Abstract
The Gram-negative opportunistic pathogen, Pseudomonas aeruginosa, has multiple multidrug efflux pumps. MexT, a LysR-type transcriptional regulator, functions as a transcriptional activator of the MexEF-OprN efflux system. MexT consists of an N-terminal DNA-binding domain and a C-terminal regulatory domain (RD). Little is known regarding MexT ligands and its mechanism of activation. We elucidated the crystal structure of the MexT RD at 2.0 Å resolution. The structure comprised two protomer chains in a dimeric arrangement. MexT possessed an arginine-rich region and a hydrophobic patch lined by a variable loop, both of which are putative ligand-binding sites. The three-dimensional structure of MexT provided clues to the interacting ligand structure. A DNase I footprinting assay of full-length MexT identified two MexT-binding sequence in the mexEF-oprN promoter. Our findings enhance the understanding of the regulation of MexT-dependent activation of efflux pumps.
Keywords: antibiotic resistance, crystal structure, lysR-type transcriptional regulator, MexT, Pseudomonas aeruginosa
INTRODUCTION
The opportunistic human pathogen Pseudomonas aeruginosa is clinically important due to its resistance to multiple antibiotics, which can be intrinsic or acquired (Piddock, 2006). In addition to its poor outer membrane permeability, chromosome-encoded efflux pumps are the major determinants of antibiotic resistance in P. aeruginosa (Sobel et al., 2005). The resistance-nodulation-cell division (RND) family efflux pumps are key contributors to the multidrug resistance of P. aeruginosa and are the most common efflux pumps found in Gram-negative bacteria (Piddock, 2006; Poole and Srikumar, 2001). To date, four RND types of efflux pumps have been identified and characterized in P. aeruginosa—MexAB-OprM, MexXY-OprM, MexCD-OprJ, and MexEF-OprN (Hinchliffe et al., 2013; Piddock, 2006). Studies of MexAB-OprM pumps and associated molecular mechanisms have demonstrated their importance for multidrug resistance (Jeong et al., 2016; Kim et al., 2015; Lister et al., 2009; Xu et al., 2012). For example, MexEF-OprN exports several types of antimicrobial compounds such as chloramphenicol, fluoroquinolones, and trimethoprim (Galie et al., 2018). In several laboratory strains and clinical isolates, MexEF-OprN affects the expression of MexAB-OrpM, indicating the existence of regulatory interplay between the two efflux pumps (Horna et al., 2018).
The expression level of MexEF-OprN in wild-type strains under laboratory conditions is typically negligible. However, treatment with the fluoroquinolone antibiotic norfloxacin leads to generate certain types of spontaneous mutant strains, such as nfxC-type mutants. And these showed constitutive expression of mexT transcriptionally (Fargier et al., 2012; Hirai et al., 1987). Activation of MexT induces overexpression of MexEF, leading to increased resistance to antibiotics (Kohler et al., 1997). In the P. aeruginosa genome, mexT is located in the upstream of mexEF-oprN operon. MexT reportedly promotes the transcription of the operon (Kohler et al., 1997; 1999). Additionally, the broad regulatory scope including multiple target genes of MexT in P. aeruginosa is being revealed adding weight on its potential as a global regulator (Tian et al., 2009). Indeed, MexT is linked to several genes associated with pathogenesis and the host–pathogen interaction (Fargier et al., 2012; Fetar et al., 2011; Jin et al., 2011; Juarez et al., 2018).
We conducted the experiments to evaluate the molecular mechanism of MexT by understanding its structure. We investigated the mechanism by which MexT regulates the expression of MexEF-OprN by producing the full-length (FL) recombinant protein in Escherichia coli. We analyzed the crystal structure of the MexT regulatory domain (RD), which encompasses putative ligand-binding sites.
