Abstract
PURPOSE
Data sharing creates potential cost savings, supports data aggregation, and facilitates reproducibility to ensure quality research; however, data from heterogeneous systems require retrospective harmonization. This is a major hurdle for researchers who seek to leverage existing data. Efforts focused on strategies for data interoperability largely center around the use of standards but ignore the problems of competing standards and the value of existing data. Interoperability remains reliant on retrospective harmonization. Approaches to reduce this burden are needed.
METHODS
The Cancer Imaging Archive (TCIA) is an example of an imaging repository that accepts data from a diversity of sources. It contains medical images from investigators worldwide and substantial nonimage data. Digital Imaging and Communications in Medicine (DICOM) standards enable querying across images, but TCIA does not enforce other standards for describing nonimage supporting data, such as treatment details and patient outcomes. In this study, we used 9 TCIA lung and brain nonimage files containing 659 fields to explore retrospective harmonization for cross-study query and aggregation. It took 329.5 hours, or 2.3 months, extended over 6 months to identify 41 overlapping fields in 3 or more files and transform 31 of them. We used the Genomic Data Commons (GDC) data elements as the target standards for harmonization.
RESULTS
We characterized the issues and have developed recommendations for reducing the burden of retrospective harmonization. Once we harmonized the data, we also developed a Web tool to easily explore harmonized collections.
CONCLUSION
While prospective use of standards can support interoperability, there are issues that complicate this goal. Our work recognizes and reveals retrospective harmonization issues when trying to reuse existing data and recommends national infrastructure to address these issues.
INTRODUCTION
Data sharing in biomedical research creates potential cost savings by reusing data, supports data aggregation, and facilitates reproducibility. As the number of participants per study can vary from as many as 10,000 to as few as 10, data from multiple studies often must be pooled to explore a research question. The ability to combine diverse data types and perform cross-domain analysis can lead to new discoveries in cancer prevention, treatment, and diagnosis, and supports the goals of precision medicine and the Cancer Moonshot. However, most patient data flow from heterogeneous systems using different software, file formats, and data models. Corwin et al1 recently reported that, because of the variability in study and data design, including the variables chosen for the study, variable names and definitions, values assigned and permitted, and value coding, merging data may not be possible and “hypothesis testing across studies may be prohibited.”(p248)
CONTEXT
Key Objective
How can we lessen the burden of retrospective data harmonization to better leverage data for cancer prevention, treatment, and diagnosis, and in support of precision medicine?
Knowledge Generated
We identified 13 retrospective harmonization challenges and potential solutions. We recommend an infrastructure and process to lesson this burden and help researchers transform their data into suitable formats for national repositories to leverage analytic tools.
Relevance
We developed recommendations for reducing the burden of retrospective data harmonization. This can greatly reduce the burden for the scientific community when trying to perform specific disease–cohort comparisons.
The National Cancer Institute (NCI) is expanding efforts to encourage data sharing by collecting multimodal data through data commons initiatives (https://datascience.cancer.gov/data-commons), the first two of which are the Genomic Data Commons (GDC) and the Proteomics Data Commons. An imaging data commons is also being planned, which will implement additional data standardization and cloud-based analysis capabilities for data from The Cancer Imaging Archive (TCIA) and other sources. Still, each commons has its own data model. Data contributors must harmonize their data to a given commons standards for data submission, and since the commons and many other databases currently do not require the use of prospective standards, the result is silos of unharmonized data that require retrospective harmonization, and some fields simply cannot be reused. To address these issues for the commons, NCI is funding a Center for Cancer Data Harmonization to provide assistance with harmonization across commons, vocabularies, and ontologies, and metadata submission.
Most researchers focus on data integration within their own communities or compliance with government or contractual reporting. Some have adopted one or more data models—the Observational Medical Outcomes Partnership,2 the National Patient-Centered Outcomes Research Network,3 NCI SEER,4 and the North American Association of Central Cancer Registries5 —whereas others define custom data models or data sets. Using multiple or competing data models inhibits uniform data collection and requires manual harmonization for analyses.
