Abstract
Checkpoint blockade immunotherapy is now a first-line treatment option for patients with melanoma. Despite achieving objective responses in about half of patients, the exact immune mechanisms elicited and those required for therapeutic success have not been clearly identified. Insight into these mechanisms is key for improving outcomes in a broader range of cancer patients. We used a murine melanoma model to track responses by different subsets of tumor-infiltrating lymphocytes (TIL) during checkpoint blockade immunotherapy. Tumors from treated mice had increased frequencies of both CD4+ and CD8+ T cells, which also showed evidence of functional reinvigoration and elevated effector cytokine production after immunotherapy. We predicted that increased T cell numbers and function within tumors reflected either infiltration by new T cells or clonal expansion by a few high-affinity tumor-reactive T cells. To address this, we compared TIL diversity before and after immunotherapy by sequencing the complementarity determining region 3 (CDR3) of all T cell receptor beta (TCRβ) genes. While checkpoint blockade effectively slowed tumor progression and increased T cell frequencies, the diversity of intratumoral T cells remained stable. This was true when analyzing total T cells and when focusing on smaller subsets of effector CD4+ and CD8+ TIL as well as regulatory T cells. Our study suggests that checkpoint blockade immunotherapy does not broaden the T cell repertoire within murine melanoma tumors, but rather expands existing T cell populations and enhances effector capabilities.
Keywords: Checkpoint blockade, Immunotherapy, T cell, TCR diversity, Melanoma
Introduction
The peripheral T cell receptor (TCR) repertoire in humans and mice is theoretically able to recognize a staggering number (> 1 × 1013) of self and non-self antigens, which is key for broad immune surveillance of pathogens and potentially malignant cells [1]. This enormous diversity among possible TCR specificities is achieved during T cell development in the thymus, where T cells undergo somatic recombination of gene segments encoding the variable regions of each α and β TCR chain. Random addition and deletion of nucleotides at the junctions between gene segments further expand the possible number of unique T cell specificities, resulting in the formation of the hypervariable complementarity determining region 3 (CDR3) that shapes TCR specificity for a given antigen presented in the context of a major histocompatibility complex (MHC) molecule. While this diversity is clearly beneficial for the recognition of a wide range of pathogens, it is unknown how such an abundance of T cell specificities impacts immunity against tumors and whether the diversity of tumor-infiltrating T cells is affected during cancer immunotherapy.
Immune checkpoint blockade (ICB) relies on the infusion of antibodies that target inhibitory receptors such as PD-1 and CTLA-4 on T cells, thereby disrupting inhibitory signaling pathways and boosting endogenous immune responses. The role of checkpoint blockade in the future of cancer treatment is beyond debate [2, 3], but the exact immune mechanisms elicited during treatment are not completely defined. The success of ICB as a cancer immunotherapy depends largely on the activity of T cells within tumors, as the frequency and function of these tumor-infiltrating lymphocytes (TIL) correlate with patient outcomes [4–6]. In animal models and in human patients, treatment with ICB increases TIL frequency and boosts T cell effector functions [7]. But beyond these metrics of immune activity, the relative diversity of T cell specificities within tumors is now being evaluated as a contributing factor in immunotherapy outcomes [5, 8–11]. This concept is still controversial and it remains undetermined whether greater TCR diversity within the TIL population or greater cellular clonality correlates with better clinical responses. Either scenario seems plausible. For example, a highly focused clonal T cell response (low diversity) strongly reactive against a prominent tumor-associated antigen could provide robust antitumor immunity. On the other hand, a wider variety of T cell specificities (high diversity) could enable the immune system to respond to multiple neo-antigens expressed by a greater number of tumor cells.
Recent advances in DNA sequencing techniques have allowed the diversity of polyclonal T cells to be measured by sequencing all the TCRβ CDR3 regions present within a heterogeneous cellular population. However, there are conflicting reports as to how cancer therapy influences TIL diversity and whether this correlates with outcomes. In a mouse model of triple-negative breast cancer, TCR diversity was shown to increase among TIL after treatment with anti-CTLA-4 but was unaltered with the addition of anti-PD-1 [12]. A murine melanoma study showed that radiation therapy broadened the TIL repertoire but diversity was unaffected by anti-CTLA-4 checkpoint blockade [5]. Another preclinical study thoroughly explored how TIL diversity was influenced by a plethora of immune modulatory treatments such as anti-PD-1, anti-CTLA-4 and anti-4-1BB to treat B16 melanoma [13]. Here, total T cell diversity was barely affected during immunotherapy. Instead, tumor-reactive clones were enriched by treatments that were more successful at slowing tumor growth. Likewise, in human breast cancer patients, treatment with ipilimumab (anti-CTLA-4) alone increased TIL density but did not affect TCR diversity [14]. Clearly, the field lacks a true consensus on this issue. One limitation of these previous studies is the relatively unfocused approach of TCR analysis on total T cells or even unfractionated whole tumor tissue without considering possible changes in diversity among distinct T cell subsets. Several T cell subsets contribute to the control of tumor progression, including CD8+ cytotoxic T lymphocytes (CTL) and interferon gamma (IFNγ) producing CD4+ T helper-1 cells [15]. In contrast, Foxp3+ T regulatory (Treg) cells suppress the effector activity of CD4+ and CD8+ T cells, impeding tumor immunity [16]. How the clonality or diversity of these individual TIL subsets is influenced during cancer immunotherapy has not been reported. Here, we have taken a different approach, focusing on individual TIL subsets. Our results demonstrated that ICB treatment increased the frequency and functionality of effector CD4+ and CD8+ T cells in melanoma tumors but did not significantly alter the TCR diversity within the TIL repertoire.
