Abstract
Aroma is an important parameter for table grapes and wines; terpenes are typical compounds in Muscat-type grape cultivars and can be easily perceived by humans because of their low olfactory threshold. Volatile terpenes contribute directly to the aroma character, while glycoside-bound terpenes are potential aromatic compounds and can be changed to their volatile forms via hydrolysis. With gas chromatography-mass spectrometry and a solid-phase microextraction method, an automatic data analysis platform was constructed; terpene compounds were identified and quantified from three table grape cultivars at three stages during berry development, and the raw data were deposited in MetaboLights. Terpene metabolite accumulation profiles are presented in this article for integrative analysis with the transcriptome data and phenotypic data to elucidate the important candidate genes and mechanism for terpene biosynthesis. Our method has applications in the identification and quantification of terpene compounds with very low or trace concentrations.
Subject terms: Plant breeding, Plant sciences
| Measurement(s) | Terpene Compound |
| Technology Type(s) | gas chromatography-mass spectrometry • solid-phase micro-extraction |
| Factor Type(s) | cultivar • developmental stage • biological replicate |
| Sample Characteristic - Organism | Vitis vinifera |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.10327715
Background & Summary
The aroma character of Muscat grape cultivars has been thoroughly studied; monoterpenes are the main compounds in Muscat grapes, in the form of terpene alcohols, alkenes, aldehydes, and their oxides. The dominant terpenes in grapes are linalool, geraniol, nerol, terpineol and citronellol. Terpenes exist in two forms: the free form is volatile and directly contributes to flavor, while the glycoside form is non-volatile but can be transformed into the volatile form via hydrolysis1–6. Generally, bound terpenes are more abundant than the free form7. In grapes, terpenes mainly exist in the vacuoles of pericarp cells and in the flesh in some varieties8; their contents are affected by the genotype9–11, development stage12,13, environment and management14–17.
Numerous methods have been used for the detection and analysis of aromatic compounds. The extraction strategies mainly include liquid-liquid extraction, steam distillation, solid-phase extraction, supercritical fluid extraction, and static headspace extraction18–21. Among these, solid-phase microextraction (SPME) has been widely used for the extraction of volatile compounds since 199622. SPME has several advantages over other sample preparation techniques: no organic solvents are needed, the process is easily automated, and small sample sizes are required23. The target compounds are not compromised, and it is easy to set up the automation system and connect to downstream analytical instruments24,25. These features make SPME particularly well suited for studies of a large number of samples. However, the selection of fiber type and model is complicated in the SPME system and should be determined by the extracted compounds and preliminary experiments; additionally, SPME is not applicable when the compounds are not easily volatilized.
Data analysis is a critical step when handling large amounts of metabolic data, and efficient software for peak identification and deconvolution should be optimized to obtain as much information as possible from a sample26,27. Fragment ions from isomers are highly overlapped between different compounds, and this coelution is one of the difficulties in separating compounds, especially for complicated samples. The automated mass spectral deconvolution and identification system (AMDIS) was developed by the American National Standards Institute and is very powerful when dealing with background noise, single peak extraction and shift correction; the detection limit can reach 10−12 g/l28.
In this study, an automatic data analysis platform was constructed, and the contents of the free and bound forms of terpenes were analyzed in 27 table grape samples. Terpene compounds were identified by comparing their retention indices and mass spectra with reference standards and the NIST11 library. Quantification was carried out with standard curves. A total of 28 terpenes were identified. Regarding their structure and category, these terpenes included 5 cyclic alkenes: β-myrcene, (Z)-β-ocimene, (E)-β-ocimene, (E,Z)-allo-ocimene, and (Z)-allo-ocimene; 6 cyclic alcohols: nerol, geraniol, β-citronellol, linalool, γ-geraniol, and isogeraniol; 3 cyclic aldehydes: geranial, neral, and citronellal; 6 cyclic ethers: nerol oxide, cis-rose oxide, trans-rose oxide, cis-furan linalool oxide, trans-furan linalool oxide, and cis-pyran linalool oxide; 1 cyclic acid: geranic acid; 3 acyclic alkenes: limonene, γ-terpinene, and terpinolene; and 2 acyclic alcohols: α-terpineol and 4-terpineol.
The contents of these terpenes could be used for correlation analysis with transcriptome data. The data will be useful for elucidating the mechanism of terpene biosynthesis in table grapes and will provide information for future breeding.
Methods
Overview of the experimental design
The berries of three genotypes were collected at three developmental stages. Approximately 300 grape berries were randomly collected for each replicate, and three replicates were harvested for each stage. The experimental design and analysis pipeline are shown in Fig. 1.
Fig. 1.
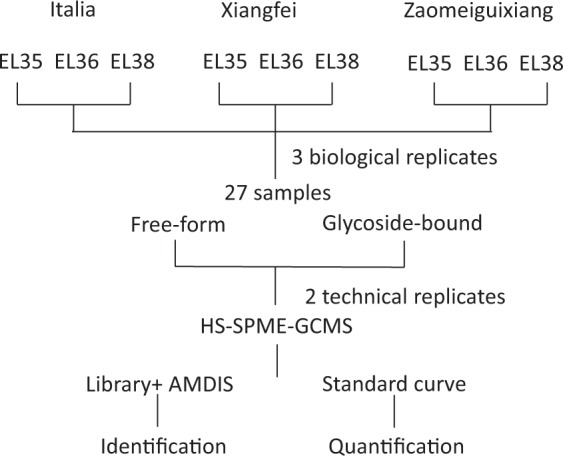
flowchart of the experimental design.
