Abstract
Introduction
Reliable in situ diagnosis of diminutive (≤ 5 mm) colorectal polyps could allow for “resect and discard” and “diagnose and leave” strategies resulting in one billion dollars in cost savings per year in the U.S. alone. Current methodologies have failed to consistently meet the Preservation and Incorporation of Valuable endoscopic Innovations (PIVI) initiative thresholds. Convolutional neural networks (CNN) have the potential to predict polyp pathology and achieve PIVI thresholds in real-time.
Methods
We developed a CNN-based optical pathology (OP) model using Tensorflow and pre-trained on ImageNet, capable of operating at 77 frames per second. 6223 images of unique colorectal polyps of known pathology, location, size, and light source (white light [WL] or narrow band imaging [NBI]) underwent 5-fold cross training (80%) and validation (20%). Separate fresh validation was performed on 634 polyp images. Surveillance intervals were calculated, comparing OP vs. true pathology (TP).
Results
In the original validation set, the negative predictive value (NPV) for adenomas was 97% among diminutive rectum/rectosigmoid polyps. Results were independent of use of NBI or WL. Surveillance interval concordance comparing OP and TP was 93%. In the fresh validation set, NPV was 97% among diminutive polyps in the rectum and rectosigmoid and surveillance concordance was 94%.
Conclusion:
This study demonstrates the feasibility of in situ diagnosis of colorectal polyps using CNN. Our model exceeds PIVI thresholds for both “resect and discard” and “diagnose and leave” strategies independent of NBI use. Point-of-care adenoma detection rate and surveillance recommendations are potential added benefits.
Keywords: Deep Machine Learning, Colorectal Cancer Screening, PIVI, Optical Pathology
INTRODUCTION
Colorectal cancer (CRC) is the 2nd leading cause of cancer deaths in the United States [1]. The majority of colorectal cancers start as benign precancerous polyps, such as adenomas [2]. Colonoscopy remains the gold standard for finding adenomas and is the only intervention capable of removing adenomas short of surgery. The National Polyp Study (NPS) showed that 70%-90% of colorectal cancers are preventable with polyp removal [3]. The same NPS colonoscopy cohort had a 53% decrease in CRC mortality compared to the SEER population, in whom an unknown percentage had colonoscopy [2].
Approximately 80% of polyps removed during colonoscopy are diminutive (≤ 5 mm) and rarely demonstrate advanced pathology [4]. Among 36,107 diminutive polyps in a large prospective database, only 0.3% of colonic polyps demonstrate high grade dysplasia and none were cancers [5][6]. Pathologic analysis of diminutive polyps contributes $1 billion annually to colonoscopy costs in the United States [7]. A reliable means to ‘optically’ diagnose diminutive polyps in situ could allow for “resect and discard” and “diagnose and leave” strategies resulting in significant cost savings, while also providing surveillance interval recommendations and calculation of adenoma detection rate (ADR) at point of care.
In 2011, the American Society for Gastrointestinal Endoscopy (ASGE) initiative titled “Preservation and Incorporation of Valuable endoscopic Innovations” (PIVI), developed performance thresholds required to “resect and discard” and/or “diagnose and leave” diminutive polyps [8]. “Resect and discard” requires an optical biopsy system with >90% concordance in recommended surveillance intervals compared to true histology for all diminutive polyps throughout the colon [8]. “Diagnose and leave” applies to diminutive non-adenomatous polyps in the rectosigmoid and requires > 90% negative predictive value (NPV) for adenomas. Numerous attempts have been made to reach both PIVI thresholds however these studies have had a number of limitations.
Deep machine learning has the potential to diagnose colorectal polyps in situ and meet optical biopsy PIVI criteria independent of setting, operator motivation, skill or training. Our goal was to create a deep learning algorithm capable of in situ diagnosis of diminutive colorectal polyps and to test its performance against PIVI thresholds under both NBI and white light (WL) without uninterpretable “low confidence” polyps.
