Abstract
Abstract
Genetic variation and genotype of Hepatitis B virus (HBV) are related to the efficiency of interferon alpha (IFN-α)-based antiviral therapy. However, the correlation of variation in interferon-stimulated response element (ISRE) and HBV genotype response to IFN-α therapy remains elusive.
Differences of ISRE between genotype B and C HBV were explored using the HBV sequences retrieved from GenBank, and further investigated by ISRE region cloning and sequencing from 60 clinical samples post-IFN-α therapy. Additionally, ISRE mutants were constructed and their relation to responsiveness of IFN-α was evaluated by real-time PCR and Southern blot analysis.
ISRE pattern between genotype B and C were found based on both clinical sample sequencing and full-length sequence alignment. The primary difference is the fourth base within the ISRE region, with T and C for genotype B and C, respectively. HBV with genotype C-type ISRE had a higher replicative capability as compared to HBV with genotype B-type ISRE after IFN-α treatment in huh7 cells.
Conclusion:
Preference of ISRE between genotype B and C HBV are distinct. Single nucleotide difference (C to T) within the HBV ISRE region may link to the efficacy of IFN-α therapy to genotype B and C HBV. Therefore, this study provides a clue for the determination of IFN-α therapy response to HBV treatment.
Keywords: hepatitis B virus, IFN-α therapy, interferon-stimulated response element, mutation
1. Introduction
Hepatitis B virus (HBV) is the causative agent of hepatitis B. More than 350 million people are chronically infected worldwide.[1,2] The genome of HBV is an enveloped partially double-stranded circular DNA molecule with 3.2 kb in length, encoding 4 overlapping genes (P, C, S, and X). Viral variation is an important characteristic of HBV, due to the lack of proofreading activity of polymerase during replication.[3] Genetic variations of HBV have been declared to be related to the severity of the liver disease, clinical outcomes, and efficacy of antiviral therapy.[1,2]
Interferon alpha (IFN-α) is the first approved and generally used drug for HBV infection, but its efficacy is far less than satisfactory. About one-third of HBeAg-positive CHB patients achieved HBeAg seroconversion after the IFN-α therapy,[4] and less efficacy was observed in HBeAg negative patients.[4] The efficacy was further compromised by accompanying side-effects. Lots of reports have been addressing on efficacy-related factors.[5–9] Among them, the virus itself is a focus. Data showed that genotype A and B are associated with a significantly higher sustained response to IFN-α than genotype C and D, respectively.[5,10–12] In vivo, associations of viral variants with the clinical outcome of therapy of pegylated (PEG) -IFN-α were confirmed.[13–16] And mutations within the HBV precore and/or basal core promoter (PC and/or BCP) region were reported to be related to the efficacy of IFN-α antiviral therapy in HBeAg-positive chronic hepatitis B patients.[6]
An IFN-stimulated response element (ISRE) is involved in the process of IFN-α action by transcriptional enhancement of IFN-stimulated genes (ISGs). ISRE has been identified in the HBV enhancer I/X gene promoter region[17] and the functional role of ISRE was not detected in the initial exploration.[18] Recently, by employing in vitro replication models and in vivo infection system, Belloni et al demonstrated that ISRE mediated IFN-α transcriptional repression of HBV transcription. ISRE mutation (TTTCACTTC to CTTTACCTTC) reduced HBV DNA through the reduction of transcription.[19] Recently, Liu reported that ISRE mutations may reflect the response of interferon treatment for hepatitis B patients.[20] However, the association of ISRE mutation to IFN-α efficacy remains elusive and deserves further investigation. In the study, the pattern of ISRE mutation and its association to efficacy were investigated.
2. Materials and methods
2.1. Patients
Sixty patients with chronic HBV infection admitted in the First and Second Affiliated Hospital of Chongqing Medical University between September 2009 and February 2014 were enrolled in this study. The inclusion criteria were as follow:
-
1.
All the patients were adult;
-
2.
Patients were all positive for HBsAg for more than 6 months;
-
3.
Patients were HBsAb negative;
-
4.
Patients had elevated serum median alanine aminotransferase (ALT) of >2 × ULN (Upper limit of normal) but <10 × ULN and detectable HBV DNA of >100 000 copies/ml.
