Abstract
MicroRNA-223-3p (miR-223-3p) is one of the potential microRNAs that have been shown to alleviate inflammatory responses in pre-clinical investigations and is highly encased in exosomes derived from bone mesenchymal stem cells (MSC-exosomes). MSC-exosomes are able to function as carriers to deliver microRNAs into cells. Autoimmune hepatitis is one of the challenging liver diseases with no effective treatment other than steroid hormones. Here, we examined whether MSC-exosomes can transfer miR-223-3p to treat autoimmune hepatitis in an experimental model. We found that MSC-exosomes were successfully incorporated with miR-223-3p and delivered miR-223-3p into macrophages. Moreover, there was no toxic effect of exosomes on the macrophages. Furthermore, treatments of either exosomes or exosomes with miR-223-3p successfully attenuated inflammatory responses in the liver of autoimmune hepatitis and inflammatory cytokine release in both the liver and macrophages. The mechanism may be related to the regulation of miR-223-3p level and STAT3 expression in the liver and macrophages. These results suggest that MSC-exosomes can be used to deliver miR-223-3p for the treatment of autoimmune hepatitis.
Keywords: autoimmune liver disease, exosomes, immunomodulatory, mesenchymal stromal cells
INTRODUCTION
Autoimmune hepatitis (AIH) is an inflammatory condition of the liver for which there is no effective treatment other than steroid hormones. A certain proportion of AIH patients experience end-stage liver diseases and require liver transplantation (Manns et al., 2010). AIH can occur in people of all ages and sex, and it has recently been recognized as a global disease (Heneghan et al., 2013). The histopathology of AIH usually presents as interface hepatitis, which features a dense portal mononuclear cell infiltration consisting of lymphocytes, monocytes/macrophages and plasma cells (Longhi et al., 2010). Although the administration of corticosteroids or corticosteroids associated with azathioprine is the conventional treatment strategy for AIH, not all patients with AIH respond well to these treatments and patients who do respond to these treatments may experience strong side effects or a relapse after steroid withdrawal (Selvarajah et al., 2012). Thus, there is a need for the development of new immunosuppressive agents or novel alternative treatments for AIH.
Mononuclear cells are the major component of the periportal cellular infiltrates in interface hepatitis associated with AIH, and these cells play an important role in the pathogenesis of AIH (Longhi et al., 2009; 2010). Previous studies have indicated that the dysfunction of Kupffer cells and peripheral blood monocyte cells (PBMCs) may be involved in the pathogenesis of AIH (Lin et al., 2016). Cytokines such as interleukin (IL)-1, IL-6, and tumor necrosis factor alpha (TNF-α) from monocytes can contribute to the loss of immune tolerance, promote the differentiation of naïve T lymphocytes into Th17 cells, and induce chronic inflammation in patients with AIH (Bettelli et al., 2006; Longhi et al., 2009). It has been confirmed that higher expression levels of IL-17, ROR-rt, IL-6, and IL-1β in the liver of patients with AIH are associated with severe inflammation and fibrosis in the liver (Zhao et al., 2011).
Exosomes are nanometer-sized membrane vesicles (40–100 nm) released from various cell types upon fusion of multivesicular bodies with the cell membrane (Raposo and Stoorvogel, 2013). Recently, exosomes have been the topic of great interest in medical research (Colombo et al., 2014; Costa-Silva et al., 2015). Importantly, numerous experiments have demonstrated exosomes as modulators of inflammation and immunity (Sadallah et al., 2011). Moreover, several recent studies have confirmed exosomes as key effectors of the paracrine function of bone marrow-derived mesenchymal stem cells (BMSCs) and demonstrated that exosomes derived from bone mesenchymal stem cells (MSC-exosomes) are able to improve recovery in animal models of graft-versus-host disease, drug-induced liver injury (Kordelas et al., 2014; Tan et al., 2014). Furthermore, MSC-exosomes have been shown to suppress Th17 differentiation and reduce IL-17 secretion in experimental models of autoimmune uveoretinitis (Shigemoto-Kuroda et al., 2017). However, whether MSC-exosomes also contribute to the protection of the liver against AIH remains to be clarified.
Current understanding is that the function of an exosome depends on its contents, for especially microRNAs (miRNAs) (Ailawadi et al., 2015). MiR-223-3p (previously referred to as miR-223 or the guide strand) is the most highly expressed miRNA in both PBMCs and BMSCs. Moreover, miR-223-3p is highly encased in exosomes derived from PBMCs and MSCs (Taibi et al., 2014; Wang et al., 2014; 2015b). Furthermore, several studies have documented that miR-223-3p can negatively regulate the expression of many inflammatory genes (i.e., STAT3 and IL-6) (Chen et al., 2012; Taibi et al., 2014; Wang et al., 2015b). Therefore, in the current study, we examined whether exosomes with miR-223-3p from MSCs could attenuate inflammatory and immune responses in an experimental model of AIH.
MATERIALS AND METHODS
Cell preparation
MSCs from C57BL/6 mice were purchased from Cyagen Biosciences (China). The identification of phenotypic properties was performed by flow cytometric analysis: CD44 100%, Sca-1 98%, CD29 94.5%, CD31 0.86% and CD117 0.28% (Meirelles Lda and Nardi, 2003). The differentiation of the cells to osteocytes, adipocytes and chondrocytes was confirmed using classical methods. The same BMSC line was used in all experiments when cells were at the 6th passage. MSCs were cultured in α-MEM (Sigma, USA) containing 10% fetal bovine serum (FBS; Sigma), 100 U/ml penicillin and 0.1 mg/ml streptomycin (Sigma). Murine macrophage RAW264.7 cells were purchased from Sigma-Aldrich (USA; product No. 91062702), and cultured in Dulbeco’s Modified Eagle’s Media (DMEM; Gibco, USA) containing 10% FBS, 2 mM glutamine (Gibco), 100 U/ml penicillin and 0.1 mg/ml streptomycin. All cells were kept in a humidified incubator at 37°C with 5% CO2.
Autoimmune hepatitis induction via hepatic S100 injection
Seventy male specific pathogen-free C57BL/6 mice at 5 to 6 weeks were purchased from the Shanghai Laboratory Animal Center (China), and were fed under specific pathogen-free conditions. The animal experiments were approved by the institutional animal committee of Wenzhou Medical University (approval No. wydw2014-0029). Out of the 70 mice, six were randomly chosen for the control group. The hepatic cytosolic S100 fraction from sixteen mice was collected as previously described (Chen et al., 2014; Lohse et al., 1990). The S-100 protein was then emulsified with an equal volume of complete Freund’s adjuvant (CFA; Sigma). The remaining 48 mice were intraperitoneally injected with this mixture (1 ml per animal) on day 0 and a repeat injection on day 7. Ten mice died in the process of modeling experimental autoimmune hepatitis (EAH). Eight mice were used to evaluate the model of EAH on day 21 by histology and blood biochemistry assay.
MSC-exosomes preparation
For the preparation of the exosomes from MSCs, the culture medium of MSCs was replaced with a-MEM medium and incubated for 24 h. The culture medium was collected and briefly centrifuged to eliminate large dead cells and cellular debris via sequential centrifugations with increasing speeds (300g for 10 min, 2,000g for 10 min, 10,000g for 30 min). After the final centrifugation, the supernatant was centrifuged again at 100,000g for 70 min to precipitate the exosomes. All centrifugations were performed at 4°C. After ultracentrifugation, the pellet was collected and washed in 50 ml phosphate-buffered saline (PBS) to remove contaminating proteins and then centrifuged again at 100,000g for 70 min (Thery et al., 2006). The pellet was then suspended in 200 μl PBS and stored at −80°C until use. Electron microscope and Western blotting analyses (with antibodies against CD63, TSG101, CD9, CD81, and cytochrome c) were employed to identify the MSC-exosomes (Lyu et al., 2015) as previously described. The total protein content of exosomes was determined using the Micro-BCA assay (Beyotime Biotechnology, China).
