Abstract
Pleomorphic adenoma is the most common salivary gland neoplasm with a variety of histologic appearances. Due to this diversity, precise preoperative diagnosis through fine needle aspiration cytology is difficult.
This study sought to identify the differentially expressed genes in pleomorphic adenoma to aid precise diagnosis and clarify the mechanism of tumorigenesis.
Suppressive subtractive hybridization was performed on pleomorphic adenoma tissues and the corresponding normal salivary gland tissues to screen of the differential expression of genes in pleomorphic adenoma.
Four known genes (microfibrillar associated protein 4 [MFAP4], dystonin [DST], solute carrier family 35 [SLC35], and potassium channel tetramerization domain containing 15 [KCTD15]) were differentially expressed in the tumors compared with the genes in normal tissues. The expression profiles were further confirmed in 15 pleomorphic adenoma and corresponding normal salivary gland tissues by quantitative real-time reverse transcription-polymerase chain reaction.
MFAP4, DST, SLC35, and KCTD15 gene expression could be potential biomarkers of pleomorphic adenoma for precise diagnosis.
Keywords: DST, KCTD15, MFAP4, pleomorphic adenoma, SLC35
1. Introduction
Pleomorphic adenoma is a benign neoplasm of the salivary glands, which is histologically extremely heterogenous and has various clinical behaviors. Its name derives from the architectural pleomorphism seen by light microscopy. Histologically, pleomorphic adenoma is highly variable in appearance, even within individual tumors, which are characterized by an admixture of polygonal epithelial and spindle-shaped myoepithelial elements in a variable background stroma that may be mucoid, myxoid, cartilaginous, or hyaline.[1,2] The tumor can extend through normal glandular parenchyma in the form of finger-like pseudopodia, leading to a high recurrence rate after surgery. Besides these histologic diversities, there is a potential for malignant transformation into carcinoma ex pleomorphic adenoma, as well as locally aggressive behavior, which makes the tumor an interesting target for molecular analysis. Recent reports have suggested that the tumor suppressor gene pleomorphic adenoma gene 1 (PLAG1) is recurrently rearranged in pleomorphic adenoma and proposed that these genes could be candidate molecules for understanding the mechanism of tumorigenesis.[3–5] But there is relatively little information in the literature, failure to grow cell lines, and the lack of a phenotypic progression model of their evolution.[6]
The suppression subtractive hybridization (SSH) method is based on the suppression polymerase chain reaction (PCR). The method combines normalization and subtraction in a single step. Normalization equalizes the abundance of complementary DNA (cDNA) with the target population and subtraction excludes the common molecules between the tester and the driver population. As a result, a 1000-fold enrichment for differentially expressed cDNAs can be easily achieved. Proper application of this method is very efficient to screen and clone large amounts of differentially expressed genes from specific diseases, such as tumors.[7]
The aim of this study was to detect characteristic gene expression patterns between pleomorphic adenoma and normal salivary gland. As a first step, we used the SSH method for the screening of differentially expressed genes of the most common type of salivary gland tumor, pleomorphic adenomas, and normal salivary gland tissue.
2. Materials and methods
2.1. Human tissue
Human salivary gland tissue samples (n = 15) were collected from patients who underwent surgery for pleomorphic adenoma in the parotid and submandibular glands at the Inha University Hospital. Adult human salivary gland tissues were obtained from the surgically removed and discarded tissue after the diagnostic tests were completed. The request for acquisition of the salivary gland tissues was approved by the Institutional Review Board, Human Subjects Protection Office, at the Inha University Hospital.