MATERIALS AND METHODS
Protein expression and purification
The native MexT RD (residues 95–304) and FL MexT (residues 1–304) were expressed as described previously (Kim et al., 2018). For the selenomethionyl (SeMet)-labeled MexT RD, the host E. coli B834(DE3) was cultured in M9 medium supplemented with L-(+)-selenomethionine, 100 μg/ml ampicillin, and other cofactors. To induce production of SeMet-labeled MexT RD, 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added, followed by incubation at 16°C for 14 h with shaking. The cells were harvested by centrifugation at 1,380g for 10 min and resuspended in 50 ml of lysis buffer (20 mM Tris-hydrochloride [HCl; pH 8.5], 300 mM sodium chloride [NaCl], 5% [v/v] glycerol, and 2 mM 2-mercaptoethanol). The cells were disrupted using a continuous French press (Beijer, Sweden) at a pressure of 23 kpsi and the lysate was clarified using centrifugation at 10,000g for 30 min. MexT was subsequently purified using TALON affinity chromatography and the RD protein was purified using anion-exchange chromatography with a HiTrap Q column (GE Healthcare, USA). We removed the hexahistidine tag using TEV protease, and further purified the proteins using HiTrap Q anion-exchange chromatography. Additionally, we carried out size-exclusion chromatography. The proteins were concentrated using Vivaspin (30 kDa molecular-weight cutoff; Millipore, USA) in buffer containing 20 mM Tris-HCl (pH 8.5), 300 mM NaCl, 5% (v/v) glycerol, and 2 mM 2-mercaptoethanol. The final protein concentration was 12 mg/ml.
After TALON affinity chromatography, FL MexT was directly loaded onto a size-exclusion chromatography column (HiLoad Superdex 16/60 200; GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.5), 500 mM NaCl, 10% (v/v) glycerol, and 2 mM 2-mercaptoethanol. The purified proteins were aliquoted and stored frozen at −173°C.
Crystallization and data collection
MexT RD was crystallized using the hanging-drop vapor-diffusion method at 14°C as described previously (Kim et al., 2018). The precipitation solution comprised 0.5 M ammonium sulfate, 1.0 M sodium citrate tribasic dehydrate (pH 5.6), 0.1 M lithium sulfate, and 2 mM ethylenediaminetetraacetic acid (EDTA). Glycerol (30% [v/v], cryoprotectant) was added to the precipitation solution. The X-ray diffraction data were collected in a flash-cooled liquid nitrogen stream at −173°C. SeMet-MexT RD was crystallized under the same crystallization conditions as for native MexT RD. The single-wavelength anomalous diffraction (SAD) dataset was collected from beamline 5C at the Pohang Accelerator Laboratory (Korea) and processed using HKL2000 software (Otwinowski and Minor, 1997).
Structural analysis of MexT RD
The 2.0 Å resolution dataset of the native crystal belongs to space group P212121 with unit-cell dimensions of a = 65.7 Å, b = 108.7 Å, and c = 109.2 Å. The initial model was built based on phase information from the SAD dataset for SeMet-substituted protein at 2.3 Å resolution using AutoSol software (Terwilliger et al., 2009). The structure was refined against the 2.0-Å-resolution native dataset using COOT and Phenix.refine softwares (Adams et al., 2010; Emsley et al., 2010).
Size-exclusion chromatography with multi-angle light scattering (SEC-MALS)
The sizes of FL MexT and MexT RD were assayed using SEC-MALS. A high-performance liquid chromatography pump (Agilent, USA) was connected to a Superdex-200 10/300 GL gel filtration column (GE Healthcare) and a MALS instrument (Wyatt Dawn Heleos, USA). The size-exclusion chromatography column was pre-equilibrated with buffer comprising 20 mM Tris-HCl (pH 8.5), 500 mM NaCl, and 2 mM 2-mercaptoethanol for FL MexT; and 20 mM Tris-HCl (pH 8.5), 300 mM NaCl, and 2 mM 2-mercaptoethanol for MexT RD. Bovine serum albumin (2 mg/ml) was used as the standard. MexT RD and FL MexT samples (2 mg/ml) were injected onto the column and eluted at a flow rate of 0.2 ml/min. The datasets were evaluated using the Debye model for fitting static light-scattering data, and refractive index peaks were presented in EASI graphs created using Astra V software (Wyatt Dawn Heleos).
DNase I footprinting assay
A DNA fragment of 630 bp (the mexT-mexT intergenic region [230 bp] + 200 bp upstream + 200 bp downstream) was amplified using polymerase chain reaction (PCR) from P. aeruginosa chromosomal DNA. For this, we used the following primers: 5′-6-fluorescein amidite-GGCGCCCGACATCATCGCCACCGTGC-3′ and 5′-GGCGCAGCTCCACCGACTCCGGGGCC3-3′.
