Abstract
Most clinical antibiotics do not have efficacy against Gram-negative pathogens, mainly because these cells are protected by the permeability barrier comprising the two membranes with active efflux. The emergence of multidrug resistant Gram-negative strains threatens the utility even of “last resort” therapeutic treatments. Significant efforts at different levels of resolution are currently focused on finding a solution to this non-permeation problem and developing new approaches to optimization of drug activities against multidrug resistant pathogens. The exceptional efficiency of the Gram-negative permeability barrier is the result of a complex interplay between the two opposing fluxes of drugs across the two membranes. In this review, we describe the current state of understanding of the problem and the recent advances in theoretical and empirical approaches to characterization of drug permeation and active efflux in Gram-negative bacteria.
Keywords: Gram-negative bacteria, antibiotic resistance, multidrug efflux, permeability barrier
Gram-negative pathogens are intrinsically resistant to most antibiotics.
Multiple antibiotic resistance in bacterial pathogens has a broad societal impact and threatens not only the most vulnerable patients such as children and the immunocompromised population but modern medicine as a whole. The susceptibility to antibiotics varies broadly depending on the specific pathogen, its clinical manifestations and the class of antibiotics. However, the majority of approved clinical antibiotics do not have efficacy against Gram-negative bacteria and are classified as “Gram-positive” only. This intrinsic resistance of Gram-negative bacteria to antibiotics is well-recognized and characterized, and was dealt with during the “golden era” of antibiotics and thereafter. The current urgency arises due to emergence in clinics of Gram-negative pathogens that are resistant to even recommended antibiotics. Gram-negative enterobacteria such as Klebsiella spp. and Enterobacter spp., Pseudomonas aeruginosa, and Acinetobacter baumannii form the core of the ESKAPE pathogens that are notorious for their multidrug resistance1–2.
Both the intrinsic and acquired antibiotic resistance in Gram-negative bacteria is enabled by two-membrane permeability barriers that comprise the inner and outer membranes with different chemical structures and compositions and active efflux pumps acting across both membranes (Fig. 1)3–4. However, these permeability barriers did not evolve equal and Gram-negative bacteria vary dramatically in their antibiotic susceptibilities (Table 1). Fluoroquinolones and aminoglycosides are examples of the broad-spectrum antibiotics, which until recently, were successfully used in treatments of P. aeruginosa infections. This species is by orders of magnitude more resistant than Escherichia coli to most clinical antibiotics. Burkholderia spp. and multidrug resistant A. baumannii strains are intrinsically resistant to aminoglycosides and are even more resistant than P. aeruginosa to many other antibiotics.
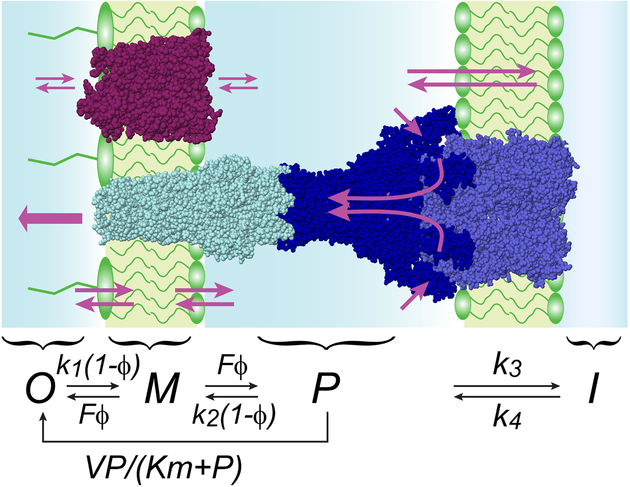
Figure 1. Schematics and kinetic model of compound permeation in the context of the two membranes with active efflux.
Small molecules traverse the outer membrane via facilitated or passive diffusion and can be extruded from the periplasmic space by active transporters. The kinetic scheme explicitly considers four compartments, outside the cell (O), within the outer membrane (M), in the periplasm (P) and in the cytoplasm (I). Active efflux is approximated as a Michaelis-Menten process. The binding to the membrane is postulated saturable, with the maximal flux F. The degree of saturation is denoted as ϕ; k1 through k4 are microscopic rate constants.
Table 1.
Minimal inhibitory concentrations (μg/mL) of select antibiotics against Gram-negative bacteria.
| Antibiotics | E. coli K-12a | P. aeruginosa PAO1b | B. cepacia ATCC 25416 b | A. baumannii AYE c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | +Pore | ΔEfflux | WT | +Pore | ΔEfflux | WT | +Pore | WT | ΔEfflux | |
| Tetracycline | 0.5 | 0.25 | 0.125 | 4 | 0.5 | 2 | >8 | 8 | 32–64 | 2–4 |
| Ciprofloxacin | 0.016 | 0.004 | 0.002 | 0.06 | 0.031 | 0.016 | 1 | 0.03 | 64 | 1 |
| Rifampin | 4 | 0.25 | 4 | 16 | 0.5 | 16 | 16 | 0.06 | 10 | 5 |
| Gentamicin | 4 | 2 | 4 | 4 | 4 | 4 | 128 | 16 | 1024 | 16 |
| Polymyxin B | 1 | 1 | 1 | 1.5 | 1.5 | 1.5 | >1024 | 512 | 2 | ND |
| Carbenicillin | 16 | 2 | 4 | 32 | 2 | 1 | >1024 | 128 | >2048 | 1024 |
These four species (Table 1) have similar yet different outer membranes (OMs). The similarity comes from the overall architecture of these membranes, as all are asymmetric bilayers composed of lipopolysaccharides (LPS) in the outer leaflet and glycerophospholipids in the inner leaflet5–8. The major features of the LPS structure, such as the presence of lipid A, the core, and O-antigen chains, are also conserved among various species but specific chemical modifications vary broadly. The species-specific OM bilayers differ in the size and numbers of LPS molecules, thicknesses, surface charge distributions, and dynamics. These differences, in turn, translate into the differences in permeability properties of OM bilayers.
In addition, a variety of OM proteins support the structure of the asymmetric bilayer and enable selective uptake of nutrients and efflux of toxic compounds and metabolites across the OM. In E. coli, antibacterial activities of large and polar antibiotics exceeding the size of general porins OmpF/OmpC (>600 Da in E. coli) are usually the most restricted by OM and the rules for permeation through enterobacterial porins are the best understood and generalized9. In contrast, the P. aeruginosa OM possesses only substrate-specific porins and its permeability properties are subject of intense investigations10–14. Recently, structures and properties of several porins in the OM of A. baumannii have also been characterized15–17. Among them, DcaP was identified as a highly abundant OM porin in an A. baumannii strain during infection15. DcaP is selective for negatively charged substrates such as succinates and also contributes to uptake of β-lactamase inhibitors such as sulbactam and tazobactam. Porins of Burkholderia spp. are still awaiting their characterization. Interestingly, some of these porins including OpcC are found to be essential in Burkholderia genomes but not in other bacterial essential genomes identified so far18.