Materials and Methods
Sample collection
Three Vitis vinifera cultivars were used for volatile metabolite analysis: ‘Xiangfei’ was registered by our team in 2000 and has a strong Muscat flavor with a green to golden skin color, ‘Italia’ is a famous mid-late-season table grape cultivar originated in Italy and has moderate Muscat flavor, and ‘Zaomeiguixiang’ has a purple-reddish color and a strong Muscat flavor.
The vines were planted in the experimental field at the Beijing Academy of Forestry and Pomology Sciences in China (39°58′N and 116°13′E) under a plastic cover and trained into a two-wire vertical trellis system with a 2.5-m row space and a 0.75-m plant space. Berry samples from three vines in 2017 were harvested at the development stages corresponding to EL35, EL36, and EL3829. The berry begins to color and soften at EL 35, the completely colored version with intermediate Brix occurs at EL 36, and the berry reaches harvest ripeness at EL38. At each stage, three replicates were harvested; for each replicate, approximately 300 grape berries were randomly collected from ten vines. The berries were frozen in liquid nitrogen and stored at −80 °C.
Extraction of volatile compounds
The extraction of volatile compounds from the grape berries followed Wen’s published method with minor modifications30. The grape berries (approximately 100 g), after discarding seeds and pedicels, were ground and then blended with 1 g of polyvinylpolypyrrolidone (PVPP) under liquid nitrogen. The resultant mixture was macerated at 4 °C for 4 hours and then centrifuged at 8,000 rpm for 15 min at 4 °C to obtain a clear juice. Five milliliters of the liquid were mixed with 1 g of sodium chloride and 10 µL of 1.00808 g/L 4-methyl-2-pentanol in a 20 mL vial (Agilent, 5188-2753 Santa Clara, California, United States) capped with a silicone septum (Agilent, 8010-0139). Two technical replicates were performed for each sample.
Extraction of glycoside-bound compounds
Ten milliliters of methanol and 10 milliliters of water were added in advance to the Cleanert PEP-SPE resins (Bonna-agela Technologies, China). Two milliliters of the clear juice were passed through the Cleanert PEP-SPE column. Water-soluble compounds were eluted with 2 mL of water, free volatiles were washed out with 10 mL of dichloromethane, and then the bound terpenes were eluted with 20 mL of methanol and collected in a round flask. The flow rate was approximately 2 mL/min. The methanol eluate was concentrated to dryness by a rotary evaporator under a vacuum at 30 degrees and then redissolved in 10 mL of a 2 M citrate-phosphate buffer solution (pH 5.0). Subsequently, 4.9 mL of the solution was transferred to a tube, and 100 μL of AR 2000 was added for incubation at 40 degrees for 16 hours. Afterwards, the liquid (5 mL) was mixed with 1 g sodium chloride and 10 µL of 1.00808 g/L 4-methyl-2-pentanol in a 15-mL vial capped with a PTFE-silicone septum.
GC-MS conditions
The volatile compounds were absorbed using headspace solid-phase microextraction (HS-SPME) and then analyzed using an Agilent 7890B-5977A gas chromatograph-mass spectrometer (GC-MS). The autosampler (CTC-PAL RSI85, CTC Analytics, Zwingen, Switzerland) was operated in SPME mode with an SPME fiber (57348-U, Supelco, Bellefonte, PA, USA). The sample vial was initially incubated at 40 °C for 30 min under agitation at 250 rpm, and then the preconditioned SPME fiber was inserted into the headspace of the vial to extract volatiles for 30 min at 40 °C under the same agitation conditions. Then, the SPME fiber was immediately inserted into the GC injection port at 250 °C for 8 min to desorb the volatiles. A 60 m × 0.25 mm HP-INNOWAX capillary column with a 0.25 μm film thickness (J&W Scientific, Folsom, CA, USA) was used to separate the volatile compounds under a 1 mL/min flow rate of helium (carrier gas). The oven temperature program was set as follows: 50 °C for 1 min, increase to 220 °C at 3 °C/min and hold at 220 °C for 5 min. The ion source was maintained at 250 °C with an MSD transfer line temperature of 250 °C. Mass scans were performed from m/z 30-350 with an ionization voltage of 70 eV.
Identification of the compounds
The identification of volatile compounds was based on the retention index of reference standards and mass spectra matching using the standard NIST 11 library. A comparison of the retention index with those reported in the literature was used when standards were not available. Automated mass spectral deconvolution and identification system software (AMDIS, version 2.69, NIST, Washington, DC, USA) was used for peak deconvolution. The parameters for deconvolution were set as follows: component width = 20; adjacent peak subtraction = 2; resolution = high; sensitivity = low; and shape requirements = medium. A self-built library was used to search and match target compounds. Parameters for peak detection were set with default values. The identification information is listed in Table 1.
Table 1.
