Abstract
Gene fusions frequently contribute to the oncogenic phenotype of many different types of cancer. Additionally, the presence of certain fusions in samples from cancer patients often directly influences diagnosis, prognosis, and/or therapy selection. As a result, the accurate detection of gene fusions has become a critical component of clinical management for many disease types. Until recently, clinical gene fusion detection was predominantly accomplished through the use of single-gene assays. However, the ever-growing list of gene fusions with clinical significance has created a need for assessing fusion status of multiple genes simultaneously. Next generation sequencing (NGS)-based testing has met this demand through the ability to sequence nucleic acid in massively parallel fashion. Multiple NGS-based approaches that employ different strategies for gene target enrichment are now available for use in clinical molecular diagnostics, each with its own strengths and weaknesses. This manuscript describes the use of anchored multiplex PCR (AMP)-based target enrichment and library preparation followed by NGS to assess for gene fusions in clinical solid tumor specimens. AMP is unique among amplicon-based enrichment approaches in that it identifies gene fusions regardless of the identity of the fusion partner. Detailed here are both the wet-bench and data analysis steps that ensure accurate gene fusion detection from clinical samples.
Keywords: gene fusion, precision medicine, molecular diagnostics, next-generation sequencing, anchored multiplex PCR, bioinformatics
SHORT ABSTRACT
This manuscript details the use of an anchored multiplex polymerase chain reaction-based library preparation kit followed by next-generation sequencing to assess for oncogenic gene fusions in clinical solid tumor samples. Both wet-bench and data analysis steps are described.
INTRODUCTION
The fusion of two or more genes into a single transcriptional entity can occur as the result of large scale chromosomal variations including deletions, duplications, insertions, inversions, and translocations. Through altered transcriptional control and/or altered functional properties of the expressed gene product, these fusion genes can confer oncogenic properties to cancer cells.1 In many cases, fusion genes are known to act as primary oncogenic drivers by directly activating cellular proliferation and survival pathways.
The clinical relevance of gene fusions for cancer patients first became apparent with the discovery of the Philadelphia chromosome and the corresponding BCR-ABL1 fusion gene in chronic myelogenous leukemia (CML).2 The small molecule inhibitor imatinib mesylate was developed to specifically target this fusion gene and demonstrated remarkable efficacy in BCR-ABL1-positive CML patients.3 Therapeutic targeting of oncogenic gene fusions has also been successful in solid tumors, with inhibition of ALK and ROS1 fusion genes in non-small cell lung cancer serving as primary examples.4,5 Recently, the NTRK inhibitor larotrectinib was FDA approved for NTRK1/2/3 fusion-positive solid tumors, regardless of disease site.6 Beyond therapy selection, gene fusion detection also has roles in disease diagnosis and prognosis. This is particularly prevalent in various sarcoma and hematologic malignancy subtypes that are diagnostically defined by the presence of specific fusions and/or for which presence of a fusion directly informs prognosis.7–11 These are but a few of the examples of the clinical application of gene fusion detection for cancer patients.
Due to the critical role in clinical decision making, accurate gene fusion detection from clinical samples is of vital importance. Numerous techniques have been applied in clinical laboratories for fusion or chromosomal rearrangement analysis including: cytogenetic techniques, reverse transcription PCR (RT-PCR), fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), and 5’/3’ expression imbalance analysis (among others).12–15 Presently, the rapidly expanding list of actionable gene fusions in cancer has resulted in the need to assess fusion status of multiple genes simultaneously. Consequently, some traditional techniques that can only query one or a few genes at a time are becoming inefficient approaches, especially when considering that clinical tumor samples are often very finite and not amenable to being divided among several assays. Next generation sequencing (NGS), however, is an analysis platform that is well suited for multi-gene testing, and NGS-based assays have become common in clinical molecular diagnostic laboratories.
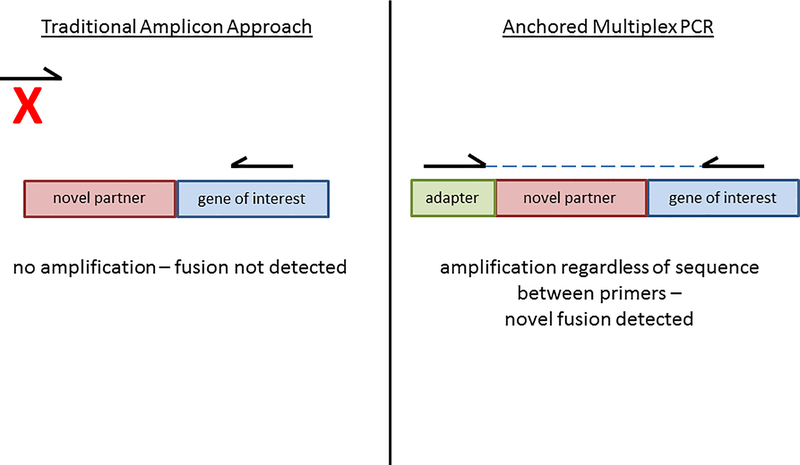
Currently used NGS assays for fusion/rearrangement detection vary in regard to the input material used, the chemistries employed for library preparation and target enrichment, and the number of genes queried within an assay. NGS assays can be based on RNA or DNA (or both) extracted from the sample. Although RNA-based analysis is hampered by the tendency of clinical samples to contain highly degraded RNA, it circumvents the need to sequence large and often repetitive introns that are the targets of DNA-based fusion testing but have proven to be difficult for NGS data analysis.16 Target enrichment strategies for RNA-based NGS assays can be largely divided into hybrid capture or amplicon-mediated approaches. While both strategies have been successfully utilized for fusion detection, each contains advantages over the other.17,18 Hybrid capture assays generally result in more complex libraries and reduced allelic dropout, whereas amplicon-based assays generally require lower input and result in less off-target sequencing 19. However, perhaps the primary limitation of traditional amplicon-based enrichment is the need for primers to all known fusion partners. This is problematic since many clinically important genes are known to fuse with dozens of different partners, and even if primer design allowed for detection of all known partners, novel fusion events would remain undetected. A recently described technique termed anchored multiplex PCR (or AMP for short) addresses this limitation.20 In AMP, a ‘half-functional’ NGS adapter is ligated to cDNA fragments that are derived from input RNA. Target enrichment is achieved by amplification between gene specific primers and a primer to the adapter. As a result, all fusions to genes of interest, even if a novel fusion partner is involved, should be detected (see Figure 1). This manuscript describes the use of the ArcherDx FusionPlex Solid Tumor kit, an NGS-based assay that employs AMP for target enrichment, for the detection of oncogenic gene fusions in solid tumor samples (see Supplementary Table 1 for complete gene list). The wet-bench protocol and data analysis steps have been rigorously validated in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory.
