Supplemental Digital Content is available in the text.
Keywords: BRAF, KIT, Melanoma, meta-analysis, mutation, NRAS
Abstract
The presence of mutations of BRAF, NRAS, and KIT genes is recognized as playing a role during carcinogenesis. Our study aims to evaluate and review other studies that present the frequency of mutations of BRAF, NRAS, and KIT genes for different populations, and analyse correlation to their clinical-pathological characteristics and to the demographics of melanoma. Thirty-two articles were selected from a collection of published literature studying 6299 patients. The parameters for correlation to different variables were calculated by odds ratio, for random and single effects. 38.5% of patients present BRAF gene mutations, 16.4% in NRAS, and 10% in KIT. Mutations of the BRAF gene were correlated to superficial spreading melanoma (odds ratio = 1.31), localization in the torso (odds ratio = 1.42) and presence of metastases. Mutations in NRAS were correlated to nodular melanoma (odds ratio = 1.57), localized in the limbs (odds ratio = 1.31). Mutations of the KIT gene were correlated to mucosal melanoma (odds ratio = 1.59). Populations in Brazil, the US, Sweden, Italian, and Australia were found to be correlated to mutations of BRAF and melanoma. Populations in Italy, Sweden, Spain, and the US were found to be correlated to mutations of NRAS. Populations in Japan, China, Turkey, Canada, and Russia were found to be correlated to mutations of KIT. Data correlated to the presence of melanoma and population type is due to the amount of studies performed across of globe.
Introduction
In the last few years there has been an increase of melanoma incidence in all populations [1]. Presently, this represents 5.3% of all skin cancers, and approximately 1.5% (9320) of patients die from this disease [2]. In 2020 it is projected that 279 938 new cases will be diagnosed, and it is estimated that around 67 809 will die as a result [3].
The incidence rate of cutaneous melanoma is greater in white population groups compared to Hispanic, African-American, Indo-American, and Asian population groups [4]. The risk of melanoma increases with age and has been described as having a 1.5 incidence rate greater for males than for females [5]. Other risk factors include exposure to ultraviolet radiation, the presence of many nevus, skin colour, hair colour, eye colour, altitude, and latitude. All these factors contribute to a change in the molecular machinery of melanocytes, which transforms a normal cell into a tumourous cell [6].
Signalling pathways altered during carcinogenesis of melanoma include the protein kinase activated by mitogen (MAPK), phosphoinositide 3-kinase (PI3K), tumour suppressor retinoblastoma, and p53 protein pathways [7]. Pathways act either collectively or individually to regulate growth, proliferation, and apoptosis. Prolonged activation of these pathways relates to the proliferation, invasion, metastasis, and survival of melanomas [7].
Mutations of BRAF, NRAS, and KIT genes have been correlated to the development of melanoma. These mutations have been described as being mutually excluding [8,9]. The tyrosine-protein kinase Kit acts as a cell surface-cells receptor, participates in the normal growth of melanocytes and intervenes in the activation of MAPK/PI3K pathways [8]. KIT gene mutations are present in 39% of mucosal melanomas (MM), 36% of acral lentiginous melanomas (ALM), and in 28% of skin with chronic solar damage. The majority of reported mutations are found in exons 9, 11, 13, and 17, and represent between 5 and 10% of mutations of diagnosed melanomas [10,11].
NRAS oncogene is a member of the superfamily of p21 GTPase proteins. These proteins cause intrinsic GTPase activity, functioning as a molecular switch for transmission in the regulator signals. They participate in the activation of MAPK/PI3K pathway, during cellular proliferation and survival [1,12]. The most common NRAS gene mutations are Q61R and Q61K and represent around 25% of mutations for biopsies of patients with melanoma [12].
Another gene implicated in the pathogenesis of melanoma is oncogene BRAF. This gene encodes a serine-threonine protein kinase of the RAF (rapidly accelerated fibrosarcoma) family. This protein plays an important role in the regulation of MAPK pathways signalling, causing changes in cell cycle, differentiation, and apoptosis [13]. The frequencies of BRAF mutation ranged from 20 to 80% [14]. The most common BRAF alteration detected in human cancers includes the V600E mutation, which is located in the activation segment of the kinase. This mutation represents between 60 and 80% of all mutations for this gene [15].
Regardless of the conclusions stated previously, it is not known whether the frequency and correlations to mutations of these genes and melanoma are the same for all populations, or if differences exist for some of the populations analysed. Additionally, how the influence of ethnic variability affects histological and clinical presentation for tumours is not known. For this reason, the aim of revising all studies evaluating the frequency of mutations of BRAF, NRAS, and KIT genes are to determine the correlation of the clinical-pathological characteristics of melanomas, and the demographic characteristics of the analysed populations.
Materials and methods
Criteria for eligibility and data recollection
The following databases were consulted: Medline, accessed through PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), Science Direct (http://www.sciencedirect.com/), and to consult literature from Latin America Scielo (http://www.scielo.cl/) was accessed. The strategy used to search for data was a combination of the words ‘melanoma AND mutations OR mutation OR mutat* AND BRAF OR B-RAF AND NRAS OR N-RAS AND C-KIT OR KIT’, ‘melanoma AND mutations OR mutation OR mutat* AND BRAF OR B-RAF’, ‘melanoma AND mutations OR mutation OR mutat* AND NRAS OR N-RAS’, ‘melanoma AND mutations OR mutation OR mutat* AND C-KIT OR KIT’. Studies included those published between January 1st 2005 and November 30th 2017, both in English and Spanish, as well as observational studies (Supplemental digital content 1, http://links.lww.com/MR/A157 lists the strategy for all queries in database search).