Identification information of the compounds.
| Compounds | Retention time | Retention Index | Molecular formula | Standard for quantification | Fragmentation | Ion for identification | Calibration Curves |
|---|---|---|---|---|---|---|---|
| β-Myrcene | 13.977 | 1173 | C10H16 | β-Myrcene | 41/69/93 | 41/93 | y = 3350.8x − 1.0946 |
| Phellandrene | 13.756 | 1165 | C10H16 | Limonene | 77/93/91/136 | 77/93 | y = 306.8x + 0.9404 |
| β-trans-Ocimene | 16.662 | 1251 | C10H16 | β-Myrcene | 79/93/121 | 79/93 | y = 3350.8x − 1.0946 |
| γ-Terpinene | 17.184 | 1253 | C10H16 | Terpinolene | 91/93/121/136 | 93/121 | y = 300.19x + 1.3797 |
| β-cis-Ocimene | 17.386 | 1242 | C10H16 | β-Myrcene | 79/93/121 | 79/93 | y = 3350.8x − 1.0946 |
| Terpinolene | 18.747 | 1291 | C10H16 | Terpinolene | 91/93/121/136 | 93/121 | y = 300.19x + 1.3797 |
| Cis Rose oxide | 21.629 | 1338 | C10H18O | Rose oxide | 69/139/154 | 139 | y = 37.312x + 3.259 |
| trans-Rose oxide | 22.289 | 1375 | C10H18O | Rose oxide | 69/139/154 | 139 | y = 37.312x + 3.259 |
| (E,Z)-Allo-Ocimene | 22.518 | 1381 | C10H16 | β-Myrcene | 105/121/136 | 105/121 | y = 3350.8x − 1.0946 |
| Allo-Ocimene | 23.471 | 1397 | C10H16 | β-Myrcene | 105/121/136 | 121/136 | y = 3350.8x − 1.0946 |
| nerol oxide | 26.414 | 1480 | C10H16O | Nerol | 41/67/68/83 | 68/83 | y = 930.79x + 8.9527 |
| Citronellal | 26.776 | 1765 | C10H18O | Citronellal | 41/69/95/121 | 41/69 | y = 70680x + 43.97 |
| Neral | 34.916 | 1755 | C10H16O | citral | 41/69/109 | 41/69 | y = 52.961x + 2.6933 |
| geranial | 37.006 | 1680 | C10H16O | citral | 41/69/84 | 41/69 | y = 52.961x + 2.6933 |
| cis-isogeraniol | 39.415 | 1818 | C10H18O | Geraniol | 41/81/109/121 | 41/81/109 | y = 1476.8x + 0.392 |
| Geranic acid | 56.744 | 2340 | C10H16O2 | Geranic acid | 41/69/100 | 41/69/100 | y = 6818.4x + 7.8939 |
| Linalool | 29.627 | 1581 | C10H18O | Linalool | 55/71/93/121 | 93 | y = 396.8x + 3.2904 |
| α-Terpineol | 35.647 | 1710 | C10H18O | α-Terpineol | 59/93/121/136 | 136 | y = 958.5x + 5.9227 |
| Nerol | 39.384 | 1797 | C10H18O | Nerol | 41/69/93 | 69 | y = 930.79x + 8.9527 |
| Geraniol | 41.043 | 1857 | C10H18O | Geraniol | 41/69 | 69 | y = 1476.8x + 0.392 |
| Citronellol | 37.966 | 1766 | C10H20O | β-Citronellol | 41/69/82/95 | 69 | y = 1323.9x + 0.3414 |
| Limonene | 15.223 | 1205 | C10H18O2 | Limonene | 68/93/121/136 | 68/93 | y = 306.8x + 0.9404 |
| cis-furan linalool oxide | 25.175 | 1448 | C10H18O2 | Linalool | 41/43/59/68 | 59/94 | y = 396.8x + 3.2904 |
| trans-furan linalool oxide | 26.567 | 1477 | C10H18O2 | Linalool | 41/43/59/68 | 59/94 | y = 396.8x + 3.2904 |
| Linalool oxide pyranoside | 37.143 | 1691 | C10H18O2 | Linalool | 41/43/59/68 | 68 | y = 396.8x + 3.2904 |
| trans-isogeraniol | 39.751 | 1829 | C10H18O | Geraniol | 41/81/109/121 | 121 | y = 1476.8x + 0.392 |
Quantification of the compounds
The quantification procedure used in this study was based on previous work by Cai31. A synthetic matrix was prepared in distilled water containing 7 g/l tartaric acid and 200 g/l glucose. The pH was adjusted to 3.3 with a 5 M NaOH solution. The standard solutions contained thirteen terpenes and had fifteen levels, each of which was the halve concentration of the previous level. These solutions were extracted and analyzed under the same conditions as those of the grape samples. An internal standard (4-methyl-2-pentanol, 10 mL, 1.0018 g/l in water) was added to both the standard solutions and samples before aroma extraction and analysis. The calibration curves for analytes were established with regression coefficients above 98%. The concentrations of volatile compounds for which it was not possible to establish calibration curves were estimated on the basis of the equations of compounds with the same functional groups and/or similar numbers of C atoms. Data management and analysis were performed using ChemStation software (Agilent Technologies, Santa Clara, California, United States).
Data Records
A total of 108 raw metabolite data files were deposited in MetaboLights32, including the free-form and glycoside-bound data of 27 samples (two technical replicates). The relative peak areas, the concentration of all samples and the statistically analyzed results were deposited in figshare33.
Technical Validation
There were three biological replicates and two technical replicates for each sample. The final concentrations of the free and bound forms of the compounds were analyzed by statistics and are listed in Online-only Table 1 and Online-only Table 2.
Online-only Table 1.