Figure 1 – Schematic representation of AMP approach.
Traditional amplicon-mediated approaches for target enrichment are limited by the fact that primers are needed for all fusion partners. Thus, novel fusion partners will not be detected. In AMP, the opposing primer is specific to the adapter, thus novel partners are detected.
PROTOCOL
1. LIBRARY PREPARATION AND SEQUENCING
1.1. General Assay Considerations and Pre-Assay Steps
1.1.1.
Assay runs typically consistent of seven clinical samples and one positive control (although the number of samples per library preparation run can be adjusted as necessary). Use a positive control that contains at least several gene fusions (that the assay targets) that have been confirmed by a manufacturer and/or have been confirmed by an orthogonal methodology. A non-template control (NTC) must be included as an additional sample in every assay run, but is only carried through second strand cDNA synthesis and Pre-Seq QC (to ensure no cDNA synthesis). Use sample diluent (10mM Tris HCl pH8.0) for the NTC.
1.1.2.
Perform total nucleic acid (TNA) extraction of formalin-fixed paraffin-embedded (FFPE) tissue.
1.1.3.
Quantify the RNA component of the TNA using a fluorometric assay.
1.1.4.
Prevent RNA degradation in the samples by limiting freeze-thaw cycles, keeping thawed nucleic acid at low temperature (chilled block, ice, or alternative), and preventing RNase contamination (wearing gloves, using an RNase decontaminator spray).
1.2. Random Priming
1.2.1.
The desired input for the assay is 200ng RNA (based on fluorometric quantitation of the TNA). Lower inputs can be used if 200ng cannot be achieved. If the sample fails post-sequencing QC, repeating with higher input may result in acceptable QC metrics.
1.2.2.
Dilute TNA in 10mM Tris HCl pH8.0 to achieve desired RNA concentration. For each sample, transfer 20μl of the dilution into the Random Priming reagent strip tubes (placed in pre-chilled aluminum block) and mix by pipetting up down 6–8 times. Briefly spin down and transfer the entire volume to 96 well PCR plate and seal with RT film.
1.2.3.
Insert plate in thermocycler block, cover with compression pad, and close lid. Incubate at 65°C for 5 minutes (heated lid).
1.2.4.
Remove plate from thermocycler and place on ice for 2 minutes.
1.3. First Strand cDNA Synthesis
1.3.1.
Transfer the entire volume of the random priming product into the First Strand reagent strip tubes (placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times. Briefly spin down, transfer entire volume to a 96 well PCR plate and seal with RT film.
1.3.2.
Insert plate in thermocycler block, cover with compression pad, and close lid. Run thermocycler program: 25°C 10min, 42°C 30 min, 80°C 20 min, 4°C hold (heated lid).
1.4. Second Stand cDNA Synthesis
1.4.1.
In a new set of PCR strip tubes create a 1:10 dilution of the first strand product by adding 1μl of first strand product to 9μl nuclease free water. Set the dilution aside to be used in Pre-Seq QC assay.
1.4.2.
Add 21μl nuclease free water to the remaining first strand product. Transfer 40μl of the first strand product and nuclease free water to the Second Strand reagent strip tube (placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times. Spin down, transfer entire volume to a 96 well PCR plate and seal with RT film.
1.4.3.
Insert plate in thermocycler block, cover with compression pad, and close lid. Run thermocycler program: 16°C 60min, 75°C 20 min, 4°C hold (heated lid).
1.4.4.
This is an acceptable stopping point. The plate can be stored at −20°C.
1.5. Pre-Seq QC
1.5.1.
Note that this quality control (QC) assay is used primarily for verification of no cDNA synthesis from the NTC. However, the data may also be informative in assay troubleshooting. For example, if a sample has a good Pre-Seq QC value but poor assay metrics (see below) then it might be an indication of a problem in the assay run, necessitating a repeat.
1.5.2.
Each sample and the NTC are run in duplicate. Also included in the Pre-Seq QC run will be a reaction NTC, which is nuclease free water (also run in duplicate). To each applicable well of an optical 96 well plate add 5μl of the assay master mix, 1μl 10X VCP Primer, and 4μl of the 1:10 dilution of the first strand product that was created in step 1.4.1.
1.5.3.
Seal the plate with RT film, spin down, and load into a qPCR instrument. Run the program: pre-amp: 1 cycle 95°C 20sec, amp: 35 cycles 95°C 3sec (4.4°C/sec ramp rate) - 60°C 30sec (2.2°C/sec ramp rate).
1.5.4.
Confirm lack of a Ct value for both the assay NTC and reaction NTC. If there is no Ct observed in the assay NTC it will no longer be carried forward in the assay. Observation of a Ct value in the reaction NTC necessitates a repeat of the Pre-Seq QC assay. Observation of a Ct in the assay NTC suggests sample contamination at some point prior in the assay, thus requiring that the entire assay (all samples) be started over
1.5.5.
