Abstract
Objective:
Enteric bacterial pathogens cause diarrheal disease and mortality at significant rates throughout the world, particularly in children younger than 5 years. Our ability to combat bacterial pathogens has been hindered by antibiotic resistance, a lack of effective vaccines, and accurate models of infection. With the renewed interest in bacteriophage therapy, we sought to use a novel human intestinal model to investigate the efficacy of a newly isolated bacteriophage against Shigella flexneri.
Methods:
An S. flexneri 2457T-specific bacteriophage was isolated and assessed through kill curve experiments and infection assays with colorectal adenocarcinoma HT-29 cells and a novel human intestinal organoid-derived epithelial monolayer model. In our treatment protocol, organoids were generated from intestinal crypt stem cells, expanded in culture, and seeded onto transwells to establish 2-dimensional monolayers that differentiate into intestinal cells.
Results:
The isolated bacteriophage efficiently killed S. flexneri 2457T, other S. flexneri strains, and a strain of 2457T harboring an antibiotic resistance cassette. Analyses with laboratory and commensal Escherichia coli strains demonstrated that the bacteriophage was specific to S. flexneri, as observed under co-culture conditions. Importantly, the bacteriophage prevented both S. flexneri 2457T epithelial cell adherence and invasion in both infection models.
Conclusions:
Bacteriophages offer feasible alternatives to antibiotics for eliminating enteric pathogens, confirmed here by the bacteriophage-targeted killing of S. flexneri. Furthermore, application of the organoid model has provided important insight into Shigella pathogenesis and bacteriophage-dependent intervention strategies. The screening platform described herein provides proof-of-concept analysis for the development of novel bacteriophage therapies to target antibiotic-resistant pathogens.
Keywords: bacteriophage, enteric pathogens, human intestinal organoid-derived epithelial infection model, Shigella flexneri
Children in the developing world are constantly exposed to enteric pathogens (1), an exposure that often leads to recurrent diarrhea, significant morbidity, and death. Repeated episodes of infectious diarrhea can cause environmental enteropathy, a condition defined by severe damage to the gastrointestinal (GI) tract, inadequate establishment of a healthy microbiome, stunted growth, and delayed development (2,3). Pathogenic Escherichia coli, Shigella, and Salmonella are prominent bacterial pathogens infecting children younger than 5 years of age throughout the globe (1,4,5). Unfortunately, vaccine development against these pathogens has not been highly effective, and antibiotic resistance causes significant treatment challenges (6–10). The World Health Organization recently included these bacteria on a list of antibiotic-resistant priority pathogens whose threat to human health demands new therapies (11). Thus, novel solutions are urgently needed to stop antibiotic-resistant infections and prevent the terrible consequences of recurrent childhood diarrhea.
Bacteriophages (phages), one of the most abundant and diverse entities present throughout the environment and the mammalian GI tract (12), have been increasingly recognized as potential tools to control pathogens. Phages can inhibit the growth of foodborne pathogens, including Shigella (13–15), while also preserving beneficial bacteria (16). More importantly, phages have shown promise in treating bacterial infections, from the first therapeutic use to treat Shigella infections to the renewed interest in phage therapy following the rise of antimicrobial resistance (12,17–20). Given phage specificity and the potential protection of the microbiome often not afforded by antibiotic treatment (21), we sought to investigate the efficacy of phages for targeting enteric pathogens. Herein, we provide proof-of-concept analysis of a Shigella-targeting phage tested in a novel human intestinal organoid-derived epithelial infection model. Insights gained from this work could facilitate the development of phage therapies for infectious diarrhea and environmental enteropathy.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Bacterial strains were routinely cultured in Luria-Bertani (LB) broth, tryptic soy broth (TSB), or TSB supplemented with 0.4% weight/volume (w/v) bile salts as previously described (22). The Shigella strains analyzed include Shigella flexneri serotype 2a strain 2457T, serotype 5a strain M90T, 2 additional clinical isolates of S. flexneri (serotypes 2a and 3a), a mutant of S. flexneri 2457T harboring a chloramphenicol resistance cassette (2457T/lpfA::cat constructed by the lambda red linear recombination method (22,23); S. dysenteriae strain 1617, and S. sonnei strain 53G. E. coli strains analyzed include the laboratory strain MC4100 and the commensal isolate HS (24). Plating for colony forming units was performed using TSB or LB plates with 1.5% agar and 0.025% Congo Red (CR, Sigma C6277). Chloramphenicol was used at 5 μg/mL where indicated.
Isolation of Bacteriophage Against S. flexneri
A bacteriophage-specific for S. flexneri 2457T was isolated from the undefined “Intesti Bacteriophage” cocktail obtained from the Eliava Institute (Tbilisi, Georgia) and purified by 3 serial plaque passages on S. flexneri 2457T. Where indicated, phage preparations were concentrated from sterile-filtered lysates of S. flexneri 2457T by polyethylene glycol (PEG)/NaCl. In brief, phages were precipitated by adding NaCl to approximately 1 mol/L and PEG-8,000 to 10% (w/v) and incubating solutions at 4°C overnight, then harvested at 12,000 × g for 60 minutes and resuspended in SM buffer (50 mmol/L Tris-HCl [pH 7.5], 100 mmol/L NaCl, 10 mmol/L MgSO4). A bacteriophage forming large plaques and able to rapidly clear a liquid culture of S. flexneri 2457T, subsequently referred to as Φ2457T, was chosen for further characterization.