Although the inner membrane (IM) of Gram-negative bacteria is a formidable barrier for large hydrophilic molecules, it is relatively permeable for the majority of amphiphilic drugs19–20. These drugs however, are countered by multidrug efflux pumps that actively expel various compounds from cells and operate and affect drug concentrations in all bacterial cell compartments4, 21. Efflux transporters, such as those belonging to the Small Multidrug Resistance or Multidrug and Toxic compound Extrusion families of proteins transport drugs across the IM and affect cytoplasmic drug accumulation22–23. However, the most efficient drug efflux pumps, such as those belonging to the Resistance-Nodulation-cell Division (RND) superfamily, bind various substrates on the periplasmic side of the IM and translocate them across the low permeability barrier of OM and into the external medium24–25. This transport is enabled by additional proteins located in the periplasm and in the OM, which together with the transporter form a trans-envelope protein conduit spanning the two membranes and the periplasm (Fig. 1)26–28. Inactivation of such trans-envelope efflux pumps increases bacterial susceptibility to various antibiotics (Table 1), whereas their overexpression is a recognized cause of the clinical antibiotic resistance29–31.
The exceptional efficiency of trans-envelope efflux pumps is the result of a complex interplay between the two opposing fluxes of drugs across the two membranes. Current efforts to quantitatively analyze this interplay and to develop approaches for the optimization of drug permeation into Gram-negative bacteria are in the focus of this review article.
Permeability barrier of the outer membrane.
Permeation of compounds across the OM is a key factor that defines their accumulation in bacteria. This process is not instantaneous even when specific pores exist in the cellular membrane that allow compounds a seemingly unfettered access to the periplasm. The rate of transmembrane transport is not only finite but also limited, and the magnitude of this limit can have dramatic effects on the overall performance of efflux transporters. An excellent analysis of transmembrane diffusion can be found in prior literature32–36. Here, we focus on factors that limit the rate of passive transport some of which were escaping attention until recently. We begin with the case of unfacilitated diffusion and then apply these ideas to mediated transport. The interplay between drug permeation into the cell and its active efflux is described in the next section.
Transmembrane diffusion is the central event during drug permeation. The rate of the diffusion can be expressed as a flux across the outer membrane
| (1), |
where A is the surface area of the membrane and dn/dt is the number of moles of the compound transported across the membrane in a unit time. When concentration of compounds is low enough, the flux is linearly related to the concentration gradient on the membrane ΔC:
| (2), |
where Pm is the permeability of the membrane. This relationship is based on Fick’s laws of diffusion, which have been derived without expectations of any saturation and postulate a proportionality between the flux and the concentration gradient. Substituting Equation 2 into the Fick’s laws leads to Equation (3):
| (3), |
where K is the partition coefficient between the membrane and solution, Dm is the diffusion coefficient within the membrane, and d is the thickness of the membrane.
The practical utility of Equation 3 is limited due to the lack of straightforward ways to evaluate Dm and K. Partition coefficient of a compound is frequently approximated with its oil-water partition coefficient, which is a fair mimic of the experiment only for molecules that are much smaller than the membrane and with negligible interaction with the polar head groups of the lipids. In reality, the size of typical antimicrobials is comparable to the length of lipid tails, and the interaction with the head groups can be substantial and varies even between analogs. Similar limitations apply to the estimates of the diffusion coefficient Dm.
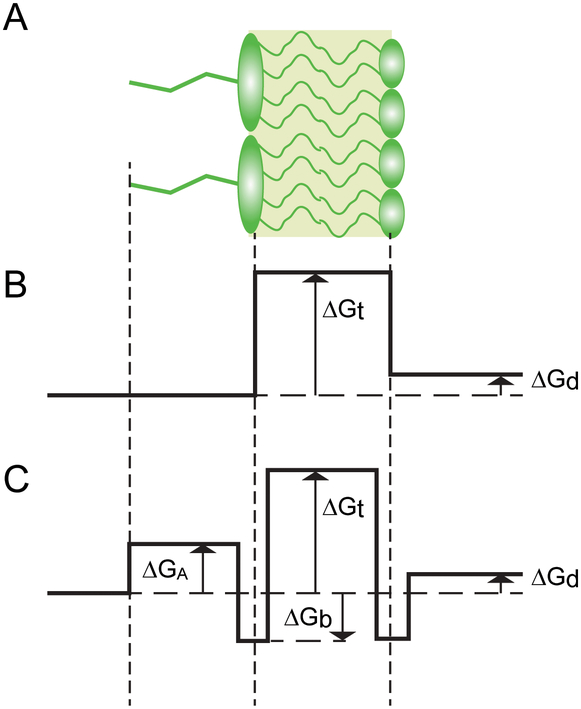
The problem can be illustrated using the energy profile of a chemical crossing the lipid bilayer (Fig. 2A). In conventional analysis, the chemical potential of a compound is postulated to change at the junction between the aqueous and lipid phases (Fig. 2B). In this case, the energetic barrier between the external medium and the periplasm is directly related to the partition coefficient via the Boltzmann equation: K = exp(−ΔGt/RT).
Figure 2. The energy profile of drug permeation across a lipid bilayer.
(A) Schematic structure of the asymmetric bilayer. (B) A simplified energy profile recognizes the Donnan equilibrium of the membrane, ΔGd, and the free energy of partitioning between the membrane and the medium, ΔGt. (C) In addition to the above, a more realistic energy profile needs to incorporate the excluded volume created by the lipid A modification, ΔGA, and the attractive interaction at the lipid-water interface, ΔGb.
A different free energy profile is expected if we recall the finite size of the traversing molecule. In this case, the lowest energy state is expected at the interface between the aqueous and lipid media, where the hydrophobic fragment of the molecule is buried within the bilayer whereas the polar moieties maximize the interaction with the head groups (Fig. 2C). One could expect in such cases that the oil-water partition coefficient would approximate well the transition state of the compound, when it is completely submerged into the non-polar phase, whereas the empirical affinity of compounds would be best represented by the interfacial region.
The energy profile depicted in Fig. 2C is not unlike that expected during enzymatic catalysis. The highest point on the energy profile plays the role of the activation energy for the process whereas the corresponding conformations of the diffusing compound represent its transition state. Unlike in catalysis, however, the transition state during diffusion is not unique but includes all possible conformations of the traversing molecule and affected lipids. Given that, it can be immediately inferred that the overall rate of transmembrane diffusion is defined by the exact shape of the energy profile, not only by its highest point. In other words, the permeability coefficient of a compound is expected to correlate with its partition coefficient and other features of its interaction with the membrane that define the shape of the transmembrane energy profile. These other features are unique to the chemical structure of a compound as well as the lipids comprising the membrane. As a result, no universal relationship between membrane permeability and partition coefficient should be expected.
Whereas the permeability of the membrane is defined by the highest region on the energy diagram, where the compound concentration is at its lowest, the apparent affinity of the membrane to compounds is decided in its most populated region, at the trenches in the energy profile. In these regions, the concentration of compounds will be enriched compared to the bulk solution by the Boltzmann factor exp(+ΔGb/RT), where ΔGb is the binding energy of the compound to the given leaflet of the membrane (Fig. 2). This region of the membrane carries no information about the energetics of the transition state. Therefore, substituting the lipid-water partition coefficient for the oil-water partition coefficient in Equation 3 should degrade, not improve the predictive power of the equation.
Saturation of transport.
An unexpected result of quantitative drug accumulation studies was the finding that the rate of transmembrane transport is saturable (37–38, see also below). In contrast, Equation 2 predicts that the transmembrane flux is linearly proportional to the concentration gradient. This idea of linearity is propagated in college textbooks and has become deeply entrenched in the field. In reality, Equation 2 holds only in the limit of low solute concentrations. At high concentrations, the molecular nature of matter must be taken into account. Specifically, one needs to recall that chemical compounds traverse the membrane by interjecting themselves between the lipids. Thus, the concentration of compounds in the membrane cannot exceed the concentration of lipids. The imposed constraint is even stricter than that because of distortions introduced into the bilayer by the diffusing molecule. Compounds that do not dissolve the membrane are probably diffusing across it without touching each other. Even at saturation, such compounds would be separated from each other by lipids.
