Abstract
The miniaturized arachnid order Palpigradi has ambiguous phylogenetic affinities owing to its odd combination of plesiomorphic and derived morphological traits. This lineage has never been sampled in phylogenomic datasets because of the small body size and fragility of most species, a sampling gap of immediate concern to recent disputes over arachnid monophyly. To redress this gap, we sampled a population of the cave-inhabiting species Eukoenenia spelaea from Slovakia and inferred its placement in the phylogeny of Chelicerata using dense phylogenomic matrices of up to 1450 loci, drawn from high-quality transcriptomic libraries and complete genomes. The complete matrix included exemplars of all extant orders of Chelicerata. Analyses of the complete matrix recovered palpigrades as the sister group of the long-branch order Parasitiformes (ticks) with high support. However, sequential deletion of long-branch taxa revealed that the position of palpigrades is prone to topological instability. Phylogenomic subsampling approaches that maximized taxon or dataset completeness recovered palpigrades as the sister group of camel spiders (Solifugae), with modest support. While this relationship is congruent with the location and architecture of the coxal glands, a long-forgotten character system that opens in the pedipalpal segments only in palpigrades and solifuges, we show that nodal support values in concatenated supermatrices can mask high levels of underlying topological conflict in the placement of the enigmatic Palpigradi.
Keywords: arthropods, Arachnida, phylotranscriptomics, evolutionary rate, gene tree
1. Introduction
The basal phylogenetic relationships of the chelicerate arthropods (e.g. spiders, scorpions and sea spiders) remain uncertain, in part owing to limitations in taxonomic sampling, as well as long-branch attraction artefacts [1–4]. Molecular phylogenetic analyses of Chelicerata have supported a basal split between Pycnogonida (sea spiders) and the remaining chelicerates, as well as the monophyly of Arachnopulmonata (arachnid orders with book lungs, such as spiders, scorpions and vinegaroons) [2,5], but other relationships within Chelicerata have proven remarkably unstable, in spite of treatment with genome-scale datasets. Among the culprits of this topological instability are the fast-evolving orders Acariformes (mites), Parasitiformes (ticks), Pseudoscorpiones (pseudoscorpions) and possibly also Solifugae (camel spiders; [5]). A recent phylogenomic assessment of the group additionally concluded that Xiphosura (horseshoe crabs) constitute a derived group of marine arachnids, rather than the sister group to a monophyletic Arachnida [6]. This unexpected result was shown to be unattributable to long-branch attraction, as sequential deletion of fast-evolving lineages had no effect on the derived placement of horseshoe crabs within the arachnids. Several works have shown that certain partitions of data matrices can indeed recover arachnid monophyly with maximal nodal support [5–7], but arachnid monophyly remains unstable and unreproducible across phylogenomic datasets, and furthermore, the backbone of chelicerate phylogeny remains intractably unresolved.
One of the limitations of previous studies, however, is the omission of a key lineage whose phylogenetic affinities within the chelicerates have been uncertain for more than a century. Palpigradi (micro-whip scorpions) is the least understood of the chelicerates and constitutes a miniaturized order of cryptic arachnids. The last of the arachnid orders to be described, palpigrades are unique among arachnids in that their first pair of walking legs are used to palpate the environment and their uplifted flagellum moves laterally; their unmodified palps are instead used for walking [8]. Morphological cladistic works have previously recovered Palpigradi as the sister group of Tetrapulmonata (a group of arachnid orders with an ancestral condition of two pairs of book lungs, such as spiders; [9]) or as the sister group of the remaining arachnids [10,11]. By contrast, molecular phylogenetic studies have variably placed them in a clade including Acari and Solifugae (Cephalosomata; [12]), sister group to mites [2], sister group to ticks [5], or in an unresolved position [13]. These molecular datasets were nevertheless either based on a small number of genes [2,12,13] or had poor representation of Palpigradi [5], owing to re-use of legacy Sanger-sequenced datasets. Palpigrades have thus, to our knowledge, never been represented in phylotranscriptomic datasets, an omission partly reflecting their small body size (typically less than 2 mm, including the posterior flagellum), cryptic habitat and difficulty of maintenance in captivity.
To address the phylogenetic position of Palpigradi, we sampled a population of the cave-inhabiting species Eukoenenia spelaea (Peyerimhofi, 1902) from the Ardovská Cave in Slovakia. We produced a single transcriptome from six living specimens and inferred the phylogenetic placement of palpigrades using a dense phylogenomic matrix consisting of 1450 loci, drawn from high-quality transcriptomic libraries and complete genomes. In addition, we added two other lineages also omitted in previous phylogenomic analyses of Chelicerata (but less controversial in phylogenetic placement than palpigrades): a newly discovered species of the erstwhile order Opilioacariformes (presently considered the putative sister group of the remaining Parasitiformes; [12,14]) and an exemplar of Schizomida (short-tailed whip scorpions), another miniaturized order of arachnids that constitutes the sister group of Thelyphonida [13,15].
Here, we show that Palpigradi are recovered as closely related to Solifugae (camel spiders) in the most complete data matrices, a result consistent with the putatively homologous architecture of the coxal glands (excretory organs) of Palpigradi and Solifugae. However, this placement is highly sensitive to dataset construction and analytical approach. We further show that high nodal support values in supermatrices mask systemic conflicts in phylogenetic signal, which can be elicited by dataset subsampling methods.