MSC-exosomesmiR-223-3p and MSC-exosomesmiR-223-3p(i) preparation
We packaged the lentiviruses that contain the vectors of LentimiRa-GFP-mmu-mir-223 Vector (mm12144; Applied Biological Materials [Canada; http://www.abmgood.com], pre-miR-223 inserted for miR-223 knockin), pLenti-III-mir-GFP-Blank (m001; Applied Biological Materials, vector for miR-223 knockin control), miRZip-223 anti-miR-223 miRNA construct (MZIP223-PA-1; System Biosciences [USA; http://www.systembio.com], miR-223 inhibitor inserted for miR-223 knockdown), and pGreenPuro Scramble Hairpin Control–Construct (MZIP000-PA-1; System Biosciences, vector for miR-223 knockdown control), respectively, according to the manufacturer’s suggested protocol. Then, we infected MSCs with these lentiviruses, respectively. The expression of green fluorescent protein (GFP) was used to monitor transfection efficiency, and stably transfected cells were selected with puromycin. Exosomes were respectively harvested from MSCs and transfected MSCs to determine the expression levels of miR-223-3p and miR-223-5p via quantitative real-time polymerase chain reaction (qRT-PCR). We defined MSCs and MSC-exosomes overexpressing miR-223-3p as MSCsmiR-223-3p and MSC-exosomesmiR-223-3p. MSCs and MSC-exosomes with miR-223-3p knockdown were defined as MSCsmiR-223-3p(i) and MSC-exosomesmiR-223-3p(i). MSCsmiR-223-3p-C°N and MSCsmiR-223-3p(i)-C°N were negative controls of MSCsmiR-223-3p and MSCsmiR-223-3p(i), respectively. Similarly, MSC-exosomesmiR-223-3p-C°N and MSC-exosomesmiR-223-3p(i)-C°N were negative controls of MSC-exosomesmiR-223-3p and MSC-exosomesmiR-223-3p(i), respectively.
Cell proliferation assay
Cell proliferation was determined using CCK-8 dye (Beyotime Inst Biotech, China) according to the manufacturer’s instructions. Briefly, MSCs and transfected MSCs (2 × 103 cells) were seeded in a 96-well plate in 100 μl medium per well, grown at 37°C for 24 h. After 10 μl CCK-8 dye was added to each well, cells were incubated at 37°C for 1 h and the absorbance was finally determined at 450 nm.
Cytotoxicity assay
Briefly, macrophages (5 × 103 cells) were seeded into a 96-well plate in 100 μl medium per well for 24 h. The cells were then respectively treated with MSC-exosomes, MSC-exosomesmiR-223-3p-C°N, MSC-exosomesmiR-223-3p(i)-C°N, MSC-exosomesmiR-223-3p, and MSC-exosomesmiR-223-3p(i) (2 μg/ml of exosomal proteins) for 24 h. After treatment, the medium was changed to a fresh medium and the CCK-8 reagent (10 μl) was added to each well and incubated for 1 h at 37°C. After incubation, the optical density was measured at 450 nm and the value was compared to that of control cells.
Treatment of macrophages with MSC-exosomes
RAW264.7 cells were respectively co-incubated with MSC-exosomes, MSC-exosomesmiR-223-3p or MSC-exosomesmiR-223-3p(i) (2 μg/ml) in DMEM with antibiotics and without FBS for one hour followed by the addition of lipopolysaccharide (LPS) (250 ng/ml) for 24 h. After treatment, the culture medium was collected for cytokine assays, and the cells were harvested for Western blotting and qRT-PCR.
MSC-exosomes uptake experiments
For the MSC-exosomes uptake experiments, MSC-exosomes were labelled with the PKH67 Green Fluorescent Cell Linker Kit (Sigma) according to the manufacturer’s protocol. Briefly, MSC-exosomes (10 μg) diluted in PBS (100 μl) were added to 2 ml diluent C (Sigma) with 4 μl PKH67 dye and incubated for 4 min. After washing with PBS, the MSC-exosomes were centrifuged at 100,000g for 70 min at 4°C to remove unbound dye. The MSC-exosomes pellet was re-suspended in 100 μl PBS (Bang et al., 2014). For uptake experiments, the green fluorescent dye PKH67-labelled exosomes were co-cultured with macrophage RAW264.7 cells for 6 h. The cells were then examined and photographed using a confocal microscope system (Olympus FV1200; Olympus, Japan).
Treatment of mice with experimental autoimmune hepatitis with MSC-exosomes
Thirty mice were randomly divided into five AIH groups: model (n = 6), prednisolone & azathioprine (n = 6), MSC-exosomes (n = 6), MSC-exosomesmiR-223-3p (n = 6), and MSC-exosomesmiR-223-3p(i) (n = 6). The mice in each of these groups were injected with S100/CFA as described in the AIH section above. Another 6 mice were intraperitoneally injected with 1.0 ml physiological saline as the control group (n = 6). Mice in the model group were treated with 200 μl PBS through tail vein injection on days 21 and 35. Mice in the prednisolone & azathioprine group were intraperitoneally given prednisolone (5 mg in 100 μl of PBS per animal) and azathioprine (5 mg in 100 μl of PBS per animal) on days 21, 28, and 35 (Lohse et al., 1998). Mice in the MSC-exosomes, MSC-exosomesmiR-223-3p, and MSC-exosomesmiR-223-3p(i) groups were intravenously injected with MSC-exosomes, MSC-exosomesmiR-223-3p, and MSC-exosomesmiR-223-3p(i) respectively (2 μg/g body weight in 200 μl of PBS per animal) on days 21 and 35. All mice were sacrificed on day 42. The liver, the spleen, and blood were collected for further analyses.
miRNA assay
For the determination of expression of miR-223-3p and miR-223-5p in MSC-exosomes, cells and tissues, the miRcute miRNA isolation kit, the miRcute Plus miRNA First-strand cDNA Synthesis kit and the miRcute Plus miRNA qPCR Detection kit (Tiangen Biotech, China; http://www.tiangen.com/productShow/t1/3/id/112.html) were respectively employed to isolate total RNA, synthesize first strand cDNA and perform qRT-PCR according to the manufacturer’s protocols. U6 snRNA was used as the internal control. The primers for real-time PCR for miR-223-3p and miR-223-5p were designed by Tiangen Biotech.
Histological staining of the liver
The liver samples were fixed in 10% neutral formalin and embedded in paraffin. The paraffin blocks were then cut into 4 μm sections and stained with H&E. The experienced liver pathologist blinded to the identity of the tissue samples performed assessment of liver tissues with code. Three sections per liver were examined. The histological scores were evaluated according to the Ishak grading system.
Serum biochemistry assay
The serum from mouse blood was isolated by centrifugation at 1,300 rpm for 10 min. The automatic biochemistry analyzer (Abbott Laboratories, USA) was used to evaluate the serum levels of alanine transaminase (ALT) and aspartate transaminase (AST) according to the manufacturer’s protocol.
Determination of cytokine concentration
The concentration of cytokines in serum or culture medium was measured using an ELISA kit (eBioscience, USA) according to the manufacturer’s protocol. Each sample was evaluated three times, and the concentrations of cytokines were determined based on standard curves.