Exclusion and inclusion criteria
The studies revised met the following criteria: (1) Studies published between 2005 and 2017. (2) Studies written in English or Spanish. (3) Studies with the frequency of BRAF or B-RAF, NRAS or N-RAS and KIT or C-KIT gene mutations. (4) Studies reporting types and subtypes of melanoma. (5) Studies describing the population where mutations were analysed.
The exclusion criteria were: publication before 2005, duplicate articles, case reports, languages other than Spanish and English, letters to the editor, reviews, studies in cell lines, studies that did not include clinical data, or did not report the location of the analysed population, abstracts, clinical studies or clinical trials and titles. Additionally, studies that reported double findings for the same population were excluded according to the hospital or reference center, date of publication, group of authors, country and bibliographical reference, or for data published previously (Fig. 1).
Fig. 1.
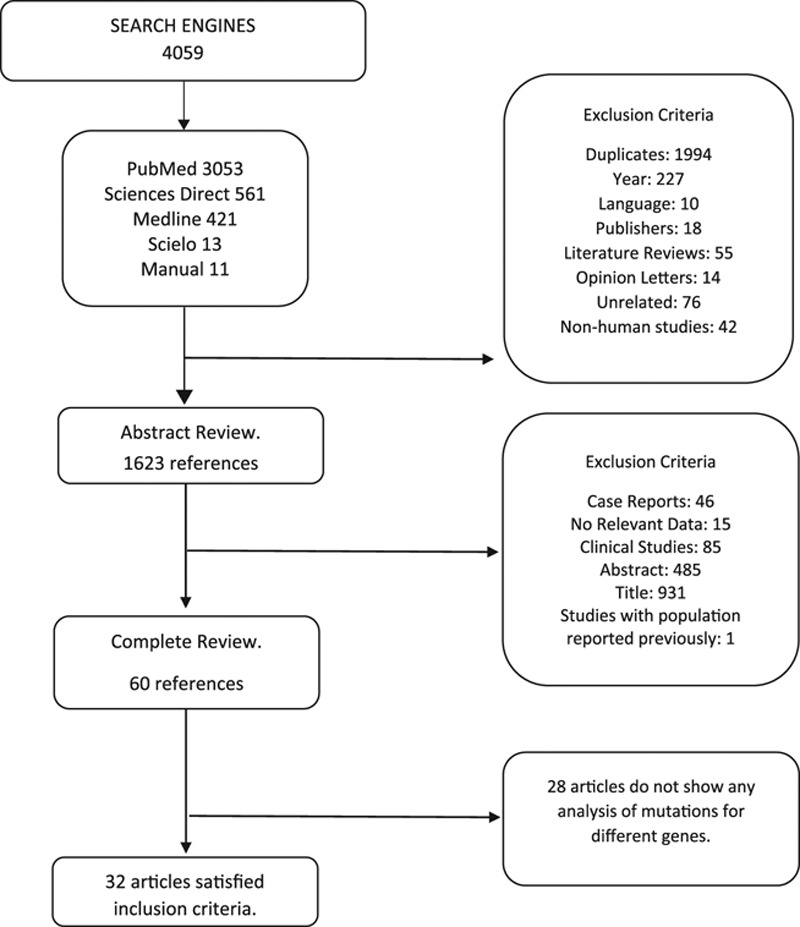
Flow diagram illustrating how studies were selected, inclusion and exclusion criteria.
Data extraction
Two reviewers independently evaluated published literature in order to preserve homogeneity and to eliminate subjectivity for the extraction of data. In the case of disagreement, the issue was resolved through a group discussion involving both reviewers. From each selected study the following information was obtained: name of the article, original author, year of publication, melanoma subtype, number of patients, country of origin of analysed population, molecular analysis and sequencing method, anatomical location of melanoma, mutations in BRAF, NRAS, and KIT gene, sex, age, chronic solar exposure, metastasis, and ulceration.
Statistical analysis
Multiple regressions were used to review the influence of the variables extracted from each one of these studies. A review of the correlation to mutations and the place of origin of the patient was proposed using the odds ratio (OR) with a confidence interval (CI) of 95%. These calculations were performed using Open Meta-Analyst software free version. For fixed effects, the Mantel–Haenszel method was used. The choice of the method of analysis was taken according to Q value in order to observe the heterogeneity between analysed studies, following the chi-square distribution with k − 1 degrees of freedom for homogenous studies. The inconsistency of the studies was quantified with the I2 statistic, that allowed us to obtain a total estimation of the variability or inconsistency of analysed studies and the value that oscillates between 0 and 100%. In terms of heterogeneity, the meta-analysis was performed using the Binary Random-Effects model, and the Random-Effects (DerSimonian-Laird) method of meta-analysis for random effects. In order to determine the bias for publication related to negative results, the comprehensive meta-analysis V.13 (free version) software was used since they are cited less frequently, thus, making them scarcer in database searches. A funnel plot analysis was performed and presented asymmetry parameter values of P = <0.05, which indicated presence of bias.