METHODS
Colonoscopy and Polyp Image Dataset
We utilized an anonymized version of our prospectively collected Colonoscopy Quality Database inclusive of all GIQuiC quality measures (GI Quality Improvement Consortium, Ltd.), polyp location, size, morphology, and linked images. The database includes over 180,000 polyp and non-polyp images acquired from Olympus 190 (90%), Olympus 180 (7%) -and Pentax i10 series colonoscopes (3%). The polyp images are categorized by light source (WL vs NBI) and image quality (excellent, adequate, poor). Only adequate-to-excellent quality polyp images linked to specimens with a single pathology were included. A total of 6223 high quality images of unique diminutive polyps (one image per polyp) with known unique pathology were identified.
Convolutional Neural Network Architecture
We developed and designed our CNN model architecture from two primary modules: Feature Extraction and Classification. The ‘base’ module of our algorithm is responsible for automated feature extraction and borrows from the Inception-ResNet-v2 algorithm developed by Google AI that achieves “state-of-the-art” accuracy on popular image classification benchmarks. The ‘head’ module of our algorithm is designed for transforming extracted features from the base layers (50mm+ parameters) into a graded scale that allows for pathologic classification. The sigmoid activation function maps the model’s output logits into a float value ranging between 0 - 1. An Nvidia Gtx 1070 GPU was used for algorithm development. Running inference on a frame requires 13 ms (77 frames per second), essentially ‘real time’.
Binary Classification: Adenomatous vs Serrated Lesions
We developed a binary CNN classifier consisting of: 1) adenomatous polyps (tubular adenoma, tubulovillous adenomas, villous adenomas and flat adenomas); vs 2) serrated polyps (hyperplastic and sessile serrated polyps). Our rationale for using this binary classification was based on three factors:
the vast majority of colorectal polyps can be categorized as either adenomas or serrated lesions [9];
pathologists (our “ground truth”) demonstrate excellent agreement distinguishing adenomatous versus serrated polyps but have fair to poor agreement separating hyperplastic polyps (non-neoplastic/surveillance-irrelevant lesions [SILs]) and sessile serrated polyps (precancerous/surveillance-relevant lesions [SRLs]) [10];
there were insufficient numbers of other polyp types in our data set to develop a robust “multiclass” model.
After excluding 945 images of polyps that did not meet our binary classification, the training dataset consisted of 5278 diminutive polyp images (3310 adenomatous polyps and 1968 serrated polyps).
Convolutional Neural Network Training and Validation
Convolutional neural network (CNN) training and validation proceeded in three stages: pre-processing, training, and inference. In the pre-processing stage, data was formatted for input in the algorithm by resizing images, normalizing pixels, and then using transfer learning to help initiate the base layer imaging weights. The base layer imaging weights were pre-trained on ImageNet, which is a large visual database designed for object recognition software. In the training stage, the CNN was trained with Tensorflow, an open-source machine learning framework. 5278 polyp images were partitioned into 5 equal-sized subsamples for 5-fold cross validation with training (80%) and validation (20%). Data augmentation provided standardization for training data “on-the-fly” including various image adjustments. In the Inference stage, the data produced by the training were used to generate test set predictions. Various augmentations to the test set generated an additional layer of predictions. Predictions are then averaged to create a composite probability, and then fed into an Adam optimizer to produce a binary result between 0-0.5 and 0.5-1 for final polyp classification data. Any value ≥ 0.5 is classified as adenoma. Any value < 0.5 is classified as serrated.
Conditional Validations:
The CNN model was then tested for performance under three separate conditions:
NBI vs WL performance: Optical Pathology (OP) was compared to True Pathology (TP) for all adenomatous vs serrated polyps of all sizes and locations.
“Diagnose and leave” performance: OP compared to TP among all diminutive polyps in the rectum and rectosigmoid.
“Resect and discard” performance: Surveillance intervals were compared based on OP vs TP of all diminutive polyps throughout the colon on a per procedure basis.
Surveillance intervals were calculated based on U.S. Multi-Society Task Force guidelines, accounting for procedure indication and polyp number, size, location and pathology (OP vs TP). We adopted the concept that serrated polyps proximal to the rectosigmoid are “surveillance relevant” (see Table 1 for calculations) [9]. The number of cases for which surveillance intervals were lengthened, shortened or unchanged by OP were analyzed.