The concentration of free-form terpenes.
| terpenes | X | Y | Z | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EL35 | SE | EL36 | SE | EL38 | SE | EL35 | SE | EL36 | SE | EL38 | SE | EL35 | SE | EL36 | SE | EL38 | SE | |
| β-Myrcene | 5.55c | 0.52 | 181.88a | 16.93 | 66.79b | 8.82 | 3.18c | 0.12 | 6.32c | 3.57 | 19.67c | 6.92 | 7.73c | 1.56 | 20.08c | 14.50 | 70.36b | 12.11 |
| phellandrene | 1.4c | 0.20 | 25.85a | 2.12 | 10.73b | 1.33 | 0.59c | 0.03 | 1.19c | 0.61 | 3.53c | 1.02 | 1.26c | 0.22 | 3.31c | 1.83 | 10.94b | 2.07 |
| β-trans-Ocimene | nd | N/A | 52.92a | 3.02 | 22.37b | 4.30 | 3.41ef | 0.06 | 3.37ef | 1.06 | 7.51d | 1.85 | 3.91def | 0.33 | 6.93de | 3.29 | 16.73c | 2.43 |
| γ-Terpinene | nd | N/A | 3.9a | 0.62 | 2.93b | 0.38 | nd | N/A | 1.24e | 0.14 | 1.69cd | 0.23 | 1.35de | 0.02 | 1.42de | 0.17 | 1.97c | 0.17 |
| β-cis-Ocimene | 3.55ef | 0.17 | 105.61a | 6.19 | 46.92b | 8.81 | nd | N/A | 4.56ef | 2.31 | 13.54d | 4.24 | 5.54def | 0.68 | 11.94de | 6.85 | 32.81c | 5.10 |
| Terpinolene | 1.29c | 0.02 | 16.65a | 0.94 | 15.34a | 7.17 | 1.13c | 0.01 | 1.8c | 0.96 | 5.97b | 1.34 | 2.11c | 0.09 | 4.28b | 0.83 | 6.16b | 0.68 |
| Cis-Rose oxide | 1.17d | 0.03 | 2.45b | 0.10 | 1.2d | 0.03 | nd | N/A | 1.01d | 0.02 | 1.06d | 0.03 | 1.15d | 0.04 | 1.98c | 0.39 | 5.48a | 0.72 |
| trans-Rose oxide | nd | 0.00 | 1.31b | 0.03 | nd | 0.00 | nd | N/A | nd | N/A | nd | N/A | nd | 0.01 | 1.16c | 0.11 | 2.17a | 0.23 |
| (E,Z)-Allo-Ocimene | nd | N/A | 28.99a | 1.97 | 17.03b | 2.05 | 2.55ef | 0.02 | 3.1de | 0.73 | 5.9d | 1.34 | 3.48de | 0.22 | 5.34de | 2.17 | 12.04c | 1.56 |
| Allo-Ocimene | nd | N/A | 11.51a | 0.60 | 7.58b | 0.69 | 2.54e | 0.02 | 2.74de | 0.24 | 3.7d | 0.44 | 2.83de | 0.08 | 3.46de | 0.71 | 5.94c | 0.57 |
| nerol oxide | 23.81cde | 4.04 | 202.76a | 13.86 | 60.58b | 7.13 | 8.1e | 0.66 | 12.83e | 1.21 | 16.56de | 1.05 | 39.42bce | 3.81 | 44.86bc | 11.02 | 208.69a | 25.86 |
| Neral | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | 0.07 | 2.71b | 0.55 | 7.83a | 1.05 |
| geranial | 2.27ab | 0.18 | 9.38b | 0.53 | 2.57ab | 1.59 | nd | N/A | 1.83ab | 0.11 | 2.03ab | 0.57 | 3.07ab | 1.20 | 4.22ab | 6.43 | 49.18a | 6.32 |
| cis-isogeraniol | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 43.33a | 7.20 |
| Geranic acid | 42.73cd | 3.44 | 67.02bcd | 5.26 | 75.23bc | 25.84 | 13.7d | 3.01 | 40.9cd | 1.81 | 32.04cd | 8.94 | 102.6ab | 30.68 | 40.85cd | 36.64 | 144.59a | 54.50 |
| Linalool | 8.43e | 1.63 | 1473.24a | 85.83 | 407.39b | 50.65 | 3.87e | 0.19 | 26.27e | 22.51 | 118.76d | 35.07 | 23.33e | 1.93 | 118.88d | 43.87 | 213.87c | 15.84 |
| 4-Terpineol | 1.32e | 0.27 | 9.52a | 0.36 | 7.11b | 0.81 | 1.07e | 0.09 | 1.05e | 0.65 | 4.96c | 0.74 | 1.7de | 0.10 | 2.04de | 0.32 | 2.92d | 0.24 |
| α-Terpineol | 10.7e | 0.62 | 181.57a | 11.42 | 118.04b | 16.39 | 9.05e | 0.21 | 16.85e | 13.66 | 99.24c | 13.21 | 23.63e | 1.16 | 53.25d | 5.86 | 59.13d | 6.90 |
| Nerol | 17.76b | 1.78 | 149.16b | 9.59 | 33.62b | 15.91 | 2.7b | 1.00 | 16.12b | 2.82 | 26.58b | 5.88 | 23.52b | 10.26 | 43.46b | 76.11 | 1572.38a | 283.38 |