Materials and methods
Mice
C57BL/6 T-bet-ZsGreen reporter (TbetZsG) mice were obtained from Taconic Farms and described previously [17, 18]. These were crossed with Foxp3-IRES-mRFP (FIR) mice from Daniel Hawiger’s laboratory (Saint Louis University) to generate double-reporter mice (TbetZsG-Foxp3RFP). Briefly, TbetZsG mice contain the coding region of ZsGreen downstream of the translational starting site of the genomic fragment encoding T-bet (T-box expressed in T cells) [18]. FIR mice contain an internal ribosomal entry site-linked monomeric red fluorescent protein (IRES-mRFP), which was placed between the translation stop codon (UGA) and the polyadenylation signal (A2UA3) of the Foxp3 gene [19]. The generation of T-bet-ZsGreen and Foxp3-RFP double-reporter mice allowed for the sorting of T cell subsets without permeabilization or fixation. Mice were screened by flow cytometry for the presence of ZsGreen and RFP.
Tumor immunotherapy
To generate established tumors, 1 × 106 B16-F0 cells were injected subcutaneously into both flanks of C57BL/6 mice. Tumors were established for 5–8 days mice prior to treatment with ICB. Combination ICB consisted of anti-PD-1 (RMP1-14), anti-CTLA-4 (9D9) and anti-lymphocyte activation gene 3 (LAG-3) (C9B7 W) administered intraperitoneally at 5 mg/kg on days 5, 10 and 15 for tumor growth studies, or on days 8, 11 and 14 for day 16 analysis of TIL. Tumors were measured using digital calipers and tumor volume was determined using the equation (L × W2)/2. For TIL analysis, tumors were excised from mice and mechanically disrupted with a sterile 3-ml syringe plunger and filtered through a 40 µM strainer. All isolation steps were performed in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Flow cytometry and cell sorting analysis
Fluorochrome-conjugated antibodies were purchased from BD Biosciences, eBioscience, and Biolegend. Aqua fluorescent reactive dye (live/dead) was purchased from Invitrogen (Thermo Fisher Scientific). Flow cytometric analysis was performed on LSR II FACS analyzer (BD Biosciences); cells were sorted using BD FACS Aria III (BD Biosciences) at the Saint Louis University Flow Cytometry Core Facility. Live lymphocytes were identified as staining negative for live/dead stain and positive for CD45. These were segregated into CD4+ and CD8+ T cells or CD4/8-negative NK1.1+ natural killer cells. CD4+ T cells were further segregated into conventional FoxP3-negative and Foxp3+ regulatory cells. Flow cytometry data were analyzed using FlowJo v.10 software (Tree Star Inc.). Intracellular cytokine staining was performed using the Cytofix/Cytoperm Plus kit (BD Biosciences) per the manufacturer’s instructions. In brief, TIL were incubated with PMA and Ionomycin (Iono) for 4 h in the presence of GolgiPlug (brefeldin A). Cells were first stained with live/dead and cell-surface markers with anti-CD45 (30-F11), anti-CD8 (53-7.6), anti-CD4 (GK1.5) and anti-NK1.1 (PK136). Cells were then fixed, permeabilized, and stained with anti-IFNγ (XMG1.2) and anti-TNF (MP6-XT22). Intracellular staining of cytoplasmic and nuclear associated proteins was performed using the eBioscience cellular perm kit per the manufacturer’s instructions. Briefly, TIL were processed and stained directly ex vivo with live/dead and cell-surface markers including anti-PD-1 (29F.1A12). Cells were fixed, permeabilized, and stained with antibodies specific for Eomes (Dan11mag), T-bet (eBio4B10), Foxp3 (FJK-16s), GzmB (GB11) and Ki67 (16A8). For cell sorting of the four T cell subsets, TIL from double-reporter mice were stained with anti-CD45, anti-CD44 (IM7), anti-CD8, anti-CD4, and anti-NK1.1.
Immunohistochemistry
Whole tumors were placed in formalin and imbedded in paraffin for sectioning. Slides were deparaffinized using xylene and rehydrated with ethanol. Antigen retrieval was performed using sodium citrate. Slides were then treated with 3% hydrogen peroxide, blocked with 5% donkey serum, and then stained with CD8 primary antibody (Cell Signaling Technologies cat#98941) overnight. Secondary HRP-conjugated antibody was added followed by diaminobenzidine (DAB) substrate. Slides were then counterstained with hematoxylin.
Next-generation high-throughput sequencing
T cell subsets were sorted on a BD FACSAria III to at least 95% purity based on differentially expression of CD8, CD4, CD44, T-bet and Foxp3. Purified T cells were pelleted and stored at negative 80 °C prior to shipping. DNA was extracted and next-generation sequencing was performed by Adaptive Biotechnologies, Seattle, WA, USA, as previously described [20].
Statistical analyses
Statistical analyses to compare multiple treatment groups were performed using unpaired, two-tailed nonparametric Mann–Whitney U tests. A log-rank (Mantel–Cox) test was used to compare recipient survival. Statistical calculations were performed using Prism 7.0, GraphPad 15 Software. All error bars represent standard deviation and indicate the mean of distribution. Whenever possible, exact P values are indicated. BLOcks Substitution Matrix 62 (BLOSUM62) analysis was completed as previously described [21]. Briefly, TCR clonotypes were aligned against previously published TCR sequences specific for gp100 antigen. Similarity in the CDR3 sequences was determined by the degree of overlap in CDR3 sequence to published sequences in addition to how various specific amino acid variations impact potential antigen specificity. Venn diagrams were generated by summing the unique clonotypes per treatment group and determining how many of those clonotypes overlap between treatment groups. Shannon diversity index was calculated using the following equation:
Results
The utility of ICB is well established as a treatment for human melanoma patients [22–24]. These clinical outcomes have been further improved with combination immunotherapy approaches (anti-PD-1/CTLA-4), though long-term remission is still only seen in a subset of patients [25]. Similarly, in animal models, resistance to combination checkpoint blockade has been demonstrated in melanoma-bearing mice treated with combination (anti-PD-1/CTLA-4) immunotherapy regimens [15]. To overcome this resistance, we have employed an aggressive combination approach using anti-PD-1/CTLA-4 plus anti-LAG-3, as anti-LAG-3 is currently being explored in clinical trials as a monotherapy and in combination with anti-PD-1 [NCT02460224]. We previously demonstrated this triple combination to be superior compared to monotherapies or even dual-antibody combinations for restoring CD8+ TIL function and improving survival of mice with disseminated leukemia [17, 26]. To determine whether this immunotherapy strategy also improved responses against melanoma, we tested different combinations in B6 mice bearing established subcutaneous melanoma tumors, again showing improved outcomes such as smaller tumors with slower growth rates and more durable responses in mice treated with anti-PD-1/CTLA-4/LAG-3 compared to other modalities (data not shown). Based on this body of work, we conducted all subsequent experiments here using this triple combination ICB to treat B6 mice bearing B16 melanoma tumors. This melanoma line has a broad mutational landscape encoding numerous neo-epitopes similar to human melanoma [27, 28], allowing for meaningful assessment of diverse T cell immune responses against bona fide tumor antigens.