Table 1.
Surveillance Intervals Calculations Based on US Multi-Society Task Force Guidelines. Final surveillance interval was defined as the lowest surveillance number combining personal history and findings at colonoscopy, comparing SRL status determined by true path (TP) vs optical path (OP) of polyps ≤ 5 mm. SRL: Surveillance Relevant Lesions. FHx: Family History. Hx: History. Lynch: Lynch syndrome. FAP: Familial Adenomatous Polyposis Syndrome. PJ: Peutz-Jeghers Syndrome.
| Personal History | Findings at Colonoscopy | |||
|---|---|---|---|---|
| Risk Category | Years | # of SRLs | Years | |
| Avg. Risk | 10 | 0 | 10 | |
| Hx Adenoma(s) | 5 | 1-2 | 5 | |
| FHx CRC | 5 | 3-9 | 3 | |
| Lynch/FAP/PJ | 1 | >9 | 1 | |
| Size (mm) | ||||
| 1-9 | 5 | |||
| 10-19 | 3 | |||
| >19 | 1 | |||
Validations against new polyp images:
To address validity of our model against fresh datasets, we utilized images of polyps with known pathology previously unexposed to the CNNs. These included 542 polyp images from 286 colonoscopies in our colonoscopy quality database, and 92 polyp images from 50 colonoscopies performed at an independent ambulatory surgical center (ASC).
RESULTS
CNN-predicted Optical Pathology vs True Pathology
Among the original validation set, the distribution of raw CNN OP outputs vs TP is shown in Figure 1. 96% of true adenomas were predicted by OP to be adenomas (prediction bins ≥ 0.5) and the 90% of true serrated polyps were predicted by OP to be serrated (prediction bins <0.5). Figure 1 also shows the distribution of “other” polyps (i.e. non-adenomatous non-serrated polyps) across raw CNN OP outputs. The majority of surveillance-relevant lesions (SRLs) such as traditional serrated adenomas (27/37), hamartomas (9/11), lymphomas (5/5), and juvenile polyps (5/6) were predicted by OP to be adenomatous lesions.
Figure 1.
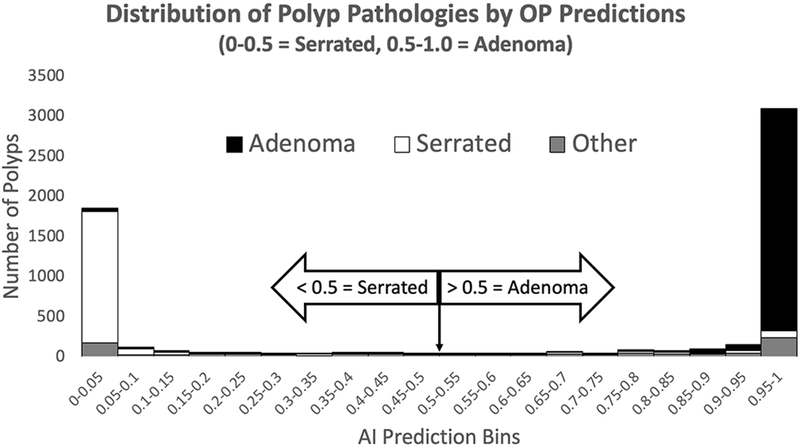
Histogram depicting distribution of polyp pathologies by OP Predictions.
Effect of NBI vs WL on Optical Pathology Performance
Among the original validation set, performance of OP across all polyp sizes and locations was compared between NBI and WL. Overall accuracy of OP was 93.6% (92.9-94.2%[95%CI])% with an NPV of 92.6% (91.5-93.8%[95%CI]) for adenomatous polyps. Positive predictive value was 94.1% (93.3-94.9%[95%CI]); sensitivity was 95.7% (95.1-96.4%[95%CI]) and specificity was 89.9% (88.6-91.3%[95%CI]). OP accuracy was 91.9% (90.2-93.6%[95%CI]) under WL and 94.0% (93.2-94.7%[95%CI]) under NBI (Figure 2). Among the fresh set of polyp images, under WL, accuracy was 92.8% (91.5-94.1% [95%CI]) and under NBI, accuracy was 93.1% (91.6-94.6% [95%CI]). A Pearson’s chi squared test showed that OP performance under WL and NBI was not significantly different in the original or fresh validation sets.
