Abstract
The preprotein binding molecular chaperone SecB functions by preventing the premature folding of the preprotein in the cytosol, and targeting it to the peripheral subunit SecA of the translocase at the cytoplasmic membrane. The nature of the interaction of SecB with soluble SecA was studied by fluorescence anisotropy spectroscopy of Ru(bpy)2(dcbpy)-labeled SecA in the presence of increasing concentrations of SecB. A more than 50-fold difference in affinity for the cytosolic SecA compared to translocase associated SecA seems to prevent unproductive binding of SecB to the cytosolic SecA and stresses its targeting function.
Keywords: Anisotropy, Preprotein targeting, SecA, SecB
1. Introduction
In Escherichia coli, preprotein synthesis and translocation across the cytoplasmic membrane are largely uncoupled [1]. The first step in preprotein translocation, therefore, is the targeting of the preprotein to the multi-subunit translocation complex or translocase at the cytoplasmic membrane. Three integral subunits SecY, -E, and -G constitute a high affinity site (KD~10 nM [2,3]) for the dissociable subunit SecA of the translocase (for a review see [4]). The homodimeric [5] SecA (204 kDa) binds the leader sequence and the mature part of the preprotein and couples the hydrolysis of ATP to the translocation of preproteins across the membrane [6]. A subset of preproteins is dependent on the cytosolic chaperone SecB for efficient translocation across the cytoplasmic membrane by the translocase (for a review see [7]). The homotetrameric [8,9] SecB (69 kDa) is a molecular chaperone that binds the mature part of the preprotein [10] and maintains it in a translocation competent state that is lacking the stable conformation characteristic of the mature folded species [11–13]. SecB has a high affinity (KD about 30–40 nM) for the membrane associated SecA [2,3]. In the presence of a preprotein such as proOmpA, the affinity of SecB for membrane bound SecA increases to 10 nM [3]. It has been suggested that a second function of SecB could be the targeting of the preprotein to the membrane associated SecA [2,3,10]. The number of SecB tetramers per cell is estimated to be approximately 1200 [8]. The number of SecA dimers is estimated to be between 1250 and 2500 copies per cell [14]. With 25–50% of the SecA found in the cytosol depending on the amount of integral translocase subunits present and on the growth conditions [15–18], a large number of SecB tetramers could be titrated in the cytosol and therefore be unable to reach the translocase at the cytoplasmic membrane. A preferential binding of SecB to the membrane bound SecA would enhance the efficiency of targeting and prevent unproductive cytosolic binding. However, whether SecB binds the cytosolic SecA with a similar affinity as it binds the membrane associated SecA is still a matter of debate [2,19,20]. In none of these studies has a measure of the affinity for the cytosolic SecA-SecB interaction been reported. Therefore, we investigated the affinity of SecB for SecA in solution using purified components. SecA was labeled with the fluorophore Ru(bpy)2(dcbpy) with a fluorescence lifetime of about 400 ns [21,22]. Since information on the rotational motion is available over a time range extending to about 3 times the fluorescence lifetime, this probe is suitable to study the rotational dynamics of large proteins, such as SecA, and protein complexes with rotational correlation times up to 1.2 μs. SecB appeared to have a dramatically lower affinity for soluble SecA than that reported for the membrane bound SecA. The implications for the targeting function of SecB are discussed.
2. Materials and methods
2.1. Bacterial strains and growth media
Unless indicated otherwise, strains were grown in Luria-Bertani (LB) broth or on LB agar supplemented with 50 μg of ampicillin/ml, 0.5% (w/v) glucose, or 1 mM isopropyl-1-thio-β-D-galactopyranoside, as required. SecYEG overproduction was done in SF100 freshly transformed with pET340 (YEG+) [15], SecA in NO2947 (MC1061 Δlac, recA56 Srl: :Tn10, Amersham International plc, UK) containing pMKL18 [23], SecB in NO2947 containing pAK330 [24], and proOmpA in E. coli strain MM52 [25] transformed with pTAQpOA (a gift of N. Nouwen).