What would be the highest concentration that a compound can reach within the membrane? Assuming a hexagonal arrangement of lipids within the bilayer, we can postulate that each solute molecule must be surrounded by at least six lipids. Given that each lipid occupies the area of 0.6 nm2, this translates into one solute molecule per 3.6 nm2 or the concentration of Cmax ≈ 0.5 M within a leaflet of the membrane. This number is probably an overestimate because it is based solely on geometric considerations and ignores the context of the membrane in live bacteria, the high density of membrane proteins and physicochemical changes in the membrane caused by its saturation with a foreign chemical.
Even with these reservations in mind, the value of Cmax appears well above compound concentrations used in a typical antimicrobial experiment. Indeed, minimal inhibitory concentrations of antibiotics stay in sub- to micromolar range, and even solubilities of some drugs do not exceed several mM. For such compounds, the saturation on the membrane will be reached if their affinity for the membrane can compensate the gap between Cmax and their working concentration. This happens when the working concentration of a compound exceeds its dissociation constant. Thus, compounds with a dissociation constant of 1 μM or less are likely to saturate the membrane during a typical microbiology experiment. Such affinity requires the compound-membrane binding energy of 30 kJ/mol or greater. This energy is fairly modest as far as bioactive molecules are concerned. For example, ciprofloxacin has a total van-der-Waals surface area of 3 nm2, 0.7 nm2 of which are polar. Given the 0.07 N/m surface tension of water, intercalation of the hydrophobic core of ciprofloxacin into the lipid bilayer should release 90 kJ/mol of the free energy of hydration. Thus, most of amphiphilic compounds are likely to exhibit saturation during transmembrane diffusion.
Facilitated diffusion.
Highly polar or charged compounds cannot cross the bilayer with any reasonable rate and rely on facilitated diffusion to penetrate into the cell. In particular, beta-lactam and fluoroquinolone antibiotics hijack the major porins OmpF and OmpC of E. coli to reach their periplasmic target. During translocations, beta-lactams form a low affinity complex with OmpF/C38–40. Single channel conductance measurements revealed a fairly low residence time of 0.14 ms for ampicillin within OmpF38. Other conductance measurements reported residence times ranging from tens of microseconds to several milliseconds. OmpF is a highly abundant protein. Its copy number in growing E. coli cells was estimated at 100,00041. Given that, the maximal flux of ampicillin via OmpF can be calculated as Vpore = 0.03 mmol∙m−2∙s−1. For comparison, a compound with the permeability constant of 10−3 cm/s would require a concentration gradient of 3 mM to achieve the same flux. Such conditions are rarely met in experiment. For practical purposes, compounds that utilize highly abundant porins for permeation will likely operate below saturation.
The high maximal flux supported by porins is offset by their low on-rate and concomitantly low affinity to the substrate. The measured on-rates were orders of magnitude below the diffusion limit38 suggesting that drug binding site within the channel represents its transition state during translocation42. Because of the low affinity, only a fraction of porins is occupied by the drug during a typical experiment. The overall flux across the pore will be expected then to follow Equation 2, with the permeability constant
| (4), |
where KM is the Michaelis constant of the pore. For ampicillin in 1 M KCl solution38, Equation 4 yields Pm = 7∙10−6 cm/s. Although this value of Pm is rather high, it is not infinite. As a consequence, even compounds that penetrate the membrane via abundant porins do not necessarily flood the cells and are subject to efficient efflux by multidrug transporters.
Bulk diffusion of compounds through the medium rarely affects the rate of their permeation into the cell. This, however, becomes a factor for compounds with the permeability constant greater than 10−3 cm/s43. In such cases, diffusion must be explicitly considered to model drug uptake rates. This also applies to methods that rely on steady state measurements such as the comparison of MICs for resistant and susceptible strains in cases where the mechanism of resistance involves drug modification. Conversely, if the mechanism of resistance is solely the active drug efflux, all transport processes in the system will reach the steady state and the external drug concentration will be at equilibrium throughout the medium.
Bifurcation kinetics of efflux.
The permeability barrier of the cellular envelope would offer bacteria little protection from antibiotics were it not for their depletion inside the cell. Active drug efflux is one of key processes that achieves that. Recently, a quantitative model of drug permeation into Gram-negative bacteria with active efflux was constructed and matched to experiment37. The model was analytically solved for a system with a transporter acting across the OM under steady state conditions. The solution revealed that the system does not conform to Michaelis-Menten kinetics or the laws of diffusion. Rather, the system behaves in a highly non-linear manner, controlled by two kinetic parameters. The first parameter, the Efflux constant KE, describes the efficiency of drug efflux by the transporter (Fig. 1). This is a unitless parameter that relates the rates of active efflux and transmembrane diffusion at low drug concentrations, when all fluxes are far from equilibrium. For the reaction mechanism illustrated in Fig. 1, the efflux constant is given the equation:
| (5), |
where V and Km are the maximal velocity and Michaelis constant of the transporter, and 0.5k2 is the rate of transmembrane diffusion. The factor 0.5 accounts for the equal probabilities of the membrane-bound compound in Fig. 1 to dissociate into the periplasm and external space. In general, the microscopic rate constants k1 and k2 are not equal but reflect the Donnan equilibrium on the membrane for a given compound. The rate of transmembrane diffusion can be related to the permeability coefficient with a simple formula:
| (6), |
where dP is the average depth of the periplasm. Depending on the physiological state of the cell, dP varies between 10 nm and 20 nm44.
The value of the Efflux constant denotes the rate of active efflux compared to passive diffusion. As such, this value also reports the fold-reduction of the internal drug concentration at steady state due to the transporter in question. For Hoechst 33342, this value was evaluated at 42037. This number is a humbling reminder of the high efficiency of active drug efflux in protecting bacteria from external toxins.
Importantly, active efflux becomes inefficient when the substrate concentration in the periplasm exceeds its Michaelis constant. This happens when active efflux is unable to keep up with passive diffusion. If such conditions can be realized, active efflux becomes inconsequential, and the intracellular drug concentration approaches equilibrium Fig. 3. Thus, one strategy to overcome drug efflux would be to focus on compounds with high permeability through the outer membrane and working concentration that exceeds their Km for the transporter. In contrast, permeation of compounds with low diffusional flux requires a low Efflux constant, which means low V/Km values (see Equation 5). This latter mechanism is known as efflux avoidance and involves a reduction of the affinity of compounds for the transmembrane pump.
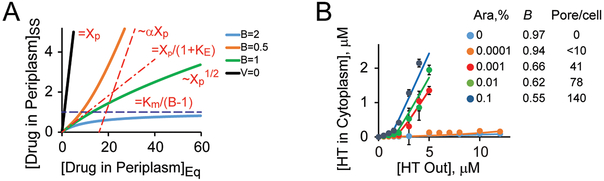
Figure 3. Bifurcation kinetics in simulation and live cells.
