Abstract
Epithelial resident memory T (eTRM) cells serve as sentinels in barrier tissues to guard against previously encountered pathogens. How eTRM cells are generated has significant implications for efforts to elicit their formation through vaccination or prevent it in autoimmune disease. Here we show that during immune homeostasis, the cytokine TGF-β epigenetically conditions resting naive CD8+ T cells and prepares them for the formation of eTRM cells in a mouse model of skin vaccination. Naive T cell conditioning occurs in lymph nodes (LNs), but not in the spleen, through MHC I-dependent interactions with peripheral tissue-derived migratory dendritic cells (DCs) and depends on DC-expression of TGF-β-activating αV-integrins. Thus, the pre-immune T cell repertoire is actively conditioned for a specialized memory differentiation fate through signals restricted to LNs.
One Sentence Summary
Naive T cells are trained in lymph nodes in their resting state to stand guard in tissues as local memory cells in the future.
T cell immune responses contract following successful control of an infection, but various types of memory cells persist and provide enhanced protection from re-encounters with the respective pathogen. Some memory cells continually recirculate through lymphoid and non-lymphoid tissues via the blood and the lymph, whereas so-called tissue-resident memory T (TRM) cells adopt states of more permanent local residence (1). This latter population includes CD8+ cells that co-express the tissue residency markers CD69 and CD103/αE-integrin and populate the epithelial layers of environmental barrier tissues, such as the skin (2, 3). These epithelial TRM (eTRM) cells form a highly sensitive sentinel system and respond to re-encounter with their cognate pathogen-derived antigen with direct antiviral or antimicrobial effector activities. Additionally, eTRM cells trigger local inflammatory responses that efficiently recruit circulating memory and other immune cells to rapidly contain the infection (4–6). eTRM cells are thought to develop locally at their site of residence from uncommitted memory precursors, which acquire the ability to respond to TGF-β through coordinated downregulation of the T-box factors T-bet and Eomesodermin (Eomes). TGF-β, in turn, induces the expression of Cd103 and other tissue residency-associated genes and enables long-term persistence of eTRM cells in the epithelium (7–11).
TGF-β is a pleiotropic cytokine with a broad range of functions in the immune system. It is widely expressed and secreted in its latent form. As such, it is abundant in most tissues where it is bound to cell surfaces and extracellular matrix via “milieu” factors such as glycoprotein-A repetitions predominant protein (GARP) or latent TGF-β binding proteins (LTBPs), respectively. The cytokine acquires its biological activity only upon simultaneous binding by integrins, which allows for the generation of force to distort the TGF-β prodomain. This, in turn, triggers the release of the growth factor domain that binds to TGF-β receptors (12). TGF-β activity in the immune system is enabled by αV-integrins expressed both by hematopoietic and non-hematopoietic cells (13). Keratinocyte-expressed αVβ6 and αVβ8 integrins, for instance, activate the pool of TGF-β that maintains the stable, long-term residence of Langerhans cells and eTRM cells in skin (14). However, the relevant microanatomical sites of CD8+ T cell exposure to TGF-β as well as the cellular mechanisms underlying its activation, which serve to initiate and drive eTRM cell differentiation during the formation of T cell memory, remain unknown.
Efficient eTRM cell formation in skin requires dendritic cell-expression of αV-integrins
In order to test whether αV-expressing dendritic cells (DCs) activate TGF-β to facilitate eTRM cell differentiation, we crossed mice with loxP-flanked Itgav alleles (15) to Cd11cCre BAC-transgenic mice, where Cre recombinase is active in >95% of all conventional DCs (16). In the resulting Cd11cCre × Itgavfl/fl mice (hereafter referred to as “αV-ΔDC” mice), αV protein was absent from the majority of DCs (Fig. S1A–B). The deletion of αV did not disrupt DC homeostasis, since the proportion of various DC populations in skin and skin-draining LNs was unchanged compared to Cd11cCre × Itgav+/+ littermate control (“WT”) mice (Fig. S1C–D). Mice whose DCs lack the β8 integrin that pairs with αV to form the primary TGF-β-activating αVβ8 heterodimer expressed in immune cells showed signs of immune activation, possibly resulting from the impaired formation of peripheral regulatory T (Treg) and T helper 17 (Th17) cells in the intestine (17, 18). Similarly, young αV-ΔDC mice showed moderate hypercellularity, expansion of CD44hi CD62Llo CD8+ T cells, and enhanced cytokine expression by CD4+ cells in spleen, but not in LNs. There was also an increase in serum IgE and IgG in these mice (Fig. S1E–J). However, no histological signs of inflammation were observed in the colon and skin (Fig. S1K), and mice displayed no signs of disease, such as weight loss, until at least 6 months of age.
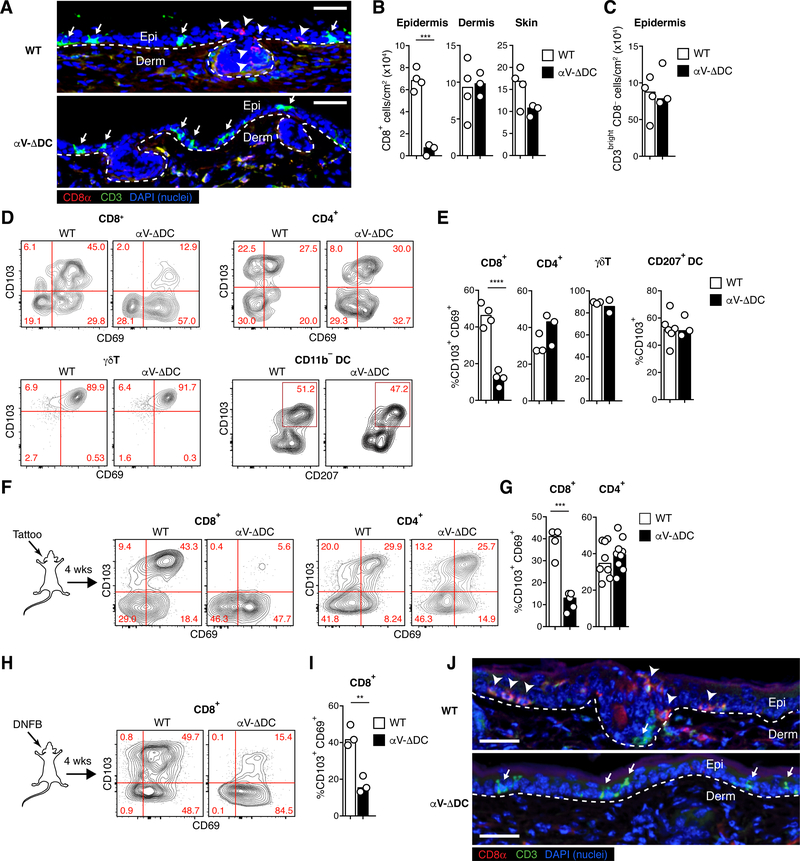
Strikingly, already in young αV-ΔDC mice, histological examination revealed a pronounced and selective loss of CD8+ T cells in the epidermis, but not in the dermis, whereas the density of CD3bright CD8− dendritic epidermal T cells (DETCs) in the skin epithelium was unchanged (Fig. 1A–C). The frequency of CD4+ T cells, which are only found within the dermis in mice (19), was also unchanged (Fig. S2A). Accordingly, there was a selective decrease in the frequency of skin CD8+ T cells co-expressing CD69 and CD103/αE-integrin, indicative of epidermal residence. By contrast, CD4+ and γΔT cells co-expressing CD69 and CD103 as well as DCs expressing CD103 were present at frequencies similar to control mice (Fig. 1D–E and fig. S2B). An even more pronounced defect in eTRM cell differentiation was evident following skin inflammation induced by either mechanical skin irritation using a tattooing device (20) or treatment with the contact sensitizer dinitrofluorobenzene (DNFB) (Fig. 1F–J and fig. S2C–D). Thus, the expression of TGF-β-activating αV-integrins on DCs is critical for the efficient formation of CD8+ eTRM cells in the stratified epithelium, but not other local CD103+ immune cells in skin.
Fig. 1. Defective epidermal TRM cell formation in absence of αV integrins on DCs.
