Abstract
Activated cancer associated fibroblasts (CAFs) play a major role in poor outcome in many diseases including pancreatic cancer. Normally quiescent with high lipid content and low proliferative capacity, CAFs receiving cues from cancer cells in the tumor microenvironment become activated and transformed into a lipid-deprived and highly proliferative myofibroblast type phenotype. Therefore, reversal of activated fibroblasts to quiescence state is an important area of investigation that may help therapeutic management of a number of diseases including pancreatic cancer. Here, we describe a unique biological function of gold nanoparticles (GNPs) and demonstrate that GNP may be used to transform activated CAFs to quiescence and provide insights into the underlying molecular mechanisms. Using immortalized and primary patient derived CAFs, we demonstrate that GNPs enhanced lipid content in the cells by inducing expression of lipogenesis genes such as FASN, SREBP2, FABP3. Using pharmacological inhibitors of lipolysis, lipophagy and fatty acid oxidation we further demonstrate that CAFs utilized GNP-induced endogenously synthesized lipid to maintain the quiescent phenotype. Consequently, treatment with GNP sensitizes CAF to FASN inhibitor or FASN siRNA. Hence, GNPs may be used as a tool to probe mechanisms of quiescence in CAFs and help device strategies to target stromal compartment exploiting the mechanisms of lipid utilization.
Keywords: gold nanoparticle, lipid droplets, quiescent cancer-associated fibroblast, activated cancer-associated fibroblasts, lipid utilization
INTRODUCTION
Fibroblasts play a major role in normal tissue homeostasis (1–2). Under the conditions of tissue injury, inflammation, environmental stress or cues from the cancer cells in the tumor microenvironment quiescent fibroblasts with high lipid content (mainly vitamin A) and low proliferative capacity become activated and transformed into a lipid-deprived highly proliferative myofibroblast type cells (3–7). In pancreatic ductal adenocarcinoma (PDAC) these activated cancer associated fibroblasts (CAFs) make it highly dense, hugely fibrotic with poor vascularity, impermeable to chemotherapeutics and consequently therapy-resistant (8–16). In addition, it is also not clear whether the high lipid content in non-activated fibroblasts plays any key role to keep them into a quiescent state. Therefore, devising strategies to transform activated fibroblasts to quiescent state and understanding molecular mechanisms are important areas of research that may be useful for therapeutic management of a number of diseases including PDAC. However, such studies are rare. Here, we demonstrate how gold nanoparticles may be used to transform activated CAFs to quiescence and provide insight into the molecular mechanisms.
We previously demonstrated self-therapeutic property of 20 nm gold nanoparticles (GNPs) that inhibited tumor growth in two preclinical orthotopic models of ovarian cancer through the inhibition of mitogen-activated protein kinase (MAPK)-activation and reversal of epithelialmesenchymal transition (EMT) via downregulation of a number of heparin binding growth factors (17–18). Furthermore, exploiting self-therapeutic property of GNPs we demonstrated disruption of bidirectional crosstalk between pancreatic cancer cells and pancreatic cancer associated fibroblasts (CAFs) that reprogrammed tumor microenvironment in pancreatic cancer leading to the inhibition of tumor growth in an orthotopic model (19). Here, we demonstrate how GNPs regulate lipid content in CAFs to transform them to quiescence and the underlying molecular mechanisms.
MATERIALS AND METHODS
Materials.
Gold (III) chloride trihydrate (520918), Sodium citrate tribasic trihydrate (S4641), Diethylumbelliferyl phosphate (DEUP) (D7692), 3-Methyladenine (3-MA) (M9281), Etomoxir (E1905), fatty acid synthase siRNA (EHU130711) and all trans ratinoic acid (R2625) were bought from Sigma-Aldrich (St. Louis, MO, USA). C75 (10005270) was purchased from Cayman chemicals (Ann Arbor, Michigan, USA). BODIPY 493/503 (D3922), 1-pyrenedecanoic acid (P31) and lipofectamine RNAiMAX transfection reagent (13778150) were bought from Invitrogen (Waltham, Massachusetts, USA). Cell culture media such as Dulbecco’s modified eagle’s medium (DMEM) (10–014-CV) and DMEM/Ham’s F12 (10–090) were obtained from Corning Inc. (Corning, NY, USA). Fetal Bovine Serum (16000–044) and Penn-Strep (15140–122) were purchased from Life technologies (Grand Island, NY, USA).
Synthesis and characterization of 20 nm GNPs.
20 nm gold nanoparticles (GNPs) were synthesized as previously described in endotoxin-free water from G Biosciences (St. Louis, MO, USA) (17). Briefly, a stock solution of gold (III) chloride trihydrate (10 mM) was diluted 40-fold with water, heated up at 340°C until boiling, added 1% pre-warmed filtered sodium citrate tribasic trihydrate and was then allowed it to boil for 12–15 mins until the color becomes dark purple. Finally, the hot GNPs were removed from the hot plate and stirred it overnight on the cooled plate. The synthesized GNPs were characterized using UV-Visible spectroscopy (Spectrostar Nano, BMG Labtech) and dynamic light scattering (Malvern Zetasizer Nano ZS). The nanoparticles were concentrated by centrifugation before treatments and the gold contents in the preparations were ascertained from the absorbance of the surface plasmon resonance (SPR) band centered at 522 nm and by measuring the content using instrumental neutron activation analysis (INAA).
Cells, cell culture and transfection.
The primary pancreatic tumor-associated fibroblasts (PTAF2 and PTAF3) were isolated from freshly resected pancreatic tissue of patients undergoing surgical resection using outgrowth methods (19, 20) at the University of Oklahoma Health Sciences Center according to the approved IRB protocol, whereas a cell line (CAF19) of activated tumor-associated fibroblast was obtained from Prof. Michael Goggins (Johns Hopkins, Baltimore, USA). These cells were herein collectively termed as activated tumor-associated fibroblasts (CAFs). CAF19 was cultured in DMEM media with a composite of 10% FBS and 1% Penn-Strep. A mixture of DMEM and Ham’s F12 media supplemented with 20%FBS and 1% Penn-Strep was used for the culture of PTAF2 and PTAF3. FASN siRNA transfection was carried out by using lipofectamine RNAiMAX transfection reagent according to the manufacterer’s protocol. In briefly, CAFs were cultured in 6 cm dish containing 1×105 cells. Post 48 h, cells were transfected with scrambled control siRNA (cat. SIC001, Sigma) and FASN siRNA at a dose of 133 nM. The effective silencing (at a protein or mRNA level) was obtained after 48 h of transfection.
Quantification of 20 nm GNPs uptake and its mechanism.