2.2. RNA extraction and cDNA synthesis
Total RNA was extracted using an Easy-spin RNA Extraction kit (iNtRON, Seoul, Korea) according to the manufacturer's instructions. The purity of RNA was assessed by comparing the absorption values at 260 and 280 nm (the values of the ratio of A260/A280 of 1.9–2.1 were considered acceptable), and by ethidium bromide staining of the 18S and 28S RNA resolved by gel electrophoresis. The RNA concentrations were determined from the A260. Two micrograms of total RNA were reverse-transcribed in a 20 μL reaction mixture that contained 50 units of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), 5 μM dithiothreitol, 40 units of RNaseOUT recombinant ribonuclease inhibitor, 0.5 μM of random hexanucleotide primers and 500 μM of deoxyribonucleotide triphosphate (dNTP) mixture. The reverse transcription reaction was carried out at 50°C for 60 minutes. Subsequently, the mixture was heated at 70°C for 15 minutes to terminate the reaction, and the cDNA was stored at −20°C.
2.3. SSH
SSH was performed with the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA) according to the manufacturer's protocol. The cDNA from human pleomorphic adenomas were used as the tester and cDNA from the corresponding normal tissues as the driver. The cDNA was amplified by long-distance PCR and was purified using a CHROMA SPIN-1000 DEPC (Clontech). Next, the tester and driver cDNAs were digested with RsaI restriction enzyme. The RsaI-digested cDNA was further purified using a QIAquick PCR Purification Kit (Qiagen, Chatsworth, CA). Two tester populations were prepared using adaptor 1 and adaptor 2R, which were independently ligated to the tester cDNA. Tester cDNA (0.25 ng) was hybridized with an excess amount (150 ng) of driver cDNA in 1 mL of hybridization mixture at 65°C for 16 hours. The second hybridization solution and 150 ng of fresh driver cDNA were then mixed and incubated at 65°C for 8 hours, so that the remaining equalized single-stranded tester cDNA was hybridized with excess driver cDNA. Thus, only the tester-specific cDNA formed the double-stranded cDNA with different adaptors on each end. They were selectively amplified by suppression PCR followed by the nested PCR. Thirty cycles of PCR were then performed using PCR primer 1 to initially amplify the differentially expressed sequences. A subsequent PCR was then performed for 14 cycles using the nested PCR primers 1 and 2R. This second PCR further enriched the differentially expressed sequences and suppressed the background sequences.[7]
2.4. Quantitative real-time reverse transcription (RT)-PCR
Overexpression of the obtained genes was confirmed by quantitative real-time RT-PCR. All real-time PCR analyses were performed on an ABI Step One real-time PCR system (Applied Biosystems, Foster City, CA). Each reaction included a 20 μL reaction mixture containing 0.1 μM of each primer, 10 μL of 2× SYBR Green PCR master mix (Applied Biosystems, including AmpliTaq Gold DNA polymerase with buffer, dNTPs mix, SYBR Green I dye, Rox dye and 10 mM MgCl2), and 1 μL of the template cDNA. The typical amplification program included activation of the enzyme at 94°C for 10 minutes, followed by 40 cycles of denaturation at 94°C for 15 seconds, and annealing and extension at 60 °C for 1 minute. The cycle threshold (CT) value for each gene was determined by the automated threshold analysis function of the ABI instrument, and the CT value for each gene was normalized to CT(GAPDH) to obtain dCT (= CT(GAPDH) − CT(test); GAPDH denotes glyceraldehyde 3-phosphate dehydrogenase). The difference of n between the 2 CT or dCT values indicates a 2n-fold difference in the amount of the target sequence between the 2 cDNA samples being compared. The primers used in the quantitative PCR are shown in Table 1.
Table 1.
Primer sequences for real-time RT-PCR experiments.

2.5. Cloning and sequencing
Ten ng of PCR products were cloned into the plasmids pGEM-T Easy Vector (Promega, Madison, WI) and transferred into Escherichia coli (E coli) DH5α competent cells for transformation. Fifty colonies were randomly picked and sequenced using the PRISM dye termination kit (Applied Biosystems). BLAST Search 2.0 (www.ncbi.nlm.nih.gov/blast/blast.cgi) was used to analyze the sequence homologies in the gene database.
2.6. Statistical analysis
Data analysis was performed using the Graph Pad Prism 5 package (GraphPad Software Inc, La Jolla, CA). The statistical significance of differences between the experiments was evaluated using Student t test. P-values less than.05 were considered statistically significant.