2.2. Materials
SecA [26], SecB [27], and proOmpA [28] were isolated as described. Protein concentration was determined by the Bradford assay [29] and by quantitative amino acid analysis (Eurosequence, Groningen, The Netherlands)
2.3. Ru(bpy)2(dcbpy) labeling of SecA
An NHS ester of Ru bis(2,2′-bipyridine)(2,2′-bipyridine-4,4′-dicarboxylic acid) (Ru(bpy)2(dcbpy) in DMF was synthesized as described[21] and used to covalently label SecA amines. While vortexing, 1 μl Ru-complex (0.23 M) was added to 200 μl SecA (5 mg/ml) in 0.2 M carbonate buffer pH 8.35 followed by a dowsing for 4 h on ice and purification of the labeled protein by gel filtration chromatography on Sephadex G25 (Pharmacia, Uppsala, Sweden), using 50 mM HEPES, 50 mM KCl, pH 7.5 for elution. The activity of the labeled protein was assayed by its in vitro preprotein translocation ATP hydrolyzing capacity.
2.4. SecA translocation ATPase assay
Translocation ATPase activity of SecA using urea treated vesicles [26] was measured as described [30] except that the reactions were supplied with 32 μg/ml SecB.
2.5. Steady state anisotropy measurements
Protein at a final concentration of 0.09 μM (dimer) in 50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM ATP was titrated with SecB. The fluorescence emission between 600 and 700 nm (slit width 8 nm) was monitored in a quartz cuvette equipped with a magnetic stirrer at an excitation wavelength of 460 nm (slit width 4 nm) and a scan rate of 1 nm/s at 25°C using calcite prism polarizers in both the vertical (I‖) and horizontal (I⊥) optical channels of a SLM-Aminco 4800C spectrofluorophotometer (SLM-Aminco, Urbana, IL, USA). Spectra were corrected for the volume increase and background fluorescence. The anisotropy was obtained by measuring the I‖ and I⊥ components of the emission
| (1) |
From the anisotropy (r) the rotational correlation time (θ) and the apparent hydrodynamic volume (V in m3/molecule) of the protein can be calculated,
| (2) |
assuming the lifetime (τ) of the protein bound fluorophore to be 350 ns, the anisotropy in the absence of rotational diffusion (r0) to be 0.26 [22], and the solvent viscosity (η) to be 1.019×10−3 Ns/m2. R is the gas constant and T is the temperature in K.
2.6. Light scattering experiments
Light scattering experiments were performed using a DynaPro-801-TC molecular sizing instrument (Protein Solutions Incorporated, High Wycombe, UK). DLS of SecB (2 mg/ml) in 50 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, and 2 mM nucleotide was measured at 25°C. Ten or more independent measurements were made from each sample and the reported values are calculated arithmetic means. The output from the dynamic light scattering is the translational diffusion coefficient DT (m2/s) of the particles in solution. Under the assumption of Brownian motion and spherical particles, this coefficient is converted to the hydrodynamic radius RH in m, of the particles using the Strokes-Einstein equation:
| (3) |
where kb is Boltzmann’s constant, T is the temperature in Kelvin, and η is the solvent viscosity and assumed to be 1.019×10−3 Ns/m2. A measure of the molecular asymmetry is the ratio RH/RS, where RS is the geometric radius for a sphere, i.e.
| (4) |
is the partial specific volume of the protein, which is 0.74 ml/g for an average protein, Mp is molecular mass of the protein, and NA is Avogadro’s number. The ratio RH/RS may be greater than unity for spherical particles that are solvated, i.e.
| (5) |
where accounts for the volume of the solvation shell (ml/g) [31].