Figure 2.
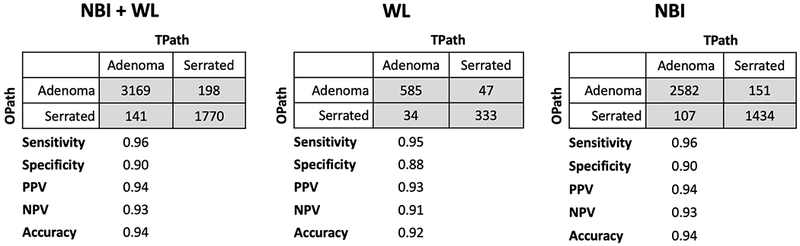
Confusion matrices of adenomas and serrated polyps under WL white light and NBI.
Optical Pathology Performance Against “Diagnose and Leave” Strategy
The PIVI threshold to “diagnose and leave” non-adenomatous polyps requires a >90% NPV for adenomatous polyps ≤ 5 mm in the rectosigmoid. Under these size and location stringencies, OP achieved an NPV for adenomas of 97.3% (95.8-98.8%[95%CI]) under both WL and NBI with an overall accuracy of 91.5% (89.3-93.8%[95%CI]) in the original validation set (see Table 2). In the fresh set of polyp images, NPV was 96.5% (94.8-98.2%[95%CI]) with an accuracy of 88.9%(86.6-91.3%[95%CI]) (see Table 3).
Table 2.
Confusion matrix for “diagnose and leave” criteria comparing diminutive adenomas to non-adenomas in the rectosigmoid and rectum.
| True Path | |||
|---|---|---|---|
| Adenoma | Non-Adenoma | ||
| Optical Pathology | Adenoma | 107 | 38 |
| Serrated Polyp | 12 | 434 | |
| Sensitivity | 0.90 | ||
| Specificity | 0.92 | ||
| PPV | 0.74 | ||
| NPV | 0.97 | ||
| Accuracy | 0.92 | ||
Table 3.
Confusion matrix for “diagnose and leave” criteria comparing diminutive adenomas to non-adenomas in the rectosigmoid and rectum with fresh data.
| True Path | |||
|---|---|---|---|
| Adenoma | Non-Adenoma | ||
| Optical Pathology | Adenoma | 167 | 60 |
| Serrated Polyp | 16 | 443 | |
| Sensitivity | 0.91 | ||
| Specificity | 0.88 | ||
| PPV | 0.74 | ||
| NPV | 0.97 | ||
| Accuracy | 0.89 | ||
Optical Pathology Performance Against “Resect and Discard” Strategy
The PIVI threshold for “resect and discard” requires 90% concordance for recommended surveillance intervals between OP and TP of polyps ≤ 5 mm throughout the colon. Among all polyps ≤ 5 mm (regardless of location or use of WL or NBI) surveillance interval concordance between TP and OP was 93.4% (92.4-94.4% [95%CI]) in the original validation set, and 94.1% (91.3-96.9% [95%CI]) in the fresh set of polyp images. Among screening and surveillance cases in the original validation set, surveillance interval concordance was 90.8% (89.3-92.3% [95% CI]% and 96.4% (95.3-97.5% [95%CI]) respectively, and was 90.8% (86.2-95.4% [95%CI] and 98.3% (96.0-100% [95%CI]), respectively in the fresh validation set.
Table 4 shows the effect of OP on recommend surveillance interval based on the 2499 procedures in our original validation dataset. Among the 192 colonoscopies for which OP shortened surveillance intervals, 167 were attributed to right-sided surveillance irrelevant polypoid lesions (154 normal, 27 lymphoid aggregates, 12 inflammatory polyps, and 1 mucosal prolapse) classified by OP as surveillance-relevant serrated polyps (44%) or adenomas (56%). The remaining 25 cases for which OP shortened surveillance intervals were attributed to OP misclassification of diminutive rectosigmoid hyperplastic polyps as adenomas. The 8 cases for which OP produced longer surveillance intervals were attributed to rectosigmoid adenomas or SSPs misclassified by OP as surveillance-irrelevant serrated polyps. Table 5 shows the effect of OP on recommend surveillance interval based on the 272 procedures in our fresh validation dataset with similar results.