3. Results
3.1. Patient demographics and presentation
Fifteen patients were identified with a histologic diagnosis of pleomorphic adenoma. All patients underwent surgical resection successfully immediately after diagnosis. The clinical characteristics of the patients are shown in Table 2. The majority of patients showed an isolated, asymptomatic mass which was mobile, firm and nontender on physical examination. None of the patients showed facial weakness. There were no patients with bilateral tumors. Presenting symptoms and morphological aspects of the patients are shown in Table 3.
Table 2.
Clinical characteristics of the patients.

Table 3.
Presenting symptoms and morphological aspects of the patients.

3.2. Identification of differentially expressed genes in human pleomorphic adenoma
The SSH method was used to examine the differential gene expression using human pleomorphic adenoma (n = 15) as the tester and the adjacent normal salivary tissues as the driver, specifically enriching for the expressed sequences that were upregulated in tumor tissues. After SSH, 50 subtractive colonies with cDNA inserts were arbitrarily picked for sequencing and identification by a basic local alignment search tool. Figure 1 shows the classification of genes from SSH libraries. Four clones of known genes were identified: microfibrillar-associated protein 4 (MFAP4), dystonin (DST), solute carrier family 35 (SLC35), and potassium channel tetramerization domain containing 15 (KCTD15) (Table 4).
Figure 1.
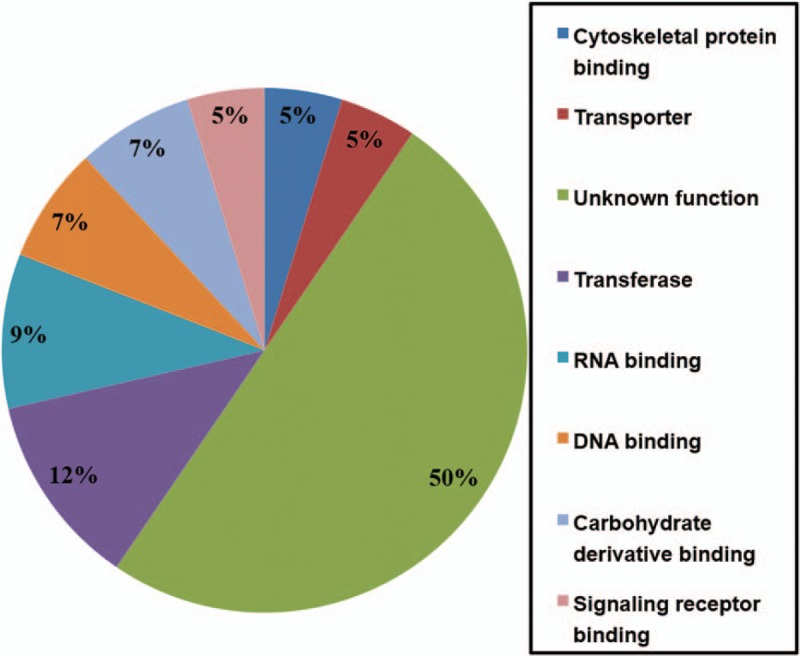
Classification of genes from SSH libraries. Putative functional classification of 50 genes from the SSH library for which identity was could be inferred. Information on function from the Mouse Genome Informatics database. SSH = suppression subtractive hybridization.
Table 4.
Representative upregulated genes in pleomorphic adenoma.

3.3. Quantitative analysis of the upregulated genes in human salivary gland pleomorphic adenoma
To evaluate the reliability of the upregulated genes in human pleomorphic adenoma, the expression levels of mRNA of the aforementioned 4 genes were examined by quantitative real-time RT-PCR analysis for 15 pairs of human pleomorphic adenoma samples and their corresponding normal tissues. To accurately quantify the expression of the 4 genes, GAPDH was amplified and the mean data from each case was used to normalize the result. The mean of the fold-changes across the 15 paired samples for MFAP4, DST, SLC35, KCTD15 were 4.3 (1.4–10.8), 1.9 (1.1–3.7), 3.1 (1.1–9.3), and 2.4 (1.3–12.5), respectively (all P < .05; Fig. 2A–D).