For visualization of Φ2457T, PEG-purified samples were loaded onto Formvar/carbon-coated nickel grids (FCF200-Ni, Electron Microscopy Sciences, Hatfield, PA) and stained with 2% uranyl acetate for transmission electron microscopy (TEM) with an FEI Tecnai Spirit Transmission Electron Microscope operating at 80 kV.
For sequencing of the phage genome, purified phage preparations were treated with DNase and RNase to remove bacterial nucleic acid contamination, and then DNA was extracted using the ZR Viral DNA Kit with IC-XL columns (Zymo Research, Irvine, CA). Purified phage genomic DNA was submitted to the MIT BioMicro Center (Cambridge, MA) for Nextera library preparation for 150 bp paired-end read sequencing on a single Illumina MiSeq lane, yielding approximately 9.7 million paired-end reads. Sequencing reads were assembled and analyzed using Geneious R9 (25). The Velvet Assembler plug-in was initially run on 1% of reads with an optimized k-mer value of 93 to yield a maximum length contig of 50,311 bp. This contig was run through NCBI BLAST (26) to generate a list of related phages, and progressiveMauve (27) was used to align full genomes and predict gene identities. For alignment to phage T1, the draft genome was permuted so that the beginning of the sequence corresponded with that of the T1 genome from NCBI (GenBank AY216660). The annotated genome has been deposited on the NCBI repository with the accession number of MH917278.
Analysis of Φ2457T in Broth Cultures
Single colonies of Shigella or E. coli strains were used to inoculate LB or TSB for overnight growth at 37°C. The following day, bacteria were subcultured with a 1:50 dilution into the specified media and grown to mid-log phase, corresponding respectively, for E. coli and Shigella, to an optical density (OD600) of 0.5 and 0.7 and to culture titers of approximately 1.0 to 2.0 × 108 CFU/mL. To generate kill curves, different concentrations of Φ2457T (or a matched volume of SM buffer) were added to the mid-log cultures and OD600 was monitored for several hours. For coculture experiments in which S. flexneri 2457T and E. coli HS were simultaneously exposed to Φ2457T, a chloramphenicol-resistant S. flexneri mutant and E. coli HS were grown separately overnight in LB as described above and subsequently either subcultured separately or co-inoculated and grown to OD600 approximately 0.7. The killing assay analysis proceeded as described above. To confirm specificity of Φ2457T in these co-culture experiments, viable bacterial titers were determined by dilution plating the samples at various time points onto CR plates with and without chloramphenicol to select for S. flexneri and total bacteria, respectively. Plates were incubated overnight at 37°C; bacteria were then enumerated. S. flexneri colonies were chloramphenicol-resistant, smaller, and red on CR plates relative to the larger, white E. coli colonies on CR plates. Control experiments verified that E. coli HS was sensitive to the chloramphenicol concentration used.
Characterization of Φ2457T Efficacy With Infected Epithelial Cells
The human colorectal adenocarcinoma HT-29 cells were maintained and the adherence and invasion infection assays were performed as previously described (22,28). S. flexneri 2457T was grown in the same conditions as mentioned above and applied to each well of the HT-29 cells, at a multiplicity of infection (MOI) of 100:1, without centrifugation of the bacteria onto the cells. For infection with Φ2457T, 4.0 × 109 plaque forming units (MOI 40:1 relative to S. flexneri titers) were applied immediately after the addition of the infecting bacterial inoculum. Infection proceeded for 3 hours. As previously described for adherence, cells were washed with 1X PBS and lysed with 0.5% Triton X-100 to enumerate the recovery bacterial titers. For invasion, cells were incubated for 45 minutes in media with 50 μg/mL gentamicin, after which cells were washed and lysed with 0.5% Triton X-100 (22,28). Bacterial recovery titers were calculated as percent recovery relative to the infecting titers.
For analyses with the human intestinal organoid-derived epithelial monolayer (HIODEM) model, the isolation, preparation of human intestinal epithelial cells, and seeding and differentiation on transwells were performed as previously described (29– 32). Human sample collection was approved by the institutional review board protocol 2015P002725, Massachusetts General Hospital, Boston, MA. Donor biopsies were obtained from consenting patients undergoing routine colonoscopies for clinical evaluations by a licensed physician. Following the isolation and propagation of intestinal crypt-derived organoids, organoid-derived cell monolayers were generated as previously described (29–31). Monolayers were used for infection analysis once the cultures reached confluence as determined by transepithelial electrical resistance monitoring and microscopic observation. At 48 hours before each experiment, the basolateral media were replenished. In the apical chamber, media were replaced with complete dulbecco’s modified eagle medium (DMEM) plus 5 μM of the γ-secretase inhibitor IX (N-[(3,5-Difluorophenyl)acetyl]-L-alanyl-2-phenylglycine-1,1-dimethylethyl ester; Calbiochem, San Diego, CA) to promote cell differentiation (29,31). On the day of each experiment, monolayers were washed with 1X PBS, both apical and basolateral media were replaced with DMEM without phenol red, and monolayers were incubated for at least 2 hours before the initiation of the experiment. S. flexneri 2457T was subcultured in TSB + 0.4% bile salts and prepared for adherence and invasion analyses as described above for the HT-29 cells (22,28). The bacterial MOI for the control infection was 150:1 while the phage MOI relative to the bacteria was 100:1.