2. Material and methods
We assembled a dataset of 45 chelicerate and 14 outgroup taxa, with datasets comprising 14 genome projects and the rest of Illumina transcriptomes (29 of the 45 transcriptomes were previously generated by us [5,6]). Electronic supplementary material, table S1 lists all species, collecting data, and sequence accession data for this study. A pipeline for RNA extraction, sequencing, transcriptome assembly with Trinity v. 2.5 [16], homology searches and phylogenetically informed orthology inference with the UPhO pipeline [17] is provided in the electronic supplementary material, Text S1 and follows our recent approaches [17,18]. Briefly, translated protein coding sequences were identified using Transdecoder v. 5.0.1 [16] and queried using blastp against sequences in the 5815 homologous clusters previously identified by us [6]. Sequences from new libraries were added into the group with the best average e-value score, and aligned and cleaned as previously described [6]. After multiple sequence alignment and trimming of ambiguously aligned regions, matrices were assembled with minimal taxon occupancy thresholds of 51 (matrix 1), 45 (matrix 2), 40 (matrix 3) and 30 (matrix 4) species. A decisive dataset of 237 loci was obtained by enforcing the presence of Palpigradi, Tetrapulmonata, Opiliones and Solifugae in each locus (matrix 5). To assess the effect of denser sampling of acarine orders, we added orthologues of the palpigrade, the opilioacarid, and the schizomid to the 233-locus and 3982-locus datasets of Lozano-Fernández et al. (‘matrix A’ and ‘matrix B’; [7]), of which the former obtained arachnid monophyly, but only under Bayesian inference and under a specific substitution model. We termed these datasets matrices 6 and 7, respectively.
Phylogenetic inference of the concatenated matrices was computed with IQ-Tree v. 1.6.1 [19], implementing the model selection of substitution and rate heterogeneity based on the Bayesian information criterion. For phylogenetic analyses using multispecies coalescent methods, species trees were estimated with Astral v. 3 [20], using the collection of orthologous gene trees from UPhO analyses as inputs, with gene tree topologies estimated using IQ-Tree v. 1.6.1 under the best substitution models inferred for concatenated approaches. PhyloBayes-mpi v. 1.7 [21] analysis was performed using two to four independent runs under the CAT + GTR + Γ model for the smaller matrices (matrices 1, 2, 5 and 6). Dayhoff-recoded matrices 1, 2 and 5 was also analysed using PhyloBayes-mpi.
Phylogenetic signal was explored using the gene-wise log-likelihood score (ΔGLS) for the unconstrained tree versus a tree constrained to recover the monophyly of Palpigradi + Solifugae (or vice versa) following Shen et al. [22]. This metric maps the relative support for each of two competing hypotheses, for every locus in the dataset; the amplitude of the log-likelihood indicates the degree of support for either hypothesis. Gene tree incongruence in the decisive dataset was visualized using quartet network mapping with SuperQ v. 1.1 [23].
3. Results
(a). Phylogenetic relationships based on concatenation
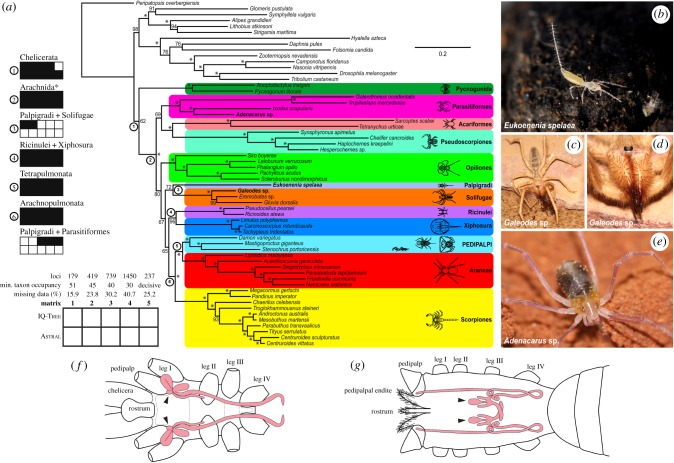
Initial homology searches used the 5815 Panarthropoda homologue clusters previously identified by us [6] and searches were conducted in the libraries of six additions to our previous analysis: E. spelaea (Palpigradi), Adenacarus sp. (Opilioacariformes), Galeodes sp. (Solifugae), Stenochrus portoricensis (Schizomida) and two additional pseudoscorpions. Properties of the primary matrices (matrices 1–5) are summarized in figure 1a and the electronic supplementary material, table S2.
Figure 1.
Phylogenomic relationships of Chelicerata. (a) Maximum-likelihood tree topology inferred from concatenated analysis of matrix 1. Number on nodes indicates bootstrap resampling frequencies. Bottom left: design of sensitivity analysis based on taxon occupancy threshold and analytical approach. Middle left: sensitivity plots for seven relationships of interest; filled squares indicate recovery of a relationship for the matrix and analysis indicated. Asterisk refers to the redefinition of Arachnida to include Xiphosura [6]. (b–d) Live habitus of key lineages sampled in this analysis. (b) The palpigrade E. spelaea. Photograph by L'. Kováč. (c) A Levantine species of the solifuge genus Galeodes. (d) Same individual as in (c), with magnification of the head. Photographs by S. Aharon. (e) A newly discovered Levantine species of the genus Adenacarus sp. Photograph by S. Aharon. (f) Schematic of palpigrade prosoma in ventral view, showing architecture of coxal gland; redrawn from [24]. (g) Schematic of solifuge prosoma in dorsal view, showing architecture of coxal gland; redrawn from [25]. Arrowheads in (f) and (g) refer to the saccule of the coxal gland, which occurs only in this pair of orders.
Maximum-likelihood (ML) analysis of all concatenated data matrices using IQ-Tree recovered tree topologies supporting the monophyly of Chelicerata, Arachnida sensu Ballesteros and Sharma (i.e. including horseshoe crabs; [6]), the clade Xiphosura + Ricinulei, Arachnopulmonata and Tetrapulmonata (figure 1a). The newly added opiliacarid and pseudoscorpion taxa were unambiguously placed within their corresponding orders. Similarly, S. portoricensis (Schizomida) was uniformly recovered with high support as the sister group of Mastigoproctus giganteus (Thelyphonida), a relationship consistent with an array of morphological and behavioural similarities of these orders [10], as well as molecular phylogenies based on two to four Sanger-sequenced genes [13,15].