The Pre-Seq QC Ct value for an adequate quality sample should be below 30 (lower Ct values correlate with more product and thus higher quality RNA). Values above 30 are not absolute indicators of failure and these sample should be continued in the assay, however all data derived from such samples should be reviewed carefully, particularly in cases for which no fusion is called.
1.6. End Repair and Bead Purification
1.6.1.
Transfer 40μl of the second strand product into the End Repair reagent strip tube (placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times, spin down and place into thermocycler block (keeping the heated lid open) and run the thermocycler program: 25°C 30min, 4°C hold.
1.6.2.
Remove purification beads from 4°C and allow to equilibrate to room temperature. Make up enough 70% ethanol to last throughout the entire library preparation. 20ml of 70% ethanol will be needed for every 8 samples that are run. Vortex beads well before use and pipet 100μl into the appropriate number of wells of a U-bottom plate.
1.6.3.
Add entire volume of the End Repair product to the beads. Pipet up and down 6–8 times to mix and incubate at room temperature for 5 minutes, followed by a 5-minute incubation on the magnet.
1.6.4.
Remove and discard the supernatant and perform two 200μl 70% ethanol washes with 30 second incubations. After the final wash remove all 70% ethanol and let air dry for 5 minutes.
1.6.5.
Remove from magnet and re-suspend beads in 22μl 10mM Tris HCl pH 8.0. Incubate off the magnet for 3 minutes followed by a 2-minute incubation on the magnet. Proceed immediately to Ligation Step 1.
1.7. Ligation Step 1 and Bead Purification
1.7.1.
Transfer 20μl from the End Repair bead purification plate (taking care not to disturb bead pellet) into the Ligation Step 1 strip tubes (placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times, spin down and transfer the entire volume into a 96 well PCR plate.
1.7.2.
Insert plate in thermocycler block, cover with compression pad and close lid. Run thermocycler program: 37°C 15min, 4°C hold (heated lid).
1.7.3.
Remove purification beads from 4°C and allow to equilibrate to room temperature. Vortex beads well before use and pipet 50μl into the appropriate number of wells of a U-bottom plate.
1.7.4.
Add entire volume of Ligation Step 1 product to the beads. Pipet up and down 6–8 times to mix and incubate at room temperature for 5 minutes, followed by a 5-minute incubation on the magnet.
1.7.5.
Remove and discard the supernatant and perform two 200μl 70% ethanol washes with 30 second incubations. After the final wash remove all 70% ethanol and let air dry for 5 minutes.
1.7.6.
Remove from magnet and re-suspend beads in 42μl 10mM Tris HCl pH 8.0. Incubate off the magnet for 3 minutes followed by a 2-minute incubation on the magnet. Proceed immediately to Ligation Step 2.
1.8. Ligation Step 2 and Bead Purification
1.8.1.
Remove MBC Adapter Strip tube reagents from 4° C storage. Proper numbering of the MBC adapter strip is critically important as sample-specific indexes are added at this point. Position the tubes horizontally with the hinges to the back and use a permanent marker to label the tubes 1, 2, 3… from left to right. It is also critical to record the sample-specific indexes for sequencing purposes (they must be entered in the sequencer worksheet).
1.8.2.
Transfer 40μl from the Ligation Step 1 bead purification plate (taking care not to disturb bead pellet) to the MBC Adapter strip tubes (placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times. Spin down and transfer the entire volume to the Ligations Step 2 reagent strip tubes (also placed in pre-chilled aluminum block). Mix by pipetting up and down 6–8 times, spin down and place in thermocycler block. With the heated lid off run the thermocycler program: 22°C 5min, 4°C hold.
1.8.3.
This is a safe stopping point and samples can be stored at −20°C.
1.8.4.
Remove Ligation Cleanup beads from 4°C storage and allow to equilibrate to room temperature. Prepare 1ml of fresh 5mM NaOH.
1.8.5.
Vortex the Ligation Cleanup beads and add 50μl to a new set of PCR strip tubes. Incubate on the magnet for 1 minute. After the 1 minute incubation remove and discard supernatant. Remove strip tubes from magnet and re-suspend in 50μl of Ligation Cleanup Buffer by pipetting up and down 6–8 times.
1.8.6.
Transfer entire volume of the Ligation Step 2 product into the Ligation Cleanup bead strip tubes. Mix the samples by vortexing and incubate at room temperature for 5 minutes. Again, mix samples by vortexing and incubate at room temperature for another 5 minutes. After the second 5 minute incubation, mix the samples by vortexing, spin down briefly and incubate on the magnet for 1 minute.
1.8.7.
Remove and discard supernatant. Add 200μl of Ligation Cleanup buffer and vortex to re-suspend. Perform a quick spin and place on the magnet for 1 minute. Perform a second wash using the Ligation cleanup buffer again.
1.8.8.
After the two washes with the Ligation cleanup buffer perform an identical wash using ultrapure water. Following the ultrapure water wash re-suspend the beads in 20μl of 5 mM NaOH and transfer to a 96 well PCR plate. Place the plate into thermocycler block with a compression pad and run the thermocycler program: 75°C 10min, 4°C hold (heated lid).
1.8.9.
Spin down the PCR plate once the samples have cooled to 4°C. Place the plate on magnet for at least 3 minutes and proceed immediately to First PCR.
1.9. First PCR and Bead Purification
1.9.1.
Remove the First PCR reagent strip tubes from 4°C Storage and place in a pre-chilled aluminum block. Also, remove the GSP1 primers from −20°C and allow to equilibrate to room temperature.
1.9.2.