Results
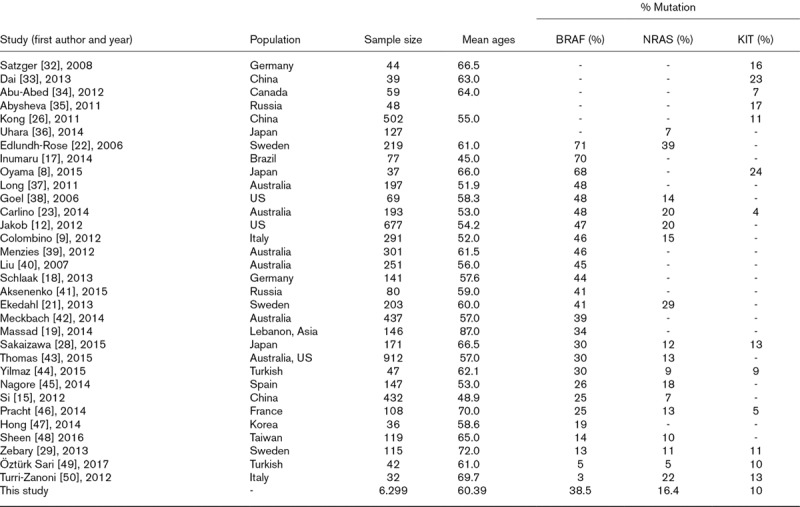
From the review of the database search, a total of 4059 articles were included. Once verified, 61 articles were considered relevant and were reviewed in their entirety. Thirty-two articles, studying 6299 patients, satisfied the inclusion criteria for clinical-pathological and demographic variable analysis (Table 1). Of the 32 articles, 26 reported mutations of the BRAF gene, 17 of the NRAS gene, and 13 of the KIT gene. The number of patients were 5480, 3904, and 1437 for analysis of mutations of BRAF, NRAS, and KIT genes, respectively. The frequency of BRAF mutations gene was 38.5% (2113), 16.4% (641) for NRAS, and 10% (150) for KIT.
Table 1.
Frequency of mutations of BRAF, NRAS, and KIT genes for studies analysed, sample size, and analysed population type
For frequency of mutations of the BRAF gene, 27 articles were reported according to sex, 15 for NRAS gene, and 8 for KIT gene. 43.6, 39, and 56% of patients who tested positive for BRAF, NRAS, and KIT gene are female, respectively. Regarding mutations in males and females for these genes, there was no correlation found between them.
A subsequent analysis was performed for the mutations reported of the BRAF gene and their relation to the histological subtype, for 25 of the 32 studies included for review. Of the 2075 patients who tested positive for BRAF mutation, 886 (42.7%) presented superficial spreading melanoma (SSM) histological subtype (OR = 1. 31, 95% CI 1.15–1.49, P < 0.001) (Fig. 2a). On the contrary, for nodular melanomas (NM), lentigo malignant melanoma (LMM), ALM, uveal melanoma (UM), MM, primary melanoma, and undifferentiated melanoma there was no relation found to BRAF gene mutations and histological subtypes.
Fig. 2.
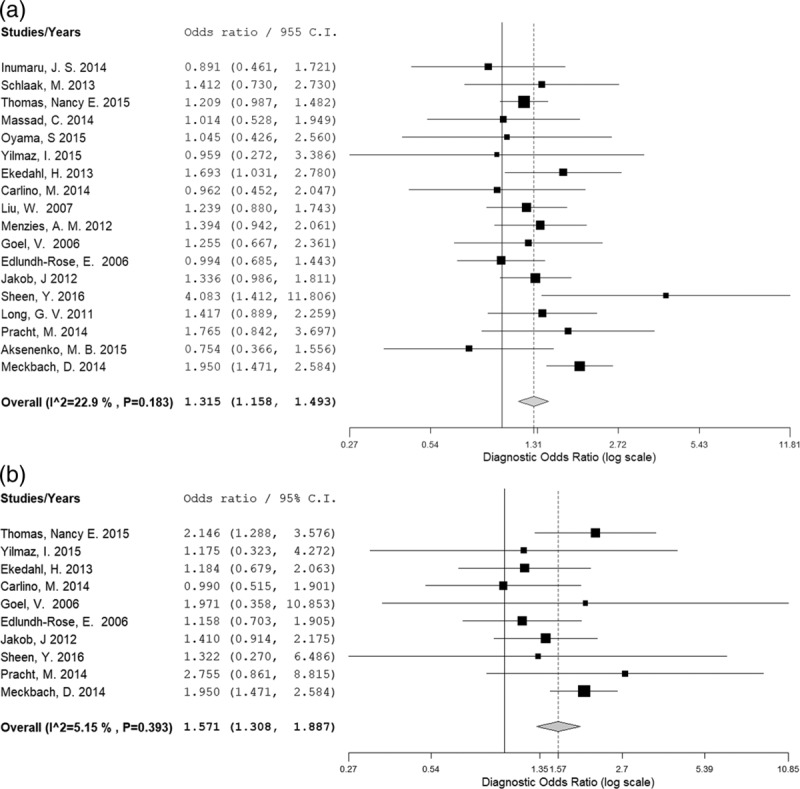
(a) Correlation to BRAF gene mutations and superficial spreading melanoma with an odds ratio value corresponding to a CI of 95% for studies performed. (b) Odds ratio with a CI of 95% for correlation to NRAS mutations and nodular melanoma. CI, confidence interval.