Radiology research studies contributing to TCIA further elucidate the multimodal data problem. They typically begin with exporting image data from specialized PACSs (picture archive and communications systems) that conform to the Digital Imaging and Communications in Medicine (DICOM) standard. This resolves many image data interoperability challenges; however, inconsistencies in the collection of nonimaging features, such as patient demographics, diagnosis, treatment regimens, or survival metrics, present additional data-sharing barriers. TCIA is growing and currently contains 100 deidentified, highly curated data sets spanning a wide variety of cancer types and imaging modalities. TCIA hosts data from clinical trials; NCI and National Institutes of Health data collection initiatives, such as The Cancer Genome Atlas (TCGA)7; and community data sets in support of government and publication data-sharing mandates. The wide variety of data types and institutions from which data sets originate was a motivator for our study.
As TCIA and its user community have evolved, researchers want to ask broader research questions that require additional supporting data. Consequently, TCIA now requests that submitters include additional data types, such as clinical, genomics, proteomics, pathology, and image analysis data where feasible. However, there is no way to query across data sets to select cohorts that are based on patient characteristics in the supporting data sets. To do so, existing data must be harmonized so that users are not required to use different search strategies for every data set. This need presented an opportunity to study retrospective harmonization while developing a pilot to determine the feasibility of querying across heterogeneous data. We used the TCIA lung and brain image collections of nonimage supporting data files for this project, using the GDC Clinical and Sample data elements as target standards. The resulting harmonized data and query approach were deployed via a software tool to allow researchers to easily explore and interact with the harmonized data.
TCIA contains data from TCGA whose clinical data are stored in the GDC; therefore, the GDC clinical data model was chosen as the standard for harmonization of non-GDC data sets. The GDC Data Dictionary (https://gdc.cancer.gov/about-data/data-dictionary) provides definitions and permitted values, and in many cases references to the Common Data Elements (CDEs) in the NCI Cancer Data Standards Registry and Repository (caDSR).8
After TCIA approves an imaging data set for submission, TCIA staff and curators use customized software to deidentify and transfer the data to its servers, including performing an extensive review of DICOM headers, pixel data, and any supporting nonimage data, taking the burden of complex Health Insurance Portability and Accountability Act compliance off the shoulders of individual investigators, but does not make an attempt to harmonize or transform data into standards.
Standards Dilemma
Use of common standards or a common data dictionary at the outset could simplify or eliminate backend harmonization if adopted, but the diversity of needs across the research community continues to drive the need for retrospective harmonization. Contributing factors include the existence of multiple standards, which vary on the basis of cancer type, such as for tumor staging (Ann Arbor, International Federation of Gynecology and Obstetrics, American Joint Committee on Cancer [AJCC9]), complexity of the standard, cost and licensing (ie, Systemized Nomenclature of Medicine Clinical Terms)10, different versions of a standard (AJCC version 7 v AJCC version 8), difficulty assessing the quality or suitability for purpose, different standards favored by different communities, and different repositories requiring the use of different standards.
Some standards are formally developed by standards development organizations, such as DICOM (https://www.dicomstandard.org/), Health Level 7,9 and the Clinical Data Interchange Standards Consortium.11 However, the use of proprietary or standardized terminology can require extra steps or cost or may be difficult to understand and use because of overlapping terms and a mix of terminology, data elements, and questionnaires, such as Systemized Nomenclature of Medicine Clinical Terms and Logical Observation Identifiers Names and Codes.12 In some cases, no standards match the researcher’s requirements. Prospectively, the existence of data that can be aggregated with a new study to strengthen research findings may influence standard selection—an example of this is the reuse of TCGA CDEs in the GDC Data Dictionary.
At the time of this writing, there are > 230 CDEs in the Clinical and Sample portion of the GDC Data Dictionary, drawn from > 1,400 TCGA CDEs used across and stratified into different cancer types. TCGA has large amounts of data already coded with these standards, and the GDC has > 356,000 files, and two petabytes of data. Two of the nine randomly selected TCIA research data sets were based on TCGA lung adenocarcinoma and lung squamous cell carcinoma data standards, illustrating that some standards are being reused, but even these data sets required harmonization as a result of slight variations in the way data were stored.