Table 4.
“Resect and discard” surveillance concordance data.
| Entire Colon (All polyps <6mm) | |||||
|---|---|---|---|---|---|
| Indications | Opath shortens interval | Unchanged | Opath lengthens interval | Totals | % Concordance |
| Screening | 151 | 1223 | 7 | 1347 | 90.8% |
| Surveillance | 41 | 1110 | 1 | 1152 | 96.4% |
| Totals | 192 | 2333 | 8 | 2499 | 93.4% |
Table 5.
“Resect and discard” surveillance concordance for fresh data.
| Entire Colon (All polyps <6mm) | |||||
|---|---|---|---|---|---|
| Indications | Opath shortens interval | Unchanged | Opath lengthens interval | Totals | % Concordance |
| Screening | 9 | 139 | 5 | 153 | 90.8% |
| Surveillance | 1 | 117 | 1 | 119 | 98.3% |
| Totals | 10 | 256 | 6 | 272 | 94.1% |
DISCUSSION
Reliable in situ diagnosis of polyp pathology could reduce pathology costs, provide patients with immediate surveillance recommendations, streamline physician workflow and improve cost-effective patient care. We created a CNN-based deep learning model with an overall accuracy of 94% for distinguishing adenomas and serrated polyps. Performance was unaffected by use of WL or NBI. Among diminutive polyps in the rectum/rectosigmoid, the NPV was 97% for adenomas, which is well over the “diagnose and leave” PIVI threshold of 90%. When considering all diminutive polyps removed throughout the colorectum under the indications of screening or surveillance, the surveillance interval concordance between OP and TP was 93%, for original data, and 94% for the fresh data, which are greater than the “resect and discard” PIVI threshold of 90%. To our knowledge, this methodology is the first to successfully achieve and surpass both PIVI thresholds independently of light source, human-based visual criteria or expertise.
There have been numerous attempts to achieve PIVI guideline thresholds using different optical modalities [11][12]. The NICE Criteria, created by the ‘Colon Tumor NBI Interest Group’, separates hyperplastic polyps (Type 1), tubular adenomas (Type 2), and invasive carcinoma (Type 3) based on color, vessels, and surface pattern under NBI without magnification [13]. In 2015, the ASGE performed a meta-analysis assessing the ability of NICE criteria to achieve PIVI standards [7]. Overall an NPV of 91% for adenomas was obtained, however post-polypectomy surveillance interval agreement was only 89%. Performance depends on the practice setting (community vs. academic), operator experience (novice vs. experienced endoscopists), and confidence level. For example, expert endoscopists from academic settings with high confidence could obtain an NPV of 95% with a post-polypectomy surveillance interval agreement of 93% [7], whereas community physicians trained on the NICE criteria were unable to reach PIVI thresholds [11][14].
Other real-time advanced imaging techniques applied to predict polyp histology include magnifying NBI, endocytoscopy, and laser-induced autofluorescence spectroscopy [15]. These technologies are limited by requirements for specialized training, access to expensive equipment, and inability to reach both PIVI thresholds [16][17][18][19]. Failure to consistently achieve PIVI thresholds is multifactorial and includes the subjectivity intrinsic to human-based criteria, e.g., NICE, and the numerous polyps that do not fit into human-based criteria [20].
Deep learning provides a new approach to overcome these limitations. CNNs determine their own criteria based on training sets of polyp images with known pathology. The resulting CNN provides a consistent ‘objective’ output independent of colonoscopist’s training, experience, and skill. Fatigue, loss of motivation, and inability to concentrate are not problems for a deep learning system.