CAFs were cultured in a 10 cm dish at a density of 5×105 cells/dish for 48 h with 5% CO2 in the presence of serum. The cells either SCM or SFM were treated with GNPs (20 μg/ml) at various time points, including 15, 30, 60, 120, 180 and 360 mins, centifuged, colleted cell pelletes after centifugation. The GNPs inside cells were quantified by using flow cytometry (21) and instrumental neutron activation analysis (INAA) (17). To disclose the mechanism of GNPs uptake, cells were pretreated with 3 types of inhibitors, including chloroquine (10 μM) (22), amiloride (125μM) and methy β-cyclodextrin (500 μM) for 2h and were then treated with GNPs (20 μg/ml) for 1h either in SCM or in SFM. The cells were washed, tripsinized, centrifuged and the GNP content in cell pelletes was measured by INAA
Fluorescence microscopy.
CAFs (CAF19, PTAF2 and PTAF3) were prepared in both serum free media (SFM) and serum containing media (SCM) from non-treated (NT) or GNPs-treated as follows: On day 0, 1 × 104 cells/well were grown on cover glass in a 12 well plate in the presence of SCM. After 48 h, on day 2, the media was replaced with either SFM or SCM. On day 3 (24 h interval), the media was again replaced with either fresh SFM or SCM and were treated with 20 ug/ml freshly prepared 20 nm GNPs or remained NT. After 48h treatment, on day 5, media was again replaced with either SFM or SCM containing BODIPY 495/505 at a dilution of 1:1000. After 30 mins incubation, the cells were washed, fixed with 4%PFA, mounted with mounting media and were observed under fluorescence microscope (Carl Zeiss Axioplan, Germany).
For the observation of lipid droplets in CAFs after combined treatments of GNPs and inhibitors, CAFs were prepared in the same way, as described above. On day 3, the media was again replaced with either fresh SFM or SCM and were then treated with GNPs (20 ug/ml), GNPs + DEUP (50 μM), GNPs + 3-MA (5 mM) and GNPs + etomoxir (50 μM) or remained NT. In parallel, the only DEUP, 3-MA and etomoxir were also used as the corresponding controls for the combined treatment groups. Post 48h, cells were incubated with SFM or SCM containing BODIPY 495/505 at a dilution of 1:1000 for 30 mins to stain the neutral lipids. The cells were washed, fixed with 4%PFA, mounted with DAPI containing mounting media and were observed under fluorescence microscope (Carl Zeiss Axioplan, Germany).
For the observation of lipid droplets in CAFs after post-treatment of inhibitors, CAFs were prepared in the same way, as described above. On day 3, cells were treated with GNPs (20 ug/ml) or remained NT for 48h. The media of GNPs treated CAFs was replaced with either DEUP (50 μM), 3-MA (5 mM), etomoxir (50 μM), SFM or SCM and were then incubated for 24h. Post 72h, cells were stained with BODIPY 495/505 at a dilution of 1:1000 for 30 mins and were observed, as described above.
On day 3, CAFs were pretreated with C75 for 2h and were then treated with GNPs for 48h. Moreover, CAFs were also pretreated with FASN siRNA (133 nM) for 48h and were then treated with GNPs for 48h. Neutral lipids and nuclei were stained with BODIPY 495/505 and DAPI respectively and images were captured.
For α-SMA staining, CAFs were prepared, as described above. On day 3, CAFs were treated with GNPs or remained NT for 48h, followed a fixation by 4% PFA and were then incubated with mouse anti-α-SMA antibody (ab7817, abcam, Cambridge, USA) (1:100) for overnight, followed by anti-ms alexa Fluor 546-conjugated secondary antibody. The images were captured under fluorescence microscope.
1 × 104 CAFs per well containing cover glass were plated into a 12 well plate in the presence of SCM. After 48 h, the media was replaced with the SCM and SFM and were then either treated with GNPs (20 ug/ml) or remained NT for 4 and 48 hrs. The old media were further replaced with the fresh corresponding media and were then treated with 1-pyrenedecanoic acid (P31 340/376, 50 μM) for 2-hr. The cells were washed, fixed with 4% PFA and were then observed under fluorescence microscope (Carl Zeiss Axioplan, Germany).
Oil red O staining.
Oil red O staining was performed, as described previously (23). Briefly, CAFs (2×104 cells/well) were seeded on a 6-well plate and were grown for 48 hr in the presence of SCM. The preparation of cells was done, as described above. For the evaluation of lipid content in various CAFs (CAF19, PTAF2 and PTAF3), cells were treated with GNPs or remained NT for 48h. Post 48h, cells were washed with PBS (3), fixed with10% formalin for 10 min at room temperature (RT) and followed by a further incubation with 10% formalin for at least 1 h. Cells were washed with 60% isopropanol for 5 min at RT, rinsed with water (5) and were then dried under chemical hood overnight. The Oil Red O working solution was added onto the dried formalin-fixed cells and were incubated for 10 min at RT. After the incubation, cells were further washed with water (5) and were dried under hood. The Oil Red O dye was eluted by adding 100% isopropanol, along with a gentle shaking for 10 mins. The optical density (OD) of the eluted dye was measured at 500 nm using 100% isopropanol as blank. In a separate experiment, the cell counts for GNPs-treated and non-treated groups after trypsinization and centrifugation were performed using hemocytometer. The data were expressed as OD/104 cells. In addition, the images for oil red o staining were also captured by using Nikon Eclipse Ni microscope.
For the measurement of lipid content in CAFs after combined treatments of GNPs and inhibitors, CAFs were prepared, as described above. On day 3, the media was again replaced with either fresh SFM or SCM and were then treated with GNPs (20 ug/ml), GNPs + DEUP (50 μM), GNPs + 3-MA (5 mM) and GNPs + etomoxir (50 μM) or remained NT. In parallel, the only DEUP, 3-MA and etomoxir were also used as the corresponding controls for the combined treatment groups. Post 48h, the lipid content/104 CAFs were measured, as described above.
For the estimation of lipid content in CAFs after post-treatment of inhibitors, CAFs were prepared, as described above. On day 3, cells were treated with GNPs (20 ug/ml) or remained NT for 48h. The media of GNPs treated CAFs was replaced with either DEUP (50 μM), 3-MA (5 mM), etomoxir (50 μM), SFM or SCM and were then incubated for 24h. Post 72h, the lipid content/104 CAFs were measured, as described above.
Real-time PCR (qRT-PCR).