Harmonization Challenges
Identifying and manually harmonizing and transforming the 31 fields common across 3 or more of the 9 TCIA files took a total 329.5 hours, or 2.3 months, extended over the course of 6 months. Several factors contributed to the difficulty: fatigue—it is difficult to focus on detailed harmonization for sustained periods of time; field names are short, often 8 characters and/or abbreviated because of database or software restrictions and syntax rules; a lack of data dictionaries that explain the meaning of each field and data value; and in some cases the data owner could not be found due to insufficient or inactive contact information.
Another hurdle is the lack of tools to support the effort. GDC provides downloadable tab separated values templates for data submission that consist only of short headings for each field but no data validation information. While the GDC Data Dictionary is HTML based, it is difficult to search, and without validation tools it is difficult to create harmonized data. Data validation applied only when the data are submitted results in multiple iterations to make it conform to the repository’s standard. As new groups submit data to the GDC, this iterative process leads to manual data dictionary extensions, adding new permitted values, and new data elements.
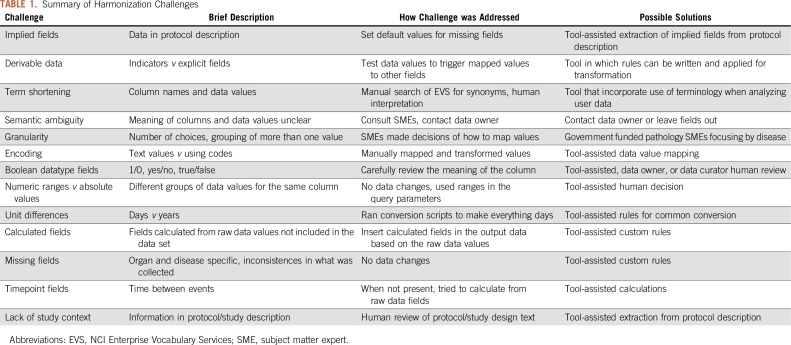
Harmonization issues are summarized in Table 1 along with potential solutions, and additional details are in the Data Supplement. Retrospective harmonization is left up to the harmonizer or repository owner, resulting in inconsistencies in analysis and reporting when reusing the same data. Some of these issues are straightforward to resolve, but they increase the time and effort required to reuse existing data, even when target data elements, such as those in the GDC Data Dictionary or the caDSR, are available to support the manual effort. These factors contribute to complexity and introduce the opportunity for errors in mappings and incorrect scientific conclusions.
TABLE 1.
Summary of Harmonization Challenges
METHODS
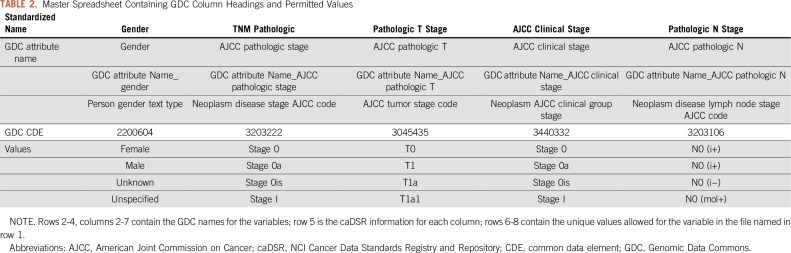
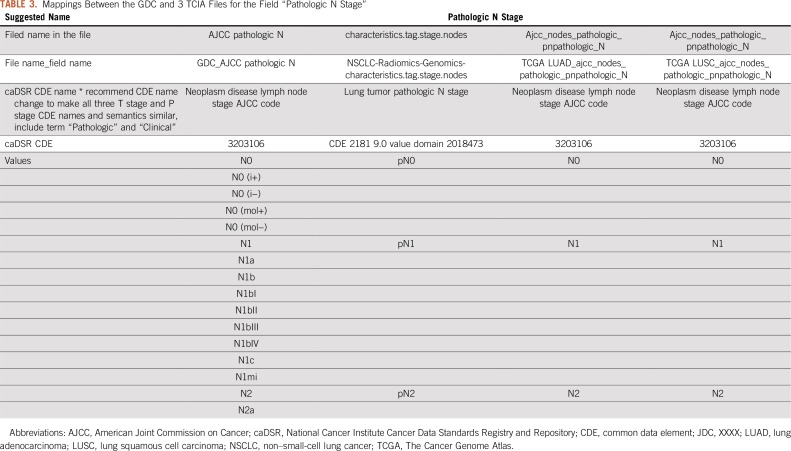
We selected 5 lung and 4 brain cancer data sets from the TCIA data portal with a total of 659 fields, of which 313 were unique. We created a master spreadsheet using the GDC CDE names as columns and one row per permissible value (enumerations) as shown in Table 2. Then, for each TCIA file, we compared the file’s column headings and inspected the unique set of data values to determine the meaning of each field. We added multiple rows for each TCIA file to capture the column heading and a row for each distinct data value. We then created a separate worksheet for each of the GDC CDEs for which there were common TCIA fields, also with one row per permitted value. In each worksheet, we aligned each TCIA file, field, and/or data value to GDC CDE permitted data values. Two files that were based on TCGA studies—lung adenocarcinoma and lung squamous cell carcinoma—included an additional row that contained the NCI caDSR CDE identifier in addition to the column headers, making it easier to determine the meaning of the data in these columns and whether the field matched the GDC CDE (Table 3).