Deep learning for polyp pathology prediction is rapidly emerging in the literature. In 2017, Byrne et al published their CNN deep learning model trained on NICE classification of polyps. When tested on 106 polyp videos, they achieved an accuracy of 94%, and NPV of 97%. Surveillance concordance was not published. Notable limitations included a dataset collected by one expert endoscopist, dependency on NBI, and exclusion of 19 polyps due to lack of high confidence results from their NICE-based deep learning model [21]. In 2018, Mori et al. published data on the use of deep learning combined with endocytoscopy and reported an NPV >90% for left colon adenomas after exclusion of low confidence polyps [22]. Their data was collected by experts in the use of the endocytoscopy which limits applicability in the community setting and bypasses the cost saving potential of deep learning.
We challenged our deep learning algorithm to differentiate adenomatous and serrated polyps on unaltered endoscopic images at standard magnification using conventional colonoscopes inclusive of WL and NBI imaging. The resulting model performed equally well under both light settings, and therefore independent of Olympus NBI technology. Because our data set was collected from endoscopists at various levels of training ranging from first year GI fellows to endoscopists with 30+ years’ experience, we expect that it’s performance will be user-independent as well as applicable in both community and academic settings. Our validation subset at a community ASC supports this conjecture.
An optimal OP technology will reliably distinguish surveillance relevant lesions (SRLs) from surveillance-irrelevant lesions (SILs). CNNs, however, can only be as good as the “ground truth” on which they are trained [23]. Our CNN was based on pathology, derived from our academic pathologists who cluster tightly in their classification rates of adenomas and serrated polyps but vary widely in their classification rates of the two primary sub-classifications of serrated polyps (HPs [SILs] and SSPs [SRLs]) [24][25]. This is in line with several studies showing poor agreement among pathologists distinguishing hyperplastic (SILs) and sessile serrated polyps (SRLs) [26]. This current limitation of pathology coupled with data suggesting that diminutive proximal HPs are associated with synchronous advanced neoplasia [27], supports suggestions that any serrated polyp proximal to the sigmoid should be considered an SRL [28].
We therefore trained our CNN on the best “ground truth” pathology (adenoma vs serrated), and adopted the rule that any serrated polyp proximal to the sigmoid is considered a SRL. Our binary approach (serrated vs adenoma) and surveillance model means that any polyp, including normal and lymphoid aggregates proximal to the sigmoid will be categorized as an SRL (serrated vs. adenoma) by our CNN. This strategy therefore biases toward OP-shortened surveillance intervals and discordance with TP. In fact, 84% of surveillance interval discordance by our OP was attributed to 167 SILs classified as SRLs by OP, 154 of which were diagnosed as “normal” colonic mucosa by our pathologists. Even so, our CNN exceeded the PIVI “resect and discard” threshold of 90% concordance. It is especially interesting that this level of concordance is achieved by combining OP for rectosigmoid polyps with a simple OP-independent rule: all diminutive polyps proximal to the sigmoid are SRLs.
Development of an OP system to further reduce surveillance discordance will require separate categories of SILs (HPs, normal, lymphoid aggregates, inflammatory, mucosal prolapse). We are currently generating a multiclass CNN to accomplish this, realizing that its performance will be limited by poor “ground truth” for separating proximal serrated polyps into HP and SSP categories. Furthermore, the category of “normal” may include a significant number of diminutive SRLs, such as adenomas, that were missed on tissue sectioning [23].
A key limitation of our study is that it is retrospective and based on static images. We currently are able to run multiple CNNs in real time during live colonoscopies without noticeable frame drops or lag. Additionally, we are optimizing live OP feedback during colonoscopy in conjunction with our polyp detection algorithm in real time [29]. Future prospective multicenter validation studies during colonoscopy are being planned.
In summary, we demonstrate the promise of real-time in situ diagnosis of colorectal polyps using deep learning. We show that in situ diagnosis is possible without dependency on specific light source, potentially applicable across multiple scope manufacturers. Deployment does not require specialized scopes or processors and only needs an off-the shelf computer with standard graphic processing unit with connections to the video source and viewing monitor. The future will bring a new deep learning assistant to the endoscopy suite to find and diagnose polyps in real time. For the patient, this means that polyps in view are found, removed, and diagnosed without unnecessary pathology fees, followed by accurate recommendations for follow-up at point-of care. Deep learning cannot replace the colonoscopist, who must drive the scope effectively and safely, clean away debris, inspect all mucosal surfaces, make sound judgements on what pathologies they find, and apply their knowledge and skill during intervention. Deep machine learning technology will continually learn from the endoscopists, leading to continual improvements in quality patient care delivery.