Total RNA was extracted from NT/GNP-treated CAFs using Quick-RNA Plus (Zymo research, Irvine, CA, USA) according to manufacturers’ protocol. iScript cDNA synthesis kit (Bio- Rad, Hercules, CA, USA) was used to retrotranscribed isolated RNA (1000 ng) and then by following the suppliers protocol for iTaq SYBR Green (Biorad), qRT-PCR was performed. The relative abundance of mRNA from GNPs-treated, compared with that of NT, where 18s is an internal control, was calculated by using the comparative Ct method (24). The details of the primer’s sequences (forward and reverse) are as shown below;
FASN: 5′-CAC AGG GAC AAC CTG GAG TT −3′ 5′- ACT CCA CAG GTG GGA ACA AG-3′
SREBP1a: 5′-CGG CGC TGC TGA CCG ACA TC −3′ 5′- CCC TGC CCC ACT CCC AGC AT-3′
SREBP2: 5′-CAA GCT TCT AAA GGG CAT CG −3′ 5′- AGT AGG GAG AGA AGC CAG CC-3′
PLIN1: 5′-TGG AAA CTG AGG AGA ACA AG-3′ 5′- ATG TCA CAG CCG AGA TGG-3′
PLIN4: 5′-GAG TCA CTG GTG CCG TAA AT −3′ 5′- CCA GTA GTC ACT GCA TCC TTA G-3′
FAB3: 5′-GAA GCT AGT GGA CAG CAA GAA −3′ 5′- GAT TGT GGT AGG CTT GGT CAT-3′
FAB4: 5′- GGA AAG TCA AGA GCA CCA TAA C −3′ 5′- GCA TTC CAC CAC CAG TTT ATC-3′
18S: 5′-TGA CTC AAC ACG GGA AAC C −3′ 5′- TCG CTC CAC CAA CTA AGA AC-3′
Immunoblotting.
Immunoblotting was performed using CAFs lysates containing a 100:1 v/v dilution of RIPA buffer and proteinase inhibitor (Pierce, Appleton, WI, USA). The cell lysate was digested with lamellae buffer containing β-mercaptoethanol at 100°C for 10 mins, followed a separation using a 10% tris-glycine SDS-PAGE gel that was transferred to PVDF membrane. The PVDF membrane was blocked in 5% BSA in TBS for at least 30 mins at room temperature and was then incubated with rab α-SMA (cat. 19245, CST) and rab FASN (cat. 3180, CST) primary monoclonal antibodies in 5% BSA. Primary antibodies was used in dilutions recommended by the manufacturer (1:1000) and secondary antibody was used at a concentration of 1:10,000. GAPDH and α-tubulin were used as loading control to confirm the equity.
Transmission Electron microscopy (TEM).
TEM was done according to the protocol, as described previously (19). Briefly, freshly prepared GNPs were loaded into a 300 Cu mesh formvour carbon coated grids and were then visualized under TEM. For the observation of internalized GNPs into CAFs, cells were treated with GNPs or remained NT for 4h in SCM or SFM. After the incubation, cells were washed with PBS and were collected the cell pellets with a brief trypsinization after centrifugation at 14,000×g for 10 min, followed by the fixation with 4% Paraformaldehyde (EM grade), 2% glutaraldehyde (EM grade), in 0.1M sodium cacodylate buffer overnight at 4°C. According to the previous described protocol, various treatments were done before the preparation of the ultrathin sections. The prepared sections which were stained with lead citrate and uranyl acetate, were viewed under a Hitachi H7600 Transmission Electron Microscope at 80 kV equipped with a 2k × 2k.
Cell viability assay.
1×104 CAFs were plated into a 12 well plate in the presence of SCM. After 48 h, cells were treated with various inhibitors ((DEUP, 3-MA and etomoxir for 24h) and FASN for 6h)) at various doses (DEUP: 6.25, 12.5, 25, 50 and 100 μM; 3-MA: 0.5, 1.0, 2.0, 4.0 and 8.0 mM; etomoxir: 0.25, 0.5, 1.0, 12.5, 25.0, 50.0 and 100 μM; C75: 12.5, 25, 50 and 100 μM; Amiloride (EIPA): 0, 31.25, 62.5, 125, 250 and 500 μM and methyl β-cyclodetrin (MβCD): 0, 0.44, 0.88, 1.75 and 3.5 mM). The cell viability was determined using MTT assay (Sigma, Ann Arbor, Michigan, USA) according to the manufacturer’s protocol. The viability of CAFs was also assessed by using cell counting method. CAFs were pretreated with C75 for 2h, washed with either SCM or SFM and were then treated with GNPs for 48h in the presence of their corresponding media. Next, the cell counts for GNPs, FASN, GNPs + FASN and NT control after trypsinization and centrifugation were performed using hemocytometer. The cell viability was expressed as a percentage (%) of cells that remained live in the SCM or SFM.
Statistical analysis.
All numerical results are reported as mean±SD. The data were presented here from a minimum of three independent experiments unless stated. The statistical significance of the difference between non-treated and treated groups were performed by using unpaired student’s t-test and one way anova followed by multiple comparison test via prism pad software.
RESULTS AND DISCUSSION
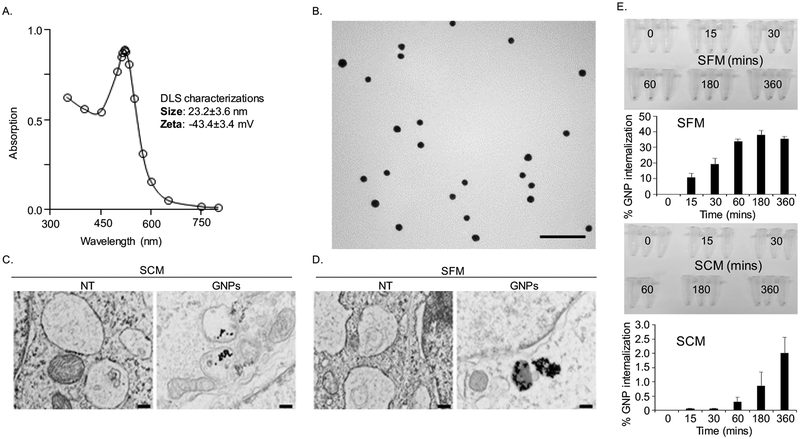
We first synthesized 20 nm GNPs by citrate reduction method and physico-chemically characterized them by UV-Visible Spectroscopy (UV-Vis), Dynamic Light Scattering (DLS), Zeta potential measurements and Transmission Electron Microscopy (TEM). The presence of surface plasmon resonance (SPR) band around 522 nm in the UV-Vis spectrum is indicative of the formation of GNPs (Figure 1A) (25). Hydrodynamic diameter (HD) and surface charge of the GNPs were determined by DLS and zeta potential measurements, respectively. DLS measurements demonstrate that GNPs of HD nearly 23 nm were formed by this method with a net negative charge of ~−43 mV as determined by zeta potential measurements (inset of Figure 1A). The size and shape of GNPs were further confirmed by TEM that demonstrated that spherical GNP of ~ 20 nm was formed by this method (Figure 1B). Since cells generally utilize lipids according to their nutritional demand particularly under nutrient deficient conditions (26) we next wanted to investigate internalization of GNPs in activated fibroblast cell line CAF19 grown both in serum containing media (SCM) mimicking abundance of nutrients and serum free media (SFM) mimicking nutrient deficient environment. TEM micrographs clearly demonstrated that CAF19 cells internalized GNPs in double membrane bound vesicles and multivesicular bodies when treated with GNPs with or without the presence of serum (Figure 1D and Figure S1A), that remains consistent with the previous report (27). To quantitatively measure the