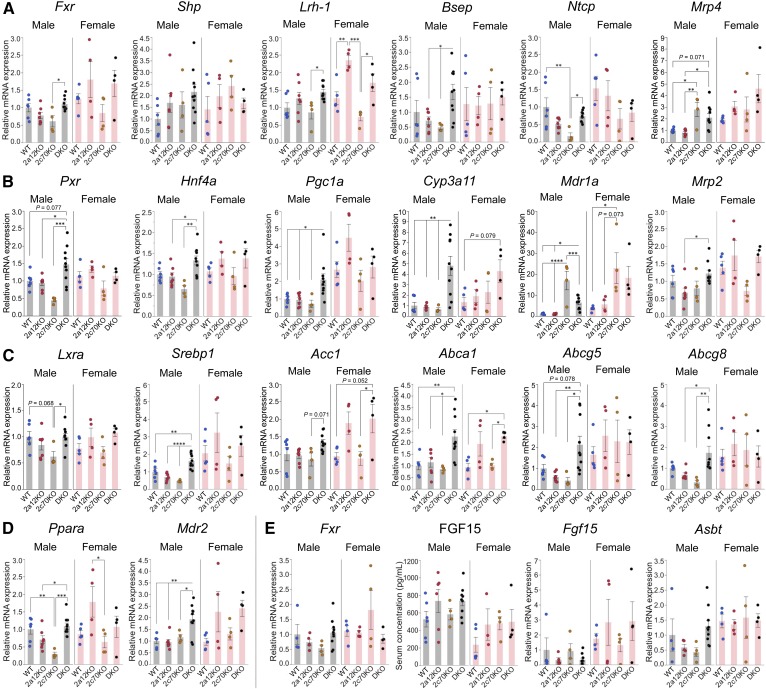
Effects of BA composition on the activities of nuclear receptors
It is established that FXR is a BA receptor controlling hepatic BA biosynthesis through the expression of a small heterodimer partner (SHP; NR0B2) in the liver and the induction of FGF15 in the small intestine (35). Because CDCA is the most potent physiological agonist (8, 9) and MCAs are antagonists of FXR (10), marked activation of FXR was expected in 2c70KO and DKO mice. However, Shp and bile salt export pump (Bsep), which must be upregulated by the activation of FXR in the liver (36), were not significantly changed in either type of mouse (Fig. 6A). Sodium-taurocholate cotransporting polypeptide (Ntcp) and multidrug resistance-associated protein (Mrp)4, which should have been downregulated by the activation of FXR (37, 38), were not suppressed in DKO mice; however, Ntcp was suppressed only in male 2c70KO mice (Fig. 6A). In addition, serum FGF15 concentrations and the expression levels of intestinal Fgf15 and another intestinal FXR target gene, apical sodium-dependent BA transporter (Asbt) (39), did not change significantly among the groups (Fig. 6E).
Fig. 6.
Downregulation of BA biosynthesis is not caused by the activation of FXR in 2c70KO and DKO mice. Serum, liver, and terminal ileum were collected from 44 mice (n = 6, 7, 4, 10, 5, 4, 4, and 4 for male WT, 2a12KO, 2c70KO, DKO, female WT, 2a12KO, 2c70KO, and DKO, respectively) at 11.1 ± 0.8 (mean ± SD) weeks of age. RNA was extracted from the liver and the terminal ileum. A: Hepatic mRNA expression levels of Fxr and related genes. B: Hepatic mRNA expression levels of Pxr and related genes. C: Hepatic mRNA expression levels of Lxra and related genes. D: Hepatic mRNA expression levels of Ppara and a related genes. E: Serum concentrations of FGF15 and mRNA expression levels of Fxr and related genes in the terminal ileum. mRNA expression levels were standardized to those of Gapdh, and the mean expression levels in male WT mice were set to 1.0. Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significantly different by the Tukey-Kramer test.
Other nuclear receptors, such as pregnane X receptor (PXR; NR1I2), PPARα (NR1C1) and LXRα, are also known to regulate Cyp7a1 expression. Hepatocyte nuclear factor 4α (HNF4α; NR2A1) complexed with PPARγ coactivator 1α (PGC1α) is necessary to transactivate Cyp7a1. Activated PXR stimulates dissociation of PGC1α from HNF4α and results in the suppression of Cyp7a1 expression (40, 41). Cyp3a11, multidrug resistance protein (Mdr)1a, and Mrp2 are all target genes for PXR and the constitutive androstane receptor (CAR; NR1I3), and Mrp4 is a target gene for CAR (42). The expression levels of these four genes (Fig. 6A, B) and the serum 4β-hydroxycholesterol concentration (supplemental Fig. S7A) strongly suggest that PXR (and maybe CAR) are activated, at least in DKO mice. In addition, the activated PXR trans-represses Shp expression (41, 43). This is probably one of the reasons why Shp is not significantly upregulated in DKO mice (Fig. 6A).
PPARα is another nuclear receptor that downregulates Cyp7a1 by interfering with the transactivation of Cyp7a1 by HNF4α (44). Mdr2 is a target gene for PPARα, and the expression was upregulated in male DKO mice (Fig. 6D), in accordance with an increased phospholipid proportion in the bile (Fig. 4C).