3. Results and discussion
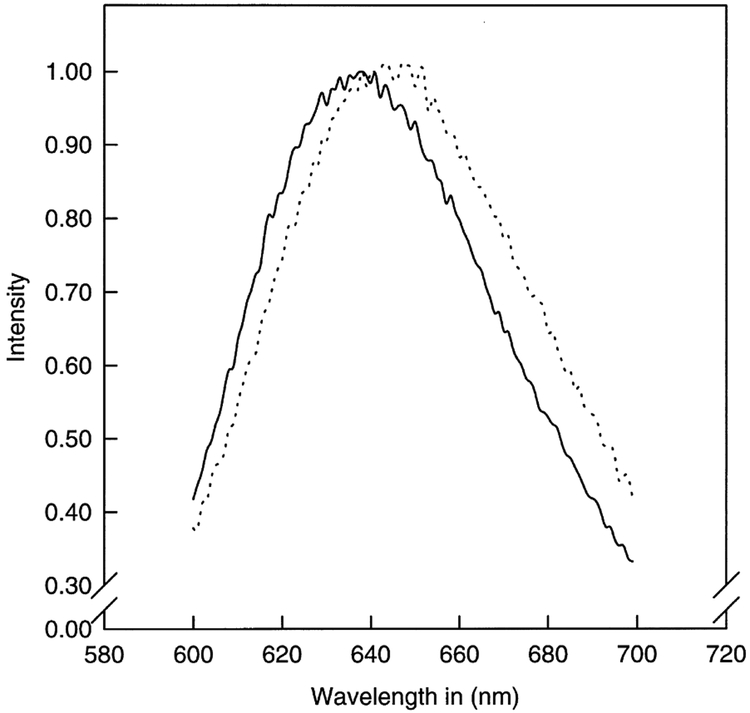
The interaction between SecB and SecA in solution by anisotropy fluorescence spectroscopy was studied using Ru-complex labeled SecA to be able to discriminate between the bound and unbound SecB. The physical basis of these measurements is the polarization or anisotropy of the emitted light when the sample is excited with vertically polarized light. In vitrified solution, i.e. without rotation, part of the polarized light will be emitted as polarized light (r0). In any other solution the molecules will rotate which causes a deviation of the polarization plane of the light or a decrease in the polarization or anisotropy (r). The degree of depolarization depends on the lifetime of the fluorophore (i.e. the time between photon absorption and emission). Information on the rotational motion is available over a time range extending to about 3 times the fluorescence lifetime, after which there is too little signal for accurate anisotropy measurements. Because the lifetimes of typical fluorophores range from 1 to 10 ns, it is difficult to determine the rotational hydrodynamics of larger proteins or membrane bound proteins. The asymmetrical Ru-complex (Ru(bpy)2(dcbpy)) has a high anisotropy in the absence of rotational motions [21]. Upon excitation with 460 nm light, the Ru-complex emits light at 650 nm with a fluorescence lifetime of about 400 ns. The covalent coupling of the Ru-complex to a protein shifts its emission wavelength approximately 10 nm (Fig. 1) to lower energy and its lifetime to 350 ns which enables the measurement of rotational correlation times up to 1.2 μs. This corresponds to the rotation of a spherical complex as big as 4000 kDa.
Fig. 1.
Emission spectrum of E. coli SecA (Ru(bpy)2(dbpy)) and free probe. Dotted line, emission spectrum of SecA (Ru(bpy)2(dbpy)); solid line, emission spectrum of the free probe in 50 mM HEPES pH 7.5, 50 mM KCl. Excitation at 460 nm.
3.1. Anisotropy of Ru-labeled SecA in the absence and presence of nucleotides
Using the labeling conditions as described in Section 2, SecA was moderately labeled and fully active as judged by its preprotein translocation ATPase activity (results not shown). Like other proteins, the Ru-labeled SecA shows a shift of about 10 nm in absorbance spectrum compared to the free Ru-complex (Fig. 1). A rotational correlation time, θ, of Ru-labeled SecA of 50 ns and a hydrodynamic volume of V = 202 nm3/molecule were calculated from the anisotropy using Eq. 2. A theoretical volume V0 = 318 nm3/molecule can be calculated using the partial specific volume of 0.74 ml/g protein [30], a hydration of 0.2 g H2O/g protein [22], and a molecular mass of 204 kDa for the dimer in the equation . Introducing this value into Eq. 2 gives a rotational correlation time θ0 of 73 ns. The fact that the apparent θ is smaller than θ0 indicates that the protein either contains domains with an individual freedom of motion, or that some of the anisotropy is lost due to fast motions of the bound Ru-complexes.
Addition of 5 mM MgCl2 and 2 mM ATP increased the θ and V of the protein by 40% each (Table 1). This results in a θ of 82 ns which is somewhat larger than the theoretical θ0 of 73 ns. An increase in hydrodynamic volume of Bacillus subtilis SecA in the presence of Mg2+ and nucleotide compared to the nucleotide free protein was also observed by dynamic light scattering (DLS) measurements [32]. SecA consists of at least two domains of similar size and the interaction between these domains is stabilized by the presence of nucleotides [32]. It is possible that these domains are connected by a flexible linker region which gives the domains a largely individual rotational freedom of movement. In the absence of nucleotides the protein would be expected to behave as a protein of about half its actual size with a θ0 of about 38 ns. This correlates with the observed apparent θ of 49 ns of SecA (Ru(bpy)2(dcbpy)) in the absence of nucleotides (Table 1). The increase of θ to 82 ns in the presence of nucleotides (Table 1) therefore indicates a decrease in the motional freedom of the SecA domains.
Table 1.