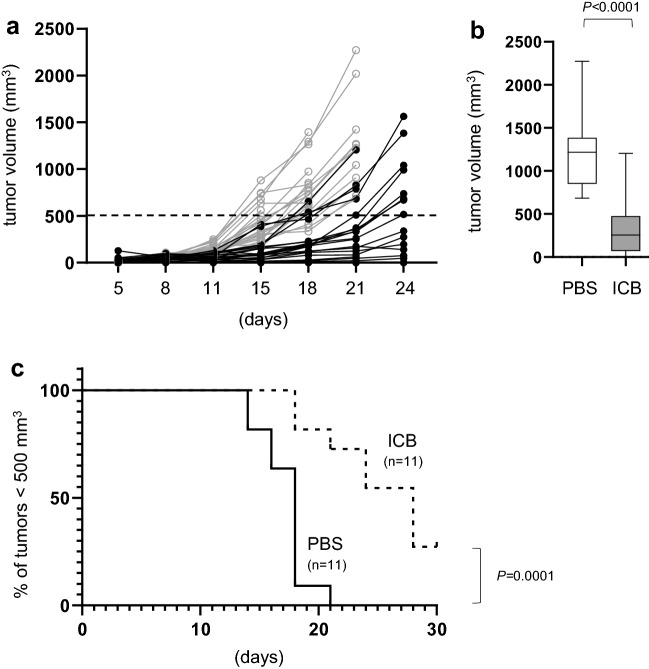
Recipients with established melanoma tumors were treated with ICB (5 mg/kg) or vehicle control (PBS) on days 4, 8, 12 and 16 post-tumor initiation. Previous work in our laboratory and others has demonstrated that isotype control antibodies do not promote antitumor immune responses [26, 29] and thus were not included here. In contrast, immunotherapy with the anti-PD-1/CTLA-4/LAG-3 combination was effective in delaying melanoma tumor progression (Fig. 1a). For example, by day 21 when a majority of mice in both groups were still alive, the average tumor volume in mice receiving ICB was significantly smaller than mice treated with PBS (Fig. 1b). In a subset of ICB-treated mice, tumors remained exceptionally small even beyond day 24 of the study. Because we do not use death as an endpoint for these studies, we instead used an arbitrary tumor volume of 500 mm3 as a surrogate threshold to compare treatment durability rather than actual survival. Whereas all mice treated with PBS crossed this threshold by day 21, those receiving ICB demonstrated resisted tumor progression with several (3/11) surviving out to day 30 (Fig. 1c). These data highlight the overall efficacy of ICB for treatment of B16 melanoma but also show the range of responses, recapitulating the diverse clinical outcomes observed in melanoma patients treated with combination checkpoint blockade [30].
Fig. 1.
Combination ICB inhibits melanoma tumor progression. Mice received bilateral subcutaneous injections of 1 × 106 B16-F0 melanoma cells. Mice were then treated with immune checkpoint blockade or PBS control on days 4, 8, 12, and 16 post-tumor injection. a Right and left tumors were measured on the days indicated and tumor volumes were determined from mice receiving PBS (gray open squares) versus ICB (closed circles). Each point represents data from an individual tumor from 11 mice per treatment group pooled from 3 independent experiments. b Comparison of PBS and ICB tumor volumes on day 21 post-tumor injection. c Kaplan–Meier survival curve using a tumor volume of 500 mm3 as a threshold for survival instead of death. P values are indicated between each treatment group
Combination ICB increases TIL abundance and function
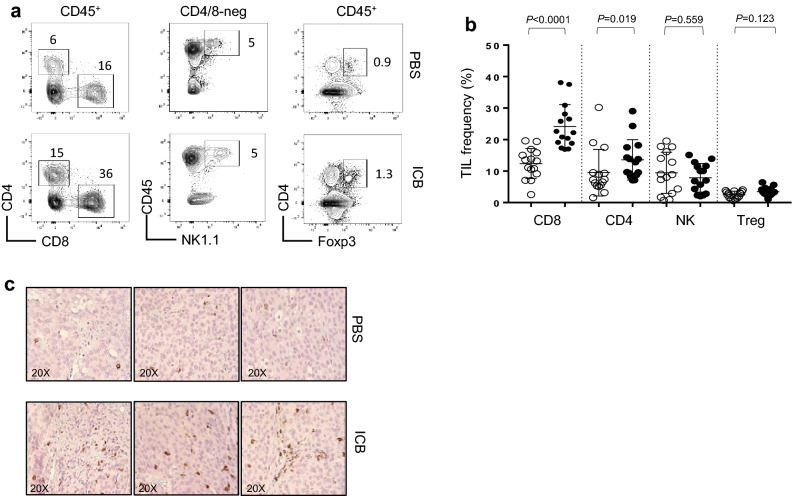
The immune mechanisms responsible for the therapeutic success of combination checkpoint blockade in melanoma are not well defined, but likely involve increased numbers and enhanced functions of tumor-infiltrating lymphocytes. To determine whether ICB immunotherapy promoted tumor infiltration by immune cells or the deletion of regulatory cells, TIL populations were quantified within established melanomas at day 16 after tumor injection. To ensure large enough tumors for sufficient TIL isolation at this abbreviated time point, the ICB schedule was delayed slightly to allow additional time for tumor growth prior to immunotherapy. Here, mice were treated with ICB or PBS on days 8, 11, and 14 relative to tumor initiation and sacrificed on day 16. At this time, tumors from treated mice showed increased percentages of both CD4+ and CD8+ T cells compared to other CD45+ lymphocytes (Fig. 2a). There were no detectable changes in the frequencies of natural killer (NK) cells or CD4+ Foxp3+ Tregs after ICB (Fig. 2a, b). Immunohistochemistry (IHC) confirmed an increase in CD8+ TIL after immunotherapy, which were widely distributed throughout tumor tissues (Fig. 2c). These data suggest that effector CD4+ and CD8+ T cells are likely responsible for the antitumor immunity elicited by this combination ICB regimen.