| Geraniol | 49.91d | 5.06 | 335.44b | 24.83 | 90.86cd | 67.12 | 25.84d | 5.20 | 45.67d | 6.52 | 65.83d | 19.63 | 137.19cd | 60.46 | 187.85c | 369.05 | 1035.58a | 387.88 |
| Citronellol | 3.86c | 0.54 | 13.63b | 0.90 | 2.02c | 1.27 | 1.8c | 0.05 | 2.03c | 0.06 | 1.94c | 0.18 | 3.6c | 0.87 | 3.94c | 5.18 | 40.86a | 6.09 |
| Limonene | 2.08e | 0.22 | 82.01a | 4.38 | 21.67bc | 6.38 | 1.13e | 0.06 | 3.69e | 3.08 | 16.07cd | 4.46 | 4.83e | 0.40 | 12.54d | 4.33 | 25.64b | 3.89 |
| α-muurolene | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 3.09a | 0.09 |
| cis-furan linalool oxide | 3.35bc | 0.34 | 12.86a | 0.60 | 3.71b | 0.15 | nd | 0.00 | 0.5d | 0.54 | 3.44bc | 0.17 | 2.8c | 0.13 | 3.43bc | 0.33 | 4.05b | 0.15 |
| trans-furan linalool oxide | 1.73c | 0.31 | 10.02a | 0.46 | 3b | 0.24 | nd | 0.00 | 0.25d | 0.25 | 1.74c | 0.24 | 1.85c | 0.15 | 1.44c | 0.25 | 2.7b | 0.18 |
| Linalool oxide pyranoside | 0.45d | 0.20 | 8.12a | 0.44 | 3.68b | 0.28 | nd | N/A | 0.42d | 0.44 | 2.79c | 0.28 | 0.27d | 0.06 | 2.51c | 0.23 | 3.65b | 0.13 |
| trans-isogeraniol | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 39.26a | 6.24 |
X, Y, and Z represent the cultivars Xiangfei, Italia and Zaomeiguixiang, respectively. Different letters represent significance levels of P < 0.05, and nd stands for nondetected. N/A stands for not available and SE stands for standard error.
Online-only Table 2.
The concentration of the bound-form terpene.
| terpenes | X | Y | Z | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EL35 | SE | EL36 | SE | EL38 | SE | EL35 | SE | EL36 | SE | EL38 | SE | EL35 | SE | EL36 | SE | EL38 | SE | |
| β-Myrcene | nd | N/A | 226.64cd | 2.85 | 463.98c | 12.03 | nd | N/A | nd | 1.62 | 168.21cd | 6.48 | nd | N/A | 2465.33a | 79.12 | 1262.42b | 26.56 |
| phellandrene | nd | N/A | 5.79c | 0.01 | 9.51bc | 0.33 | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 20.10a | 0.50 | 12.42b | 0.15 |
| β-trans-Ocimene | nd | N/A | 41.87d | 1.44 | 115.39c | 3.38 | nd | N/A | 14.67d | 0.46 | 29.71d | 1.34 | 36.72d | 0.95 | 601.16a | 19.26 | 279.24b | 6.18 |
| γ-Terpinene | nd | N/A | nd | N/A | 8.79c | 0.07 | nd | N/A | nd | N/A | nd | N/A | 7.82c | 0.02 | 26.43a | 0.69 | 15.75b | 0.20 |
| β-cis-Ocimene | 36.92d | 2.32 | 90.13d | 2.91 | 227.08c | 6.60 | 2.12d | 0.26 | 44.46d | 0.79 | 62.31d | 2.62 | 75.26d | 2.77 | 1176.79a | 37.71 | 567.18b | 11.97 |
| Terpinolene | 7.51d | 0.03 | 8.71c | 0.05 | 11.89c | 0.14 | nd | N/A | 7.48c | 0.02 | 7.98c | 0.04 | 8.34c | 0.03 | 41.82a | 1.14 | 20.90b | 0.29 |
| cis Rose oxide | 16.64d | 0.01 | 16.68cd | 0.01 | 16.95c | 0.02 | nd | N/A | 16.44d | 0.01 | 16.55d | 0.01 | 16.68cd | 0.01 | 22.74a | 0.18 | 19.24b | 0.04 |
| trans-Rose oxide | 16.41cd | 0.00 | 16.43cd | 0.00 | 16.52c | 0.01 | nd | N/A | 16.35d | 0.00 | 16.39d | 0.00 | 16.42cd | 0.00 | 18.50a | 0.06 | 17.32b | 0.01 |
| (E,Z)-Allo-Ocimene | nd | N/A | 12.13d | 0.13 | 39.53c | 1.34 | nd | N/A | 4.04d | 0.18 | 8.00d | 0.05 | 12.19d | 0.21 | 202.00a | 6.49 | 94.72b | 1.98 |
| Allo-Ocimene | nd | N/A | 4.47cd | 0.08 | 18.92c | 0.73 | nd | N/A | nd | N/A | 2.36d | 0.02 | 4.54cd | 0.13 | 125.31a | 4.10 | 53.04b | 1.17 |