In contrast to FXR, PXR, and PPARα, LXRα is a positive regulator of Cyp7a1 expression in mice. Srebp1, acetyl-CoA carboxylase 1 (Acc1), Abca1, Abcg5, and Abcg8 are all target genes for LXRα. The expression levels of these five genes (Fig. 6C) and increased biliary proportion of cholesterol (Fig. 4C) suggest that LXRα is activated in male DKO mice.
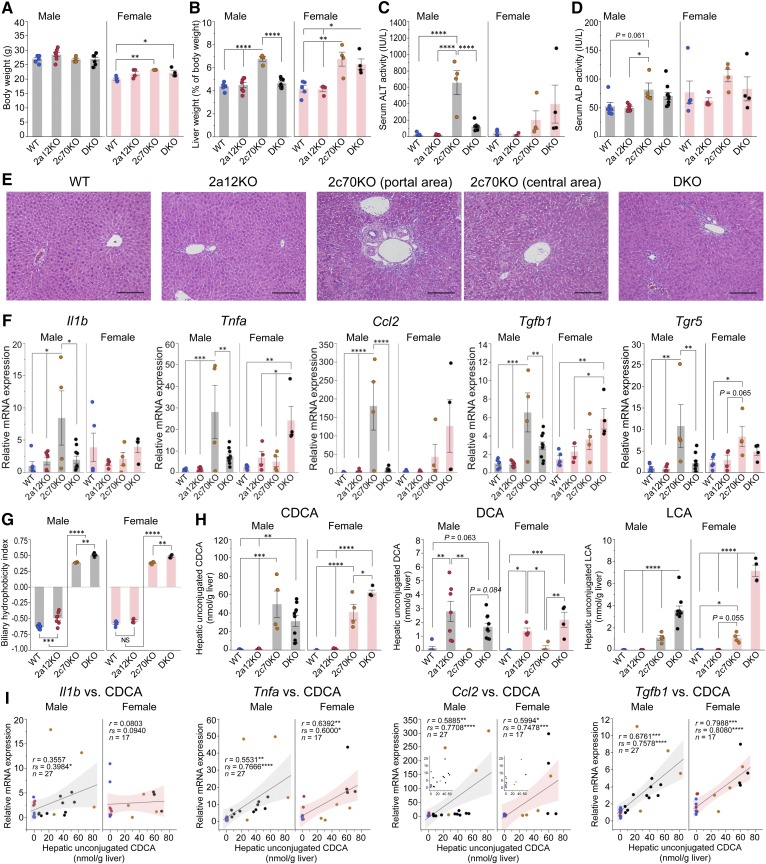
Inflammatory cytokines inhibit BA biosynthesis in 2c70KO and DKO mice
The fertility of 2c70KO mice was markedly low and that of DKO mice was slightly low compared with that of WT mice. Furthermore, unexpected death within a week after weaning (4 weeks old) was observed in 20–25% of both types of mice. However, the growth of surviving animals was good at least until 11 weeks of age (Fig. 7A). Liver weights were significantly increased in male 2c70KO mice and female 2c70KO and DKO mice (Fig. 7B), and serum ALT and ALP activities showed changes that were similar to changes in the liver weights (Fig. 7C, D). Histologic sections of liver from male 2c70KO mice revealed diffuse bile ductular reaction and necrosis of hepatocytes (∼10%) with notable infiltration of lymphocytes and neutrophils, suggesting chronic hepatocyte damage and inflammation (Fig. 7E). In male DKO mice, a slight bile ductular reaction was seen, but necrosis of hepatocytes was not observed. In female 2c70KO and DKO mice, the degree of these histologic findings was variable among individual mice depending on serum ALT levels. In contrast, liver histology showed no significant findings in 2a12KO mice.
Fig. 7.
CDCA concentrations correlate with mRNA expression levels of inflammatory cytokines and liver injury. A total of 44 mice (n = 6, 7, 4, 10, 5, 4, 4, and 4 for male WT, 2a12KO, 2c70KO, DKO, female WT, 2a12KO, 2c70KO, and DKO, respectively) were euthanized at the age of 11.1 ± 0.8 (mean ± SD) weeks, and serum and the liver were analyzed. A–D: Comparison of body weight (A), liver weight (B), and serum activities of ALT (C) and ALP (D) among genotypes. E: Representative histopathologic features of the livers from male WT, 2a12KO, 2c70KO, and DKO mice. Hematoxylin/eosin stain. Scale bars, 100 μm. F: Hepatic mRNA expression levels of inflammatory cytokines and Tgr5. mRNA expression levels were standardized to those of Gapdh, and the mean expression levels in male WT mice were set to 1.0. G: Hydrophobicity indices of total BAs in gallbladder bile. H: Concentrations of unconjugated hydrophobic BAs (CDCA, DCA, and LCA) in the liver. I: Correlations between unconjugated CDCA concentrations and mRNA expression levels of inflammatory cytokines (Il1b, Tnfa, Ccl2, and Tgfb1) in the liver. Shaded areas represent 95% confidence intervals of regression lines. Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significantly different by the Tukey-Kramer test, or significant by Pearson’s correlation coefficient (r) or Spearman’s correlation coefficient (rs).
Hepatic mRNA expression levels of the inflammatory cytokines interleukin 1β (Il1b), Tnfa, chemokine (C-C motif) ligand 2 (Ccl2), and transforming growth factor β1 (Tgfb1) were all higher (3.9, 2.5, 3.2, and 1.5 times, respectively) in female WT mice than in male WT mice (Fig. 7F). In 2c70KO and DKO mice, these cytokines were upregulated depending on serum ALT levels. It has been suggested that the activation of macrophage TGR5 regulates the production of inflammatory cytokines (11, 45–48). The expression level of hepatic Tgr5 was significantly elevated in both sexes of 2c70KO mice (Fig. 7F), but this could be due to monocyte infiltration.