Western blotting
Protein samples were extracted from MSC-exosomes, cells or tissues according to a previously described protocol (Wang et al., 2009). Equal amounts of protein were subjected to 10% SDS-PAGE and immobilized on PVDF membranes. The membranes were blocked in skimmed milk for 1 h at room temperature and then incubated with different primary antibodies overnight. The source and dilutions of antibodies are as follows: rabbit anti-CD63 (1:500 dilution; Abcam, UK), rabbit anti-CD9 (1:400 dilution; Abcam), rabbit anti-CD81 (1:400 dilution; Abcam), rabbit anti-cytochrome c (1:1,000 dilution; Abcam), rabbit anti-TSG101 (1:400 dilution; Abcam), rabbit anti-p-STAT3 (1:50,000 dilution; Abcam), rabbit anti-STAT3 (1:1,000 dilution; Abcam), and anti-Sema3A (1:1,000 dilution; Abcam). The antibody against GAPDH (1:1,000 dilution; Sigma-Aldrich) was used as an internal control. After incubation with secondary antibodies (goat anti-rabbit antibody, 1:5,000 dilution; Biosharp, USA), the proteins on membranes were visualized by incubation with a chemiluminescent reagent (Millipore Corporation, USA) and exposure on Kodak films.
Flow cytometric analyses of lymphocytes
Splenic mononuclear cells were isolated from fresh mouse spleen using lymphocyte separation medium (TBDscience, China). For the analysis of Treg cells, the mononuclear cells were labelled with anti-mouse CD4 antibody conjugated with FITC (eBioscience) and CD25 antibody conjugated with PE (eBioscience) for 30 min, incubated with fixation/permeabilization reagent for 35 min and then stained with anti-mouse Foxp3 antibody conjugated with PE-Cyanine5.5 (eBioscience) for 30 min. For the analysis of Th17 cells, the lymphocytes were stained with anti-mouse CD4 antibody conjugated with FITC, and incubated with a combination of PMA/Ionomycin mixture (eBioscience) and BFA/Monensin mixture (eBioscience) for 4 to 6 h at 37°C, followed by staining with anti-mouse IL-17 antibody conjugated with PE (eBioscience). The stained cells were examined using a BD FACSCalibur platform (BD Bioscience, USA) following the manufacturer’s instruction. Flow cytometric data were analyzed using FlowJo software (FlowJo, USA).
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated with the RNA extraction kit (Aidlab Biotechnologies, China) and the first-strand cDNA was reverse-transcribed with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) according to the instruction manual. The qRT-PCR was performed at 95°C for 15 min following by 40 cycles of denaturing at 95°C for 10 sec, annealing at different melting temperature (listed in Supplementary Table S1) for 32 sec and extending at 72°C for 10 sec. The fluorescence signal was recorded at the end of the annealing step using a real-time fluorescence PCR spectrometer (7500 Real-Time PCR System; ABI, USA). ΔCt depicted the different Ct values of the target gene and GAPDH. It was analyzed with 2−ΔCt as relative contents.
Statistical analysis
The data were expressed as the mean ± SD. A one- or two-way ANOVA and an least significant difference test were used to determine the significance between multiple groups. IBM SPSS Statistics 19.0 software (IBM, USA) and GraphPad Prism 5 (GraphPad Software, USA) were used to analyze the statistical differences. P < 0.05 was considered statistically significant.
RESULTS
Identification and characterization of exosomes and miR-223-3p
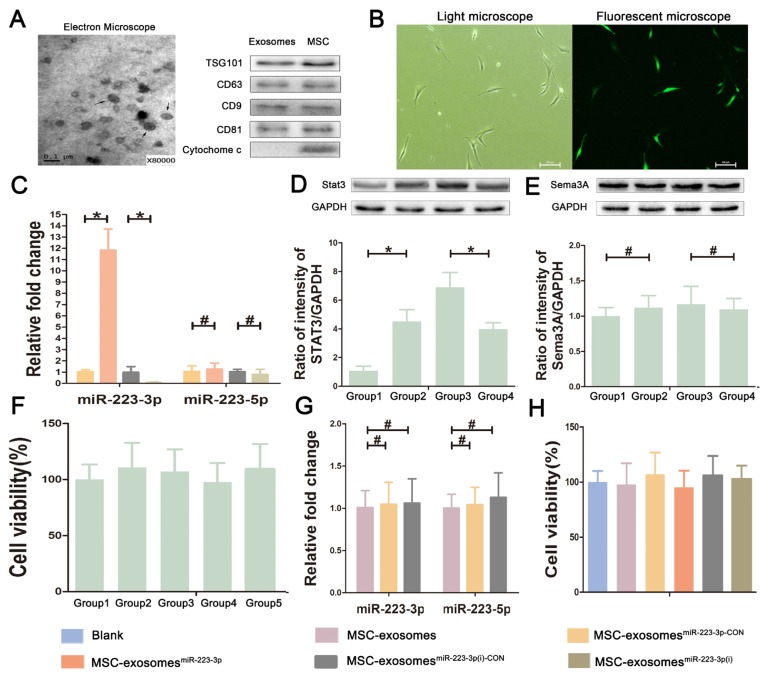
Exosomes were isolated from MSCs and identified using both transmission electron microscope (TEM) and Western blotting. As shown in Figure 1A, TEM revealed that exosomes are cup-shaped membrane vesicles of 40 to 100 nm in diameter. Western blot analysis indicated that the exosomes were positive for CD9, CD63, CD81, and TSG101, but negative for cytochrome c as previously reported (Lyu et al., 2015). The exosomes with miR-223-3p were further analyzed after transfection of MSCs with lentivirus-encoding pre-miR-223 and lentivirus-encoding anti-miR-223. The expression of GFP was used to monitor the infection efficiency (Fig. 1B). According to the GFP signal, over 90% of cells were successfully transfected. In order to verify the transfection effect, the expression levels of miR-223-3p and miR-223-5p in exosomes derived from transfected MSCs were determined via qRT-PCR. As shown in Figure 1C, the level of miR-223-3p was significantly higher in MSC-exosomesmiR-223-3p than that in MSC-exosomesmiR-223-3p-C°N, while the expression of miR-223-3p was dramatically down-regulated in MSC-exosomesmiR-223-3p(i) when compared to MSC-exosomesmiR-223-3p(i)-C°N. Compared with miR-223-3p, the level of miR-223-5p was not significantly changed in MSC-exosomesmiR-223-3p and MSC-exosomesmiR-223-3p(i). MiR-223-3p and miR-223-5p could negatively regulate the expression of Stat3 and Sema3A, respectively. To further verify the results of transfection, the expression of Stat3 and Sema3A in transfected MSCs was analyzed via Western blotting. As shown in Figures 1D and 1E, the results showed that the expression of Stat3 was obviously lower in MSCsmiR-223-3p than that in MSCsmiR-223-3p-C°N, while the expression of Stat3 was significantly up-regulated in MSCsmiR-223-3p(i) when compared to MSCsmiR-223-3p(i)-C°N. Moreover, Western blot results also showed no significant difference in the expression of Sema3A, the downstream target of miRNA-223-5p, in each group. In summary, based on the above PCR and Western blot results, we found that the expression level of miR-223-3p was significantly changed after transfection of lentivirus containing pre-miR-223 and miR-223 inhibitor, but miR-223-5p was not obviously changed, which may be related to miR-223-3p as the guide strand of miR-223 and the advantage of miR-223-3p expression over miR-223-5p (see the database available at http://www.mirbase.org/index.shtml).
Fig. 1. Characterizations of exosomes derived from bone mesenchymal stem cells.