(A) The main regimes of compound accumulation in Gram-negative bacteria calculated for a model in Fig. 137. The steady state (SS) concentration of a drug in the periplasm is plotted against its equilibrium concentration (EQ), which takes into account the Donnan equilibrium. The black line is for compounds without active efflux, the three other lines are for compounds with the Efflux constant of 10 and the indicated values of the Barrier constant. At low drug concentration, the steady state is reduced compared to equilibrium by a factor of 1+KE in all cases. At high concentrations, the result depends on the value of B. If B < 1, the steady state asymptotically approaches the equilibrium modified by a factor α = (1-B)/(1+B). If B > 1, the intracellular drug level cannot exceed its threshold. (B) Experimental observation of a bifurcation in accumulation of Hoechst 33342 (HT) in E. coli cells harboring and inducible pore. The expression level of the pore and the resulting B-values are indicated. Reproduced from37.
The second kinetic parameter that controls the permeation of compounds across the double-membrane cell envelope is called the Barrier constant. For the mechanism depicted in Fig. 1, it is defined according to Equation 7:
| (7). |
Similar to the Efflux constant, this parameter is unitless and defines the ratio of active drug efflux and passive diffusion. Unlike the Efflux constant, B relates the maximal fluxes, which would be attainable at high substrate concentrations. As argued in the previous section, the maximal diffusional flux of compounds into the cell is finite regardless of the mechanism of transmembrane permeation. Even when diffusion is facilitated by an abundant channel protein, the value of B could be reasonably small given the high efflux rates of RND transporters.
The solution to the reaction depicted in Fig. 3 undergoes a bifurcation controlled by the value of B. When B is less than 1, the solution is qualitatively similar to the one derived without any consideration for possible saturation of influx. Although active efflux remains highly efficient at low drug concentrations, it can be ultimately overrun by increasing the external drug concentration (Fig. 3). Not so when B > 1. In this case, the desired drug concentration cannot be reached, and the transporter would not be saturated. The internal drug concentration will remain below a threshold no matter how much of it is added to the medium Fig. 3. If this threshold happens to be below the efficacy level of the drug, bacteria will be completely insensitive to this antibiotic. In principle, this mechanism of resistance, which is achieve through an increase of B above 1, could result in an infinitely large increase in MIC. The only way to sensitize bacteria to such compounds would be by bringing B down to below 1, either by increasing the maximal influx or by decreasing the maximal efflux rate (Table 1).
Experimental separation of active efflux and outer membrane contributions in activities of antibiotics.
How should compounds be modified to increase their intracellular accumulation and hence their effective intracellular concentrations? The permeation of each class of antibiotics is affected by a slow uptake and active efflux to its own degree. Establishing whether increased uptake or reduced efflux is the most efficient way to increase the potency of a specific class of compounds is one of the tasks in a drug optimization process. However, difficulties in separating the contributions of diffusion and active efflux in permeation and antibacterial activities of compounds significantly complicate this task45–46. Both small molecule potentiators and genetic modifications are used to manipulate the permeability of the OM. Polymyxins are often used to permeabilize the OM of E. coli, but these cationic peptides do not work in all strains, alter the physical structure of the outer membrane bilayer, and have significant interactions with other antibiotics47. Mutant strains with defects in the biosynthesis of LPS are also hypersusceptible to antibiotics, but as in the case of polymyxin, these modifications increase cell surface hydrophobicity and facilitate predominantly the penetration of hydrophobic antibiotics48. To address the penetration of hydrophilic compounds, bacterial strains and species producing mutant or larger porins are used45, 49. However, such porins are still highly selective according to the properties of the compounds.
To sidestep most of these difficulties, Krishnamoorthy et al. developed a hyperporination approach37, 50–51. In this approach, a modified siderophore uptake channel is expressed from the chromosome and used to create a large 2.4 nm nonspecific pore in the OM of Gram-negative bacteria. The pore is large enough for passage of small proteins52 and does not discriminate between compounds on the basis of their hydrophilicity50. When expressed at high levels, it bypasses the permeability barrier of the OM and reduces the Barrier factor below 1, enabling permeation even for such large antibiotics as vancomycin or rifampin (Table 1). The activities of antibiotics, the intracellular penetration of which is mostly limited by the OM barrier are affected by hyperporination to the largest extent, whereas substrates of efflux pumps are affected by both the OM barrier and active efflux (Table 1). Hyperporination of efflux-proficient cells and their efflux-deficient variants creates a set of strains with variable OM barriers and efflux efficiencies that report on contributions of these two factors in the intracellular accumulation of compounds and their antibacterial activities (Table 1).
Comparative analyses of such strains constructed for E. coli, P. aeruginosa, A. baumannii, B. cepacia, and B. thailandensis led to several unexpected findings. Kinetic measurements of the intracellular accumulation of fluorescent probes demonstrated that in all these species, the permeability barriers are synergistic and undergo kinetic bifurcations (Fig. 3B)50–51. As predicted by kinetic modeling, both hyperporination and efflux inactivation change the kinetics of intracellular accumulation of fluorescent probes (Fig. 3B). However, these changes are also species-specific and highlight the species-specific differences in the structure and composition of their OM, and in the activities and repertoires of the corresponding trans-envelope efflux pumps.
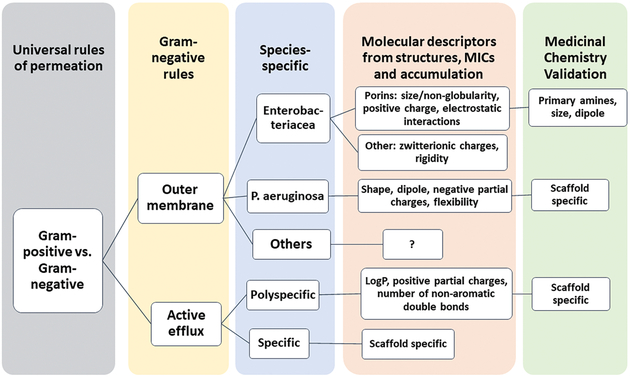
Growth inhibitory activities of antibiotics are in agreement with kinetic uptake data. Correlation analyses of antibiotic activities measured in different strains and species showed that synergism between active efflux pumps and passive barriers universally protect bacteria from structurally diverse antibiotics, even those previously thought to be privileged in efflux avoidance (Table 1)51. Antibiotics form four distinct clusters that are separated based on the magnitude of the effects caused by efflux inactivation and hyperporination. Notably, no obvious correlation was found between the consequences of transporter inactivation and hyperporination. These results suggest that the two effects are mechanistically independent of each other and that the two barriers “filter” antibiotics based on different properties (Fig. 4). The species-specific variabilities in activities of antibiotics are clustered at the lower level, reflecting the contribution of structural particularities of the permeability barriers (Fig. 4).
Figure 4. Rules of permeation and their hierarchy.
Summarized views and results discussed here and in9–10, 51, 55.
Despite the biological diversity of permeability barriers and the lack of apparent chemical similarities between antibiotic classes, antibiotics within each of the identified clusters are expected to share certain structural characteristics that are recognized by either the OM or the active efflux barrier51. Identification of these structural characteristics could lead to the more efficient optimization of antibacterial compounds against Gram-negative bacteria.
Molecular determinants of antibiotic activities and permeation.