Figure 2.
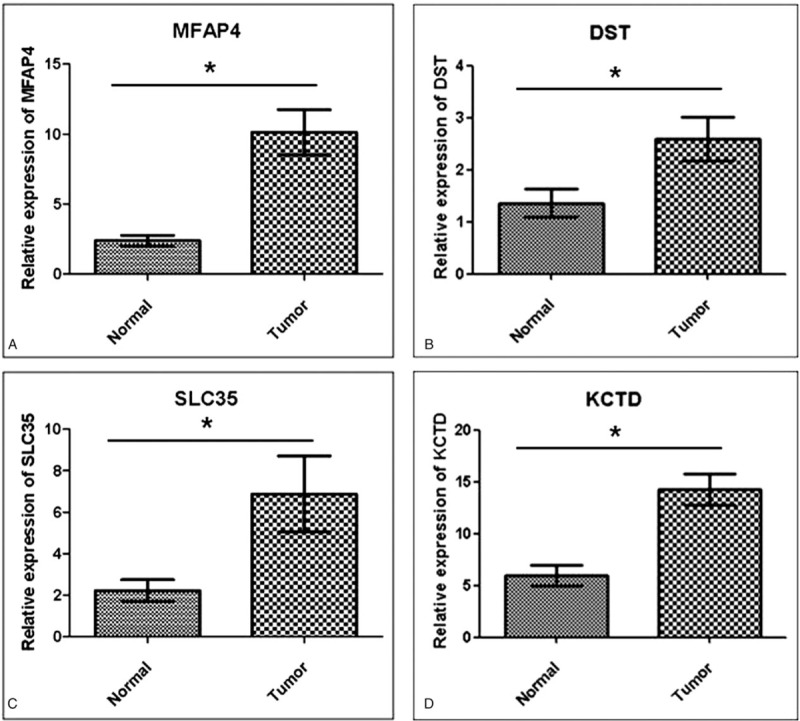
Quantitative real-time PCR analysis of the differentially expressed genes (MFAP4, DST, SLC35, and KCTD15) on 15 pleomorphic adenoma samples and corresponding normal salivary gland tissues. The graph shows the mean and standard deviation. Four expressed genes were statistically significant (∗P < .05). (A) MFAP4 on pleomorphic adenoma versus corresponding normal salivary gland tissue. (B) DST on pleomorphic adenoma versus corresponding normal salivary gland tissue. (C) SLC35 on pleomorphic adenoma versus corresponding normal salivary gland tissue. (D) KCTD15 on pleomorphic adenoma versus corresponding normal salivary gland tissue. DST = dystonin, KCTD15 = potassium channel tetramerization domain containing 15, MFAP4 = microfibrillar associated protein 4, PCR = polymerase chain reaction, SLC35 = solute carrier family 35.
4. Discussion
Pleomorphic adenoma is the most common benign neoplasm of the salivary glands, which accounts for more than 50% of cases. However, occasionally, some types of tumor have aggressive features. The rate of recurrence is relatively high even after surgery, and the malignant transformation rate in pleomorphic adenoma has been estimated at 6%. The underlying mechanism of the high recurrence rate and malignant transformation of pleomorphic adenoma is still not clear. One explanation for the diversity of salivary gland tumors is myoepithelial differentiation. It is generally accepted that the myoepithelial cell is the primary proliferating cell in pleomorphic adenoma.[8] The neoplastic myoepithelium of pleomorphic adenoma displays a wide range of cellular modifications that give rise to histologic diversity. Myoepithelial cells are ectodermal in origin and envelop the glandular/acinar and ductal elements of salivary, mammary, sweat, lacrimal, mucous, and mucoserous glands of the aerodigestive tract.[9] In salivary glands, the myoepithelial cells are associated with the acini and intercalated ducts, and have a dual epithelial and smooth muscle phenotype reflected in their desmosomes between adjacent cells, intermediate size filaments, endocytic vesicles, and microfilaments.[2] They have diverse functions that include extracellular matrix synthesis, a paracrine role, and a tumor-suppressor role.[10]
The other explanation is the altered regulation of genes, which is known to be related to tumor features such as invasiveness, recurrence, and metastasis. If the mechanism of these features in pleomorphic adenoma could be found, we can expect a better prognosis in treatment. In this study, we identified 4 genes (MFAP4, DST, SLC35, and KCTD15) that were differentially expressed between the pleomorphic adenoma and the normal salivary gland tissue.
