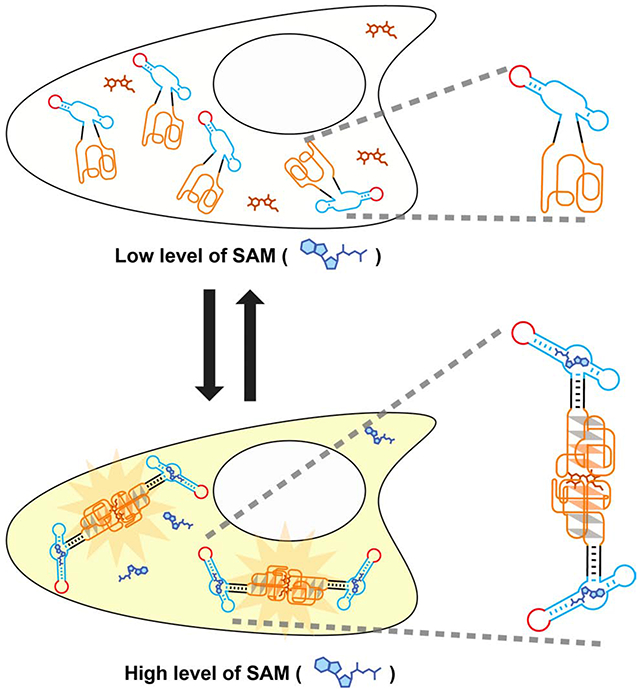
Summary
Corn is a fluorogenic RNA aptamer that forms a high-affinity quasi-symmetric homodimer. The Corn dimer interface binds DFHO, resulting in highly photostable yellow fluorescence. Because of its photostability, Corn would be useful in RNA-based small molecule biosensors, where quantitative accuracy would be affected by photobleaching. Here we describe a strategy for converting the constitutive Corn dimer into a small-molecule regulated fluorescent biosensor that detects S-adenosyl methionine (SAM) in vitro and in living cells. We fused the Corn aptamer into a helical stem that was engineered by circularly permuting the SAM aptamer from the SAM-III riboswitch. In the absence of SAM, the Corn portion of this fusion RNA is unable to dimerize. However, upon binding SAM, the RNA dimerizes and binds DFHO. This RNA-based biosensor enables detection of SAM dynamics in living mammalian cells. Together, these data describe a class of RNA-based biosensor based on small-molecule regulated dimerization of Corn.
Graphical Abstract
eTOC Blurb
Corn is a fluorogenic RNA aptamer that forms a high-affinity quasi-symmetric homodimer and exhibits remarkable photostability. Kim et al. describe a Corn-based sensor which enables detection of metabolite in live mammalian cells with reduced photobleaching. Upon binding its target metabolite, this Corn-based sensor undergoes dimerization which leads to its fluorescence.
Introduction
Although most genetically encoded small molecule sensors are engineered proteins, sensors can also be designed using RNA. RNA-based sensors comprise an analyte-binding aptamer and a fluorogenic RNA aptamer that becomes fluorescent only when the target is present (Paige et al., 2012). It is potentially easier to develop new sensors using RNA rather than protein since it is relatively easy to generate target-binding RNA aptamers using the SELEX (systematic evolution of ligands by exponential enrichment) technique (Ellington and Szostak, 1990; Tuerk and Gold, 1990).
Until recently, genetically encoded RNA-based sensors were exclusively used in bacterial cells (Kellenberger et al., 2013; Litke and Jaffrey, 2019; Paige et al., 2012; Su et al., 2016; You et al., 2015). RNA-based sensors could not be used in mammalian cells since small engineered RNAs are rapidly degraded and do not accumulate to levels needed to generate a detectable fluorescence signal (Filonov et al., 2015; Litke and Jaffrey, 2019). However, the recently developed Tornado (Twister-optimized RNA for durable overexpression) expression system allows RNA aptamers and RNA-based sensors to be expressed as highly stable circular RNAs that accumulate to micromolar levels, similar to protein-based sensors (Litke and Jaffrey, 2019). Using this new expression approach, RNA-based sensors can generate sufficient fluorescence signals needed for imaging small molecules in mammalian cells.
Here we describe a class of RNA-based sensors that function by undergoing target-induced RNA dimerization. These sensors use the Corn fluorogenic aptamer, which binds DFHO (3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-oxime), its fluorogenic ligand, to induce yellow fluorescence. Corn has an unusual structure, which involves two Corn aptamers binding to form an exceptionally tight binding dimer (Kd < 1 nM) (Warner et al., 2017). Dimerization is independent of DFHO, but the dimer interface provides the binding site for DFHO. To create RNA-based sensors that utilize Corn, we fused Corn into a stem-loop structure of the SAM aptamer, in a way that disrupts Corn fluorescence. Upon SAM binding, the Corn-based sensor switches from an RNA monomer to an RNA dimer, resulting in Corn fluorescence. We show that SAM-regulated Corn dimerization enables simple detection of SAM levels in mammalian cells with reduced photobleaching. Overall, these results demonstrate a class of RNA-based sensor in which metabolite binding controls RNA dimerization.
Results
Strategy for designing metabolite sensors using Corn
To design an RNA-based sensor, current approaches require an “entry point” in the fluorogenic aptamer (Paige et al., 2012). For example, Spinach contains a stem-loop structure near its fluorophore-binding pocket (Paige et al., 2011) that serves as an entry point into which a metabolite-binding aptamer is inserted (Figure S1A).
However, Corn lacks an entry point. Structural analysis of Corn (Warner et al., 2017) shows that its only stem provides its 5’ and 3’ ends at its base (Figure 1A). Insertion of a metabolite-binding aptamer elsewhere into Corn is not possible since mutation anywhere results in a near complete loss of fluorescence (Song et al., 2017; Warner et al., 2017). Thus, it is not clear how to insert a metabolite-binding aptamer into Corn to turn it into an RNA-based biosensor.
Figure 1. Strategy for designing metabolite sensors using Corn.
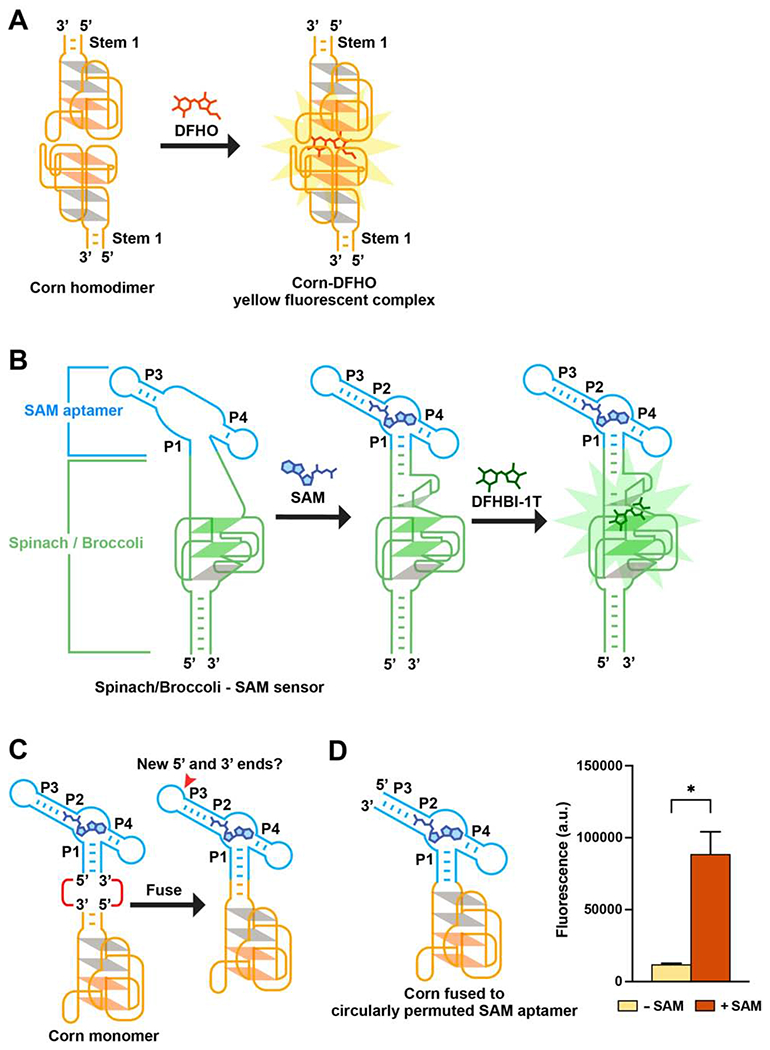
(A) Corn contains one helical stem (Stem 1) unlike Spinach or Broccoli. DFHO binds the Corn dimer interface, between the apical G-quadruplex in each Corn monomer. Orange parallelograms indicate the two G-quartets. Grey parallelograms indicate two mixed-sequence tetrads.