The goal of any discovery efforts is to obtain the most potent antibacterial activity by maximizing the affinity to the target and OM permeation and by minimizing active efflux of a compound. Two general strategies can be exploited to identify structural properties of compounds that are associated with permeation and efflux. The first strategy utilizes bacterial strains with efflux- proficient and deficient genetic backgrounds and variable permeability of the OM to trace the changes in intracellular activities of compounds as related to chemical modifications and physico-chemical properties of compounds (the structure-activity relationship or SAR). The second strategy utilizes similar sets of strains but follows the intracellular accumulation of compounds with the goal to define the structure-accumulation relationship, which is independent on the biological activity of compound. Both strategies can be integrated with various biochemical and computational approaches to define affinities to the specific targets, efflux pumps and/or affinities and permeation through specific OM porins. A variety of statistical methods and machine learning techniques are used at different stages to analyze relationships and to generate predictive models that eventually must be validated in medicinal chemistry programs (Fig. 4).
Studies of antibacterial activities.
Previous SAR studies of antibacterial compounds in wild-type and efflux-deficient strains led to partial characterization of the specificity of efflux pumps45–46, 53. Studies of Haemophilus influenzae, E. coli, and P. aeruginosa strains revealed that the antibacterial activities of the very polar and low-molecular-weight (MW) compounds on the one hand, and of zwitterionic and high-MW compounds on the other, tend to be the least affected by efflux pumps, suggesting that such compounds are poor substrates for multidrug transporters45–46. In contrast, an increase in hydrophobicity was found to correlate with the increased propensity of a compound to be a substrate of efflux pumps in studies of Salmonella enterica serovar Typhimurium53. However, the extent to which the OM barrier biases these conclusions remained unclear.
Recently, the relative potencies of fluoroquinolones and beta-lactams (as measured by MIC) were analyzed in hyperporinated, efflux-deficient and wild-type strains of E. coli and P. aeruginosa and the dependence of the antibacterial activities on efflux, the OM barrier, or both (as measured by MIC ratios) in these species10. A random forest (RF) classification was then applied to measured MICs and MIC ratios to identify the most important physicochemical properties. The top 20 descriptors for classifying the MICs or MIC ratios for each species were identified and showed that: 1) charge descriptors dominate the MIC fingerprints of both species and are important for both permeation and antibacterial activity; 2) physical properties, chemical structure and shape descriptors are selective for MIC ratios and describe contributions of active efflux and OM barriers and their synergistic interactions; and 3) the molecular shape descriptors are characteristic of the OM barriers. These studies further suggested that E. coli efflux pumps are optimized to expel compounds that permeate the OM, whereas in P. aeruginosa the relationships are more complex, with efflux pumps and the OM barrier selecting for different properties of compounds (Fig. 4)10.
Permeation through porins.
As discussed above hydrophilic small beta-lactams cross the OM of E. coli and other Enterobacteriaceae species through the water-filled porins. The permeation through these porins is characterized in a great detail by a combination of structural, electrophysiological and computational approaches. Together this multidisciplinary approach generated a scoring function that can be used to predict the permeation of compounds through enterobacterial porins9. The generalized “rules” of permeation through porins emphasize size of the molecule and electrostatic interactions of the diffusing molecule with the porin through its charge and dipole (Fig. 4). These properties are also identified as top descriptors of antibacterial activities of beta-lactam and fluoroquinolones and their dependence on permeation in E. coli (see above)10 and in intracellular accumulation experiments discussed below54–55.
Whole cell studies of compound permeation and accumulation.
Although studies of antibiotic activities are well-standardized and generate data that are comparable across different studies, the approaches to establish structure-accumulation relationships are more complicated and their specific outcomes are sensitive to the experimental design and methods. The detection methods for measuring changes in compound concentrations are very diverse and include radiometry,56–60 intrinsic fluorescence61 and fluorescent probes,58–59, 62–70 Raman spectroscopy,71 enzyme kinetics (e.g., for β-lactamase, peptidases),49, 72–74 Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS)75 and LC-MS/MS59, 76–80. Among these, MS- and fluorescence-based techniques are the most broadly used and versatile.
The intracellular accumulation of environment-sensitive dyes is the most sensitive approach to compare and analyze differences in permeability barriers in laboratory and clinical isolates and in genetically altered variants57, 74, 81–89. The fluorescence of these dyes is enhanced when they bind to membranes, proteins or nucleic acids. As discussed above, time-dependent changes in fluorescence provide kinetic information about permeation, efflux, and intracellular accumulation (Fig. 3A). Such assays could be used in a high throughput format or can be adapted to microfluidics and microscopy for single-cell analyses90–91. The fluorescence enhancement of the probe is often specific to cellular compartments, enabling assessment of intracellular localization. These studies however are limited to specific chemical classes of compounds.
At present, LC-MS/MS could probably be considered as the gold standard in efflux and permeation measurements due to its broad applicability, high accuracy and versatility.59, 79–80, 92–93 However, the method has certain limitations, as cells must be separated from the external solution without a loss of the intracellular compound. Depending on the properties of compounds, significant experimental error can be present due to non-specific binding to filter, plastic tubes or cell surfaces or washing the compound out of cells94. Some of these problems are alleviated by measuring relative changes in intracellular concentration by using sets of genetically modified strains with varying efflux capacities and/or OM permeabilities. The throughput of the assay can be increased by changing to a 96-well format and taking advantage of 9 s/sample solid phase extraction (SPE)–MS80. Yet, the conclusions based on LC-MS/MS measurements depend on specifics of the experiment.
Davis et al. used principal component analysis to identify physical properties associated with high accumulation of sulfonyl adenosines in E. coli and Gram-positive bacteria as measured by LC-MS/MS at a single external concentration of compounds. This revealed significant differences between compounds with similar logP values, indicating that hydrophobicity alone is insufficient to predict accumulation accurately. Additional positive correlations for the intracellular accumulation in E. coli were found with ring content and size, and negative correlations with ring complexity, H-bonding, heteroatoms, and three dimensionality77. These analyses also helped rationalize differences observed across the other bacteria. Using a similar experimental design and a single-point measurements, Richter et al. analyzed the accumulation of ~180 diverse compounds in E. coli and applied RF classification to 297 molecular descriptors to identify properties that are most important for the intracellular accumulation55. Molecules that are most likely to accumulate include primary or secondary amines, are rigid, have low globularity, and are amphiphilic. Notably, charge, molecular weight, and clogD7.4 showed no correlative relationship with accumulation.
Both these studies successfully used the discovered heuristics to design molecules with improved intracellular accumulation and antibacterial activities. Focused on physico-chemical properties of compounds, they did not address the question whether the changes in intracellular accumulation of compounds correlate with their antibacterial activities. To address this specific question, Iyer et al.54 analyzed the intracellular accumulation of a set of over a hundred compounds (inhibitors of NAD-dependent DNA ligase, LigA) in the wild type and efflux-deficient E. coli cells. The set included compounds with a range of antibacterial and biochemical potencies as measured by IC50 and MIC values. In agreement with the above studies, either hydrophobicity or positive charge of compounds increased the amount of the total accumulated compound and accumulation was affected by active efflux. Interestingly, these changes in the accumulation failed to predict whole-cell antibacterial activities of compounds, leading to the conclusion that the single-point measurements of total accumulation are not necessarily predictive of the amount of compound that will be available to the intracellular target. Indeed, the kinetics of compound accumulation in Gram-negative bacteria varies depending on the intracellular compartment (Fig. 3 and see above) and several concentrations and time points have to be analyzed to establish functional correlations.
Together these studies demonstrated the conceptual and methodological feasibility of establishing species-specific “rules” of permeation. At present we have a practical understanding of how compounds permeate the protective barriers of Enterobacteriacea (Fig. 4). Further studies are needed to achieve similar levels of understanding for other Gram-negative bacteria.