internalized GNPs, we first determined time course (0, 15, 30, 60, 120 and 180 mins) of internalization of GNPs into CAFs by using flow cytometry in the presence of serum free media (SFM) and serum containing media (SCM). The results depicted in Figure S1B showed that internalization of GNPs resulted in an increase in the geometric mean intensity of side scattering (SSC) population in a time-dependent manner. It is reported that the intensity of SSC represents the inner density or granularity of each cell, whereas that of forward scattering (FSC) is related to the cellular volume (21). Therefore, it is likely that GNPs inside cells increased the granularity, resulting in a time-dependent increase in the intensity of SSC population. In order to further confirm the results, we measured internalized GNPs by INAA at various time points (0, 15, 30, 60, 180 and 360 mins) after culturing CAFs in SFM and SCM. In SFM, the uptake of GNPs into cells increased in a time-dependent manner. Maximum uptake (~40 % of the treated dose) was observed at 3 h time point, beyond which internalization reached a plateau. However, in case of SCM, the uptake is slow (~ 2 % of treated dose) and continuously increasing with time (Figure 1E). This is likely due to the fact that the presence of abundant nutrients in SCM reduces the uptake of the exogenous materials. However, we do not rule out the possibility of other mechanisms such as the corona formation between GNPs and serum containing proteins. In addition, we quantified amount of internalized GNP by INAA as a readout of pathways involvement. To disclose the mechanism of GNPs uptake, we employed 3 widely used chemical inhibitors such as methyl β-cyclodextrin (MβCD to inhibit caveolae-mediated endocytosis, amiloride (EIPA) to inhibit macropinocytosis and chloroquine to inhibit clathrin-mediated endocytosis (CME). At first, we optimized the concentration of MβCD and EIPA at which most of the cells remained alive (Figure S1C), whereas we used chloroquine (CQ) at a previously reported dose of 10 μM (22). Inhibitors pretreated CAFs were incubated with GNPs at a dose of 20 μg/ml for 1 h in the presence of SCM and SFM and then the internalized GNPs were quantified by INAA. The results depicted in Figure S1D demonstrated that MβCD blocked GNPs uptake around 27% in SFM and 80% in SCM. Similarly, EIPA inhibited gold uptake around 31% in SFM and 80% in SCM. Interestingly, however, CQ (a vesicular disruption by elevating the intracellular pH of endosomes and lysosomes), only marginally inhibited uptake (~8 % of treated dose) of GNP in SFM, while inhibition is ~ 76 % in SCM. The possibility for the predominant inhibition of GNPs uptake in SCM is that GNPs coated with proteins, known as corona is prone to take up by cells through the receptors-mediated endocytosis (28), while large particles are mainly followed up another route of cellular entry (e.g. large particles via macropinocytosis). Collectively, these results suggest that GNPs are internalized into cells by multiple pathways, caveolae-mediated, clathrin-mediated and macropinocytosis in SCM, whereas uptake is mainly through caveolar and micropinocytosis in SFM.
Figure 1.
GNPs characterizations and its cellular uptake. (A) The UV-Visible spectrum of as synthesized GNPs exhibits SPR band around 522 nm. (B) TEM micrographs of as synthesized GNPs showing spherical particles of ~ 20 nm diameter was formed by this method. Scale bar 100 nm. (C-D) Observation of internalized GNPs into CAFs through TEM. Treatment of CAFs with GNPs (20 μg/ml) was performed with or without the presence of serum for 4h. Cells without GNP treatment were used as controls (NT). Cells were then trypsinized, fixed, sectioned and then visualized under TEM. Scale bar 200 nm. (E) Quantification of internalized GNPs into CAFs via instrumental neutron activation analysis (INAA). CAFs were treated with GNPs at various time points, including 15, 30, 60, 180 and 360 mins in the presence or absence of serum and were then collected the cell pelletes by centrifugation with a brief tripsinization. The cells pelletes were captured with a digital camara and were then quantified by INAA.
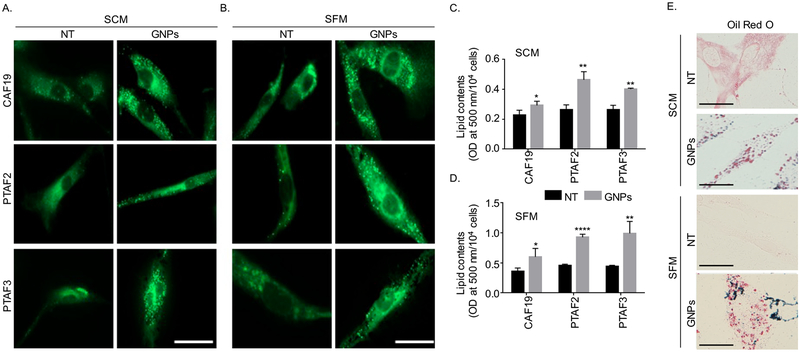
To test whether GNPs can transform activated fibroblasts to quiescent characterized by high lipid content, we next visualized lipid content in immortalized activated CAFs (CA19) and primary patient derived CAFs (PTAF2 and PTAF3) isolated and characterized as we described previously (19, 20). We first treated the cells with GNPs either under serum free (SFM) or serum containing (SCM) condition for 48 h and labeled lipid droplets by a fluorescent dye BODIPY (10) and visualized lipid droplets using fluorescence microscope. Compared to non-treated (NT) control, treatment with GNPs either in SCM or SFM condition significantly increased lipid content in both immortalized and primary CAFs as visualized by BODIPY fluorescence (green) (Figure 2A, B). Interestingly, however, GNPs did not increase lipid content in pancreatic cancer cell line PANC1 under the same conditions (Figure S4). In separate experiments, we quantified lipid content of the cells under the same experimental conditions as above but using Oil Red O staining method as described previously (23). Oil O Red dye was extracted from the cells using 100 % isopropanol and optical density (OD) was measured at 500 nm. As well, the oil red o stained lipid droplets inside cells were visulalized. Compared to NT control, treatment with GNP significantly increased lipid content in CAF19 under both serum containing and serum free media (Figure 2C, D, E and Figure S2, S3), however, the effect was much prominent under serum free condition as expected. Together, these results indicate that GNP has the potential to transform activated CAFs to quiescence through the modulation of lipid contents in the cells. However, mechanisms through which GNP increased lipid content in CAFs were not known that we wanted to investigate next.