Study Highlights.
WHAT IS KNOWN:
The Preservation and Incorporation of Valuable endoscopic Innovations (PIVI) guidelines provide criteria for adoption of “resect and discard” and “diagnose and leave” strategies for optical pathology.
Convolutional Neural Networks (CNNs) show promise in achieving accurate real-time optical pathology.
WHAT IS NEW HERE:
We demonstrate that our CNN-based optical pathology exceeds both PIVI thresholds when applied to polyp images obtained during colonoscopy.
Accuracy was achieved using images from standard colonoscopes performed by novice and expert colonoscopists using white light or NBI.
ACKNOWLEDGEMENTS
Preliminary results of this study were presented as an oral abstract at the “American College of Gastroenterology Annual Meeting and Postgraduate Course” in Philadelphia on October 9th, 2018. We would like to thank all UCI and Monterey Bay GI Consultants Medical Group physicians, nurses, technicians and staff who assisted in capturing and annotating images in the colonoscopy database.
Financial Support: None
Ethics: The research dataset is not considered human-subjects research for IRB purposes as determined by the University of California, Irvine (UCI) Human Research Protections Program (HS#2015-1939 [eAPP# 8814] dated 1/13/2016). The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR001414. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Potential Competing Interests: WK, JS, and AN are co-founders of Docbot, Inc., the company responsible for CNN development and implementation.
REFERENCES
- [1].Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Mortality-All COD, Aggregated with State, Total US (1969-2014) <Katrina/Rita Population Adjustment>. Bethesda, MD: National Cancer Institute; 2016. Updated September 10, 2018. [Google Scholar]
- [2].Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic Polypectomy and Long-Term Prevention of Colorectal-Cancer Deaths. N Engl J 2012; 366:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Winawer SJ, Zauber AG, Ho MN, et al. Prevention of Colorectal Cancer by Colonoscopic Polypectomy. N Engl J Med 1993; 329:1977–81. [DOI] [PubMed] [Google Scholar]
- [4].Lee J. Resection of Diminutive and Small Colorectal Polyps: What is the Optimal Technique? Clin Endosc 2016; 49:355–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ponugoti PL, Cummings OW, and Rex DK. Risk of cancer in small and diminutive colorectal polyps. Dig Liver Dis 2017; 49:34–7. [DOI] [PubMed] [Google Scholar]
- [6].Kaminski MF, Hassan C, Bisschops R, et al. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy Guideline. Endoscopy 2014; 46:435–57. [DOI] [PubMed] [Google Scholar]
- [7].Abu Dayyeh BK, Thosani N, Konda V, et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc 2015; 81:502e1–16. [DOI] [PubMed] [Google Scholar]
- [8].Rex DK, Kahi C, O’Brien M, et al. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc 2011; 73:419–22. [DOI] [PubMed] [Google Scholar]
- [9].Rex DK. Serrated Polyps in the Colon. Gastroenterol Hepatol 2014; 10:671–4. [PMC free article] [PubMed] [Google Scholar]
- [10].Wong NA, Hunt LP, Novelli MR, et al. Observer agreement in the diagnosis of serrated polyps of the large bowel. Histopathology 2009; 55:63–6. [DOI] [PubMed] [Google Scholar]
- [11].Ladabaum U, Fioritto A, Mitani A, et al. Real-time optical biopsy of colon polyps with narrow band imaging in community practice does not yet meet key thresholds for clinical decisions. Gastroenterology 2013; 144:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Byrne MF, Shahidi N, and Rex DK. Will computer-aided detection and diagnosis revolutionize colonoscopy? Gastroenterology 2017; 153:1460–4e1. [DOI] [PubMed] [Google Scholar]
- [13].Hewett D, Kaltenbach T, Sano Y, et al. Validation of a simple classification system for endoscopic diagnosis of small colorectal polyps using narrow-band imaging. Gastroenterology 2012; 143:599–607. [DOI] [PubMed] [Google Scholar]