An analysis of mutations in the NRAS gene and the histological characteristics of melanoma were reported in 17 articles. Of the 641 patients who presented mutations in NRAS gene, 21.1% (135) presented NM, and 19.1% (127) presented the histological subtype of undifferentiated melanoma. A statistical analysis found a correlation to mutations of NRAS gene and nodular subtype (OR = 1.57, 95% CI 1.30–1.88, P < 0.001) (Fig. 2b), and with undifferentiated (OR = 1.35, 95% CI 1.06–1.71). An analysis of the mutations of the KIT gene and their relation to different melanoma subtypes was not found to be correlated to histological subtypes. With ALM, an analysis of fixed effects was performed according to the homogeneity of collected data (OR = 1.45, 95% CI 0.89–2.11, Q = 7.97, df = 6, P = 0.240, I2 = 24.74).
The correlation of mutations and anatomical location of melanoma was also analysed. Thirty-four percent (674/1978) of patients with melanoma were reported to have presented a correlation to melanomas located in the torso with mutation of the BRAF gene (OR = 1.41, 95% CI 1.21–1.63, P< 0.001) (Fig. 3). The correlation between mutations of the NRAS gene and the location of melanoma in the limbs was significant. 34.7% of patients (212/610) had mutations in limbs (OR = 1.31, 95% CI 1.01–1.70, P = 0,036). For anatomical locations in head, neck, torso, mucosa, feet, and hands no correlation was found for mutations of NRAS gene.
Fig. 3.
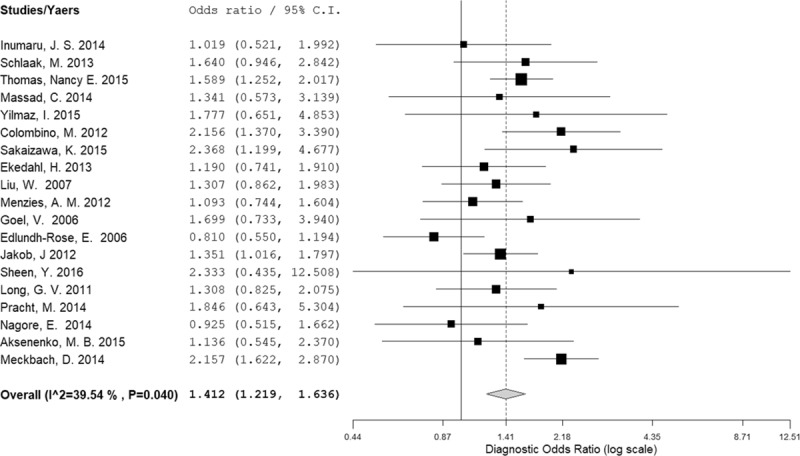
Correlation to BRAF mutations and anatomical localization of melanoma (torso).
Thirteen of all studies analysed reported the melanoma localization and the presence of KIT gene mutations. A correlation was shown for localization in mucosa and presence of KIT gene mutations (OR = 1.59, 95% CI 1.10–2.31, P < 0.013, Q = 12.4, df = 8).
Five of the 26 studies, that analysed BRAF gene mutations report the relation to metastatic sites. The presence of mutations in BRAF gene, and their relationship to metastases is significant (OR = 1.95, 95% CI 1.13–3.35, Q = 18.8, df = 5, P = 0.016 and I2 = 78), but for mutations on NRAS and KIT gene this relationship was not found.
Correlated risk factors
There was no correlation found to chronic solar exposure and presence of mutations in BRAF (OR = 0.941, 95% CI 0.729–1.21, Q = 8.391, df = 9, P = 0.495 and I2 = 0), or NRAS (OR = 0.362, 95% CI 0.06–1.97, P = 0.24), and KIT (OR = 1.00, 95% CI 0.323–3.12, P = 0.99). A correlation to ulceration presence and NRAS gene mutations was found (OR = 2.90, 95% CI 1.24–6.73, P = 0.003). Such a factor is not found to be correlated to the presence of BRAF gene mutations, nor for KIT gene mutations.
Patient origin country
One aspect that has not been found to be related to the presence of mutations in the population’s origin country. According to this statement, an analysis was performed to determine if a relationship exists between mutations of BRAF, NRAS, and KIT genes, and population type according to country of origin. No global relationship was found for the studied populations and the presence of BRAF mutations (OR = 1.04, 95% CI 0.99–1.10, P < 0.090). Additionally, a significant statistic was found for the heterogeneity of the collected data in the analysed articles (Q = 209.42, df = 12, P < 0.001). Given the observation of bias, an independent analysis was performed for each population.
The correlation to population and the presence of BRAF gene mutations was found by independent analysis; in Brazil (OR = 2.91, 95% CI 1.47–2.97), Australia (OR = 1.13, 95% CI 1.02–1.25), Italy (OR = 1.25, 95% CI 1.02–1.54), US (OR = 1.33, 95% CI 1.18–1.49), and Sweden (OR = 1.41, 95% CI 1.20–1.65). Populations analysed from other sources did not demonstrate such correlations.