(A) The morphology of MSC-exosomes under an electron microscope and proteins expression in exosomes and MSCs. Arrows indicate exosomes as cup-shaped membrane vesicles of 40 to 100 nm in diameter. Western blot analysis indicates that exosomes are positive for CD9, CD63, CD81, and TSG101, but negative for cytochrome c. (B) The transfected cells under a light microscope and a fluorescent microscope at a magnification of ×200. Scale bars = 100 μm. After transfection, MSCs, which transfected successfully, displayed green fluorescence in fluorescent microscope image. (C) The expression levels of miR-223-3p and miR-223-5p in transfected MSC-exosomes. (D and E) The protein levels of Stat3 and Sema3A, the downstream target of miR-223-3p and miR-223-5p respectively, in transfected MSCs via Western blotting. Group 1, MSCsmiR-223-3p group; Group 2, MSCsmiR-223-3p-C°N group; Group 3, MSCsmiR-223-3p(i) group; Group 4, MSCsmiR-223-3p(i)-C°N group. (F) The effect of lentivirus, miR-223-3p knockin or knockdown on the proliferation viability of MSCs. Group 1, MSCs group; Group 2, MSCsmiR-223-3p-C°N group; Group 3, MSCsmiR-223-3p group; Group 4, MSCsmiR-223-3p(i)-C°N group; Group 5, MSCsmiR-223-3p(i) group. The results showed no difference. (G) The effect of lentivirus itself on the expression of miR-223-3p and miR-223-5p in MSC-exosomes. (H) The effect of MSC-exosomes and transfected MSC-exosomes on the viability of macrophages. The results showed no difference. Data are presented as mean ± SD from three independent experiments. *P < 0.05, #P > 0.05.
To test whether lentivirus, miR-223-3p knockin or knockdown affects the cell characteristics of MSCs, we compared the proliferation characteristic between MSCs and transfected MSCs using the CCK-8 kit. The results suggested no significant difference in the proliferation characteristic of MSCs and transfected MSCs (Fig. 1F). Moreover, in order to test the effect of lentivirus itself on the MSC-exosomes, we compared the expression levels of miR-223-3p and miR-223-5p between MSC-exosomes and MSC-exosomes which transfected the negative control lentivirus. The results showed that lentivirus itself did not affect the expression of miR-223-3p and miR-223-5p in MSC-exosomes (Fig. 1G). In addition, by cytotoxicity assay, we found that MSC-exosomes and transfected MSC-exosomes did not affect the viability of macrophages (Fig. 1H), suggesting that lentivirus and MSC-exosomes may have no significant toxicity on cells.
Attenuation of autoimmune hepatitis in mice by MSC- exosomes and MSC-exosomesmiR-223-3p
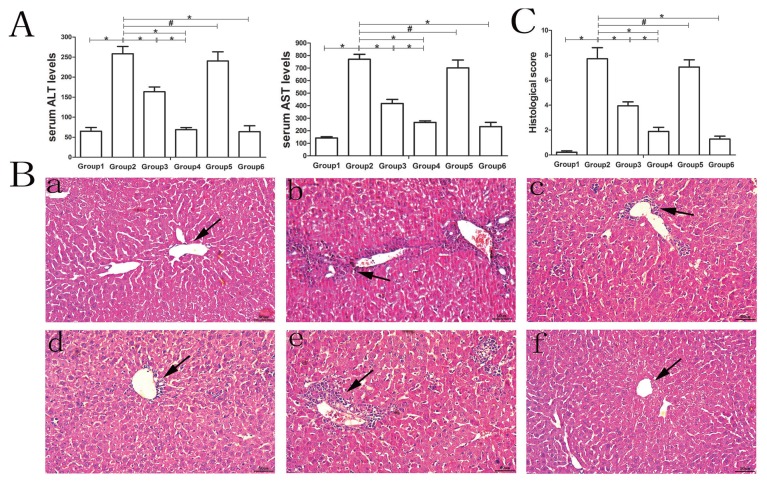
AIH was successfully established in mice with the administration of S100/CFA as shown in Figure 2. In the model group, there was a significant increase in the level of both ALT and AST. In the liver section, there were typical features of interface hepatitis characterized by the infiltration of mononuclear cells in the centrilobular or portal areas as well as intralobular inflammatory lesions and necrosis. However, treatment with either MSC-exosomes or MSC-exosomesmiR-223-3p significantly reduced the elevated levels of both ALT and AST (Fig. 2A), especially with MSC-exosomesmiR-223-3p, which reduced the level of both ALT and AST to that in the control group and to a similar extent as treatment with prednisolone and azathioprine. Moreover, treatment with MSC-exosomes, MSC-exosomesmiR-223-3p, as well as prednisolone and azathioprine significantly improved inflammatory lesions and reduced the infiltration of mononuclear cells into the centrilobular or portal regions (Fig. 2B). In contrast, MSC-exosomesmiR-223-3p(i) significantly increased the level of transaminase and inflammatory lesions compared to MSC-exosome treatment, suggesting a role of miR-223-3p carried by MSC-exosomes in attenuating inflammatory responses and liver injury. In addition, the liver sections were assessed by experienced pathology and scored with the Ishak grading system. A significantly higher histological hepatitis score (Fig. 2C) was found in the model group and MSC-exsomesmiR-223-3p(i) group compared with that in the control group. After treatment with MSC-exosomes, MSC-exosomesmiR-223-3p or the drugs, the score declined significantly.
Fig. 2. Liver injury in hepatic S100/CFA-induced AIH mice.
The automatic biochemistry analyzer was used to evaluate the serum levels of ALT and AST. (A) The serum levels of ALT and AST in the different groups of mice. Data are presented as mean ± SD from six mice. Group 1, control group; Group 2, model group; Group 3, MSC-exosomes-treated group; Group 4, MSC-exosomesmiR-223-3p-treated group; Group 5, MSC-exosomesmiR-223-3p(i)-treated group; Group 6, drug-treated group (the drug is defined as steroids and azathioprine). (B) The typical liver section stained with H&E at a magnification of ×200. Scale bars = 50 μm. Arrows indicate the infiltration of mononuclear cells in the centrilobular as well as intralobular inflammatory lesions and necrosis. Label a, control group; b, model group; c, MSC-exosomes-treated group; d, MSC-exosomesmiR-223-3p-treated group; e, MSC-exosomesmiR-223-3p(i)-treated group; f, drug-treated group. (C) The histological scoring of each group according to the Ishak grading system. Three histological sections per animal were examined. Group 1, control group; Group 2, model group; Group 3, MSC-exosomes-treated group; Group 4, MSC-exosomesmiR-223-3p-treated group; Group 5, MSC-exosomesmiR-223-3p(i)-treated group; Group 6, drug-treated group. Data are presented as mean ± SD from six mice. *P < 0.05, #P > 0.05.
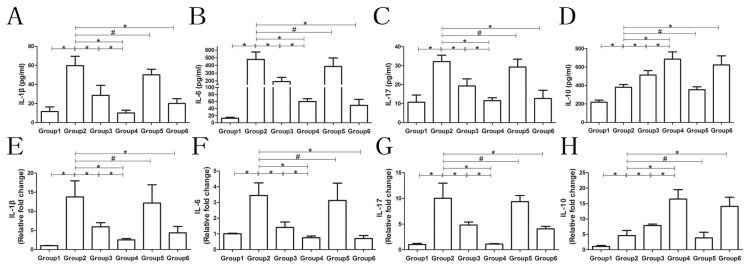
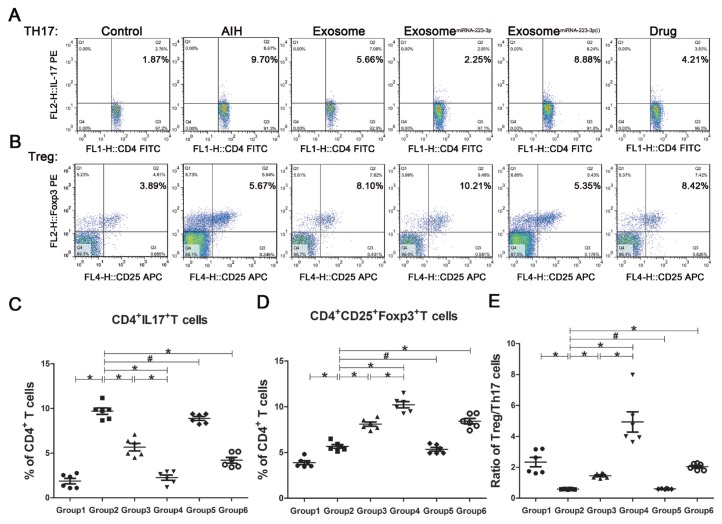
Figures 3 and 4 show the regulation of inflammatory cytokines and immune regulatory cells in AIH mice with and without exosomes treatment. As shown in Figure 3, treatment with MSC-exosomes, MSC-exosomesmiR-223-3p, as well as prednisolone and azathioprine significantly reduced the serum levels of IL-1β, IL-6, and IL-17 but significantly increased the serum level of IL-10 compared to PBS or MSC-exsomesmiR-223-3p(i) treatment. Moreover, the mRNA levels of these cytokines in the liver exhibited a similar trend in response to these treatments. Regulation of immune regulatory cells-Treg and Th17-cells is shown in Figure 4. There was a significant increase in the proportion of Th17 cells in the model mice but not much increase in Treg cells compared with those in the control mice. Both MSC-exosomes and MSC-exosomesmiR-223-3p were able to reduce the proportion of Th17 cells and increase the proportion of Treg cells. The same change was observed after the administration of prednisolone and azathioprine. However, MSC-exosomesmiR-223-3p(i) significantly increased the proportion of Th17 cells and reduced the proportion of Treg cells compared to MSC-exosome treatment. The ratio of Treg/Th17 is the most important parameter. In the model mice, the ratio was significantly decreased but treatment with MSC-exosomes reverted the ratio to the normal level. Moreover, treatment of MSC-exosomesmiR-223-3p or the drugs significantly elevated the ratio. In contrast, MSC-exosomesmiR-223-3p(i) treatment remarkably reduced the ratio compared to MSC-exosome treatment.