(B) Schematic diagram of the Spinach-based SAM sensor. SAM induces folding of P1, which serves as a transducer domain. P1 folding enables Spinach folding and binding to DFHBI-1T (right).
(C) Since the P1 stem has the 5’ and 3’ ends of the SAM aptamer, fusing Corn to the SAM aptamer P1 stem would result in an RNA lacking 5’ and 3’ ends. The P3 stem contains the new 5’ and 3’ ends.
(D) Corn fused to the circularly permuted SAM aptamer exhibits SAM-induced fluorescence. In vitro transcribed RNA (1 μM) was incubated with 0.1 mM SAM or water for 1 h (37°0 and 1 mM MgCl2). Addition of SAM resulted in a 7.4-fold increase in fluorescence (ex 505 nm; em 545 nm). Mean and SEM values are shown (n = 3). * P = 0.0384
We therefore considered another strategy for sensor design. Rather than using an entry point in the fluorogenic aptamer, we asked if an entry point in the metabolite-binding aptamer could be used. In this way, the fluorogenic aptamer can be inserted into the metabolite-binding aptamer at its entry point.
To test this, we focused on the SAM-binding aptamer from the SAM-III riboswitch (Lu et al., 2008). SAM binding induces a conformational change in the SAM-binding aptamer (Smith et al., 2010), which we reasoned could be used to regulate the folding of Corn.
We first identified a potential entry point in the SAM aptamer. The SAM aptamer folds into a three-way junction, with SAM binding at the junction (Lu et al., 2008; Wilson et al., 2011) (Figure S1B). When SAM binds, the P1 stem is stabilized by additional π-stacking interactions with the adenosine portion of SAM (Lu et al., 2008). In earlier studies, we fused the P1 stem into the entry point in Spinach, and showed that SAM binding to its aptamer induced Spinach folding and fluorescence (Figure 1B). Therefore, it may be possible to fuse the P1 stem into the stem of Corn, thus enabling SAM-dependent regulation of Corn fluorescence.
However, fusing the P1 stem with the base helix of Corn would be problematic since this fusion RNA would no longer have 5’ and 3’ ends (Figure 1C). Therefore, a new set of 5’ and 3’ ends are needed. Thus, we decided to create a new set of 5’ and 3’ ends in the SAM aptamer without disrupting its ability to bind SAM. Based on the overall structure of Corn fused into the SAM aptamer, we considered inserting new 5’ and 3’ ends at the end of the P3 stem (Figure 1C).
Since the P3 stem end does not show high sequence conservation (Figure S1C) (Appasamy et al., 2013; Fuchs et al., 2006) and the P3 stem also does not undergo SAM-induced structural changes (Wilson et al., 2011), we reasoned that it would tolerate mutations. Thus, we designed a circularly permuted SAM aptamer containing a new set of 5’ and 3’ ends at the end of the P3 stem (Figure S1D).
To test whether the circularly permuted SAM aptamer can still bind SAM, we fused it to Broccoli and asked if it exhibits SAM-dependent fluorescence. We fused Broccoli to the SAM aptamer P1 stem, but in this case we used the circularly permuted SAM aptamer. Since the circularly permuted SAM aptamer has the new 5’ and 3’ ends of the sensor, we needed to remove the 5’ and 3’ ends in Broccoli to make this a single contiguous RNA. We therefore linked the Broccoli 5’ and 3’ ends by encoding a four nucleotide-long connecting loop (Figure S1E). This SAM aptamer-Broccoli fusion RNA exhibited a 2.4-fold increase in fluorescence in response to SAM (Figure S1F). The data suggests the circularly permuted SAM aptamer retains the ability to bind SAM. Additionally, these data show that the P1 stem of the circularly permuted SAM aptamer can be an entry point for a fluorogenic aptamer.
Design of a Corn-based SAM sensor
To create a Corn-based SAM sensor, we fused the 26-nt core sequence of the Corn aptamer to the P1 stem of the circularly permuted SAM aptamer (Figure S1G). Addition of SAM resulted in a 7.4-fold increase in fluorescence (Figure 1D). Thus, this design allows Corn to function as a SAM sensor.
To maintain low fluorescence in the absence of SAM, the transducer needs to be unfolded, thus disrupting the Corn structure. However, addition of SAM needs to cause the P1 stem to become stabilized, which should induce dimerization of Corn and subsequent fluorescence (Figure 2A). We therefore tested different transducer sequences with different lengths and predicted thermodynamic stabilities (Figure 2B). Transducer 2 and 4 exhibited ~20-fold increases in fluorescence while the Transducer 5 exhibited a 10-fold increase in fluorescence. However, Transducer 5 was substantially brighter, achieving ~50% of the maximum brightness as defined by the control tRNA-Corn (Figure 2B). We therefore used Transducer 5 for all subsequent experiments.
Figure 2. Design of a Corn-SAM sensor.
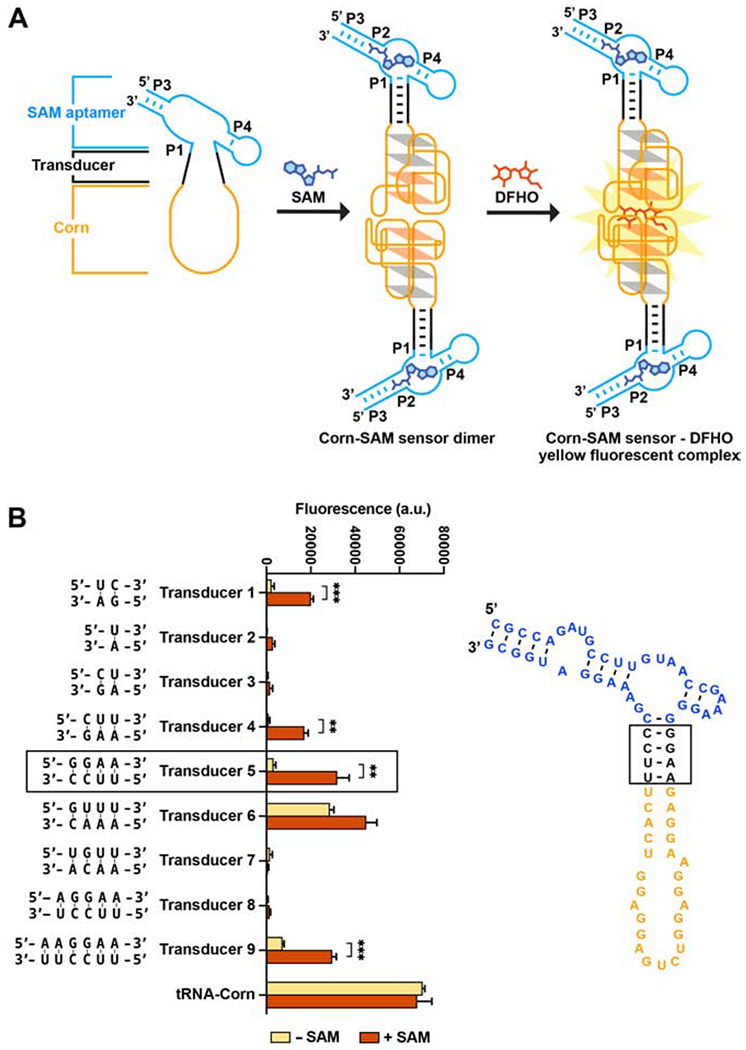
(A) Schematic of SAM binding to the SAM aptamer, which induces folding, dimerization, and fluorogenic activation of Corn.