ML analyses of matrices 1 and 2 recovered Palpigradi as the sister group of Solifugae (bootstrap resampling frequency (BS) = 72% and 46%, respectively), whereas the analysis of matrices 3 and 4 placed Palpigradi as the sister group of Parasitiformes (BS = 61% and 74%, respectively). The decisive dataset also recovered Palpigradi + Parasitiformes with modest support (BS = 78%) (electronic supplementary material, figure S1).
(b). Assessment of topological instability using taxon deletion
Three orders of arachnids (Acariformes, Parasitiformes and Pseudoscorpiones) have previously been shown to exhibit long-branch attraction artefacts [5–7], and there is some evidence that Solifugae may be prone to this phenomenon as well [6]. To assess whether Palpigradi is affected by the removal of long-branch orders, we undertook the sequential deletion of long-branch orders in ML analyses for matrix 1 (179 genes) and matrix 4 (1450 genes), starting with all outgroup taxa, then deleting Acari (mites and ticks), then Pseudoscorpiones, and lastly Solifugae (i.e. in decreasing order of ordinal mean patristic distance). Consistent with previous results, the position of mites, ticks and pseudoscorpions changed upon deletion of outgroups and/or outgroups as well as Acari (electronic supplementary material, figure S3). Specifically, the removal of outgroups, mites and ticks resulted in the sister group relationship of pseudoscorpions with scorpions with support (BS = 65% for matrix 1, BS = 90% for matrix 5; electronic supplementary material, figure S3), a recurring relationship across phylogenomic analyses [5,6,26]. Deletion of outgroups and long-branch taxa similarly affected Solifugae, destabilizing its placement across analyses and pushing this order closer to the base of the arachnid tree (electronic supplementary material, figure S3). For palpigrades, the removal of outgroups had the greatest effect on their placement, resulting in topologies where palpigrades were recovered as sister group to the remaining arachnids (regardless of the matrix analysed). No other orders exhibited subtree pruning and regrafting moves, or branch rearrangements, upon taxon deletion. Palpigradi are therefore part of the subset of chelicerate orders that exhibit topological instability upon perturbation of branch length distributions.
(c). Ordered phylogenomic subsampling
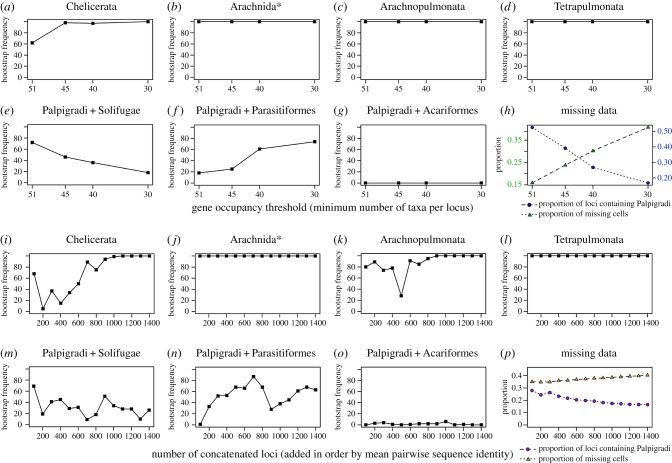
To assess how nodal support for the placement of Palpigradi is affected by matrix completeness, bootstrap resampling pseudoreplicates from matrices 1–4 were evaluated for seven topological hypotheses. We evaluated support for the monophyly of Chelicerata, Arachnida (including Xiphosura), Arachnopulmonata and Tetrapulmonata, which are robustly supported and stable across molecular phylogenetic datasets [5,6]. Separately, we evaluated support for three alternative placements of Palpigradi: sister group to Solifugae, sister group to Acariformes (proposed by van der Hammen [27], recovered by Regier et al. [2]) and sister group to Parasitiformes (previously recovered by Sharma et al. [5]).
The four stable nodes exhibited either maximal support across datasets (Arachnida, Arachnopulmonata and Tetrapulmonata) or monotonic increase to maximal nodal support upon addition of genes (Chelicerata) (figure 2a–d). Support for Palpigradi + Solifugae was moderate in the smallest and most complete matrix and decreased monotonically upon addition of genes (figure 2e), whereas the inverse trend was observed for Palpigradi + Parasitiformes (figure 2f). Low/negligible bootstrap support was observed for Palpigradi + Acariformes (figure 2g). We separately observed that as matrix size increased, the proportion of loci sampling Palpigradi decreased monotonically, indicating that the distribution of missing data is non-random with respect to E. spelaea (figure 2h).
Figure 2.
Dissection of phylogenetic signal using ordered phylogenomic subsampling. (a–g) Bootstrap resampling frequencies for selected phylogenetic hypotheses inferred from sequential addition of loci in order of completeness (taxon occupancy threshold, i.e. minimum number of taxa per locus). Labels over panels indicate the hypothesis assessed. (h) Proportion of missing sequence data (green) and proportion of loci including E. spelaea (blue) as a function of taxon occupancy threshold. (i–o) Bootstrap resampling frequencies for selected phylogenetic hypotheses inferred from sequential addition of matrices in order of mean pairwise sequence identity, starting with the most conserved loci. Labels over panels indicate the hypothesis assessed. (p) Proportion of missing sequence data (orange) and proportion of loci including E. spelaea (purple) as a function of taxon occupancy threshold. Asterisk refers to the redefinition of Arachnida to include Xiphosura [6].