Fig. 3. The levels of inflammatory cytokines in mouse serum and liver.
ELISA and qRT-PCR were employed to investigate serum and liver levels of inflammatory cytokines. (A-D) The serum levels of IL-1β, IL-6, IL-17, and IL-10, respectively. (E-H) The liver levels of IL-1β, IL-6, IL-17, and IL-10, respectively. For the liver, the data are expressed as fold changes relative to the control group. All histograms are presented as mean ± SD, n = 6. *P < 0.05, #P > 0.05. Group 1, control group; Group 2, model group; Group 3, MSC-exosomes-treated group; Group 4, MSC-exosomesmiR-223-3p-treated group; Group 5, MSC-exosomesmiR-223-3p(i)-treated group; Group 6, drug-treated group (the drug is defined as steroids and azathioprine).
Fig. 4. Typical flow cytometric plots and different phenotypes of lymphocytes.
Flow cytometry was employed to investigate the differentiation of CD4+ lymphocytes. (A and B) The representative flow cytometric plots of CD4+CD25+Foxp3+ Treg and CD4+IL-17+ Th17 cells, respectively, labelled with the corresponding percentage of CD4+ T cells. (C-E) The ratios of Th17/CD4+, ratio of Treg/CD4+ and ratio of Treg/Th17 respectively. Data are presented as mean ± SD, n = 6. *P < 0.05, #P > 0.05. Group 1, control group; Group 2, model group; Group 3, MSC-exosomes-treated group; Group 4, MSC-exosomesmiR-223-3p-treated group; Group 5, MSC-exosomesmiR-223-3p(i)-treated group; Group 6, drug-treated group.
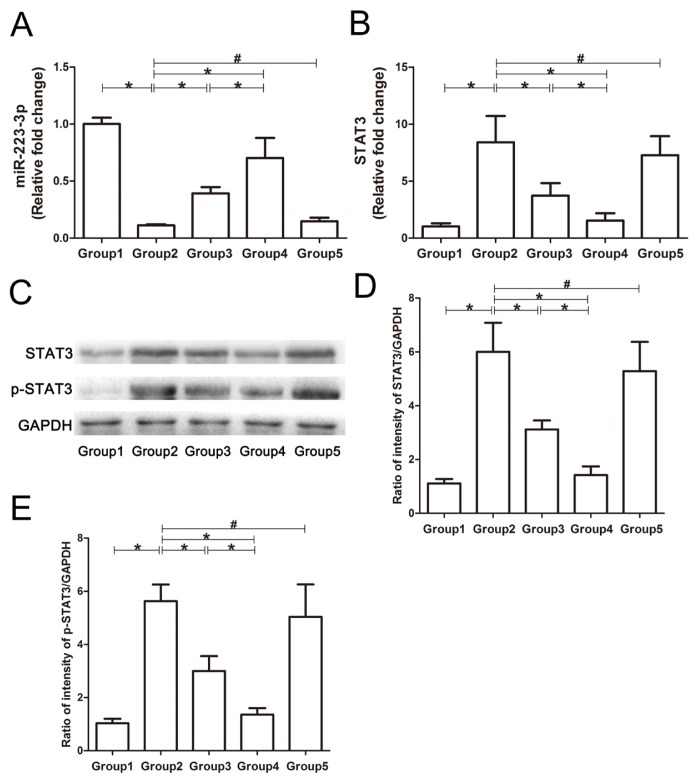
The mechanism by which MSC-exosomes and MSC-exosomesmiR-223-3p modulate immune regulatory cells was further investigated. As shown in Figure 5, the expression of miR-223-3p was significantly reduced in the livers of model or MSC-exosomesmiR-223-3p(i) therapy mice. However, MSC-exosomes or MSC-exosomesmiR-223-3p treatment was able to increase the level of miR-223-3p to lower than the normal level. In addition, the expression of STAT3 and p-STAT3 was significantly elevated in the livers of model or MSC-exosomesmiR-223-3p(i) therapy mice, but MSC-exosomes and MSC-exosomesmiR-223-3p treatments reduced both STAT3 and p-STAT3 levels.
Fig. 5. Expression of miR-223-3p, p-STAT3, and STAT3 in the livers of different groups of mice.
Western blotting and qRT-PCR were used to analyze the expression of miR-223-3p, p-STAT3 and STAT3 in the livers of mice in each group. For the PCR results, the data are expressed as fold changes relative to the control group. (A) The expression of miR-223-3p in the liver. (B) The expression of STAT3 mRNA in the liver. (C) The typical images of Western blots for p-STAT3, STAT3, and GAPDH. (D) The STAT3 levels normalized to GAPDH in the liver. (E) The p-STAT3 levels normalized to GAPDH in the liver. Group 1, control group; Group 2, model group; Group 3, MSC-exosomes-treated group; Group 4, MSC-exosomesmiR-223-3p-treated group; Group 5, MSC-exosomesmiR-223-3p(i)-treated group. Data are presented mean ± SD, n = 6. *P < 0.05, #P > 0.05.