(A) Histological cross-sections from ear skin of 8–10 week-old αV-ΔDC and WT littermate control mice. Arrowheads indicate CD8+ CD3dim TRM cells in the epidermis of WT, but not αV-ΔDC mice, whereas arrows indicate CD3bright DETCs present in both strains. Dashed lines indicate the dermal–epidermal border. Scale bar = 50 μm. Epi: epithelium, Derm: dermis. (B, C) Density of CD8+ T cells in epidermis, dermis, and total skin (B) and of CD3bright CD8− T cells in epidermis (C). Data are means and replicates and representative of two independent experiments. Each symbol represents the mean of 3–4 values per animal obtained from different histological sections. (D, E) Expression of the tissue residence marker CD103 (as well as CD69 on T cells) on indicated immune cell subsets (Thy1+ CD8β+ T cells, Thy1+ CD4+ T cells, Thy1+ TCRd+ γΔT cells, CD11c+ MHC II+ CD11b− DCs) from skin of αV-ΔDC and WT littermate control mice. Data are means and replicates and representative of two independent experiments. (F-I) Expression of CD69 and CD103 on skin CD8+ or CD4+ T cells 4 weeks after sterile inflammation induced by mechanical irritation with a tattooing device (F, G) or topical DNFB treatment (H, I) in 8–10-week-old mice. Data are means and replicates and representative of five (F, G) or three (H, I) independent experiments. (J) Ear skin 4 weeks after DNFB treatment. Note the enrichment of CD8β+ CD3dim eTRM cells (arrow-heads) in WT mice, but their absence in the epithelium of αV-ΔDC, which is instead still populated by CD3bright DETC (arrows). Data are representative of two animals per group. Scale bar = 50 μm. **/***/****: p<0.01/p<0.001/p<0.0001 (Two-tailed unpaired Student’s t-tests in (B), (C), (E), (G), (I)).
Naive T cells are preconditioned for eTRM cell formation by DC prior to antigenic priming
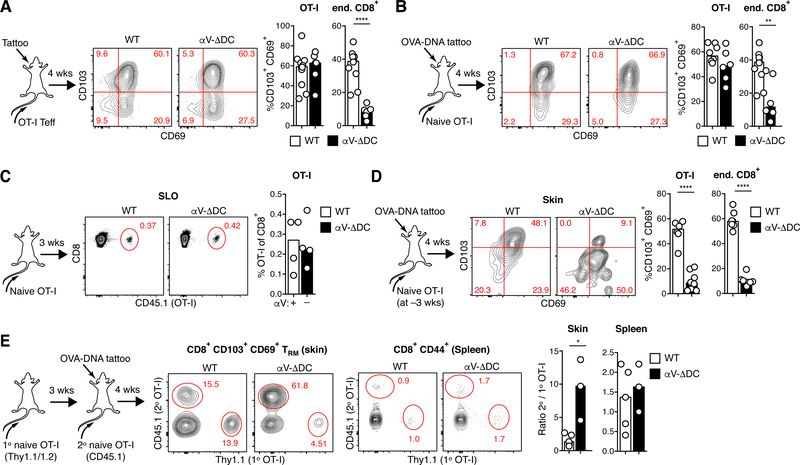
Terminal eTRM cell differentiation is thought to be initiated locally in skin from uncommitted memory precursors (7, 11). We therefore hypothesized that dermal αV-expressing DCs may enhance eTRM cell formation by generating a subepithelial pool of active TGF-β to which CD8+ memory precursor T cells would then be exposed upon extravasation into the inflamed skin. However, when we intravenously injected ex vivo activated ovalbumin (OVA)-specific effector CD8+ T (TEFF) cells from OT-I TCR transgenic mice, they differentiated into CD69+ CD103+ eTRM cells in the inflamed skin of αV-ΔDC and WT hosts with similar efficiencies (Fig. 2A and fig. S2E). Thus, the local activation of TGF-β by αV-expressing DCs in skin is not required for eTRM cell formation.
Fig. 2. DC-expressed αV-integrins condition naive CD8+ T cells for epithelial TRM cell formation prior to vaccine-induced activation.
(A) 106 Thy1.1/2 congenic ex vivo activated OT-I effector cells were intravenously injected into Thy1.2 αV-ΔDC or WT recipients, whose ears were simultaneously inflamed through tattoo injury. After 4 weeks, CD69 and CD103 expression was assessed on transferred OT-I and host CD8+ T cells in skin. Data are means and replicates and representative of six independent experiments. (B) 105 CD44lo CD62Lhi naive OT-I cells were adoptively transferred into αV-ΔDC or WT recipients, whose ears were simultaneously tattooed with OVA-encoding plasmid DNA to prime OT-I in skin-draining LNs. After 4 weeks, CD69 and CD103 expression was assessed on transferred OT-I and host CD8+ T cells in skin. Data are means and replicates and representative of five independent experiments. (C, D) 106 CD44lo CD62Lhi naive OT-I cells were adoptively transferred into αV-ΔDC or WT recipients. Three weeks later, OT-I cell frequency in pooled LNs and spleen (SLO) was determined in some animals (C), while the remaining animals were vaccinated by ear tattoo with OVA-encoding plasmid DNA (D). After 4 weeks, CD69 and CD103 expression was assessed on transferred OT-I and host CD8+ T cells in skin. Data are means and replicates and representative of four independent experiments. (E) 106 Thy1.1/2 congenic CD44lo CD62Lhi naive OT-I cells (1° OT-I) were adoptively transferred into αV-ΔDC or WT recipients. Three weeks later, a second batch of 105 CD45.1 congenic OT-I cells (2° OT-I) was transferred into the same recipients and mice were vaccinated by ear tattoo with OVA-encoding plasmid DNA. After 4 weeks, the ratios of OT-I derived from the second and the first injected batch was assessed in the pools of CD69+ CD103+ TRM cells in skin and of CD44+ CD8+ T cells in spleen. Data are means and replicates and representative of two independent experiments. */**/****: p<0.05/p<0.01/p<0.0001 (Two-tailed unpaired Student’s t-tests in (A-E)).
Alternatively, we hypothesized that CD8+ T cells may be exposed to TGF-β activated by DCs during antigenic priming in skin-draining LNs, which might endow them with enhanced eTRM cell differentiation or skin homing capacity (e.g. through the TGF-β-dependent induction of T cell ligands for selectins expressed on inflamed skin endothelium (21)). However, the transfer of naive OT-I T cells and vaccination with OVA-expressing plasmid DNA also generated similar frequencies of eTRM cells in the skin of αV-ΔDC and WT hosts (Fig. 2B and fig. S2F).
Since TGF-β-activation by αV-expressing DCs was important for eTRM cell formation, but was not critical during T cell priming in draining LNs or during terminal differentiation at the effector site, we considered other contexts in which T cells may be exposed to TGF-β. Resting T cells depend on interactions with self-antigen presented by DCs for their maintenance (22). Thus, we hypothesized that αV-expressing DCs may at the same time, during immune homeostasis, expose naive CD8+ T cells in secondary lymphoid organs (SLOs) to active TGF-β and thereby precondition them for enhanced eTRM cell formation upon eventual foreign antigen encounter. We therefore transferred naive OT-I cells into αV-ΔDC mice where such preconditioning might be lost, or into WT mice where it would be sustained. After 3 weeks, at which time OT-I cell frequency among naive CD8+ T cells in SLOs was comparable in both strains (Fig. 2C), we vaccinated mice with OVA-DNA via the ear skin. Four weeks following this regimen, we observed a reduction in the frequency of CD103+ OT-I eTRM cells at vaccinated skin sites in αV-ΔDC (but not in WT hosts), which matched the reduction of eTRM cells derived from the hosts’ endogenous polyclonal CD8+ T cell populations (Fig. 2D). To control for potentially variable T cell priming efficiency in individual animals, we also transferred two batches of naive OT-I cells into either αV-ΔDC or WT mice. The first and second batches were injected 3 weeks and immediately prior to OVA skin vaccination, respectively (Fig. 2E). Cells from both batches formed eTRM cells at nearly equal efficiencies in WT mice, suggesting that their capacity to form eTRM cells had not changed over the course of 3 weeks. By contrast, cells from the first batch were tenfold less efficient at forming eTRM cells than those from the second batch in αV-ΔDC hosts. Thus, the capacity of naive CD8+ T cells to form eTRM cells declines over time when DCs do not express αV-integrins. No such bias in favor of more recently transferred cells was observed among circulating memory T cells in spleen (Fig. 2E).
Naive cell preconditioning is required for optimal eTRM cell-mediated antiviral protection
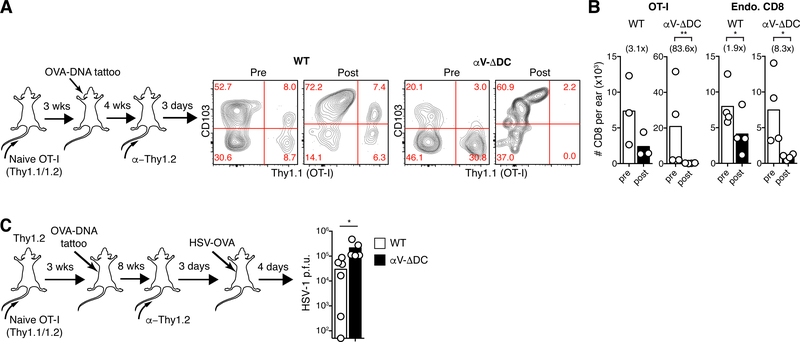
A hallmark of eTRM cells is their resistance to antibody-mediated depletion (10, 19, 23). Accordingly, when we injected T cell-depleting antibodies into WT or αV-ΔDC animals seeded with skin memory cells derived from naive OT-I cells transferred 3 weeks prior to skin vaccination, we observed a loss of CD103− dermal TRM cells and a corresponding enrichment of CD103+ eTRM cells in all animals, as expected (Fig. 3A). The total numbers of remaining endogenous and OT-I CD8+ T cells in skin were each only moderately (2–3-fold) reduced in WT hosts. By contrast, the decrease in endogenous CD8+ T cell numbers was much more pronounced (eightfold) and OT-I cells were essentially eliminated in αV-ΔDC hosts (Fig. 3B). Thus, even the few eTRM cells that had formed from “deconditioned” naive precursors in αV-ΔDC hosts were less stably resident within the epithelium. Consequently, eTRM cells derived from deconditioned precursors were less effective at protecting animals from challenge with OVA-expressing HSV-1 (Fig. 3C). Thus, continual exposure to DCs expressing TGF-β-activating αV-integrins sustained the capacity of naive CD8+ T cells to efficiently form protective epidermal TRM cells. By contrast, the formation of circulating memory cells appeared to be unaffected.