(B) Each indicated transducer sequence was tested for its ability to mediate SAM-induced fluorescence. In vitro transcribed Corn-SAM sensor RNA (1 μM) was incubated with 10 μM DFHO and 0.1 mM SAM for 1 h at 37°0. Fluorescence was measured (ex 505 nm; em 545 nm). The optimal transducer is indicated in a black-lined box. Mean and SEM values are shown in the plot (n = 3). *** P = 0.0004 (Transducer 1), ** P = 0.0010 (Transducer 2), ** P = 0.0067 (Transducer 5), *** P = 0.0004 (Transducer 9)
The Corn-based sensor approach can be applied to other metabolites
To test if this new Corn-based sensor design can be applied to detect other metabolites, we fused Corn to the cyclic di-GMP (cdiGMP) and S-adenosyl homocysteine (SAH) aptamers (Kellenberger et al., 2013; Su et al., 2016). Both aptamers contain both a base stem and a stem-loop structure, which could potentially be modified to become an entry point for Corn (Figures S2A and S2B). Additionally, both of these aptamers have been previously used in allosteric Spinach-based sensors (Kellenberger et al., 2013; Su et al., 2016).
We tested different transducer domains in the Corn-cdiGMP and Corn-SAH sensors and identified optimized sensors, which each exhibit more than a ~150-fold increase in fluorescence upon addition of the cognate metabolite (Figures S2C and S2D). The high fold-increase in fluorescence is due to the low background fluorescence in the absence of metabolite. These data demonstrate that diverse metabolite sensors can be developed using this approach.
SAM induces the dimerization of the Corn-SAM sensor
To determine if SAM induces dimerization of the Corn-based sensor, which would thus allow DFHO binding and fluorescence, we measured RNA dimerization by native polyacrylamide gel electrophoresis (PAGE). As a control, we in vitro transcribed the Corn aptamer expressed within a tRNA folding scaffold, which improves its folding (Song et al., 2017). tRNA-Corn migrated predominantly as a dimer, and only the band that corresponded to the dimer exhibited fluorescence when DFHO was soaked into the gel, as previously described (Song et al., 2017; Warner et al., 2017). However, the Corn-SAM sensor migrated exclusively as a monomer and did not activate DFHO fluorescence (Figure 3A). When the Corn-SAM sensor RNA was premixed with SAM, and then loaded on the gel, the Corn-based sensor migrated as a dimer in a SAM concentration-dependent manner (Figure 3A). Overall, these data suggest that SAM induces sensor dimerization.
Figure 3. SAM induces the dimerization of the Corn-SAM sensor.
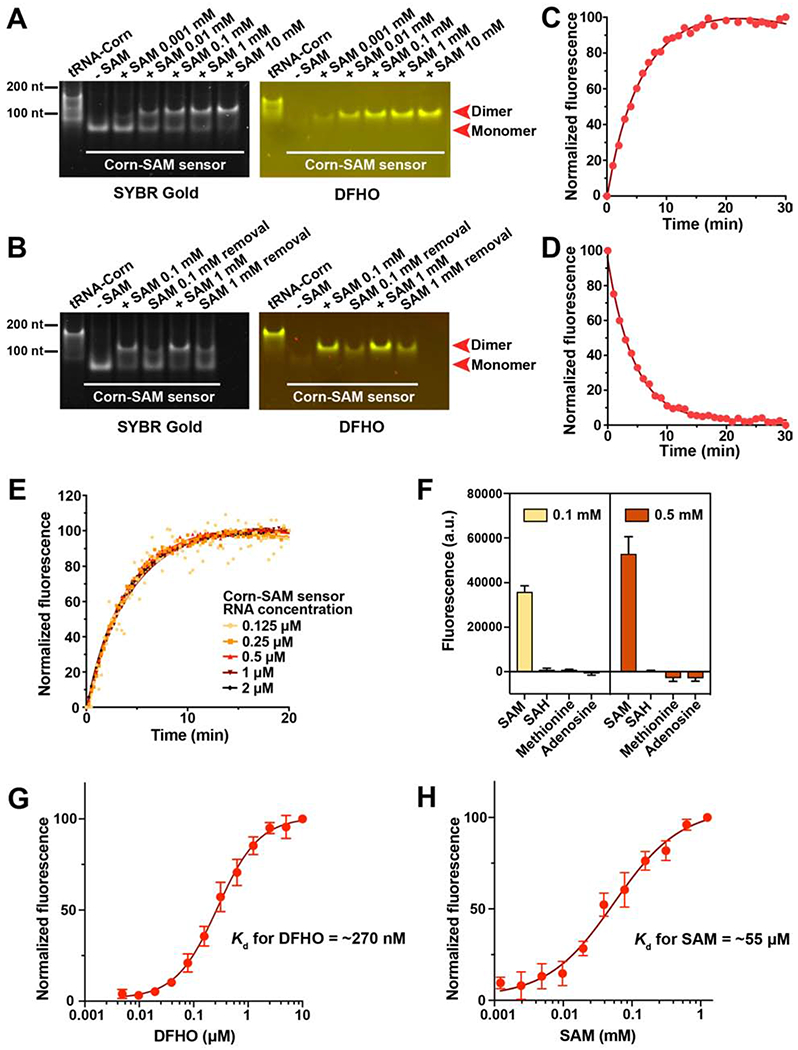
(A) Native PAGE analysis of the Corn-SAM sensor shows increased dimerization upon addition of SAM. Corn-SAM sensor RNA (0.5 μM) was resolved by native PAGE after incubation with SAM for 1 h at 25°C. The gel was stained with 10 μM DFHO, to visualize fluorescence-competent dimeric Corn, followed by SYBR Gold staining to visualize total RNA. tRNA-Corn RNA was used as a positive control for dimerization.
(B) The Corn-SAM dimer becomes a monomer after SAM removal by rapid gel filtration.
(C) Time course of Corn-SAM sensor activation by 0.1 mM SAM. The fluorescence signal of 1 μM Corn-SAM sensor RNA (37°C and 0.5 mM MgCl 2) was recorded every minute.
(D) Time course of Corn-SAM sensor deactivation upon removal of SAM by gel filtration.
(E) Effect of Corn-SAM sensor RNA concentration on sensor activation rate. Fluorescence was recorded every 15 sec for 20 min after the addition of 0.1 mM SAM. The activation rate was similar for all RNA concentrations.
(F) Corn-SAM sensor is only activated by SAM and not related molecules. Mean and SEM values are shown (n = 3).
(G) In vitro transcribed Corn-SAM sensor RNA (0.05 μM) was incubated with 100 μM SAM for 2 h (25°C and 5 mM MgCl 2) followed by the incubation with the indicated concentration of DFHO. Half-maximal fluorescence was reached at ~270 nM. The Hill coefficient calculated using GraphPad Prism was ~1. Mean and SEM values are shown (n = 3).
(H) Measurement of the EC50 of the Corn-SAM sensor. The Hill coefficient calculated using GraphPad Prism was ~1. Mean and SEM values are shown (n = 3).
The Corn-based sensor shows reversibility, specificity and sensitivity needed for imaging SAM in mammalian cells
For use in cells, the Corn-SAM sensor should exhibit reversibility, kinetics of activation and deactivation that correspond with endogenous SAM dynamics, and high specificity and sensitivity for SAM detection. To assess reversibility of the Corn-SAM sensor, we used gel filtration to remove the SAM and assessed sensor deactivation by measuring loss of dimerization by nondenaturing gel electrophoresis. After SAM removal, the sensor was primarily monomeric, suggesting reversibility of the sensor (Figure 3B).
We next measured the activation kinetics of the Corn-based sensor at 37°C with 0.5 mM MgCl 2, a concentration that is more physiologic than the 5 mM used in the gel experiments (Figure 3C). In these experiments, >80% of the maximal fluorescence activation was seen in 10 min (Figure 3C). Deactivation rates were detected by removing SAM by gel filtration. SAM removal resulted in >80% sensor deactivation within 10 min (Figure 3D). Thus, the Corn-based sensor exhibits rapid fluorescence activation and deactivation kinetics.
Since the activation rate of these dimeric sensors could be affected by RNA concentration, we varied the concentration of the Corn-SAM sensor RNA and added 0.1 mM SAM and 20 μM DFHO (Figure 3E). This titration showed that the Corn-SAM sensor activation rate is not affected by the concentration of the Corn-SAM sensor RNA in this concentration range.
The Corn-SAM sensor appeared to be specific for SAM since it was not appreciably activated by metabolites that resemble SAM (Figure 3F).
The Kd for DFHO (~270 nM) of the fully-SAM bound Corn-SAM sensor is ~4-fold larger than the Kd value (70 nM) reported for tRNA-Corn (Song et al., 2017) (Figure 3G). This likely reflects a slight alteration of the Corn structure by the SAM aptamer. Based on the Kd, the Corn-SAM sensor will have sufficient DFHO for fluorescence since imaging conditions utilize 10 μM DFHO.
