Abstract
Objective:
We developed and validated a nomogram predicting the likelihood of occult lymph node metastases in surgically resectable esophageal cancers.
Background:
Patients with esophageal cancer with positive lymph nodes benefit from neoadjuvant therapy, but limitations in current clinical staging techniques mean nodal metastases often go undetected preoperatively.
Methods:
The National Cancer Database was queried for patients with clinical T1–3N0M0 cancer undergoing upfront esophagectomy from 2004 to 2014. Multivariable logistic regression was used to develop the risk model using both statistical significance and clinical importance criteria for variable selection. Predictive accuracy was assessed and bootstrapping was used for validation. A nomogram was constructed for presentation of the final model.
Results:
Of 3186 patients, 688 (22%) had pathologic lymph node involvement (pN+) and 2498 (78%) had pN0 status. Variables associated with pN+ status included histology [adenocarcinoma vs squamous: odds ratio (OR) 1.75], tumor stage (T1: reference, T2: OR 1.90, T3: OR 2.17), tumor size (<1 cm: reference, 1–2 cm: OR 2.25, 2–3 cm: OR 3.82, 3–4 cm: OR 5.40, 4–5 cm: OR 5.66, ≥5 cm: OR 6.02), grade (1: reference, 2: OR 2.62, 3: OR 4.39, 4: OR 4.15, X: OR 1.87), and presence of lymphovascular invasion (absent: reference, present: OR 4.70, missing: OR 1.87), all P < 0.001. A nomogram with these variables had good predictive accuracy (Brier score: 0.14, calibration slope: 0.97, c-index: 0.77).
Conclusions:
We created a nomogram predicting the likelihood of pathologic lymph node involvement in patients with esophageal cancer who are clinically node negative using a generalizable dataset. Risk stratification with this nomogram could improve delivery of appropriate perioperative care.
Keywords: esophageal cancer, lymph nodes, nomogram
The treatment of esophageal cancer varies by stage. Patients with esophageal cancer with positive lymph nodes experience a survival benefit with neoadjuvant chemoradiation before surgical resection.1 National guidelines have incorporated this evidence and recommend multidisciplinary treatment for patients with demonstrated locoregional disease, whereas patients with early stage localized cancer may be candidates for surgical resection alone.2,3 Clinical staging is typically performed with endoscopic ultrasound (EUS) and positron emission tomography-computed tomography, but limitations in these techniques mean lymph node metastases often go undetected preoperatively. Prior institutional studies have shown rates of occult nodal metastases can range from 16% to 39%, even for clinical early stage disease.4,5 This indicates that current staging practices alone may not be adequate for appropriately risk-stratifying patients preoperatively.
Given the inaccuracies in clinical staging for identifying nodal disease, physicians often consider additional clinical data when deciding whether to prescribe induction chemoradiation for clinically node-negative patients. Efforts to identify clinical variables associated with occult nodal metastases have revealed that multiple tumor-related variables can be predictive.6,7 Development of a usable clinical tool for predicting which patients are at high risk of occult nodal metastases would improve patient selection for appropriate preoperative therapy.
In this study, we aimed to develop and validate a nomogram using readily available clinical variables from a broadly generalizable database to predict the likelihood of occult lymph node metastases in surgically resectable esophageal cancers.
METHODS
Data Source
This study selected eligible patients from the National Cancer Database (NCDB) Participant Use File for esophageal cancer. The NCDB is a retrospective dataset from the American Cancer Society and the American College of Surgeons that captures greater than 70% of all new nationwide cancer diagnoses from more than 1500 Commission on Cancer accredited hospitals. This dataset was selected because its broad representation of patients with esophageal cancer would allow the resulting nomogram to be generalizable to the United States population. This study was exempt from Washington University’s Institutional Review Board approval because the dataset is deidentified.