| nerol oxide | 65.11d | 0.51 | 66.88d | 0.59 | 96.93c | 1.50 | 48.89d | 0.37 | 53.89d | 0.90 | 56.89d | 0.23 | 100.79c | 1.34 | 469.91a | 14.54 | 218.50b | 2.58 |
| Citronellal | 292.93d | 0.92 | 372.55d | 3.09 | 471.13bc | 6.56 | 236.55d | 1.31 | 306.75d | 3.15 | 347.35d | 4.49 | 365.23d | 5.29 | 1230.16a | 31.75 | 676.88b | 7.85 |
| Neral | 14.64de | 0.05 | 14.67de | 0.03 | 18.75c | 0.16 | 13.70e | 0.02 | 14.18de | 0.09 | 14.39de | 0.02 | 16.33d | 0.11 | 43.87a | 0.73 | 24.21b | 0.16 |
| geranial | 14.27e | 0.03 | 14.71de | 0.03 | 17.55c | 0.12 | 13.78e | 0.01 | 14.41e | 0.08 | 14.73de | 0.04 | 15.71d | 0.04 | 30.87a | 0.47 | 19.83b | 0.09 |
| cis-isogeraniol | nd | N/A | 8.38cd | 0.13 | 11.89bc | 0.25 | nd | N/A | nd | N/A | 6.22cd | 0.05 | 6.56cd | 0.07 | 40.45a | 1.15 | 18.46b | 0.22 |
| Geranic acid | 579.99d | 14.63 | 367.87d | 21.20 | 2921.57b | 109.64 | 185.55d | 15.29 | 308.82d | 28.46 | 450.35d | 8.47 | 1775.95c | 15.06 | 4971.88a | 134.20 | 967.30cd | 15.75 |
| Linalool | 22.12c | 0.10 | 175.18a | 5.19 | 179.63a | 4.87 | 17.57c | 0.06 | 27.94c | 0.81 | 71.82b | 1.96 | 28.53c | 0.28 | 204.30a | 6.24 | 102.95b | 1.34 |
| α-Terpineol | 31.96c | 0.03 | 34.06c | 0.10 | 39.58b | 0.31 | 30.85c | 0.05 | 32.64c | 0.20 | 33.07c | 0.10 | 33.84c | 0.15 | 66.02a | 1.15 | 42.13b | 0.23 |
| Nerol | 228.83c | 3.79 | 506.78c | 13.28 | 824.18c | 21.56 | 86.67c | 0.65 | 205.77c | 2.90 | 308.27c | 11.35 | 298.33c | 7.02 | 4215.95a | 129.51 | 2267.67b | 37.62 |
| Geraniol | 146.64d | 1.56 | 223.76d | 4.15 | 442.51c | 10.72 | 96.09d | 0.61 | 181.65d | 0.72 | 230.27d | 6.39 | 257.38d | 2.88 | 1130.06a | 31.58 | 642.51b | 8.88 |
| Citronellol | 12.48e | 0.95 | 36.26d | 0.98 | 64.26c | 1.10 | 17.44de | 0.48 | 14.32e | 0.33 | 22.21de | 0.16 | 32.69de | 0.34 | 190.26a | 4.70 | 116.94b | 1.51 |
| Limonene | 7.20d | 0.22 | 12.71cd | 0.23 | 22.03c | 0.41 | 8.35cd | 0.11 | 7.62cd | 0.08 | 9.45cd | 0.04 | 12.55cd | 0.08 | 112.07a | 3.38 | 59.24b | 1.08 |
| α-muurolene | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A |
| cis-furan linalool oxide | nd | N/A | 18.90b | 0.03 | 18.24c | 0.03 | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 21.03a | 0.02 | 18.38c | N/A |
| trans-furan linalool oxide | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A |
| Linalool oxide pyranoside | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 19.77a | 0.01 | nd | N/A |
| trans-isogeraniol | nd | N/A | 3.74cd | 0.01 | 4.97bc | 0.02 | nd | N/A | nd | N/A | nd | N/A | nd | N/A | 17.38a | 0.52 | 7.97b | 0.10 |
X, Y, Z represent the cultivars Xiangfei, Italia and Zaomeiguixiang. Different letters represent significant levels of P < 0.05, and nd stands for nondetected. N/A stands for not available, SE stands for standard error.
Usage Notes
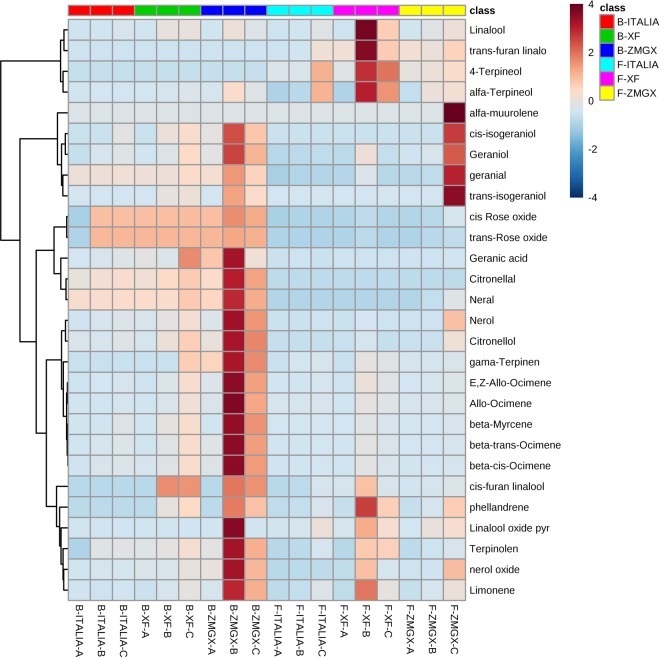
The final concentration of all the samples were presented in excel files and deposited in the figshare. Figure 2 shows the heatmap of terpene concentration for three cultivars at three stages. Figure 3 shows the PCA analysis results.
Fig. 2.
Heatmap of terpene concentration in table grapes during three developmental stages.
Fig. 3.