Figure 2.
GNP treatment increased neutral lipid contents in activated cancer-associated fibroblasts (CAFs). (A-B). CAFs (CAF19, PTAF2 and PTAF3 were plated at a density of 104 cells/well in a 12 well-plate with cover glass) followed treatment with GNPs (20 ug/ml) or remained non-treated (NT) in SCM or SFM condition. After 48h, neutral lipids were stained with BODIPY (green, λex./em.=493/503 nm) for 30 mins and were visualized under fluorescence microscope. Representative images of cells were shown here from three independent experiments. Scale bar 20 μm. (C-D). Quantification of lipid contents in CAFs cultured in SCM and SFM using Oil Red O. CAFs (2×104 cells/well in 6-well plate) were prepared and were treated with GNPs or remained NT. Post 48h, lipids were stained with Oil Red O dye and that dye was extracted by using 100% isopropanol. In separate experiment, cells were counted by using hemocytometer. The optical density (OD) value of Oil Red O dye was measured at 500 nm and was expressed as OD/104 cells. Statistical significance was determined by student’s unpaired t-test(NT vs GNPs, n=3, mean±s.d., *p<0.05, **p<0.01 and ****p<0.0001). (E) Images after Oil red o staining CAFs were prepared, stained with oil red o dye and were then visualized by Nikon Eclipse Ni microscope. Scale bar 20 μm.
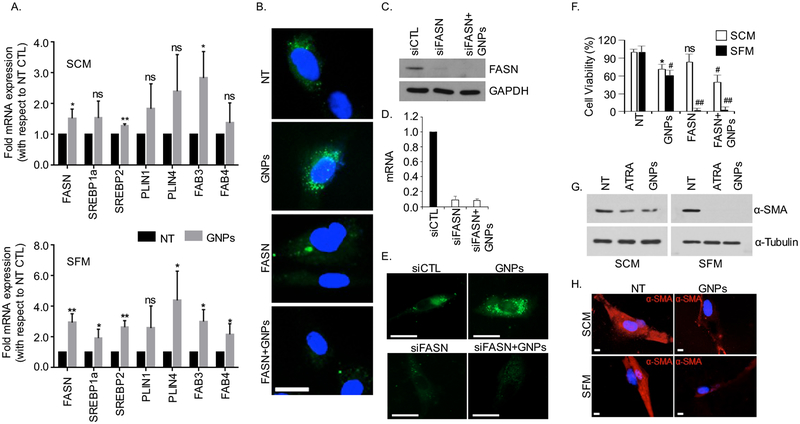
GNPs may enhance lipid content of the cells either by increasing lipid biosynthesis by upregulating lipogenic genes or by enhancing extracellular lipid uptake or interfering with lipid utilization by the cells or may be by regulating all of these three possibilities. Lipids are usually stored in the cells in the form of lipid droplets (LDs) and usually associated with the upregulation of several fatty acid-related genes, including fatty acid binding proteins (FABP3, FABP4), perilipin family (PLIN1, PLIN4), sterol regulatory element binding proteins (e.g. SREBP1, SREBP2) and fatty acid synthatase (FASN) (19, 23). Therefore, we wanted to investigate if GNPs regulated expression of these genes using quantitative real time PCR (qRT-PCR) as one of the mechanisms of enhanced lipid content of the cells by GNP treatment. In the presence of serum, treatment of CAF19 with GNP enhanced mRNA expression of FASN, SREBP2 and FABP3 whereas under serum free conditions GNP significantly increased mRNA levels of all of the lipogenic genes tested when compared with non-treated control. Importantly, mRNA levels of FASN, SREBP2 and FABP3 were significantly enhanced both in SCM and SFM condition upon GNP treatment when compared with non-treated controls (Figure 3A). These findings indicate that enhanced expression of lipogenic genes such as FASN, SREBP2 and FABP3 may be responsible for enhanced lipid content of the cells upon GNP treatment. Since FASN is a key lipogenic enzyme which helps cells to produce long-chain fatty acids that are stored in the in cytoplasm as LDs for cells to utilize under environmental stress (29), we next wanted to investigate if inhibition of FASN activity would impair GNP-induced enhancement of the lipid content in the cells. We first carried out a dose dependent viability assay with CAF19 cells to determine the optimum dose of C75, a pharmacological inhibitor of FASN (30), to be used to interfere with the lipid synthesis but without hampering cell viability (Figure S5). Pre-treatment with C75 or by knocking down FASN using FASN siRNA completely abolished GNP induced increase in lipid content CAF19 cells in SCM, confirming that FASN played a critical role in GNP-induced increase in lipid content in CAFs (Figure 3B–E). Afterwards we tested the effect of the combination treatment on the viability of CAFs. As expected non-toxic of dose of C75 did not alter viability of CAF19 whereas GNP treatment decreased viability by 30–40 % in SCM as well as in SFM condition. However, combination treatment with GNP and C75 further reduced the viability of CAF19 by ~60 %. These results indicate that CAFs utilized lipids to remain viable when treated with GNP by enhancing endogenous lipid synthesis. Therefore, combination treatment with FASN inhibitor and GNP decreased cell viability significantly when compared to GNP treatment or C75 treatment alone. Furthermore, C75 completely abolished viability of CAF19 cells when treated in SFM condition, indicating that cells relied on lipids to survive under starving condition (Figure 3F). We thereupon wanted to investigate whether GNP induced increase in lipid content in CAFs transformed them from activated to quiescent type phenotype. Expression of α-SMA is a marker for activated CAFs that is expected to decrease when transformed into quiescent phenotype (31). Therefore, we probed expression of α-SMA using immunoblot (Figure 3G) and immunofluorescence (Figure 3H) experiments. With concomitant increase in lipid content, treatment with GNP reduced α-SMA expression in CAF19 cells, indicating reversal of activated fibroblast phenotype to quiescence (Figure 3G–H). Collectively, these results support that GNPs enhanced lipid content in activated CAFs by increasing expression of lipogenic genes mainly FASN and transformed activated fibroblast towards quiescence.
Figure 3.