Unconjugated and hydrophobic BAs in the enterohepatic circulation repress CYP7A1 via the production of regulatory cytokines by macrophages (49). Biliary BA hydrophobicity indices were calculated (50), and we found that 2c70KO and DKO mice had much higher values than WT and 2a12KO mice (Fig. 7G). However, the hydrophobicity indices were not sufficient enough to explain the difference in cytokine production among the groups. Subsequently, we compared the hepatic concentration of each unconjugated hydrophobic BA (CDCA, DCA, and LCA) among the groups and found that CDCA reflected the synthesis of cytokines the most (Fig. 7H). Indeed, hepatic unconjugated CDCA concentrations were positively correlated with mRNA expression levels of Il1b in male mice and those of Tnfa, Ccl2, and Tgfb1 in male and female mice (Fig. 7I).
DISCUSSION
In this study, we demonstrated that BA 7α-rehydroxylation in mice is primarily catalyzed by CYP2A12 and showed that the disruption of Cyp2a12 and/or Cyp2c70 results in a dramatic change in the BA composition in mice. Although the conjugated amino acids (mainly glycine conjugated in humans) and proportion of each BA are still different from those in humans, the major components and hydrophobicity of BAs in DKO mice are quite similar to those of humans.
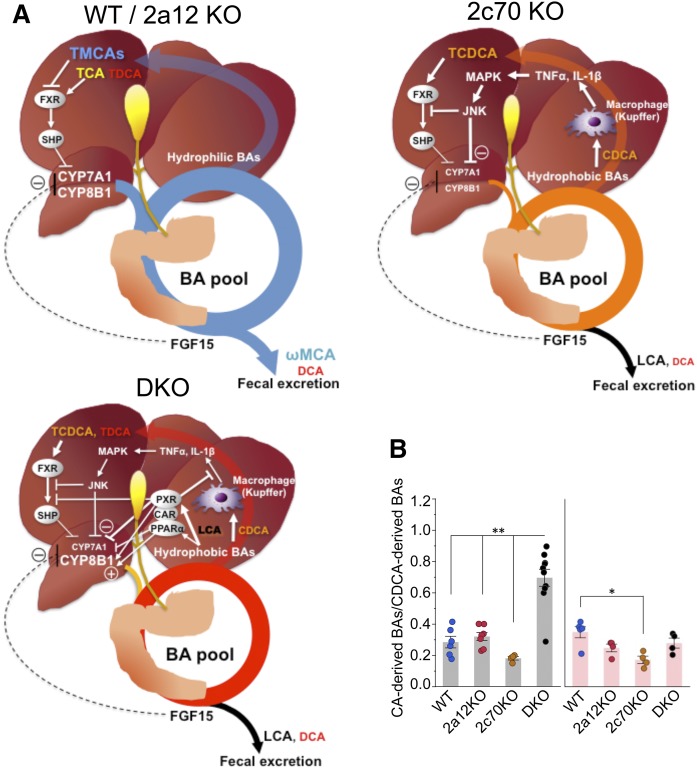
Except for the BA composition, the reduced BA pool size was characteristic of 2c70KO and DKO mice. Because CDCA is the most potent physiological agonist (8, 9) and MCAs are antagonists (10) of FXR, we expected FXR to be activated in 2c70KO and DKO mice. However, no evidence for the activation of FXR in these mice was obtained because hepatic expression levels of Shp and Bsep, ileal expression levels of Fgf15 and Asbt, and serum concentrations of FGF15 were not significantly different from those of WT mice. Instead of the activation of FXR, stimulation of the cytokine/c-Jun N-terminal kinase (JNK) signaling pathway was suggested as the major mechanism behind the inhibition of BA biosynthesis in 2c70KO mice (Fig. 8A). This FXR/SHP-independent and cytokine-mediated regulation pathway was first reported by Miyake, Wang, and Davis (49). They showed that unconjugated CDCA, but not conjugated CDCA, induces the expression of inflammatory cytokines such as IL-1β and TNFα in hepatic macrophages (Kupffer cells). These cytokines are secreted into the sinusoids and recognized by cytokine receptors on hepatic parenchymal cells, thus causing activation of MAPK and the JNK signaling pathway. The activation of the JNK pathway downregulates Cyp7a1 and Cyp8b1 by inhibiting HNF4α and/or PGC-1α by activated c-Jun (51, 52). Moreover, inflammation-activated cellular signaling pathways modulate FXR activity by posttranslational modifications (phosphorylation, acetylation, and SUMOylation) (53–55), which may deactivate FXR target genes including Shp, Bsep, and Fgf15 in 2c70KO and DKO mice. These results suggest that the inflammation-activated signaling pathways are the predominant mechanism, preferred over the FXR/SHP and FXR/FGF15 pathways, for regulating BA biosynthesis under pathological conditions.
Fig. 8.