TABLE 2.
Master Spreadsheet Containing GDC Column Headings and Permitted Values
TABLE 3.
Mappings Between the GDC and 3 TCIA Files for the Field “Pathologic N Stage”
Mapping TCIA fields to the GDC CDEs did not always result in one-to-one matches. In some cases, we had to derive the meaning of a field from the protocol description or make a best guess on the basis of the TCIA file’s data value. We attempted to contact the original researcher/data owner to help interpret the data when the meaning was ambiguous, but this was not always successful. Mapping was particularly difficult for histologic type as a result of differences in abbreviations and granularity across studies. We consulted a pathologist to help determine which histologic types were appropriate to group together. Assessing disease progression and disease recurrence was also difficult due to differences in field naming and different timepoint fields across data sets.
We manually aligned and transformed the existing TCIA data with standard GDC permitted values for each field. Finally, we categorized the types of harmonization challenges and developed recommendations and guidelines for better supporting retrospective data harmonization to help address this time-consuming manual effort.
RESULTS
On the basis of our analysis of 9 of 100 data sets available on TCIA, there was only 6% to 15% overlap in terms and data values used by the different submitters, and 76% of those required transformation because the data values for the same field were different due to the lack of a dominant standard. Many researchers still complete their entire study and consider data sharing as an afterthought; thus, support for retrospective harmonization will persist.
Pilot Deployment
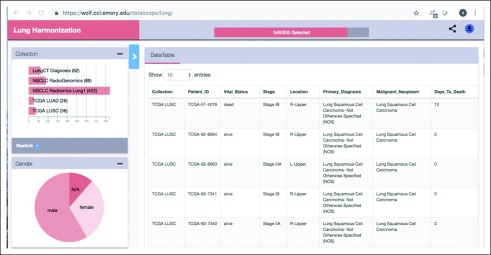
We developed and piloted a tool with which to easily explore and interact with harmonized TCIA data sets. Harmonized and curated data were loaded into MongoDB, an open source, document-oriented, NoSQL database (https://www.mongodb.com). Disease-specific instances of MongoDB were instantiated, thereby allowing a researcher to query across the various TCIA lung or breast cancer collections (Fig 1). A set of REST API end points were deployed using the Bindaas middleware to query and access the data via a pilot portal at Emory University. This REST API was also leveraged to develop a graphic dashboard that allowed users to easily explore and interact with the data set. The combination of a visual and a programmatic interface would help researchers create cohorts that draw data from multiple collections.
FIG 1.
Dashboard of pilot. Users can search across files to create a specialized cohort across data sets that can be leveraged in data exploration using a subset of The Cancer Imaging Archive data sets. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; N/A, not applicable; NSCLC, non–small-cell lung cancer; TCGA, The Cancer Genome Atlas.