Patient Selection
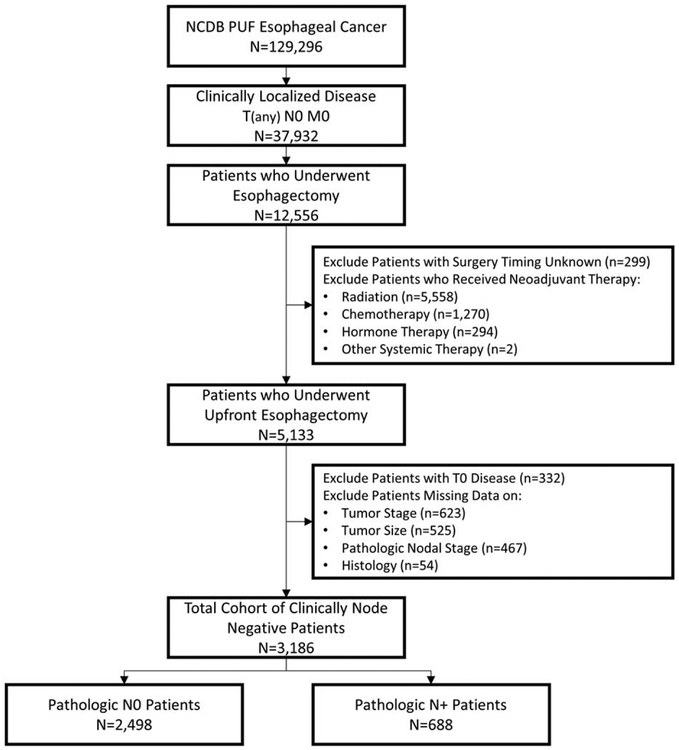
Individuals who had clinically localized esophageal cancer (cT1–3N0M0) and underwent esophagectomy from 2004 to 2014 were included in the study. Patients were excluded if they underwent any type of neoadjuvant therapy (including radiation, chemotherapy, hormone, therapy, or other systemic therapy); they were documented as having clinical T0 (no evidence of a primary tumor) or Tis (high-grade dysplasia) disease; or they were missing data on the timing of their surgery, tumor stage, tumor size, pathologic lymph node staging, or histology. This cohort was then divided into 2 groups for analysis: patients who were pathologically node negative (pN0) and patients who were pathologically node positive (pN+), including pN1–3 disease. A summary of study selection criteria can be seen in our Consolidated Standards of Reporting Trials diagram (Fig. 1).
FIGURE 1.
Consolidated Standards of Reporting Trials (CONSORT) diagram.
Development of the Prediction Model
We abstracted and categorized the following demographic and tumor variables: age, sex, race (white vs non-white), insurance status (private vs public), median annual income by zip code (lowest quartile of <$38,000 vs >$38,000), education status by zip code (lowest quartile of >21% without a high school degree vs the remainder), population by zip code (>250,000 vs <250,000), Charlson Deyo Score (0, 1, ≥2), treatment center type (academic vs nonacademic), year of diagnosis, histology (squamous cell vs adenocarcinoma), tumor stage, tumor size, grade, and presence of lymphovascular invasion.
Baseline characteristics were assessed for the entire cohort and compared between pN0 and pN+ patients using t tests and chisquare tests for continuous and categorical variables, respectively. Univariable analysis was performed to identify variables associated with pN+ status. We then used multivariable logistic regression to develop the risk model. We selected predictors using both statistical significance (P < 0.05) and clinical importance criteria. All statistical analyses were performed using SAS version 9.4 (SAS Institute, Inc, Cary, NC) and R Package Regression Modeling Strategies.
Evaluation of the Predictive Performance of the Model
The predictive accuracy of the model was assessed using 3 measures: (1) Brier score for overall performance, (2) calibration slope for calibration, and (3) c-index for discrimination. In addition to these numeric measures, we also used the calibration plot and receiver operating characteristic curve to display the calibration and discrimination aspects of our final model.
Bootstrapping using 500 repetitions was used for internal validation of our model and to obtain bias-corrected predictive accuracy measures of the final model. For each bootstrap sample, each predictive accuracy measure was assessed in the bootstrap sample and in the original data set. The mean difference between the assessment in bootstrap sample and in the original data was defined as the “optimism.” The “optimism” of each accuracy measure was subtracted from the final model to yield the bias-corrected predictive accuracy measures of the final model.8
We also performed additional analyses, creating and assessing our model with 3 clinically relevant variations. First, we excluded the variable lymphovascular invasion because it is not always available preoperatively. Second, we excluded the cT3N0 patients, because many such individuals are prescribed induction chemoradiation based on depth of invasion. Finally, we divided the cT1N0 patients into T1a, T1b, and T1 NOS (not otherwise specified) populations. We then examined the predictive accuracy of our model in these contexts.