The PCR product was purified from agarose gel electrophoresis. We mixed 400 ng of DNA and 200 ng of purified FL MexT for 30 min at room temperature. Next, 0.04 units of DNase I (Fermentas) was added, and the reaction mixture (10 mM Tris [pH 8.0], 50 mM KCl, 8 mM MgCl2, 50 ng/μl BSA, 5% glycerol) was incubated for 2 min at room temperature. The reactions were terminated by adding an equal volume of stopping buffer (200 mM NaCl, 1% sodium dodecyl sulfate, 30 mM EDTA), and DNase I was inactivated by heating at 75°C for 10 min. The DNA fragments were recovered via phenol extraction and alcohol precipitation and fragment-sequences were analyzed using an ABI3730xl DNA Analyzer (Applied Biosystems, USA). The size distribution was scanned using Peak Scanner software (ver. 1.0; Applied Biosystems).
RESULTS AND DISCUSSION
Overall structure of the MexT regulatory domain
The FL MexT (residues 1–304) and its RD (residues 95–304) were produced in E. coli and purified until homogeneity was attained. FL MexT and MexT RD were homotetrameric and homodimeric in solution, respectively, as determined using SEC-MALS (Fig. 1).
Fig. 1. Molecular size of the purified proteins.
(A) FL MexT. (B) MexT RD. Primary y-axis, molar mass determined using multi-angle light scattering (MALS; black line); secondary y-axis, concentration. Protein concentration was assessed by measuring the absorbance at 280 nm (red), light scattering (LS; green), and the refractive index (RI; purple). X-axis, elution time from size exclusion chromatography.
To obtain phase information, we acquired the diffraction dataset using the crystal of the SeMet-labeled protein, which was crystallized in the presence of a high concentration of ammonium sulfate. The structure was determined via the SAD method using the anomalous signals from SeMet, which was refined to the 2.0-Å-resolution native dataset with an R factor of 0.21 and Rfree of 0.25 (Table 1). The obtained crystals belonged to the spacegroup P212121, and the asymmetric unit contained four protomers or two homodimers (Fig. 2). Its overall structure shows the features, typical of the RDs of LysR-type transcriptional regulators (LTTRs). The extensive dimeric interaction in the crystal structure is consistent with the results of SEC-MALS (Fig. 1).
Table 1.
Statistics for X-ray data collection and refinement
| Native MexT RD | Se-Met MexT RD | |
|---|---|---|
| Data collection | ||
| Beamline | PAL 5C | PAL 5C |
| Wavelength (Å) | 0.97960 | 0.97940 |
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 65.7, 108.7, 109.2 | 65.9, 108.7, 109.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50-2.00 (2.03-2.00)a | 50-2.30 (2.34-2.30)a |
| Rmergeb | 0.042 (0.50) | 0.10 (0.83) |
| I/σI | 23.76 (2.91) | 13.07 (2.08) |
| Completeness (%) | 97.1 (93.6) | 98.8 (94.4) |
| Redundancy | 5.6 (3.7) | 7.9 (3.5) |
| Refinement (PDB code : 6L33) | ||
| Resolution (Å) | 31.8-2.0 | |
| No. of reflections | 50,630 | |
| Rwork/Rfreec | 0.21/0.25 | |
| No. of total atoms | 6,365 | |
| Wilson B-factor (Å) | 22.18 | |
| RMSD | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.67 | |
| Ramachandran plot | ||
| Favored (%) | 97.0 | |
| Allowed (%) | 3.0 | |
| Outliers (%) | 0.0 | |
| PDB ID | 6L33 | |
RMSD, root mean square deviation.
Values in parentheses are for the highest-resolution shell.
Rmerge = ΣhklΣi|Ii(hkl) − [I(hkl)]|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of the ith observation of reflection hkl and [I(hkl)] is the average intensity of the i observations.
Rfree calculated for a random set of 10% of reflections not used in the refinement.