Rotational correlation times of Ru-labeled SecA and its derived apparent molecular volume and radius
3.2. Determination of the affinity of SecB for SecA and the stoichiometry of the complex
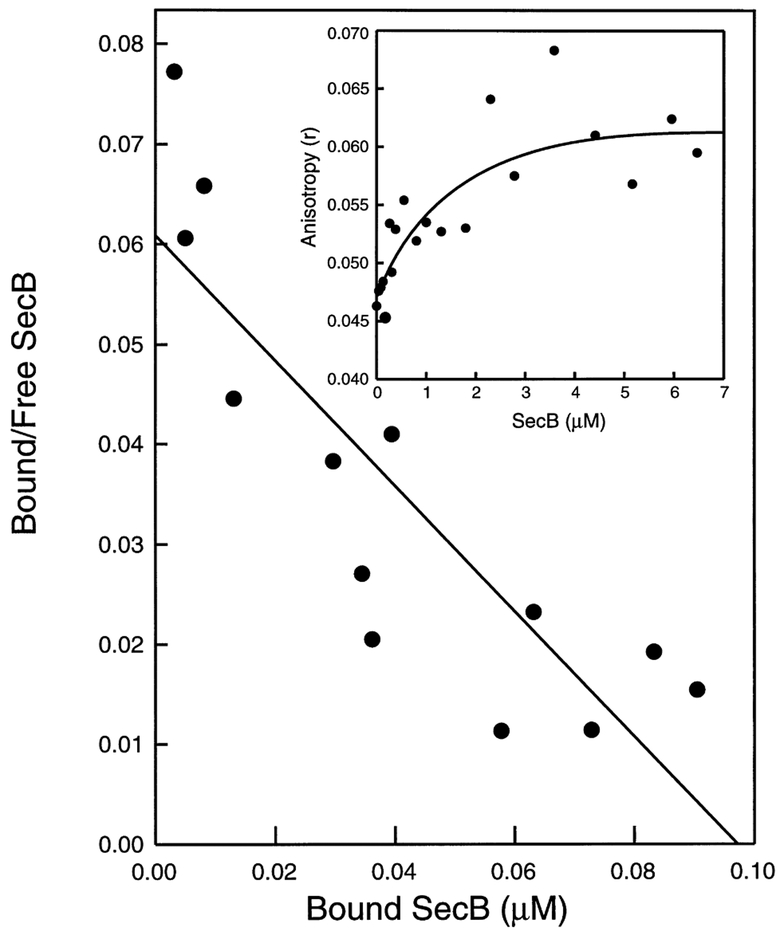
It is assumed that SecA in vivo will be in a nucleotide bound state. Therefore, Ru-labeled SecA was titrated with SecB in the presence of 5 mM MgCl2 and 2 mM ATP (being the most abundant adenosine in the cell). The anisotropy was measured as above at an excitation wavelength of 460 nm and an emission of 620–660 nm. The best fit (S.D. ± 25%) for the Scatchard [33] analysis of the anisotropy data was obtained when a stoichiometry of one SecB tetramer binding site per SecA dimer was assumed (Fig. 2). A similar binding stoichiometry was recently demonstrated for the binding of SecB to the SecYEG bound SecA [3]. However, the determined apparent KD of 1.6 ± 0.5 μM for the binding of SecB to SecA in solution is at least 50 times higher than the reported KD of about 30 nM for the binding of SecB to membrane bound SecA [3]. This huge difference in affinity, in combination with the increased concentration of SecA at the membrane (due to the localization), will specifically direct the cytosolic SecB to the membrane bound SecA. It seems feasible that this might function as a mechanism to enhance the efficiency of preprotein targeting by SecB to the membrane. Membrane associated SecA has a higher affinity (KD 10 nM [3]) for SecB in the presence than in the absence of preproteins. The Ru-labeled SecA complexed with SecB was able to bind proOmpA as indicated by an increase in θ of about 30%. Because stable proOmpA-SecB complexes can only be maintained in the presence of an excess of SecB due to the high association and dissociation rates of these complexes [12], it is not possible to determine the affinity of proOmpA-SecB complexes for soluble SecA by equilibrium anisotropy measurements. Although preprotein-SecB complexes can readily be isolated from cytosolic fractions [10], binary SecA-SecB complex have not yet been isolated in vivo. The ternary complex of preprotein, SecB and SecA has only been isolated in a cell free export system [20]. Therefore, it seems unlikely that the affinity of preprotein-SecB complexes would be higher for the soluble SecA than for the membrane associated SecA.
Fig. 2.