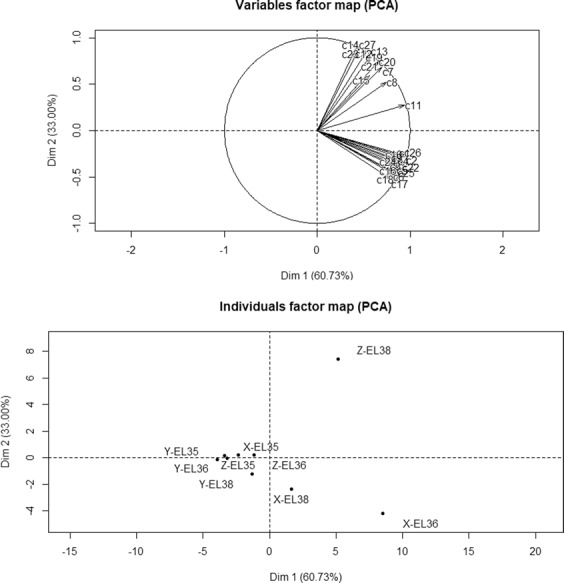
PCA analysis of terpene concentration at three stages.
The B represents the glycoside-bound form. F represents the free volatile form. The A, B, and C at the bottom of the X axis represent the berry developing stages EL35, EL36 and EL38, respectively. XF and ZMGX represent the cultivars Xiangfei and Zaomeiguixiang, respectively.
X stands for Xiangfei, Y stands for Italia, Z stands for Zaomeiguixiang, the terpene names of C1-C26 were shown in Table 1.
Supplementary information
Acknowledgements
This work was supported by the Beijing Academy of Agricultural and Forestry Sciences (KJCX20180411) and the China Agriculture Research System (CARS-29).
Online-only Tables
Author contributions
L.S. designed the experiments and wrote the manuscript, B.Q.Z. analyzed the data, X.Y.Z. collected the samples and performed the GC-MS experiment. G.J.Z., A.L.Y., H.L.W. and X.Y.W. review the manuscript, H.Y.X. designed the experiments, reviewed the manuscript and supervised the study.
Code availability
The code for the heatmap and PCA analysis is available in Supplementary file 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-019-0321-1.
References
- 1.Wilson B, Strauss CR, Williams PJ. The distribution of free and glycosidically-bound monoterpenes among skin, juice, and pulp fractions of some white grape varieties. Amer. J. Enol.Viticult. 1986;37:107–111. [Google Scholar]
- 2.Hjelmeland AK, Ebeler SE. Glycosidically bound volatile aroma compounds in grapes and wine: a review. Amer. J. Enol. Viticult. 2015;66:1–11. doi: 10.5344/ajev.2014.14104. [DOI] [Google Scholar]
- 3.Voirin SG, Baumes RL, Bitteur SM, Gunata ZY, Bayonove CL. Novel monoterpene disaccharide glycosides of vitis vinifera grapes. J. Agric. Food Chem. 1990;38:1373–1378. doi: 10.1021/jf00096a016. [DOI] [Google Scholar]
- 4.Cabrita MJ, Costa-Freitas AM, Laureano O, Stefano SD. Glycosidic aroma compounds of some Portuguese grape cultivars. J. Sci. Food Agric. 2006;86:922–931. doi: 10.1002/jsfa.2439. [DOI] [Google Scholar]
- 5.Wu Y, et al. Aroma characterization based on aromatic series analysis in table grapes. Sci. Rep. 2016;6:31116. doi: 10.1038/srep31116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilc T, Werck-Reichhart D, Navrot N. Meta-analysis of the core aroma components of grape and wine aroma. Front. Plant. Sci. 2016;7:1472. doi: 10.3389/fpls.2016.01472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenoll J, Manso A, Hellin P, Ruiz L, Flores P. Changes in the aromatic composition of the Vitis vinifera grape Muscat Hamburg during ripening. Food Chemistry. 2009;114:420–428. doi: 10.1016/j.foodchem.2008.09.060. [DOI] [Google Scholar]
- 8.Luan F, Wust M. Differential incorporation of 1-deoxy-d-xylulose into (3S)-linalool and geraniol in grape berry exocarp and mesocarp. Phytochemistry. 2005;60:451–459. doi: 10.1016/S0031-9422(02)00147-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, et al. The free and enzyme-released volatile compounds of distinctive Vitis amurensis var. Zuoshanyi grapes in China. Eur. Food Res. Technol. 2015;240:985–997. doi: 10.1007/s00217-014-2403-9. [DOI] [Google Scholar]
- 10.Yang C, Wang Y, Wu B, Fang J, Li S. Volatile compounds evolution of three table grapes with different flavour during and after maturation. Food Chem. 2009;128:823–830. doi: 10.1016/j.foodchem.2010.11.029. [DOI] [Google Scholar]
- 11.Yang C, et al. Volatiles of grape berries evaluated at the germplasm level by headspace-SPME with GC-MS. Food Chem. 2011;114:1106–1114. doi: 10.1016/j.foodchem.2008.10.061. [DOI] [Google Scholar]
- 12.Kalua CM, Boss PK. Evolution of volatile compounds during the development of Cabernet Sauvignon grapes (Vitis vinifera L.) J. Agric. Food Chem. 2009;57:3818–3830. doi: 10.1021/jf803471n. [DOI] [PubMed] [Google Scholar]
- 13.Kalua CM, Boss PK. Comparison of major volatile compounds from Reisling and Cabernet Sauvignon grapes (Vitis vinifera L.) from fruitset to harvest. Aus. J. Grape and Wine Res. 2010;16:337–348. doi: 10.1111/j.1755-0238.2010.00096.x. [DOI] [Google Scholar]
- 14.Bureau SM, Razungles AJ, Baumes RL. The aroma of Muscat of Frontignac grapes: effect of the light environment of vine or bunch on volatiles and glycoconjugates. J. Sci. Food Agric. 2000;80:2012–2020. doi: 10.1002/1097-0010(200011)80:14<2012::AID-JSFA738>3.0.CO;2-X. [DOI] [Google Scholar]