The global relation between the analysed population and the presence of mutations in NRAS gene was shown to be correlated (OR = 1.34, 95% CI 1.23–1.47, P < 0.001). The analysis for the presence of mutations of this gene, and the specific population, was shown to be significant for Italy (OR = 1.52, 95% CI 1.11–2.07), Sweden (OR = 2.87, 95% CI 2.36–3.49), Spain (OR = 1.80, 95% CI 1.18–2.74), and the US (OR = 1.89, 95% CI 1.61–2.23). The analysis of this relation to the global population and the presence of mutations in KIT gene was shown to be correlated as well (OR = 2.08, 95% CI 1. 73–2.49, P < 0.001). The populations in which the greatest correlation was found are China (OR = 2.71, 95% CI 2.01–3.67), Japan (OR = 3.88, 95% CI 2.60–5.80), Turkey (OR = 3.77, 95% CI 1.79–7.92), Canada (OR = 2.84, 95% CI 1.02–7.93), and Russia (OR = 2.62, 95% CI 1.26–5.45) (Fig. 4a).
Fig. 4.
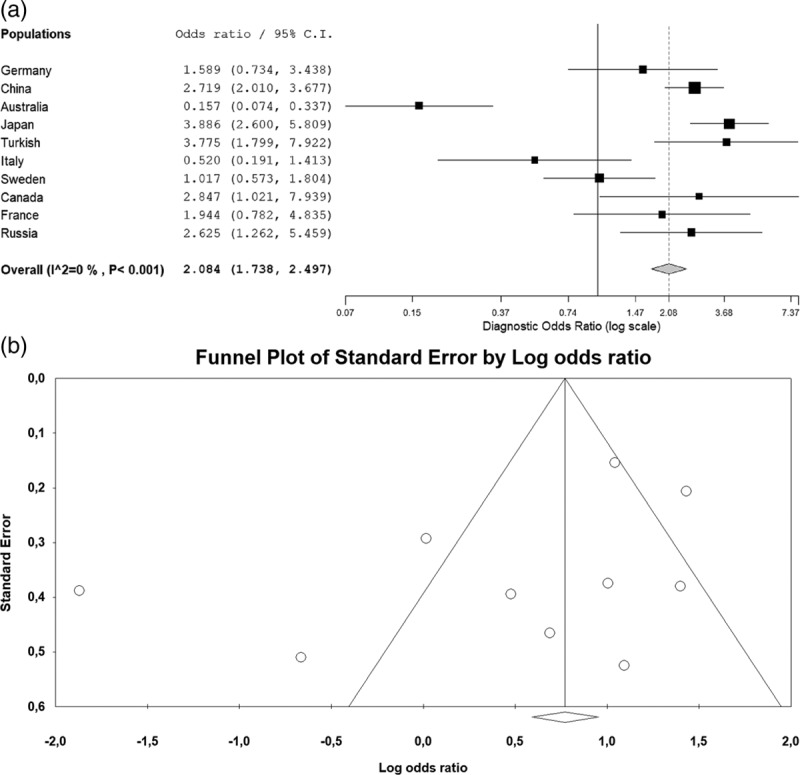
(a) Correlation to KIT gene mutations and population type. (b) Funnel plot of KIT gene mutations and population type.
Funnel plot analysis demonstrates no bias in published studies which analyse mutations in BRAF, NRAS, and KIT genes for patients with melanoma, according to their sex, melanoma subtype, localization of melanoma, chronic solar exposure, and ulceration. Nevertheless, a published bias is present in the analysis of genes, the presence of metastasis, and population type (Fig. 4b).
Discussion
Melanoma has been understood according to the identification of mutations and genetic changes, thus, allowing for the development of a more effective approach to the disease, especially in advanced cases. The knowledge of the frequencies of mutations in the BRAF, KIT, and NRAS genes seen along the study in the different geographical regions with the pathological and clinical characteristics, is important for treatment planning and the sifting of evidence.
For the present meta-analysis, correlations to patients with BRAF, NRAS, and KIT mutations, clinical-pathological characteristics and risk factors correlated to the development of melanoma were grouped together. Such information may help to guide development of therapeutic strategies and also, aid in the identification of specific risk factors for each population.
Various population studies demonstrate correlation to clinical-pathological characteristics, histological subtypes, and presentation of BRAF gene mutations; nevertheless, these results are not conclusive and vary according to the population and samples analysed. Different studies have reported that BRAF mutations are more frequent in skin that has been exposed to sunlight intermittently, which is correlated to a melanoma subtype with surface spreading and localization in the torso [16]; nonetheless, we do not find any correlation between the presence of BRAF mutations and solar exposure. Likewise, some studies performed on populations from Italy (Libra et al. [13]) and Brazil (Inumaru et al. [17]) found that the presence of the V600 mutation, of BRAF gene, is not correlated to factors such as sex, melanoma type, chronic solar exposure, presence of metastases, and ulceration. This differs from a study performed on a German population where a correlation to the presence of BRAF mutations and localization in the torso was found (<0.0001) [18]. Another study of 146 patients in Lebanon (Asia) also reported the presence of subcutaneous metastatic melanoma correlated to the presence BRAF mutations [19]. In this meta-analysis, we found a correlation to the presence of BRAF mutations for the histological subtype of SSM with localization in the torso and the presence of metastases, but not for solar exposure.
BRAF gene mutations are considered as driver mutations due to their presence in nevus, correlated to the development of melanoma and PMs and metastasic outgrowths [20]. In the present meta-analysis, a correlation to the presence of BRAF mutations and the presence of metastatic melanomas was found. Edlundh-Rose et al. and Ekedahl et al. found the presence of mutations for this gene in PM and metastases in samples from such patients [21,22]. Conversely, Carlino et al. [23] reported no correlation to the presence of BRAF and NRAS gene mutations and the progression of metastatic disease.