The Corn-SAM sensor detected SAM throughout the concentration range (0.01-0.1 mM) of SAM seen in several mammalian cells lines (Sitz et al., 1983), with half-maximal fluorescence at ~55 μM (Figure 3H). Binding showed a Hill coefficient of ~1, suggesting that there is no cooperative binding of SAM (Figure 3H). DFHO does not substantially affect SAM binding affinity (Figures S3A and S3B). Overall, these data suggest that the SAM sensor has binding specificity and sensitivity which would make it suitable for use in mammalian cells.
The Corn-SAM sensor exhibits improved photostability in living cells compared to other sensors
To monitor SAM dynamics in live HEK293T cells we used the Tornado expression system to express the sensor (Figure 4A; Table S1) (Litke and Jaffrey, 2019). In this method, an RNA of interest is first expressed as a linear transcript flanked by Twister ribozymes in cells. Upon autocatalytic ribozyme cleavage, the 5’ and 3’ ends of the RNA contain termini that are ligated by the endogenous RNA ligase RtcB. Unlike linear RNA-based sensors and aptamers which accumulate to low nanomolar concentrations in mammalian cells, circular RNAs expressed using the Tornado expression system achieve micromolar levels and therefore generate a fluorescence signal sufficient for detection using fluorescence microscopy (Litke and Jaffrey, 2019).
Figure 4. The Corn-SAM sensor enables detection of SAM dynamics in living cells.
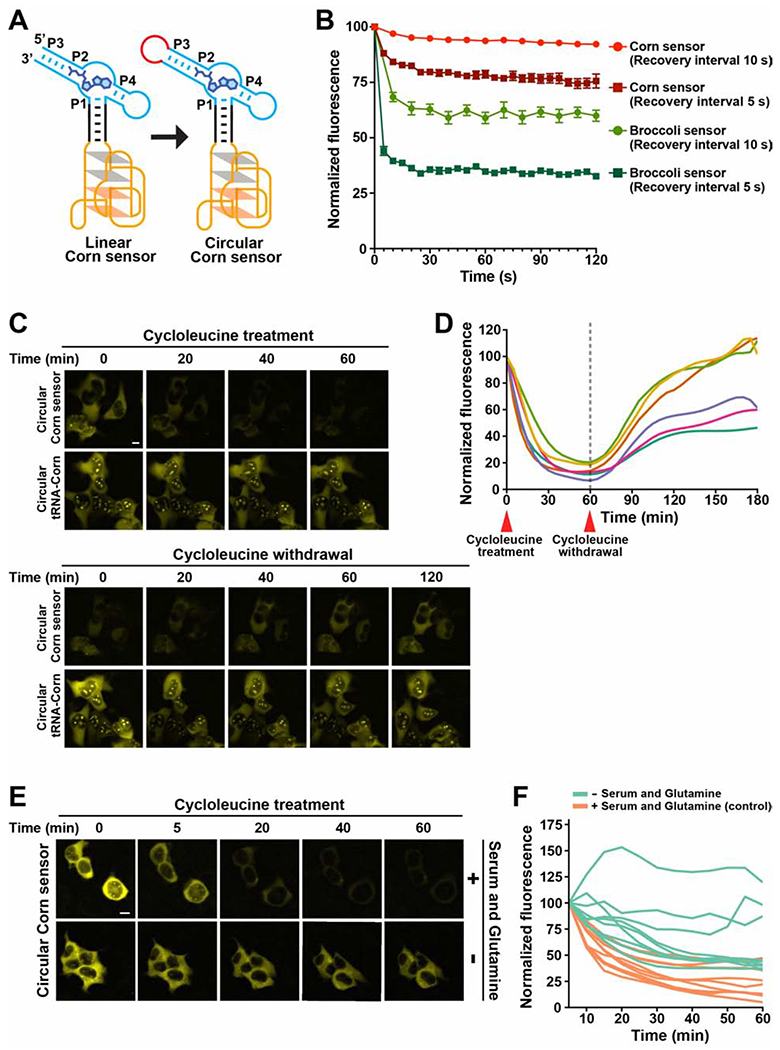
(A) The Tornado expression system was used to express the sensor as a circle. The Tornado system leaves a small RNA loop, indicated in red.
(B) Photostability comparison of the Corn- and Broccoli-SAM sensor. Cells expressing each sensor were imaged (1000 ms) and then the shutter was closed for 5 s or 10 s between each image acquisition. The mean fluorescence intensity in individual cells and SEM were obtained from cells in independently acquired images (n = 3). The Broccoli-SAM sensor showed more photobleaching compared to the Corn-SAM sensor. See also Figure S3F.
(C) Single-cell imaging in HEK293T cells showed a drop in Corn-SAM sensor fluorescence after cycloleucine treatment, and recovery after cycloleucine was washed away. Circular tRNA-Corn, which is constitutively fluorescent, was used as a control. ex 500 ± 12 nm; em 542 ± 13.5 nm. Scale bar 10 μm.
(D) SAM trajectory plots of 6 individual cells from 2 repeated experiments. Each trajectory plot was generated based on the mean fluorescence intensity in single cells during the time-course imaging experiment. Fluorescence was measured at 13 time points over 1 h after the addition of 25 mM cycloleucine, and at 24 time points over the 2 h after the withdrawal of cycloleucine.
(E) Single-cell imaging of the circular Corn-SAM sensor fluorescence after cycloleucine treatment in the serum- and glutamine-deprived conditions. Prior to imaging, the cell culture media was changed to either serum and glutamine-containing media or serum- and glutamine-deprived media and then cycloleucine was added to a final concentration of 25 mM. Scale bar 10 μm.
(F) SAM trajectory plots were generated based on E. Mean cell fluorescence intensities were calculated at 12 time points over a 55 min after the addition of cycloleucine (25 mM). n = 9 cells per condition from two biological replicates.
To confirm cellular expression of the circular Corn-SAM sensor, total cellular RNA was resolved on a gel and stained with DFHO and SAM (Figure S3C; Table S1). When the linear form of the sensor was expressed, no band was detected (Figure S3C). However, the circular Corn-SAM sensor generated a readily detectable band (Figure S3C). Treatment of cells with the transcription inhibitor actinomycin D for 6 h, which typically causes a complete loss of linear aptamers due to their rapid instability (Filonov et al., 2015), did not affect the expression level of the circular Corn-based sensor, as expected for circular RNA (Figure S3C). We further confirmed that transfection and expression of the circular Corn-SAM sensor does not affect cell viability (Figure S3D).
To measure the photostability of the sensor, we imaged transfected cells continuously using a 400 ms image acquisition time, and measured the cellular fluorescence. In these experiments, the Broccoli fluorescence was rapidly photobleached, while the Corn fluorescence photobleached much more slowly (Figure S3E).
We next imaged the Corn-based and Broccoli-based SAM sensors using “pulsed illumination.” In this protocol, a fluorescence image is acquired, and then the shutter is closed for a specific amount of time to allow the photobleached aptamer to recover fluorescence by rebinding fluorophore (Han et al., 2013; Wang et al., 20l3). Cellular fluorescence was imaged using a 1 s image acquisition time followed by recovery for 5 s or 10 s (Figures 4B and S3F). Using either recovery interval, the Corn-based sensor showed substantially higher and consistent fluorescence levels compared to the Broccoli-SAM sensor (Figures 4B and S3F). The plateau of fluorescence is due to the equilibrium of light-induced fluorophore ejection and fluorophore rebinding (Han et al., 2013; Wang et al., 2013). Overall, these data show that the Corn-based sensor is more resistant to photobleaching.
The Corn-SAM sensor enables detection of SAM dynamics in living cells
To determine whether the Corn-SAM sensor can detect endogenous SAM levels, we used cycloleucine, which inhibits SAM biosynthesis (Lombardini and Talalay, 1971). Flow cytometry analysis of HEK293T cells expressing the Corn-SAM sensor in individual cells showed a marked decrease in fluorescence 1 h after addition of 25 mM cycloleucine (Figure S4A). Cells expressing circular tRNA-Corn, which is constitutively fluorescent, were not affected by cycloleucine. Thus, the Corn-SAM sensor can detect changes in the intracellular level of SAM in mammalian cells.