To assess how nodal support for the placement of Palpigradi is affected by an evolutionary rate, 14 matrices were assembled wherein orthologues were concatenated in order by their mean pairwise sequence identity (MPSI), a proxy for rate of evolution [5,6]. Genes were thus added to a phylogenetic matrix at increments of 100, starting with the slowest-evolving genes. Bootstrap resampling frequencies were scored for the same seven nodes. For the four stable nodes, nodal support trajectory either remained consistently at 100% (Arachnida and Tetrapulmonata) or trended towards maximal support in the largest matrices (Chelicerata and Arachnopulmonata) (figure 2i–l). Notably, support for Arachnopulmonata decreased to 28% in the analysis of the 500 slowest-evolving genes, owing to the nested placement of Pseudoscorpiones within Arachnopulmonata as the sister group of scorpions (figure 2k). This phenomenon reflects our previous finding that Pseudoscorpiones, probably a member of Arachnopulmonata, is highly prone to long-branch attraction artefacts, a result not mitigated by the addition of exemplars in this analysis [5,6].
By contrast, the placement of Palpigradi + Solifugae was better supported than Palpigradi + Parasitiformes only in two of the 14 matrices (figure 2m,n). Contrary to the expectation of robustly supported or robustly rejected nodes, support for neither hypothesis trended towards a maximal value, but rather, fluctuated between values that reflected modest support. Support for Palpigradi + Acariformes never exceeded 6% (figure 2o). The matrices assembled by MPSI did not exhibit dramatically different proportions of missing data, nor representation of Palpigradi orthologues (figure 2p).
(d). Coalescent-based species tree methods
Coalescent-based analyses with all five matrices using Astral recovered Palpigradi as part of a grade with the long-branch orders Acariformes, Parasitiformes and Pseudoscorpiones, with variable support for the position of palpigrades (electronic supplementary material, figure S4a). To assess support for competing hypotheses generated by concatenation-based analyses, we reran Astral using gene trees of each matrix (matrices 1–4) while constraining the species tree to one of two topologies: the ML tree topology inferred for matrix 1 (figure 1a; Palpigradi + Solifugae) and the ML tree topology inferred for matrix 4 (Palpigradi + Parasitiformes). Local posterior probabilities (PP) for either placement of Palpigradi were highest for the unconstrained topology (PP = 0.57–0.98 across matrices 1–4), intermediate for the topology with the Palpigradi + Parasitiformes constraint and lowest for the topology with the Palpigradi + Solifugae constraint (electronic supplementary material, figure S4b). However, normalized quartet scores obtained under either constrained topology were nearly equal in value to each other, as well as to the quartet scores obtained under the unconstrained tree, with maximum difference in scores never exceeding 0.5% across matrices 1–4 (electronic supplementary material, figure S4c). These patterns were also observed for the decisive dataset of 237 genes, wherein Palpigradi is represented in 100% of loci (maximum difference in normalized quartet scores of 0.6%).
(e). Gene-wise log-likelihood score and gene tree discordance
For some problematic cases in systematics, aberrant tree topologies have been shown to result from a small number of anomalous genes [22]. To investigate this possibility, we computed the ΔGLS for the two competing topologies yielded by concatenated analyses of matrices 1 and 4. This analysis provides a comparison of the support for one of two competing tree topologies, for every locus in the dataset [22]. As constraints, we enforced only the nodes Palpigradi + Parasitiformes (T1) and Palpigradi + Solifugae (T2). ML trees were estimated from the decisive dataset to rule out the effects of loci missing focal lineages. These analyses revealed nearly equal numbers of gene trees supporting either hypothesis (electronic supplementary material, figure S4d), consistent with the incidence of a short internode in this part of the arachnid tree. Separately, we visualized the dominant bipartitions among the ML gene tree topologies of the decisive dataset by constructing supernetworks using the SuperQ method [23]. Briefly, such supernetworks are a visualization of gene tree conflict, where edge lengths are proportional to the numbers of loci supporting a given bipartition. Reticulations correspond to gene tree conflict—the larger and denser the reticulation, the greater the conflict between loci. We observed an unresolved star topology with pronounced reticulations corresponding to basal divergences of Arachnida (electronic supplementary material, figure S4e). The only relationships exhibiting the signature of unambiguous resolution were a small subset of superordinal relationships consistently recovered by all analyses (e.g. Arachnopulmonata; Tetrapulmonata; Pedipalpi), and the monophyly of all non-Acari chelicerate orders. As indicated by this supernetwork visualization, the position of Palpigradi is entirely ambiguous across the gene trees (note the short terminal edge leading to E. spelaea in electronic supplementary material, figure S4e; all terminal edges in the SuperQ network are identical in length, as they do not convey information beyond the locations of the tips in the network). Consistent with this ambiguous distribution of signal, runs of PhyloBayes-mpi [21] never reached stationarity for any of our analyses after exceeding 10 000 cycles. In addition to not recovering a clear placement of Palpigradi, the resulting tree topologies from PhyloBayes-mpi analyses consistently recovered the non-monophyly of Arachnida (electronic supplementary material, figure S5).