BA metabolism in mouse models with hydrophobic BA composition. A: Compared with WT and 2a12KO mice, 2c70KO and DKO mice had reduced BA synthesis and smaller BA pool sizes. FXR was not significantly activated in 2c70KO and DKO mice. In 2c70KO mice, unconjugated CDCA induced inflammatory cytokines (TNFα, IL-1β, etc.) in hepatic macrophages (Kupffer cells). The released cytokines inhibit CYP7A1 and CYP8B1 in hepatocytes through signaling pathways, including the JNK pathway. In DKO mice, LCA directly or indirectly activated PXR, CAR, and PPARα, which coordinately downregulates CYP7A1, upregulates CYP8B1, and inhibits cytokine production in Kupffer cells. B: The ratio of CA-derived BAs (CA and DCA) to CDCA-derived BAs (CDCA, LCA, and MCAs) in enterohepatic circulation. Each column and error bar represents the mean and SEM. *P < 0.01 and **P < 0.0001 were considered significantly different by the Tukey-Kramer test.
Cyp2c-cluster-null mice lacking MCAs have already been reported (16), but the regulation of BA metabolism appears to be very different from that observed in our 2c70KO mice. In Cyp2c-cluster-null mice, hepatic mRNA expression levels of Cyp7a1 and Cyp8b1 were not significantly reduced, and the hepatic CA/CDCA ratio was much higher than that in 2c70KO mice. Fecal BA analysis showed that the deconjugation of TCDCA in Cyp2c-cluster-null mice was impaired as compared with the 2c70KO mice. Therefore, hepatic concentrations of unconjugated CDCA were higher in the 2c70KO mice than in the Cyp2c-cluster-null mice, which induces pro-inflammatory cytokines, downregulates Cyp7a1 and Cyp8b1, and reduces the CA/CDCA ratio in 2c70KO mice. Thus, the difference of BA metabolism between Cyp2c-cluster-null mice and the current 2c70KO mice may be explained by different gut microbiomes between these two mouse models.
One might think that the changes in BA composition may alter the structure of the gut microbiome that allows more uptake of intestinal LPS and pro-inflammatory cytokines from the gut. However, serum LPS and TNFα concentrations in 2c70KO mice were not significantly elevated compared with those in WT mice (supplemental Fig. S8). These results suggest that the hepatic inflammation in 2c70KO mice was not the consequence of the pro-inflammatory cytokines derived from the gut.
Very recently, De Boer et al. (56) generated an acute hepatic Cyp2c70 KO mouse model with CRISPR/Cas9-mediated somatic genome editing. In this mouse model, the liver inflammation was not severe, downregulation of Cyp8b1 was little, and the CA/CDCA ratio was high compared with our 2c70KO mice. A major difference between the acute hepatic Cyp2c70 KO mice and our whole-body 2c70KO mice is that the former have significant amounts of MCAs. As mentioned in (56), some functional CYP2C70 appears to still be present in the liver due to incomplete inactivation of the Cyp2c70 gene. Extrahepatic expression of CYP2C70 may also contribute to the production of MCAs to some extent, although Cyp2c70 is expressed almost exclusively in the liver (57).
In comparison to 2c70KO mice, DKO mice had decreased CDCAs and increased proportions of DCAs and LCAs. LCA is an agonist of PXR and regulates the detoxification of LCA (58, 59). In DKO mice, PXR appears to be activated because hepatic mRNA expression levels of the target gene Cyp3a11 and of its serum surrogate marker, 4β-hydroxycholesterol, were both elevated. Along with PXR, CAR also plays a central role in LCA detoxification (60, 61) although there is no current evidence that LCA is an agonist of CAR. On the other hand, LCA may be an agonist of PPARα (62), and the expression levels of Mdr2, a target gene of PPARα, were upregulated in DKO mice. It is known that the activation of PXR (40) and PPARα (44) downregulates CYP7A1, whereas the activation of PXR (63), CAR (64), and PPARα (65) upregulates CYP8B1 and an increase in the ratio of CA to CDCA+βMCA (65). In fact, CYP8B1 activity (Fig. 5) and the ratio of CA-derived BAs to CDCA-derived BAs (Fig. 8B) were higher in DKO mice than in 2c70KO mice. In addition, SHP is suppressed by LCA depending on PXR and CAR (61), and the activation of PXR may suppress the immune response in the liver (66). These data suggest that BA biosynthesis in DKO mice is primarily regulated by the activation of PXR/CAR/PPARα (Fig. 8A).
Apart from the basic characteristics of the regulation of BA pool size in 2c70KO and DKO mice, sex-related differences in BA metabolism (67–69) were also found in our KO mice. The most notable sex difference was that liver damage, as evaluated by liver weight, serum ALT activity, liver histopathology, and hepatic expression levels of cytokines, was obvious in male 2c70KO mice but significantly attenuated in male DKO mice. In contrast, liver damage in female 2c70KO mice was not as much as that observed in male 2c70KO mice; however, the damage was not attenuated in female DKO mice. One of the reasons for this difference is that female DKO mice have higher hepatic unconjugated CDCA concentrations than male DKO mice (Fig. 7H). Because the upregulation of CYP8B1 in female DKO mice is not as much as that in male DKO mice (Fig. 5), the production of CDCA is not inhibited like it is in male DKO mice (Fig. 8B, supplemental Fig. S4).