To determine how SAM levels change in individual cells over time in response to cycloleucine, we used fluorescence microscopy to quantify fluorescence in individual HEK293T cells. Cells were treated with 25 mM cycloleucine and images were acquired every 5 min for 1 h (Figure 4C). We generated a SAM “trajectory” in each cell by plotting the fluorescence at each time point. In these experiments, all cycloleucine-treated cells showed a uniform and substantial loss of fluorescence by 30 min (Figure 4D). Importantly, Corn-SAM sensor RNA levels did not change during these experiments (Figures S4B–S4D).
However, when cycloleucine was washed out, cellular heterogeneity was observed. Approximately half the cells completely recovered their SAM levels by 30 to 60 min, but another subset of cells recovered only ~50% of their SAM levels (Figure 4D).
Since SAM levels drop very quickly after inhibiting SAM biosynthesis with cycloleucine, it is clear that SAM is rapidly consumed. Since SAM is used for diverse metabolic pathways linked to cell growth, such as methylation of lipids, proteins, and nucleic acids (Chiang et al., 1996; Sutter et al., 2013; Ye et al., 2017), we asked if these pathways account for the rapid SAM consumption seen in cells. To test this, we inhibited new SAM synthesis with cycloleucine and examined SAM consumption rates in serum- and glutamine-deprived culturing conditions, which we expected would globally downregulate cellular metabolism (Figure 4E). Compared to the control, serum- and glutamine-deprivation markedly reduced SAM consumption rate in individual cells, although the cells showed heterogeneity in SAM consumption rates (Figure 4F). Overall, these data support the idea that the metabolic state is linked to the SAM consumption rate.
To determine if RNA methylation accounts for rapid utilization of SAM in cells, we compared the SAM consumption rate in cells treated with actinomycin D to inhibit transcription by all three RNA polymerases (Figures S4E and S4F). No noticeable difference was detected in cells that were treated with actinomycin D compared to the control. Thus, RNA methylation is not the major consumer of SAM in these experiments.
Overall, these data demonstrate that the Corn-SAM sensor enables SAM dynamics to be monitored in individual mammalian cells in real time.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Samie R. Jaffrey (srj2003@med.cornell.edu).
Experimental Model and Subject Details Cell Lines and Transfection
HEK293T/17 (human embryonic kidney, female, ATCC CRL-11268) cell line was cultured in full DMEM (Thermo Fisher Scientific 11995-065) with 10% FBS, 100 U/ml penicillin and 100μg/ml of streptomycin under standard tissue culture conditions (at 37°C and 5% CO 2). Cells were detached for splitting and plating using TrypLE Express (Life Technologies) according to the manufacturer’s instructions. Cells were screened for mycoplasma contamination before passaging using Hoechst 33258, according to ATCC recommendations. The cell line was recently purchased prior to use, but not additionally authenticated. To transfect cells with plasmids, FuGENE HD (Promega 2311) was used as transfection reagent. 1 d before transfection, cells were seeded onto 24-well plates (Corning CLS3527).
Method Details
Cloning of the Corn-SAM sensors
To prepare pAV-Corn-SAM sensor, DNA templates containing the sequence of Corn-SAM sensor were prepared with flanking Sall and Xbal restriction sites. These constructs were cloned downstream of a U6+27 promoter and upstream of a U6 terminator in a pAV vector that contains the SV40 origin (Paul et al., 2002). This U6 promoter includes the first 27 nucleotides of U6 RNA as described previously.
In the case of preparing pAV-Tornado-Corn-SAM sensor, DNA templates containing the sequence of Corn-SAM sensor were prepared with flanking Notl and SacII restriction sites. The vector used for cloning is the pAV-Tornado vector which contains a U6+27 promoter, a U6 terminator, SV40 origin and a Tornado expression cassette (Litke and Jaffrey, 2019; Paul et al., 2002). The Corn-SAM sensor sequence was inserted between Tornado expression cassette (Litke and Jaffrey, 2019) on a pAV vector through cloning using NotI and SacII restriction sites.
Preparation of RNA in vitro
Double-stranded DNA templates were designed to contain a 5’ T7 promoter to be used for in vitro transcription. Double-stranded DNA templates were prepared from single-stranded DNA oligos (Integrated DNA Technologies). DNA templates were amplified by PCR using Taq DNA polymerase (NEB M02373) and checked for quality using 1% agarose gel electrophoresis. PCR reactions were purified with the QIAquick PCR purification kit (Qiagen 28104).
In vitro transcription reactions using the AmpliScribe™ T7-Flash™ transcription kit (Lucigen ASF3507) were carried out at 37°0 for >4 h. Transcription reactions were terminated by treating with RNase-Free DNase I (Lucigen ASF3507) at 37°C for at least 15 min. RNAs were then purified from reactions by using Micro Bio-Spin Columns with Bio-Gel P-30 (Bio-Rad 7326223) followed by phenol-chloroform extraction with premixed acid phenol:chloroform:IAA (Invitrogen AM9732). Purified RNAs were then precipitated using isopropanol, and dissolved in nuclease-free water (Growcells UPW100012). Quality of samples was checked by running on a precast 6% TBE-Urea Gel (Life Technologies EC68655) at 180 V for 30 min. After staining with SYBR Gold (ThermoFisher S11494) diluted 1:10,000 in TBE buffer, RNA bands were imaged using a ChemiDoc MP (Bio-Rad) with a preset channel (ex 302 nm; em 590/110 nm).
Measurements of RNA fluorescence in vitro
Measurements of Broccoli fusion RNAs
Purified RNAs dissolved in nuclease-free water were heated up to 75°C for 5 min and cooled down to 4°C for 5 min before incubating with the in dicated buffer. RNAs were diluted to a final concentration of 1 μM in buffer solution containing 100 mM KCl, 1 mM MgCl2, 40 mM K-HEPES (pH 7.5) and 10 μM DFHBI-1T (synthesized (Song et al., 2014) or Lucerna 410-1mg). To test if the circularly permuted SAM aptamer-Broccoli fusion RNA enables to sense SAM, a final concentration of 0.1 mM SAM (Sigma-Aldrich A7007) was added to the prepared RNA sample. After 1 h incubation at 37°0, fluorescence signal of the sample was measured at 37°C using a Fluoromax-4C (Horiba Scientific) with 470 nm excitation and 505 nm emission, 5 nm slit widths, and 0.1 s integration time. Background signal was subtracted from the signal obtained from each RNA sample measurement. To obtain background signal, an RNA-free sample containing 10 μM DFHBI-1T in 100 mM KCl, 1 mM MgCl2, 40 mM K-HEPES (pH 7.5) was used, and was subjected to fluorescence measurement at 37°C.
Corn sensors with different transducers
Purified RNAs dissolved in nuclease-free water were heated up to 75°C for 5 min and cooled down to 4°C for 5 min before incubating with the in dicated buffer. Corn-SAM sensor RNAs, Corn-SAH sensor RNAs or Corn-cdiGMP sensor RNAs were diluted to a final concentration of 1 μM in buffer solution containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5) and 10 μM DFHO (synthesized (Song et al., 2017) or Lucerna 500-1mg). In the case of the Corn-cdiGMP sensor measurements, 3 mM MgCl2 was used in buffer solution instead of 0.5 mM MgCl2 (Kellenberger et al., 2013). To test the ability of each indicated transducer sequence to mediate analyte-induce fluorescence, 0.1 mM of the following analyte, SAM (Sigma-Aldrich A7007), cdiGMP (Sigma-Aldrich SML1228) or SAH (Sigma-Aldrich A9384), was added to each cognate sensor RNA sample solution. After 1 h incubation at 37°0, fluorescence signal of each sample was measured at 37°C using a Fluoromax-4C (H oriba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. Background signal was subtracted from the signal obtained from each RNA sample measurement. To obtain background signal, an RNA-free sample containing 10 μM DFHO in 100 mM KCl, 0.5 mM MgCl2 (or 3 mM MgCl2 in the case of the Corn-cdiGMP sensors), 40 mM K-HEPES (pH 7.5) was used, and was subjected to fluorescence measurement at 37°C.
Activation and deactivation rates
To measure the activation rate of the Corn-SAM sensor, RNAs were diluted to 1 μM in buffer solution containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5) and 10 μM DFHO. 0.1 mM SAM was then rapidly added to the stirring RNA solution, and fluorescence emission was recorded over a 30-min period at 1 min interval at 37°C. The fluorescence signal of the sample was measured using a Fluoromax-4C (Horiba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. The fluorescence measurement was normalized to the intensity at 30 min (100) and the intensity at 0 min (0).