GNPs treatment increased lipogenic gene expression and induced quiescence in CAFs. (A) GNPs upregulate the mRNA levels of lipid metabolism genes in CAFs analyzed by qRT-PCR. CAFs (2×105 cells/10 cm dish) were cultured and were then treated with GNPs for 48 h in SCM and SFM condition separately. mRNA levels of important lipid metabolism genes (FASN, SREBP1a, SREBP2, PLIN1, PLIN4, FABP3 and FABP4) were determined using qRT-PCR, where 18s was used as internal control. Results were expressed as fold mRNA expression, with respect to non-treated controls (NT) and represented as mean±s.d. of three individual experiments (n=3). Statistical analyses were performed using student’s unpaired t-test (NT vs GNPs, ns = non-significant, *p<0.05, and **p<0.01). (B). Inhibition of fatty acid synthatase (FASN) by C75 reduced lipid droplets formation. CAFs were pretreated with C75 at a dose of 50 μM for 2h in the presence of SCM and followed by treatment with GNPs for 48h. The cells were stained with BODIPY (green, λex./em.=493/503 nm), fixed and were visualized under microscope after mounting with DAPI (blue, λex./em.=340/380 nm). Scale bar 20 μm. (C-E) Knock-down of FASN by siRNA reduced the lipid droplets formation. CAFs were transfected with FASN siRNA at a dose of 133 nM. Post 48h, the mRNA and protein expression were determined by RT-qPCR and immunoblotting, where 18s and GAPDH were used controls respectively. Neutral lipids into control and knock-down cells were also visualized by staining BODIPY (green, λex./em.=493/503 nm) under microscope. Scale bar 20 μm (F) CAFs were pretreated with C75 at a dose of 50 μM either in SCM or SFM, post 48h, cells were counted by using hemocytometer. Results were expressed as the viable cells (%) with respect to NT from three individual experiments (n=3) and statistical analyses were performed using one way anova, followed by multiple comparison test (NT vs treated, mean±s.d., ns=non-significant, *p<0.05, #p<0.001 and ##p<0.0001). (G) Western blot for α-smooth muscle actin (α-SMA) showing decreased expression of α-SMA upon 48h GNPs and ATRA treatment, where GAPDH was used as the loading control. (H) Immunofluorescence for α-SMA showing reduced fluorescence (red) upon GNP treatment. Scale bar 5 μm.
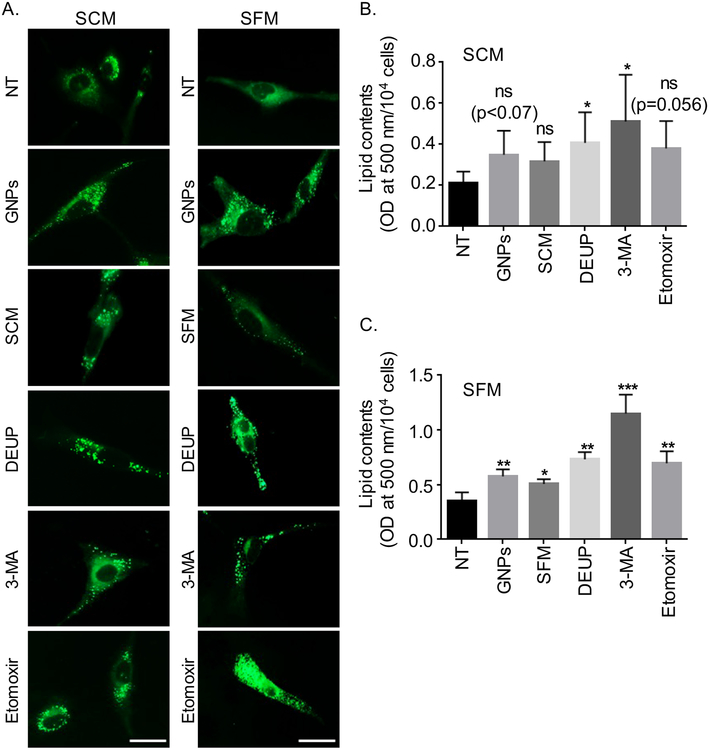
As discussed above treatment of CAF19 cells with FASN inhibitor under serum free condition completely abolished their viability indicating that cells utilized lipids to survive under serum free condition. Therefore, we wanted to investigate whether cells utilized lipid droplets (LDs) induced by GNP treatment to remain viable in quiescent state. LDs first needs to be broken down to free fatty acids (FFAs) in order for cells to utilize them in cellular energy homeostasis (31). There are mainly two major pathways by which cells break down lipids; i) lipolysis: break down of lipids by the action of various lipases; ii) lipophagy: break down of lipids by autophagic pathways (28). Finally FFAs thus formed needs to be transported to the mitochondrion by carnitine palmitoyl transporter 1A (CPT1A) to be used as fuels for fatty acid oxidation (FAO). To discern the effect of lipolysis in LD breakdown we used diethylumbelliferyl phosphate (DEUP, a selective inhibitor for lipase function) (32). We also used 3-methyladenine (3-MA, an inhibitor for blocking autophagosome formation) (33) to decipher a role of lipophagy in LD breakdown and etomoxir, an inhibitor of CPT1A that prevents FFAs transport to the mitochondria and inhibits FAO (34). We first determined toxicity profile of each of the inhibitors on CAF19 cells to select a non-toxic dose of the inhibitors to be used in the study (Figure S5). We next investigated effects of these inhibitors on LD utilization by CAFs. We induced LD formation by GNP treatment under both SCM and SFM condition and then monitored utilization of these LD by CAFs in the presence of different inhibitors. Similar to the observations described above treatment of CAF19 cells with GNP under SCM and SFM condition increased lipid content (Figure 4A). When SCM and SFM were replaced with fresh SCM and SFM respectively, and visualized after 24 h, no significant change in LD fluorescence was observed in SCM, however, a significant drop in LD fluorescence was observed in SFM, indicating that cells utilized LDs that were induced by GNP treatment to remain viable. When cells were treated with DEUP and 3-MA under the same conditions, a significant increase in LD fluorescence was observed under both SCM and SFM condition although the effects were more prominent under SFM condition, as expected. Interestingly, treatment with Etomoxir did not significantly change LD fluorescence in SCM, however, LD fluorescence increased significantly in SFM condition. These results clearly demonstrated that cells partially utilized GNP-induced lipid droplet both under SCM and SFM condition, however, the effect was more prominent under starving condition where cells predominantly rely on lipid store to survive. We next quantified lipid contents of the cells using Oil O Red staining as we described above. The results clearly support the same conclusions drawn from the LD visualization experiments (Figure 4B, C). Next we investigated whether treatment with a mixture of GNP and inhibitors would have similar effect. Combination treatment exhibited similar effect on LDs accumulation and utilization as determined by LD visualization by fluorescence and quantification by Oil O Red staining as described above (Figure S6A–C). We then wanted to investigate whether GNP treatment impacted lipid uptake in CAFs. We used a fluorescence fatty acid analog P31 (1-pyrenedecanoic acid, P31) either in SFM or in SCM as exogenous lipid source to determine the effect of GNP on lipid uptake. Fluorescence images clearly demonstrated that treatment of CAFs with GNP for either 4h or 48 h did not alter the uptake of P31 as compared to non-treated controls both under SCM and SFM condition. In addition, the presence of partial co-localization (yellow) between P31 and LDs with and without GNP treatment clearly demonstrated that GNP did not interfere with lipid uptake and induced endogenous lipogenesis. (Figure S7A, B).