Microscopy
HIODEM monolayers were fixed in 4% paraformaldehyde at room temperature for 30 minutes followed by storage in 70% ethanol until paraffin embedding. For immunofluorescence analysis, sections were stained using mouse anti-EpCAM for enterocytes and rabbit-anti-Shigella-biotin conjugated antibodies (Thermo Fisher, Waltham, MA). Fluorescent-conjugated secondary monoclonal antibodies or streptavidin-biotin conjugates were used for detection. 4’,6-Diamidino-2-phenylindole (Thermo Fisher) was used to stain the nuclei. Samples were imaged using a Nikon A1SiR confocal microscope. For TEM analysis, samples were fixed in 2% paraformaldehyde with 2.5% glutaraldehyde in 0.1 mol/L sodium cacodylate, mounted on grids, and imaged using a JEOL transmission electron microscope.
Statistical Analyses
All experiments were performed at least 2 independent times with technical triplicates for each experiment. All data plotted represent the average ± the standard error of the mean (SEM). Statistical significance was determined using a Student t test or an analysis of variance. A P value of <0.05 was considered significant.
RESULTS
Φ2457T Is Highly Specific for S. flexneri Strains
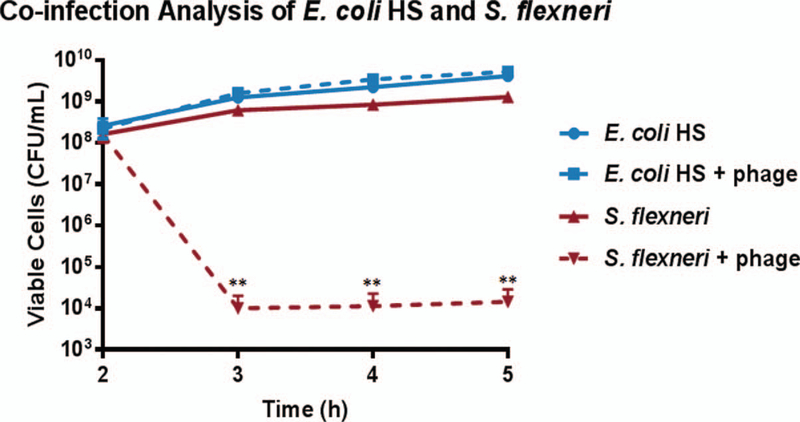
The Φ2457T phage (Fig. 1A) was found to be related to the well-characterized E. coli phage T1 (33), sharing approximately 70% nucleotide identity and having similar genomic structure (Supplementary Digital Content 1, http://links.lww.com/MPG/B515). We repeatedly observed efficient and specific binding of Φ2457T to S. flexneri 2457T (Fig. 1A) and killing of this strain in LB broth (Fig. 1B), and in TSB and TSB supplemented with bile salts (data not shown), which mimics in vivo conditions as the bacteria transit the GI tract (22). We also observed efficient killing of 2457T in tissue culture media (DMEM) following 2-hour growth in the 3 media types listed above (data not shown). Φ2457T killed S. flexneri serotypes 2a and 3a but not 5a (Fig. 1B). Interestingly, Φ2457T efficiently killed S. dysenteriae strain 1617 at higher concentrations but did not observably kill S. sonnei strain 53G (Fig. 1C). Φ2457T did not kill an E. coli laboratory strain or commensal E. coli strain HS in liquid cultures (Fig. 1D). The specificity of Φ2457T for S. flexneri was confirmed in coculture experiments of E. coli HS and 2457T in which plating was performed on differential media: only 2457T was susceptible to the phage (Fig. 2). In all, the data demonstrate that Φ2457T is highly specific for S. flexneri 2457T.
FIGURE 1.