Conclusions
In the past few years, significant advances have been made in rationalization of drug permeation into Gram-negative pathogens possessing two membranes with active efflux. In combination, kinetic modeling, mechanistic analyses and experimental measurements of compound accumulation and activities in cells are posed to solve the riddle of Gram-negative permeability barriers. The methodological developments in controlling the permeability barrier of the outer membrane and in separating contributions of efflux and the low permeation in activities and accumulation of compounds are expected to reveal molecular properties of compounds selected by different barriers. Increasing the chemical diversity of analytes and incorporation of integrated kinetic approaches in analyses of intracellular accumulation and target engagement could further stimulate medicinal chemistry programs aimed at optimization of antibacterial agents against multidrug resistant Gram-negative bacteria.
Acknowledgements
This work was supported by National Institutes of Health/ National Institute of Allergy and Infectious Diseases Grants AI136795 and AI136799 and Defense Threat Reduction Agency grant HDTRA1-14-1-0019. The contents of this article do not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.
Footnotes
Competing Interests. Authors declare no competing interests.
References
- 1.Tommasi R; Brown DG; Walkup GK; Manchester JI; Miller AA, ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 2015, 14 (8), 529–42. [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW; Scheld M; Bartlett J; Talbot GH; Bradley JS; Spellberg B; Edwards JE; Gilbert D; Rice LB, Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clinical Infectious Diseases 2009, 48 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H, Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 1994, 264 (5157), 382–388. [DOI] [PubMed] [Google Scholar]
- 4.Zgurskaya HI; Rybenkov VV; Krishnamoorthy G; Leus IV, Trans-envelope multidrug efflux pumps of Gram-negative bacteria and their synergism with the outer membrane barrier. Res Microbiol 2018, 169 (7–8), 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneleit P; Kleber H-P, The outer membrane of Acinetobacter: structure-function relationships In The Biology of Acinetobacter, Towner KJ; Bergogne-Berezin E; Fewson CA, Eds. Plenum Press: New York and London, 1991; pp 259–272. [Google Scholar]
- 6.Kocincova D; Lam JS, Structural diversity of the core oligosaccharide domain of Pseudomonas aeruginosa lipopolysaccharide. Biochemistry Moscow 2011, 76 (7), 755–760. [DOI] [PubMed] [Google Scholar]
- 7.Pier GB, Pseudomonas aeruginosa lipopolysaccharide: A major virulence factor, initiator of inflammation and target for effective immunity. International Journal of Medical Microbiology 2007, 297 (5), 277–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinion-Dubiel AD; Goldberg JB, Review: Lipopolysaccharide of Burkholderia cepacia complex. Journal of Endotoxin Research 2003, 9 (4), 201–213. [DOI] [PubMed] [Google Scholar]
- 9.Acosta-Gutiérrez S; Ferrara L; Pathania M; Masi M; Wang J; Bodrenko I; Zahn M; Winterhalter M; Stavenger RA; Pagès J-M; Naismith JH; van den Berg B; Page MGP; Ceccarelli M, Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins. ACS Infectious Diseases 2018, 4 (10), 1487–1498. [DOI] [PubMed] [Google Scholar]
- 10.Cooper SJ; Krishnamoorthy G; Wolloscheck D; Walker JK; Rybenkov VV; Parks JM; Zgurskaya HI, Molecular Properties That Define the Activities of Antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect Dis 2018, 4 (8), 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eren E; Parkin J; Adelanwa A; Cheneke B; Movileanu L; Khalid S; van den Berg B, Toward Understanding the Outer Membrane Uptake of Small Molecules by Pseudomonas aeruginosa. Journal of Biological Chemistry 2013, 288 (17), 12042–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Berg B, Structural Basis for Outer Membrane Sugar Uptake in Pseudomonads. Journal of Biological Chemistry 2012, 287 (49), 41044–41052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier S; Bouffartigues E; Bodilis J; Maillot O; Lesouhaitier O; Feuilloley MGJ; Orange N; Dufour A; Cornelis P, Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev 2017, 41 (5), 698–722. [DOI] [PubMed] [Google Scholar]
- 14.Isabella VM; Campbell AJ; Manchester J; Sylvester M; Nayar AS; Ferguson KE; Tommasi R; Miller AA, Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa. Chem Biol 2015, 22 (4), 535–47. [DOI] [PubMed] [Google Scholar]
- 15.Bhamidimarri SP; Zahn M; Prajapati JD; Schleberger C; Söderholm S; Hoover J; West J; Kleinekathöfer U; Bumann D; Winterhalter M; van den Berg B, A Multidisciplinary Approach toward Identification of Antibiotic Scaffolds for Acinetobacter baumannii. Structure 2019, 27 (2), 268–280.e6. [DOI] [PubMed] [Google Scholar]
- 16.Zahn M; Bhamidimarri Satya P.; Baslé A; Winterhalter M; van den Berg B, Structural Insights into Outer Membrane Permeability of Acinetobacter baumannii. Structure 2016, 24 (2), 221–231. [DOI] [PubMed] [Google Scholar]
- 17.Zahn M; D’Agostino T; Eren E; Baslé A; Ceccarelli M; van den Berg B, Small-Molecule Transport by CarO, an Abundant Eight-Stranded β-Barrel Outer Membrane Protein from Acinetobacter baumannii. Journal of Molecular Biology 2015, 427 (14), 2329–2339. [DOI] [PubMed] [Google Scholar]
- 18.Gislason AS; Turner K; Domaratzki M; Cardona ST, Comparative analysis of the Burkholderia cenocepacia K56–2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia. Microbial Genomics 2017, 3 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silver LL, A Gestalt approach to Gram-negative entry. Bioorg Med Chem 2016, 24 (24), 6379–6389. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido H; Thanassi DG, Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother 1993, 37 (7), 1393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido H; Zgurskaya HI, Antibiotic efflux mechanisms. Curr Opin Infect Dis 1999, 12 (6), 529–36. [DOI] [PubMed] [Google Scholar]
- 22.Bay DC; Rommens KL; Turner RJ, Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim Biophys Acta 2007. [DOI] [PubMed] [Google Scholar]
- 23.Omote H; Hiasa M; Matsumoto T; Otsuka M; Moriyama Y, The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 2006, 27 (11), 587–93. [DOI] [PubMed] [Google Scholar]
- 24.Eicher T; Cha HJ; Seeger MA; Brandstatter L; El-Delik J; Bohnert JA; Kern WV; Verrey F; Grutter MG; Diederichs K; Pos KM, Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 2012, 109 (15), 5687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruggerone P; Murakami S; Pos KM; Vargiu AV, RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem 2013, 13 (24), 3079–100. [DOI] [PubMed] [Google Scholar]