Fig. 3. Residual eTRM cells formed in absence of DC-expressed αV-integrins are unstable and provide reduced antiviral protection.
(A-B) 106 CD44lo CD62Lhi naive OT-I cells were adoptively transferred into αV-ΔDC or WT recipients. Three weeks later, animals were vaccinated by ear tattoo with OVA-encoding plasmid DNA. After an additional 4 weeks, circulating T cells were depleted with α-Thy1.2 mAbs, and then, 3 d later, the expression of CD103 (A) and absolute numbers in skin of total CD8+ T cells (B) derived from OT-I (left) or from polyclonal endogenous cells (right) were determined. Number in parentheses indicate fold decrease in αV-ΔDC mice Data are means and replicates and representative of two independent experiments. (C) Mice were seeded with OT-I eTRM cells as in (A), but in the flank. Eight weeks later, circulating T cells were depleted, and an additional 3 d later, animals were exposed at vaccinated skin sites with HSV-OVA. Four days later, local viral titers were determined in skin. Data are medians and biological replicates, each replicate representing the mean of a technical triplicate, and representative of two independent experiments. */**: p<0.05/p<0.01 (Two-tailed paired Student’s t-tests in (B), *: p<0.05 (Mann–Whitney U test in (C)).
αV-expressing DCs regulate chromatin accessibility in preconditioned naive CD8+ T cells
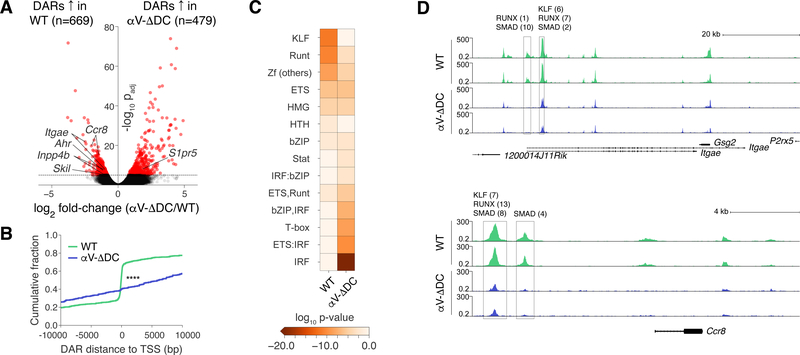
We hypothesized that exposure to active TGF-β may induce a permissive epigenetic state in resting CD8+ T cells that enhances their capacity to form eTRM cells upon activation. For instance, TGF-β may prepare genes relevant to eTRM cell formation for more rapid expression. We therefore purified CD44lo naive CD8+ T cells from young WT and αV-ΔDC mice and investigated their genome-wide chromatin accessibility through the assay for transposase accessible chromatin using high-throughput sequencing (ATAC-seq). Among the 13 Mb of detected accessible chromatin (<0.04% of the mouse genome), 6.8% was differentially accessible, with 496 kb (3.8%) being more accessible in naive CD8+ T cells from WT and 391 kb (3.0%) being more accessible in cells from αV-ΔDC mice. Peak calling and merging yielded low numbers of differentially accessible regions (DARs) of DNA both in cells from WT and from αV-ΔDC hosts (Fig. 4A and table S1). Interestingly, DARs in WT cells clustered around gene transcription start sites (TSSs), indicating that these were potentially expressed or ready to be expressed. By contrast, DARs in αV-ΔDC cells were more frequently located distal to the TSS of the most proximal gene (Fig. 4B). These latter DARs were enriched in binding motifs for transcription factors (TFs) associated with cellular activation, such as interferon regulatory factors (IRFs) and T-box factors. By contrast, DARs in T cells from WT mice were enriched in binding motifs for the Krüppel-like family of TFs (KLFs) and other zinc finger (Zf) DNA-binding proteins, as well as for Runx family TFs, including Runx3 (Fig. 4C and fig. S3). Runx3 has recently been identified as an important positive regulator of eTRM cell differentiation (11). Of note, Runx TFs interact with and enable the functions of TGF-β-regulated Smad TFs (24). Thus, the role of Runx3 during eTRM cell differentiation may, at least in part, be based on its requirement for TGF-β-mediated gene regulation.
Fig. 4. Resting naive CD8+ T cells are epigenetically conditioned by TGF-β signals in secondary lymphoid tissues.
(A) Volcano plot of chromatin accessibility changes between CD44lo naive CD8+ T cells from WT and αV-ΔDC mice, considering merged DARs. (B) Cumulative distributions of distances from DARs in cells from WT (green) and αV-ΔDC (blue) animals to the closest transcription start site. The two-sample Kolmogorov–Smirnov was used to compare cumulative distributions. (C) Enrichment of indicated transcription factor binding motif families in DARs. (D) Normalized chromatin accessibility near the Itgae (top) and Ccr8 (bottom) loci. The rectangles mark detected DARs. All analyses were performed on 2 mice per group with similar results. ****: p<0.0001 (Two-sample Kolmogorov–Smirnov test in (B)).
Enhanced accessibility of eTRM cell genes in preconditioned naive CD8+ T cells
The presence of Runx motifs in regions accessible in naive cells from WT but not αV-ΔDC mice also suggests that some epigenetic changes driving eTRM cell differentiation may already occur in resting T cells prior to activation. Indeed, among a set of 21 signature genes upregulated during eTRM cell differentiation in skin (7, 25), five were proximal to DARs in WT CD8+ T cells, including Itgae (encoding CD103), Ahr, and Ccr8 (Fig. 4A). By contrast, the tissue egress-promoting gene S1pr5, which is downregulated in eTRM cells (7, 25), contained a DAR upstream of its TSS in cells from αV-ΔDC mice (fig. S4A). DARs proximal to eTRM cell genes mostly contained multiple clustered Runx, KLF, and Smad motifs (Fig. 4D and fig. S4A). Since KLFs also interact with Smad TFs and mediate TGF-β signaling (26), this pattern of motif enrichment suggested the involvement of TGF-β in the epigenetic changes we observed in naive CD8+ T cells. Accordingly, genes proximal to DARs in WT cells were most strongly associated with the TGF-β pathway and with cell cycle arrest among biological processes (fig. S4B, C).
The preconditioned state is reversible and actively maintained in secondary lymphoid organs
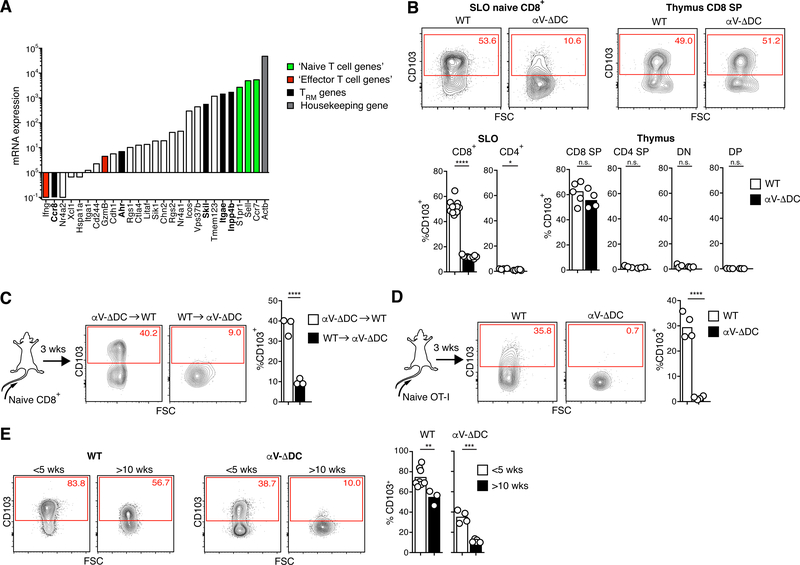
Based on our analysis of publicly available gene expression data sets deposited through the Immunological Genome Project (www.immgen.org), and using the transcript levels of well characterized naive and effector cells genes as a reference, some eTRM cell genes proximal to DARs in WT cells were expressed in naive CD8+ T cells (Itgae, Skil, Inpp4b), whereas others were not (Ahr, Ccr8) (Fig. 5A). Among the former, Itgae was highly expressed as integrin αE/CD103 protein on naive CD8+, but not on CD4+ T cells in WT mice (Fig. 5B). Since CD103 expression by naive CD8+ T cells has been shown to depend on TGF-β signals (27), we used it as a surrogate marker of TGF-β-mediated conditioning. As predicted from our ATAC-seq analysis, CD103 expression was substantially reduced on naive CD8+ T cells in αV-ΔDC mice, but not on CD8 single-positive (SP) thymocytes from the same animals (Fig. 5B). Since thymocyte expression of CD103 also depends on the cooperation of Runx3 and TGF-β signals (28), normal thymocyte expression may reflect alternative local mechanisms of TGF-β-activation, such as αV-integrins expressed by thymic epithelial or other stromal cells.