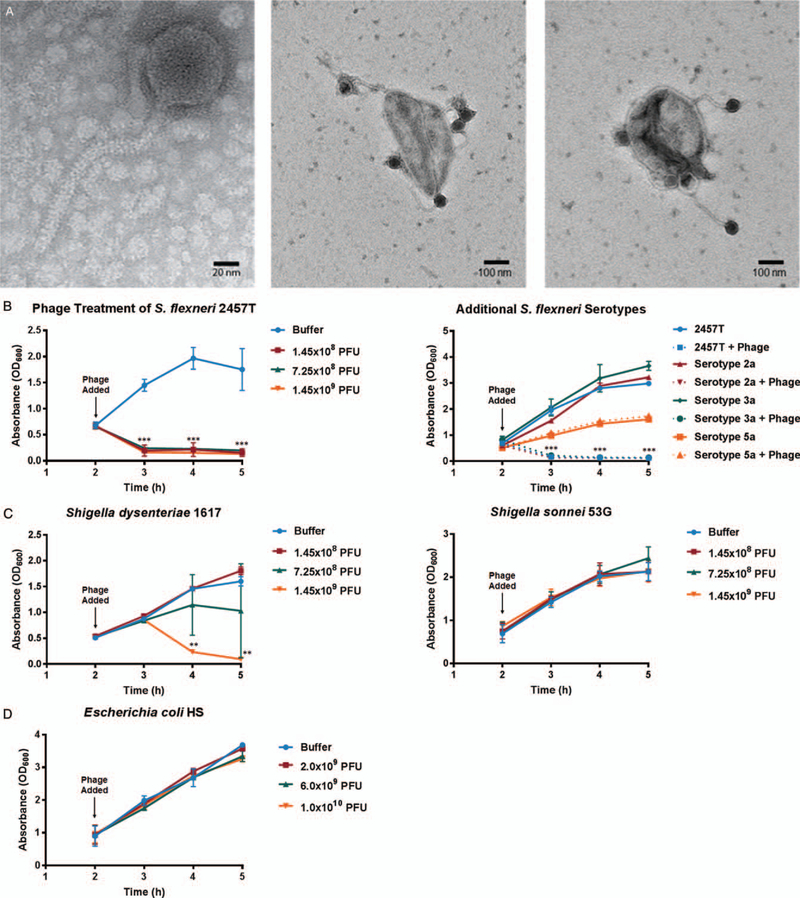
Efficacy of Φ2457T against Shigella flexneri 2457T. A, Transmission electron microscopy (TEM) image of Φ2457T phage (left) and the phage attached to S. flexneri 2457T (center and right). B, Kill curve analysis for S. flexneri 2457T in LB broth using various concentrations of Φ2457T is provided on the left. Efficient killing (red, green, and yellow lines, ***, P < 0.001) was observed when phage was applied to the cultures at 2 hours (arrow) compared to continuous growth of S. flexneri in control media (blue line). Data represent the average of 3 independent experiments ± SEM. Additional analysis of S. flexneri serotypes 2a, 3a, and 5a are provided on the right. Killing was detected for clinical isolates 2a and 3a (***, P < 0.001); however, no killing was detected for S. flexneri 5a laboratory strain M90T (yellow line). Data represent the average of 2 independent experiments ± SEM. C, Analysis of S. dysenteriae strain 1617 (left) and S. sonnei strain 53G (right). Killing was detected for S. dysenteriae at higher phage concentrations (yellow line, **, P ≤ 0.01 at 4 and 5 hours relative to buffer). No killing was detected for S. sonnei at phage concentrations tested. Data represent the average of 2 independent experiments ± SEM. D, Analysis of E. coli HS (commensal). No killing was detected at any concentration tested. Data represent the average of 2 independent experiments ± SEM. PFU = plaque forming units.
FIGURE 2.
Analysis of Φ2457T in co-cultures of S. flexneri 2457T and E. coli HS. Φ2457T (1.45 × 108 PFU) or control buffer was added to co-cultures at 2 hours, followed by bacterial enumeration (CFU/mL) for each strain. An average of 2.5 to 3.2 × 109 CFU/mL of E. coli HS was recovered from cocultures ± phage treatment. An average of 3.6 × 108 CFU/mL of S. flexneri 2457T was recovered from cocultures without phage while significantly fewer (1.0 × 104, **, P < 0.01) colonies were recovered with phage. Data represent an average of 3 independent experiments ± SEM.
Characterization of Φ2457T Efficacy in S. flexneri Infection
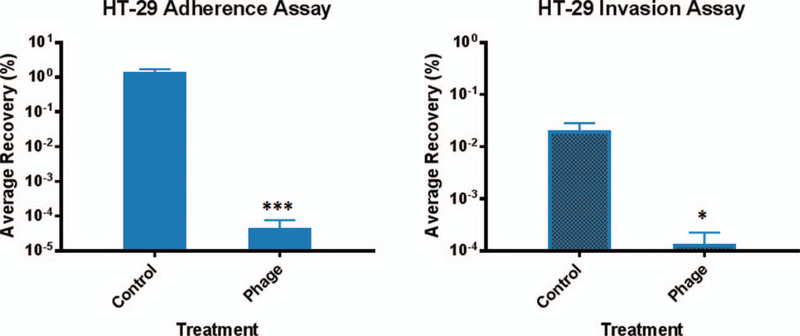
As an initial test, we analyzed Φ2457T during S. flexneri 2457T infection of human colonic HT-29 cells by measuring bacterial adherence and invasion in the presence and absence of the phage. The control bacterial MOI for the HT-29 cell infection was 100:1 and the phage-to-bacteria MOI was 40:1. Following growth of S. flexneri 2457T in LB, Φ2457T repeatedly prevented both bacterial adherence to and invasion of the epithelial cells (Fig. 3). Similar results were obtained when the bacteria were subcultured in TSB or TSB supplemented with bile salts before infection (data not shown). The few 2457T colonies that were recovered from the phage-treated wells were irregular in shape and white on CR plates, indicating loss of virulence (34). A screen verified that these colonies were not phage resistant (data not shown), indicating that the phage treatment effectively lysed S. flexneri 2457T, thereby preventing both adherence to and invasion of HT-29 cells.