(f). Assessment of tree topology with denser sampling of Acari
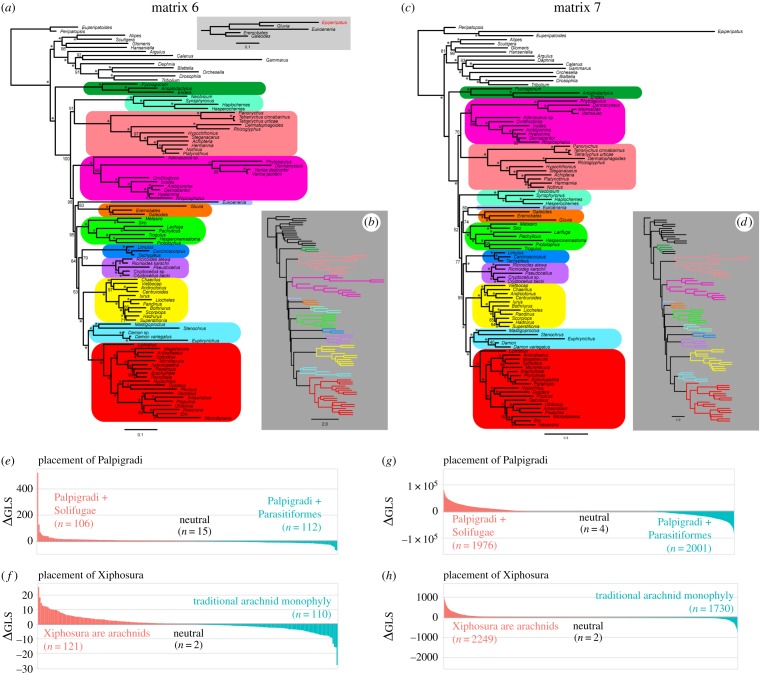
To evaluate the impact of richer sampling of Acariformes and Parasitiformes on the placement of Palpigradi, we added three key lineages (palpigrade, schizomid and opilioacarid) to a 233-locus phylogenomic matrix of curated genes (matrix A) recently published by Lozano-Fernández et al. [7], to test how this augmented dataset (matrix 6) would support or refute the hypothesis of arachnid monophyly that was previously recovered by matrix A under the CAT model. But upon reanalysing their matrix, we encountered some anomalies that impinge on the reproducibility of their matrix A [7]. To circumvent these anomalies, we then added the three terminals to the 3982-locus matrix also published by Lozano-Fernández et al. [7] (matrix B), whose orthology was established using the Orthologous MAtrix algorithm (matrix 7). Thereupon, we encountered additional anomalies in the contruction of matrix B. These anomalies and associated caveats in interpretation are detailed in the electronic supplementary material, Text S2.
ML analysis of matrix 6 under posterior mean site frequency (PMSF) (LG + C20 + F + Γ), a mixture model alternative to the CAT implementation in PhyloBayes, did not recover the monophyly of Arachnida (figure 3a); Xiphosura was recovered as sister group to Ricinulei with modest support (BS = 83%). Moreover, this analysis recovered the diphyly of Onychophora, owing to the nested placement of Epiperipatus sp. within Solifugae + Palpigradi. We deduced that this anomalous placement was the result of extensive missing data for Epiperipatus sp. (87.83% missing data). Upon removal of this terminal, we obtained: (i) the same placement of Xiphosura as derived arachnids (BS = 79%); (ii) the monophyly of Uropygi (=Schizomida + Thelyphonida) (BS = 100%); and (iii) the placement of Opilioacariformes at the base of Parasitiformes (BS = 95%); and the sister group relationship of Palpigradi and Solifugae (BS = 83%). Other relationships were as shown in figure 3a. As with analyses of all other matrices, Astral analysis of the same dataset recovered a grade of the unstable arachnid orders at the base of the species tree (figure 3b). Analyses of matrix 7 recovered the same placements for Xiphosura, Schizomida and Palpigradi as matrix 6; Opilioacariformes was recovered as nested within Parasitiformes, with support (figure 3c,d). Consistent with the discordant resolution implied by Astral, interrogation of phylogenetic signal for both palpigrade placement and arachnid monophyly using a ΔGLS approach showed that both these datasets lacked signal for key basal relationships, like arachnid monophyly (figure 3e–h). The recovery of similar ΔGLS distributions for palpigrade placement across three different approaches to orthology inference suggests that conflicting signal in the placement of Palpigradi is systemic.
Figure 3.
(a,b) ML tree topology of matrix 6 under the PMSF (CAT approximation) model (a) and Astral counterpart (b). (c,d) ML tree topology of matrix 7 under the PMSF (CAT approximation) model (c) and Astral counterpart (d). Numbers on nodes correspond to bootstrap resampling frequencies. (e,f) Ranked distribution of ΔGLS for matrix 6 evaluating alternative placements of Palpigradi (e) and arachnid monophyly (f). (g,h) Ranked distribution of ΔGLS for matrix 7 evaluating alternative placements of Palpigradi (e) and arachnid monophyly (f).
4. Discussion
(a). The phylogenetic placement of Palpigradi is not resolved by phylotranscriptomics
The backbone phylogeny of Chelicerata has remained obdurately recalcitrant to stable resolution, despite the application of thousands of orthologous loci drawn from complete genomes and high-quality transcriptomes [6,7]. Beyond the incidence of multiple long-branch orders and marked asymmetry in extant diversity, phylogenomic efforts have also faced limitations in taxonomic sampling of small-bodied and/or rare lineages, such as palpigrades and numerous groups of mites. Only one previous molecular phylogenetic analysis of chelicerates sampled all ordinal lineages with more than two loci; the 62 protein-encoding locus analysis by Regier et al. [2] recovered Palpigradi as the sister group of Acariformes with low support in a subset of analyses, with this clade at the base of an unstable Arachnida, together with Parasitiformes and Pseudoscorpiones. Analysis of larger datasets has shown that up to thousands of genes may be required to resolve with confidence the short internodes that constitute the backbone of chelicerate phylogeny, but these works lacked transcriptomic representation of Palpigradi, as well as the small-bodied order Schizomida and the rare lineage Opilioacariformes [5–7]. Inclusion of the palpigrade terminal from the Regier et al. [2] dataset in the phylogenomic matrix of Sharma et al. [5] resulted in the representation of Palpigradi by only three genes in a 500-locus matrix, and consequently, an unsupported placement of palpigrades as a sister group to Parasitiformes in that study.