Attenuation of LPS-induced cytokine release from macrophages by exosomes and miR-223-3p
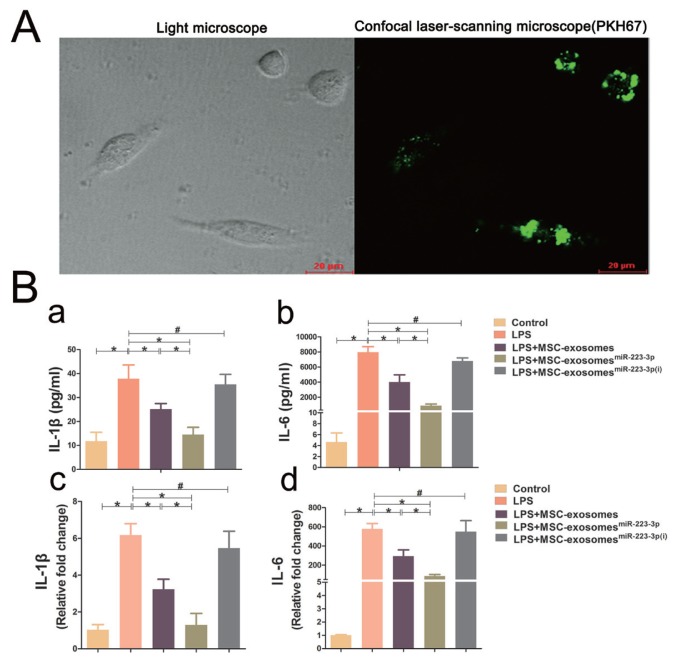
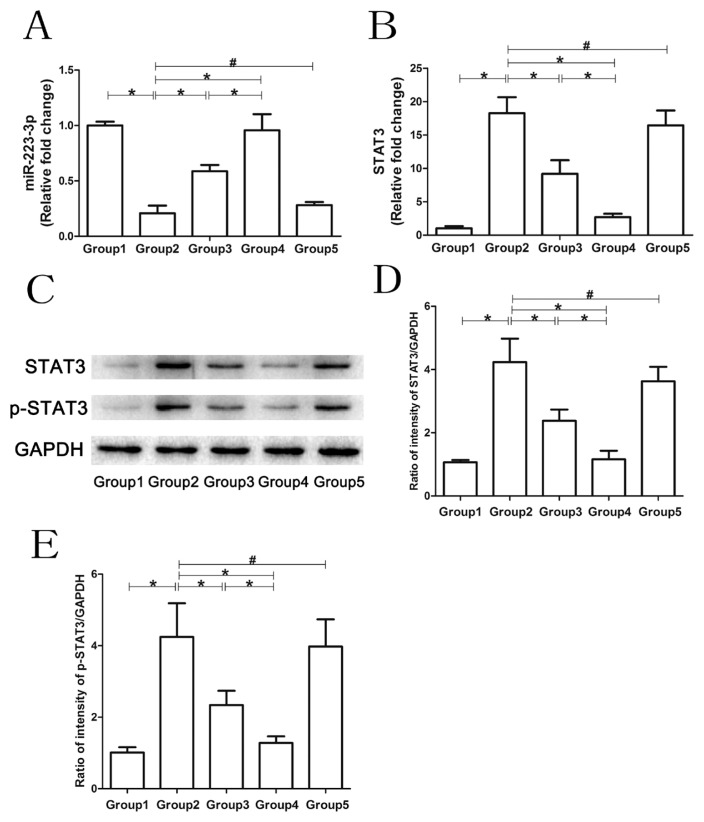
The macrophage cell line RAW264.7 was employed to validate the effect of MSC-exosomes, MSC-exosomesmiR-223-3p and MSC-exosomesmiR-223-3p(i) on cytokine release. As shown in Figure 6A, MSC-exosomes labelled with GFP were internalized into macrophages. Moreover, treatment with both MSC-exosomes and MSC-exosomesmiR-223-3p was able to attenuate the LPS-induced increase in IL-1β and IL-6 at both mRNA and protein levels. However, treatment with MSC-exosomesmiR-223-3p(i) could not attenuate the elevation of LPS-induced inflammatory cytokines (Fig. 6B). The mechanism by which exosomes and miR-223-3p affect cytokine release from macrophages was further investigated and depicted in Figure 7. LPS treatment significantly reduced the miR-223-3p level, while in addition to MSC-exosomesmiR-223-3p(i) treatment, treatment with both MSC-exosomes and MSC-exosomesmiR-223-3p was able to bring the miR-223-3p level back to normal. Moreover, LPS treatment also significantly increased STAT3 and p-STAT3 levels in macrophages. Treatment with both MSC-exosomes and MSC-exosomesmiR-223-3p could reduce the LPS-induced elevation of STAT3 and p-STAT3 in macrophages. However, compared with macrophages treated with LPS, treatment with MSC-exosomesmiR-223-3p(i) could not reduce the expression of STAT3 and p-STAT3 at the mRNA and protein levels.
Fig. 6. Internalization of MSC-exosomes into macrophages and expression of cytokines in macrophages.
(A) Confocal laser-scanning microscopy images of macrophages with internalized green dye-labelled MSC-exosomes (×400 magnification). Scale bars = 20 μm. Macrophages and supernatants were collected for cytokine expression analysis via ELISA and qRT-PCR. For the PCR results, the data are expressed as fold changes relative to the control group. (B) The mRNA and protein levels of IL-1β and IL-6. Labels ‘a’ and ‘c’ indicate IL-1β protein in the culture medium and IL-1β mRNA in macrophages, respectively. Labels ‘b’ and ‘d’ indicate IL-6 protein in the culture medium and IL-6 mRNA in macrophages, respectively. Data represent the mean ± SD from six independent experiments. *P < 0.05, #P > 0.05.
Fig. 7. Regulation of gene expression in macrophages by MSC-exosomes.
Western blotting and qRT-PCR were used to analyze the expression of miR-223-3p, p-STAT3 and STAT3 in macrophages in each group. For the PCR results, the data are expressed as fold changes relative to the control group. (A) The expression of miR-223-3p in macrophages in response to different treatments. (B) The STAT3 mRNA levels in macrophages in response to different treatments. (C-E) The protein levels of p-STAT3 and STAT3 in macrophages in response to different treatments. Group 1, control group; Group 2, LPS group; Group 3, LPS+MSC-exosomes group; Group 4, LPS+MSC-exosomesmiR-223-3p group; Group 5, LPS+MSC-exosomesmiR-223-3p(i) group. Data are presented mean ± SD, n = 6. *P < 0.05, #P > 0.05.
Overall animal health and adverse events
There were no differences in overall health issures related to the treatment. At the end of treatment, the body weights of all mice were increased but there was no significant difference between any two groups of mice. Although there was fatty deposition in part of the liver parenchyma and injection site of S100 in CFA, that could be due to the lipid component of the CFA. Moreover, no mice died in any group during treatment. Major adverse events (such as injection site reactions, emboli, proliferation) were not observed after exosomes or steroid treatment.
DISCUSSION
In several models of experimental autoimmune diseases, the administration of MSCs has been documented to suppress inflammation and autoimmunity (Le Blanc and Mougiakakos, 2012). Our previous research demonstrated that transplantation of MSCs was effective in reducing hepatic inflammation in an experimental model of AIH in mice (Chen et al., 2014). Although there are numerous reports describing the efficacy of cell-based therapies for liver diseases (Du et al., 2013; Zhao et al., 2016), exogenous MSCs could distribute to different organs in the body and cause emboli or proliferation (Wang et al., 2012). MSC-exosomes are nano-size particles released from MSCs that have been considered key carriers of paracrine factors from MSCs. Moreover, MSC-exosomes are gaining attention in the treatment of various diseases, because the MSC-exosome-mediated intercellular delivery of bioactive substances affects cellular function (Wang et al., 2015b; Xin et al., 2013). Tan et al. (2014) demonstrated that MSC-exosomes can promote hepatic regeneration in a drug-induced liver injury model. The application of MSC-exosomes could minimize the potential adverse effects of administering MSCs such as toxicity, immune response, emboli formation, and tumourigenesis, etc. In addition, MSC-exosomes can carry functional molecules such as miRNAs, which can be regulated to enhance the therapeutic benefit of the exosomes (Xin et al., 2013). Our current study shows that MSC-exosomes carrying miR-223-3p could attenuate the development of hepatic S100/CFA-induced AIH in mice.