Creation of the Nomogram
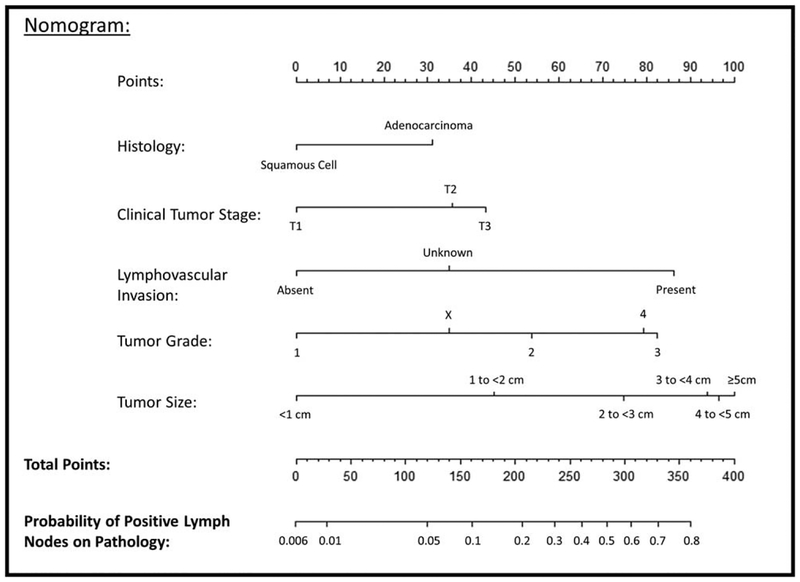
A nomogram was developed as the graphical representation of our final model.9 The nomogram has a reference line on the top for scoring points for each predictor from 0 to 100. The predictive variables are displayed below with bars that scale their effect size, demonstrating visually the relative weight of each variable and allowing for points to be assigned to each significant clinical characteristic.10 The summation of points from each predictor and the corresponding predicted probability of positive lymph nodes can be read from the bottom 2 lines.
RESULTS
Baseline Clinical Characteristics
After application of selection criteria, 3186 patients were available for analysis: 688 (22%) had pathologic lymph node involvement (pN1–3) and 2498 (78%) had pN0 status (Fig. 1). A summary of patient demographic and tumor characteristics are displayed in Table 1. The mean age was 65 years, and the majority of patients were men (80%), white (92%), had a Charlson Deyo Score of 0 (68%), were treated in an academic center (62%), and had adenocarcinoma (79%), with no significant differences in these measures by pathologic nodal status after adjusting for multiple comparisons. Patients with pathologic lymph node involvement had higher rates of more advanced clinical tumor stage, larger tumor size, higher tumor grade, and presence of lymphovascular invasion (all P < 0.001).
TABLE 1.
Patient and Tumor Characteristics of the Overall Cohort of Clinically Node Negative Patients, and Characteristics by Pathologic Node Status
| Overall Cohort (n = 3186) | Path NO (n = 2498) | Path N+ (n = 688) | P | |
|---|---|---|---|---|
| Age | 65.0 ± 10.1 | 65.0 ± 9.9 | 65.2 ± 10.7 | 0.655 |
| Sex (male) | 2545 (79.9%) | 1980 (79.3%) | 565 (82.1%) | 0.098 |
| Race (white) | 2925 (91.8%) | 2286 (91.5%) | 639 (92.9%) | 0.248 |
| Insurance (private) | 1272 (39.9%) | 996 (39.9%) | 276 (40.1%) | 0.908 |
| Population (>250,000) | 2266 (71.1%) | 1789 (71.6%) | 477 (69.3%) | 0.241 |
| Median income (<$38,0O0) | 472 (14.8%) | 365 (14.6%) | 107 (15.6%) | 0.539 |
| >21% Without high school diploma | 399 (12.5%) | 324 (13.0%) | 75 (10.9%) | 0.147 |
| Charlson Deyo score | ||||
| 0 | 2158 (67.7%) | 1699 (68.0%) | 459 (66.7%) | 0.482 |
| 1 | 806 (25.3%) | 632 (25.3%) | 174 (25.3%) | |
| ≥2 | 222 (7.0%) | 167 (6.7%) | 55 (8.0%) | |
| Academic center | 1975 (62.0%) | 1562 (62.5%) | 413 (60.0%) | 0.231 |
| Diagnosis year | ||||
| 2004 | 201 (6.3%) | 163 (6.5%) | 38 (5.5%) | 0.151 |
| 2005 | 211 (6.6%) | 157 (6.3%) | 54 (7.9%) | |
| 2006 | 238 (7.5%) | 184 (7.4%) | 54 (7.9%) | |
| 2007 | 275 (8.6%) | 208 (8.3%) | 67 (9.7%) | |
| 2008 | 404 (12.7%) | 306 (12.3%) | 98 (14.2%) | |
| 2009 | 411 (12.9%) | 334 (13.4%) | 77 (11.2%) | |
| 2010 | 344 (10.8%) | 273 (10.9%) | 71 (10.3%) | |
| 2011 | 322 (10.1%) | 253 (10.1%) | 69 (10.0%) | |
| 2012 | 278 (8.7%) | 209 (8.4%) | 69 (10.0%) | |
| 2013 | 264 (8.3%) | 221 (8.9%) | 43 (6.3%) | |
| 2014 | 238 (7.5%) | 190 (7.6%) | 48 (7.0%) | |
| Histology: | ||||
| Squamous | 675 (21.2%) | 548 (21.9%) | 127 (18.5%) | 0.048 |
| Adenocarcinoma | 2511 (78.8%) | 1950 (78.1%) | 561 (81.5%) | |
| Tumor stage | ||||
| T1 | 1840 (57.8%) | 1602 (64.1%) | 238 (34.6%) | <0.001 |