- 15.Wang Y, et al. Effects of cluster thinning on vine photosynthesis, berry ripeness and flavonoid composition of Cabernet Sauvignon. Food Chem. 2018;248:101–110. doi: 10.1016/j.foodchem.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Xu XQ, et al. Differences in volatile profiles of Cabernet Sauvignon grapes grown in two distinct regions of China and their responses to weather conditions. Plant Physiol. Biochem. 2015;89:123–133. doi: 10.1016/j.plaphy.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Koundouras S, Marinos V, Gkoulioti A, Kotseridis Y, Van LC. Influence of vineyard location and vine water status on fruit maturation of non-irrigated cv. Agiorgitiko (Vitis vinifera L.). effects on wine phenolic and aroma components. J. Agric. Food Chem. 2006;54:5077–5086. doi: 10.1021/jf0605446. [DOI] [PubMed] [Google Scholar]
- 18.Ortega-Heras M, Gonzalez-SanJose ML, Beltran S. Aroma composition of wine studied by different extraction methods. Analytica Chimica Acta. 2002;458:85–93. doi: 10.1016/S0003-2670(01)01526-4. [DOI] [Google Scholar]
- 19.Falque-Lepez E, Fernandez-Gomez E. Comparison of solvents for determination of monoterpenes in wine using liquid-liquid extraction. Chromatographia. 2000;52:798–802. doi: 10.1007/BF02491007. [DOI] [Google Scholar]
- 20.Caven-Quantrill DJ, Buglass AJ. Comparison of micro-scale simultaneous distillation–extraction and stir bar sorptive extraction for the determination of volatile organic constituents of grape juice. J Chromatogr A. 2006;1117:121–131. doi: 10.1016/j.chroma.2006.03.091. [DOI] [PubMed] [Google Scholar]
- 21.Pascual-Martia MC, Salvador A, Chafer A, Berna A. Supercritical fluid extraction of resveratrol from grape skin of Vitis vinifera and determination by HPLC. Talanta. 2001;54:735–740. doi: 10.1016/S0039-9140(01)00319-8. [DOI] [PubMed] [Google Scholar]
- 22.Steffen A, Pawliszyn J. Analysis of flavor volatiles using headspace solid-phase microextraction. J. Agric. Food Chem. 1996;44:2187–2193. doi: 10.1021/jf950727k. [DOI] [Google Scholar]
- 23.Atapattu SN, Rosenfeld JM. Solid phase analytical derivatization as a sample preparation method. J Chromatogr A. 2013;1296:204–213. doi: 10.1016/j.chroma.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Abdulra’uf LB, Tan GH. Chemometric approach to the optimization of HS-SPME/GC–MS for the determination of multiclass pesticide residues in fruits and vegetables. Food Chem. 2015;177:267–273. doi: 10.1016/j.foodchem.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Da Silva GC, et al. Method development by GC–ECD and HS-SPME–GC–MS for beer volatile analysis. Food Chem. 2015;167:71–77. doi: 10.1016/j.foodchem.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 26.Boccard J, Veuthey JL, Rudaz S. Knowledge discovery in metabolomics: An overview of MS data handling. J. Sep Sci. 2010;33:290–304. doi: 10.1002/jssc.200900609. [DOI] [PubMed] [Google Scholar]
- 27.Dunn WB, Ellis DI. Metabolomics: Current analytical platforms and methodologies. Trends Anal Chem. 2005;24:285–294. doi: 10.1016/j.trac.2004.11.021. [DOI] [Google Scholar]
- 28.Stein SE. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am Soc Mass Spectrom. 1999;10:770–781. doi: 10.1016/S1044-0305(99)00047-1. [DOI] [Google Scholar]
- 29.Coombe BG. Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1995;1:100–110. doi: 10.1111/j.1755-0238.1995.tb00086.x. [DOI] [Google Scholar]
- 30.Wen YQ, et al. Free and glycosidically bound aroma compounds in cherry (Prunus avium L.) Food Chem. 2014;152:29–36. doi: 10.1016/j.foodchem.2013.11.092. [DOI] [PubMed] [Google Scholar]
- 31.Cai, et al. Influence of pre-fermentation cold maceration treatment on aroma compounds of Cabernet Sauvignon wines fermented in different industrial scale fermenters. Food Chem. 2014;154:217–229. doi: 10.1016/j.foodchem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Sun L. 2019. The accumulation profiles of Terpene metabolites of three Muscat table grape cultivars through HS-SPME-GCMS. MetaboLights. MTBLS968 [DOI] [PMC free article] [PubMed]
- 33.Sun L. 2019. The accumulation profiles of Terpene metabolites of three Muscat table grape cultivars through HS-SPME-GCMS. figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Sun L. 2019. The accumulation profiles of Terpene metabolites of three Muscat table grape cultivars through HS-SPME-GCMS. MetaboLights. MTBLS968 [DOI] [PMC free article] [PubMed]
- Sun L. 2019. The accumulation profiles of Terpene metabolites of three Muscat table grape cultivars through HS-SPME-GCMS. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The code for the heatmap and PCA analysis is available in Supplementary file 1.