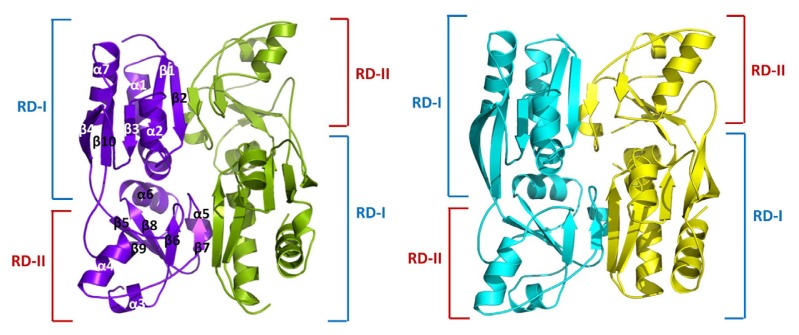
Fig. 2. Structure of MexT RD dimer.
Chains A–D of the MexT RD dimers are colored purple, green, cyan, and yellow, respectively, with secondary structures labeled accordingly. RD-I comprises three α-helices (α1, α2, and α7), and five β-strands (β1, β2, β3, β4, and β10). RD-II comprises four α-helices (α3, α4, α5, and α6), and five β-strands (β5, β6, β7, β8, and β9). Red line, RD-I; blue line, RD-II.
Structure of RD protomers
MexT RD protomers are composed of 10 β-strands and 7 α-helices, as in the typical LTTR RD (Jo et al., 2015). Each protomer can be divided into two subdomains—RD-I (residues 95–172, 274–304) and RD-II (residues 173–273). The folding pattern of the two subdomains was similar to Rossman-fold topology; i.e., a central β-sheet bound by helices and loops. The RD-I subdomain comprised three α helices (α1, α2, and α7) and five β-strands (β1, β2, β3, β4, and β10), whereas the RD-II subdomain comprised four α helices (α3, α4, α5, and α6) and five β-strands (β5, β6, β7, β8, and β9). The RD-I and RD-II subdomains were connected by two loops—one connects β4 of RD-I to β5 of RD-II, and the other connects β9 of RD-II to β10 of RD-I (Fig. 2).
Dimeric arrangement of MexT RD
The four protomers (chains A–D) in the asymmetric unit are arranged as two of dimers (chains A:B and C:D; Fig. 2). The overall arrangement of the MexT RD dimer was similar to those of other LTTRs. Within crisscrossing protomer dimer, RD-I of one protomer directly interacted with RD-II of the other protomer. The major interactions of two protomers were intermolecular hydrogen bonds between residues in the RD-1 β2 backbone and the RD-II β7 backbone at the dimeric interface. Hydrophobic interactions were found at the dimeric interface of RD-1 α1 and RD-II α5, and likely stabilize the dimer (Supplementary Fig. S1).
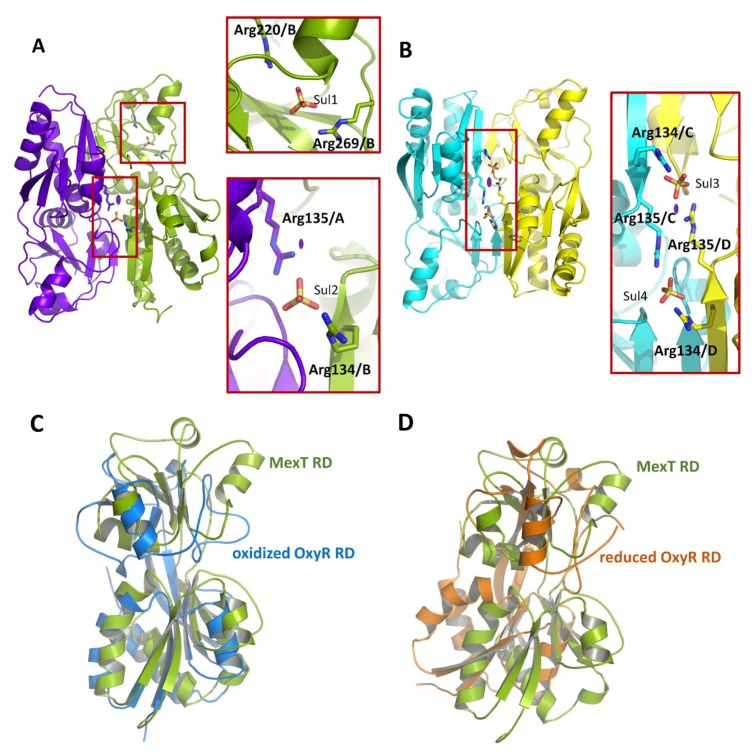
The asymmetric unit contained five sulfate ions, which may originate from the crystallization solution (Fig. 3 and Supplementary Fig. S2). Of the five sulfate ions, three were found at the dimeric interfaces; the sulfate ions interacted with Arg134 and Arg135 near the molecular pseudo-2-fold axis—one in the A:B chain dimer and two in the C:D chain dimer (Fig. 3). The fourth sulfate ion is found in the putative ligand-binding site (Fig. 4A). The fifth sulfate ion is located outside of the A:B chain dimer, interacting with Arg120 and Arg121 of chain A (Supplementary Fig. S2).