Here, we overcame limitations in sampling Palpigradi at the phylogenomic level and assembled a data matrix with representation of all chelicerate orders with up to 1450 loci. The palpigrade library we generated was considerably smaller and far less complete than those of other species (14.7% of benchmarked universal single-copy orthologues (BUSCOs) for arthropods recovered), a result of limitations in RNA preservation and extraction of rare, minute specimens in a challenging fieldwork theatre. Nevertheless, we were able to assess palpigrade placement with up to 237 loci in the decisive dataset, whose construction was predicated on the presence of sequence data for Palpigradi and other key lineages.
Analyses of our largest matrices recovered Palpigradi as the sister group to Parasitiformes, with moderate support (BS = 74%, matrix 4; electronic supplementary material, figure S1). The modest support for this relationship precipitated an investigation of interpartition conflict, using ordering phylogenomic subsampling techniques. Intriguingly, we established that in the most complete matrix, Palpigradi was recovered as the sister group of Solifugae (figure 2e) with comparable nodal support to the competing topology favouring Palpigradi + Parasitiformes in the largest matrix (BS = 72%). Acariformes was never supported as a part of this clade (contrary to a previous analysis based on two nuclear ribosomal genes [12]), nor as a sister group to Palpigradi, as suggested by a 62-locus analysis [2]. Addition of orthogroups with greater amounts of missing data resulted in the degradation of signal supporting Palpigradi + Solifugae, suggesting that the relationship Palpigradi + Parasitiformes may be driven by data incompleteness and the cumulatively decreasing proportion of loci that sample Palpigradi (figure 2e,h).
We also explored the effect of evolutionary rate using sequential addition of faster-evolving genes in concatenated matrices. This analysis revealed varying levels of support for the relationship Palpigradi + Solifugae as well as Palpigradi + Parasitiformes. While analysis of the 100 slowest-evolving loci recovered moderate support for Palpigradi + Solifugae (BS = 69%), nodal support did not trend towards zero or 100% for either competing hypothesis, suggesting pervasive interpartition conflict throughout the orthogroups we examined, as previously reported in chelicerate phylogenomic datasets [6]. This inference was corroborated by species tree coalescent-based approaches, which revealed nearly equal distributions of quartets and gene trees (electronic supplementary material, figure S4b–d) supporting any of the putative placements of Palpigradi recovered in this study.
Taken together, these results suggest high levels of noise with respect to the position of Palpigradi for the orthologues presently available for E. spelaea. Moreover, the deletion analyses we performed suggest that Palpigradi is prone to topological instability as a function of taxonomic sampling of long-branch orders, thereby further complicating a stable resolution of chelicerate phylogeny. Apropos, addition of palpigrades to phylogenomic matrices containing the most comprehensive sampling of chelicerates to date, recovered the clade Palpigradi + Solifugae with modest support but only in concatenated analyses (figure 3a,c). Astral analyses of the same datasets show little mitigation of gene tree conflict upon denser sampling of the long-branch orders Acariformes and Parasitiformes in matrix 6, both for the placement of palpigrades and for the monophyly of Arachnida (figure 3b); the corresponding analysis for matrix 7 supported both a basally branching placement of Palpigradi, as well as the non-monophyly of Arachnida (figure 3d).
(b). The relationship Palpigradi + Solifugae is consistent with the architecture of the coxal glands
The unexpected recovery of Palpigradi as sister group to Solifugae in the most complete matrix merits scrutiny, because this relationship has not been supported in recent analyses of morphological or molecular datasets. Paralleling molecular phylogenetic datasets, morphological phylogenies of Chelicerata have historically exhibited significant discordance with respect to the placement of Palpigradi, owing to various interpretations of the plesiomorphic versus derived nature of arachnid character systems [9–11]. As examples, the palpigrade flagellum (annulated posterior appendage resembling a ‘tail’) closely resembles flagella found in extant Thelyphonida (vinegaroons) and Schizomida and invites the inference of their relationship to Pedipalpi [28]. However, comparable structures also occur in Uraraneida (an extinct order that is sister group to modern spiders; [29,30]) as well as some extinct Pycnogonida [31]. Palpigradi lack eyes altogether, obviating the applicability of characters pertaining to arachnid median versus lateral eyes. The mouth of Palpigradi, consisting of an anteriorly projecting cone-shaped structure incorporating the labrum as its upper lobe, is comparable to that of Pycnogonida, Solifugae and Acari [28]. The ‘pulmonary sacs’ found on the ventral opisthosoma (abdomen) of large palpigrade species were held to be remnants of book lungs, but similar opisthosomal protuberances also occur in embryos of Solifugae [8,12]. Pepato et al. [12] proposed that palpigrades were related to Solifugae and Acariformes (with the latter two orders forming the clade Poecilophysida [32]), a result obtained with weak support in combined analyses of morphological and molecular data. They termed the clade formed by this trio of orders Cephalosomata, based upon the patterns of sclerotization of the head segments anterior to leg III, but this relationship was never supported in any of our analyses. Separately, no morphological phylogenies or homology schema have ever united palpigrades with Parasitiformes.