Anisotropy of E. coli SecA (Ru(bpy)2(dbpy)) in the presence of increasing SecB concentrations. Scatchard plot with a KD of 1.6 μM for the SecB tetramer assuming a complete ligandation of the SecA dimer. Inset, fitted line showing the increase in anisotropy plotted against the SecB (as tetramer) concentration in μM.
3.3. Comparison of the size of SecB as determined by DLS and anisotropy spectroscopy
Upon binding of SecB, the apparent hydrodynamic volume of Ru-labeled SecA increased by 40 ± 6%. Based on the molecular mass of 69.2 kDa of SecB an increase of 34% was expected. However, DLS experiments show that SecB behaves as a particle with an apparent molecular mass of 80 kDa in 50 mM Tris-HCl. In the presence of salt and Mg2+ATP, its apparent molecular mass increases to 90 kDa (Table 2). As an unhydrated solid sphere SecB is expected to have a radius of 2.73 nm (Eq. 4), whereas the hydrated radius seems to be 4 nm (Table 2). Introducing both radii into Eq. 5 yields a volume for its hydration shell of 1.68 ml/g protein, which clearly indicates that the protein contains much more water accessible surface than expected for a spherical protein. It can be concluded that shape asymmetry contributes to the RH/RS ratio, and this asymmetry seems to be more pronounced at higher ionic strengths. Similar ionic strength conditions increased the binding of ANS, which has an affinity for hydrophobic amino acid clusters, to SecB [34]. This also indicates that the ionic strength of the solvent influences the conformation of SecB. It could for instance be ellipsoid or spherical with a cavity. Using this molecular mass, an increase in apparent hydrodynamic volume of 40% is to be expected, which correlates well with the data obtained by the anisotropy measurements. A similar increase in θ of Ru-labeled SecA upon binding of SecB in the presence of ATP as in the presence of ADP was observed.
Table 2.
Determination of the apparent molecular weight of E. coli SecBa at 25°C by dynamic light scatter measurementsb
| Buffer 50 mM | DTc (10−13 m2/s) | RHd (nM) | MW (kDa) | Polydispersity (nm) |
|---|---|---|---|---|
| A | 615 (1) | 3.90 (0.00) | 79.5 (0.3) | 0.14 (0.09) |
| B | 579 (2) | 4.07 (0.01) | 90.3 (1.0) | 1.62 (0.01) |
SecB concentrations were 1 and 3 mg/ml, respectively.
The data are based on at least 10 measurements.
Translational diffusion coefficient.
Hydrodynamic radius of gyration. A: 50 mM Tris-HCl pH 8.0; B: 50 mM HEPES KOH pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM ATP.
The standard deviation is indicated in parentheses.
The results presented here provide a measure of the affinity of the SecB-SecA interaction in solution. The affinity of interaction is 50-fold poorer than that of the binding of SecB to membrane associated SecA [3], further underscoring the proposed targeting function of SecB [2]. Moreover, the best fit of the binding data by Scatchard analysis suggests that the binding stoichiometry is one tetrameric SecB per SecA dimer. This value is identical to that reported for the membrane bound SecA [3]. SecB binds to the carboxyl-terminal region of SecA [3,35], and studies with refolded heterodimers of wild-type SecA and SecAN880, a deletion mutant that lacks the highly conserved SecB binding domain, demonstrate that both carboxyl termini of the SecA dimer are needed to form a genuine SecB binding site [3]. Given the low SecB binding affinity of soluble SecA, it thus appears that the SecA conformation is triggered for SecB binding once it has associated with the SecYEG complex at the membrane. The changes in the apparent θ of Ru-labeled SecA in the absence and presence of nucleotides suggest that the two thermodynamically discernable domains of SecA [32] are possibly connected by a flexible linker region.
Acknowledgements:
The expert technical assistance of Tjaard Pijning is gratefully acknowledged. We wish to thank Peter Fekkes for critically reading the manuscript. This work was supported by a pioneer grant of the Netherlands Organization for Scientific Research (N.W.O.).
Abbreviations:
- BSA
bovine serum albumin
- DLS
dynamic light scattering
- DTT
dithiothreitol
- Ru(bpy)2(dcbpy)
Ru bis(2,2′-bipyridine)(2,2′-bipyridine-4,4′-dicarboxylIC acid)
Footnotes
This article is dedicated to the memory of Mies Held.