To study the effect of Corn-SAM sensor RNA concentration on sensor activation rate, we varied the concentration of the Corn-SAM sensor RNA and measured the activation rates. In this experiment, RNAs were diluted to 0.125 μM, 0.25 μM, 0.5 μM, 1 μM or 2 μM in buffer solution containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5) and 20 μM DFHO. 0.1 mM SAM was then rapidly added to each stirring RNA solution, and fluorescence emission was recorded over a 20-min period at 15 sec interval at 37°C. The fluorescence signal of the sample was measured using a Fluoromax-4C (Horiba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. The fluorescence measurement was normalized to the intensity at 20 min (100) and the intensity at 0 min (0).
To measure the deactivation rate of the Corn-SAM sensor, 1 μM RNA sensor and 10 μM DFHO was first incubated with 0.1 mM SAM (Sigma-Aldrich A7007) in buffer solution containing 100 mM KCl, 0.5 mM MgCl2 and 40 mM K-HEPES (pH 7.5) for 1 h at 37°C. When the sensor reached the maximal fluorescence, the solution was transferred to a Micro Bio-Spin Columns with Bio-Gel P-30 (Bio-Rad 7326223). This column was buffer-exchanged with buffer solution containing 100 mM KCl, 0.5 mM MgCl2 and 40 mM K-HEPES (pH 7.5). The sample was loaded on the buffer-exchanged column and spun at 1,000 × g for 4 min. The flow-through after gel filtration contains the sensor RNA in SAM-free buffer. 10 μM DFHO was newly added to the collected flow-through. Then, the fluorescence emission of the flow-through was recorded over a 30-min period at 1 min interval at 37°C. The fluo rescence signal of the sample was measured using a Fluoromax-4C (Horiba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. The fluorescence measurement was normalized to the intensity at 0 min (100) and the intensity at 30 min (0).
Each fluorescence emission measurement was plotted as a dot against time. One site-Total model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve for the activation rate measurements. Dissociation-One phase exponential decay model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve for the deactivation rate measurements.
Measurements with different SAM analogs
The Corn-SAM sensor RNAs were diluted to 1 μM in buffer solution containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5) and 10 μM DFHO. To test if fluorescence signal of the Corn-SAM sensor is specifically activated by SAM, 0.1 mM or 0.5 mM of the following analyte, SAM (Sigma-Aldrich A7007), SAH (Sigma-Aldrich A9384), adenosine (Sigma-Aldrich 9251) or methionine (Sigma-Aldrich M9625), was added to the prepared RNA samples. Then, the samples were incubated at 37°C for 1 h. The fluores cence signal of each sample was measured at 37°C using a Fluoromax-4C (Horiba Scientific) wi th 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. Background signal was subtracted from the signal obtained from each RNA sample measurement. To obtain background signal, an RNA-free sample containing 10 μM DFHO in 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5) was used, and was subjected to fluorescence measurement at 37°C.
Dose-response curve measurements
To measure the EC50 for SAM of the Corn-SAM sensor, a dose-response curve for the Corn-SAM sensor in response to SAM was determined by measuring the increase in fluorescence as a function of SAM concentration in the presence of a fixed concentration of RNA sensor (0.5 μM or 0.1 μM) and a fixed concentration of DFHO (10 μM). Corn-SAM sensor RNAs were incubated with a range of concentrations of SAM (Sigma-Aldrich A4377) in buffer solution (100 mM KCl, 0.5 mM MgCl2 or 5 mM MgCl2, 40 mM K-HEPES, pH 7.5 and 10 μM DFHO) at 37°C for 1 h. 0.5 μM of RNA was used for the 0.5 mM MgCl2 condition and 0.1 μM of RNA was used for the 5 mM MgCl2 condition. After 1 h incubation, the fluorescence signal of each sample was measured at 37°C using a Fluoromax-4C (Horiba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time.
To measure the EC50 for DFHO of the Corn-SAM sensor, the increase in fluorescence was measured as a function of DFHO concentration in the presence of a fixed concentration of RNA sensor (0.05 μM) and a fixed concentration of SAM (100 μM). 0.05 μM sensor RNA was incubated with 100 μM SAM (Sigma-Aldrich A7007) in buffer solution (100 mM KCl, 5 mM MgCl2, 40 mM K-HEPES, pH 7.5) at 25°C for 2 h. Then, the samples were incubated with a range of concentrations of DFHO. The fluorescence signal of each sample was measured at 25°C using a Fluoromax-4C (Horiba Scientific) with 505 nm excitation and 545 nm emission, 5 nm slit widths, and 0.1 s integration time. For each concentration of DFHO measured, a background signal for DFHO alone was also measured and subtracted from the signal measured for RNA and DFHO together.
Agonist vs. response-variable slope (four parameters) model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve and calculate a Hill coefficient from the measurements.
Native PAGE imaging analysis
Native PAGE conditions were prepared by casting 10×10 cm 10% PAGE gels using 30% acrylamide/bis-acrylamide (29:1) solution (Sigma-Aldrich A3574), 10% ammonium persulfate, tetramethylethylenediamine, and the Native PAGE buffer. The final concentration of the native PAGE buffer in 10% PAGE gel was 40 mM K-HEPES (pH 7.5), 100 mM KCl and 5 mM MgCl2. After polymerization, the samples were run at 40 V for 2 h in a water-cooled PAGE chamber to keep the buffer temperature at ~25°2. For staining, gels were incubated in the native PAGE buffer containing 10 μM DFHO for 15 min. DFHO-stained gels were analyzed on a ChemiDoc MP imaging station (Bio-Rad) at 470/30 nm excitation and 530/28 nm emission (green channel) and 530/28 nm excitation and 605/50 nm emission (red channel). Each presented image is an overlay of both scans. Next, counterstaining with SYBR Gold (ThermoFisher S11494) 1:10,000 diluted in the native PAGE buffer was performed for 15 min followed by gel imaging to detect all RNA species in each lane. SYBR Gold-stained bands were imaged using UV excitation (302 nm) and 590/110 nm emission on the ChemiDoc MP imaging station.
Denaturing PAGE imaging analysis
2 d after the transfection of the corresponding plasmids, total cellular RNA was harvested from cultured cells by removing media followed by TRIzol™ LS Reagent (Invitrogen 10296010) addition. RNA was purified from the TRIzol LS mixtures according to the manufacturer’s instructions.
1.5 μg of total RNA was loaded on each lane of precast 6% TBE-Urea Gels (Life Technologies EC68655). Gels were run at 180 V in TBE buffer for 35 min. Gels were washed 3× 5 min with water and then additionally washed for 2 h. Then, gels were stained for 1 h with 10 μM DFHO and 0.1 mM SAM (Sigma-Aldrich A7007) in buffer containing 40 mM K-HEPES (pH 7.5), 100 mM KCl and 5 mM MgCl2. Corn bands were then imaged using a ChemiDoc MP (Bio-Rad) with 470/30 nm excitation and 532/28 nm emission (green channel) and 530/28 nm excitation and 605/50 nm emission (red channel). Each presented image is an overlay of both scans. Gels were washed additionally with water and stained with SYBR Gold (ThermoFisher S11494) 1:10,000 diluted in TBE buffer. SYBR Gold-stained bands were imaged using UV excitation (302 nm) and 590/110 nm emission on the ChemiDoc MP imaging station.
Isothermal titration calorimetry (ITC)
Corn-SAM sensor RNA solution and SAM solution were prepared in the same buffer (40 mM HEPES, pH 7.5, 100 mM KCl, 5 mM MgCl2). In vitro transcribed Corn-SAM sensor RNA was dissolved to a final concentration ~107.5 μM in 330 μl solution. The concentration of RNA was determined using denaturing PAGE analysis followed by SYBR Gold staining and comparing SYBR Gold signal to RNA standards (Thermo Fisher Scientific SM1831). We cannot exclude the possibility that the RNA preparation includes some misfolded RNA which is incapable of binding SAM. This may give the appearance of a lower molar ratio of SAM to sensor RNA at each SAM injection. In the condition of DFHO presence, 400 μM DFHO was included in the final RNA solution (0.56% DMSO in final). The RNA solution was loaded in the cell of a Nano ITC microcalorimetry device (TA Instruments) and 50 μL of 877 μM SAM (Sigma-Aldrich A4377) were loaded in the titration syringe. For the condition of DFHO presence, 0.56% DMSO was included in the final SAM solution. 3 μL of titrant was added into the reaction chamber for the first injection, and 2 μL per injection was added for the following 24 injections. Between the injections, there was a 1200 s interval for the first 7 injections and a 1000 s interval for the following 18 injections. Measurements were performed at 37°C and the syringe rotation speed was set to 250 rpm. The NanoAnalyze Software (TA Instruments) was used to model SAM binding to the Corn-SAM sensor.