A sister group relationship of the miniaturized palpigrades to the typically large-bodied, fast-moving and predatory Solifugae obtained by the most complete matrices (figures 1 and 3) seems immediately counterintuitive. Yet, surprisingly, this hypothesis was proposed over a century ago in an obscure work by Buxton [24]. One historically overlooked character system that unites palpigrades and solifuges is the architecture of the coxal glands, a pair of excretory organs unique to arachnids. In most arachnids, the coxal gland consists of a terminal saccule and a collecting tubule leading into the labyrinth, which is lined with excretory epithelium. At the distal end of the labyrinth lies a collecting bladder and a short exiting tubule leading to the coxal gland pore, which typically opens at the base of the coxae of leg I (in addition to leg III in some orders). In Palpigradi and Solifugae (figure 1f,g), the coxal gland is distinguished from those of other arachnid orders in that (i) an additional large sac lined with secretory epithelium lies between the collecting tubule and the labyrinth, and (ii) the coxal gland exit pore opens on the pedipalpal segment [24]. Nevertheless, this character was subsequently coded in a series of morphological phylogenetic matrices as autapomorphic in Solifugae [9,10,12]. A review of the pertinent literature [25,28,33] corroborates the observation of Buxton [24] that the palpigrade coxal gland is more similar to its solifuge counterpart than those of any other chelicerates. Only the position of the coxal gland opening on the pedipalpal segment of solifuges is autapomorphic, as it occurs in a dorsolateral position, rather than on the ventral surface of the pedipalpal coxa like in palpigrades [24,34].
We add the caveat that identifying morphological characters consistent with a favoured hypothesis is a practice prone to post hoc interpretation and investigator bias. The congruence of a single character system with the relationship of Palpigradi + Solifugae does not safeguard phylogenetic accuracy, especially given the odd combination of plesiomorphic and apomorphic morphological traits exhibited by Palpigradi. Rather, the shared condition of the coxal glands of these two otherwise morphologically disparate orders underscores the incidence of noise inherent in the base of Arachnida across all available phylogenetic data classes, including morphology. Given the incidence of this character, we conclude only that a grouping of Palpigradi with Solifugae is plausible and cannot be ruled out by available data. Historical hypotheses that we can rule out with confidence include the phylogenetic proximity of Palpigradi to Pedipalpi [28], to Tetrapulmonata [8] or to Acariformes [27]. Future investigations of chelicerate phylogeny should emphasize the sampling of basally branching groups within each of the poorly sampled orders of Chelicerata, such as prokoeneniid palpigrades and austrodecid sea spiders.
Supplementary Material
Acknowledgements
We are indebted to Scott V. Edwards, Jordi Paps, and two anonymous reviewers, whose feedback improved a previous draft of this work. Assistance in the field was provided by Shlomi Aharon and Shemesh Ya'aran. Eitan Rechet provided guidance on the taxonomy of Opilioacariformes. Access to computing nodes for intensive tasks was provided by the Center for High Throughput Computing (CHTC) and the Bioinformatics Resource Center (BRC) of the University of Wisconsin–Madison.
Ethics
The collection of specimens of E. spelaea adhered to the conditions of License 3102/2009 of the Ministry of Environment of the Slovak Republic. Specimens of Oplioacariformes and Solifugae were collected under permit 2017/41718, issued by the Israel National Parks Authority.
Data accessibility
Illumina raw read data: NCBI SRA accession numbers SRR8080645-647. Trinity assemblies, multiple sequence alignments (Matrices 1-7), partition files, phylogenetic trees: Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.t1g1jwsz4 [35].
Authors' contributions
Specimens were collected by L.K. and E.G.-R. Molecular work was performed by P.P.S. and C.E.S.L.; C.E.S.L., J.A.B. and P.P.S. performed bioinformatic and phylogenetic analyses. J.A.B., C.E.S.L. and P.P.S. wrote the paper. All the authors edited and approved the final content of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by National Science Foundation (grant no. IOS-1552610) to P.P.S. L'.K. was supported from research grants APVV-17-0477 and VEGA 1/0346/18. J.A.B. was supported by the M. Guyer postdoctoral fellowship. C.E.S.L. was supported by a postdoctoral CONACYT grant (reg. 207146/454834). Fieldwork in Israel was supported by a National Geographic Society Expeditions Council grant no. NGS-271R-18 to J.A.B.
References
- 1.Roeding F, Borner J, Kube M, Klages S, Reinhardt R, Burmester T. 2009. A 454 sequencing approach for large scale phylogenomic analysis of the common emperor scorpion (Pandinus imperator). Mol. Phylogenet. Evol. 53, 826–834. ( 10.1016/j.ympev.2009.08.014) [DOI] [PubMed] [Google Scholar]
- 2.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079–1083. ( 10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 3.Meusemann K, et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol. Biol. Evol. 27, 2451–2464. ( 10.1093/molbev/msq130) [DOI] [PubMed] [Google Scholar]
- 4.Borner J, Rehm P, Schill RO, Ebersberger I, Burmester T. 2014. A transcriptome approach to ecdysozoa phylogeny. Mol. Phylogenet. Evol. 80, 79–87. ( 10.1016/j.ympev.2014.08.001) [DOI] [PubMed] [Google Scholar]
- 5.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 6.Ballesteros JA, Sharma PP. 2019. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst. Biol. 68, 896–917. ( 10.1093/sysbio/syz011) [DOI] [PubMed] [Google Scholar]