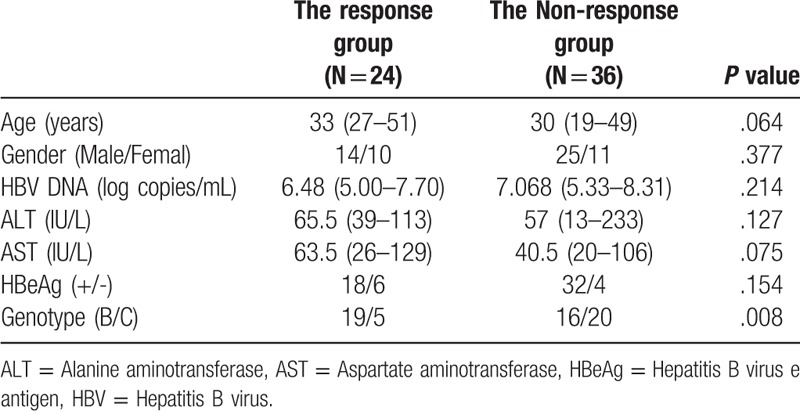
The exclusion criteria were that patients were positive for HAV, HCV, HDV or HIV and exhibited evidence of the preexistence of liver disease. The clinical data of included patients were shown in Table 1. For treatment, all the patients received subcutaneous injection of 5 MU recombinant IFN-α (IFN-α-1b, Sinogen, Kexing Biotech Co. Ltd. Shenzhen, China) 3 times per week for 24 weeks. All patients gave written informed consent and the study was approved by the ethics committee of the university.
Table 1.
Clinical features of hepatitis B patients at baseline.
2.2. Patient grouping and sample collection
The patients were divided into the response group (Rs, n = 24) and the non-response group (NRs, n = 36) according to the follow-up outcomes. HBV DNA and Serum were checked at the initial and at the end of the therapy. In Rs, HBV DNA decreased by less than 1000 copies/ml, the ALT returned to the normal level or HBeAg seroconversion was achieved (loss of HBeAg with or without anti-HBe) at the end of therapy. In NRs, patients did not achieve the criteria mentioned as above.
2.3. DNA extraction and PCR amplification
Virus DNA was extracted from 200 μl of serum using a TIANamp virus DNA kit (Tiangen Biotech Co., Beijing, China) according to the manufacturer's instructions. HBV ISRE region was amplified using primer pairs (forward 5’-GAGTCCCTTTATGCCGCTGT-3’; reverse 5’-GTAAACAAAGGACGTCCCGC-3’), which were synthesized by Beijing Genomics Institute (Beijing, China). PCR was performed using Premix PrimeSTAR HS (Takara, Dalian, China). The amplification reactions consisted of an initial denaturation step of 5 min at 98°C, followed by 35 cycles of 10 s at 98°C, 15 s at 60°C, 40 s at 72°C. The reaction was finalized with a 10 minutes extension at 72°C. The PCR products were sequenced by the Beijing Genomics Institute (Beijing, China). The HBV genotype from each sample was characterized using Genbank HBV genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi).
2.4. Analysis of the pattern of ISRE variation among sequences from GenBank
The complete genome sequences of HBV were downloaded from GenBank using HBV as the search query on May 20, 2018. Only the 3215 bp genotype B and C HBV genome were included in the study. Seven hundred eighty four genotype B and 1287 genotype C HBV were aligned and analyzed with the program Mega Version 6 (Tamura et al. 2013). The HBV ISRE region (nt1089–1098) was extracted and characterized to identify the type of ISRE sequence.
2.5. Plasmid constructs
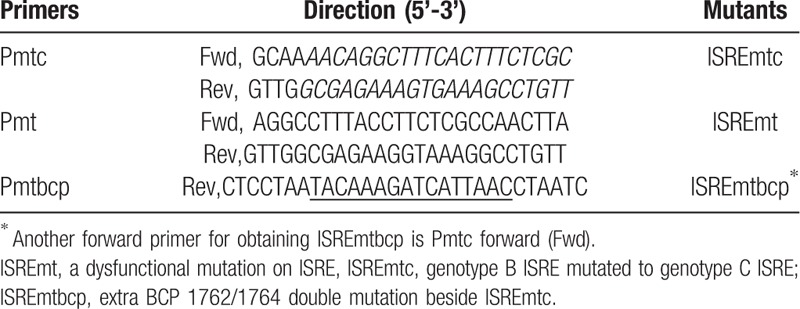
The original expression plasmid (PCH9–1.1x-HBV-B), containing a CMV-early-promoter-driven HBV genotype B full genome and a 0.1× genome fragment from genotype D, was constructed in our lab by a fragment substitution reaction.[21,22] To construct different mutant plasmid, site-directed mutagenesis was used. The target mutant region was firstly amplified by mutant primers (Table 2). Pmtc primer pair was used to generate genotype C ISRE mutation (ISREmtc), Pmtbcp primer pair was used to induce an extra BCP 1762/1764 double mutation beside genotype C ISRE mutation (ISREmtbcp), while Pmt primers were used to induce mutation dysfunctional for ISRE involving in the IFN-α signal cascade (ISREmt). The gel-purified PCR amplicons were used as a metaprimer and the original PCH9–1.1×-HBV-B genotype plasmid as a template. The plasmids with specific mutations were constructed by routine molecular cloning. ISRE regions of constructed plasmids (ISREmtc, ISREmtbcp, ISREmt) were confirmed by sequencing.