MTT assay
Cells were transfected with the indicated plasmids with FuGENE HD (Promega). Cells were subcultured onto mouse laminin-coated plates 1 d after transfection. To prepare mouse laminin-coated plates, plates were coated with Cultrex Mouse Laminin I (Thermo Fisher 340001002) for at least 1 h and rinsed once in water. 3 d after transfection, cells were incubated with 2.5 mg/mL thiazolyl blue tetrazolium bromide (MTT). After 3 h incubation at 37°0, cells were treated with 2.4 mM HCl and 0.06% NP-40 in final. MTT absorbance readings were performed at 590 nm absorbance. Signal obtained from MTT solution without cells was used for subtracting background signal. Values were normalized to the average value of the no transfection condition.
Flow cytometry analysis
HEK293T cells were transfected with the Corn-SAM sensor-expressing Tornado plasmid, the tRNA-Corn-expressing Tornado plasmid (Litke and Jaffrey, 2019) or 5S rRNA-expressing plasmid (Filonov et al., 2014). Cells were subcultured onto mouse laminin-coated plates 1 d after transfection. To prepare mouse laminin-coated plates, plates were coated with Cultrex Mouse Laminin I (Thermo Fisher 340001002) for at least 1 h and rinsed once in water. 2 d after transfection, cells were treated with 25 mM cycloleucine (Sigma-Aldrich A48105) for the indicated time. Then, cells were washed with 1× PBS once, harvested using TrypLE Express Enzyme, resuspended in the 4% FBS/1× PBS solution containing 10 μM DFHO and 5 mM MgSo4, and kept on ice until analysis on LSRFortessa™ (BD Biosciences). Populations of cells were gated to avoid cell doublets and cell debris detected by forward and side scattering. 5S rRNA expressing cells were used as a negative control for Corn fluorescence (488 nm excitation and 545 ± 17.5 nm emission). Plots were generated using FlowJo softwar (Tree Star, Inc.).
Microscopy and image processing
For imaging cells, we used glass-bottomed 24-well plates (MatTek Corporation P24G-1.5-13-F) that were coated with poly-D-lysine (Cultrex 3429-100-01) for at least 1 h and rinsed once in water. These plates were additionally coated with Cultrex Mouse Laminin I (Thermo Fisher 340001002) for at least 1 h and rinsed once in water. Cells were subcultured onto pre-treated glass-bottomed plates 1 d after transfection. Cell culture media was changed to phenol red-free DMEM (Thermo Fisher Scientific 31053028) supplemented with 10% FBS and 1× GlutaMax-I (Thermo Fisher Scientific 35050061) 16 h before imaging. HEK293T cells expressing the circular Corn-SAM sensor or the circular tRNA-Corn were treated with 10 μM DFHO (synthesized (Song et al., 2017) or Lucerna 500-1mg) 1 h before imaging. In case of imaging cells expressing the circular Broccoli-based sensor, cells were pretreated with 40 μM DFHBI-1T (synthesized (Song et al., 2014) or Lucerna 410-1mg), instead of DFHO. Live-cell fluorescence images were acquired with a CoolSnap HQ2 CCD camera through a 40× air objective (NA 0.75) mounted on a Nikon Eclipse TE2000-E microscope and analyzed with the NIS-Elements software. Conditions were maintained at 37°C and 5% CO2 during live-cell imaging. The filter set used for Broccoli detection was a filter cube with excitation filter 470 ± 20 nm, dichroic mirror 495 nm (long pass), and emission filter 525 ± 25 nm. Corn detection used a filter cube with excitation filter 500 ± 12 nm, dichroic mirror 520 nm (long pass), and emission filter 542 ± 13.5 nm. Cell mean fluorescence intensity was computed using Fiji (Schindelin et al., 2012) by measuring the total fluorescence signal in a region of interest (ROI) divided by ROI area μm2) and subtracting background based on the mean fluorescence intensity of an untransfected cell. ROI was defined as cell cytoplasm area. Cell nucleus area was excluded from ROI.
Photostability analysis
HEK293T cells were transfected with a Tornado plasmid encoding the Corn-SAM sensor or a Tornado plasmid encoding the Broccoli-SAM sensor (Litke and Jaffrey, 2019). After 1 d, transfected cells were subcultured onto coated glass-bottomed plates. 4 d after transfection, cells were imaged by using the live-cell imaging conditions as described above. For a “pulsed illumination” method, cellular fluorescence was acquired using a 1 s image acquisition time followed by the indicated recovery intervals (5 s or 10 s) for 120 s. During the recovery interval, the shutter was closed so that the cells were not exposed to light. For a “continuous illumination” method, cells were continuously illuminated throughout the experiment (10 s). Cellular fluorescence was acquired for 10 s using a 400 ms acquisition time.
Intracellular SAM imaging
HEK293T cells were transfected with a Tornado plasmid encoding the Corn-SAM sensor or a Tornado plasmid encoding the tRNA-Corn (Litke and Jaffrey, 2019). After 1 d, transfected cells were subcultured onto coated glass-bottomed plates. 3.5-4 d after transfection, cells were imaged. Using the live-cell imaging conditions described above, we imaged cells for 1 h at 5 min intervals after adding cycloleucine (Sigma-Aldrich A48105) to 25 mM. Then, we withdrew cycloleucine by changing cell culture media into fresh media and continued to image cells every 5 min for 2 additional h. Acquisition time: 500 ms.
Data and Code availability
This study did not generate or analyze datasets/code.
Quantification and Statistical analysis
In vitro experiments (Figures 1D, 2B, 3F, 3G, 3H, S1F, S2C, S2D, S3B and S3D) were performed in triplicate. The mean and SEM values are shown for the plotted graphs (n = 3). Significance was determined by unpaired t test with Welch’s correction provided in Prism 8 (Graphpad). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
The average and SEM values of cell mean fluorescence intensities obtained from cells in independently acquired images (n = 3) are shown for the plotted line graphs in Figures 4B and S3E. The average and SEM values of cell mean fluorescence intensity obtained from n = 8 cells from 5 acquired images are shown for the plotted line graph in Figure S4D. Cell mean fluorescence intensity was calculated using Fiji (Schindelin et al., 2012) by measuring the total fluorescence signal in a region of interest (ROI) divided by ROI area μm2) and subtracting background based on mean fluorescence intensity of an untransfected cell. ROI was defined as cell cytoplasm area.
To plot SAM trajectories in single cells, we obtained images of HEK293T cells expressing the circular Corn-SAM sensor from 2 repeated experiments. The mean intensity of each cell at time 0 was defined as 100% in Figures 4D and S4F. In case of Figure 4F, the signal at time point 0 was excluded for plotting trajectories due to unstably spiked signal after changing cell culture media. Thus, the mean intensity of each cell at 5 min was defined as 100% in Figure 4F. The mean fluorescence intensity at any other time point is normalized to the value at time 0 in Figures 4D and S4F. The mean fluorescence intensity at any other time point is normalized to the value at 5 min in Figure 4F. Normalized mean fluorescence of each cell was plotted as a function of time. In Figure 4D, measurements at 37 time points were fitted with a smoothed curve to generate a SAM trajectory in a single cell by using Savitzky-Golay filter (Press et al., 1992). In Figure 4F, measurements at 12 time points were fitted with a smoothed curve to generate a SAM trajectory in a single cell by using Savitzky-Golay filter. In Figure S4F, measurements at 13 time points were fitted with a smoothed curve to generate a SAM trajectory in a single cell by using Savitzky-Golay filter. R (R Core Team, 2018) was used to apply Savitzky-Golay filter (signal package) to data and generate a trajectory plot (ggplot package).