The presence of mutations of NRAS, and KIT genes has also been correlated to the subtype and localization of melanoma. Devitt et al. in Australian patients, found a correlation to NRAS mutations and the histological subtype of nodular melanoma (P = <0.001) [24]. Similarly, Jakob et al. [12] in 2012 studied 677 samples of the American population and reported a correlation to anatomical localization (P = <0.001) and to subtypes with a greater degree of correlation to PM. As would be expected, the data analysed in this study demonstrated a correlation to the presence of mutations and localization of melanoma in lower and upper limbs and NM subtype.
The mutations of the KIT gene are found with greater frequency for mucosal melanoma (30%), unlike for BRAF and NRAS genes. The mutations of this gene are heterogeneous and are found along the length of exon 11. Additionally, these mutations occur independently from mutations of BRAF and NRAS genes, indicating that the pathogenesis of cutaneous melanoma and mucosal melanoma occurs differently [25].
In cutaneous melanoma, the mutation of KIT has been found more frequently for ALM [26]. This melanoma subtype is prevalent for Asian populations and in Latin America. Despite this, we have not found studies in Latin American populations that allow us to verify the correlation between KIT and ALM mutations. Furthermore, KIT mutation has been related to chronic solar damage [27], and yet our study could not affirm said correlation (OR = 1.0, CI 0.3–3.1); such a result is possible due to population diversity and the frequency of histological subtype in the studies analysed. According to what was reported by Sakaizawa et al. [28] in Japanese patients where the correlation to melanoma type and anatomical localization was analysed (P = 0.554). Correspondingly, Zebary et al. [29] in 2013 did not observe any correlation between melanoma type, ulceration, clinical and histopathological characteristics for patients analysed with ALM (P = 0.039) for KIT mutations. In this meta-analysis, the correlation to clinical-pathological characteristics, for patients with melanoma diagnosis, was shown.
As was previously stated, several studies have identified the correlation to BRAF, NRAS, and KIT gene mutations, for different clinical-pathological characteristics and clinical risk factors. In this meta-analysis, such an existing correlation is confirmed independently of the period of time that has been measured. Although the incidence of melanoma continues to increase heterogeneously across the globe [30]. For example, Lee et al. [31], performed a meta-analysis in 2010, including 36 published articles between 1989 and 2010 where a correlation was conclusively found to SSM and NM and the presence of BRAF and N-RAS mutations. We found this correlation in published studies between 2005 and 2017. Additionally, we found a correlation to ulceration and the presence of mutations in NRAS.
On the other hand, for isolated populations various studies demonstrate the analysis of mutations and their correlation to melanoma. Consequently, it has not demonstrated whether the correlation to BRAF, NRAS, and KIT mutations gene is related to population type, as well as for the geographical characteristics of the region where the population resides. The meta-analysis presented a correlation to the presence of BRAF gene mutations for patients with melanoma and the populations of Brazil, Australia, Italy, US, and Sweden.
Hence, the presence of mutations of the NRAS gene and populations from Italy, Sweden, Spain, and the US is found to be correlated. This coincides with the greater frequency of melanoma subtype of SSM and NM for these populations and with the frequency of mutations for these two genes and subtypes, compared to the MM and ALM. However, a correlation is found for different geographical regions when the presence of KIT gene mutations are analysed. The greatest correlations were found in the populations of Japan followed by Turkey, Canada, China, and Russia, respectively. This indicates that it is possible that a correlation found by this study, for the region analysed, is due to the greater frequency of mutations for histological subtypes. These findings allow us to generate strategies for prevention and treatment, based in a molecular point of view, for populations that are at risk of developing melanoma.
The difference in the frequency of mutations of BRAF, NRAS, and KIT genes for different populations reflects existing variability. It may be more adequate, in order to homogenize the study of melanoma, to collect information on a region by region basis. In this study, correlation is only analysed for populations from certain countries and melanoma subtypes when such mutations are found.
Our study has limitations. This is a systematic review and meta-analysis of the literature of reports for frequency mutations of BRAF, NRAS, and KIT genes around the world and is more likely to be compromised by selection and enrichment bias of patients from a specific region of a country; it being the case that patient selection in the majority of studies is performed at reference centers. The combined estimate may be overestimating the frequency of mutations and therefore, may not be representing differences for separate regions in one country. Additionally, the analysis of the frequency of mutations and their correlation to the analysed population represents published bias due to the variability of incidence for disease. The majority of studies performed occur in White populations with high rates of incidence for some melanoma subtypes.
Acknowledgements
We acknowledge the Hospital Universitario–Centro Dermatológico Federico Lleras Acosta, E.S.E.
This study was supported by the Hospital Universitario—Centro Dermatológico Federico Lleras Acosta, E.S.E.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Footnotes
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.melanomaresearch.com.