It is noteworthy that liver damage was significantly attenuated in male DKO mice compared with that in male 2c70KO mice. As mentioned above, we hypothesize that the major mechanisms behind this attenuation were the reduction of hepatic unconjugated CDCA concentrations and the suppression of inflammatory cytokine production (66) through the activation of PXR/CAR/PPARα. Because LCAs and DCAs, potent agonists of TGR5 (11), increased in DKO mice, the roles of macrophage TGR5 in the regulation of cytokine production may also be important. However, there is no consensus regarding the fact that the activation of macrophage TGR5 leads to anti-inflammatory effects (11, 45–48). In our female DKO mice, liver damage and cytokine production were not reduced, although the concentrations of LCAs and DCAs were higher than in male DKO mice (Fig. 3A). Therefore, it is unlikely that the activation of TGR5 was the primary cause of the improvement in male DKO mice.
In the present study, we obtained additional new findings regarding BA metabolism. First, conjugated BAs are much better substrates for CYP2A12 and CYP2C70 than unconjugated BAs in mice. Second, CYP2C70 metabolizes CDCA to αMCA and ursodeoxycholic acid (UDCA) to βMCA, but the accumulation of UDCA is much less than that of CDCA in 2c70KO mice, suggesting that in mice, most βMCA is synthesized from CDCA via αMCA but not via UDCA. Third, MDCA and HDCA are synthesized from LCA by CYP3A. However, DKO mice have reduced pools of MDCA and HDCA, although they have a markedly enlarged LCA pool and levels of activated CYP3A. Therefore, transformations of αMCA by intestinal bacteria appear to be a major route to MDCA and HDCA in mice (supplemental Fig. S6).
BAs are biological detergents participating in the digestion and absorption of lipids and are physiological regulators of multiple receptors. The potencies of BAs as detergents and regulators are determined by their molecular structures that are controlled by the liver; they are also affected by gut microbiota. Therefore, the marked difference in the BA composition between mice and humans suggests difficulty in extrapolating the results of mouse experiments to humans. Recent studies on the roles of BAs in mousse models of colorectal (70) and liver cancers (12, 71) and metabolic diseases (72, 73) are very informative. It would be intriguing if the same conclusions can be obtained when our DKO mice are used instead of conventional mice. In addition, the development of more pathophysiologically relevant mouse models for hepatobiliary diseases, including nonalcoholic steatohepatitis (74), primary biliary cholangitis (75), and primary sclerosing cholangitis (76), are desired. The utilization of mice with human-like BA composition may serve this purpose.
In summary, we identified principal enzymes responsible for the differences in BA composition between mice and humans and established and characterized Cyp2a12/Cyp2c70 DKO mice with human-like BA composition. These DKO mice could be a useful model for investigating the roles of hydrophobic and hydrophilic BAs in various human diseases.
Supplementary Material
Acknowledgments
The authors thank Sachiko Murayama, Hiroki Maeda, Raku Kato, Hideto Takahashi, Yoshihisa Ikeda, Takahiro Machiura, and Takashi Ueda (Charles River Laboratories Japan, Inc.) for the breeding and genotyping of KO mice.
Footnotes
Abbreviations:
- Asbt
- apical sodium-dependent bile acid transporter
- ALP
- alkaline phosphatase
- ALT
- alanine transaminase
- BA
- bile acid
- Bsep
- bile salt export pump
- CA
- cholic acid
- CAR
- constitutive androstane receptor
- Ccl2
- chemokine (C-C motif) ligand 2
- CDCA
- chenodeoxycholic acid
- CYP
- cytochrome P450
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP8B1
- 7α-hydroxy-4-cholesten-3-one 12α-hydroxylase
- DCA
- deoxycholic acid
- DKO
- double KO
- FGF15
- fibroblast growth factor 15
- HCA
- hyocholic acid
- HDCA
- hyodeoxycholic acid
- HNF4α
- hepatocyte nuclear factor 4α
- Il1b
- interleukin 1β
- JNK
- c-Jun N-terminal kinase
- LCA
- lithocholic acid
- LCA-3S
- lithocholic acid 3-sulfate
- LPS
- lipopolysaccharide
- MCA
- muricholic acid
- MDCA
- murideoxycholic acid
- Mdr
- multidrug resistance protein
- Mrp
- multidrug resistance-associated protein
- Ntcp
- sodium-taurocholate cotransporting polypeptide
- PGC1α
- PPARγ coactivator 1α
- PXR
- pregnane X receptor
- SHP
- small heterodimer partner
- TCDCA
- taurochenodeoxycholic acid
- TDCA
- taurodeoxycholic acid
- Tgfb1
- transforming growth factor β1
- TGR5
- Takeda G protein-coupled receptor 5
- TLCA
- taurolithocholic acid
- TLCA-3S
- taurolithocholic acid 3-sulfate
- UDCA
- ursodeoxycholic acid
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grants 15K15300 (T.H.), 17H04167 (A.H.), 18K08017 (T.H.), and 18K07920 (J.I.). The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Thakare R., Alamoudi J. A., Gautam N., Rodrigues A. D., and Alnouti Y.. 2018. Species differences in bile acids I. Plasma and urine bile acid composition. J. Appl. Toxicol. 38: 1323–1335. [DOI] [PubMed] [Google Scholar]
- 2.Schaap F. G., Trauner M., and Jansen P. L.. 2014. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11: 55–67. [DOI] [PubMed] [Google Scholar]