- 26.Du D; Wang Z; James NR; Voss JE; Klimont E; Ohene-Agyei T; Venter H; Chiu W; Luisi BF, Structure of the AcrAB-TolC multidrug efflux pump. Nature 2014, 509 (7501), 512–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z; Fan G; Hryc CF; Blaza JN; Serysheva II; Schmid MF; Chiu W; Luisi BF; Du D, An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zgurskaya HI; Weeks JW; Ntreh AT; Nickels LM; Wolloscheck D, Mechanism of coupling drug transport reactions located in two different membranes. Front Microbiol 2015, 6, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aires JR; Kohler T; Nikaido H; Plesiat P, Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 1999, 43 (11), 2624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Kievit TR; Parkins MD; Gillis RJ; Srikumar R; Ceri H; Poole K; Iglewski BH; Storey DG, Multidrug Efflux Pumps: Expression Patterns and Contribution to Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. Antimicrob Agents Chemother 2001, 45 (6), 1761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne S; Courvalin P; Perichon B, Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 2011, 55 (3), 947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter TS; Parkin J; Khalid S, The Free Energy of Small Solute Permeation through the Escherichia coli Outer Membrane Has a Distinctly Asymmetric Profile. J Phys Chem Lett 2016, 7 (17), 3446–51. [DOI] [PubMed] [Google Scholar]
- 33.Comer J; Schulten K; Chipot C, Diffusive Models of Membrane Permeation with Explicit Orientational Freedom. J Chem Theory Comput 2014, 10 (7), 2710–8. [DOI] [PubMed] [Google Scholar]
- 34.Parisio G; Stocchero M; Ferrarini A, Passive Membrane Permeability: Beyond the Standard Solubility-Diffusion Model. Journal of Chemical Theory and Computation 2013, 9 (12), 5236–5246. [DOI] [PubMed] [Google Scholar]
- 35.Mälkiä A; Murtomäki L; Urtti A; Kontturi K, Drug permeation in biomembranes: In vitro and in silico prediction and influence of physicochemical properties. European Journal of Pharmaceutical Sciences 2004, 23 (1), 13–47. [DOI] [PubMed] [Google Scholar]
- 36.Mitragotri S; Johnson ME; Blankschtein D; Langer R, An Analysis of the Size Selectivity of Solute Partitioning, Diffusion, and Permeation across Lipid Bilayers. Biophysical Journal 1999, 77 (3), 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westfall DA; Krishnamoorthy G; Wolloscheck D; Sarkar R; Zgurskaya HI; Rybenkov VV, Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS One 2017, 12 (9), e0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nestorovich EM; Danelon C; Winterhalter M; Bezrukov SM, Designed to penetrate: Time-resolved interaction of single antibiotic molecules with bacterial pores. Proceedings of the National Academy of Sciences 2002, 99 (15), 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pages JM; James CE; Winterhalter M, The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 2008, 6 (12), 893–903. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz G; Danelon C; Winterhalter M, On Translocation through a Membrane Channel via an Internal Binding Site: Kinetics and Voltage Dependence. Biophysical Journal 2003, 84 (5), 2990–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikaido H, Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003, 67 (4), 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acosta-Gutierrez S; Ferrara L; Pathania M; Masi M; Wang J; Bodrenko I; Zahn M; Winterhalter M; Stavenger RA; Pages JM; Naismith JH; van den Berg B; Page MGP; Ceccarelli M, Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins. ACS Infect Dis 2018, 4 (10), 1487–1498. [DOI] [PubMed] [Google Scholar]
- 43.Malkia A; Murtomaki L; Urtti A; Kontturi K, Drug permeation in biomembranes: in vitro and in silico prediction and influence of physicochemical properties. Eur J Pharm Sci 2004, 23 (1), 13–47. [DOI] [PubMed] [Google Scholar]
- 44.Van Wielink JE; Duine JA, How big is the periplasmic space? Trends in Biochemical Sciences 1990, 15 (4), 136–137. [DOI] [PubMed] [Google Scholar]
- 45.Manchester JI; Buurman ET; Bisacchi GS; McLaughlin RE, Molecular determinants of AcrB-mediated bacterial efflux implications for drug discovery. J Med Chem 2012, 55 (6), 2532–7. [DOI] [PubMed] [Google Scholar]
- 46.Brown DG; May-Dracka TL; Gagnon MM; Tommasi R, Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J Med Chem 2014, 57 (23), 10144–61. [DOI] [PubMed] [Google Scholar]
- 47.Vaara M, Agents that increase the permeability of the outer membrane. Microbiol Rev 1992, 56 (3), 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuorio R; Vaara M, The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob Agents Chemother 1992, 36 (4), 826–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kojima S; Nikaido H, Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proceedings of the National Academy of Sciences 2013, 110 (28), E2629–E2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishnamoorthy G; Wolloscheck D; Weeks JW; Croft C; Rybenkov VV; Zgurskaya HI, Breaking the Permeability Barrier of Escherichia coli by Controlled Hyperporination of the Outer Membrane. Antimicrob Agents Chemother 2016, 60 (12), 7372–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnamoorthy G; Leus IV; Weeks JW; Wolloscheck D; Rybenkov VV; Zgurskaya HI, Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. MBio 2017, 8 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohammad MM; Howard KR; Movileanu L, Redesign of a plugged beta-barrel membrane protein. The Journal of biological chemistry 2011, 286 (10), 8000–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikaido H; Basina M; Nguyen V; Rosenberg EY, Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. Journal of Bacteriology 1998, 180 (17), 4686–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iyer R; Sylvester MA; Velez-Vega C; Tommasi R; Durand-Reville TF; Miller AA, Whole-Cell-Based Assay To Evaluate Structure Permeation Relationships for Carbapenem Passage through the Pseudomonas aeruginosa Porin OprD. ACS Infect Dis 2017. [DOI] [PubMed] [Google Scholar]
- 55.Richter MF; Drown BS; Riley AP; Garcia A; Shirai T; Svec RL; Hergenrother PJ, Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasdemir UO; Chevalier J; Nordmann P; Pages JM, Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. Journal of clinical microbiology 2004, 42 (6), 2701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li XZ; Zhang L; Nikaido H, Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 2004, 48 (7), 2415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piddock LJV; Johnson MM, Accumulation of 10 Fluoroquinolones by Wild-Type or Efflux Mutant Streptococcus pneumoniae. Antimicrobial Agents and Chemotherapy 2002, 46 (3), 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai H; Rose K; Liang LH; Dunham S; Stover C, Development of a liquid chromatography/mass spectrometry-based drug accumulation assay in Pseudomonas aeruginosa. Anal Biochem 2009, 385 (2), 321–5. [DOI] [PubMed] [Google Scholar]
- 60.Diver JM; Piddock LJV; Wise R, The accumulation of five quinolone antibacterial agents by Escherichia coli. Journal of Antimicrobial Chemotherapy 1990, 25 (3), 319–333. [DOI] [PubMed] [Google Scholar]
- 61.Cinquin B; Maigre L; Pinet E; Chevalier J; Stavenger RA; Mills S; Réfrégiers M; Pagès J-M, Microspectrometric insights on the uptake of antibiotics at the single bacterial cell level. Scientific reports 2015, 5, 17968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ricci V; Piddock L, Accumulation of garenoxacin by Bacteroides fragilis compared with that of five fluoroquinolones. J Antimicrob Chemother 2003, 52 (4), 605–9. [DOI] [PubMed] [Google Scholar]