- 7.Lozano-Fernández J, Tanner AR, Giacomelli M, Carton R, Vinther J, Edgecombe GD, Pisani D. 2019. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat. Commun. 10, 2295 ( 10.1038/s41467-019-10244-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giribet G, et al. 2014. The first phylogenetic analysis of Palpigradi (Arachnida)—the most enigmatic arthropod order. Invertebr. Syst. 28, 350–360. ( 10.1071/IS13057) [DOI] [Google Scholar]
- 9.Shultz JW. 1990. Evolutionary morphology and phylogeny of Arachnida. Cladistics 6, 1–38. ( 10.1111/j.1096-0031.1990.tb00523.x) [DOI] [PubMed] [Google Scholar]
- 10.Shultz J. 2007. A phylogenetic analysis of the arachnid orders based on morphological characters. Zool. J. Linn. Soc. 150, 221–265. ( 10.1111/j.1096-3642.2007.00284.x) [DOI] [Google Scholar]
- 11.Garwood RJ, Dunlop J. 2014. Three-dimensional reconstruction and the phylogeny of extinct chelicerate orders. PeerJ 2, e641 ( 10.7717/peerj.641/supp-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pepato AR, da Rocha CEF, Dunlop JA. 2010. Phylogenetic position of the acariform mites: sensitivity to homology assessment under total evidence. BMC Evol. Biol. 10, 235 ( 10.1186/1471-2148-10-235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giribet G, Edgecombe GD, Wheeler WC, Babbitt C. 2002. Phylogeny and systematic position of Opiliones: a combined analysis of chelicerate relationships using morphological and molecular data. Cladistics 18, 5–70. ( 10.1006/clad.2001.0185) [DOI] [PubMed] [Google Scholar]
- 14.Harvey M. 2002. The neglected cousins: what do we know about the smaller arachnid orders? J. Arachnol. 30, 357–372. ( 10.1636/0161-8202(2002)030[0357:TNCWDW]2.0.CO;2) [DOI] [Google Scholar]
- 15.Clouse RM, et al. 2017. First global molecular phylogeny and biogeographical analysis of two arachnid orders (Schizomida and Uropygi) supports a tropical Pangean origin and mid-Cretaceous diversification. J. Biogeogr. 44, 2660–2672. ( 10.1111/jbi.13076) [DOI] [Google Scholar]
- 16.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros JA, Hormiga G. 2016. A new orthology assessment method for phylogenomic data: unrooted phylogenetic orthology. Mol. Biol. Evol. 33, 2117–2134. ( 10.1093/molbev/msw069) [DOI] [PubMed] [Google Scholar]
- 18.Santibáñez-López C, Ontano A, Ballesteros JA, Harvey M, Sharma PP. 2018. Transcriptomic analysis of pseudoscorpion venom reveals a unique cocktail dominated by enzymes and protease inhibitors. Toxins 10, 207–212. ( 10.3390/toxins10050207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L-T, Schmidt HA, Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52. ( 10.1093/bioinformatics/btv234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 22.Shen X-X, Hittinger CT, Rokas A. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1, 0126 ( 10.1038/s41559-017-0126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruenewald S, Spillner A, Bastkowski S, Boegershausen A, Moulton V. 2013. SuperQ: computing supernetworks from quartets. IEEE/ACM Trans. Comput. Biol. Bioinform. 10, 151–160. ( 10.1109/TCBB.2013.8) [DOI] [PubMed] [Google Scholar]
- 24.Buxton BH. 1917. Notes on the anatomy of arachnids. J. Morphol. 29, 1–31. ( 10.1002/jmor.1050290102) [DOI] [Google Scholar]
- 25.Börner C. 1904. Beiträge zur Morphologie der Arthropoden: I. Ein Beitrag zur Kenntnis der Pedipalpen. Zoologica16, 1–174.
- 26.Sharma PP, Fernández R, Esposito LA, González-Santillán E, Monod L. 2015. Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal. Proc. R. Soc. B 282, 20142953 ( 10.1093/molbev/mss208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Hammen L. 1977. A new classification of Chelicerata. Zool. Meded. 51, 307–319. [Google Scholar]
- 28.Hansen HJ, Sorensen W. 1897. The order Palpigradi Thor. (Koenenia mirabilis Grassi) and its relationship to the other Arachnida. Entomol. Tidskr. 18, 223–240. [Google Scholar]
- 29.Wang B, Dunlop JA, Selden PA, Garwood RJ, Shear WA, Müller P, Lei X. 2018. Cretaceous arachnid Chimerarachne yingi gen. et sp. nov. illuminates spider origins. Nat. Ecol. Evol. 2, 614–622. ( 10.1038/s41559-017-0449-3) [DOI] [PubMed] [Google Scholar]
- 30.Huang D, Hormiga G, Cai C, Su Y, Yin Z, Xia F, Giribet G. 2018. Origin of spiders and their spinning organs illuminated by mid-Cretaceous amber fossils. Nat. Ecol. Evol. 2, 623–627. ( 10.1038/s41559-018-0475-9) [DOI] [PubMed] [Google Scholar]
- 31.Poschmann M, Dunlop JA. 2006. A new sea spider (Arthropoda : Pycnogonida) with a flagelliform telson from the lower Devonian Hunsruck Slate, Germany. Palaeontology 49, 983–989. ( 10.1111/j.1475-4983.2006.00583.x) [DOI] [Google Scholar]
- 32.Dunlop JA, Krüger J, Alberti G. 2012. The sejugal furrow in camel spiders and acariform mites. Arachnol. Mitt. 43, 8–15. ( 10.5431/aramit4303) [DOI] [Google Scholar]
- 33.Millot J. 1943. Sur l‘anatomie et l'histophysiologie de Koenenia mirabilis Grassi. Rev. Franç. Entomol. 9, 137–145. [Google Scholar]
- 34.Buxton BH. 1913. Coxal glands of the Arachnids. Zool. Jahrb. 14, 16–50. [Google Scholar]
- 35.Ballesteros JA, Santibáñez López CE, Kováč L', Gavish-Regev E, Sharma PP. 2019. Data from: Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi Dryad Digital Repository. ( 10.5061/dryad.t1g1jwsz4) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ballesteros JA, Santibáñez López CE, Kováč L', Gavish-Regev E, Sharma PP. 2019. Data from: Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi Dryad Digital Repository. ( 10.5061/dryad.t1g1jwsz4) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Illumina raw read data: NCBI SRA accession numbers SRR8080645-647. Trinity assemblies, multiple sequence alignments (Matrices 1-7), partition files, phylogenetic trees: Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.t1g1jwsz4 [35].