- 3.Rudling M. 2016. Understanding mouse bile acid formation: Is it time to unwind why mice and rats make unique bile acids? J. Lipid Res. 57: 2097–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fickert P., and Wagner M.. 2017. Biliary bile acids in hepatobiliary injury - what is the link? J. Hepatol. 67: 619–631. [DOI] [PubMed] [Google Scholar]
- 5.Li J., and Dawson P. A.. 2019. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 1865: 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schölmerich J., Becher M. S., Schmidt K., Schubert R., Kremer B., Feldhaus S., and Gerok W.. 1984. Influence of hydroxylation and conjugation of bile salts on their membrane-damaging properties–studies on isolated hepatocytes and lipid membrane vesicles. Hepatology. 4: 661–666. [DOI] [PubMed] [Google Scholar]
- 7.Kitani K., Kanai S., Sato Y., and Ohta M.. 1994. Tauro α-muricholate is as effective as tauro β-muricholate and tauroursodeoxycholate in preventing taurochenodeoxycholate-induced liver damage in the rat. Hepatology. 19: 1007–1012. [PubMed] [Google Scholar]
- 8.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., and Shan B.. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. [DOI] [PubMed] [Google Scholar]
- 9.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 10.Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H. U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., and Backhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 11.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278: 9435–9440. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto S., Loo T. M., Atarashi K., Kanda H., Sato S., Oyadomari S., Iwakura Y., Oshima K., Morita H., Hattori M., et al. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 499: 97–101. [Erratum. 2014. Nature. 506: 396.] [DOI] [PubMed] [Google Scholar]
- 13.Louis P., Hold G. L., and Flint H. J.. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12: 661–672. [DOI] [PubMed] [Google Scholar]
- 14.Meir K., Kitsberg D., Alkalay I., Szafer F., Rosen H., Shpitzen S., Avi L. B., Staels B., Fievet C., Meiner V., et al. 2002. Human sterol 27-hydroxylase (CYP27) overexpressor transgenic mouse model. Evidence against 27-hydroxycholesterol as a critical regulator of cholesterol homeostasis. J. Biol. Chem. 277: 34036–34041. [DOI] [PubMed] [Google Scholar]
- 15.Makino I., and Nakagawa S.. 1978. Changes in biliary lipid and biliary bile acid composition in patients after administration of ursodeoxycholic acid. J. Lipid Res. 19: 723–728. [PubMed] [Google Scholar]
- 16.Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J., and Gonzalez F. J.. 2016. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheer N., Kapelyukh Y., Chatham L., Rode A., Buechel S., and Wolf C. R.. 2012. Generation and characterization of novel cytochrome P450 Cyp2c gene cluster knockout and CYP2C9 humanized mouse lines. Mol. Pharmacol. 82: 1022–1029. [DOI] [PubMed] [Google Scholar]
- 18.Murakami K., and Okuda K.. 1981. Purification and characterization of taurodeoxycholate 7α-monooxygenase in rat liver. J. Biol. Chem. 256: 8658–8662. [PubMed] [Google Scholar]
- 19.Kunne C., Acco A., Hohenester S., Duijst S., de Waart D. R., Zamanbin A., and Oude Elferink R. P.. 2013. Defective bile salt biosynthesis and hydroxylation in mice with reduced cytochrome P450 activity. Hepatology. 57: 1509–1517. [DOI] [PubMed] [Google Scholar]
- 20.Nelson D. R. 2005. Gene nomenclature by default, or BLASTing to Babel. Hum. Genomics. 2: 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renaud H. J., Cui J. Y., Khan M., and Klaassen C. D.. 2011. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol. Sci. 124: 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon J. W., and Ruddle F. H.. 1981. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 214: 1244–1246. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino Y., Mizuno S., Kato K., Mizuno-Iijima S., Tanimoto Y., Ishida M., Kajiwara N., Sakasai T., Miwa Y., Takahashi S., et al. 2017. Simple generation of hairless mice for in vivo imaging. Exp. Anim. 66: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami M., Iwamoto J., Honda A., Tsuji T., Tamamushi M., Ueda H., Monma T., Konishi N., Yara S., Hirayama T., et al. 2018. Detection of Gut Dysbiosis due to Reduced Clostridium Subcluster XIVa Using the Fecal or Serum Bile Acid Profile. Inflamm. Bowel Dis. 24: 1035–1044. [DOI] [PubMed] [Google Scholar]
- 25.Honda A., Miyazaki T., Ikegami T., Iwamoto J., Yamashita K., Numazawa M., and Matsuzaki Y.. 2010. Highly sensitive and specific analysis of sterol profiles in biological samples by HPLC-ESI-MS/MS. J. Steroid Biochem. Mol. Biol. 121: 556–564. [DOI] [PubMed] [Google Scholar]
- 26.Honda A., Salen G., Matsuzaki Y., Batta A. K., Xu G., Leitersdorf E., Tint G. S., Erickson S. K., Tanaka N., and Shefer S.. 2001. Differences in hepatic levels of intermediates in bile acid biosynthesis between Cyp27–/– mice and CTX. J. Lipid Res. 42: 291–300. [PubMed] [Google Scholar]
- 27.Honda A., Mizokami Y., Matsuzaki Y., Ikegami T., Doy M., and Miyazaki H.. 2007. Highly sensitive assay of HMG-CoA reductase activity by LC-ESI-MS/MS. J. Lipid Res. 48: 1212–1220. [DOI] [PubMed] [Google Scholar]