Fig. 3. Dimeric arrangement of MexT RD.
(A) Sulfates in the A:B chain dimer. A sulfate ion (Sul1) is bound to Arg220 and Arg269 of chain B. The other sulfate ion (Sul2) interacts with Arg134 of chain A (purple) and Arg135 of chain B (green; bottom panel). (B) Sulfates in the C:D chain dimer. Two sulfate ions (Sul3 and Sul4) are bound. Red squares, sulfates bound to arginine residues; the main residues involved are marked. Purple ovals, two fold enlargements of detail. (C) Oxidized (active) OxyR RD from E. coli (PDB code, 1I6A; blue) superimposed on the MexT RD (green). (D) Reduced (inactive) OxyR RD from E. coli (PDB code, 1A69; orange) superimposed on the MexT RD (green).
Fig. 4. Putative ligand binding sites and variable (V)-loops.
(A) Conformation of the V-loop in each monomer. Red lines represent V-loops; a red dotted line represents a disordered region of chain D. A stick model was generated to show positively charged and hydrophobic residues with chain B. (B) The pocket at the putative ligand-binding site. The pocket at the putative ligand binding site in chain B is shown with a sulfate ion on the surface. A stick model was generated to show positively charged and hydrophobic residues. The hydrophobic patch is lined.
The dimeric arrangements of the two dimers were similar (Supplementary Fig. S3), but the relative position of subdomain RD-II within the overall frame showed ~5.0 Å deviation. The orientation of the protomers in the dimer can be used to distinguish active from inactive LTTRs (Choi et al., 2001; Jo et al., 2015). To examine the conformation of the RD dimer, the obtained MexT RD dimer (chains A:B) was superimposed onto the E. coli OxyR RD dimer in the reduced (inactive; PDB code, 1A69) and oxidized (active; PDB code, 1I6A) forms as a prototype LTTR (Choi et al., 2001). The conformation of the MexT RD dimer fit better to that of the oxidized OxyR RD (Fig. 3C and 3D). However, further studies are required to confirm this idea.
Putative ligand-binding site
Compared to a typical LTTR E. coli OxyR, MexT exhibits substantial variation in the conformation of the putative ligand-binding sites at the inter-subdomain junction. These structural variations of MexT appears to be beyond the subtle superficial conformational changes resulting from crystal packing contacts. We noted a loop connecting β6 and α4 in RD-II (residues 199–208), which we named the variable (V)-loop (Fig. 4A). A V-loop with two different conformations lines the putative ligand-binding sites, which are not involved in the crystal packing contacts between the symmetry-related molecules (Supplementary Fig. S4). In chains A and B, Leu205 of the V-loop forms a hydrophobic bond with Val222 of RD-II, and with Val130 and Val132 of RD-I of the other protomer, leading to the formation of a cavity at the putative ligand-binding site (Fig. 4A; chains A and B). In chain C, Leu205 in the loop forms hydrophobic bonds with Tyr156 and Tyr138 of RD-I but has no inter-protomer interaction (Fig. 4A; chain C). The electron density of the V-loop of chain D is low and disordered (residues 204–217). However, the conformation of the V-loop of chain D was similar to that of chain C (Fig. 4A; chain D). Thus, our findings suggest that the V-loop regulates the putative ligand-binding sites.