Figure 4.
Utilization of lipid droplets by CAFs. (A) CAFs were treated with GNPs or remained untreated (NT) for 48h. GNPs-treated CAFs were further incubated with SFM, SCM, DEUP (50μM), 3-MA (5 mM), etoximir (50 μM) either in SCM or SFM for 24h or remained untreated after which LDs were visualized by BODIPY fluorescence (green, λex./em.=493/503 nm). LDs were stained with BODIPY for 30 mins, fixed and then visualized under fluorescence microscope. Representative images of cells were shown here from three independent experiments (n=3). Scale bar 20 μm. (B-C). Quantification of lipid contents in CAFs using Oil Red O staining. Experiments were performed as above except staining with Oil O Red instead of BODIPY. The optical density (OD) value of Oil Red O dye was measured at 500 nm and was expressed as OD/104 cells. Statistical significance was determined by student’s unpaired t-test of three independent experiments (mean±s.d., n=3, ns=non-significant, *p<0.05, **p<0.01 and ***p<0.001, NT vs treated groups).
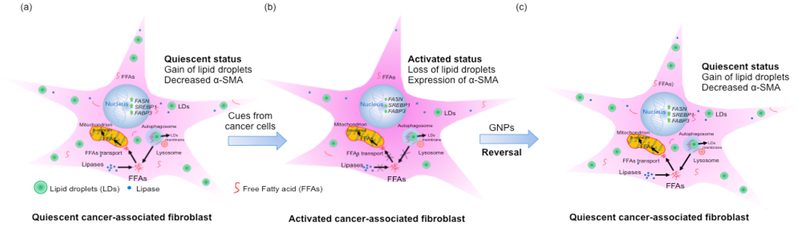
In summary, we demonstrated a unique biological function of gold nanoparticles. Gold nanoparticles reprogram activated cancer associated fibroblast to quiescent by enhancing lipid synthesis and lipid utilization (Figure 5). Since activated fibroblast plays major role in a number of pathological conditions such as fibrosis, cancer, gold nanoparticles may find wide applications in those pathological conditions where reversal of activated fibroblast to quiescence is necessary to improve pathological outcome. In addition, the current study also provides some mechanistic insight on the roles of lipid droplets in maintaining quiescent phenotype through lipid utilization which may be exploited to target stromal compartment to improve poor outcome in diseases.
Figure 5.
Gold nanoparticle transforms activated cancer-associated fibroblast to quiescence by modulating lipid content and utilization.
Supplementary Material
ACKNOWLEDGEMENT:
This work was supported by 2R01CA136494, CA213278, HL120585. We also thank the Peggy and Charles Stephenson Cancer Center at the University of Oklahoma Health Sciences Center for a seed grant and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103639 for the use of Histology and Immunohistochemistry Core, which provided immunohistochemistry and image analysis service
Footnotes
Supporting Information
The data for this research study are available within the article text and the supporting information section containing Figure S1–Figure S7 and their captions, along with a graphical abstract.
The authors declare no conflict of interest.
REFERENCES:
- (1).Dvorak HF, Tumors: Wounds That Do Not Heal. Similarities between Tumor Stroma Generation and Wound Healing. N Engl J Med 1986, 315, 1650–1659. [DOI] [PubMed] [Google Scholar]
- (2).Kalluri R, The Biology and Function of Fibroblasts in Cancer. Nat Rev Cancer 2016, 16, 582–598. [DOI] [PubMed] [Google Scholar]
- (3).Apte MV; Haber PS; Applegate TL; Norton ID; McCaughan GW; Korsten MA; Pirola RC; Wilson JS, Periacinar Stellate Shaped Cells in Rat Pancreas: Identification, Isolation, and Culture. Gut 1998, 43, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Phillips PA; McCarroll JA; Park S; Wu MJ; Pirola R; Korsten M; Wilson JS; Apte MV, Rat Pancreatic Stellate Cells Secrete Matrix Metalloproteinases: Implications for Extracellular Matrix Turnover. Gut 2003, 52, 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Apte MV; Pirola RC; Wilson JS, Pancreatic Stellate Cells: A Starring Role in Normal and Diseased Pancreas. Front Physiol 2012, 3, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Masamune A; Shimosegawa T, Signal Transduction in Pancreatic Stellate Cells. J Gastroenterol 2009, 44, 249–260. [DOI] [PubMed] [Google Scholar]
- (7).Wehr AY; Furth EE; Sangar V; Blair IA; Yu KH, Analysis of the Human Pancreatic Stellate Cell Secreted Proteome. Pancreas 2011, 40, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mahadevan D; Von Hoff DD, Tumor-stroma Interactions in Pancreatic Ductal Adenocarcinoma. Mol Cancer Ther 2007, 6, 1186–1197. [DOI] [PubMed] [Google Scholar]
- (9).Ijichi H; Chytil A; Gorska AE; Aakre ME; Bierie B; Tada M; Mohri D; Miyabayashi K; Asaoka Y; Maeda S; Ikenoue T; Tateishi K; Wright CV; Koike K; Omata M; Moses HL, Inhibiting Cxcr2 Disrupts Tumor-stromal Interactions and Improves Survival in a Mouse Model of Pancreatic Ductal Adenocarcinoma. J Clin Invest 2011, 121, 4106–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sherman MH; Yu RT; Engle DD; Ding N; Atkins AR; Tiriac H; Collisson EA; Connor F; Van Dyke T; Kozlov S; Martin P; Tseng TW; Dawson DW; Donahue TR; Masamune A; Shimosegawa T; Apte MV; Wilson JS; Ng B; Lau SL; Gunton JE; Wahl GM; Hunter T; Drebin JA; O’Dwyer PJ; Liddle C; Tuveson DA; Downes M; Evans RM, Vitamin D Receptor-mediated Stromal Reprogramming Suppresses Pancreatitis and Enhances Pancreatic Cancer Therapy. Cell 2014, 159, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Olive KP; Jacobetz MA; Davidson CJ; Gopinathan A; McIntyre D; Honess D; Madhu B; Goldgraben MA; Caldwell ME; Allard D; Frese KK; Denicola G; Feig C; Combs C; Winter SP; Ireland-Zecchini H; Reichelt S; Howat WJ; Chang A; Dhara M; Wang L; Ruckert F; Grutzmann R; Pilarsky C; Izeradjene K; Hingorani SR; Huang P; Davies SE; Plunkett W; Egorin M; Hruban RH; Whitebread N; McGovern K; Adams J; Iacobuzio-Donahue C; Griffiths J; Tuveson DA, Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science (New York, N.Y.) 2009, 324, 1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jacobetz