Fig. 5. CD103 is actively maintained on naive CD8+ T cells through αV-integrin-expressing DCs.
(A) ImmGen database RNA-seq gene expression analysis of naive CD8+ T cells. Green bars denote selected genes known to be expressed at the protein level in naive T cells, red bars denote selected genes known to be expressed at very low levels or not to be expressed in naive T cells. Black bars denote skin eTRM cell genes having greater accessibility in naive CD8+ T cells from WT as compared to αV-ΔDC mice. White bars denote other skin eTRM cell genes not differentially accessible in cells from WT compared to αV-ΔDC mice. (B) Expression of CD103 on CD44lo CD62Lhi naive CD4+ and CD8+ T cells (pooled from LNs and spleen) and on thymocyte subsets from αV-ΔDC and WT littermate control mice. Data are means and replicates and representative of four independent experiments. (C) 106 naive polyclonal CD8+ T cells from αV-ΔDC were adoptively transferred into CD45.1 congenic C57BL/6 mice or vice versa and re-isolated 3 weeks later from pooled LNs and spleens for analysis of CD103 expression. Data are means and replicates and representative of three independent experiments. (D) 106 naive OT-I T cells were adoptively transferred into αV-ΔDC or WT hosts and isolated from pooled LNs and spleens 3 weeks later for analysis of CD103 expression. Data are means and replicates and representative of three independent experiments. (E) CD103 expression on CD44lo naive CD8+ T cells from the peripheral blood of WT and αV-ΔDC mice at less than 5 or more than 10 wks of age. Data are means and replicates. */**/***/****: p<0.05/p<0.01/p<0.001/p<0.0001 (Two-tailed unpaired Student’s t-tests in (B-E)).
The defect in CD103 expression on naive CD8+ T cells from αV-ΔDC mice was not cell-intrinsic, since treatment with TGF-β in vitro induced their expression of CD103 at similar levels as in WT T cells (fig. S5B), and transfer of CD103lo cells from αV-ΔDC mice expressed CD103 upon adoptive transfer into WT hosts (Fig. 5C). Conversely, both polyclonal as well as clonal OT-I T cells from αV-sufficient mice lost CD103 expression in αV-ΔDC hosts, indicating that expression must be actively maintained following induction (Fig. 5C–D). Importantly, activated T cells transiently downregulate TGF-β receptors (29). Accordingly, CD103 expression in CD8+ T cells was first lost upon antigenic priming in LNs prior to its eventual re-expression at considerably higher levels in the eTRM cells that form in skin (fig. S5C–E). Thus, following eTRM cell egress from thymus, DC-dependent exposure to active TGF-β in secondary lymphoid organs (SLOs) renders multiple genes important in their differentiation, including Itgae, more accessible in naive CD8+ T cells. Despite the transient loss of gene expression upon T cell activation, at least of Cd103, this enhanced accessibility potentially facilitates the accelerated transcriptional activation and more efficient formation of eTRM cells.
Preconditioning occurs in lymph nodes, but not in the spleen
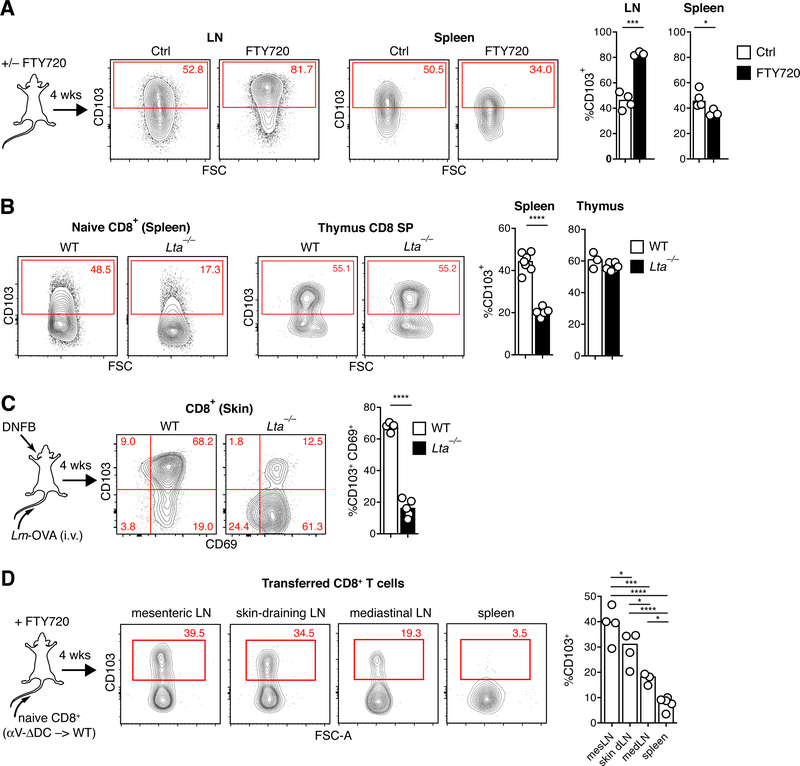
Since the majority of CD8 SP thymocytes expressed CD103, it was possible that some CD103+ naive CD8+ T cells were recent thymic emigrants (RTEs) that transiently retained CD103 expression induced in the thymus independently of αV-expressing DCs. Indeed, in WT mice younger than 5 weeks of age, in which the majority of peripheral T cells are RTEs (30), CD103 expression on naive CD8+ T cells was higher than in mice older than 10 weeks of age, when the thymic output rate has stabilized. This difference was even more pronounced in αV-ΔDC mice, in which naive CD8+ T cells included a distinct CD103hi population that was absent in adult mice (Fig. 5E). To further assess the magnitude of the contribution of RTEs to the CD103hi population of naive CD8+ T cells in adult WT mice, we treated them with the functional S1P-receptor antagonist FTY720. This blocks lymphocyte tissue egress and thereby “traps” T cells within their respective tissue locations, including the thymus (31). If CD103hi naive CD8+ T cells were predominantly RTEs, this treatment would be predicted to produce a decrease in CD103 expression in SLOs. However, CD103 expression did not decrease, but further increased on naive CD8+ T cells trapped in LNs. By contrast, CD103 expression decreased in cells trapped in the spleen (Fig. 6A). Thus, RTEs do not appear to make a large contribution to the CD103hi naive CD8+ T cell pool in adult mice. Furthermore, CD103 expression in peripheral T cells is preferentially induced and sustained in LNs, but not in the spleen. To test this hypothesis in an independent model, we analyzed lymphotoxin-α-deficient (Lta−/−) mice, which lack LNs (32). Naive CD8+ T cells in these animals expressed low levels of CD103 comparable to αV-DC mice (Fig. 6B). Expression was however normal in Lta−/− CD8 SP thymocytes and could be restored in splenic Lta−/− naive CD8+ T cells by exposure to TGF-β in vitro (Fig. 6B and fig. S6A). Thus, there was no cell-intrinsic defect obstructing TGF-β-driven induction of CD103. TEFF-like cells did not accumulate in spleens of Lta−/− mice, indicating that this feature of αV-ΔDC mice was not directly caused by a lack of exposure of naive CD8+ T cells to TGF-β-activating αV+ DC in LNs (fig. S6B).
Fig. 6. Naive CD8+ T cell preconditioning occurs in lymph nodes, but not spleens.
(A) C57BL/6 mice were treated with FTY720 to block lymphocyte egress from all lymphoid tissues. After 4 weeks of continued treatment, naive CD8+ T cells in LNs and spleen were analyzed for CD103 expression. Data are means and replicates and representative of six independent experiments. (B) CD103 expression in naive CD8+ T cells in spleens and thymi of WT and Lta−/− mice. Data are means and replicates and representative of two independent experiments. (C) Frequency of CD69+ CD103+ eTRM cells in skin 4 weeks after ‘prime and pull’ immune challenge through i.v. injection with OVA-expressing Listeria monocytogenes to produce a circulating TEFF cell pool (“prime”) followed by DNFB+ treatment of ear skin (“pull”) of WT an Lta−/− mice. Data are means and replicates and representative of four independent experiments. (D) CD103 expression of naive CD8+ T cells from αV-ΔDC donors in indicated SLOs of continuously FTY720-treated WT hosts 4 weeks following adoptive transfer. Data are means and replicates and representative of two independent experiments. */**/***/****: p<0.05/p<0.01/p<0.001/p<0.0001 (Two-tailed unpaired Student’s t-tests in (A-C), One-way ANOVA in (D)).