As mentioned above, one sulfate ion is located at the putative ligand-binding site (Fig. 3A). Arg220 and Arg269 ionically interact with the sulfate ion at the putative ligand-binding site of chain B (Fig. 3A). In particular, guanidine group of Arg269 in chain B can make two configurations by flipping. And it is the site where one sulfate ion can bind. Pocket-formation by the flipping guanidine group makes space for holding small molecules. The sulfate-binding region extends to the hydrophobic surface region decorated by Tyr138, Phe201, Phe208, and Phe229 (Fig. 4B and Supplementary Fig. S5). Highlighting the space of adjacent sulfate-binding region and hydrophobic patches in the ligand-binding site, our structure suggests that candidate ligands should have a negatively charged (or polar) moiety linked to a hydrophobic moiety.
Cinnamaldehyde upregulated the expression of MexEF-OprN. Indeed, cinnamaldehyde has a polar aldehyde moiety and a hydrophobic phenyl ring (Juarez et al., 2017). The polar aldehyde moiety likely interacts with the positively charged pocket, which is composed of Arg220 and Arg269, and the hydrophobic phenyl ring probably participates in the interaction. A molecular model showed that cinnamaldehyde fitted the pocket well (Dallakyan and Olson, 2015) (Supplementary Fig. S6). Molecules with such characteristics may activate MexT and alter its association with DNA.
Binding of MexT to the promoter region of mexEF-oprN
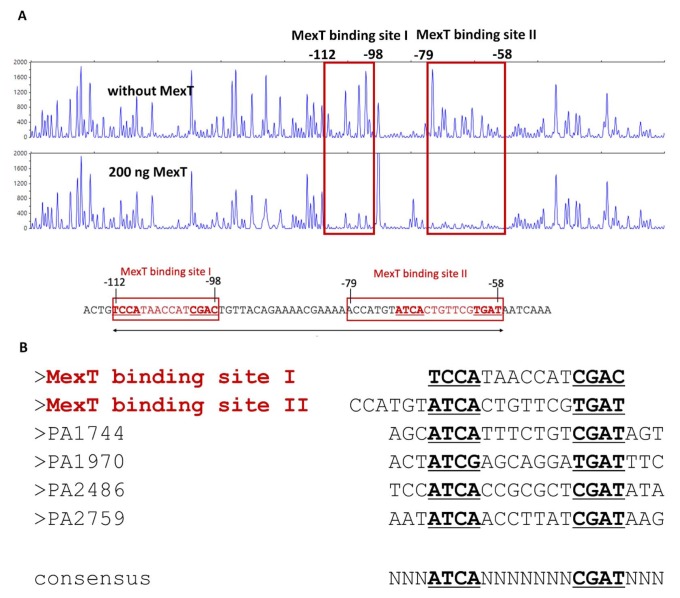
To elucidate the interaction between MexT with the promoter of mexEF-oprN, a DNase I footprinting assay was performed focusing on the region between mexT and mexE, which spans the promoter region of the mexEF-oprN operon. According to the short fragment DNA size analysis data, MexT binding protected two parts of the DNA sequence (MexT binding sites I and II) that are 17 bp apart. MexT binding site I is located between −112 bp and −89 bp, and the MexT binding site II is between −72 and −58 bp from the transcriptional initiation site of the mexEF-oprN operon, which is located 30 bp upstream of the mexE start codon (Maseda et al., 2010) (Fig. 5).
Fig. 5. MexT binding sequences identified by DNase I footprinting.
(A) A 630-bp DNA fragment containing the mexT-mexE intergenic region was labeled with 6-fluorescein amidite and then used as a DNA probe. The fluorescence-labeled DNA probe (400 ng) was incubated without or with 200 ng of purified FL MexT. The regions protected by MexT are indicated by red squares (MexT binding site I and II). Nucleotide numbers shown are the relative positions when the transcription start site mexE is designated as +1. (B) Comparison of the MexT-binding sequences. The two putative MexT-binding sequences in the mexEF-oprN regulatory region are shown at the top. The MexT-binding sequences from other promoters of MexT regulons are aligned below. Nucleotides in bold and underlined represent a partially conserved palindromic sequence, suggesting the potential consensus MexT-binding sequence, ATCA(N)7CGAT.