MFAP4 is an extracellular matrix protein that mediates cell-to-cell or cell-to-matrix interaction.[11] It repairs collectins in the extracellular compartment during inflammation.[12] From the view-point of the importance of extracellular component in pleomorphic adenoma, it was reported that matrix metalloproteinase and their tissue inhibitors are overexpressed in stroma rather than in the epithelium. Therefore, it is presumed that the extracellular matrix synthesis of myoepithelium in the stroma, which could be related to MFAP4, is crucial for the development and progression of pleomorphic adenoma.[13] Furthermore, MFAP4 stimulates ex vivo expansion of hematopoietic stem cells and is involved in stromal tumors.[11,14] Based on these findings, MFAP4 could be related to tumor growth and fibrosis.
DST encodes a member of the plakin protein family of adhesion junction plaque proteins.[15] Some isoforms are expressed in neural and muscle tissue, and function to anchor intermediate neural filaments to the actin cytoskeleton, and some isoforms are expressed in epithelial tissue.[16] Takashi et al reported that DST was overexpressed in human melanoma cell lines and suggested that auto-antibodies against DST could be a potent melanoma marker.[17] Overexpression of DST in a human epidermoid carcinoma cell line and mammary ductal carcinoma in situ has been confirmed.[18] Interestingly, downregulation of DST was also found in metastatic prostate cancer and mammary invasive ductal cell carcinoma.[19] The authors suggested that the downregulation of DST could be indicative of an invasive phenotype and metastasis. Future study of DST expression in carcinoma ex pleomorphic adenoma compared with pleomorphic adenoma could verify that theory in pleomorphic adenoma.
The solute carrier family SLC35 consists of at least 17 molecular species in humans. The family members encode nucleotide sugar transporters localizing at the golgi apparatus and/or the endoplasmic reticulum.[19] Although there has been the little study of the contributions of SLC35 to tumorigenesis, Kumamoto et al reported that the amount of SLC35A2 mRNA, one of the SLC35 family, was increased significantly in malignant colon cancer tissues.[20] It was suggested that the expression of SLC35A2 on the colon cancer cells contributes to hematogenous metastasis of the cancer cells.[19]
The KCTD15 gene encodes the potassium channel tetramerization domain containing 15 and may be associated with obesity, although its function related to tumorigenesis is unclear.[21] In the previous report, it was suggested that KCTD15 negative correlates with the Wnt/beta-catenin pathway.[22] The Wnt signaling pathway plays a central role in the regulation of cell adhesion, proliferation, differentiation, and epithelial-mesenchymal transition.[23] Recently, Zhao et al reported the increased expression of the Wnt/beta-catenin pathway in pleomorphic adenoma animal model (PLAG1 transgenic mice).[24] Contrary to this, we found KCTD15 overexpression in pleomorphic adenoma, which could suppress Wnt/beta-catenin pathway. Further study of the KCTD15 and Wnt/beta-catenin will be needed to deduce the function of KCTD15 related tumorigenesis in pleomorphic adenoma.
To the best of our knowledge, this is the first study to screen the differences in gene expression between pleomorphic adenoma and normal salivary gland tissue using the SSH method. Our studies also showed that the SSH method has a good tool to screen and identify the expression pattern of genes simultaneously.