Immune responses to skin challenge are predicted to be impaired in Lta−/− mice given their lack of LNs. Therefore, in order to assess eTRM cell formation in these animals, we resorted to a “prime and pull” approach, whereby systemic T cell activation is combined with locally induced tissue inflammation in order to elicit TRM cell formation (33, 34). Four weeks following intravenous (i.v.) injection with OVA-expressing Listeria monocytogenes and skin treatment with DNFB, the frequency of CD103+ skin eTRM cells was reduced to levels comparable to Lta−/− mice and αV-ΔDC mice (Fig. 6C). Thus, low CD103 expression reflective of impaired naive CD8+ T cell conditioning in Lta−/− mice is also linked to a profound reduction in the ability to form eTRM cells in skin upon immune challenge. To further assess whether LNs draining different tissues varied in their capacity for T cell conditioning, we transferred CD103lo naive CD8+ T cells from αV-ΔDC mice into FTY720-treated WT hosts. Four weeks later, CD8+ T cells in mesenteric LNs showed the most pronounced increase in expression of CD103, followed by CD8+ T cells in skin-draining LNs. By contrast, the induction of CD103 was moderate in lung-draining mediastinal LNs and absent in spleen (Fig. 6D). Thus, αV-expressing DCs in LNs are specialized to precondition naive CD8+ T cells to differentiate into eTRM cells.
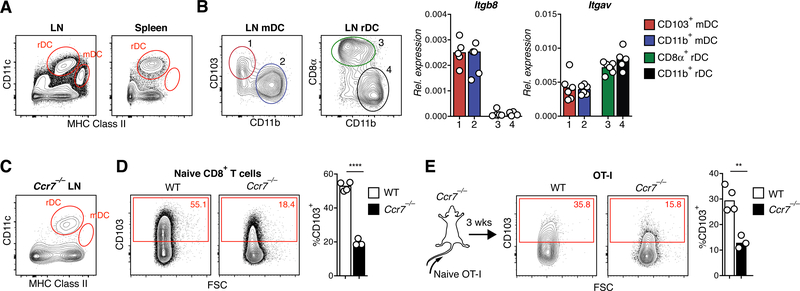
Preconditioning activity in αVβ8-expressing migratory, but not resident DCs
A key difference between LNs and spleen is the presence in the former of a CD11cint MHC IIhi migratory DC (mDC) population (Fig. 7A) and (35). These cells continually traffic from non-lymphoid tissues to draining LNs via the lymph. Although both mDCs and resident DCs (rDCs), including CD11b+, CD103+, and CD8+ subsets, expressed αV, only mDCs expressed the β8-integrin chain (Fig. 7B) to form the αVβ8 heterodimer that enables them to activate TGF-β (17). Naive CD8+ T cells expressed CD103 at similarly low levels in mice lacking β8-integrin in DCs (Cd11cCre × Itgb8fl/f, or “β8-ΔDC” mice) as in αV-ΔDC mice, although this defect was again less pronounced in very young mice with the highest frequencies of RTEs (fig. S6C–D). Reduced eTRM cell formation in the skin of β8-ΔDC mice further confirmed a role specifically for αVβ8 in T cell preconditioning (fig. S6E). However, the reduction in skin eTRM cells was less severe than in αV-ΔDC mice, suggesting that either TGF-β-independent functions of DC-expressed αV-integrins, or TGF-β-activation through other αV-heterodimers expressed by DC, such as αVβ3 and αVβ5, contribute to TGF-β activity on CD8+ T cells.
Fig. 7. αVβ8-expressing migratory DCs precondition naive CD8+ T cells for epithelial TRM cell formation in skin.
(A) CD11cint MHC IIhi migratory DCs in LNs are absent in spleens of C57BL/6 mice. (B) CD103+ and CD11b+ subsets of migratory DCs as well as CD8+ and CD11b+ subsets of resident DCs were purified by FACS and analyzed for expression of αV and β8 integrin mRNA by RT-qPCR. Data are means and replicates and representative of two independent experiments. (C) CD11cint MHC IIhi migratory DCs are absent in LNs of Ccr7−/− mice. (D) CD103 expression in naive CD8+ T cells in pooled LNs and spleen of Ccr7−/− mice. Data are means and replicates and representative of four independent experiments. (E) 106 naive OT-I T cells were adoptively transferred into Ccr7−/− or C57BL/6 hosts. CD103 expression on naive OT-I cells from pooled LNs and spleen was analyzed 3 weeks later. Data are means and replicates and representative of two independent experiments. **/****: p<0.01/p<0.0001 (Two-tailed unpaired Student’s t-tests in (D-E)).
To further test if naive T cell preconditioning through TGF-β depends on mDCs, we analyzed CCR7-deficient (Ccr7−/−) mice, in which DC trafficking from peripheral tissues to draining LNs is defective and in which mDCs are absent from both the spleen and LNs (Fig. 7C and Ref. (36)). Similar to αV-DC and Lta−/− mice, CD103 expression was reduced on naive CD8+ T cells in Ccr7−/− mice (Fig. 7D). However, since Ccr7−/− T cells may also be excluded from TGF-β-mediated preconditioning based on their impaired ability to enter LNs, we transferred CCR7-sufficient OT-I T cells that can enter LNs into Ccr7−/− hosts. These cells also lost CD103 expression (Fig. 7E), confirming the critical role for CCR7-dependent migration of αV-expressing DCs from the skin to the LNs to precondition naive CD8+ T cells to differentiate into eTRM cells in the skin.
eTRM cell preconditioning occurs through MHC I-dependent DC–T cell interactions
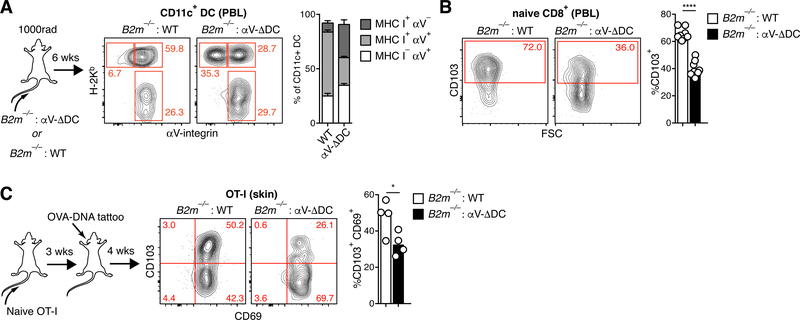
Naive CD8+ T cell survival and responsiveness depends on non-cognate, but MHC I-dependent, interactions with DCs (37). The question arose whether resting CD8+ T cells receive TGF-β-signals during MHC-I-dependent physical interactions with αV-integrin expressing DCs, or if DCs produce a pool of active TGF-β that is available indiscriminately to all T cells. We therefore generated mixed bone marrow chimeras (BMCs) from MHC I-deficient B2m−/− and αV-ΔDC donors in order to segregate expression of these two proteins on DCs and to test whether self-peptide/MHC I ligands and TGF-β-activating αV-integrins need to be co-expressed by the same DC for naive T cell preconditioning to be effective. This strategy was only partially effective since about a third of DCs still co-expressed MHC I and αV, possibly as a result of transfer of membrane proteins between DCs (38). The remaining two thirds were divided evenly between MHC I-expressing and αV-expressing subsets (Fig. 8A). However, even a partial reduction in the frequency of DCs co-expressing MHC I and αV resulted in a decrease in the frequency of CD103+ naive CD8+ T cells, as compared to control BMCs (Fig. 8B). Accordingly, the capacity of OT-I cells transferred into B2m−/−:αV-ΔDC BMCs to form eTRM cells in the skin upon vaccination was impaired (Fig. 8C). Thus, αV-expressing DCs in LNs do not produce a public pool of active TGF-β available to all T cells but rather present it to naive CD8+ T cells discretely during MHC I-dependent interactions.
Fig. 8. eTRM cell preconditioning of naive CD8+ T cells occurs through homeostatic MHC I-dependent DC interactions.
(A) Lethally irradiated CD45.1 congenic mice were injected with 1:1 mixtures of either WT or αV-ΔDC and MHC I-deficient B2m−/− bone marrow cells. Eight weeks later, the expression of H-2Kb and αV on CD45.1+ CD11+ cells in peripheral blood was assessed to determine the frequency of cells that express either one of both proteins. Data are means ± SEM (n=8 for WT and n=6 for αV-ΔDC) and representative of four independent experiments. (B) CD103 expression on naive CD8+ T cells in the peripheral blood of αV-ΔDC : B2m−/− mixed BMCs, in which expression of αV and MHC I on DCs is segregated. Data are means and replicates and representative of two independent experiments. (C) 106 naive OT-I cells were adoptively transferred into BMCs. Three weeks later, mice were vaccinated by ear tattoo with OVA-encoding plasmid DNA. After an additional 4 weeks, CD69 and CD103 expression was assessed on transferred OT-I T cells in skin. Data are means and replicates and representative of two independent experiments. */****: p<0.05/p<0.0001 (Two-tailed unpaired Student’s t-tests in (B-C)).