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| One shot™ Stbl3™ chemical competent E.coli | Thermo Fisher | C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DFHBI-1T | (Song et al., 2014) or Lucerna | Cat#410 |
| DFHO | (Song et al., 2017) or Lucerna | Cat#500 |
| S-adenosyl-methionine chloride dihydrochloride | Sigma-Aldrich | Cat#A7007 |
| S-adenosyl-methionine iodide | Sigma-Aldrich | Cat#A4377 |
| S-adenosyl-homocysteine | Sigma-Aldrich | Cat#A9384 |
| adenosine | Sigma-Aldrich | Cat#9251 |
| methionine | Sigma-Aldrich | Cat#M9625 |
| cyclic di-GMP | Sigma-Aldrich | Cat#SML1228 |
| 30 % acrylamide/bis-acrylamide (29:1) solution | Sigma-Aldrich | Cat#A3574 |
| ammonium persulfate | Sigma-Aldrich | Cat#A3678 |
| N,N,N’,N’-Tetramethylethylenediamine | Sigma-Aldrich | Cat#T9281 |
| cycloleucine | Sigma-Aldrich | Cat#A48105 |
| TRIzol™ LS Reagent | Invitrogen | Cat#10296010 |
| Thiazolyl blue tetrazolium bromide (MTT) | Abcam | Cat#ab146345 |
| actinomycin D | Sigma-Aldrich | Cat#A9415 |
| Critical Commercial Assays | ||
| AmpliScribe™ T7-Flash™ transcription kit | Lucigen | Cat#ASF3507 |
| Micro Bio-Spin Columns with Bio-Gel P-30 | Bio-rad | Cat#7326223 |
| FuGENE HD | Promega | Cat#2311 |
| Experimental Models: Cell Lines | ||
| HEK293T/17 | ATCC | CRL-11268 |
| Oligonucleotides | ||
| RiboRuler Low Range RNA Ladder | Thermo Fisher | Cat#SM1831 |
| Primers for Broccoli fused to circularly permuted SAM aptamer in vitro transcription, See Table S2 | This paper | N/A |
| Primers for Corn-SAM sensors in vitro transcription, See Table S2 | This paper | N/A |
| Primers for Corn-cdiGMP sensors in vitro transcription, See Table S2 | This paper | N/A |
| Primers for Corn-SAH sensors in vitro transcription, See Table S2 | This paper | N/A |
| Primers for tRNA-Corn in vitro transcription, See Table S2 | This paper | N/A |
| Primers for Corn-SAM sensors cloning, See Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pAV-Tornado-Broccoli-SAM sensor | (Litke and Jaffrey, 2019) | N/A |
| pAV-T ornado-tRNA-Corn | (Litke and Jaffrey, 2019) | N/A |
| pAV-Tornado-Corn-SAM sensor | This paper | N/A |
| pAV-Corn-SAM sensor | This paper | N/A |
| pAV-5S rRNA | (Paige et al., 2011) | N/A |
| Software and Algorithms | ||
| FlowJo | TreeStar | www.flowjo.com |
| ImageJ | NIH | imagej.net |
| Fiji | NIH | fiji.sc |
| NIS-Element | Nikon | www.nikon.com |
| R | The R Foundation | www.r-project.org |
| Prism 8 | GraphPad | www.graphpad.com |
| NanoAnalyze Software | TA Instruments | https://www.tainstruments.com |
SIGNIFICANCE.
Here we describe a class of genetically encoded small molecule sensor composed of a fluorogenic RNA that dimerizes and exhibits fluorescence upon binding its small molecule ligand. The constitutively dimeric Corn aptamer can be converted into a conditional dimer by fusing its base into a helical stem within the SAM aptamer. In the absence of SAM, this fusion RNA is not capable of dimerizing. However, upon binding SAM, Corn undergoes dimerization and can then bind and activate its fluorogenic ligand DFHO. In this way, small molecule binding can be coupled to cellular fluorescence, enabling imaging of SAM dynamics in living cells. These studies reveal an approach for constructing RNA-based small molecule biosensors, and demonstrate that RNA dimerization can be utilized for designing RNA devices.
Highlights.
The fluorogenic RNA aptamer Corn was converted into a photostable metabolite sensor
The constitutive Corn dimer was engineered into metabolite-regulated dimer
The sensor undergoes dimerization upon binding to SAM and activates fluorescence
The sensor detects SAM levels in live mammalian cells with improved photostability
Acknowledgements
We thank members of the Jaffrey lab for helpful comments and suggestions. We thank J.S. Paige, W. Song and G.S. Filonov for early contributions to this project, J.L. Litke, S. Suter, J. Wu, J.D. Moon, X. Li, S. Dey, Q. Hou, and L. Mo for useful comments and suggestions, and X. Li for synthesizing DFHO and DFHBI-1T. This work was supported by NIH grant R01NS064516 and R35NS111631 and the American Diabetes Association Pathway to Stop Diabetes Grant 1-18-VSN-02 to S.R.J. and by Kwanjeong Educational Foundation to H.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
S.R.J. is the co-founder of Lucerna Technologies and has equity in this company. Lucerna has licensed technology related to Spinach and other RNA–fluorophore complexes.
References
- Appasamy SD, Ramlan EI, and Firdaus-Raih M (2013). Comparative Sequence and Structure Analysis Reveals the Conservation and Diversity of Nucleotide Positions and Their Associated Tertiary Interactions in the Riboswitches. PLoS One 8, e73984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, and McCann PP (1996). S-Adenosylmethionine and methylation. FASEB J. 10, 471–480. [PubMed] [Google Scholar]
- Ellington AD, and Szostak JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Filonov GS, Moon JD, Svensen N, and Jaffrey SR (2014). Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Kam CW, Song W, and Jaffrey SR (2015). In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol 22, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, and Henkin TM (2006). The SMKbox is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol 13, 226–233. [DOI] [PubMed] [Google Scholar]
- Han KY, Leslie BJ, Fei J, Zhang J, and Ha T (2013). Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc 135, 19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Sales-Lee J, and Hammond MC (2013). RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc 135, 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke JL, and Jaffrey SR (2019). Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol 37, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardini JB, and Talalay P (1971). Formation, functions and regulatory importance of S-adenosyl-l-methionine. Adv. Enzyme Regul 9, 349–384. [DOI] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, and Ke A (2008). Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. & Mol. Biol 15, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY, and Jaffrey SR (2011). RNA mimics of green fluorescent protein. Science 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Nguyen-Duc T, Song W, and Jaffrey SR (2012). Fluorescence Imaging of Cellular Metabolites with RNA. Science 335, 1194–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, and Engelke DR (2002). Effective expression of small interfering RNA in human cells. Nat. Biotechnol 20, 505–508. [DOI] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, and Flannery BP (1992). Numerical Recipes in C: The Art of Scientific Computing. In Cambridge University Press, 650–655. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitz TO, Godbum KE, Somers KD, and Nazar RN (1983). Significance of S-Adenosylmethionine Pools in the Hypomethylation of Ribosomal RNA During the Propagation of Tissue Culture Cells and Oncogenesis. Cancer Res. 43, 5681–5686. [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, and Henkin TM (2010). The SAM-responsive SMK box is a reversible riboswitch. Mol. Microbiol 78, 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Strack RL, Svensen N, and Jaffrey SR (2014). Plug-and-play fluorophores extend the spectral properties of spinach. J. Am. Chem. Soc 136, 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Filonov GS, Kim H, Hirsch M, Li X, Moon JD, and Jaffrey SR (2017). Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol 13, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Hickey SF, Keyser SGL, and Hammond MC (2016). In Vitro and in Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl- l -homocysteine (SAH). J. Am. Chem. Soc 138, 7040–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter BM, Wu X, Laxman S, and Tu BP (2013). Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154, 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. (2018). R: A Language and Environment for Statistical Computing.
- Tuerk C, and Gold L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Wang P, Querard J, Maurin S, Nath SS, Le Saux T, Gautier A, and Jullien L (2013). Photochemical properties of Spinach and its use in selective imaging. Chem. Sci 4, 2865–2873. [Google Scholar]
- Warner KD, Sjekloa L, Song W, Filonov GS, Jaffrey SR, and Ferré-D’Amaré AR (2017). A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol 13, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, and Foster MP (2011). Tuning riboswitch regulation through conformational selection. J. Mol. Biol 405, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Sutter BM, Wang Y, Kuang Z, and Tu BP (2017). A Metabolic Function for Phospholipid and Histone Methylation. Mol. Cell 66, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Litke JL, and Jaffrey SR (2015). Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci 112, E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets/code.