Table 2.
Primers for construction of different HBV constructs using PCH9–1.1×-B PCH9 plasmids.
2.6. Transfection of Huh7 Cells with HBV expression plasmid
Huh7 hepatoma cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2 incubator. For transient transfection, Huh7 cells were seeded into 6-well plates 16 to 18 hours before transfection. When the cells were 60% to 70% confluent, the culture medium was replaced with fresh medium and 1.5 μg HBV constructs (PCH9–1.1×-HBV-B, ISREmtc, ISREmtbcp, ISREmt) was transfected into each well using TransIT-LT1 reagent (Mirus Bio, Madison) according to the instructions. After transfection for 24 hours, IFN-α-1b (Sinogen, Shenzhen, China) at a final concentration of 1000 IU/ml was added directly to the culture medium every 24 hours. Four days after transfection, the culture medium was collected and the cells were harvested. All experiments were conducted in triplicate.
2.7. Serological testing of HBsAg and HBeAg by Enzyme-linked immunosorbent assay (ELISA)
The level of HBsAg and HBeAg secreted into the culture supernatants were tested using HBsAg and HBeAg ELISA kits (Ke Hua Co., Ltd., Shanghai) according to the manufacturer's instructions. The absorbance value at the wavelength of 450 nm was read with an Elx800 universal microplate reader (Bio-Tek Instruments, USA). As recommendation, a ratio of S/CO (Signal/cutoff) value ≥2 was considered a positive response for HBsAg or HBeAg. The relative inhibitory rate was calculated using the following formula: inhibitory rate (%) = (S/COtransfectiongroup-S/COIFN-treatedgroup)/S/COtransfectiongroup × 100%. The average inhibitory rate was expressed a mean ± SD.
2.8. Quantification of HBV DNA copies by real-time quantitative PCR (qPCR)
HBV intracellular viral core DNA was extracted 4 days after transfection as previously reported.[22] Briefly, cells were rinsed twice with PBS and lysed with 500 μl of lysis buffer (10 mM Tris–HCl [pH 8.0], 1 mM EDTA, 1% NP-40, and 2% sucrose) at 37°C for 10 minutes. After centrifugation at 15,000 g for 4 minuts, the supernatant was transferred to a fresh microcentrifuge tube and 1 M MgCl2 was added to achieve a final Mg2+ concentration of 10 mM. DNase I (Promega, Oakland, USA) was added to digest the contaminating plasmid at a final concentration of 40U/ml. After 3 hours at 37°C, the reaction was terminated with 10 mM EDTA, and then 200 μl of 35% polyethylene glycol 8000 containing 1.5 M NaCl. After incubation for 1 hour on ice the viral nucleocapsids were pelleted by centrifugation at 13,000 g for 10 minutes at 4°C, and then digested overnight in 400 μl of buffer containing 1 mg/ml proteinase K (Takara, Dalian, China). The digestion mixture was extracted twice with phenol. The DNA was precipitated with ethanol and dissolved in TE buffer (10 mM Tris–HCl [pH 8.0], 1 mM EDTA). The HBV DNA was detected with real-time qPCR with SYBR Green using the PCH9–1.1×-HBV-B at different concentrations (5 × 102, 5 × 103, 5 × 104, 5 × 105, 5 × 106 and 5 × 107 copies/μl) as a template to make the standard curve. The HBV-specific primers used for amplification were as follows: 5’- CCTAGTAGTCAGTTATGTCAAC-3’ (sense) and 5’-TCTATAAGCTGGAGGAGTGCGA-3’ (antisense). The condition of amplification contained initial denaturation at 95°C for 2 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 50°C for 30 seconds and extension at 72°C for 20 seconds.
2.9. Southern blot analysis
HBV replicative intermediates were extracted from the cells as shown above and then separated on 0.8% agarose gels. The DNA samples were transferred to nylon membranes (Roche Diagnostics, Mannheim, Germany). After ultraviolet cross-linking and prehybridization, the membranes were hybridized with a digoxigenin-labeled full-length HBV DNA probe using the DIG DNA Labeling and Detection Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. The signal was detected by exposure on an X-ray film and scanning using the Versa Doc Imaging system (Bio-Rad, Hercules, CA, USA).