Discussion
In this study, we show that MHC I-dependent interactions with DCs expressing TGF-β-activating αV-integrins epigenetically precondition naive CD8+ T cells during the steady-state to efficiently form epidermal TRM cells upon foreign antigen encounter. This activity is restricted to migratory DCs found in LNs, but not in the spleen, delineating an unanticipated division of labor between these lymphoid tissues.
DCs are known imprint specific migratory patterns in the T cells they activate. For instance, DCs in skin-draining LNs induce the preferential expression of homing molecules for entry into skin, whereas DCs in mesenteric LNs elicit tropism for the small intestine (39–41). eTRM cell preconditioning is distinct in that it already occurs during immune homeostasis, whereas imprinting for tissue-selective homing occurs during T cell priming. Furthermore, αV-integrin-mediated exposure to TGF-β is similar in skin and mesenteric LNs, judging from comparable induction of CD103 in naive T cells at both sites. Preconditioning was however less pronounced in mediastinal LNs, raising the possibility that migratory DCs from different tissues possess varying capacities for TGF-β activation in draining LNs.
It is perhaps counterintuitive that the propensity for eTRM cell formation is not conferred upon T cells during antigenic priming in LNs, but rather earlier, under resting conditions. As a result, naive CD8+ T cells, by transiently retaining the preconditioned state during recirculation, remain endowed with the potential to form eTRM cells even when activated in other SLOs, such as the spleen. In this scenario, the propensity for eTRM cell differentiation does not result from activation at a specific anatomical site, but may instead be restricted to a subset of a heterogenous naive T cell pool that is competent to receive TGF-β-signals from αV-integrin-expressing mDCs at steady-state. Although the naive T cell repertoire is generally viewed as functionally homogenous and uncommitted to any particular memory or effector fate (42), the precedent for such heterogeneity comes from studies on the developmental history of individual naive T cells (43) or their self-reactivity (44). In both studies, subsets were defined with an enhanced ability to rapidly differentiate into short-lived effector cells. Similarly, cell-intrinsic properties that remain to be defined may render a subset of naive CD8+ T cells, identifiable by expression of the CD103 integrin, competent for TGF-β-mediated preconditioning for a future eTRM cell fate.
It has long remained unclear how the multitude of biological functions of a widely secreted cytokine like TGF-β on various cell types could be regulated and coordinated. Recently, LRRC33 was identified as a novel TGF-β milieu protein expressed on microglia, which enables highly localized TGF-β activity in the CNS in coordination with αVβ8-integrins on glial cells (45). Although no milieu proteins expressed by naive CD8+ T cells have yet been identified, our observation that exposure to activated TGF-β is limited to cells that engage in homeostatic, MHC-I-restricted physical interactions with αV-expressing DCs further illustrates an important general principle. Namely, that the potent biological activity of this pleiotropic cytokine can be restricted to individual cells through a two-cell mechanism and thereby “privatized”.
It will be of interest to determine if the DC-driven, TGF-β-dependent preconditioning of naive CD8+ T cells in LNs can be limiting in situations where maximal formation of eTRM cells would be desirable (e.g. during vaccination). In such cases, transiently optimizing local or systemic TGF-β activity in preparation for vaccine administration, ideally targeted to CD8+ T cells in SLOs, may allow for the manipulation of T cell memory differentiation to improve vaccine-induced protection. Conversely, approaches to disrupt naive T cell preconditioning might serve to attenuate de novo eTRM cell formation and aid in the treatment of diseases such as psoriasis, in which eTRM cells play pathogenic roles.
Materials and Methods
Mice
Mice with either floxed αV alleles (Itgavfl/fl) or floxed β8 alleles (Itgb8fl/fl) were previously described (15, 17) and crossed to CD11cCre BAC-transgenic mice (16) obtained from The Jackson Laboratory. C57BL/6J, CD45.1 or Thy1.1 congenic C57BL/6J, OT-I × Tcra−/−, Cd103−/−, B2m−/−, Ccr7−/−, and Lta−/− mice were purchased from The Jackson Laboratory and further bred in-house. Animals were housed in specific pathogen-free facilities at the Massachusetts General Hospital (MGH) and all experimental studies were approved and performed in accordance with guidelines and regulations implemented by the MGH Institutional Animal Care and Use Committee (IACUC).
Irradiation bone marrow chimeras
Recipient mice were i.p. injected with 200 μg of α-NK1.1 mAbs (clone PK136, BioXCell) 1 d prior to irradiation to transiently deplete NK cells and enhance engraftment of B2m−/− bone marrow. Bone marrow was dislodged from femurs and tibiae of donor mice by flushing with 10 ml of PBS without Ca2+/Mg2+ (“PBS” hereafter) and filtered once through a 40-μm filter. Cells from either WT or αV-ΔDC mice were mixed at a 1:1 ratio with cells from B2m−/− mice and resuspended at 5 × 107 total cells/ml. Recipients were irradiated at 1,000 rad (Cesium) in a rotating chamber and injected retro-orbitally with 5 × 106 total donor bone marrow cells/mouse in 100 μl of PBS. Mice were provided sulfamethoxazole in their drinking water for 4 weeks post-irradiation.
DNA Vaccination, mechanical skin irritation, and DNFB treatment
For DNA vaccination against OVA the 6162 bp expression plasmid pOD CAGGS was kindly provided by Dr. J.J. Moon. pOD CAGGS is derived from pCAGGS (46) and uses the CAG promoter to drive expression of full length chicken ovalbumin fused on its N-terminus to the leader sequence of the H-Kb α-chain (MVPCTLLLLLAAALAPTQTRA) and fused on its C-terminus to 44 aa of the transmembrane domain of H-2Db (HEGLPEPLTLRWEPPPSTDSYMVIVAVLGVLGAMAIIGAVVAFV) to achieve membrane incorporation. Mice were anesthetized using isoflurane. Ear or flank skin was shaved and epilated using Nair hair removal cream. A droplet of H20 containing 3 μg of pOD CAGGS DNA was then tattooed into the skin using a sterile disposable 11-needle bar mounted on a rotary tattoo device (Biotouch), as previously described (20). To create a transient inflammatory reaction through mechanical skin irritation alone, plasmid DNA was omitted from this procedure. Alternatively, mice were treated with 10 μl of 0.5% DNFB in a 4:1 acetone:olive oil emulsion to inflame ear skin.
HSV-OVA Infection
HSV-OVA was obtained from Dr. T. Gebhardt (University of Melbourne). For epicutaneous infection by scarification a small (~5 mm2) area of skin overlying the upper pole of the spleen was abraded with 20 strokes of 150-grit sandpaper attached to the end of a pencil (47). Then, 106 plaque-forming units (PFU) of HSV-OVA in 10 μl HBSS was applied to the abraded site, and mice were bandaged for 2 d as described (47, 48). To measure viral titers, a 1-cm2 area of skin surrounding the infection site was harvested into DMEM, cut into small pieces, and snap frozen in a dry ice/70% ethanol bath. The snap-frozen tissue was thawed at 37 °C in a water bath and homogenized in gentleMACS M tubes in a total volume of 2 ml using RNA_01 protocol. Samples were centrifuged for 5 min at 500 × g to remove debris and aggregates and supernatants serially diluted (ranging 10−1–10−6) in serum-free Vero Medium (DMEM, 1% HEPES, 1% GlutaMAX). Serially diluted virus suspension (200 μl) was added to Vero cells grown to 90% confluency in 24-well plates (in duplicate for each sample and dilution). Following 1 h of incubation at 37 °C with gentle rocking of the plate every 10–15 min for even distribution of virus, viral dilutions were aspirated, cells lightly washed with PBS and cultured for 2 d at 37 °C in 1 ml/well of incubation medium (DMEM, 1% HEPES, 1% GlutaMAX, 1% penicillin–streptomycin, 5% (non-fetal) bovine serum, 7.5 of μg/ml human IgG). Cells were then washed with PBS, stained with 1 ml of crystal violet solution (20% EtOH, 80% H2O, 0.5 g/100 ml of crystal violet powder) for 15 min, washed with H2O, and dried overnight. Plaques were counted manually.
Prime and pull immune challenge
The attenuated OVA-expressing Listeria monocytogenes strain L.m.-OVA ΔactA was provided by Dr. J.J. Moon (MGH) and was grown in brain heart infusion (BHI) medium containing 34 μg/ml of chloramphenicol to an absorbance of ~0.1 at 600 nm. L.m.-OVA ΔactA (2 × 107 CFU) were then injected intravenously into mice to induce a systemic TEFF cell response (“Prime”). Four days following infection, mice ears were treated with DNFB, as described above, to produce skin inflammation (“Pull”) and enable seeding by TRM cells from the circulating pool of TEFF cells.