| T2 | 926 (29.1%) | 639 (25.6%) | 287 (41.7%) | |
| T3 | 420 (13.2%) | 257 (10.3%) | 163 (23.7%) | |
| Tumor size | ||||
| <1 cm | 563 (17.7%) | 539 (21.6%) | 24 (3.5%) | <0.001 |
| l–<2 cm | 688 (21.6%) | 605 (24.2%) | 83 (12.1%) | |
| 2–<3 cm | 682 (21.4%) | 531 (21.3%) | 151 (22.0%) | |
| 3–<4 cm | 509 (16.0%) | 343 (13.7%) | 166 (24.1%) | |
| 4–<5 cm | 331 (10.4%) | 216 (8.7%) | 115 (16.7%) | |
| ≥5 cm | 413 (13.0%) | 264 (10.6%) | 149 (21.7%) | |
| Grade | ||||
| 1 | 391 (12.3%) | 371 (14.9%) | 20 (2.9%) | <0.001 |
| 2 | 1356 (42.6%) | 1103 (44.2%) | 253 (36.8%) | |
| 3 | 1153 (36.2%) | 780 (31.2%) | 373 (54.2%) | |
| 4 | 53 (1.7%) | 35 (1.4%) | 18 (2.62%) | |
| Unknown | 233 (7.3%) | 209 (8.4%) | 24 (3.5%) | |
| Lymphovascular invasion | ||||
| Yes | 945 (29.7%) | 171 (6.9%) | 155 (22.5%) | <0.001 |
| No | 326 (10.2%) | 846 (33.9%) | 99 (14.4%) | |
| Missing | 1915 (60.1%) | 1481 (59.3%) | 434 (63.1%) |
Development and Evaluation of the Predictive Model
Univariable and multivariable logistic regression was performed to assess for variables associated with pathologic lymph node involvement, and a summary of the results is displayed in Table 2. The variables independently associated with occult lymph node metastases and included in the final model were histology, tumor stage, tumor size, grade, and lymphovascular invasion.
TABLE 2.
Univariable and Multivariable Analyses of Variables Associated with Pathologically Positive Lymph Nodes
| Univariable OR (95% CI) | P | Multivariable OR (95% CI) | P | |
|---|---|---|---|---|
| Age | 1.00 (0.99–1.01) | 0.642 | ||
| Sex (female vs male) | 0.83 (0.67–1.04) | 0.098 | ||
| Race (white vs non-white) | 1.21 (0.88–1.67) | 0.248 | ||
| Insurance (private vs nonprivate) | 1.01 (0.85–1.20) | 0.908 | ||
| Population (<250,000 vs >250,000) | 1.12 (0.93–1.34) | 0.242 | ||
| Median income <$38,OOO | 0.93 (0.74–1.17) | 0.539 | ||
| High school diploma (<21% vs 21%) | 1.22 (0.93–1.60) | 0.147 | ||
| Charlson Deyo score | ||||
| 0 | (reference) | 0.483 | ||
| 1 | 1.02 (0.84–1.24) | |||
| 2 | 1.22 (0.88–1.68) | |||
| Academic center | 1.11 (0.94–1.32) | 0.232 | ||
| Diagnosis year | ||||
| 2004 | (reference) | 0.155 | ||
| 2005 | 1.48 (0.92–2.36) | |||
| 2006 | 1.26 (0.79–2.01) | |||
| 2007 | 1.38 (0.88–2.16) | |||
| 2008 | 1.37 (0.90–2.09) | |||
| 2009 | 0.99 (0.64–1.52) | |||
| 2010 | 1.12 (0.72–1.73) | |||
| 2011 | 1.17 (0.75–1.82) | |||
| 2012 | 1.42 (0.91–2.21) | |||
| 2013 | 0.84 (0.52–1.35) | |||
| 2014 | 1.08 (0.67–1.74) | |||
| Adenocarcinoma vs squamous | 1.24 (1.00–1.54) | 0.048 | 1.75 (1.38–2.21) | <0.001 |
| Tumor stage | ||||
| T1 | (reference) | <0.001 | (reference) | <0.001 |
| T2 | 3.02 (2.49–3.67) | 1.90 (1.52–2.35) | ||
| T3 | 4.27 (3.36–5.42) | 2.17 (1.65–2.85) | ||
| Tumor size | ||||
| <1cm | (reference) | <0.001 | (reference) | <0.001 |
| l–<2 cm | 3.08 (1.93–4.92) | 2.25 (1.39–3.65) | ||
| 2–<3 cm | 6.39 (4.08–9.98) | 3.82 (2.40–6.09) | ||
| 3–<4 cm | 10.87 (6.94–17.02) | 5.40 (3.37–8.66) | ||
| 4–<5 cm | 11.96 (7.49–19.08) | 5.66 (3.45–9.29) | ||
| ≥5 cm | 12.67 (8.04–19.99) | 6.02 (3.70–9.80) | ||
| Grade | ||||
| 1 | (reference) | <0.001 | (reference) | <0.001 |
| 2 | 4.26 (2.66–6.81) | 2.62 (1.61–4.26) | ||
| 3 | 8.87 (5.56–14.14) | 4.39 (2.71–7.12) | ||