- 63.Bensikaddour H; Fa N; Burton I; Deleu M; Lins L; Schanck A; Brasseur R; Dufrene YF; Goormaghtigh E; Mingeot-Leclercq MP, Characterization of the interactions between fluoroquinolone antibiotics and lipids: a multitechnique approach. Biophys J 2008, 94 (8), 3035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakashima R; Sakurai K; Yamasaki S; Nishino K; Yamaguchi A, Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 2011, 480 (7378), 565–9. [DOI] [PubMed] [Google Scholar]
- 65.Asuquo AE; Piddock LJ, Accumulation and killing kinetics of fifteen quinolones for Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa. J Antimicrob Chemother 1993, 31 (6), 865–80. [DOI] [PubMed] [Google Scholar]
- 66.Bazile S; Moreau N; Bouzard D; Essiz M, Relationships among antibacterial activity, inhibition of DNA gyrase, and intracellular accumulation of 11 fluoroquinolones. Antimicrobial Agents and Chemotherapy 1992, 36 (12), 2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bedard J; Wong S; Bryan LE, Accumulation of enoxacin by Escherichia coli and Bacillus subtilis. Antimicrobial Agents and Chemotherapy 1987, 31 (9), 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mccaffrey C; Bertasso A; Pace J; Georgopapadakou NH, Quinolone Accumulation in Escherichia-Coli, Pseudomonas-Aeruginosa, and Staphylococcus-Aureus. Antimicrobial Agents and Chemotherapy 1992, 36 (8), 1601–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piddock LJV; Jin YF; Ricci V; Asuquo AE, Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. Journal of Antimicrobial Chemotherapy 1999, 43 (1), 61–70. [DOI] [PubMed] [Google Scholar]
- 70.Chapman JS; Georgopapadakou NH, Routes of quinolone permeation in Escherichia coli. Antimicrobial Agents and Chemotherapy 1988, 32 (4), 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heidari-Torkabadi H; Bethel CR; Ding Z; Pusztai-Carey M; Bonnet R; Bonomo RA; Carey PR, “Mind the Gap”: Raman Evidence for Rapid Inactivation of CTX-M-9 β-Lactamase Using Mechanism-Based Inhibitors that Bridge the Active Site. Journal of the American Chemical Society 2015, 137 (40), 12760–12763. [DOI] [PubMed] [Google Scholar]
- 72.Zimmermann W; Rosselet A, Function of the Outer Membrane of Escherichia coli as a Permeability Barrier to Beta-Lactam Antibiotics. Antimicrob Agents Chemother 1977, 12 (3), 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikaido H; Rosenberg EY; Foulds J, Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J Bacteriol 1983, 153 (1), 232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugawara E; Nikaido H, OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J Bacteriol 2012, 194 (15), 4089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tian H; Six DA; Krucker T; Leeds JA; Winograd N, Subcellular Chemical Imaging of Antibiotics in Single Bacteria Using C60-Secondary Ion Mass Spectrometry. Analytical Chemistry 2017, 89 (9), 5050–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graef F; Gordon S; Lehr CM, Anti-infectives in Drug Delivery-Overcoming the Gram-Negative Bacterial Cell Envelope. Curr Top Microbiol Immunol 2016, 398, 475–496. [DOI] [PubMed] [Google Scholar]
- 77.Davis TD; Gerry CJ; Tan DS, General Platform for Systematic Quantitative Evaluation of Small-Molecule Permeability in Bacteria. ACS Chemical Biology 2014, 9 (11), 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhat J; Narayan A; Venkatraman J; Chatterji M, LC-MS based assay to measure intracellular compound levels in Mycobacterium smegmatis: linking compound levels to cellular potency. J Microbiol Methods 2013, 94 (2), 152–8. [DOI] [PubMed] [Google Scholar]
- 79.Zhou Y; Joubran C; Miller-Vedam L; Isabella V; Nayar A; Tentarelli S; Miller A, Thinking Outside the “Bug”: A Unique Assay To Measure Intracellular Drug Penetration in Gram-Negative Bacteria. Analytical Chemistry 2015, 87 (7), 3579–3584. [DOI] [PubMed] [Google Scholar]
- 80.Widya M; Pasutti WD; Sachdeva M; Simmons RL; Tamrakar P; Krucker T; Six DA, Development and Optimization of a Higher-Throughput Bacterial Compound Accumulation Assay. ACS Infectious Diseases 2019, 5 (3), 394–405. [DOI] [PubMed] [Google Scholar]
- 81.Masi M; Refregiers M; Pos KM; Pages JM, Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat Microbiol 2017, 2, 17001. [DOI] [PubMed] [Google Scholar]
- 82.Husain F; Nikaido H, Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol 2010, 78 (2), 320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin J; Hsieh YH; Cui J; Damera K; Dai C; Chaudhary AS; Zhang H; Yang H; Cao N; Jiang C; Vaara M; Wang B; Tai PC, Using Chemical Probes to Assess the Feasibility of Targeting SecA for Developing Antimicrobial Agents against Gram-Negative Bacteria. ChemMedChem 2016, 11 (22), 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allam A; Maigre L; Alves de Sousa R; Dumont E; Vergalli J; Pages JM; Artaud I, New amphiphilic neamine conjugates bearing a metal binding motif active against MDR E. aerogenes Gram-negative bacteria. Eur J Med Chem 2017, 127, 748–756. [DOI] [PubMed] [Google Scholar]
- 85.Coldham NG; Webber M; Woodward MJ; Piddock LJ, A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother 2010, 65 (8), 1655–63. [DOI] [PubMed] [Google Scholar]
- 86.Abdali N; Parks JM; Haynes KM; Chaney JL; Green AT; Wolloscheck D; Walker JK; Rybenkov VV; Baudry J; Smith JC; Zgurskaya HI, Reviving Antibiotics: Efflux Pump Inhibitors That Interact with AcrA, a Membrane Fusion Protein of the AcrAB-TolC Multidrug Efflux Pump. ACS Infect Dis 2017, 3 (1), 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Opperman TJ; Kwasny SM; Kim HS; Nguyen ST; Houseweart C; D’Souza S; Walker GC; Peet NP; Nikaido H; Bowlin TL, Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother 2014, 58 (2), 722–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian CD; Wu XC; Teng Y; Zhao WP; Li O; Fang SG; Huang ZH; Gao HC, Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 2012, 56 (3), 1458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaczmarek FM; Dib-Hajj F; Shang W; Gootz TD, High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother 2006, 50 (10), 3396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iino R; Sakakihara S; Matsumoto Y; Nishino K, Large-Scale Femtoliter Droplet Array for Single Cell Efflux Assay of Bacteria In Bacterial Multidrug Exporters: Methods and Protocols, Yamaguchi A; Nishino K, Eds. Springer New York: New York, NY, 2018; pp 331–341. [DOI] [PubMed] [Google Scholar]
- 91.Iino R; Nishino K; Noji H; Yamaguchi A; Matsumoto Y, A Microfluidic Device for Simple and Rapid Evaluation of Multidrug Efflux Pump Inhibitors. Frontiers in Microbiology 2012, 3 (40). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mortimer PGS; Piddock LJV, A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. Journal of Antimicrobial Chemotherapy 1991, 28 (5), 639–653. [DOI] [PubMed] [Google Scholar]
- 93.Liu HL; Sabus C; Carter GT; Du C; Avdeef A; Tischler M, In vitro permeability of poorly aqueous soluble compounds using different solubilizers in the PAMPA assay with liquid chromatography/mass spectrometry detection. Pharm Res 2003, 20 (11), 1820–1826. [DOI] [PubMed] [Google Scholar]
- 94.Six DA; Krucker T; Leeds JA, Advances and challenges in bacterial compound accumulation assays for drug discovery. Curr Opin Chem Biol 2018, 44, 9–15. [DOI] [PubMed] [Google Scholar]
- 95.CLSI, Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7th ed., CLSI document M02-A11. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA, 2012. [Google Scholar]