FIGURE 3.
Φ2457T prevents both adherence to and invasion of S. flexneri 2457T in HT-29 cells. Following subculture of 2457T in LB media, adherence (left), and invasion (right) assays were performed +/− phage. Phage treatment routinely resulted in no or very few 2457T recovery colonies (P < 0.001 (***) for adherence, P = 0.025 (*) for invasion), indicating that the presence of the phage inhibited both adherence and invasion of S. flexneri 2457T. The average percent recovery of bacterial titers from the infection is plotted ± the SEM from 6 independent experiments, each with technical triplicates.
To confirm the results with a more physiologically relevant infection model, we analyzed Φ2457T in the HIODEM model. The model is derived from stem cells isolated from intestinal tissue, propagated as organoids, and subsequently trypsinized and seeded onto transwells to generate a two-dimensional polarized, differentiated model of the intestinal epithelium in which enterocytes, mucus-producing goblet cells, and antigen sampling M cells are present (29–31). The model used was derived from the cecum, the cellular architecture of which was confirmed by TEM (Fig. 4A). Additional characterization of the model is provided in Supplementary Digital Content 2 (http://links.lww.com/MPG/B516). Following subculture of the bacteria in the in vivo–like conditions of TSB supplemented with bile salts, S. flexneri 2457T adhered to and invaded the model efficiently. The infecting bacterial MOI for the model was 150:1. Infection was, however, significantly reduced following the administration of Φ2457T at a phage-to-bacteria MOI of 100:1 (Fig. 4B). Confocal immunofluorescence analysis confirmed both the infection and the efficacy of Φ2457T treatment (Fig. 4C). As with the HT-29 infection assays, any 2457T colonies recovered from the phage treatment analyses appeared irregular in shape and white on CR plates, and remained phage sensitive upon further testing. Taken together, these results confirmed the efficacy of phage treatment with Φ2457T in in vivo–like conditions, and established HIODEM as an effective model of S. flexneri infection and a platform for the preclinical testing of therapeutics against human-specific pathogens such as Shigella.
FIGURE 4.
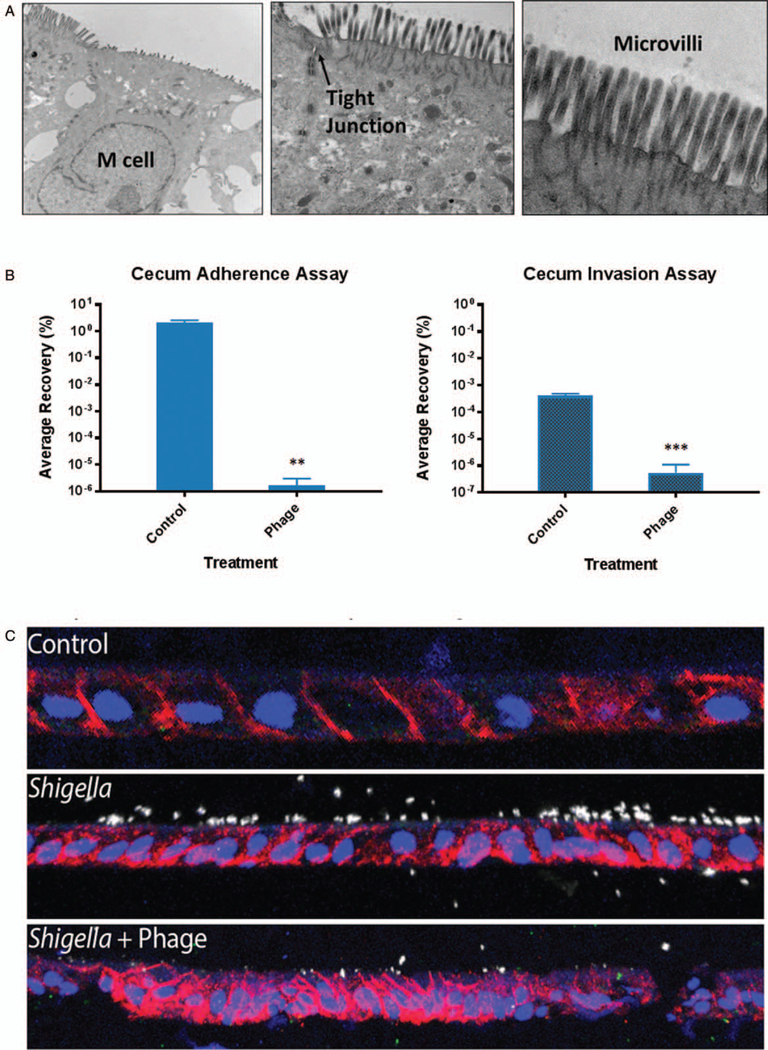
Efficacy of Φ2457T in the human intestinal organoid-derived epithelial monolayer (HIODEM) model. A, Cecum transmission electron microscopy (TEM) analysis showing M cells (absent microvilli, left, 12,000×), enterocyte tight junctions (middle, 25,000×), and mature enterocyte microvilli (right, 100,000×). B, Φ2457T prevents adherence (left, **, P = 0.01) and invasion (right, ***, P = 0.003). The average percent 2457T infection recovery is plotted ± SEM from 6 independent experiments, each with technical triplicates. C, Confocal immunofluorescence (red, EpCAM, intestinal epithelial cells; white anti-LPS Shigella; blue, 4’,6-diamidino-2-phenylindole [DAPI], nuclei) of uninfected organoids (top), 2457T infection control (middle), and 2457T infection with phage treatment (bottom). A reduction in 2457T infection (white) resulted from phage treatment.