References
- 1.Apalla Z, Nashan D, Weller RB, Castellsagué X. Skin cancer: epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol Ther (Heidelb). 2017; 7:5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Stat Facts: Melanoma of the Skin. Estados Unidos: National Cancer Institute. https://seer.cancer.gov/statfacts/html/melan.html. [Accessed 26 October 2018] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 4.Cormier JN, Xing Y, Ding M, Lee JE, Mansfield PF, Gershenwald JE, et al. Ethnic differences among patients with cutaneous melanoma. Arch Intern Med. 2006; 166:1907–1914 [DOI] [PubMed] [Google Scholar]
- 5.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014; 28:1005–1011 [PubMed] [Google Scholar]
- 6.Fabbrocini G, Triassi M, Mauriello MC, Torre G, Annunziata MC, De Vita V, et al. Epidemiology of skin cancer: role of some environmental factors. Cancers (Basel). 2010; 2:1980–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia J, Jia P, Hutchinson KE, Dahlman KB, Johnson D, Sosman J, et al. A meta-analysis of somatic mutations from next generation sequencing of 241 melanomas: a road map for the study of genes with potential clinical relevance. Mol Cancer Ther. 2014; 13:1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyama S, Funasaka Y, Watanabe A, Takizawa T, Kawana S, Saeki H. BRAF, KIT and NRAS mutations and expression of c-KIT, phosphorylated extracellular signal-regulated kinase and phosphorylated AKT in japanese melanoma patients. J Dermatol. 2015; 42:477–484 [DOI] [PubMed] [Google Scholar]
- 9.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012; 30:2522–2529 [DOI] [PubMed] [Google Scholar]
- 10.Rivera RS, Nagatsuka H, Gunduz M, Cengiz B, Gunduz E, Siar CH, et al. C-kit protein expression correlated with activating mutations in KIT gene in oral mucosal melanoma. Virchows Arch. 2008; 452:27–32 [DOI] [PubMed] [Google Scholar]
- 11.Montone KT, van Belle P, Elenitsas R, Elder DE. Proto-oncogene c-kit expression in malignant melanoma: protein loss with tumor progression. Mod Pathol. 1997; 10:939–944 [PubMed] [Google Scholar]
- 12.Jakob JA, Bassett RL, Jr, Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012; 118:4014–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libra M, Malaponte G, Navolanic PM, Gangemi P, Bevelacqua V, Proietti L, et al. Analysis of BRAF mutation in primary and metastatic melanoma. Cell Cycle. 2005; 4:1382–1384 [DOI] [PubMed] [Google Scholar]
- 14.Platz A, Egyhazi S, Ringborg U, Hansson J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008; 1:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Si L, Kong Y, Xu X, Flaherty KT, Sheng X, Cui C, et al. Prevalence of BRAF V600E mutation in chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer. 2012; 48:94–100 [DOI] [PubMed] [Google Scholar]
- 16.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012; 12:349–361 [DOI] [PubMed] [Google Scholar]
- 17.Inumaru JS, Gordo KI, Fraga Junior AC, Silva AM, Leal CB, Ayres FM, et al. Analysis of the BRAF V600E mutation in primary cutaneous melanoma. Genet Mol Res. 2014; 13:2840–2848 [DOI] [PubMed] [Google Scholar]
- 18.Schlaak M, Bajah A, Podewski T, Kreuzberg N, von Bartenwerffer W, Wardelmann E, et al. Assessment of clinical parameters associated with mutational status in metastatic malignant melanoma: a single-centre investigation of 141 patients. Br J Dermatol. 2013; 168:708–716 [DOI] [PubMed] [Google Scholar]
- 19.Massad C, Loya A, Taraif S, Saroufim M, Kibbi AG, Habib R, et al. BRAF mutation status in primary and metastatic melanomas in two regions with differing potential ultraviolet radiation exposure. Clin Exp Dermatol. 2014; 39:932–943 [DOI] [PubMed] [Google Scholar]
- 20.Mehnert JM, Kluger HM. Driver mutations in melanoma: lessons learned from bench-to-bedside studies. Curr Oncol Rep. 2012; 14:449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekedahl H, Cirenajwis H, Harbst K, Carneiro A, Nielsen K, Olsson H, et al. The clinical significance of BRAF and NRAS mutations in a clinic-based metastatic melanoma cohort. Br J Dermatol. 2013; 169:1049–1055 [DOI] [PubMed] [Google Scholar]
- 22.Edlundh-Rose E, Egyházi S, Omholt K, Månsson-Brahme E, Platz A, Hansson J, Lundeberg J. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006; 16:471–478 [DOI] [PubMed] [Google Scholar]
- 23.Carlino MS, Haydu LE, Kakavand H, Menzies AM, Hamilton AL, Yu B, et al. Correlation of BRAF and NRAS mutation status with outcome, site of distant metastasis and response to chemotherapy in metastatic melanoma. Br J Cancer. 2014; 111:292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. 2011; 24:666–672 [DOI] [PubMed] [Google Scholar]
- 25.Slipicevic A, Herlyn M. KIT in melanoma: many shades of gray. J Invest Dermatol. 2015; 135:337–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y, Si L, Zhu Y, Xu X, Corless CL, Flaherty KT, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res. 2011; 17:1684–1691 [DOI] [PubMed] [Google Scholar]
- 27.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006; 24:4340–4346 [DOI] [PubMed] [Google Scholar]