The relative inhibitory rate of HBV DNA was calculated using the following formula: inhibitory rate (%) = (S/COtransfectio group-S/COIFN-treatedgroup)/S/COtransfectiongroup × 100%. The average inhibitory rate was expressed as mean±SD. Reduction of HBV DNA levels (Log 10 copies/ml) from baseline in Huh7 cells. Data of groups were compared by one-way ANOVA and further Tukey Least Significant Difference (LSD) to find out which groups differ.
2.10. Statistical analysis
The results of continuous variables were expressed as the median and interquartile ranges. Intergroup differences between the response group and the non-response group were examined by the Chi-Squared test, the Fisher exact test, or Mann–Whitney test where appropriate. HBeAg and HBsAg levels were compared by one-way ANOVA. In all tests, P < .05 was considered statistically significant. All analysis was carried out using GraphPad Prism version 7 for Windows (GraphPad Software Inc., San Diego, CA).
3. Results
3.1. ISRE patterns in HBV sequences from patients’ sera
648-bp region including ISRE was amplified and sequenced for the characterization of ISRE difference among patients. In general, 5 types of ISREs were detected (Table 3). The 2 most common ISRE variants were TTTTACTTTC (37 cases, 61.67%; 16 responders and 21 non-responders) and TTTCACTTTC (19 cases, 31.67%; 4 responders and 15 non-responders). The 83% of ISRE variant TTTTACTTTC sequence was from genotype B and all of ISRE variant TTTCACTTTC sequence was from genotype C. Thus, we named these 2 ISRE variants as genotype B- and C- ISRE, respectively. Other 3 ISRE variants were TTTTTCGTTT (1 case, 1.67%; 1 responder), TTTTACGTTC (2 case, 3.33%; 2 responders), and TTTTACTTTT (1 case, 1.67%; 1 non-responder).
Table 3.
ISRE pattern from sera of patients between response and non-response group∗.

In total, there were 41 cases with the fourth base T in ISRE (37 cases of genotype B ISRE, 2 cases of TTTTACGTTC, 1 case of TTTTACTTTT, and 1 case of TTTTTCGTTT) and 19 cases with forth base C (all 19 cases of genotype C ISRE). In the cases with T-type ISRE, 20 cases were in the response group and 21 cases in the non-response group. While in the cases with C-type ISRE, 4 cases were in response group and 15 cases in the non-response group. Compared to the C-type ISRE patients, the responders within the patients carrying the T-type ISRE showed statistically significant using Chi-Squared test (Chi-Squared value 4.16, P = .041), demonstrating that the fourth base T in ISRE region, can predict the response to IFN treatment. In addition, we performed ANOVA analysis to investigate the relationship of HBV genotype and ISRE pattern alone or together to IFN response. These results revealed that the ISRE pattern is significantly related to the response to interferon treatment (P = .003), but not the genotype (P = .424). And The ISRE pattern and genotype together significantly relate to the response of interferon treatment (P = .003).
3.2. ISRE pattern of genotype B and C HBV from GenBank database
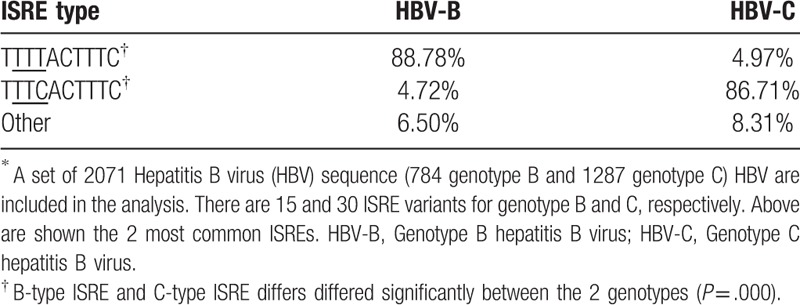
Most cases with T-type ISRE (85.37%, 35 of 41) were from genotype B HBV, while C-type ISRE (73.68%, 14/19) cases were mainly from genotype C HBV based on the above clinical data. Next, the association of ISRE differences and genotype (B and C) was further investigated by bioinformatics analysis used sequences from GeneBank. A set of 2071 HBV full-length sequences consist of 784 genotype B and 1287 genotype C were included in the analysis. There were 15 and 30 ISRE variants for genotype B and genotype C, respectively. The ISRE preference differed significantly between the 2 genotypes (P = .000). 88.78% of the genotype B HBV preferred an ISRE sequence of TTTTACTTTC, while 86.71% of the genotype C HBV was with an ISRE sequence of TTTCACTTTC (Table 4). The ISRE preferences of HBV genotype B and C from published full-length HBV showed the same as the ISRE pattern detected in the study.
Table 4.
ISRE preference of genotype B and C∗.