Fig. 2.
Immunotherapy increases CD4+ and CD8+ TIL. a Representative flow plots and b corresponding graph of the frequency of tumor-infiltrating CD4+, CD8+, NK, and Tregs in mice treated with PBS control (open circle) or ICB (closed circle). Mice received bilateral subcutaneous injections of 1 × 106 B16-F0 melanoma cells and were treated with ICB or PBS on days 8, 11, and 14 post-tumor injection (n = 15). Tumors were harvest on day 16 and analyzed by flow cytometry for immune cell frequency and c CD8+ TIL visualized by IHC. P values are indicated above each treatment group
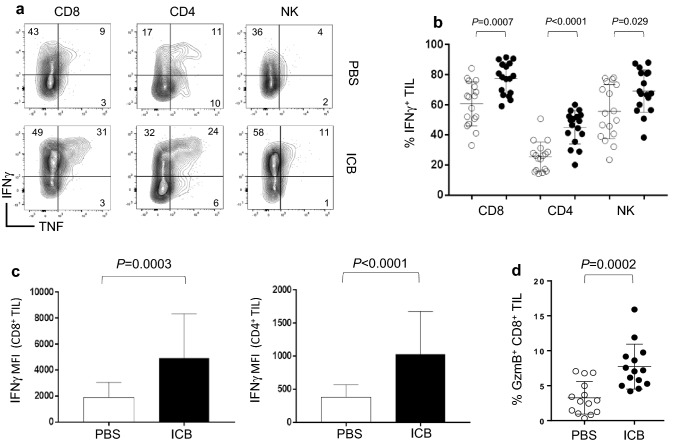
T cells are instrumental in controlling tumor progression but require differentiation and the acquisition of effector functions to mediate a therapeutic impact. Effector CD4+ T helper 1 (TH1) cells as well as CTL are important producers of effector molecules such as IFNγ, TNF, and Granzyme B (GzmB) necessary for antitumor immunity. Since combination ICB provided enhanced control of melanoma progression corresponding with increased T cell frequencies, we determined if immunotherapy also increased the functionality of different TIL populations. Melanoma tumors were implanted and recipient mice were treated with ICB on days 8, 11, and 14. On day 16, expression of intracellular cytokines and effector molecules were measured in TIL by flow cytometry. Relative to mice treated with PBS, those receiving immunotherapy had significantly higher frequencies of multifunctional CD8+ and CD4+ TIL as well as natural killer cells producing IFNγ and TNF following ex vivo restimulation (Fig. 3a, b). These CD8+ and CD4+ TIL also produced more IFNγ on a per cell basis relative to those from PBS-treated mice as indicated by increased mean fluorescence intensity (MFI) of IFNγ staining (Fig. 3c). Furthermore, CD8+ TIL from recipients treated with ICB demonstrated increased expression of GzmB directly ex vivo (Fig. 3d). These results show that immunotherapy with combination ICB promotes the accumulation of effector T cells within established melanoma tumors.
Fig. 3.
Checkpoint blockade increases TIL effector functions. Mice received bilateral subcutaneous injections of 1 × 106 B16-F0 melanoma cells and were treated with immune checkpoint blockade or PBS control on days 8, 11, and 14 post-tumor injection. Tumors were harvest on day 16 and analyzed by flow cytometry for effector molecule production. a Representative flow plots and b corresponding graph of the frequency of IFNγ and TNF producing tumor-infiltrating CD8+, CD4+, and NK cells from mice treated with PBS control (open circle) or ICB (closed circle) after ex vivo restimulation. c MFI of IFNγ for CD8+ and CD4+ TIL. d Frequency of Granzyme B producing CD8+ TIL (n = 14). For graphs b and c, n = 17 from five independent experiments. For graph c, n = 14 from four independent experiments. P values are indicated above each treatment group
Combination ICB increases CD8+ TIL reinvigoration
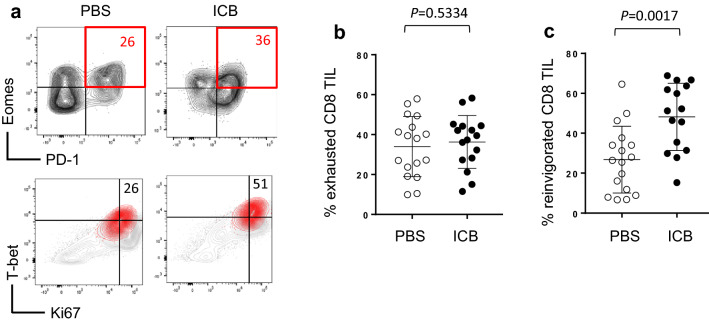
Exhaustion of T cells within the tumor microenvironment is a particularly effective form of immune evasion in both human and mouse melanoma [31, 32]. Thus, the functional reinvigoration of TIL is key to the success of immune checkpoint blockade. Exhausted T cells are identifiable by co-expression of PD-1 and the transcription factor Eomesodermin (Eomes) and are characterized by a loss of proliferative capacity and effector function [5]. Exhausted T cells that have been reinvigorated via immunotherapy or radiation therapy retain the PD-1 and Eomes profile but gain expression of the proliferation marker Ki67 and effector genes like IFNγ and GzmB [5], which are controlled by the transcription factor T-bet. We therefore define exhausted CD8+ T cells as Eomes+ PD-1+ and reinvigorated CD8+ T cells as Eomes+ PD1+ Ki67+ T-bet+. Here, CD8+ TIL from mice treated with either PBS control or ICB showed no difference in the frequency of CD8+ TIL with an exhausted Eomes+ PD-1+ phenotype (Fig. 4a, b). However, the proportion of these TIL displaying a reinvigorated Eomes+ PD1+ Ki67+ T-bet+ phenotype was significantly increased in mice treated with ICB (Fig. 4a, c). These data suggest that combination checkpoint blockade induces reinvigoration of T cells within melanoma tumors, promoting effector activities that contribute to the control of tumor progression.