miRNAs are important intracellular regulatory molecules that are becoming the focus of studies on various diseases, especially in the regulation of immune responses (Wang et al., 2015b). Li et al. (2017) demonstrated that the introduction of miR-223-3p can ameliorate alcohol-induced liver injury through the inhibition of IL-6 expression in neutrophils. Furthermore, Wang et al. (2015b) also found that exosomes derived from MSCs carrying miR-223-3p can reduce the LPS-induced production of IL-6 and IL-1β in macrophages, which is consistent with the findings of our current study. In this experiment, the expression of miR-223-3p in the MSC-exosomes was up-regulated and down-regulated by transfection of lentivirus to verify its function. To date, miR-223-3p has been confirmed to negatively modulate the expression of inflammation-related gene STAT3, which is known to be an important upstream activator of IL-1β and IL-6 (Chen et al., 2012). Both IL-1β and IL-6 have been documented to be involved in AIH (Maggiore et al., 1995; Zhao et al., 2013). In addition, STAT3 has been known to activate RORt and promote the secretion of IL-17 in Th17 cells, which play a crucial role in autoimmune diseases (Shigemoto-Kuroda et al., 2017). In clinical studies, high levels of TNF-α and IL-6 in children with AIH have been found to be related to liver damage, and IL-1β level has also been shown to be closely related to the progression of AIH (Maggiore et al., 1995; Zhao et al., 2013). Moreover, the cytokine IL-6 can promote the differentiation of naïve T lymphocytes into Th17 cells (Bettelli et al., 2006; Chen et al., 2007) which secrete IL-17 to promote the development of AIH (Hammerich et al., 2011). Furthermore, the cytokine IL-1β can convert human Treg cells into Th17cells (Deknuydt et al., 2009). Treg cells are a key immune cell population that secretes IL-10 to maintain immune homeostasis and tolerance in the liver (An Haack et al., 2015). Several studies have reported that Treg and Th17 cells as well as their secreted IL-10 and IL17 are increased in experimental AIH. More importantly, the Treg/Th17 ratio is significantly reduced in AIH (Eisenstein and Williams, 2009; Kato et al., 2001; Peiseler et al., 2012; Wang et al., 2015a; Zhao et al., 2011). All these reports are consistent with the results of the current investigation, suggesting that altered inflammatory and immune responses occur in AIH. Interestingly, our data show that MSC-exosomes are successfully able to deliver miRNA-223-3p to regulate the expression of IL-1β and IL-6 in the liver and alter the proportions of Treg and Th17 cells in the spleen of mice with hepatic S100/CFA-induced AIH. The reduced ratio of Treg/Th17 was significantly rescued by MSC-exosomes carrying miRNA-223-3p.
Although our data indicate a role of miRNA-223-3p in experimental AIH, there are several limitations in the present study. First, it is possible that the MSC-exosomes can affect other cell types such as endothelial cells in addition to monocytes/macrophages. Moreover, recent investigations indicate that MSC-exosomes injected into mice via the tail vein can be detectable in the liver, the heart, the kidney, the lung and the brain (Gatti et al., 2011; Yu et al., 2015). Thus, further studies are warranted to modify exosomes that can specifically target the liver or other organs. One such modification to dendritic cell-derived exosomes has been reported. By modifying dendritic cell-derived exosomes with adhesive molecules, Alvarez-Erviti et al. (2011) demonstrated that these exosomes can cross the blood-brain barrier and deliver functional siRNA into the mouse brain. Second, because the cellular condition can affect the production and composition of the exosomes (Eldh et al., 2010), the production and composition of exosomes may change with different passages of MSCs. For example, Xin et al. (2012) showed that MSC-exosomes exposed to ischemic tissues contain high levels of miRNAs (miR-133b). Therefore, further studies are important to identify the molecular constituents of exosomes. Third, we did not collect tissues from other organs (such as the brain and heart) to evaluate organ-specific or systemic toxicities of the treatment with MSC-exosomes. Moreover, the study was conducted in the limited time of observation; the long-term adverse effects of treatment remain to be investigated.
In conclusion, our results demonstrated that administration of MSC-exosomes attenuated liver injury in our experimental model of AIH. The attenuation of liver injury was more pronounced with the administration of MSC-exosomes carrying miRNA-223-3p. The mechanism could be related to the miRNA-223-3p-mediated regulation of STAT3 gene and inflammatory cytokines (IL-1β and IL-6) expression, and elevation of the Treg/Th17 ratio. The findings of the current study provide a novel strategy for the development of a therapeutic method for patients with AIH.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the members of the Wenzhou Key Laboratory of Hepatology. The authors also thank the Hepatology Institute of Wenzhou Medical University for technical assistance.
This work was supported by National Natural Science Foundation of China (81570514, 81600466 and 81770585), Science Foundation of Zhejiang Province (LY15H030017 and LQ15H030006); Medical and Health Technology projects of Zhejiang Province (2020KY276); Medical Award Fund, Beijing, China (YJHYXKYJJ-162); Innovation Team of the Early Warning and Intervention to End-stage Liver Disease of Wenzhou (C20150005); and National Science and Technology Major Project, China (2017ZX10202201, 2017ZX10203201, and 2018ZX10725506-001).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ailawadi S., Wang X., Gu H., Fan G.C. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim Biophys Acta. 2015;1852:1–11. doi: 10.1016/j.bbadis.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- An Haack I., Derkow K., Riehn M., Rentinck M.N., Kuhl A.A., Lehnardt S., Schott E. The role of regulatory CD4 T cells in maintaining tolerance in a mouse model of autoimmune hepatitis. PLoS One. 2015;10:e0143715. doi: 10.1371/journal.pone.0143715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A., et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Chen Q., Wang H., Liu Y., Song Y., Lai L., Han Q., Cao X., Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1beta production in macrophages by targeting STAT3. PLoS One. 2012;7:e42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen S., Liu L., Zou Z., Cai Y., Wang J., Chen B., Xu L., Lin Z., Wang X., et al. Mesenchymal stem cells ameliorate experimental autoimmune hepatitis by activation of the programmed death 1 pathway. Immunol Lett. 2014;162:222–228. doi: 10.1016/j.imlet.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Chen Z., Laurence A., O’shea J. Signal transduction pathways and transcriptional regulation in the control of Th17 differentiatio. Semin Immunol. 2007;19:400–408. doi: 10.1016/j.smim.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deknuydt F., Bioley G., Valmori D., Ayyoub M. IL-1beta and IL-2 convert human Treg into T(H)17 cells. Clin Immunol. 2009;131:298–307. doi: 10.1016/j.clim.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Du Z., Wei C., Cheng K., Han B., Yan J., Zhang M., Peng C., Liu Y. Mesenchymal stem cell-conditioned medium reduces liver injury and enhances regeneration in reduced-size rat liver transplantation. J Surg Res. 2013;183:907–915. doi: 10.1016/j.jss.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Eisenstein E.M., Williams C.B. The T(reg)/Th17 cell balance: a new paradigm for autoimmunity. Pediatr Res. 2009;65:26R–31R. doi: 10.1203/PDR.0b013e31819e76c7. [DOI] [PubMed] [Google Scholar]
- Eldh M., Ekstrom K., Valadi H., Sjostrand M., Olsson B., Jernas M., Lotvall J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One. 2010;5:e15353. doi: 10.1371/journal.pone.0015353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti S., Bruno S., Deregibus M.C., Sordi A., Cantaluppi V., Tetta C., Camussi G. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transplant. 2011;26:1474–1483. doi: 10.1093/ndt/gfr015. [DOI] [PubMed] [Google Scholar]