Next, the two MexT-binding site sequences identified by DNase I footprinting were aligned with the promoter sequences of other genes in the MexT regulon (PA1744, PA1970, PA2486, and PA2759). The MexT regulon was identified with microarray-based transcriptome profiling (Tian et al., 2009), and was further experimentally confirmed by electromobility shift assays and β-galactosidase assays using transcriptional fusion reporters (Maseda et al., 2010; Tian et al., 2009). The sequence alignment showed a palindromic consensus pattern of ATCA(N7)CGAT at each MexT-binding site. Notably, there was partial overlap between the front region ATCA(N5)GTCGAT(N4)ACYAT and the nod box sequence ATC(N9)GAT, indicating a region of degeneracy (marked as N9) in the binding motif bracketed by ATC and GAT. While the sequence ATCA(N7)CGAT is the binding motif of the DNA-binding domain (DBD) dimer of LTTR, our findings show that the DBD dimer of MexT facilitates close contact between the above two regions. Therefore, it is not surprising that the tetrameric conformation of MexT brings the DNA region (55 bp) spanning the two MexT-binding sites into close association.
The MexT-binding sequence has been predicted in previous studies (Goethals et al., 1992; Kohler et al., 1997; 1999; Maseda et al., 2010; Tian et al., 2009). We identified MexT-binding sequence with the upstream region of the mexEF-oprN operon, which partially agrees with the predicted palindromic consensus sequence (Goethals et al., 1992; Kohler et al., 1997; 1999; Maseda et al., 2010; Tian et al., 2009). Because these were in close proximity, MexT multimers likely directly interact with the sequence spanning the two binding sites, similar to other LTTRs (Maddocks and Oyston, 2008; Muraoka et al., 2003a; 2003b).
Since recombinant MexT was not tested with candidate ligands, its binding to MexT binding sites I and II likely represents its inactive status as a mexEF-oprN promoter. The effect of cognate ligand binding on protein–DNA binding is unclear. Importantly, the two MexT consensus sequence positions form an interrupted palindromic sequence compared to those seen in other LTTRs. MexT binding site II was predicted to have high similarity with the nod box (Maseda et al., 2010). However, MexT binding site I was only discovered in this study.
In conclusion, MexT is a LTTR (Kohler et al., 1997; 1999). The typical tetrameric arrangement of OxyR (Jo et al., 2015) and the atypical tetrameric arrangement of VV2_1132 and HypT have been characterized previously (Jang et al., 2018; Jo et al., 2019). Recognition of ligand binding or stimuli by the RD induces structural changes in transcriptional regulators, which alters their DNA binding (Jo et al., 2015). We elucidated the crystal structure of the RD of MexT of P. aeruginosa. Since the RD should contain the region required for sensing the signals in response to the environmental cues to turn on the efflux pumps, our study is important in understanding the molecular nature of the gene expression regulation required for the efflux pump-mediated antibacterial resistance of P. aeruginosa. Furthermore, our findings will facilitate the development of drugs with activity against P. aeruginosa by promoting the expression of multidrug efflux pumps.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Foundation of Korea (NRF-2017R1A2B2003992 to NCH). We made use of beamline 5C at the Pohang Accelerator Laboratory (Pohang, Republic of Korea).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S.E. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/S0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier E., Mac Aogain M., Mooij M.J., Woods D.F., Morrissey J.P., Dobson A.D., Adams C., O’Gara F. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J Bacteriol. 2012;194:3502–3511. doi: 10.1128/JB.06632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetar H., Gilmour C., Klinoski R., Daigle D.M., Dean C.R., Poole K. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother. 2011;55:508–514. doi: 10.1128/AAC.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galie S., Garcia-Gutierrez C., Miguelez E.M., Villar C.J., Lombo F. Biofilms in the food industry: health aspects and control methods. Front Microbiol. 2018;9:898. doi: 10.3389/fmicb.2018.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals K., Van Montagu M., Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci U S A. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P., Symmons M.F., Hughes C., Koronakis V. Structure and operation of bacterial tripartite pumps. Annu Rev Microbiol. 2013;67:221–242. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- Hirai K., Suzue S., Irikura T., Iyobe S., Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:582–586. doi: 10.1128/AAC.