DISCUSSION
While the global incidence of diarrheal disease has substantially decreased in the past 25 years owing in large part to improved nutrition, clean water, and vaccination campaigns against rotavirus (35), rates of enteric infections, especially from human-restricted bacterial pathogens, are still significant. Shigella remains the second leading cause of all diarrheal deaths and the third leading cause of diarrheal deaths in children under the age of 5 years (36). Children who survive infection risk continual exposure to pathogens, development of environmental enteropathy, and long-term effects on growth and development (2,3). Current challenges to treat diarrheal diseases include both lack of effective vaccines (37,38) and antibiotic resistance. Vaccine development has been complicated by our incomplete understanding of pathogenesis (22) and limited animal models, which do not adequately replicate human infection (39). New technologies and novel approaches are urgently needed to develop successful therapies against enteric pathogens such as Shigella.
Bacteriophage therapy is a promising alternative to antibiotics; however, the idea has been revitalized only recently so current studies are limited (12,17–20). Discrepancies between in vitro and in vivo efficacy analyses with animal models (40) are a concern. Therefore, the overall goal of this study was to test phage therapy efficacy against the human-specific pathogen S. flexneri using a preclinical human intestinal model closely resembling in vivo infection (HIODEM). Here, we have shown that a phage isolated against S. flexneri 2457T is effective at targeting the pathogen in multiple types of media. Of interest was testing phage efficacy in in vivo–like conditions to examine killing potential in environments present in the human host. Therefore, we used bile salts to mimic conditions encountered by Shigella in the small intestine before infection in the colon (22). The fact that the phage was still effective in bile salts is encouraging, particularly since bile exposure can increase the antibiotic tolerance of bacterial pathogens (22,41). Furthermore, Φ2457T effectively and specifically killed an antibiotic-resistant strain of S. flexneri used in the co-culture experiments with E. coli HS. Therefore, our data suggest that phage therapy may deliver improved targeted therapeutic efficacy over conventional antibiotics. Reports in the clinical setting have also indicated such outcomes (19–21).
In host range assays, effective killing by Φ2457T was restricted to S. flexneri 2457T and clinical isolates of S. flexneri serotypes 2a and 3a. Phage efficacy was diminished against S. dysenteriae while killing was not detected against the S. flexneri serotype 5a or S. sonnei strains tested. These results are not surprising, as the phage used here was isolated from a cocktail for activity against S. flexneri 2457T. Phage specificity and range are critical concerns for bacteriophage-based antibacterial treatments, especially when considering phage to treat human infections (42). Our future goals are to take new approaches in phage engineering (43,44) to improve phage specificity against multiple Shigella strains and to extend the specificity to additional enteric bacteria such as Salmonella and pathogenic E. coli (45). Furthermore, phage engineering would enable us to target pathogens without harming the complex symbiotic population of the GI microbiota. The fact that Φ2457T did not kill E. coli MC4100 or HS is encouraging, but additional investigations using complete bacterial communities are warranted to ensure that commensal organisms remain viable during treatment. We expect that therapeutic use of engineered phage will have improved efficacy over conventional antibiotics and may represent an innovative treatment option for malnourished children at risk of developing environmental enteropathy, for whom the microbiota is especially critical to growth and development. Finally, engineered phages could also be employed to address concerns of phage resistance in bacterial pathogens or concerns of phage recombination leading to the acquisition and spread of virulence genes (12,46).
Of equal importance is the use of reliable human infection models to evaluate preclinical therapeutic candidates. The second goal of this study was to test the use of a human-specific infection model to evaluate phage efficacy. The HIODEM model is derived from tissue samples of the cecum, the intraperitoneal pouch at the beginning of the colon. Additional HIODEM models generated from the terminal ileum of the small intestine provide high-through-put tools in which noncancerous enterocytes, M cells, and mucus-producing goblet cells are represented. These models can be used to analyze infection by bacterial pathogens and to enable a better understanding of how enteric pathogens infect the GI tract (29,30).
We demonstrated S. flexneri 2457T adherence to and invasion of the human cecum HIODEM model. Importantly, we obtained invasion following apical administration of the bacteria, without the need to artificially centrifuge the bacteria onto the cells (47), closely mimicking the conditions of human infection. This result is important because Shigella is unable to invade via the apical surface and must transit M cells or disrupt the epithelial barrier to reach the basolateral pole of the epithelium for invasion (48,49). Thus, our model more accurately reproduces natural Shigella infection than previous analyses, which required direct basolateral administration of bacteria by seeding inverted human adenocarcinoma cells in order to examine Shigella infection of polarized epithelial cells (50). Finally, the HIODEM model is expected to provide a tool for reliable screening of therapeutic candidates to help advance products into clinical development. As demonstrated in this study, the addition of Φ2457T prevented S. flexneri adherence and invasion in the HIODEM model, as was observed in HT-29 cells. Thus, we have developed an efficient, safe model to examine bacteriophage treatment of Shigella infection. We expect to apply this model to other aspects of therapeutic development.