3.3. Expression and replication of HBV from different ISRE constructs in Huh7 cells
To investigate the relationship of genotype-specific ISRE variants and antiviral response to IFN-α in vitro, 4 constructs were generated based on HBV-replicating pCH-9/3091 plasmid. Three different mutations within the ISRE region were induced into the plasmid (Fig. 1). ISREwtb represented genotype B-ISRE and wildtype BCP region in original pCH-9/3091 plasmid carrying 1.1× genotype B HBV. ISREmt represented the mutation in the ISRE region, which could abolish ISGF3 binding in EMSA assays,[18] but did not alter the HBV polymerase polypeptide sequence. ISREmtc converted genotype B ISRE to genotype C ISRE with point mutation in ISRE region. ISREmtbcp had an additional mutation in BCP region (A1762T/G1764A) except ISRE mutation as ISREmtc. Huh7 cells were transiently transfected with the genotype B pCH-9/3091 construct (ISREwt) and 3 other mutated constructs (ISREmtc, ISREmt, ISREmtbcp).
Figure 1.

Sequence around ISRE and BCP region of HBV 4 constructs used in the study. ISREwtb, wildtype genotype B ISRE; ISREmt, a dysfunctional mutation on ISRE, ISREmtc, genotype B ISRE mutated to genotype C ISRE; ISREmtbcp, extra BCP 1762/1764 double mutation beside ISREmtc.
HBsAg and HBeAg in supernatants were measured 4 days after transfection by ELISA. HBsAg was detected in all the samples transfected with different HBV ISRE constructs, while HBeAg was detected at a rather low level (data not shown). The inhibitory rate of IFN-α to different HBV ISRE constructs was around 50% and 25% for HBsAg and HBeAg, respectively (Fig. 2A and 2B). However, no statistical significance in response to IFN-α treatment between the 4 HBV constructs was shown.
Figure 2.
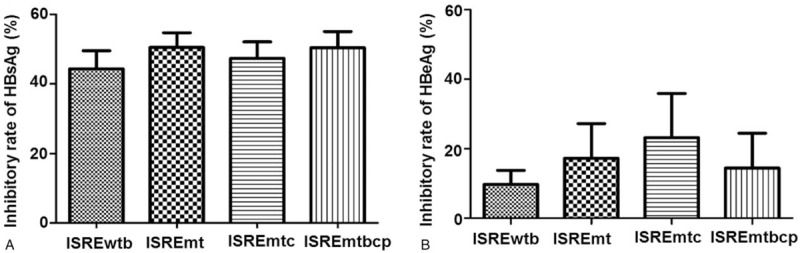
Inhibition on the expression of extracellular HBsAg and HBeAg of different constructs by IFN-α in Huh7 cells after transfection for 72 h. The relative inhibitory rate was calculated as described in methods. The average inhibitory rate was expressed a mean±SD. The inhibitory rate of IFN-α to different constructs was around 50% and 25% for HBsAg (A) and HBeAg (B), respectively. No statistical significance in response to IFN-α treatment was shown.
Intracellular HBV replicative intermediates were measured by qPCR and southern blotting, and the results showed reduced HBV DNA levels in all constructs after IFN-α treatment was given. The inhibitory rate of IFN-α to HBV constructs carrying original genotype B ISRE (ISREwtb) was 73.6 ± 8.15%, while the inhibitory rate of other 3 HBV constructs (ISREmt, ISREmtC, and ISREmtbcp) were 49.67 ± 5.03%, 52.67 ± 7.77%, 51.33 ± 7.57%, respectively. This meant a significant reduction of the inhibitory rate of IFN-α to HBV after mutation within ISRE (P < .05) and the inhibitory rate of BCP mutation beyond ISRE region had no significant difference compared to the other 2 mutations (P > .05). This result indicated BCP 1762/1764 double mutations in the study showed no impact on the HBV response to IFN-α in vitro (Fig. 3A). HBV replication was further quantified by southern blotting analysis (Fig. 3B). Distinct reductions in HBV replicative intermediates were observed in transfected Huh7 cells treated by IFN-α with different HBV plasmids as compared to the IFN-α-treatment naïve group. In contrary with the IFN-α-treatment naïve group, the mutant HBV constructs showed a higher level of replication, indicating a single ISRE mutation at the forth base (from T to C) enhanced the HBV replication. The enhancement of HBV replication could antagonize the inhibitory effect of IFN-α. The inhibition of HBV replication by IFN-α may happen at the transcriptional level, as reported previously.[23,24] The enhancement of HBV replication by ISRE mutation needs further investigation. All the results demonstrate that mutation of ISRE from genotype B-type to genotype C-type results in reduced inhibition of HBV replication by IFN-α and may relate to, at least partially, the response of genotype B and C to IFN-α in the clinical sample.