References
- [1].Randall LL (1993) Cell 33, 231–240. [DOI] [PubMed] [Google Scholar]
- [2].Hartl F-U, Lecker S, Schiebel E, Hendrick JP and Wickner W (1990) Cell 63, 269–279. [DOI] [PubMed] [Google Scholar]
- [3].Fekkes P, van der Does C and Driessen AJM (1997) EMBOJ. (in press). [Google Scholar]
- [4].Driessen AJM (1994) J. Membrane Biol 142, 145–149. [DOI] [PubMed] [Google Scholar]
- [5].Driessen AJM (1993) Biochemistry 32, 13190–13197. [DOI] [PubMed] [Google Scholar]
- [6].Lill R, Dowhan W and Wickner W (1990) Cell 60, 271–280. [DOI] [PubMed] [Google Scholar]
- [7].Kumamoto CA (1991) Mol. Microbiol 5, 19–22. [DOI] [PubMed] [Google Scholar]
- [8].Watanabe M and Blobel G (1989) Proc. Natl. Acad. Sci. USA 86, 1895–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smith VF, Schwartz L, Randall LL and Smith RD (1996) Protein Sci. 5, 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kumamoto CA and Francetić O (1993) J. Bacteriol 175, 2184–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hardy SJS and Randall LL (1991) Science 251, 439–443. [DOI] [PubMed] [Google Scholar]
- [12].Fekkes P, den Blaauwen T and Driessen AJM (1995) Biochemistry 34, 10078–10085. [DOI] [PubMed] [Google Scholar]
- [13].Zahn R, Perrett S and Fersht AR (1996) J. Mol. Biol 261, 43–61. [DOI] [PubMed] [Google Scholar]
- [14].Matsuyama S-I, Fujita Y, Sagara K and Mitzushima S (1992) Biochim. Biophys. Acta 1122, 77–84. [DOI] [PubMed] [Google Scholar]
- [15].Van der Does C, Den Blaauwen T, De Wit JG, Manting EH, Groot NA, Fekkes P and Driessen AJM (1996) Mol. Microbiol 22, 619–629. [DOI] [PubMed] [Google Scholar]
- [16].Chun S-Y and Randall LL (1994) J. Bacteriol 176, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim YJ, Rajapandi T and Oliver DB (1994) Cell 78, 845–853. [DOI] [PubMed] [Google Scholar]
- [18].Cabelli RJ, Dolan KM, Qian L and Oliver DB (1991)J. Biol. Chem 266, 24420–24427. [PubMed] [Google Scholar]
- [19].Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford JB Jr., Kumamoto C and Wickner W (1989) EMBO J. 8, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hoffschulte HK, Drees B and Müller M (1994) J. Biol. Chem 269, 12833–12839. [PubMed] [Google Scholar]
- [21].Terpetschnig E, Szmacinski H, Malak H and Lakowicz JR (1995) Biophys. J 68, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Terpetschnig E, Szbacinski H and Lakowicz JR (1995) Anal. Biochem 227, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klose M, Schimz K-L, Van der Wolk J, Driessen AJM and Freudl R (1993) J. Biol. Chem 268, 4504–4510. [PubMed] [Google Scholar]
- [24].Kumamoto CA and Nault AK (1989) Gene 75, 167–175. [DOI] [PubMed] [Google Scholar]
- [25].Oliver DB and Beckwith J (1981) Cell 25, 765–772. [DOI] [PubMed] [Google Scholar]
- [26].Cunningham K, Lill R, Crooke K, Rice M, Moore K, Wickner W and Oliver DB (1989) EMBO J. 8, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Weiss JB, Ray PH and Bassford PJ Jr. (1988) Proc. Natl. Acad. Sci. USA 85, 8978–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Crooke E, Guthrie B, Lecker S, Lill R and Wickner W (1988) Cell 54, 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bradford MM (1976) Anal. Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- [30].Lill R, Cunningham K, Brundage LA, Ito K, Oliver DB and Wickner W (1989) EMBO J. 8, 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schmitz KS (1990) An Introduction to Dynamic Light Scattering by Macromolecules, Academic Press, Boston, MA. [Google Scholar]
- [32].Den Blaauwen T, Fekkes P, De Wit JG, Kuiper W and Driessen AJM (1996) Biochemistry 35, 11994–12004. [DOI] [PubMed] [Google Scholar]
- [33].Scatchard G (1949) Ann. NY Acad. Sci 51, 660–672. [Google Scholar]
- [34].Randall LL (1992) Science 257, 241–245. [DOI] [PubMed] [Google Scholar]
- [35].Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J and de Kruijff B (1995) J. Biol. Chem 270, 2702–2709. [DOI] [PubMed] [Google Scholar]