| 4 | 9.54 (4.62–19.70) | 4.15 (1.92–8.97) | ||
| Unknown | 2.13 (1.15–3.95) | 1.87 (0.99–3.56) | ||
| Lymphovascular invasion | ||||
| No | (reference) | <0.001 | (reference) | <0.001 |
| Yes | 7.75 (5.73–10.47) | 4.70 (3.41–6.48) | ||
| Missing | 2.50 (1.98–3.16) | 1.87 (1.46–2.39) |
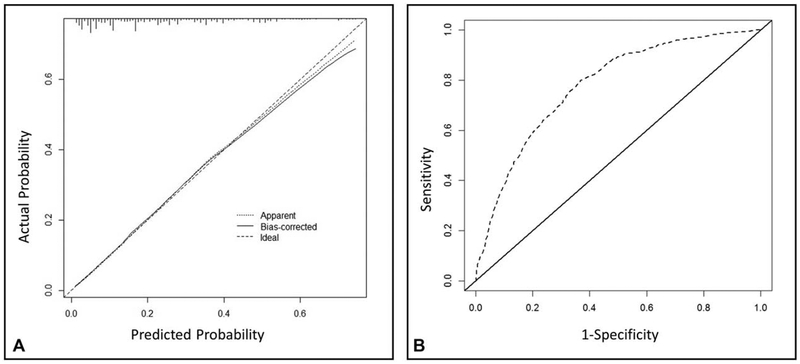
The predictive accuracy of the model was assessed using (1) Brier score for overall performance, which assesses the difference between observed and predicted values with values closer to 0 indicating better predictive ability; (2) calibration slope for calibration, which assesses the agreement between observed and predicted values with values closer to 1 indicating better performance; and (3) c-index for discrimination, which assesses how well the model distinguishes between those with and without the outcome of interest with values of 0.5 indicating a noninformative model and 1 indicating perfect discrimination. Bootstrapping with 500 repetitions was used for model validation, and the bias-corrected measures of accuracy were Brier score of 0.14, calibration slope of 0.97, and c-index of 0.77, respectively. The calibration slope and the receiver operating characteristic curve were also plotted to graphically assess calibration and discrimination, respectively, and are displayed in Figure 2. The Brier score is a measure of overall performance, and captures aspects of both calibration and discrimination. It is a representation of the difference between the predicted probability and the actual outcome. The score ranges from 0 to 1, with values closer to 0 indicating better predictive ability. The calibration slope tests the concordance between predicted values and outcomes with a perfect slope equal to 1. The c-statistic tests the discrimination of the model, or the ability to tell a patient who has pN+ disease from one who has pN0 disease. Values range from 0.5 to 1, with values closer to 1 indicating better discrimination. Together, the values we obtained for these measures indicate reasonably good predictive accuracy.
FIGURE 2.
A, Calibration slope and (B) receiving operating characteristic (ROC) curve of our model. Our model had a calibration slope of 0.97 and c-index of 0.77, respectively.
We then analyzed clinically relevant variations. First, the variable lymphovascular invasion was excluded, and model accuracy was assessed. This model had a Brier score of 0.145, calibration slope of 0.97 and c-index of 0.75, indicating slightly worse overall performance. Since the original model was built incorporating a “missing” category for lymphovascular invasion to allow use in all patients, and discrimination was better when this variable was included, we retained the original risk model. Second, we excluded cT3N0 patients. Predictive accuracy measures for this model included a Brier score of 0.13, calibration slope of 0.96, and c-index of 0.77, indicating comparable performance as the original. Finally, we divided cT1N0 patients into T1a, T1b, and T1 NOS populations. The regression coefficients for T1a (P = 0.9) and T1b (P = 0.6) were not different from 0 (reference: T1 NOS). Because none of these variations improved the model, we retained the original for creation of the nomogram.