Fig. 4.
Immunotherapy induces CD8+ TIL reinvigoration. Mice with established B16 tumors were treated with ICB or PBS control on days 8, 11, and 14 post-tumor injection. Tumors were harvest on day 16 and analyzed by flow cytometry. a Representative flow plots of CD8+ TIL from mice treated with PBS control or ICB. Red quadrant and inset numbers in the top panels are the percentage of Eomes+ PD1+ CD8+ TIL. Red contour in the bottom panels are reinvigorated (Eomes+ PD-1+ T-bet+ Ki67+) CD8+ TIL taken from the red quadrant above. Inset numbers are the percentage of reinvigorated CD8+ TIL. Pooled data for exhausted (b) and reinvigorated (c) CD8+ TIL (n = 17) are displayed from five independent experiments. P values are indicated above each treatment group
T cell receptor diversity among melanoma TIL is unaffected by ICB
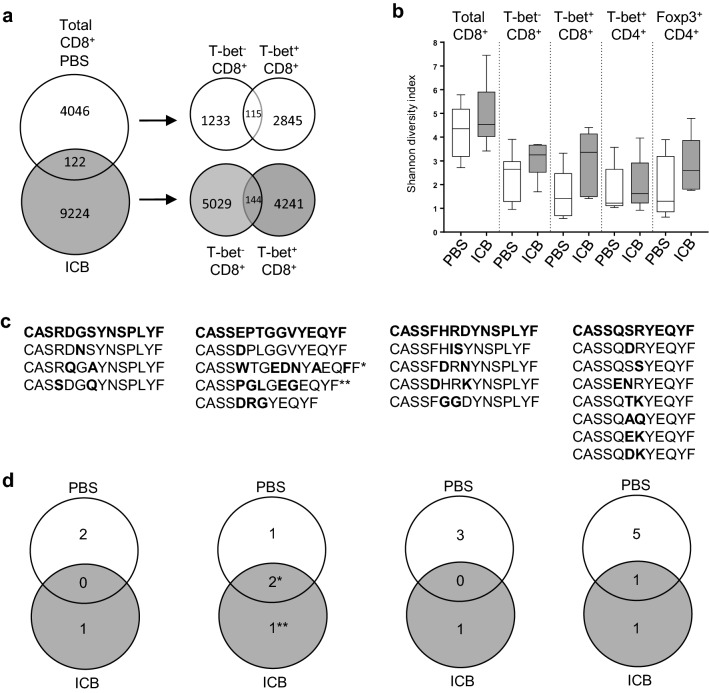
Because checkpoint blockade induced greater function and frequency of CD4+ and CD8+ T cells within tumors, we sought to determine whether this combination immunotherapy also impacted the diversity of the TIL repertoire. As described above, tumors were initiated and recipients were treated with ICB on days 8, 11, and 14. On day 16, TIL were sorted from recipient mice into four distinct subpopulations of T cells; effector CTL (CD8+ T-bet+), recently activated CD8+ T cells (CD44+ T-bet−), differentiated TH1 cells (CD4+ T-bet+), and Tregs (CD4+ Foxp3+). This is a novel approach to define TIL diversity compared to other studies that have examined unfractionated T cells or sequenced TCR genes from whole tumor tissue [12, 13]. To enable sorting of different T cell subsets without the need for permeabilization or fixation, B6 reporter mice expressing ZsGreen downstream of the T-bet promoter and RFP downstream of the Foxp3 (TbetZsG-Foxp3RFP double-reporter mice) promoter were used as tumor-bearing recipients. Genomic DNA was isolated from sorted T cells and subjected to next-generation high-throughput sequencing of the TCRβ CDR3 region. Among total CD8+ T cells, more than twice as many unique TCR clonotypes (9224) were detected among TIL from treated mice compared to control mice (4046), with only 122 overlapping TCR sequences (Fig. 5a). Similar results were observed when CD8+ T cells were segregated by expression of T-bet (Fig. 5a). Despite the increase in the number of unique TCR sequences after ICB treatment, this does not necessarily represent a change in diversity, as this analysis did not take into account the evenness of all TCR distributed across the entire population. When evenness was considered, we measured no significant change in the diversity of the TCR repertoires (as measured by the Shannon diversity index) between ICB-treated and PBS-treated control (Fig. 5b). While unanticipated, this outcome is in agreement with a recent study measuring TCR diversity in whole tumor tissue, where the Shannon diversity index was unchanged between untreated mice and those receiving a potent immunotherapy regimen of anti-PD-1 plus anti-4-1BB [13].
Fig. 5.