Adoptive T cell transfers, in vitro activation of T cells, and T cell depletion
Naive CD8+ T cells were purified from LN and spleen single-cell suspensions by immunomagnetic negative cell selection using the Miltenyi naive CD8+ T cell isolation kit and adoptively transferred into sex-matched recipients by retro-orbital injection. For the in vitro activation of T cells, OT-I × Tcra−/− splenocytes were pulsed with 1 μM SIINFEKL peptide (New England Peptide) in 1 ml of T cell medium (RPMI, 10% FCS, 1% HEPES, 1% sodium pyruvate, 1% GlutaMAX, 1% non-essential amino acids, 55 μM 2-mercaptoethanol) for 1 h at 37 °C, diluted in 9 ml of T cell medium, and cultured at 37 °C in 5% CO2. Two days later, 20 ng/ml of IL-2 was added and cell density was maintained at 106 cells/ml. OT-I cells were adoptively transferred on day 5 after activation by retro-orbital injection.
For T cell depletion, one dose of anti-Thy1.2 mAbs (3 μg, clone 30H12) was injected i.v.
FTY720 treatment
Mice received intraperitoneal injections of 1 mg/kg BW FTY720 (fingolimod) (Sigma-Aldrich) in 150 μl of H20 every 2–3 d.
Naive T cell treatment with TGF-β1
CD44lo naive CD8+ T cells were purified from LNs and spleens by immunomagnetic negative cell selection using the Miltenyi naive CD8+ T cell isolation kit. Cells were cultured in 96-well flat-bottom plates at a density of 2 × 106 cells/m in serum-free XVIVO10 medium supplemented with 100 ng/ml of rmIL-15 and 5 ng/ml of rmIL-7 (BioLegend). Titrated amounts of acid-activated recombinant mouse TGF-β1 (Cell Signaling) were added as indicated and cells were cultured at 37 °C in 5% CO2 for 72 h before analyzing CD103 surface expression.
Isolation of cells from tissues and staining for flow cytometry
For cell isolations from non-lymphoid tissues, mice were i.v. injected with 3 μg of Alexa Fluor 700- or FITC-labeled α-CD45.2, or PE-Cy7-labeled α-CD8β antibody 3 min prior to euthanasia in order to label intravascular leukocytes for exclusion from analysis. All organs were harvested into ice-cold FACS buffer (PBS with 0.5% BSA and 2 mM EDTA).
Skin was processed as previously described (49). Briefly, separated dorsal and ventral halves of ear skin or flank skin were minced into small pieces and placed in 2.5 ml of digest buffer A (DMEM; 2% FBS; 1% HEPES; 25 U/ml collagenase IV) or digest buffer B (DMEM; 2% FBS; 1% HEPES; 125 μg/ml liberase TM (Sigma-Aldrich); 0.5 mg/ml hyaluronidase Type I-S from bovine testes(Sigma-Aldrich)) for 1 h at 37 °C under agitation, then quenched with 10% FBS and 1.5 mM EDTA and blended using a gentleMACS tissue blender (Miltenyi, C tubes, m_impTumor_01 protocol).
Spleens, thymi, and LNs were minced and passed through a 40-μm cell strainer. Red blood cells were lysed with ACK lysis buffer as necessary. For the isolation of DCs, minced spleens and LNs were digested in digest buffer A for 20 min under agitation at 37 °C before passing through a 40-μm cell strainer.
Cell surface epitopes were stained in FACS buffer in the dark at 4 °C for 15 min, followed by staining with fixable viability dye (Zombie Dyes, BioLegend) at room temperature for 15 min. For detection of intracellular epitopes, cells were fixed and permeabilized (eBioscience Fixation/Permeabilization kit) and stained with antibodies for 30 min. in the dark at room temperature. All antibodies used are listed in table S2.
Histology
Mice were epilated (Nair) and excised ear skin was fixed for 15 min at room temperature in 4% PFA (freshly diluted from 16%), washed six times for 30 min each time in PBS, and stored overnight in 30% sucrose. Eighteen hours later, tissues were immersed in OCT, snap-frozen in a dry ice/2-methylbutane bath and stored at −80 °C until sectioning. Frozen cross-sections (10–20 μm) were air-dried at room temperature for 5 min and loaded into a Shandon Immunostaining Chamber with 1 ml of PBS. Tissues were fixed again with 100 μl 4% PFA for 10 min, washed with 1.2 ml of PBS and then washed with 200 μl of 0.3% PBST. Tissues were then blocked with 100 μl of blocking buffer (5% normal donkey serum, 1% BSA, 2% cold-water-fish gelatin, 0.3% Triton X-100 in PBS), for 30 min at room temperature. Sections were incubated overnight at 4 °C with primary antibodies (in blocking buffer, 100 μl total per slide), washed with 1 ml 0.3% PBST, and incubated with streptavidin for 1 h at room temperature. Slides were washed with 1 ml of 0.3% PBST and then 2 ml of PBS. Slides were then mounted with 100 μl of DAPI fluoromount (Southern Biotech) and cured overnight in the dark at room temperature. Images were acquired on a Zeiss LSM 800 confocal microscope using a 20x/0.8 N.A dry lens and processed using Imaris Software (Bitplane) and ImageJ software (NIH).
The density of immune cells in skin was determined by manual counting and extrapolation of the number of cells per cm2 skin surface based on the number of cells per section and the skin surface represented by the section (10-μm thickness × measured length of the epithelium in the field of view). Non-consecutive sections were analyzed for quantification to avoid overlapping of cells between sections.
Separate Tissues (ear skin and large intestine) from 7-week-old mice were fixed in neutral-buffered formalin (Sigma-Aldrich) overnight, paraffin-embedded, sectioned, and stained with hematoxylin and eosin.
ELISA of serum immunoglobulins
Serum immunoglobulins were measured as previously described (50). Immulon 2HB microtiter plates (DYNEX) were coated with 10 μg/ml of goat anti-mouse Ig (Southern biotech) in PBS at 4 °C overnight. Plates were then blocked with 1% BSA in PBS and incubated with serial dilutions of serum samples in PBS. Specific Ig isotypes were detected using alkaline phosphatase-conjugated isotype-specific antibodies (Southern Biotech). Alkaline phosphatase activity was developed with disodium p-nitrophenyl phosphate substrate (Southern Biotech). Antibody concentrations were calculated as titers relative to purified Ig standards (Southern Biotech).
PCR analysis of floxed Itgav alleles
Five thousand cells of each type were purified by FACS and genomic DNA was extracted using the Arcturus PicoPure DNA Extraction Kit. DNA was amplified for 35 cycles with an annealing temperature of 57 °C using Q5 High Fidelity DNA Polymerase and the following primers: Intav4avf: 5′-TTCAGGACGGCACAAAGACCGTTG-3′ and Intav5b3: 5′-CACAAATCAAGGATGACCAAACTGAG-3′. WT Itgav and floxed Itgav allele products were sized 150 bp and 400 bp, respectively, whereas the recombined floxed Itgav allele yielded no product. The ratio of WT Itgav to floxed Itgav product was determined for each sample and normalized to the ratio in a tail sample of an Itgav fl/wt control animal not expressing Cre recombinase, in order to determine the degree of deletion in each cell type.
Preparation of genomic material and ATAC-seq data analysis
LN CD44lo naive CD8+ T cells (3 × 104 per replicate) were purified by FACS into PBS containing 10% FBS in DNA loBind Eppendorf tubes. Pelleted cells were lysed in 50 μl of reaction mix (25 μl of 2X TD, 2.5 μl of Tn5 enzyme, 0.25 μl of 2% digitonin, and 22.25 μl of nuclease-free water). The mix was incubated at 37 °C for 30 min with agitation at 300 rpm. DNA was purified using a QIAgen MinElute Reaction Cleanup kit and Nextera sequencing primers ligated using PCR amplification. Agencourt AMPure XP bead cleanup (Beckman Coulter/Agencourt) was used post-PCR and library quality was verified using a Tapestation machine. Samples were sequenced on an Illumina HiSeq 2000 sequencer using paired-end 5′ bp reads.
For analysis of sequences, first, adapters were trimmed using AdapterRemoval (v. 2.2.1a) (51). Second, the paired-end ATAC-seq were aligned to the mouse reference genome (mm10) using Bowtie2 (2.3.4.1) (52) with the following parameters --no-discordant --no-unal --no-mixed -X 2000. Third, the reads were shifted (Tn5 insertion) after removing the reads from mitochondrial DNA and duplicated reads.
ATAC-seq peak regions of the pooled sample were called using HOMER (v4.9.1) (53) with the following parameters -region -size 500 -minDist 50 -o auto -tbp 0. Then, the fragment counts for each peak region for each sample were obtained using bedtools (v2.24.0) (54). Differentially accessible peak regions between conditions using the two replicates per condition (adjusted p-value ≤ 1 × 10−5) were called using DESeq (v1.30.0) (55) using the “pooled” dispersion estimation with the “local” fit. Then, overlapping differentially accessible peak regions (adjusted p-value ≤ 1 × 10−5) per condition were merged using bedtools. For each merged peak region, as illustrated in Fig. 4A, we associate adjusted p- and log2 fold-change-values using the adjusted p- and log2 fold-change-values of the region of smallest adjusted p-value (among the regions prior to merging).