Figure 3.
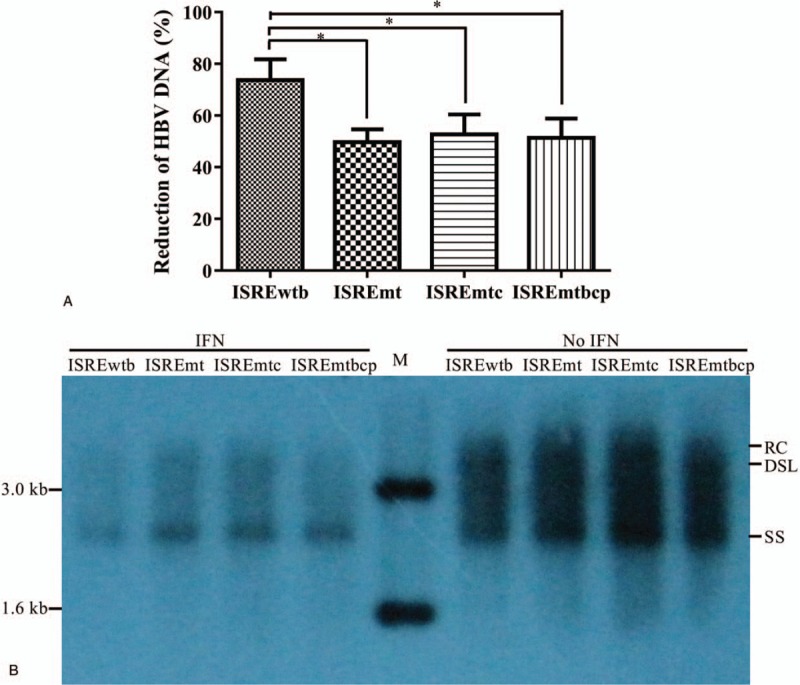
Expression of intracellular HBV replicative intermediates of different plasmid constructs with or without IFN-α treatment (1000 U/ml) in Huh7 cells. Reduction of HBV DNA level (Log10 copies/ml) from baseline in Huh7 cell measured by qPCR (A), and southern blot (B). Results were shown as mean values from 3 independent experiments. ∗ indicates a significant difference, P < .05. RC, relaxed circular HBV DNA; DSL, double strand linear HBV DNA; SS, single strand linear HBV DNA.
4. Discussion
In the study, ISRE variation among HBV genotype B and C was explored, and the association of ISRE variation with antiviral response of IFN-α was further evaluated. Results showed that a single nucleotide variation at the fourth base of ISRE for genotype B and C might explain, at least partially, the different response of IFN-α therapy among patients infected with HBV genotype B and C.
Investigation of the correlation of genetic variation to IFN-α antiviral efficacy is beneficial for us to predict the clinical outcome of antiviral drugs. IFN antiviral response is partially genotype-dependent. The PEG-IFN-α response rate of genotype B is significantly higher than that of genotype C in HBeAg-positive patients in the Asian population, within which genotype B and C HBV are dominating.[1,25] Other variations such as mutations in PreC/BCP region and ISREs are also involved in the difference of the antiviral response. Interestingly, a single mutation within ISRE is reported to relate to the outcome of IFN-α therapy.[26] However, no HBV genotyping information within the patients was offered. ISRE was reported to be associated with IFN-α-mediated suppression of viral gene expression of HBV,[17,19,20,26] probably through affecting its binding stability to ISGF3 complexes, as well as IRF3.[27] To address the effect of ISRE mutation on IFN-α, we mutated the genotype B ISRE and found the first and fourth base mutation of genotype B ISRE (ISREmt) reduced the inhibitory effect of IFN-α on HBV DNA replication in huh7 cells. The result is consistent with previous reports.[26] Further, we changed genotype B ISRE to genotype C ISRE, and found this change reduced the viral replication under the treatment of IFN-α. This pattern coincides with the clinical findings that genotype B HBV has a relatively higher response of IFN-α therapy.[1] Thus, the proportion of genotype B and C in the clinical sample may explain the difference of antiviral response of IFN-α therapy in our sample, as well as the previous report.[20]
Dynamic changes of HBV sequences could affect the outcome of therapy during the IFN-α therapy.[1,4,13] An earlier study concluded that the IFN-α resistance to HBV was related to genotype C with a higher frequency of BCP A1762T/G1764A mutation than genotype B.[28] In vitro, PC and BCP mutations resulted in IFN-α resistance.[29,30] However, clinical data reveal the inconsistency of PC and/or BCP mutations with IFN-α response.[6,11,14,15] In this study, HBV with additional BCP A1762T/G1764A mutation showed no distinct difference in levels of antigen expression and HBV replicative intermediate in vitro. In the study, 1.1× HBV full-length expressing plasmid was employed, while 3.2 kb linear HBV monomers were employed in the previous work.[29] This difference may be an explanatory factor for the inconsistency.