The underlying data management and visual analytic components are highly scalable and capable of supporting data explorations through faceted search. However, it is worth noting that as the variety of attributes that can be leveraged in search and data exploration increases, the data harmonization and curation process can be slowed.
DISCUSSION
While the goal of our project was to identify and harmonize data from existing heterogeneous TCIA data collections for the purpose of cross-study query and analysis, we learned in depth about the challenges of retrospective harmonization and documented possible solutions. With the ability to harmonize existing data in the 9 files we analyzed, we expanded the available data to > 659 fields with as many as 1,070 records—the smallest file had 26 records and the largest 187 records—and in some cases, as with TCGA data, the information on the same patient was expanded by linking different files. In theory, reusing existing data—if cost effective—saves the cost of funding new research and can accelerate scientific discovery. Although prospective use of data standards can avoid the need for retrospective harmonization, more can be done to help researchers with existing retrospective data harmonization needs.
The approach to retrospective data harmonization should not rely solely on manual effort. We feel that a national infrastructure should be developed and deployed by government agencies, such as NCI, with tools and resources made publicly and freely available to provide a scalable solution rather than submitting data in its native state for the repository owner to manually harmonize.
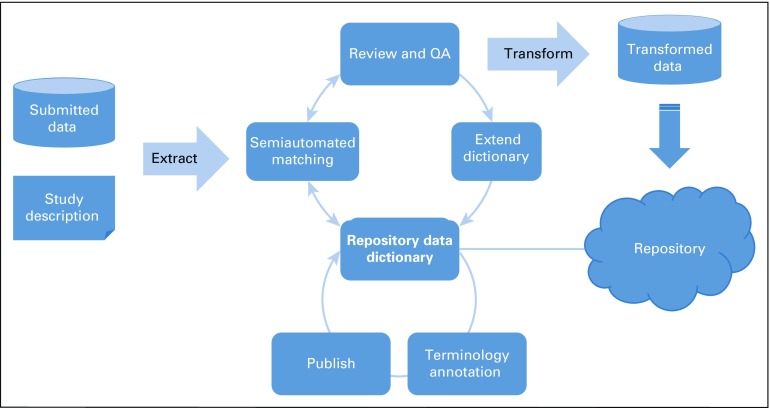
The key components of such an infrastructure are depicted in Figure 2, described in Table 4, and highlighted below.
FIG 2.
National Infrastructure for Data Harmonization consists of a data dictionary with standardized content annotated with standard terminology, published in machine-readable format, leveraged for semiautomated matching of data sets to repository standards, reviewed by subject matter experts, used to transform data into a compatible format, and used to extend the data dictionary standards for new content. QA, quality assurance.
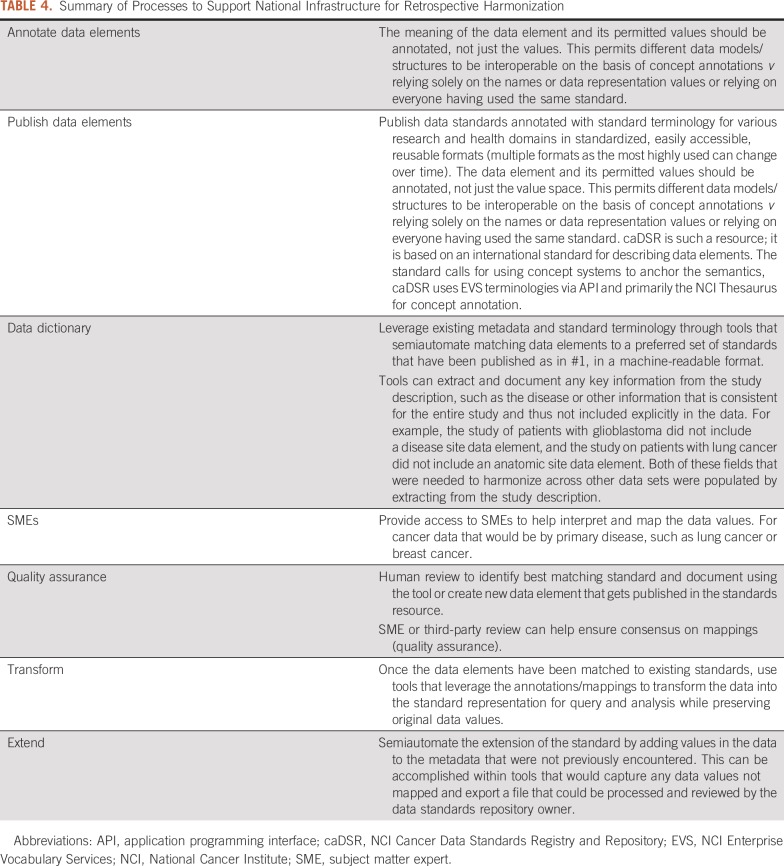
TABLE 4.