Creation and Use of the Nomogram
A nomogram displaying the predictive variables and corresponding point scales is shown in Figure 3. The steps for using the nomogram are (1) determine the patient’s value for each predictive variable, (2) draw a straight line upwards from each predictive value to the top point reference line, (3) sum the points from each predictor, (4) locate the sum on the total points reference line, and (5) draw a straight line from total points line down to the bottom probability line to find the patient’s likelihood of pathologically positive lymph nodes.
FIGURE 3.
A nomogram for predicting the likelihood of occult positive lymph nodes in clinically node negative esophageal cancer patients. To use the nomogram, the value for each predictor is determined by drawing a line upward to the point reference line, the points are summed, and a line is drawn downward from the total points line to find the predicted probability of node positivity.
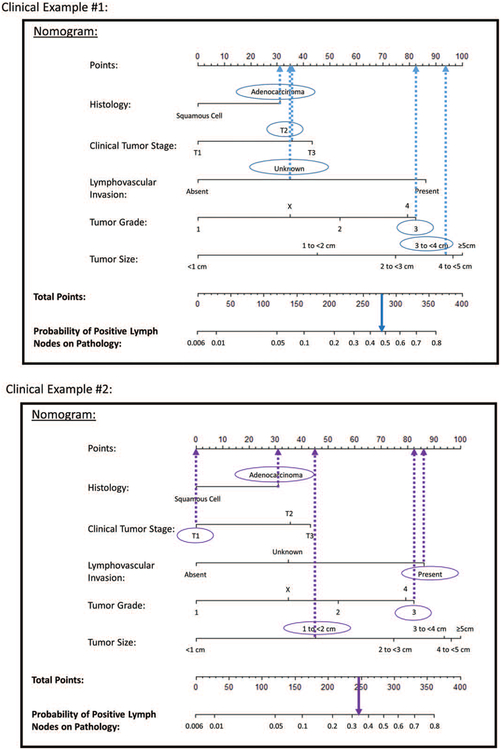
The applicability of the nomogram can be illustrated through some clinical examples (Fig. 4). In the first example, we calculate the predicted probability of pN+ for a patient who is staged with EUS as T2N0 with an estimated 3.5 cm adenocarcinoma that is grade 3, with lymphovascular invasion status unknown. Points are assigned for each feature: 31 for adenocarcinoma histology, 35 for T2 tumor stage, 35 for unknown lymphovascular invasion status, 83 for grade 3, and 94 for size 3 to 4 cm. The total of 278 points corresponds to a nearly 50% chance of pN+ for this patient. In the second example, we calculate the predicted probability of pN+ for a T1N0 patient with a 1.5 cm adenocarcinoma who underwent endoscopic mucosal resection and was found to have evidence of a grade 3 tumor and presence of lymphovascular invasion. Points are again assigned for each feature: 31 for adenocarcinoma histology, 0 for T1 tumor stage, 87 for presence of lymphovascular invasion, 83 for grade 3, and 45 for size 1 to 2 cm. The total of 246 points corresponds to an approximately 35% chance of pN+ disease. The expected likelihood of node-positive disease for individual patients based on features of their disease can be used for preoperative counseling and treatment planning.
FIGURE 4.
Clinical examples of nomogram use.
DISCUSSION
Nomograms have long been used in oncology to calculate an individual patient’s estimated prognosis based on relevant clinical parameters, and examples are available for colon, gastric, breast, bladder, renal, and prostate cancers.11 Nomograms incorporate more detailed clinical data that is not included in the standard tumor-node-metastasis staging system, and thereby can provide more refined estimates of patient probability of survival. In addition, nomograms have also been used to predict lymph node positivity in many of these same tumors, where prognosis or treatment depends on this probability.11
In this study, we developed and internally validated a nomogram to predict the likelihood of occult nodal metastases in patients with esophageal cancer, and found our model had good predictive accuracy. This tool was developed using relevant clinically available tumor factors within the NCDB, including histology, tumor stage, tumor size, grade, and presence of lymphovascular invasion. These factors have been shown to have prognostic significance12,13 but have not all been previously assembled into a usable clinical tool for predicting node disease for an individual patient. A prior study used data from 273 patients at 2 institutions, and created a nomogram including tumor length, clinical tumor stage, and clinical node stage.13 This model also demonstrated good predictive accuracy, although it included clinically node-positive patients for whom the standard of care would be neoadjuvant treatment,1 independent of data from the nomogram. Our study focused on clinically node-negative patients who may be treated with surgery alone. Patients treated with surgery alone who are pathologically upstaged due to nodal metastases have significantly worse survival,4 so this population represents an area for improvement of care.