This study has some limitations that warrant consideration. Although our results suggest the possibility of the association between these new overexpressed genes and pleomorphic adenoma, there is still little information about the relationship of these genes to pleomorphic adenoma. Many genes (such as EGF, EGFR, ErbB-2, FAS, FGF-2, PDGF, WT1, p63, calponin, beta-catenin, REG I, TGF beta-1, and etc) have previously been implicated in the pathogenesis of salivary gland pleomorphic adenoma.[1,25] Additional study is needed to find the relationships and tumorigenesis mechanism between the known genes and new expressed 4 genes. Also, the supplement study with any sort of protein expression analysis (Western blots, immunohistochemistry) to demonstrate translation of the depicted genes at the protein level and to find the location (epithelial, myoepithelial, or stromal cell) where the new 4 genes products overexpressed were not performed. Furthermore, our study had limitations including the small number of samples and the lack of the clinical comparative data of the expressed genes. So, long term study with a large sample size to know the functions of these newly expressed genes will be helpful in understanding the tumorigenesis mechanism for pleomorphic adenoma, and their functions should be confirmed.
We screened the differences in new gene expression between pleomorphic adenoma and normal salivary gland tissue using the SSH method. Four genes (MFAP4, DST, SLC35, and KCTD15) have been implicated in tumorigenesis. Their mechanism of pleomorphic adenoma in salivary gland remains to be solved.
Author contributions
Conceptualization: Byung Han Cho.
Data curation: Young-Mo Kim.
Formal analysis: Jun-Hyeog Jang.
Investigation: Hong-Ju Kim.
Methodology: Jun-Hyeog Jang.
Project administration: Young-Mo Kim.
Resources: Young-Mo Kim.
Software: Jeong-Seok Choi.
Supervision: Young-Mo Kim.
Validation: Jeong-Seok Choi.
Visualization: Jun-Hyeog Jang.
Writing – original draft: Jeong-Seok Choi, Byung Han Cho.
Writing – review and editing: Jeong-Seok Choi, Jun-Hyeog Jang.
Jeong-Seok Choi orcid: 0000-0001-9669-2141.
Footnotes
Abbreviations: DST = dystonin, KCTD15 = potassium channel tetramerization domain containing 15, MFAP4 = microfibrillar associated protein 4, PLAG1 = pleomorphic adenoma gene 1, SLC35 = solute carrier family 35, SSH = suppressive subtractive hybridization.
How to cite this article: Choi JS, Cho BH, Kim HJ, Kim YM, Jang JH. Identification of new genes of pleomorphic adenoma. Medicine. 2019;98:51(e18468).
J-SC and BHC contributed equally to this work.
Y-MK and J-HJ contributed equally to this work.
This research was supported by the Medical Research Center (MRC) (NRF-2014R1A5A2009392, NRF-2016R1A2B4008811, and NRF-2019R1H1A2102005) of the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) and an Inha University Research Grant.
The authors have no conflicts of interest to disclose.