Summary of Processes to Support National Infrastructure for Retrospective Harmonization
Data Dictionaries
Access to repositories of data element specifications in a standardized machine-readable format is critical. These dictionaries should adhere to the same FAIR principles as those described for data sharing: Findable, Accessible, Interoperable, Reusable (https://www.force11.org/group/fairgroup/fairprinciples). Details of how data dictionaries can be made FAIR are included in the Data Supplement.
Key points for data dictionaries are:
Use standard terminology to annotate data elements and models
Use a common conceptual model in which fields are described but the answer lists are not, thereby providing a basis for recording the same semantics for a particular field with different coding
Records mappings of the data owners’ data elements to this model for later use by data mapping and transformation tools that can help address some of the types of harmonization issues described in Table 1
Machine readable format
Tools
Tools should be built to leverage existing standards to semiautomate data annotations and to extract new fields from the data set’s description. The data dictionary as above should support these tools and would greatly reduce the burden of retrospective harmonization. Data repository owners should provide access to a tailored transformation tool that leverages the data elements required for the repository as the targets for semiautomated mapping and transformation of user data into valid data for submission.
Access to Subject Matter Experts
In cases in which there is semantic ambiguity, contacting the data owner may be the sole strategy for postharmonization, and if the data owner cannot be reached, sometimes fields have to be left out to ensure data quality. Subject matter experts (SMEs) should assist in determining appropriate mappings between similar terms, such as those that are broader or narrower, and serve as reviewers once the data curator has reviewed and revised the tool-driven semiautomated matching. This expert reviewer can help catch errors and recommend new data elements or extensions to existing data elements.
The research community could leverage the infrastructure by:
Annotating their data elements and models with domain-specific, standard terminology
Publishing this content in standardized data dictionaries in machine-readable format
Using tools that leverage the content for semiautomated annotation mapping and transforming to existing data elements and conceptual models, as well as extension of the data dictionary
Leveraging SMEs to ensure quality and consistency in new content to dynamically extend the dictionaries to fully describe repository data
NCI models several of these infrastructure components. caDSR is a reference repository from which abbreviated data dictionaries can be built, providing well-defined metadata that is semantically based in controlled terminologies. Its content is standardized on the ISO metadata repository standard (ISO/IEC 11179), which calls for the use of standard terminology concepts for tagging the meaning of each data element and any of its permitted data values. All versions of data element specifications are maintained over time. It holds the Biomedical Research Integrated Domain Group Model, an ISO standard conceptual model created through a collaboration between Clinical Data Interchange Standards Consortium, NCI, the US Food and Drug Administration, and Health Level 7, to which national data models, such as Observational Medical Outcomes Partnership and National Patient-Centered Outcomes Research Network, have been harmonized. Content is available in human and machine-readable formats. NCI is testing the reuse of this content in several ways—with a tool that imports caDSR information into a data mapping and transformation tool to aid retrospective data harmonization, and with algorithms to semiautomate matching new data sets to existing caDSR information. NCI has SMEs to support the creation of annotated data specifications and is also funding a center to provide SME support for data commons.