- Hammerich L., Heymann F., Tacke F. Role of IL-17 and Th17 cells in liver diseases. Clin Dev Immunol. 2011;2011:345803. doi: 10.1155/2011/345803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan M.A., Yeoman A.D., Verma S., Smith A.D., Longhi M.S. Autoimmune hepatitis. Lancet. 2013;382:1433–1444. doi: 10.1016/S0140-6736(12)62163-1. [DOI] [PubMed] [Google Scholar]
- Kato M., Ikeda N., Matsushita E., Kaneko S., Kobayashi K. Involvement of IL-10, an anti-inflammatory cytokine in murine liver injury induced by Concanavalin A. Hepatol Res. 2001;20:232–243. doi: 10.1016/S1386-6346(00)00137-6. [DOI] [PubMed] [Google Scholar]
- Kordelas L., Rebmann V., Ludwig A.K., Radtke S., Ruesing J., Doeppner T.R., Epple M., Horn P.A., Beelen D.W., Giebel B. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973. doi: 10.1038/leu.2014.41. [DOI] [PubMed] [Google Scholar]
- Le Blanc K., Mougiakakos D. Multipotent mesenchymal stromal cells and the innate immune system. Nat Rev Immunology. 2012;12:383–396. doi: 10.1038/nri3209. [DOI] [PubMed] [Google Scholar]
- Li M., He Y., Zhou Z., Ramirez T., Gao Y., Gao Y., Ross R.A., Cao H., Cai Y., Xu M., et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47phox-oxidative stress pathway in neutrophils. Gut. 2017;66:705–715. doi: 10.1136/gutjnl-2016-311861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Zhang J., Zhou L., Wang B. Altered function of monocytes/macrophages in patients with autoimmune hepatitis. Mol Med Rep. 2016;13:3874–3880. doi: 10.3892/mmr.2016.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse A., Dienes H.P., Buschenfelde K.H.M.Z. Suppression of murine experimental autoimmune hepatitis by T-cell vaccination or immunosuppression. Hepatology. 1998;27:1536–1543. doi: 10.1002/hep.510270611. [DOI] [PubMed] [Google Scholar]
- Lohse A.W., Manns M., Dienes H.P., Meyer zum Buschenfelde K.H., Cohen I.R. Experimental autoimmune hepatitis: disease induction, time course and T-cell reactivity. Hepatology. 1990;11:24–30. doi: 10.1002/hep.1840110106. [DOI] [PubMed] [Google Scholar]
- Longhi M.S., Ma Y., Mieli-Vergani G., Vergani D. Aetiopathogenesis of autoimmune hepatitis. J Autoimmun. 2010;34:7–14. doi: 10.1016/j.jaut.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Longhi M.S., Mitry R.R., Samyn M., Scalori A., Hussain M.J., Quaglia A., Mieli-Vergani G., Ma Y., Vergani D. Vigorous activation of monocytes in juvenile autoimmune liver disease escapes the control of regulatory T-cells. Hepatology. 2009;50:130–142. doi: 10.1002/hep.22914. [DOI] [PubMed] [Google Scholar]
- Lyu L., Wang H., Li B., Qin Q., Qi L., Nagarkatti M., Nagarkatti P., Janicki J.S., Wang X.L., Cui T. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J Mol Cell Cardiol. 2015;89:268–279. doi: 10.1016/j.yjmcc.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiore G., De Benedetti F., Massa M., Pignatti P., Martini A. Circulating levels of interleukin-6, interleukin-8, and tumor necrosis factor-alpha in children with autoimmune hepatitis. J Pediatr Gastroenterol Nutr. 1995;20:23–27. doi: 10.1097/00005176-199501000-00005. [DOI] [PubMed] [Google Scholar]
- Manns M.P., Czaja A.J., Gorham J.D., Krawitt E.L., Mieli-Vergani G., Vergani D., Vierling J.M. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–2213. doi: 10.1002/hep.23584. [DOI] [PubMed] [Google Scholar]
- Meirelles Lda S., Nardi N.B. Murine marrow-derived mesenchymal stem cell: isolation, in vitro expansion, and characterization. Br J Haematol. 2003;123:702–711. doi: 10.1046/j.1365-2141.2003.04669.x. [DOI] [PubMed] [Google Scholar]
- Peiseler M., Sebode M., Franke B., Wortmann F., Schwinge D., Quaas A., Baron U., Olek S., Wiegard C., Lohse A.W., et al. FOXP3+ regulatory T cells in autoimmune hepatitis are fully functional and not reduced in frequency. J Hepatol. 2012;57:125–132. doi: 10.1016/j.jhep.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadallah S., Eken C., Schifferli J.A. Ectosomes as modulators of inflammation and immunity. Clin Exp Immunol. 2011;163:26–32. doi: 10.1111/j.1365-2249.2010.04271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah V., Montano-Loza A.J., Czaja A.J. Systematic review: managing suboptimal treatment responses in autoimmune hepatitis with conventional and nonstandard drugs. Aliment Pharmacol Ther. 2012;36:691–707. doi: 10.1111/apt.12042. [DOI] [PubMed] [Google Scholar]
- Shigemoto-Kuroda T., Oh J.Y., Kim D.K., Jeong H.J., Park S.Y., Lee H.J., Park J.W., Kim T.W., An S.Y., Prockop D.J., et al. MSC-derived extracellular vesicles attenuate immune responses in two autoimmune murine models: type 1 diabetes and uveoretinitis. Stem Cell Reports. 2017;8:1214–1225. doi: 10.1016/j.stemcr.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taibi F., Metzinger-Le Meuth V., Massy Z.A., Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta. 2014;1842:1001–1009. doi: 10.1016/j.bbadis.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Tan C.Y., Lai R.C., Wong W., Dan Y.Y., Lim S.K., Ho H.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res Ther. 2014;5:76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3.22) doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Wang L., Du H., Liu Y., Wang L., Ma X., Zhang W. Chinese medicine bu xu hua yu recipe for the regulation of treg/th17 ratio imbalance in autoimmune hepatitis. Evid Based Complement Alternat Med. 2015a;2015:461294. doi: 10.1155/2015/461294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gu H., Qin D., Yang L., Huang W., Essandoh K., Wang Y., Caldwell C.C., Peng T., Zingarelli B., et al. Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis. Sci Rep. 2015b;5:13721. doi: 10.1038/srep13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang W., Yang Y., Wang Y., Peng T., Chang J., Caldwell C.C., Zingarelli B., Fan G.C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim Biophys Acta. 2014;1842:701–711. doi: 10.1016/j.bbadis.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zingarelli B., O’Connor M., Zhang P., Adeyemo A., Kranias E.G., Wang Y., Fan G.C. Overexpression of Hsp20 prevents endotoxin-induced myocardial dysfunction and apoptosis via inhibition of NF-kappaB activation. J Mol Cell Cardiol. 2009;47:382–390. doi: 10.1016/j.yjmcc.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Han Z.B., Song Y.P., Han Z.C. Safety of mesenchymal stem cells for clinical application. Stem Cells Int. 2012;2012:652034. doi: 10.1155/2012/652034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Li Y., Buller B., Katakowski M., Zhang Y., Wang X., Shang X., Zhang Z.G., Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–1564. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Li Y., Liu Z., Wang X., Shang X., Cui Y., Zhang Z.G., Chopp M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells. 2013;31:2737–2746. doi: 10.1002/stem.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Kim H.W., Gong M., Wang J., Millard R.W., Wang Y., Ashraf M., Xu M. Exosomes secreted from GATA-4 overexpressing mesenchymal stem cells serve as a reservoir of anti-apoptotic microRNAs for cardioprotection. Int J Cardiol. 2015;182:349–360. doi: 10.1016/j.ijcard.2014.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Tang Y., You Z., Wang Q., Liang S., Han X., Qiu D., Wei J., Liu Y., Shen L., et al. Interleukin-17 contributes to the pathogenesis of autoimmune hepatitis through inducing hepatic interleukin-6 expression. PLoS One. 2011;6:e18909. doi: 10.1371/journal.pone.0018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Zhou H., Su S.B. A critical role for interleukin-1β in the progression of autoimmune diseases. Int Immunopharmacol. 2013;17:658–669. doi: 10.1016/j.intimp.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Zhao X., Shi X., Zhang Z., Ma H., Yuan X., Ding Y. Combined treatment with MSC transplantation and neutrophil depletion ameliorates D-GalN/LPS-induced acute liver failure in rats. Clin Res Hepatol Gastroenterol. 2016;40:730–738. doi: 10.1016/j.clinre.2016.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.