- 28.Honda A., Salen G., Shefer S., Batta A. K., Honda M., Xu G., Tint G. S., Matsuzaki Y., Shoda J., and Tanaka N.. 1999. Bile acid synthesis in the Smith-Lemli-Opitz syndrome: effects of dehydrocholesterols on cholesterol 7α-hydroxylase and 27-hydroxylase activities in rat liver. J. Lipid Res. 40: 1520–1528. [PubMed] [Google Scholar]
- 29.Hirayama T., Honda A., Matsuzaki Y., Miyazaki T., Ikegami T., Doy M., Xu G., Lea M., and Salen G.. 2006. Hypercholesterolemia in rats with hepatomas: increased oxysterols accelerate efflux but do not inhibit biosynthesis of cholesterol. Hepatology. 44: 602–611. [DOI] [PubMed] [Google Scholar]
- 30.Honda A., Salen G., Matsuzaki Y., Batta A. K., Xu G., Leitersdorf E., Tint G. S., Erickson S. K., Tanaka N., and Shefer S.. 2001. Side chain hydroxylations in bile acid biosynthesis catalyzed by CYP3A are markedly up-regulated in Cyp27–/– mice but not in cerebrotendinous xanthomatosis. J. Biol. Chem. 276: 34579–34585. [DOI] [PubMed] [Google Scholar]
- 31.Kurose K., Tohkin M., Ushio F., and Fukuhara M.. 1998. Cloning and characterization of Syrian hamster testosterone 7α-hydroxylase, CYP2A9. Arch. Biochem. Biophys. 351: 60–65. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., and Klaassen C. D.. 2010. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51: 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., et al. 2006. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281: 27816–27826. [DOI] [PubMed] [Google Scholar]
- 34.Björkhem I., and Åkerlund J-E.. 1988. Studies on the link between HMG-CoA reductase and cholesterol 7α-hydroxylase in rat liver. J. Lipid Res. 29: 136–143. [PubMed] [Google Scholar]
- 35.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 36.Ananthanarayanan M., Balasubramanian N., Makishima M., Mangelsdorf D. J., and Suchy F. J.. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276: 28857–28865. [DOI] [PubMed] [Google Scholar]
- 37.Denson L. A., Sturm E., Echevarria W., Zimmerman T. L., Makishima M., Mangelsdorf D. J., and Karpen S. J.. 2001. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 121: 140–147. [DOI] [PubMed] [Google Scholar]
- 38.Renga B., Migliorati M., Mencarelli A., Cipriani S., D’Amore C., Distrutti E., and Fiorucci S.. 2011. Farnesoid X receptor suppresses constitutive androstane receptor activity at the multidrug resistance protein-4 promoter. Biochim. Biophys. Acta. 1809: 157–165. [DOI] [PubMed] [Google Scholar]
- 39.Neimark E., Chen F., Li X., and Shneider B. L.. 2004. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 40: 149–156. [DOI] [PubMed] [Google Scholar]
- 40.Li T., and Chiang J. Y. L.. 2005. Mechanism of rifampicin and pregnane X receptor (PXR) inhibition of human cholesterol 7α-hydroxylase gene (CYP7A1) transcription. Am. J. Physiol. Gastrointest. Liver Pathol. 288: G74–G84. [DOI] [PubMed] [Google Scholar]
- 41.Pavek P. 2016. Pregnane X Receptor (PXR)-Mediated Gene Repression and Cross-Talk of PXR with Other Nuclear Receptors via Coactivator Interactions. Front. Pharmacol. 7: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolson A. H., and Wang H.. 2010. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv. Drug Deliv. Rev. 62: 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T., and Chiang J. Y.. 2006. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4α and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab. Dispos. 34: 756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrapodi M., and Chiang J. Y.. 2000. Peroxisome proliferator-activated receptor α (PPARα) and agonist inhibit cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J. Lipid Res. 41: 514–520. [PubMed] [Google Scholar]
- 45.Högenauer K., Arista L., Schmiedeberg N., Werner G., Jaksche H., Bouhelal R., Nguyen D. G., Bhat B. G., Raad L., Rauld C., et al. 2014. G-protein-coupled bile acid receptor 1 (GPBAR1, TGR5) agonists reduce the production of proinflammatory cytokines and stabilize the alternative macrophage phenotype. J. Med. Chem. 57: 10343–10354. [DOI] [PubMed] [Google Scholar]
- 46.Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., et al. 2016. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 45: 802–816. [DOI] [PubMed] [Google Scholar]
- 47.Lou G., Ma X., Fu X., Meng Z., Zhang W., Wang Y. D., Van Ness C., Yu D., Xu R., and Huang W.. 2014. GPBAR1/TGR5 mediates bile acid-induced cytokine expression in murine Kupffer cells. PLoS One. 9: e93567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Z., Zhou J., Zhao S., Tian C., Wang P., Xu C., Chen Y., Cai W., and Wu J.. 2016. Chenodeoxycholic acid activates NLRP3 inflammasome and contributes to cholestatic liver fibrosis. Oncotarget. 7: 83951–83963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyake J. H., Wang S. L., and Davis R. A.. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J. Biol. Chem. 275: 21805–21808. [DOI] [PubMed] [Google Scholar]
- 50.Heuman D. M. 1989. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 30: 719–730. [PubMed] [Google Scholar]
- 51.Li T., Jahan A., and Chiang J. Y.. 2006. Bile acids and cytokines inhibit the human cholesterol 7α-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jahan A., and Chiang J. Y.. 2005. Cytokine regulation of human sterol 12α-hydroxylase (CYP8B1) gene. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G685–G695. [DOI] [PubMed] [Google Scholar]