- 28.Sakaizawa K, Ashida A, Uchiyama A, Ito T, Fujisawa Y, Ogata D, et al. Clinical characteristics associated with BRAF, NRAS and KIT mutations in japanese melanoma patients. J Dermatol Sci. 2015; 80:33–37 [DOI] [PubMed] [Google Scholar]
- 29.Zebary A, Omholt K, Vassilaki I, Höiom V, Lindén D, Viberg L, et al. KIT, NRAS, BRAF and PTEN mutations in a sample of swedish patients with acral lentiginous melanoma. J Dermatol Sci. 2013; 72:284–289 [DOI] [PubMed] [Google Scholar]
- 30.Dimitriou F, Krattinger R, Ramelyte E, Barysch MJ, Micaletto S, Dummer R, Goldinger SM. The world of melanoma: epidemiologic, genetic, and anatomic differences of melanoma across the globe. Curr Oncol Rep. 2018; 20:87. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011; 164:776–784 [DOI] [PubMed] [Google Scholar]
- 32.Satzger I, Schaefer T, Kuettler U, Broecker V, Voelker B, Ostertag H, et al. Analysis of c-KIT expression and KIT gene mutation in human mucosal melanomas. Br J Cancer. 2008; 99:2065–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai B, Cai X, Kong YY, Yang F, Shen XX, Wang LW, Kong JC. Analysis of KIT expression and gene mutation in human acral melanoma: with a comparison between primary tumors and corresponding metastases/recurrences. Hum Pathol. 2013; 44:1472–1478 [DOI] [PubMed] [Google Scholar]
- 34.Abu-Abed S, Pennell N, Petrella T, Wright F, Seth A, Hanna W. KIT gene mutations and patterns of protein expression in mucosal and acral melanoma. J Cutan Med Surg. 2012; 16:135–142 [DOI] [PubMed] [Google Scholar]
- 35.Abysheva SN, Iyevleva AG, Efimova NV, Mokhina YB, Sabirova FA, Ivantsov AO, et al. KIT mutations in russian patients with mucosal melanoma. Melanoma Res. 2011; 21:555–559 [DOI] [PubMed] [Google Scholar]
- 36.Uhara H, Ashida A, Koga H, Ogawa E, Uchiyama A, Uchiyama R, et al. NRAS mutations in primary and metastatic melanomas of Japanese patients. Int J Clin Oncol. 2014; 19:544–548 [DOI] [PubMed] [Google Scholar]
- 37.Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011; 29:1239–1246 [DOI] [PubMed] [Google Scholar]
- 38.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006; 126:154–160 [DOI] [PubMed] [Google Scholar]
- 39.Menzies AM, Haydu LE, Visintin L, Carlino MS, Howle JR, Thompson JF, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012; 18:3242–3249 [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Kelly JW, Trivett M, Murray WK, Dowling JP, Wolfe R, et al. Distinct clinical and pathological features are associated with the BRAF(T1799A(V600E)) mutation in primary melanoma. J Invest Dermatol. 2007; 127:900–905 [DOI] [PubMed] [Google Scholar]
- 41.Aksenenko MB, Kirichenko AK, Ruksha TG. Russian study of morphological prognostic factors characterization in BRAF-mutant cutaneous melanoma. Pathol Res Pract. 2015; 211:521–527 [DOI] [PubMed] [Google Scholar]
- 42.Meckbach D, Bauer J, Pflugfelder A, Meier F, Busch C, Eigentler TK, et al. Survival according to BRAF-V600 tumor mutations–an analysis of 437 patients with primary melanoma. PLoS One. 2014; 9:e86194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas NE, Edmiston SN, Alexander A, Groben PA, Parrish E, Kricker A, et al. ; GEM Study Group. Association between NRAS and BRAF mutational status and melanoma-specific survival among patients with higher-risk primary melanoma. JAMA Oncol. 2015; 1:359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yilmaz I, Gamsizkan M, Kucukodaci Z, Berber U, Demirel D, Haholu A, Narli G. BRAF, KIT, NRAS, GNAQ and GNA11 mutation analysis in cutaneous melanomas in turkish population. Indian J Pathol Microbiol. 2015; 58:279–284 [DOI] [PubMed] [Google Scholar]
- 45.Nagore E, Requena C, Traves V, Guillen C, Hayward NK, Whiteman DC, Hacker E. Prognostic value of BRAF mutations in localized cutaneous melanoma. J Am Acad Dermatol. 2014; 70:858–862.e1 [DOI] [PubMed] [Google Scholar]
- 46.Pracht M, Mogha A, Lespagnol A, Fautrel A, Mouchet N, Le Gall F, et al. Prognostic and predictive values of oncogenic BRAF, NRAS, c-KIT and MITF in cutaneous and mucous melanoma. J Eur Acad Dermatol Venereol. 2015; 29:1530–1538 [DOI] [PubMed] [Google Scholar]
- 47.Hong JW, Lee S, Kim DC, Kim KH, Song KH. Prognostic and clinicopathologic associations of BRAF mutation in primary acral lentiginous melanoma in Korean patients: a preliminary study. Ann Dermatol. 2014; 26:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheen YS, Liao YH, Liau JY, Lin MH, Hsieh YC, Jee SH, Chu CY. Prevalence of BRAF and NRAS mutations in cutaneous melanoma patients in taiwan. J Formos Med Assoc. 2016; 115:121–127 [DOI] [PubMed] [Google Scholar]
- 49.Öztürk Sari Ş, Yilmaz İ, Taşkin OÇ, et al. BRAF, NRAS, KIT, TERT, GNAQ/GNA11 mutation profile analysis of head and neck mucosal melanomas: a study of 42 cases. Pathology. 2016; 49:55–61 [DOI] [PubMed] [Google Scholar]
- 50.Turri-Zanoni M, Medicina D, Lombardi D, Ungari M, Balzarini P, Rossini C, et al. Sinonasal mucosal melanoma: molecular profile and therapeutic implications from a series of 32 cases. Head Neck. 2013; 35:1066–1077 [DOI] [PubMed] [Google Scholar]