MA; Chan DS; Neesse A; Bapiro TE; Cook N; Frese KK; Feig C; Nakagawa T; Caldwell ME; Zecchini HI; Lolkema MP; Jiang P; Kultti A; Thompson CB; Maneval DC; Jodrell DI; Frost GI; Shepard HM; Skepper JN; Tuveson DA, Hyaluronan Impairs Vascular Function and Drug Delivery in a Mouse Model of Pancreatic Cancer. Gut 2013, 62, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Straussman R; Morikawa T; Shee K; Barzily-Rokni M; Qian ZR; Du J; Davis A; Mongare MM; Gould J; Frederick DT; Cooper ZA; Chapman PB; Solit DB; Ribas A; Lo RS; Flaherty KT; Ogino S; Wargo JA; Golub TR, Tumour Micro-environment Elicits Innate Resistance to RAF Inhibitors Through HGF Secretion. Nature 2012, 487, 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wilson TR; Fridlyand J; Yan Y; Penuel E; Burton L; Chan E; Peng J; Lin E; Wang Y; Sosman J; Ribas A; Li J; Moffat J; Sutherlin DP; Koeppen H; Merchant M; Neve R; Settleman J, Widespread Potential for Growth-factor-driven Resistance to Anticancer Kinase Inhibitors. Nature 2012, 487, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Erkan M; Hausmann S; Michalski CW; Fingerle AA; Dobritz M; Kleeff J; Friess H, The Role of Stroma in Pancreatic Cancer: Diagnostic and Therapeutic Implications. Nat Rev Gastro Hepat 2012, 9, 454–467. [DOI] [PubMed] [Google Scholar]
- (16).Qu C; Wang Q; Meng Z; Wang P, Cancer-Associated Fibroblasts in Pancreatic Cancer: Should They Be Deleted or Reeducated? Integr Cancer Ther 2018, 17, 1016–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Arvizo RR; Saha S; Wang E; Robertson JD; Bhattacharya R; Mukherjee P, Inhibition of Tumor Growth and Metastasis by a Self-therapeutic nanoparticle. Proc Natl Acad Sci U S A 2013, 110, 6700–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Xiong X; Arvizo RR; Saha S; Robertson DJ; McMeekin S; Bhattacharya R; Mukherjee P, Sensitization of Ovarian Cancer Cells to Cisplatin by Gold Nanoparticles. Oncotarget 2014, 5, 6453–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Saha S; Xiong X; Chakraborty PK; Shameer K; Arvizo RR; Kudgus RA; Dwivedi SK; Hossen MN; Gillies EM; Robertson JD; Dudley JT; Urrutia RA; Postier RG; Bhattacharya R; Mukherjee P, Gold Nanoparticle Reprograms Pancreatic Tumor Microenvironment and Inhibits Tumor Growth. ACS nano 2016, 10, 10636–10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bachem MG; Schneider E; Gross H; Weidenbach H; Schmid RM; Menke A; Siech M; Beger H; Grunert A; Adler G, Identification, Culture, and Characterization of Pancreatic Stellate Cells in Rats and Humans. Gastroenterology 1998, 115, 421–432. [DOI] [PubMed] [Google Scholar]
- (21).Park J; Ha MK; Yang N, and Yoon TH, Flow Cytometry-Based Quantification of Cellular Au Nanoparticles. Anal. Chem 2017, 89, 2449–2456 [DOI] [PubMed] [Google Scholar]
- (22).Conde J; Oliva N; Atilano M; Song SH; and Artzi N, Self-Assembled RNA-Triple-Helix Hydrogel Scaffold for MicroRNA Modulation in the Tumour Microenvironment. Nat Mater. 2016, 15, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chakraborty PK; Xiong X; Mustafi SB; Saha S; Dhanasekaran D; Mandal NA; McMeekin S; Bhattacharya R; Mukherjee P, Role of Cystathionine Beta Synthase in Lipid Metabolism in Ovarian Cancer. Oncotarget 2015, 6, 37367–37384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Schmittgen TD; Livak KJ, Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat Protoc 2008, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- (25).Wu Y; Zhang Q; Ruan Z; Yin Y, Intrinsic Effects of Gold Nanoparticles on Proliferation and Invasion Activity in SGC-7901 Cells. Oncol Rep 2016, 35, 1457–1462. [DOI] [PubMed] [Google Scholar]
- (26).Li Y; Zong WX; Ding WX, Recycling the Danger via Lipid Droplet Biogenesis After Autophagy. Autophagy 2017, 13, 1995–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Canton I, Battaglia G Endocytosis at the Nanoscale. Chem. Soc. Rev 2012, 41 (7), 2718–39. [DOI] [PubMed] [Google Scholar]
- (28).Ng CT; Tang FM; Li JJ; Ong C; Yung LL; and Bay BH, Clathrin-Mediated Endocytosis of Gold Nanoparticles In Vitro. Anat Rec (Hoboken) 2015, 298, 418–427 [DOI] [PubMed] [Google Scholar]
- (29).Flavin R; Peluso S; Nguyen PL; Loda M, Fatty Acid Synthase as a Potential Therapeutic Target in Cancer. Future Oncol 2010, 6, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Menendez JA; Lupu R, Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nat Rev Cancer 2007, 7, 763–777. [DOI] [PubMed] [Google Scholar]
- (31).Ferdek PE; Jakubowska MA, Biology of Pancreatic Stellate Cells-More Than Just Pancreatic Cancer. Pflug Arch Eur J Phy 2017, 469, 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Singh R; Kaushik S; Wang Y; Xiang Y; Novak I; Komatsu M; Tanaka K; Cuervo AM; Czaja MJ, Autophagy Regulates Lipid Metabolism. Nature 2009, 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rambold AS; Cohen S; Lippincott-Schwartz J, Fatty Acid Trafficking in Starved Cells: Regulation by Lipid Droplet Lipolysis, Autophagy, and Mitochondrial Fusion Dynamics. Dev Cell 2015, 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Blommaart EF; Krause U; Schellens JP; Vreeling-Sindelarova H; Meijer AJ, The Phosphatidylinositol 3-Kinase Inhibitors Wortmannin and LY294002 Inhibit Autophagy in Isolated Rat Hepatocytes. Eur J Biochem 1997, 243, 240–246. [DOI] [PubMed] [Google Scholar]
- (35).Lopaschuk GD; Wall SR; Olley PM; Davies NJ, Etomoxir, a Carnitine Palmitoyltransferase I Inhibitor, Protects Hearts From Fatty Acid-Induced Ischemic Injury Independent of Changes in Long Chain Acylcarnitine. Circ Res 1988, 63, 1036–1043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.