In conclusion, this study demonstrates that a wild-type phage specifically killed the pathogenic strain S. flexneri 2457T without affecting the commensal strain E. coli HS. Furthermore, Φ2457T was effective against S. flexneri 2457T following exposure to in vivo–like culture conditions, which have been shown to increase the antibiotic resistance potential of the pathogen. Efficacy of Φ2457T was demonstrated in both traditional assays and the novel, human-specific model of the human GI epithelium, which was shown to accurately recapitulate the natural S. flexneri 2457T infection process. Future directions include applying this infection model to newly isolated or engineered phages to improve specificity, minimize effects on the microbiota, and circumvent the development of phage resistance in bacterial pathogens. Ultimately, it is expected that phage therapy will enhance our ability to combat infectious diarrhea, help to overcome the burdens of antibiotic resistance, and provide prophylactic and treatment options for people exposed to various enteric pathogens, particularly malnourished children in the developing world.
Supplementary Material
What Is Known
Infectious diarrhea causes a significant global health burden, particularly in children in the developing world.
Bacteriophage therapy has the potential to become a promising treatment alternative for diarrhea-causing bacterial pathogens in an era of rampant antimicrobial resistance.
What Is New
Bacteriophages can specifically kill Shigella flexneri in numerous growth and human coculture infection conditions, potentially without harming commensal bacteria.
Combined with a novel human-specific infection model, this work serves as the basis for future development of bacteriophage therapy against enteric bacterial pathogens.
Acknowledgments:
The authors would like to thank Dr. Brett Swierczewski, Walter Reed Army Institute of Research, for the clinical isolates of S. flexneri used in this study; Ms. Diane Capen, Microscopy Core Facility at the Center for Systems Biology, MGH, for TEM sample processing; Dr. Mark Mimee in Dr. Lu’s lab for assistance with the sequence analysis; and Dr. Karen Pepper in Dr. Lu’s laboratory for critical reading of the manuscript.
Funding was provided through the Bill and Melinda Gates Foundation grant OPP1139856. Additional financial support was provided by the National Institute of Allergy and Infectious Diseases grants U19-AI082655 (Cooperative Center on Human Immunology to A.F.) and K22AI104755 (to C.S.F.). The Lu lab acknowledges additional financial support by the National Science Foundation (MCB-1350625, 1521925), the National Institutes of Health (5-P50-GM098792–05 and 4-R33-AI121669–03), the Office of Naval Research (N00014–13-l-0424), the Army Research Office (W911NF-ll-1–0281), the Broad Institute, the Koch Institute, the Defense Threat Reduction Agency (HDTRAl-14–1-0007), the Kenneth Rainin Foundation (2016–3066), the Ellison Foundation (AG-NS-0948–12), the Wertheimer Fund, the Institute for Soldier Nanotechnologies (W9111NF-13-D-0001), J-WAFS, Novartis (14081955), the Desphande Center at MIT, the Singapore-MIT Alliance for Research and Technology, the Center for Microbiome Informatics and Technology (15127713), and Excet, Inc (17033835). The TEM core at MGH is supported by National Institute of Neurological Disorders and Stroke P30NS045776. Support for the Philly Dake Electron Microscope Facility was provided by the National Institutes of Health grant 1S10RR023594S10 and by funds from the Dake Family Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in the study decision, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
T.K.L. is a co-founder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, and Eligo Biosciences. T.K.L. also holds financial interests in nest.bio, Ampliphi, IndieBio, and MedicusTek.
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013;382:209–22. [DOI] [PubMed] [Google Scholar]
- 2.Lee G, Paredes Olortegui M, Penataro Yori P, et al. Effects of Shigella-,Campylobacter- and ETEC-associated diarrhea on childhood growth. Pediatr Infect Dis J 2014;33:1004–9. [DOI] [PubMed] [Google Scholar]
- 3.Prendergast AJ, Kelly P. Interactions between intestinal pathogens, enteropathy and malnutrition in developing countries. Curr Opin Infect Dis 2016;29:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015;12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marks F, Von Kalckreuth V, Aaby P, et al. Incidence of invasive Salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study. Lancet Glob Health 2017;5:e310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya D, Bhattacharya H, Sayi DS, et al. Changing patterns and widening of antibiotic resistance in Shigella spp. over a decade (2000–2011), Andaman Islands, India. Epidemiol Infect 2015;143:470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013;26:822–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lokken KL, Walker GT, Tsolis RM. Disseminated infections with antibiotic-resistant non-typhoidal Salmonella strains: contributions of host and pathogen factors. Pathog Dis 2016;74:pii: ftw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump JA, Sjolund-Karlsson M, Gordon MA, et al. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 2015;28:901–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puzari M, Sharma M, Chetia P. Emergence of antibiotic resistant Shigella species: a matter of concern. J Infect Public Health 2018; 11:451–4. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017; Press Release. Available at: http://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed Accessed October 30, 2018.