- [14].Kuiper T, Marsman WA, Jansen JM, et al. Accuracy for optical diagnosis of small colorectal polyps in nonacademic settings. Clin Gastroenterol Hepatol 2012; 10:1016–20. [DOI] [PubMed] [Google Scholar]
- [15].Mori Y, Kudo SE, Berzin TM, et al. Computer-aided diagnosis for colonoscopy. Endoscopy 2017; 49:813–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tischendorf JJ, Schirin-Sokhan R, Streetz K, et al. Value of magnifying endoscopy in classifying colorectal polyps based on vascular pattern. Endoscopy 2010; 42:22–27. [DOI] [PubMed] [Google Scholar]
- [17].Iwatate M, Ikumoto T, Hattori S, et al. NBI and NBI Combined with Magnifying Colonoscopy. Diagn Ther Endosc 2012; 2012:173269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Misawa M, Kudo S, Mori Y, et al. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology 2016; 150:1531–2. [DOI] [PubMed] [Google Scholar]
- [19].Rath T, Tontini GE, Vieth M, et al. In vivo real-time assessment of colorectal polyp histology using an optical biopsy forceps system based on laser-induced fluorescence spectroscopy. Endoscopy 2016; 48:557–62. [DOI] [PubMed] [Google Scholar]
- [20].Kumar S, Fioritto A, Mitani A, et al. Optical biopsy of sessile serrated adenomas: do these lesions resemble hyperplastic polyps under narrow-band imaging? Gastrointest Endosc 2013; 78:902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Byrne MF, Chapados N, Soudan F, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019; 68:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mori Y, Kudo SE, Misawa M, et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med 2018; 169:357–366. [DOI] [PubMed] [Google Scholar]
- [23].Ponugoti P, Rastogi A, Kaltenbach T, et al. Disagreement between high confidence endoscopic adenoma prediction and histopathological diagnosis in colonic lesions ≤3 mm in size. Endoscopy 2019; DOI: 10.1055/a-0831-2348 [DOI] [PubMed] [Google Scholar]
- [24].Turkiewicz J, Clary M, and Karnes WE. Variable classification of right-sided serrated colon polyps among pathologists. Gastrointest Endosc 2017; 87:AB465–66. [Google Scholar]
- [25].Flores SL, Ziogas A, Albers G, et al. Variable recognition of sessile serrated adenomas among colonoscopists and pathologists - a compounded roadblock to reducing interval colon cancers. Gastrointest Endosc 2014; 79:AB170. [Google Scholar]
- [26].Rex DK, Alikhan M, Cummings O, et al. Accuracy of pathologic interpretation of colorectal polyps by general pathologists in community practice. Gastrointest Endosc 1999; 50:468–74. [DOI] [PubMed] [Google Scholar]
- [27].Hamoudah T, Ma K, Esteban M, et al. Patients with small and diminutive proximal hyperplastic polyps have higher rates of synchronous advanced neoplasia compared with patients without serrated lesions. Gastrointest Endosc 2018; 87:1518–26. [DOI] [PubMed] [Google Scholar]
- [28].Rex DK. Small proximal hyperplastic polyps are associated with synchronous advanced neoplasia. NEJM Journal Watch. [https://www.jwatch.org/na45974/2018/02/01/small-proximal-hyperplastic-polyps-are-associated-with] Published February 1, 2018. Accessed February 1, 2019.
- [29].Urban G, Tripathi P, Alkayali T, et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018; 155:1069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer Clin J 2017; 67:177–93. [DOI] [PubMed] [Google Scholar]
- [31].Karkanis SA, Iakovidis DK, Maroulis DE, et al. Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed 2003; 7:141–52. [DOI] [PubMed] [Google Scholar]
- [32].Apel D, Jakobs R, Schilling D, et al. Accuracy of high-resolution chromoendoscopy in prediction of histologic findings in diminutive lesions of the rectosigmoid. Gastrointest Endosc 2006; 63:824–28. [DOI] [PubMed] [Google Scholar]