- 53.Kemper J. K. 2011. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Biochim. Biophys. Acta. 1812: 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balasubramaniyan N., Luo Y., Sun A. Q., and Suchy F. J.. 2013. SUMOylation of the farnesoid X receptor (FXR) regulates the expression of FXR target genes. J. Biol. Chem. 288: 13850–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byun S., Jung H., Chen J., Kim Y. C., Kim D. H., Kong B., Guo G., Kemper B., and Kemper J. K.. 2019. Phosphorylation of hepatic farnesoid X receptor by FGF19 signaling-activated Src maintains cholesterol levels and protects from atherosclerosis. J. Biol. Chem. 294: 8732–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Boer J. F., Verkade E., Mulder N. L., de Vries H. D., Huijkman N. C., Koehorst M., Boer T., Wolters J. C., Bloks V. W., van de Sluis B., et al. A human-like bile acid pool induced by deletion of Cyp2c70 modulates effects of farnesoid X receptor activation in mice. J. Lipid Res. Epub ahead of print. September 10, 2019; doi:10.1194/jlr.RA119000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graves J. P., Gruzdev A., Bradbury J. A., DeGraff L. M., Edin M. L., and Zeldin D. C.. 2017. Characterization of the tissue distribution of the mouse Cyp2c subfamily by quantitative PCR analysis. Drug Metab. Dispos. 45: 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., Mackenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. 2001. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 98: 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie W., Radominska-Pandya A., Shi Y., Simon C. M., Nelson M. C., Ong E. S., Waxman D. J., and Evans R. M.. 2001. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. USA. 98: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J., Huang W., Qatanani M., Evans R. M., and Moore D. D.. 2004. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J. Biol. Chem. 279: 49517–49522. [DOI] [PubMed] [Google Scholar]
- 61.Uppal H., Toma D., Saini S. P., Ren S., Jones T. J., and Xie W.. 2005. Combined loss of orphan receptors PXR and CAR heightens sensitivity to toxic bile acids in mice. Hepatology. 41: 168–176. [DOI] [PubMed] [Google Scholar]
- 62.Bock K. W. 2012. Human UDP-glucuronosyltransferases: feedback loops between substrates and ligands of their transcription factors. Biochem. Pharmacol. 84: 1000–1006. [DOI] [PubMed] [Google Scholar]
- 63.Honda A., Ikegami T., Nakamuta M., Miyazaki T., Iwamoto J., Hirayama T., Saito Y., Takikawa H., Imawari M., and Matsuzaki Y.. 2013. Anticholestatic effects of bezafibrate in patients with primary biliary cirrhosis treated with ursodeoxycholic acid. Hepatology. 57: 1931–1941. [DOI] [PubMed] [Google Scholar]
- 64.Beilke L. D., Aleksunes L. M., Holland R. D., Besselsen D. G., Beger R. D., Klaassen C. D., and Cherrington N. J.. 2009. Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metab. Dispos. 37: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gafvels M., Einarsson C., and Alexson S. E.. 2000. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. J. Biol. Chem. 275: 28947–28953. [DOI] [PubMed] [Google Scholar]
- 66.Sun M., Cui W., Woody S. K., and Staudinger J. L.. 2015. Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug Metab. Dispos. 43: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turley S. D., Schwarz M., Spady D. K., and Dietschy J. M.. 1998. Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets. Hepatology. 28: 1088–1094. [DOI] [PubMed] [Google Scholar]
- 68.Fu Z. D., Csanaky I. L., and Klaassen C. D.. 2012. Gender-divergent profile of bile acid homeostasis during aging of mice. PLoS One. 7: e32551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uppal H., Saini S. P., Moschetta A., Mu Y., Zhou J., Gong H., Zhai Y., Ren S., Michalopoulos G. K., Mangelsdorf D. J., et al. 2007. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 45: 422–432. [DOI] [PubMed] [Google Scholar]
- 70.Fu T., Coulter S., Yoshihara E., Oh T. G., Fang S., Cayabyab F., Zhu Q., Zhang T., Leblanc M., Liu S., et al. 2019. FXR regulates intestinal cancer stem cell proliferation. Cell. 176: 1098–1112.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., Agdashian D., Terabe M., Berzofsky J. A., Fako V., et al. 2018. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 360: eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., Liu J., Deng Y., Xia J., Chen B., et al. 2018. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 24: 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang C., Xie C., Li F., Zhang L., Nichols R. G., Krausz K. W., Cai J., Qi Y., Fang Z. Z., Takahashi S., et al. 2015. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 125: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farrell G., Schattenberg J. M., Leclercq I., Yeh M. M., Goldin R., Teoh N., and Schuppan D.. 2019. Mouse models of nonalcoholic steatohepatitis: toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 69: 2241–2257. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka A., Leung P. S. C., and Gershwin M. E.. 2018. Evolution of our understanding of PBC. Best Pract. Res. Clin. Gastroenterol. 34–35: 3–9. [DOI] [PubMed] [Google Scholar]
- 76.Nakamoto N., Sasaki N., Aoki R., Miyamoto K., Suda W., Teratani T., Suzuki T., Koda Y., Chu P. S., Taniki N., et al. 2019. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 4: 492–503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.