We illustrate (Fig. 4) how using this tool could make a difference in clinical management. For all esophageal cancers, but particularly for patients with clinical T2N0 cancers, there is a demonstrated lack of reliability in EUS and positron emission tomography-computed tomography staging.12,14–16 Consequently, some providers choose to routinely administer induction therapy while others do not. Similarly, in T3N0 tumors, some centers are choosing to perform upfront minimally invasive esophagectomy with possible postoperative administration of adjuvant therapy17 instead of standard induction chemoradiation followed by surgery.1 This model could improve patient selection for neoadjuvant treatment in this setting by identifying those who are most likely to benefit and sparing those with low risk of nodal disease the additional cost and risks of chemoradiation. In our first clinical example of a patient with T2N0 cancer, the probability of occult nodal disease was approximately 50%. In a previously published decision analysis examining the role of induction therapy for T2N0 esophageal cancer,18 we demonstrated that a survival benefit could be expected with induction chemoradiation if there was a 48% probability of upstaging. Based on our nomogram, the patient in this example could be a candidate for evidence-based neoadjuvant treatment.
With the increasing application of endoscopic mucosal resection and endoscopic submucosal dissection for T1 cancers, the ability to reliably predict nodal disease in T1 cancers has become especially important. Currently, guidelines recommend endoscopic treatment for T1a tumors and esophagectomy for T1b tumors if the patient is a surgical candidate, or endoscopic treatment or chemoradiation if the patient is not.2 Our second clinical example illustrates how the nomogram could be helpful for risk stratifying these early stage patients to decide on treatment or surveillance strategies: the T1N0 patient has a 35% of occult nodal disease after discovery of high-risk pathologic features on endoscopic mucosal resection. This information may better inform referral for esophagectomy for complete resection and pathologic staging, or it may help guide additional nonoperative therapy or a more intensive surveillance strategy if the patient is a poor surgical candidate. Beyond stratification into T1a and T1b, the information gained from this nomogram could help both patients and clinicians quantify individual risk based on their clinical data.
Our study has some limitations that merit discussion. First, a retrospective dataset was used to identify patients undergoing upfront esophagectomy for inclusion into the study. This avoids treatment effect on pathologic lymph node assessment, but it is possible that treatment selection bias is present within our sample—the clinical features of patients with T2N0 or T3N0 disease that do not receive induction therapy may be different than the whole population of patients with this stage disease. In addition, patients who do not share the typical demographics of patients with esophageal cancer nationwide may not be as well represented by this model. Finally, our model has not been externally validated. We did, however, use a broadly representative dataset that captures the majority of nationwide esophageal cancer diagnoses to derive our model. Furthermore, we used bootstrapping, which has been shown to provide the best and least biased estimates,19 to internally validate our clinical model, which provided good optimism adjusted estimates of its predictive capability.
These limitations are balanced by the strengths of our study and the utility of the nomogram. By using a robust sample size of more than 3000 clinically node-negative patients who underwent an upfront operation and pathologic exam of their lymph nodes, and incorporating multiple variables into the instrument, our nomogram expands on previously published models examining lymph node positivity in esophageal cancer.13 We incorporated potentially available and clinically relevant variables. Tumor size and stage are routinely assessed with endoscopy and EUS. Histology and grade are frequently available from a biopsy. Identification of the presence of lymphovascular invasion preoperatively depends on both the quality of a biopsy and use of staining techniques,12,20,21 but can be determined without surgical resection22,23 and when available, provides very useful information. We included categories for missing information for grade and lymphovascular invasion to allow general applicability of our instrument even when these data for an individual are not available. This easily usable tool can provide additional clinical information in settings in which clinical staging is known to be frequently inaccurate. The probability of nodal disease from the nomogram can be used in conjunction with other clinical data for shared decision making or treatment selection to improve the care of patients with esophageal cancer.
Acknowledgments
This work was supported by a Barnes Jewish Hospital Foundation Grant (T.R.S.); NIH grants 2T32HL7776-21 (T.R.S., M.S.) and K07CA178120 (V.P.); and the Division of Cardiothoracic Surgery.
Footnotes
Presented at the American Association for Thoracic Surgery, 99th Annual Meeting, May 4–7, 2019, Toronto, Canada.
The authors report no conflicts of interest.