Add 2μl of the GSP1 primers to each well of the First PCR reagent strip tubes. Transfer 18μl of the Ligation 2 cleanup product to the First PCR reagent strip tubes and mix by pipetting up and down 6–8 times. Spin down and transfer to 96 well PCR plate. Place plate into thermocycler block with a compression pad and run the thermocycler program: 95°C 3min, 15 cycles 95°C 30sec - 65°C 5 min (100% ramp rate), 72°C 3min, 4°C hold (heated lid).
1.9.3.
This is a safe stopping point and samples can be stored at 4°C overnight or at −20°C for long term storage.
1.9.4.
Remove purification beads from 4°C and allow to equilibrate to room temperature. Vortex beads well before use and pipet 24μl into the appropriate number of wells of a U-bottom plate.
1.9.5.
Transfer 20μl of First PCR product to the beads. Pipet up and down 6–8 times to mix and incubate at room temperature for 5 minutes, followed by a 2-minute incubation on the magnet.
1.9.6.
Remove and discard the supernatant and perform two 200μl 70% ethanol washes with 30-second incubations. After the final wash remove all 70% ethanol and let air dry for 2 minutes.
1.9.7.
Remove from magnet and re-suspend the beads in 24μl 10 mM Tris HCl pH8.0. Incubate off the magnet for 3 minutes followed by a 2-minute incubation on the magnet. Proceed immediately to Second PCR.
1.10. Second PCR and Bead Purification
1.10.1.
Remove the Second PCR reagent strip tubes from 4°C and place in a pre-chilled aluminum block. Proper numbering of the Second PCR strip tubes is critically important as sample-specific indexes are added at this point. Position the tubes horizontally with the hinges to the back and use a permanent marker to label the tubes 1, 2, 3… from left to right. It is also critical to record the sample-specific indexes for sequencing purposes (they must be entered in the sequencer worksheet). Also, remove the GSP2 primers from −20°C and allow to equilibrate to room temperature.
1.10.2.
Add 2μl of the GSP2 primers to each well of the Second PCR reagent strip tubes. Transfer 18μl of the First PCR cleanup product to the Second PCR reagent strip tubes and mix by pipetting up and down 6–8 times. Spin down and transfer to 96 well PCR plate. Place plate into thermocycler block with a compression pad and run the thermocycler program: 95°C 3min, 18 cycles 95°C 30sec - 65°C 5 min (100% ramp rate), 72°C 3min, 4°C hold (heated lid).
1.10.3.
This is a safe stopping point and samples can be stored at 4°C overnight or at −20°C for long term storage.
1.10.4.
Remove purification beads from 4°C and allow to equilibrate to room temperature. Vortex beads well before use and pipet 24μl into the appropriate number of wells of a U-bottom plate.
1.10.5.
Transfer 20μl of Second PCR product to the beads. Pipet up and down 6–8 times to mix and incubate at room temperature for 5 minutes, followed by a 2-minute incubation on the magnet.
1.10.6.
Remove and discard the supernatant and perform two 200μl 70% ethanol washes with 30 second incubations. After the final wash remove all 70% ethanol and let air dry for 2 minutes.
1.10.7.
Remove from magnet and re-suspend the beads in 24μl 10 mM Tris HCl pH8.0. Incubate off the magnet for 3 minutes followed by a 2-minute incubation on the magnet. Transfer 20μl of Second PCR cleanup product to a new 96 well PCR plate.
1.10.8.
This is a safe stopping point and libraries can be stored at −20°C for long term storage or proceed immediately to Library Quantification.
1.11. Library Quantitation
1.11.1.
Remove the library quantitation master mix and standards from −20°C storage and allow to equilibrate to room temperature.
1.11.2.
Perform 1:5 dilution of all libraries (Second PCR product) using 10mM Tris HCL pH8.0 followed by a serial dilution of 1:199, 1:199, and 20:80 using 10mM Tris HCl pH 8.0 0.05% Tween. The final two dilutions of 1:200,000 and 1:1,000,000 respectively will be run in triplicate.
1.11.3.
Add 6μl of master mix to each well of an optical 96-well plate followed by 4μl of appropriate dilution or Standard. Spin down the plate and load it onto the qPCR instrument. Use the following cycling conditions: 1 cycle 95°C 5min, 35 cycles 95°C 30sec (4.4°C/sec ramp rate) - 60°C 45sec (2.2°C/sec ramp rate).
1.11.4.
After library quantification is complete and library concentrations are determined (through averaging both tested dilutions), normalize all libraries to 2nM using 10mM Tris HCl pH8.0. Make the library pool by combining 10μl of each normalized library into one 1.5ml micro centrifuge tube.
1.12. Sequencing of Libraries
1.12.1.
Remove sequencer reagent cartridge from −20°C storage and place in DI water up to the fill line for at least one hour. Also, remove sequencer reagent kit from 4°C and allow to equilibrate to room temperature for at least one hour. The maximum number of libraries that can be sequenced on this sequencing platform is 8 (one of which must be the positive control).
1.12.2.
Make the Denatured Amplicon Library (DAL) pool by combining 10μl of the library pool with 10μl of 0.2N NaOH and incubate for 5 minutes at room temperature. After the 5-minute incubation add 10μl of 200 mM Tris HCl pH 7.0, followed by 970 μl of HT1 buffer.
1.12.3.
Make the final load tube by combining 300μl of HT1, 25μl of 20pM PhiX and 675μl of the DAL pool. Vortex and spin down the load tube. Add entire volume of the load tube to the sample well of the sequencer reagent cartridge and load cartridge into the sequencer running 2×151 bp reads with 2×8 index reads.
2. DATA ANALYSIS
2.1. Sequencing Data Handling and Analysis
2.1.1.