Extensive genetic heterogeneity exists within individual HBV patient. One limitation of our study is that the plasmid represents only a single variation. An unbiased method for overall phenotype achievement might be transfected with all kinds of variants. It is theoretically feasible but unrealistic. Recently developed colony methods might provide an alternative way for HBV population phenotype analysis.[31] Further study on genetic correlates of IFN-α resistance may consider genetic heterogeneity.
Author contributions
Conceptualization: Yanan Guo, He Lu, Jieli Hu, Ailong Huang.
Data curation: Yanan Guo, He Lu, zeng tu.
Formal analysis: Yanan Guo, He Lu.
Funding acquisition: zeng tu.
Investigation: Yanan Guo, Nur Fazleen Binti Idris, Yimin Li, Jieli Hu, Ailong Huang.
Methodology: Yanan Guo, He Lu, Lei Xu, Yimin Li, Ailong Huang, zeng tu.
Project administration: Ailong Huang, zeng tu.
Resources: Ailong Huang.
Supervision: Ailong Huang, zeng tu.
Validation: Ailong Huang, zeng tu.
Writing – original draft: Yanan Guo, He Lu, Lei Xu, Nur Fazleen Binti Idris, zeng tu.
Writing – review & editing: Lei Xu, Nur Fazleen Binti Idris, Jieli Hu, Ailong Huang, zeng tu.
Footnotes
Abbreviations: (PEG-) IFN-α = pegylated-interferon alpha, ALT = alanine aminotransferase, BCP = basal core promoter, DSL = double strand linear HBV DNA, ELISA = enzyme-linked immunosorbent assay, HBeAg = hepatitis B e antigen, HBsAg = hepatitis B virus surface antigen, HBV = hepatitis B virus (HBV), ISGs = IFN-stimulated gene, ISRE = interferon-stimulated response element, ISREmt = dysfunctional mutation on ISRE, ISREmtbcp = extra BCP 1762/1764 double mutation beside ISREmtc, ISREmtc = ISRE mutation to genotype C, IU = international unit, NRs = No response group, PC = precore promoter, qPCR = real-time qualitative PCR, RC = relaxed circular HBV DNA, Rs = response group, S/CO value = signal/cutoff, SS = single strand linear HBV DNA.
How to cite this article: Guo Y, Lu H, Xu L, Idris NF, Li Y, Hu J, Huang A, TU Z. The response of hepatitis B virus genotype to interferon is associated with a mutation in the interferon-stimulated response element. Medicine. 2019;98:51(e18442).
This work was supported in part by Postdoctoral Science Foundation (Grant No. 2013M542264 and Xm2014006), Chongqing Research Program of Basic Research and Frontier Technology (Grant No. cstc2015jcyjA10006 and cstc2014jcyjA10024) and the Natural Science Foundation of China (Grant No. 81501751 and 81201948, 2014.05, 2015.02).
The authors have no conflicts of interest to declare.