References
- [1].Langman G, Andrews CL, Weissferdt A. WT1 expression in salivary gland pleomorphic adenomas: a reliable marker of the neoplastic myoepithelium. Mod Pathol 2011;24:168–74. [DOI] [PubMed] [Google Scholar]
- [2].Savera AT, Zarbo RJ. Defining the role of myoepithelium in salivary gland neoplasia. Adv Anat Pathol 2004;11:69–85. [DOI] [PubMed] [Google Scholar]
- [3].Kandasamy J, Smith A, Diaz S, et al. Heterogeneity of PLAG1 gene rearrangements in pleomorphic adenoma. Cancer Genet Cytogenet 2007;177:1–5. [DOI] [PubMed] [Google Scholar]
- [4].Asp J, Persson F, Kost-Alimova M, et al. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer 2006;45:820–8. [DOI] [PubMed] [Google Scholar]
- [5].Martins C, Fonseca I, Roque L, et al. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol 2005;18:1048–55. [DOI] [PubMed] [Google Scholar]
- [6].Williams MD, Chakravarti N, Kies MS, et al. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin Cancer Res 2006;12:7353–8. [DOI] [PubMed] [Google Scholar]
- [7].Diatchenko L, Lau YF, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A 1996;93:6025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dardick I, Burford-Mason AP. Current status of histogenetic and morphogenetic concepts of salivary gland tumorigenesis. Crit Rev Oral Biol Med 1993;4:639–77. [DOI] [PubMed] [Google Scholar]
- [9].Riva A, Valentino L, Lantini MS, et al. 3D-structure of cells of human salivary glands as seen by SEM. Microsc Res Tech 1993;26:5–20. [DOI] [PubMed] [Google Scholar]
- [10].Sternlicht MD, Safarians S, Rivera SP, et al. Characterizations of the extracellular matrix and proteinase inhibitor content of human myoepithelial tumors. Lab Invest 1996;74:781–96. [PubMed] [Google Scholar]
- [11].Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, et al. Array comparative genomic hybridization reveals distinct DNA copy number differences between gastrointestinal stromal tumors and leiomyosarcomas. Cancer Res 2006;66:8984–93. [DOI] [PubMed] [Google Scholar]
- [12].Toyoshima T, Ishida T, Nishi N, et al. Differential gene expression of 36-kDa microfibril-associated glycoprotein (MAGP-36/MFAP4) in rat organs. Cell Tissue Res 2008;332:271–8. [DOI] [PubMed] [Google Scholar]
- [13].Lausen M, Lynch N, Schlosser A, et al. Microfibril-associated protein 4 is present in lung washings and binds to the collagen region of lung surfactant protein D. J Biol Chem 1999;274:32234–40. [DOI] [PubMed] [Google Scholar]
- [14].Zhang X, Wang Y, Yamamoto G, et al. Expression of matrix metalloproteinases MMP-2, MMP-9 and their tissue inhibitors TIMP-1 and TIMP-2 in the epithelium and stroma of salivary gland pleomorphic adenomas. Histopathology 2009;55:250–60. [DOI] [PubMed] [Google Scholar]
- [15].Molleken C, Sitek B, Henkel C, et al. Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology 2009;49:1257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kasperkiewicz M, Zillikens D. The pathophysiology of bullous pemphigoid. Clin Rev Allergy Immunol 2007;33:67–77. [DOI] [PubMed] [Google Scholar]
- [17].Brown A, Bernier G, Mathieu M, et al. The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat Genet 1995;10:301–6. [DOI] [PubMed] [Google Scholar]
- [18].Young KG, Kothary R. Dystonin/Bpag1–a link to what? Cell Motil Cytoskeleton 2007;64:897–905. [DOI] [PubMed] [Google Scholar]
- [19].Shimbo T, Tanemura A, Yamazaki T, et al. Serum anti-BPAG1 auto-antibody is a novel marker for human melanoma. PLoS One 2010;5:e10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee CW. An extract of cultured A431 cells contains major tissue antigens of autoimmune bullous diseases. Br J Dermatol 2000;143:821–3. [DOI] [PubMed] [Google Scholar]
- [21].Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res 2006;66:5278–86. [DOI] [PubMed] [Google Scholar]
- [22].Vanaja DK, Cheville JC, Iturria SJ, et al. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res 2003;63:3877–82. [PubMed] [Google Scholar]
- [23].Ishida N, Kawakita M. Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35). Pflugers Arch 2004;447:768–75. [DOI] [PubMed] [Google Scholar]
- [24].Kumamoto K, Goto Y, Sekikawa K, et al. Increased expression of UDP-galactose transporter messenger RNA in human colon cancer tissues and its implication in synthesis of Thomsen-Friedenreich antigen and sialyl Lewis A/X determinants. Cancer Res 2001;61:4620–7. [PubMed] [Google Scholar]
- [25].Queimado L, Obeso D, Hatfield MD, et al. Dysregulation of Wnt pathway components in human salivary gland tumors. Arch Otolaryngol Head Neck Surg 2008;134:94–101. [DOI] [PubMed] [Google Scholar]