Download sequencing metrics from the NGS instrument.
2.1.2.
Ensure that sequencing data falls into the below ranges (specific for the kit used in this example). Sequencing runs not meeting these metrics are subject to repeat.
cluster Density (Density (K/mm2)): 945K – 1800K
% of reads passing filter (Clusters PF (%)): >90%
quality scores (%>=Q30): >85%
PhiX alignment (Aligned %): 1.5% - 6.0%
PhiX error rate (Error Rate (%)): <1.0%
reads passing filter (Reads PF (M)): >22 million
2.1.3.
Review the % of reads attributed to each sample (i.e. each sample-specific index). For a typical run in which the PhiX spike-in was ~5% and 8 samples were sequenced, each sample should ideally account for approximately 11.9% of the reads. The acceptable range for each sample is 5–25%. If any samples do not fit within this range, repeating library quantitation and sequencing is necessary.
2.2. Submission of raw sequence data to the analysis algorithm (version 4.1.1.7 of ArcherDx Analysis described here)
2.2.1.
In this example, the analysis software is deployed as a ‘virtual machine’ run on an internal server. One CPU and 12GB of RAM are required for each sample to be analyzed concurrently (typically 8 samples are processed concurrently requiring 8 CPU and 96GB RAM). Processing typically takes 3–8hrs.
2.2.2.
Default analysis settings within the analysis system are used, with the exception of MIN_AVERAGE_UNIQUE_RNA_START_SITES_PER_GSP2_CONTROLS (this is changed from 10 to 20) and READ_DEPTH_NORMALIZATION (this is set to 0 to prevent down-sampling).
2.2.3.
To start an analysis job, click on ‘Perform Analysis’ in the user interface, submit a name for the job, and then select the necessary FASTQ files (ensuring both reads (R1 and R2) are selected for each sample).
2.2.4.
Ensure ‘RNA Fusion’ is selected for RNA Analysis Types, ‘Illumina (paired)’ is selected for platform, and ‘FusionPlex Solid Tumor Panel’ is selected for Target Region. Then click on Submit Analysis.
2.3. Assay Data Interpretation
2.3.1.
Open the user interface of the analysis system and select the desired job. Select the positive control sample. Ensure that all expected fusions and oncogenic isoforms have been detected and are listed in the Strong Evidence tab.
2.3.2.
If any of the tracked fusions in the positive control are not detected for a given run of the assay, it may be an indication of a problem in that run, which necessitates a repeat (from beginning of the assay).
2.3.3.
Examine the read statistics of each clinical sample in the run by clicking on the Read Statistics tab. Each sample should be represented by at least 1 million Total Fragments. If less than 1 million, the library must be re-sequenced with considerations for re-quantitation to ensure the initial quantitation was not aberrantly high.
2.3.4.
Good quality samples will generally display on ‘On Target %’ >85%, and the RNA ‘Molecular Bins’ should be >20,000 and comprise >30% of the reads. However, failure to meet these metrics does not automatically trigger sample failure.
2.3.5.
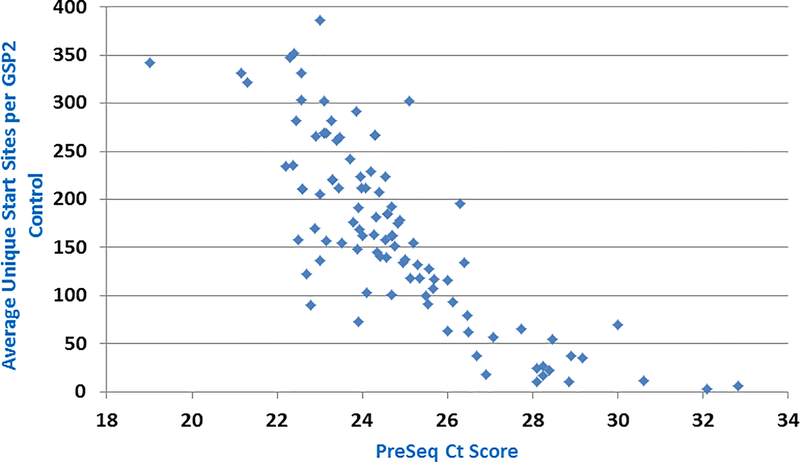
Inspect the ‘Average Unique Start Sites per GSP2 Control’ value. It is a measure of sequencing complexity from primers to four housekeeping genes. This metric is a critical determinant of RNA quality in the sample (if sequencing of genes expected to be expressed in the sample is poor, then confidence in the ability to detect an expressed gene fusion is low). The cutoff for this metric is 20. If any sample displays a value less than 20, then results for the sample must be reported as ‘uninformative’ if no high confidence fusion is found. The status of this metric can also be determined in the summary page of the run (status will be listed as ‘Fusion QC: Pass’ or ‘ Fusion QC: Number of Unique RNA GSP2 Control Start Sites Low’ depending on whether the value is greater than or less than 20). A high confidence fusion in a sample that did not pass this QC metric can still be reported if it is determined to be real (see below). Generally, higher QC scores using this metric correlate with lower PreSeq Ct scores, as expected (Fig 2).
Figure 2 – Correlation between PreSeq QC score and post-sequencing QC.
The PreSeq Ct and Average Unique Start Sites per GSP2 Control values are correlated for a series of 100 samples. Generally, as expected, low PreSeq scores correlate with higher SS/GSP2 values (both are indicators of good quality RNA).
2.4. Fusion Call Interpretation
2.4.1.