References
- [1].Lazarus JV, Block T, Bréchot C, et al. The hepatitis B epidemic and the urgent need for cure preparedness. Nat Rev Gastroenterol Hepatol 2018;15:517. [DOI] [PubMed] [Google Scholar]
- [2].McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology 2009;49(S5):S45–55. [DOI] [PubMed] [Google Scholar]
- [3].Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res 2010;40:14–30. [DOI] [PubMed] [Google Scholar]
- [4].Lok AS, McMahon BJ, Brown RS, et al. Antiviral therapy for chronic hepatitis B viral infection in adults: a systematic review and meta-analysis. Hepatology 2016;63:284–306. [DOI] [PubMed] [Google Scholar]
- [5].Buster EHCJ, Hansen BE, Lau GKK, et al. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-Alfa. Gastroenterology 2009;137:2002–9. [DOI] [PubMed] [Google Scholar]
- [6].Tseng TC, Yu ML, Liu CJ, et al. Effect of host and viral factors on hepatitis B e antigen-positive chronic hepatitis B patients receiving pegylated interferon-alpha-2a therapy. Antivir Ther 2011;16:629–37. [DOI] [PubMed] [Google Scholar]
- [7].Fan HB, Guo YB, Zhu YF, et al. Hepatitis B virus genotype B and high expression of interferon alpha receptor beta subunit are associated with better response to pegylated interferon alpha 2a in Chinese patients with chronic hepatitis B infection. Hepat Mon 2012;12:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Song J, Zhou Y, Li S, et al. Susceptibility of different hepatitis B virus isolates to interferon-alpha in a mouse model based on hydrodynamic injection. PLoS One 2014;9:e90977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Y, Wu Y, Ye S, et al. The response to interferon is influenced by hepatitis B virus genotype in vitro and in vivo. Virus Res 2013;171:65–70. [DOI] [PubMed] [Google Scholar]
- [10].Wai CT, Chu CJ, Hussain M, et al. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology 2002;36:1425–30. [DOI] [PubMed] [Google Scholar]
- [11].Erhardt A, Blondin D, Hauck K, et al. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut 2005;54:1009–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rijckborst V, Hansen BE, Cakaloglu Y, et al. Early on-treatment prediction of response to peginterferon alfa-2a for HBeAg-negative chronic hepatitis B using HBsAg and HBV DNA levels. Hepatology 2010;52:454–61. [DOI] [PubMed] [Google Scholar]
- [13].Jansen L, Welkers MR, van Dort KA, et al. Viral minority variants in the core promoter and precore region identified by deep sequencing are associated with response to peginterferon and adefovir in HBeAg negative chronic hepatitis B patients. Antiviral Res 2017;145:87–95. [DOI] [PubMed] [Google Scholar]
- [14].Yang H-C, Chen C-L, Shen Y-C, et al. Distinct evolution and predictive value of hepatitis B virus precore and basal core promoter mutations in interferon-induced hepatitis B e antigen seroconversion. Hepatology 2013;57:934–43. [DOI] [PubMed] [Google Scholar]
- [15].Sonneveld MJ, Rijckborst V, Zeuzem S, et al. Presence of precore and core promoter mutants limits the probability of response to peginterferon in hepatitis B e antigen-positive chronic hepatitis B. Hepatology 2012;56:67–75. [DOI] [PubMed] [Google Scholar]
- [16].Erhardt A, Reineke U, Blondin D, et al. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology 2000;31:716–25. [DOI] [PubMed] [Google Scholar]
- [17].Rang A, Gunther S, Will H. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J Hepatol 1999;31:791–9. [DOI] [PubMed] [Google Scholar]
- [18].Alcantara FF, Tang H, McLachlan A. Functional characterization of the interferon regulatory element in the enhancer 1 region of the hepatitis B virus genome. Nucleic Acids Res 2002;30:2068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Belloni L, Allweiss L, Guerrieri F, et al. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest 2012;122:529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lu JJ, Chen EQ, Yang JH, et al. A mutation in the interferon regulatory element of HBV may influence the response of interferon treatment in chronic hepatitis B patients. Virol J 2012;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tu Z, Hu Y, Xu L, et al. An improved method for simple and efficient hepatitis B virus genome cloning. J Virol Methods 2014;205:75–80. [DOI] [PubMed] [Google Scholar]
- [22].Hu J-L, Cui J, Guo J-J, et al. Phenotypic assay of a hepatitis B virus strain carrying an rtS246T variant using a new strategy. J Med Virol 2012;84:34–43. [DOI] [PubMed] [Google Scholar]
- [23].Mao R, Nie H, Cai D, et al. Inhibition of hepatitis b virus replication by the host zinc finger antiviral protein. PLoS Pathog 2013;9:e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hao J, Jin W, Li X, et al. Inhibition of alpha interferon (IFN-()-Induced MicroRNA-122 Negatively Affects the Anti-Hepatitis B Virus Efficiency of IFN-(. J Virol 2013;87:137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lau GKK, Piratvisuth T, Luo KX, et al. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med 2005;352:2682–95. [DOI] [PubMed] [Google Scholar]
- [26].Liu F-J, Chen E-Q, Zhou Q-L, et al. Functional characterization of interferon regulation element of hepatitis B virus genome in vivo. Ind J Virol 2012;23:278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsuchida T, Kawai T, Akira S. Inhibition of IRF3-dependent antiviral responses by cellular and viral proteins. Cell Res 2009;19:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kao JH, Wu NH, Chen PJ, et al. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 2000;33:998–1002. [DOI] [PubMed] [Google Scholar]
- [29].Wang Y, Wei L, Jiang D, et al. In vitro resistance to interferon-alpha of hepatitis B virus with basic core promoter double mutation. Antiviral Res 2007;75:139–45. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, Wei L, Jiang D, et al. In vitro resistance to interferon of hepatitis B virus with precore mutation. World J Gastroenterol 2005;11:649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Qin Y, Tang X, Garcia T, et al. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol 2011;85:10167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]