The TCR diversity among melanoma TIL subsets is not significantly altered during checkpoint blockade immunotherapy. Reporter mice were injected subcutaneously with B16-F0 melanoma cells and treated on days 8, 11, and 14 post-tumor initiation. On day 16, TIL from individual mice were sorted independently and sent to Adaptive for deep sequencing. a Venn diagram representing the sum of all unique clonotypes from CD8+ TIL of 12 separate mice treated with PBS or 11 separate mice treated with ICB. The number of unique and shared clonotypes between differently treated groups is inset. b Shannon diversity index comparing PBS- and ICB-treated mice for each T cell subset. c Four amino acid sequences (bold) from predicted Gp100-specific TCRβ. Variations of each consensus sequence found in our data sets are listed with individual amino acid differences in bold. d Venn diagrams show distribution of each of the four Gp100-specific TCR among PBS- and ICB-treated animals with the number of unique and shared sequences inset
It is not yet possible to reliably derive antigen specificity directly from TCRβ CDR3 amino acid sequences, making it difficult to predict how many of these TCR may be tumor specific. However, to gain some insight into the specificity of TCR in our CD8+ TIL populations, we interrogated our data set for the presence of four published TCRβ sequences reported to be specific for the melanoma antigen Gp100 (Pmel) [33, 34]. No identical matches to the four published Gp100-specific TCRβ sequences were identified in the CD8+ TIL TCR repertoires. However, using both the basic local alignment search tool (BLAST) and BLOSUM62, we identified 18 distinct TCR clonotypes that are predicted to share antigen specificity based on amino acid sequence similarity within the CDR3 region (Fig. 5c). The BLOSUM is used to score alignments between protein sequences and has been previously used to mathematically predict TCR epitope specificity to hepatitis C antigens [21]. These Gp100-specific TCR were distributed across TIL populations from 4/11 mice (36%) treated with ICB and 6/12 mice (50%) receiving PBS control. Of these TCR sequence variations, 11 were found at very low frequencies among TIL only in untreated mice, 4 were found exclusively in mice treated with ICB, and 3 were shared between both groups of mice (Fig. 5d). A caveat to this distribution pattern is that two TCR clonotypes (marked with asterisks in the second column) were present at exceptionally high frequencies in individual ICB-treated mice (25%* and 18%** of total TCR sequences compared to less than 1% PBS-treated mice). Therefore, Gp100-specific TIL were slightly more common in untreated mice but the number of these melanoma-reactive TIL was far greater in 2/11 mice treated with ICB. While such analysis on TIL specificity is clearly limited at this stage, TIL are assumed to contain a heterogeneous population of T cells reactive against both tumor and non-tumor antigens alike. However, the exact specificity of these TIL does not impede our ability to describe the overall T cell diversity within tumors. We interpret our studies to suggest that immunotherapy with immune checkpoint blockade does not significantly alter the T cell repertoire within murine melanomas.
Discussion
Checkpoint blockade immunotherapy has dramatically improved outcomes for patients with melanoma [23, 35]. Subsequent combination blockade approaches have extended this clinical success to an even greater proportion of patients, eliciting long-term (at least 3 years) overall survival in more than half of patients treated [25]. However, a complete understanding into the immune mechanisms operative during treatment has been elusive. An analysis of human melanoma TIL revealed that CTLA-4 and PD-1 likely play distinct roles in suppression of immune function, and blockade of these molecules during immunotherapy induces expansion of unique CD4+ and CD8+ T cell subsets [36]. It remains to be determined whether expansion of these individual subsets alters the diversity of tumor-infiltrating T cells and whether this contributes to antitumor immunity.
Whether or not the diversity of tumor-reactive T cells influences patient outcomes remains a controversial topic. In a clinical study of 42 patients with prostate cancer, treatment with anti-CTLA-4 (ipilimumab) induced modest diversification of peripheral blood T cell clonotypes in both responding and non-responding patient cohorts and was associated with immune-related adverse events [37]. In melanoma patients, a higher density of CD4+ and CD8+ TIL of a more clonal nature (reduced diverse) correlated with better responses to PD-1 blockade (pembrolizumab) [11]. In contrast, assessment of a small group of melanoma patients treated with ipilimumab associated TIL diversity with some clinical benefit although no impact on overall survival was noted [38]. Thus, clinical responses have so far been inconsistent and difficult to interpret.
To gain insight into how checkpoint blockade immunotherapy influences the breadth of T cells infiltrating tumors, we explored TIL diversity in the B16 murine melanoma tumor model. B16 tumors have a well-characterized mutational landscape necessary for recognition by a broad range of neoantigen-specific T cells [28]. Treatment of tumor-bearing mice with combination checkpoint blockade immunotherapy resulted in significantly smaller tumors over time—reflecting improved immune responses against the cancer. Indeed, recipients treated with immunotherapy displayed elevated frequencies of CD4+ and CD8+ T cells within tumors, and these T cells showed increased expression of effector molecules, indicative of enhanced functionality.
Because these TIL populations expanded after immunotherapy, we anticipated that the diversity of these subsets would also be impacted; either reduced due to clonal expansion of a small number of tumor-reactive T cells or increased due to activation and infiltration by a broader population of endogenous T cells. However, sequencing of the genes encoding all TCRβ chains within the tumor microenvironment revealed no significant changes in the overall diversity of T cells responding to immunotherapy. This is in agreement with a previous study in B16 melanoma TIL where radiation therapy induced a broader TCR repertoire, which was unaltered by the addition of checkpoint blockade antibodies [5]. In our study this was also true for individual TIL subsets such as T-bet+ CD8+ effector CTL and for T-bet+ CD4+ TH1 cells. The analysis of the TCRβ repertoires of TIL revealed a diverse heterogeneous T cell population. We did not identify any previously published tumor antigen-specific clonotypes within the TIL TCR repertoires. This is not particularly surprising, as the TCR repertoire is vastly diverse and antigen-specific TCR clonotypes shared among multiple individuals account for only a small percentage of the repertoire [39–41]. A more focused assessment of melanoma-reactive TCR clonotypes showed a wide distribution of predicted Gp100-specific TCR in TIL regardless of immunotherapy. However, these data are limited to a very small subset of TCR specific for a single melanoma antigen. As increases in technology allow for more accurate prediction of TCR specificity from CDR3 sequencing data, a more comprehensive assessment of how checkpoint blockade influences tumor-reactive TIL will be possible.