Carefully scrutinize every called fusion under the Strong Evidence tab (the majority of Strong Evidence fusions called by the system are artifactual). Also scan the Weak Evidence fusion bin, although the presence of a non-artifactual fusion in this bin is an extremely rare event. To assess validity of a fusion, first note the ‘Reads (#/%)’ and ‘Start Sites’ metrics. Generally, confidence in a called fusion increases the higher these metrics are. Fusions that are supported by less than 5 start sites and by less than 10% of the reads from the supporting primer should be considered as highly suspicious as being artifactual.
2.4.2.
Make note of the icons displayed in the ‘Filters’ column. Several of these icons alert the user of a potential source of an artifactual call. Frequent among these (and primarily found in the Weak Evidence bin of called fusions) are instances where the partners are known paralogs or show similarity to each other in sequence (indicating likely mispriming or erroneous alignment). Another common source of artifactual fusion calls occurs when the reads supporting the fusion contain a high error rate that is indicative of poor mapping to the reference genome, and this is associated with a distinct icon. Transcriptional readthrough events are identified via another icon. These are common and are generally ignored. Several other icons are also used in the user interface, but a full description of all is beyond the scope of this manuscript. Common artifacts that can generally be ignored without further investigation include: ADCK4-NUMBL, SYN2-PPARG, DGKG-ETV5, ETV6-AXL, ETV1-ERG, ETV4-ETV1, FGFR3-PDGFRB, EGFR-NTRK1, RAF1-RET, ALK-TFEB, NOTCH1-RET, RELA-RET, NTRK3-ALK, BRAF-ADAMTS8, and FCGR2C-MAST.
2.4.3.
Visualize the supporting reads for each potential fusion. This can be achieved by clicking on the ‘Visualize’ link in the user interface, which takes the user to a web-based JBrowse view of pileups of individual fusion-supporting reads. Confirm that the reads are generally free of mismatch (although some ‘noise’ is to be expected), that a significant proportion (ideally >30 bases) of the reads align to the fusion partner, and that sequences immediately adjacent to the breakpoint between the genes and the primer binding sites are free of insertions or deletions. Confirm that aligned sequence includes the 3’ portion of the primer binding sequences for primers that support the fusion reads (if 3’ portions are not included, it can be an indication of mis-priming).
2.4.4.
If coding sequences of both genes are involved in the fusion, ensure that the 3’ partner remains in-frame. Click on the ‘Translation’ link in the interface (which will give a ‘True’ or ‘False’ status for each reference sequence combination) and also manually confirm that fusion is in frame through inspection of the called breakpoints in a genome browser. The called breakpoints can be determined by hovering the mouse over the red dots in the fusion schematic.
2.5.5.
Since most (but not all) genomic breakpoints for legitimate fusions occur in intronic regions and result in splicing together of whole exons, fusions in which exon boundaries comprise the breakpoints for both partners are generally viewed with a higher degree of confidence. However, since there are many published examples of mid-exonic breakpoints in fusions, this should not be used as absolute criteria in the interpretation of a fusion.
2.5.6.