- 12.Goodridge LD. Bacteriophages for managing Shigella in various clinical and non-clinical settings. Bacteriophage 2013;3:e25098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anany H, Chen W, Pelton R, et al. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in meat by using phages immobilized on modified cellulose membranes. Appl Environ Microbiol 2011;77:6379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagens S, Loessner MJ. Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr Pharm Biotechnol 2010;11:58–68. [DOI] [PubMed] [Google Scholar]
- 15.Shahin K, Bouzari M. Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J Food Sci Technol 2018;55:550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffey B, Mills S, Coffey A, et al. Phage and their lysins as biocontrol agents for food safety applications. Annu Rev Food Sci Technol 2010;1:449–68. [DOI] [PubMed] [Google Scholar]
- 17.Reardon S. Phage therapy gets revitalized. Nature 2014;510:15–6. [DOI] [PubMed] [Google Scholar]
- 18.Pallavali RR, Degati VL, Lomada D, et al. Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS One 2017;12:e0179245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalifa L, Gelman D, Shlezinger M, et al. Defeating antibiotic- and phage-resistant Enterococcus faecalis using a phage cocktail in vitro and in a clot model. Front Microbiol 2018;9:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua Y, Luo T, Yang Y, et al. Phage therapy as a promising new treatment for lung infection caused by Carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol 2017;8:2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng M, Liang J, Zhang Y, et al. The bacteriophage EF-P29 efficiently protects against lethal vancomycin-resistant Enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Front Microbiol 2017;8:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickerson KP, Chanin RB, Sistrunk JR, et al. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun 2017;85:pii: e01067–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000;97:6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faherty CS, Harper JM, Shea-Donohue T, et al. Chromosomal and plasmid-encoded factors of Shigella flexneri induce secretogenic activity ex vivo. PLoS One 2012;7:e49 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 27.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010;5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faherty CS, Wu T, Morris CR, et al. The synthesis of OspD3 (ShET2) in Shigella flexneri is independent of OspC1. Gut Microbes 2016;7:486–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senger S, Ingano L, Friere R, et al. Human fetal-derived enterospheres provide insights on intestinal development and a novel model to study necrotizing enterocolitis (NEC). Cell Mol Gastroenterol Hepatol 2018;5:549–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickerson KP, Senger S, Zhang Y, et al. Salmonella typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EBioMedicine 2018;31:92–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015;64:911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build cryptvillus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- 33.Roberts MD, Martin NL, Kropinski AM. The genome and proteome of coliphage T1. Virology 2004;318:245–66. [DOI] [PubMed] [Google Scholar]
- 34.Maurelli AT, Blackmon B, Curtiss R 3rd. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun 1984;43:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 2017;17:909–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotloff KL. Shigella infection in children and adults: a formidable foe. Lancet Glob Health 2017;5:e1166–7. [DOI] [PubMed] [Google Scholar]
- 37.Barry EM, Pasetti MF, Sztein MB, et al. Progress and pitfalls in Shigella vaccine research. Nat Rev Gastroenterol Hepatol 2013;10: 245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitisuttithum P, Islam D, Chamnanchanunt S, et al. Clinical trial of an oral live Shigella sonnei vaccine candidate, WRSS1, in Thai adults. Clin Vaccine Immunol 2016;23:564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim YJ, Yeo SG, Park JH, et al. Shigella vaccine development: prospective animal models and current status. Curr Pharm Biotechnol 2014;14:903–12. [DOI] [PubMed] [Google Scholar]
- 40.Maura D, Morello E, Du Merle L, et al. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ Microbiol 2012;14:1844–54. [DOI] [PubMed] [Google Scholar]
- 41.Sistrunk JR, Nickerson KP, Chanin RB, et al. Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev 2016;29:819–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardy P, Pantucek R, Benesik M, et al. Genetically modified bacteriophages in applied microbiology. J Appl Microbiol 2016;121: 618–33. [DOI] [PubMed] [Google Scholar]
- 43.Citorik RJ, Mimee M, Lu TK. Bacteriophage-based synthetic biology for the study of infectious diseases. Curr Opin Microbiol 2014;19:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 2014;32:1141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ando H, Lemire S, Pires DP, et al. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst 2015;1:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abedon ST, Kuhl SJ, Blasdel BG, et al. Phage treatment of human infections. Bacteriophage 2011;1:66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hale TL, Formal SB. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect Immun 1981;32:137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 2008;21:134–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maldonado-Contreras A, Birtley JR, Boll E, et al. Shigella depends on SepA to destabilize the intestinal epithelial integrity via cofilin activation. Gut Microbes 2017;8:544–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCormick BA, Siber AM, Maurelli AT. Requirement of the Shigella flexneri virulence plasmid in the ability to induce trafficking of neutrophils across polarized monolayers of the intestinal epithelium. Infect Immun 1998;66:4237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.