In conclusion, our work demonstrates that data-sharing approaches for retrospective data harmonization and transformation currently require intensive human intervention. It highlights a need for infrastructure and support for the use of shared data standards to better support both retrospective and prospective data harmonization. As part of the TCIA team, we developed a recommendation for the adoption of standards for future TCIA submissions to avoid complex retrospective harmonization, but we have also described harmonization issues and proposed solutions to these challenges.
While our experience reinforces the conclusions of Corwin et al1 that the use of common standards before data submission is the best approach for data sharing, we outline these solutions to streamline and improve retrospective data harmonization.
ACKNOWLEDGMENT
We thank Anthony Kervalage, Edward Helton, and Ulrike Wagner for support and discussions on this study.
Footnotes
Funded in whole or in part with federal funds from the National Cancer Institute under Contracts No. HHSN261200800001E and HHSN316201200117W and Grants No. 5U01CA187013 and 1U24CA215109. A.B. was a White House Presidential Innovation Fellow and detail at the National Cancer Institute for the duration of this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
AUTHOR CONTRIBUTIONS
Conception and design: Amrita Basu, Denise Warzel, Justin S. Kirby, Ashish Sharma
Financial support: Amrita Basu
Administrative support: Amrita Basu, Aras Eftekhari
Provision of study material or patients: Amrita Basu
Collection and assembly of data: Amrita Basu, Denise Warzel, Aras Eftekhari, Justin S. Kirby, John Freymann, Ashish Sharma
Data analysis and interpretation: Amrita Basu, Denise Warzel, Aras Eftekhari, Janice Knable, Ashish Sharma, Paula Jacobs
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Amrita Basu
Employment: Leidos Health
Travel, Accommodations, Expenses: Leidos Health
Justin S. Kirby
Employment: Leidos Biomedical Research
Janice Knable
Employment: Science Application International Corporation
Ashish Sharma
Patents, Royalties, Other Intellectual Property: A pending patent application describing computational staining for detecting tumor-infiltrating lymphocytes (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Corwin EJ, Moore SM, Plotsky A, et al. Feasibility of combining common data elements across studies to test a hypothesis. J Nurs Scholarsh. 2017;49:249–258. doi: 10.1111/jnu.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reisinger SJ, Ryan PB, O’Hara DJ, et al. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. J Am Med Inform Assoc. 2010;17:652–662. doi: 10.1136/jamia.2009.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pletcher MJ, Forrest CB, Carton TW. PCORnet’s collaborative research groups. Patient Relat Outcome Meas. 2018;9:91–95. doi: 10.2147/PROM.S141630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altekruse S.F., Rosenfeld GE, Carrick DM, et al. SEER cancer registry biospecimen research: Yesterday and tomorrow. Cancer Epidemiol Biomarkers Prev. 2014;23:2681–2687. doi: 10.1158/1055-9965.EPI-14-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang L.J., Altekruse S, Adamo M, et al. SEER and NAACCR data completeness methods: How do they impact data quality in central cancer registries? J Registry Manag. 2012;39:185–186. [PubMed] [Google Scholar]
- 6.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [Erratum: Nature 494:506, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warzel DB, Andonaydis C, McCurry B, et al. Common data element (CDE) management and deployment in clinical trials. AMIA Annu Symp Proc. 2003;2003:1048. [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Costa C, Schulz S. HL7 FHIR: Ontological reinterpretation of medication resources. Stud Health Technol Inform. 2017;235:451–455. [PubMed] [Google Scholar]
- 10.Miñarro-Giménez JA, Martínez-Costa C, López-García P, et al. Building SNOMED CT post-coordinated expressions from annotation groups. Stud Health Technol Inform. 2017;235:446–450. [PubMed] [Google Scholar]
- 11.Hume S, Sarnikar S, Becnel L, et al. Visualizing and validating metadata traceability within the CDISC standards. AMIA Jt Summits Transl Sci Proc. 2017;2017:158–165. [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: A 5-year update. Clin Chem. 2003;49:624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 13.Dueck AC, Mendoza TR, Mitchell SA, et al. Validity and reliability of the US National Cancer Institute’s Patient-Reported Outcomes Version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE) JAMA Oncol. 2015;1:1051–1059. doi: 10.1001/jamaoncol.2015.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]