For each fusion, manually ensure that sequences adjacent to the breakpoint are not homologous to the next expected contiguous bases for each partner. This will require inspection in a genome browser, making sure to inspect all possible contiguous exons. A common source of artifactual fusion is misalignment or mis-priming due to homology between two gene partners, and this is not always called by the system via the icons described above.
REPRESENTATIVE RESULTS
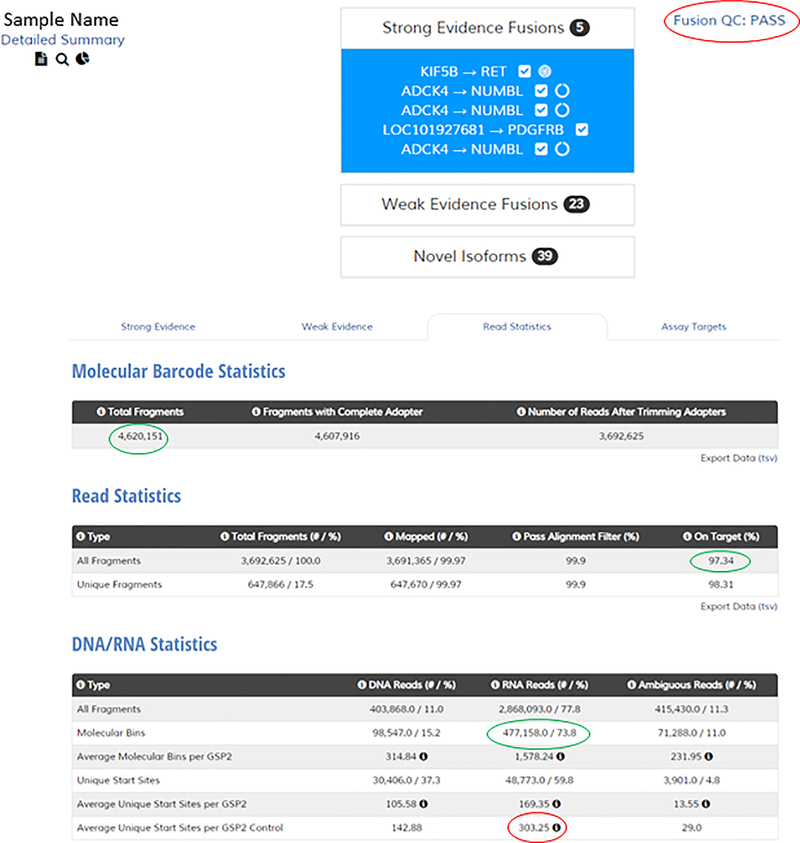
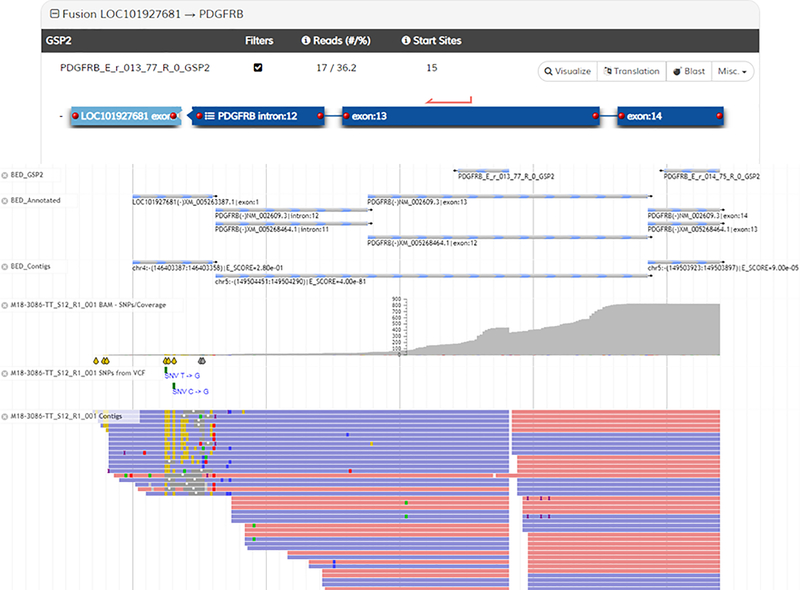
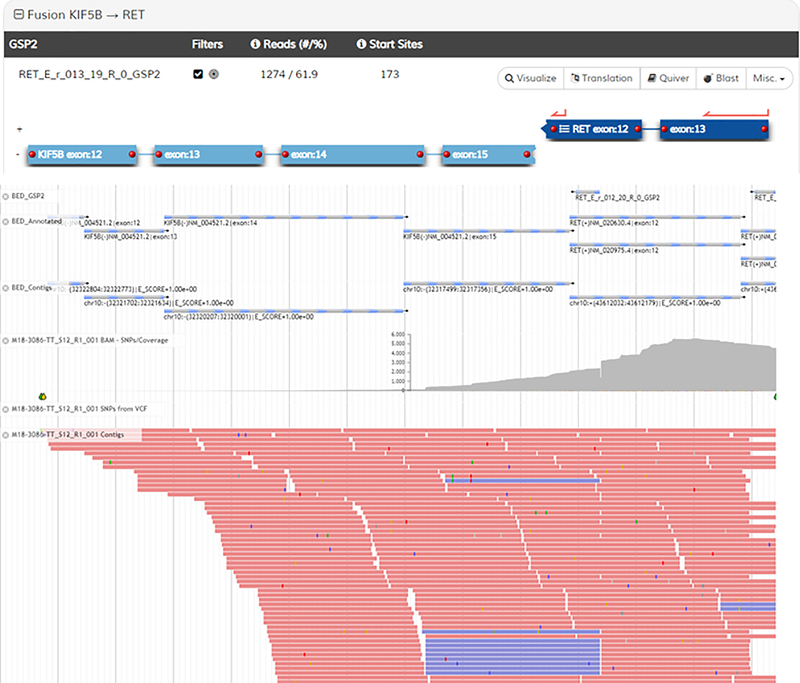
Shown in Representative Results Figures 3–5 are screenshots from the analysis user interface demonstrating results from a lung adenocarcinoma sample. In Figure 3, the sample summary is shown (top) that lists the called Strong Evidence fusions, as well as the QC status (circled in red). The ADCK4-NUMBL fusion (of which 3 isoforms are listed) is immediately ignored because it is a persistent transcriptional readthrough event (noted by the broken circle icons next to the listings). The bottom of Figure 3 is a screenshot of the Read Statistics page. Particularly informative metrics are circled in green. The critical QC metric that determines the pass/fail nature of the sample is highlighted in red (this is the metric that dictates pass/fail status in the summary above). If this value is below 20, a negative fusion result is deemed ‘uninformative’. Figures 4 and 5 are screenshots of the fusion schematics (top) and JBrowse views of fusion supporting reads (bottom) for the two fusions that warranted further investigation. The KIF5B-RET fusion (Fig 4) demonstrates a high number of supporting reads, a high % of reads from the primer supporting the fusion, and a high number of start sites. Additionally, several contiguous exons of the fusion partner (KIF5B) are included in the alignment, the fusion was verified to be in frame, and the reads supporting the fusion are generally free of mismatch. For these reasons, the fusion is deemed real and reportable. The LOC101927681-PDGFRB fusion (Fig 5) demonstrates lower supporting metrics. Furthermore, the portion of the reads mapping to the partner are relatively short and contain a high error rate, which strongly suggests misalignment and an artifactual fusion call. Finally, intronic sequence of PDGFRB is included, further suggesting an artifact (most, but not all, legitimate fusions are comprised solely of sequence that maps to coding regions of both genes). For these reasons, this fusion is deemed an artifact and not reportable.
Figure 3 – Example sample summary and read statistic views.
Top: view of a sample summary in the user interface with strong evidence fusions listed. Bottom: view of the read statistics page in the user interface.
Figure 5.
Figure 4.
DISCUSSION
Anchored multiplex PCR-based target enrichment and library preparation followed by next-generation sequencing is well suited for multiplexed gene fusion assessment in clinical tumor samples. By focusing on RNA input rather than genomic DNA, the need to sequence large and repetitive introns is avoided. Additionally, since this approach amplifies gene fusions regardless of the identity of the fusion partner, novel fusions are detected. This is a critical advantage in the clinical realm, and there have been many examples of actionable novel gene fusions identified through AMP reported in the literature 21–25.