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horna G., Lopez M., Guerra H., Saenz Y., Ruiz J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci Rep. 2018;8:16463. doi: 10.1038/s41598-018-34694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y., Choi G., Hong S., Jo I., Ahn J., Choi S.H., Ha N.C. A novel tetrameric assembly configuration in VV2_1132, a LysR-type transcriptional regulator in Vibrio vulnificus. Mol Cells. 2018;41:301–310. doi: 10.14348/molcells.2018.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Kim J.S., Song S., Shigematsu H., Yokoyama T., Hyun J., Ha N.C. Pseudoatomic structure of the tripartite multidrug efflux pump AcrAB-TolC reveals the intermeshing cogwheel-like interaction between AcrA and TolC. Structure. 2016;24:272–276. doi: 10.1016/j.str.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Jin Y., Yang H., Qiao M., Jin S. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J Bacteriol. 2011;193:399–410. doi: 10.1128/JB.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo I., Chung I.Y., Bae H.W., Kim J.S., Song S., Cho Y.H., Ha N.C. Structural details of the OxyR peroxide-sensing mechanism. Proc Natl Acad Sci U S A. 2015;112:6443–6448. doi: 10.1073/pnas.1424495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo I., Kim D., No T., Hong S., Ahn J., Ryu S., Ha N.C. Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator. Proc Natl Acad Sci U S A. 2019;116:3740–3745. doi: 10.1073/pnas.1811509116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez P., Broutin I., Bordi C., Plesiat P., Llanes C. Constitutive activation of MexT by amino acid substitutions results in MexEF-OprN overproduction in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2018;62:e02445–17. doi: 10.1128/AAC.02445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez P., Jeannot K., Plesiat P., Llanes C. Toxic electrophiles induce expression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa through a novel transcriptional regulator, CmrA. Antimicrob Agents Chemother. 2017;61:e00585–17. doi: 10.1128/AAC.00585-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Jeong H., Song S., Kim H.Y., Lee K., Hyun J., Ha N.C. Structure of the tripartite multidrug efflux pump AcrAB-TolC suggests an alternative assembly mode. Mol Cells. 2015;38:180–186. doi: 10.14348/molcells.2015.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Ahn J., Ha N. Purification and preliminary analysis of the regulatory domain of MexT from Pseudomonas aeruginosa, a LysR-type transcriptional activator of the MexEF-OprN multidrug efflux pump. Biodesign. 2018;6:42–45. [Google Scholar]
- Kohler T., Epp S.F., Curty L.K., Pechere J.C. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6300–6305. doi: 10.1128/JB.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Michea-Hamzehpour M., Henze U., Gotoh N., Curty L.K., Pechere J.C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S.E., Oyston P.C. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154(Pt 12):3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- Maseda H., Uwate M., Nakae T. Transcriptional regulation of the mexEF-oprN multidrug efflux pump operon by MexT and an unidentified repressor in nfxC-type mutant of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2010;311:36–43. doi: 10.1111/j.1574-6968.2010.02063.x. [DOI] [PubMed] [Google Scholar]
- Muraoka S., Okumura R., Ogawa N., Nonaka T., Miyashita K., Senda T. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol. 2003a;328:555–566. doi: 10.1016/S0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- Muraoka S., Okumura R., Uragami Y., Nonaka T., Ogawa N., Miyashita K., Senda T. Purification and crystallization of a LysR-type transcriptional regulator CBNR from Ralstonia eutropha NH9. Protein Pept Lett. 2003b;10:325–329. doi: 10.2174/0929866033478942. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Piddock L.J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K., Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- Sobel M.L., Neshat S., Poole K. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J Bacteriol. 2005;187:1246–1253. doi: 10.1128/JB.187.4.1246-1253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z.X., Fargier E., Mac Aogain M., Adams C., Wang Y.P., O’Gara F. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res. 2009;37:7546–7559. doi: 10.1093/nar/gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Moeller A., Jun S.Y., Le M., Yoon B.Y., Kim J.S., Lee K., Ha N.C. Assembly and channel opening of outer membrane protein in tripartite drug efflux pumps of Gram-negative bacteria. J Biol Chem. 2012;287:11740–11750. doi: 10.1074/jbc.M111.329375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.