REFERENCES
- 1.Van Hagen P, Hulshof MCCM, van Lanschot JJB, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–2084. [DOI] [PubMed] [Google Scholar]
- 2.Farjah F, Gerdes H, Gibson M, et al. NCCN Guidelines Version 2.2018 Esophageal and Esophagogastric Junction Cancers NCCN Evidence BlocksTM Available from: https://www.nccn.org/professionals/physician_gls/pdf/esophageal_blocks.pdf. 2018. Accessed October 17, 2018. [Google Scholar]
- 3.Little AG, Lerut AE, Harpole DH, et al. The Society of Thoracic Surgeons practice guidelines on the role of multimodality treatment for cancer of the esophagus and gastroesophageal junction. Ann Thorac Surg. 2014;98:1880–1885. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree TD, Yacoub WN, Puri V, et al. Endoscopic ultrasound for early stage esophageal adenocarcinoma: implications for staging and survival. Ann Thorac Surg. 2011;91:1509–1516. [DOI] [PubMed] [Google Scholar]
- 5.Shin S, Kim HK, Choi YS, et al. Clinical stage T1-T2N0M0 oesophageal cancer: accuracy of clinical staging and predictive factors for lymph node metastasisy†. Eur J Cardio-Thoracic Surg. 2014;46:274–279. [DOI] [PubMed] [Google Scholar]
- 6.Dubecz A, Kern M, Solymosi N, et al. Predictors of lymph node metastasis in surgically resected T1 esophageal cancer. Ann Thorac Surg. 2015;99:1879–1885. discussion 1886. [DOI] [PubMed] [Google Scholar]
- 7.Weksler B, Kennedy KF, Sullivan JL. Using the National Cancer Database to create a scoring system that identifies patients with early-stage esophageal cancer at risk for nodal metastases. J Thorac Cardiovasc Surg. 2017;154:1787–1793. [DOI] [PubMed] [Google Scholar]
- 8.Harrell F Regression Modeling Strategies with applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. 2nd ed, Switzerland: Springer; 2015. [Google Scholar]
- 9.Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. 1st ed, New York: Springer; 2009. [Epub ahead of print]. [Google Scholar]
- 10.Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364–1370. [DOI] [PubMed] [Google Scholar]
- 11.Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16:e173–e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samson P, Puri V, Robinson C, et al. Clinical T2N0 esophageal cancer: identifying pretreatment characteristics associated with pathologic upstaging and the potential role for induction therapy. Ann Thorac Surg. 2016;101:2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaur P, Sepesi B, Hofstetter WL, et al. A clinical nomogram predicting pathologic lymph node involvement in esophageal cancer patients. Trans Meet Am Surg Assoc. 2010;128:215–222. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree TD, Kosinski AS, Puri V, et al. Evaluation of the reliability of clinical staging of T2 N0 esophageal cancer: a review of the Society of Thoracic Surgeons database. Ann Thorac Surg. 2013;96:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofstetter W Treatment of clinical T2N0M0 esophageal cancer. Ann Surg Oncol. 2014;21:3713–3714. [DOI] [PubMed] [Google Scholar]
- 16.Speicher PJ, Ganapathi AM, Englum BR, et al. Induction therapy does not improve survival for clinical stage T2N0 esophageal cancer. J Thorac Oncol. 2014;9:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahoor H, Luketich JD, Levy RM, et al. A propensity-matched analysis comparing survival after primary minimally invasive esophagectomy followed by adjuvant therapy to neoadjuvant therapy for esophagogastric adenocarcinoma. J Thorac Cardiovasc Surg. 2015;149:538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenkovich TR, Panni RZ, Hudson JL, et al. Comparative effectiveness of upfront esophagectomy versus induction chemoradiation in clinical stage T2N0 esophageal cancer: a decision analysis. J Thorac Cardiovasc Surg. 2018;155:2221.e1–2230.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steyerberg EW, Harrell FE, Borsboom GJ, et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774–781. [DOI] [PubMed] [Google Scholar]
- 20.Saad RS, Lindner JL, Liu Y, et al. Lymphatic vessel density as prognostic marker in esophageal adenocarcinoma. Am J Clin Pathol. 2009;131:92–98. [DOI] [PubMed] [Google Scholar]
- 21.Goscinski MA, Nesland JM, Giercksky K-E, et al. Primary tumor vascularity in esophagus cancer. CD34 and HIF1-( expression correlate with tumor progression. Histol Histopathol. 2013;28:1361–1368. [DOI] [PubMed] [Google Scholar]
- 22.Cen P, Hofstetter WL, Lee JH, et al. Value of endoscopic ultrasound staging in conjunction with the evaluation of lymphovascular invasion in identifying low-risk esophageal carcinoma. Cancer. 2008;112:503–510. [DOI] [PubMed] [Google Scholar]
- 23.Cen P, Hofstetter WL, Correa AM, et al. Lymphovascular invasion as a tool to further subclassify T1b esophageal adenocarcinoma. Cancer. 2008; 112: 1020–1027. [DOI] [PubMed] [Google Scholar]