Since the assay is RNA based, it is critical to preserve RNA quality in samples during processing. It is also critical to determine which samples produced RNA sequencing that was too poor to trust negative fusion results. This is achieved by assessment of sequencing data from primers to four housekeeping genes. If sequencing of these genes is poor, then negative fusion results are deemed uninformative. In addition, given the complexity and multitude of wet-bench steps in the assay, it is important to include a fusion-positive control in every assay run. By doing so, compromised assay runs become apparent through analysis of expected fusion events in the control.
As with all amplicon-based approaches, AMP is highly reliant on individual primer performance. When assessing multiple exons of multiple genes, it is inevitable that some primers will not perform as well as others. Therefore, it is critical for users to know where the assay underperforms due to primer inefficiency. Additionally, NGS-based assays require complex bioinformatic data analysis. If the algorithms employed are not thoughtfully designed, false-negative and false-positive results are likley. It is very important that all gene fusions called by analysis algorithms be manually inspected by the user.
With an ever growing list of actionable gene fusions that should be assessed in clinical tumor specimens, use of multiplexed assays like AMP will continue to increase in clinical laboratories. Future applications of the technique will likely focus on combining fusion and mutation assessment within a single assay. Regardless of the molecular assay approach, users must always be aware of assay limitations and should always establish quality control metrics to guide data interpretation.
Supplementary Material
Supplementary Table 1 – List of genes for which gene-specific primers are included in the assay (to various exons and introns)
ACKNOWLEDGEMENTS
This work was supported by the Molecular Pathology Shared Resource of the University of Colorado (National Cancer Institute Cancer Center Support Grant No. P30-CA046934) and by the Colorado Center for Personalized Medicine.
Footnotes
DISCLOSURES
D.L.A. has received consulting fees from Bayer Oncology, Genentech, AbbVie, and Bristol Myers Squibb
K.D.D has received sponsored travel from ArcherDx.
REFERENCES
- 1.Mitelman F, Johansson B & Mertens F The impact of translocations and gene fusions on cancer causation. Nature Reviews Cancer. 7 (4), 233–245, (2007). [DOI] [PubMed] [Google Scholar]
- 2.Daley GQ, Van Etten RA & Baltimore D Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 247 (4944), 824–830, (1990). [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. New England Journal of Medicine. 344 (14), 1038–1042, (2001). [DOI] [PubMed] [Google Scholar]
- 4.Solomon BJ et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. New England Journal of Medicine. 371 (23), 2167–2177, (2014). [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. New England Journal of Medicine. 371 (21), 1963–1971, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drilon A et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. New England Journal of Medicine. 378 (8), 731–739, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai A et al. SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. New England Journal of Medicine. 338 (3), 153–160, (1998). [DOI] [PubMed] [Google Scholar]
- 8.de Alava E et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. Journal of Clinical Oncology. 16 (4), 1248–1255, (1998). [DOI] [PubMed] [Google Scholar]
- 9.Pierron G et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nature Genetics. 44 (4), 461–466, (2012). [DOI] [PubMed] [Google Scholar]
- 10.Papaemmanuil E et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New England Journal of Medicine. 374 (23), 2209–2221, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arber DA et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 127 (20), 2391–2405, (2016). [DOI] [PubMed] [Google Scholar]
- 12.Sanders HR et al. Exon scanning by reverse transcriptase-polymerase chain reaction for detection of known and novel EML4-ALK fusion variants in non-small cell lung cancer. Cancer Genetics. 204 (1), 45–52, (2011). [DOI] [PubMed] [Google Scholar]
- 13.Camidge DR et al. Optimizing the Detection of Lung Cancer Patients Harboring Anaplastic Lymphoma Kinase (ALK) Gene Rearrangements Potentially Suitable for ALK Inhibitor Treatment. Clinical Cancer Research. 16 (22), 5581–5590, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mino-Kenudson M et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clinical Cancer Research. 16 (5), 1561–1571, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suehara Y et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clinical Cancer Research. 18 (24), 6599–6608, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies KD et al. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. Journal of Thoracic Oncology. 13 (10), 1474–1482, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeser JW et al. Validation of a Targeted RNA Sequencing Assay for Kinase Fusion Detection in Solid Tumors. Journal of Molecular Diagnostics. 19 (5), 682–696, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beadling C et al. A Multiplexed Amplicon Approach for Detecting Gene Fusions by Next-Generation Sequencing. Journal of Molecular Diagnostics. 18 (2), 165–175, (2016). [DOI] [PubMed] [Google Scholar]
- 19.Mamanova L et al. Target-enrichment strategies for next-generation sequencing. Nature Methods. 7 (2), 111–118, (2010). [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature Medicine. 20 (12), 1479–1484, (2014). [DOI] [PubMed] [Google Scholar]
- 21.Davies KD et al. Dramatic Response to Crizotinib in a Patient with Lung Cancer Positive for an HLA-DRB1-MET Gene Fusion. JCO Precision Oncology. 2017. (1), (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vendrell JA et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Scientific Reports. 7 (1), 12510, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agaram NP, Zhang L, Cotzia P & Antonescu CR Expanding the Spectrum of Genetic Alterations in Pseudomyogenic Hemangioendothelioma With Recurrent Novel ACTB-FOSB Gene Fusions. American Journal of Surgical Pathology. 42 (12), 1653–1661, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skalova A et al. Molecular Profiling of Mammary Analog Secretory Carcinoma Revealed a Subset of Tumors Harboring a Novel ETV6-RET Translocation: Report of 10 Cases. American Journal of Surgical Pathology. 42 (2), 234–246, (2018). [DOI] [PubMed] [Google Scholar]
- 25.Guseva NV et al. Anchored multiplex PCR for targeted next-generation sequencing reveals recurrent and novel USP6 fusions and upregulation of USP6 expression in aneurysmal bone cyst. Genes Chromosomes and Cancer. 56 (4), 266–277, (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 – List of genes for which gene-specific primers are included in the assay (to various exons and introns)