ATAC-seq coverage tracks were generated using deepTools (v.3.0.2) (56). The size factors estimated by DESeq were used to normalize ATAC-seq coverage tracks visualized in Figs. 4D and fig. S4A.
HOMER was used to annotate the merged differentially accessible regions per condition and to calculate distances from DARs to the closest transcription start sites in order to generate Fig. 4B. Additionally, HOMER was used to find enriched motifs in the merged differentially accessible regions of each condition with the following parameters (-size given -mask -nomotif) and using the default motif collection (364 motifs). To generate Fig. 4C, we used the motif family information included in the HOMER motif database and discarded the families with the “?” symbol in their identifier. Moreover, we separated the KLF motifs from the Zf family: the KLF family contains all the KLF motifs and the Zf (others) family contains all other Zf motifs. The Genomic Regions Enrichment of Annotations Tool (GREAT) was used for gene ontology and molecular signature enrichment analysis (57). DARs were inputted into the GREAT and gene associations were defined by “basal plus extension” (basal as 5 kb upstream of and 1kb downstream from the TSS, and extension to estimate gene regulatory regions 1 Mb upstream and downstream of the TSS.)
Reverse transcription qPCR
Dendritic cells from each indicated subset (2 × 105 per subset) were purified by FACS into TriZOL for RNA extraction and RNA was further purified using RNeasy Plus Micro or Mini Kit (Qiagen). RNA was reverse transcribed using a High Capacity cDNA Transcription Kit (Life Technologies) and RT-qPCR was performed using SYBR Green detection (Roche LightCycler 480 kit). Primer sequences used for amplification are as follows: Itgav (Forward: 5′-CGGGTCCCGAGGGAAGTTA-3′, Reverse: 5′-TGGATGAGCATTCACATTTGAG-3′) and Itgb8 (Forward: 5′-AGTGAACACAATAGATGTGGCTC-3′, Reverse: 5′-TTCCTGATCCACCTGAAACAAAA-3′).
Statistical analysis
Two-tailed, paired or unpaired Student’s t-tests (for normally distributed data) or Mann–Whitney U tests (for non-normally distributed data) was used for comparisons between two groups. One-way ANOVA test was used for comparison between multiple groups. The two-sample Kolmogorov–Smirnov test was used to compare cumulative distributions. All statistical tests were performed with GraphPad Prism software, and p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. S. Pillai, A. Wagers, U. von Andrian, S. Beyaz, and N. Giovannone for helpful discussions and critical feedback on the manuscript. Funding: This work was supported by the Bob and Laura Reynolds MGH Research Scholar Award and NIH grant R21 AR070981 (to T.R.M), R01 AI040618 (to A.D.L), T32 CA207201 (to V.M.), T32 AR007258 (to E.C.), R01 AI121546 (to S.K.B.), R01 AI107087 (to K.L.J), and by core funding from the Wellcome Trust (203128/Z/16/Z) to The Wellcome Centre for Cell-Matrix Research at University of Manchester.
Footnotes
Competing Interests:
The authors declare no competing interests
Data and materials availability
ATAC-seq data are available under GEO accession number GSE133504. All other data needed to evaluate the conclusions in this paper are available in the main text or the supplementary materials.
Supplementary Materials:
Table S1-S6
Table S1 (as separate *.txt-file)
Table S2
References and Notes
- 1.Masopust D, Soerens AG, Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 37, 521–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller SN, Mackay LK, Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Gebhardt T, Palendira U, Tscharke DC, Bedoui S, Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol. Rev. 283, 54–76 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gebhardt T et al. , Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Schumacher TN et al. , Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science (80-. ). 346, 101–105 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Schenkel JM et al. , T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacKay LK et al. , The developmental pathway for CD103+ CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Masopust D et al. , Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laidlaw BJ et al. , CD4+ T Cell Help Guides Formation of CD103+ Lung-Resident Memory CD8+ T Cells during Influenza Viral Infection. Immunity. 41, 633–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackay LK et al. , T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. 43, 1101–1111 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Milner JJ et al. , Runx3 programs CD8 + T cell residency in non-lymphoid tissues and tumours. Nature. 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson IB, Rifkin DB, Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb. Perspect. Biol. 8, 1–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Travis MA, Sheppard D, TGF-β activation and function in immunity. Annu. Rev. Immunol. 32, 51–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammed J et al. , Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol. 17, 414–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacy-Hulbert A et al. , Ulcerative colitis and autoimmunity induced by loss of myeloid v integrins. Proc. Natl. Acad. Sci. 104, 15823–15828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caton ML, Smith-Raska MR, Reizis B, Notch–RBP-J signaling controls the homeostasis of CD8 − dendritic cells in the spleen. J. Exp. Med. 204, 1653–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travis MA et al. , Loss of integrin αvβ8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 449, 361–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acharya M et al. , αv Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. J. Clin. Invest. 120, 4445–4452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebhardt T et al. , Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 477, 216–219 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Ariotti S et al. , Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc. Natl. Acad. Sci. 109, 19739–19744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagers AJ, Kansas GS, Potent induction of alpha(1,3)-fucosyltransferase VII in activated CD4+ T cells by TGF-beta 1 through a p38 mitogen-activated protein kinase-dependent pathway. J. Immunol. 165, 5011–5016 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Hochweller K et al. , Dendritic cells control T cell tonic signaling required for responsiveness to foreign antigen. Proc. Natl. Acad. Sci. 107, 5931–5936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schenkel JM, Fraser KA, Vezys V, Masopust D, Sensing and alarm function of resident memory CD8 + T cells. Nat. Immunol. 14, 509–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito Y, Miyazono K, RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr. Opin. Genet. Dev. 13, 43–47 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Mackay LK et al. , Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 26.McConnell BB, Yang VW, Mammalian Krüppel-Like Factors in Health and Diseases. Physiol. Rev. 90, 1337–1381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang N, Bevan MJ, TGF-β signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat. Immunol. 13, 667–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grueter B et al. , Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J. Immunol. 175, 1694–1705 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Sanjabi S, Mosaheb MM, Flavell RA, Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 31, 131–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ, Continued maturation of thymic emigrants in the periphery. Nat. Immunol. 5, 418–425 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Cyster JG, Schwab SR, Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30, 69–94 (2012). [DOI] [PubMed] [Google Scholar]
- 32.De Togni P et al. , Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 264, 703–707 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Mackay LK et al. , Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl. Acad. Sci. 109, 7037–7042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin H, Iwasaki A, A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 491, 463–467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henri S et al. , The dendritic cell populations of mouse lymph nodes. J. Immunol. 167, 741–8 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Ohl L et al. , CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 21, 279–288 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Takada K, Jameson SC, Self–class I MHC molecules support survival of naive CD8 T cells, but depress their functional sensitivity through regulation of CD8 expression levels. J. Exp. Med. 206, 2253–2269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakim LM, Bevan MJ, Cross-dressed dendritic cells drive memory CD8+T-cell activation after viral infection. Nature. 471, 629–631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson-Lindbom B et al. , Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J. Exp. Med. 198, 963–969 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Andrian UH et al. , Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 424, 88–93 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Dudda JC, Simon JC, Martin S, Dendritic cell immunization route determines CD8+ T cell trafficking to inflamed skin: role for tissue microenvironment and dendritic cells in establishment of T cell-homing subsets. J. Immunol. 172, 857–863 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Buchholz VR, Schumacher TNM, Busch DH, T Cell Fate at the Single-Cell Level. Annu. Rev. Immunol. 34, 65–92 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Smith NL et al. , Developmental Origin Governs CD8 + T Cell Fate Decisions during Infection. Cell. 174, 117–130.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Fulton RB et al. , The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat. Immunol. 16, 107–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin Y et al. , A Milieu Molecule for TGF-beta Required for Microglia Function in the Nervous System. Cell. 174, 156–171.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niwa H, Yamamura K, Miyazaki J, Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Goel N, Docherty JJ, Fu MM, Zimmerman DH, Rosenthal KS, A modification of the epidermal scarification model of herpes simplex virus infection to achieve a reproducible and uniform progression of disease. J. Virol. Methods. 106, 153–158 (2002). [DOI] [PubMed] [Google Scholar]
- 48.van Lint A et al. , Herpes simplex virus-specific CD8+ T cells can clear established lytic infections from skin and nerves and can partially limit the early spread of virus after cutaneous inoculation. J. Immunol. 172, 392–397 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Shade K-TC et al. , A single glycan on IgE is indispensable for initiation of anaphylaxis. J. Exp. Med. 212, 457–467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acharya M et al. , αv Integrins combine with LC3 and atg5 to regulate Toll-like receptor signalling in B cells. Nat. Commun. 7, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schubert M, Lindgreen S, Orlando L, AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes. 9, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Huber W, Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T, DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, 187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLean CY et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.