Our results indicate that successful immunotherapy of murine B16 melanoma does not require tumor infiltration by new T cells with broader specificities, as the breadth of T cell specificities present after expansion essentially mirrored that prior to expansion. This conclusion aligns with another study where the diversity index of TCR within whole tumor tissues was unaltered even after treatment with a successful immunotherapy combination of anti-PD-1/4-1BB [13]. In contrast, increased TIL diversity was observed in mice treated with a low-efficacy monotherapy of anti-4-1BB, which failed to control melanoma tumor growth. Together with our studies, these data raise questions about the utility of TIL diversity as a predictor or driver of therapeutic success. Results from our study also suggest successful immunotherapy was not reliant on the select expansion of certain T cell clones, but rather induced relatively uniform expansion of the majority of tumor-infiltrating T cells, thus leaving overall TIL diversity unaffected. The stability of population diversity among Foxp3+ regulatory T cells was not surprising given these cells did not expand during treatment, but to our knowledge this is the first report of TCRβ chain diversity in tumor-infiltrating regulatory CD4+ T cells. Of course, our observations here are limited to one particular murine melanoma model. Whether the success of immunotherapy in human cancer patients is also independent of changes in TIL diversity has yet to be determined. It is likely that different therapeutic approaches and distinct cancers could yield new conclusions.
Acknowledgements
The authors thank Sherri Koehm and Joy Eslick (Saint Louis University) for technical assistance with flow cytometry and cell sorting.
Abbreviations
- ATCC
American Type Culture Collection
- BLAST
Basic local alignment search tool
- BLOSUM62
BLOck substitution matrix 62
- CDR3
Complementarity determining region
- DAB
Diaminobenzidine
- Eomes
Eomesodermin
- FIR
Foxp3-IRES-mRFP
- GzmB
Granzyme B
- ICB
Immune checkpoint blockade
- Iono
Ionomycin
- IRES
Internal ribosome entry site
- LAG-3
Lymphocyte-activation gene-3
- RFP
Red fluorescent protein
- T-bet
T-box expressed in T cells
- TbetZsG
T-bet-ZsGreen
- TH1
T helper 1 cell
- Treg
T regulatory cell
Author contributions
LMK helped develop methodology, performed all experiments, analyzed and interpreted data, and helped write the manuscript. RMT was the study supervisor, designed the study, and helped write the manuscript. KW and RJD assisted in translating and interpreting the TCR sequencing results as well as writing the manuscript. JZ performed the immunohistochemistry staining and analysis.
Funding
This work was supported by a grant from the Alvin J. Siteman National Cancer Institute Comprehensive Cancer Center and The Foundation for Barnes-Jewish Hospital (Grant no. P30 CA091842) to Ryan M. Teague.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All mice were maintained under specific pathogen-free conditions and used in accordance with protocols established by the Institutional Animal Care and Use Committee of the Department of Comparative Medicine, Saint Louis University School of Medicine. All work with mice was performed according to protocol #2437 approved by the Animal Care and Use Program at Saint Louis University (Office of Laboratory Animal Welfare Assurance number D16-00141) which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALACI).
Animal source
TbetZsG original breeder mice were obtained from Taconic Farms, and the FIR mice were a gift from Daniel Hawiger’s laboratory (Saint Louis University).
Cell line authentication
The B16-F0 cell line was obtained commercially from and authenticated by American Type Culture Collection (ATCC) (Cat# CRL-6322). The cell line was free of mycoplasma, and correct morphology was confirmed by microscopy.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4(2):123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 2.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 5.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, Herati RS, Mansfield KD, Patsch D, Amaravadi RK, Schuchter LM, Ishwaran H, Mick R, Pryma DA, Xu X, Feldman MD, Gangadhar TC, Hahn SM, Wherry EJ, Vonderheide RH, Minn AJ. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, Adamow M, Kuk D, Panageas KS, Carrera C, Wong P, Quagliarello F, Wubbenhorst B, D’Andrea K, Pauken KE, Herati RS, Staupe RP, Schenkel JM, McGettigan S, Kothari S, George SM, Vonderheide RH, Amaravadi RK, Karakousis GC, Schuchter LM, Xu X, Nathanson KL, Wolchok JD, Gangadhar TC, Wherry EJ. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017 doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, Zitvogel L. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. 2016;44(6):1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH, Mardis ER, Linette GP. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, Fong L. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med. 2014;6(238):238ra270. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert L, Tsoi J, Wang X, Emerson R, Homet B, Chodon T, Mok S, Huang RR, Cochran AJ, Comin-Anduix B, Koya RC, Graeber TG, Robins H, Ribas A. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res. 2014;20(9):2424–2432. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosby EJ, Wei J, Yang XY, Lei G, Wang T, Liu CX, Agarwal P, Korman AJ, Morse MA, Gouin K, Knott SRV, Lyerly HK, Hartman ZC. Complimentary mechanisms of dual checkpoint blockade expand unique T-cell repertoires and activate adaptive anti-tumor immunity in triple-negative breast tumors. Oncoimmunology. 2018;7(5):e1421891. doi: 10.1080/2162402X.2017.1421891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoi A, Takeda K, Nagaoka K, Iino T, Matsushita H, Ueha S, Aoki S, Matsushima K, Kubo M, Morikawa T, Kitaura K, Suzuki R, Kakimi K. Increased diversity with reduced “diversity evenness” of tumor infiltrating T-cells for the successful cancer immunotherapy. Sci Rep. 2018;8(1):1058. doi: 10.1038/s41598-018-19548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page DB, Yuan J, Redmond D, Wen YH, Durack JC, Emerson R, Solomon S, Dong Z, Wong P, Comstock C, Diab A, Sung J, Maybody M, Morris E, Brogi E, Morrow M, Sacchini V, Elemento O, Robins H, Patil S, Allison JP, Wolchok JD, Hudis C, Norton L, McArthur HL. Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after immunotherapy. Cancer Immunol Res. 2016;4(10):835–844. doi: 10.1158/2326-6066.CIR-16-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA. 2010;107(9):4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao JL, Savage PA. Unlocking the complexities of tumor-associated regulatory T cells. J Immunol. 2018;200(2):415–421. doi: 10.4049/jimmunol.1701188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berrien-Elliott MM, Yuan J, Swier LE, Jackson SR, Chen CL, Donlin MJ, Teague RM. Checkpoint blockade immunotherapy relies on T-bet but not Eomes to induce effector function in tumor-infiltrating CD8+ T cells. Cancer Immunol Res. 2015;3(2):116–124. doi: 10.1158/2326-6066.CIR-14-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, Feigenbaum L, Zhao K, Paul WE. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37(4):660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci USA. 2005;102(14):5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf KJ, Emerson RO, Pingel J, Buller RM, DiPaolo RJ. Conventional and regulatory CD4+ T cells that share identical TCRs are derived from common clones. PLoS One. 2016;11(4):e0153705. doi: 10.1371/journal.pone.0153705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbe C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, Smylie M, Dummer R, Hill A, Hogg D, Haanen J, Carlino MS, Bechter O, Maio M, Marquez-Rodas I, Guidoboni M, McArthur G, Lebbe C, Ascierto PA, Long GV, Cebon J, Sosman J, Postow MA, Callahan MK, Walker D, Rollin L, Bhore R, Hodi FS, Larkin J. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berrien-Elliott MM, Jackson SR, Meyer JM, Rouskey CJ, Nguyen TL, Yagita H, Greenberg PD, DiPaolo RJ, Teague RM. Durable adoptive immunotherapy for leukemia produced by manipulation of multiple regulatory pathways of CD8+ T-cell tolerance. Cancer Res. 2013;73(2):605–616. doi: 10.1158/0008-5472.CAN-12-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome I. Consortium IBC. Consortium IM-S. PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, Koslowski M, Kuhn AN, Britten CM, Huber C, Tureci O, Sahin U. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 29.Huang RY, Francois A, McGray AR, Miliotto A, Odunsi K. Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology. 2017;6(1):e1249561. doi: 10.1080/2162402X.2016.1249561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, Lao CD, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Hill A, Hogg D, Marquez-Rodas I, Jiang J, Rizzo J, Larkin J, Wolchok JD. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19(11):1480–1492. doi: 10.1016/S1470-2045(18)30700-9. [DOI] [PubMed] [Google Scholar]
- 31.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DSM, Pauken KE, Huang AC, Gangadhar TC, Amaravadi RK, Schuchter LM, Feldman MD, Ishwaran H, Vonderheide RH, Maity A, Wherry EJ, Minn AJ. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540e1512–1554e1512. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, Adamow M, Kuk D, Panageas KS, Carrera C, Wong P, Quagliarello F, Wubbenhorst B, D’Andrea K, Pauken KE, Herati RS, Staupe RP, Schenkel JM, McGettigan S, Kothari S, George SM, Vonderheide RH, Amaravadi RK, Karakousis GC, Schuchter LM, Xu X, Nathanson KL, Wolchok JD, Gangadhar TC, Wherry EJ. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, Dellemijn TA, Antony PA, Spiess PJ, Palmer DC, Heimann DM, Klebanoff CA, Yu Z, Hwang LN, Feigenbaum L, Kruisbeek AM, Rosenberg SA, Restifo NP. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198(4):569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzuto GA, Merghoub T, Hirschhorn-Cymerman D, Liu C, Lesokhin AM, Sahawneh D, Zhong H, Panageas KS, Perales MA, Altan-Bonnet G, Wolchok JD, Houghton AN. Self-antigen-specific CD8+ T cell precursor frequency determines the quality of the antitumor immune response. J Exp Med. 2009;206(4):849–866. doi: 10.1084/jem.20081382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB, Powderly JD, Leming PD, Lipson EJ, Puzanov I, Smith DC, Taube JM, Wigginton JM, Kollia GD, Gupta A, Pardoll DM, Sosman JA, Hodi FS. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32(10):1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, Allison JP. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170(6):1120e1117–1133e1117. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh DY, Cham J, Zhang L, Fong G, Kwek SS, Klinger M, Faham M, Fong L. Immune toxicities elicted by CTLA-4 blockade in cancer patients are associated with early diversification of the T-cell repertoire. Cancer Res. 2017;77(6):1322–1330. doi: 10.1158/0008-5472.CAN-16-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postow MA, Manuel M, Wong P, Yuan J, Dong Z, Liu C, Perez S, Tanneau I, Noel M, Courtier A, Pasqual N, Wolchok JD. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J Immunother Cancer. 2015;3:23. doi: 10.1186/s40425-015-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carpenter SM, Nunes-Alves C, Booty MG, Way SS, Behar SM. A higher activation threshold of memory CD8+ T cells has a fitness cost that is modified by TCR affinity during tuberculosis. PLoS Pathog. 2016;12(1):e1005380. doi: 10.1371/journal.ppat.1005380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunes-Alves C, Booty MG, Carpenter SM, Rothchild AC, Martin CJ, Desjardins D, Steblenko K, Kloverpris HN, Madansein R, Ramsuran D, Leslie A, Correia-Neves M, Behar SM. Human and murine clonal CD8+ T cell expansions arise during tuberculosis because of TCR selection. PLoS Pathog. 2015;11(5):e1004849. doi: 10.1371/journal.ppat.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas N, Best K, Cinelli M, Reich-Zeliger S, Gal H, Shifrut E, Madi A, Friedman N, Shawe-Taylor J, Chain B. Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics. 2014;30(22):3181–3188. doi: 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]