Abstract
Chronic alcohol consumption induces adipose tissue atrophy. However, the mechanisms for how alcohol induces lipodystrophy and its impact on liver steatosis and injury are not fully elucidated. Autophagy is a highly conserved lysosomal degradation pathway, which regulates cellular homeostasis. Mice with autophagy deficiency in adipose tissue have impaired adipogenesis. However, whether autophagy plays a role in alcohol-induced adipose atrophy and how altered adipocyte autophagy contributes to alcohol-induced liver injury remain unclear. To determine the role of adipose autophagy and mechanistic target of rapamycin (mTOR) in alcohol-induced adipose and liver pathogenesis, we generated adipocyte-specific Atg5 knockout (KO), adipocyte-specific mTOR KO, adipocyte-specific Raptor KO, and adipocyte-specific tuberous sclerosis complex 1 KO mice by crossing floxed mice with Adipoq-Cre. The KO mice and their matched wild-type mice were challenged with chronic-plus-binge alcohol mouse model. Chronic-plus-binge alcohol induced adipose atrophy with increased autophagy and decreased Akt/mTOR signaling in epididymal adipose tissue in wild-type mice. Adipocyte-specific Raptor KO mice experienced exacerbated alcohol-induced steatosis, but neither adipocyte-specific mTOR nor adipocyte-specific tuberous sclerosis complex 1 KO mice exhibited similar detrimental effects. Adipocyte-specific Atg5 KO mice had increased circulating levels of fibroblast growth factor 21 and adiponectin and were resistant to alcohol-induced adipose atrophy and liver injury. In conclusion, autophagy deficiency in adipose tissue leads to reduced sensitivity to alcohol-induced adipose atrophy, which ameliorates alcohol-induced liver injury in mice.
Alcoholic liver disease (ALD) is a worldwide health issue claiming 2 million lives per year. The pathogenesis of ALD is characterized by steatosis, alcoholic hepatitis, fibrosis, cirrhosis, and eventually hepatocellular carcinoma.1, 2, 3 The only known curable treatment for late-stage ALD is liver transplantation. Currently, no other curable treatments are available, which has generated a need for novel treatments, especially in the early stages of ALD. Alcohol consumption damages multiple organs, including liver, pancreas, lung, kidney, adipose, and brain.4 In recent years, increasing evidence implicates the role of adipose-liver axis in the pathogenesis of ALD.5,6 Chronic alcohol consumption leads to increased lipolysis, adipose tissue atrophy, and proinflammatory adipokine secretion.7,8 Moreover, chronic ethanol increases cytochrome P450 2E1 activity in adipose tissues, which leads to BH3 interacting domain death agonist–mediated apoptosis and activation of complement via C1q, resulting in adipose tissue inflammation.9 However, the mechanisms for how alcohol-induced adipose injury contributes to ALD remain largely unknown.
Autophagy is a highly conserved, genetically programmed lysosomal degradation pathway that functions to maintain cellular homeostasis.10 Emerging evidence suggests that autophagy affects the mass, differentiation, and physiological functions of adipose tissue.11, 12, 13 Interestingly, up-regulated autophagy proteins and genes have also been observed in human adipose tissue samples in obesity.14 However, the role of autophagy in alcohol-induced adipose tissue atrophy has not been studied.
Mechanistic target of rapamycin (mTOR), a highly conserved serine-threonine protein kinase, is a key regulator of cell growth and metabolism.15 The tumor suppressor proteins tuberous sclerosis complex 1 (TSC1) and TSC2 form a complex and negatively regulate mTOR activation.16,17 Pharmacologic and genetic evidence has proved that mTOR is required for adipogenesis and adipose maintenance in vitro and in vivo.18, 19, 20 Activation of mTOR suppresses lipolysis, stimulates lipogenesis, and promotes fat storage.21 In a recent study, Crowell et al22 reported that mice fed with a Lieber-DeCarli ethanol diet for 24 weeks had altered mTOR signaling in epididymal white adipose tissue. Nevertheless, the exact role of mTOR in alcohol-induced autophagy and adipose atrophy has not been elucidated.
In this study, we investigated the effect of chronic-plus-binge (Gao-binge) alcohol on mTOR and autophagy pathways in mouse adipose tissue. We further determined the role of mTOR and autophagy in alcohol-induced adipose atrophy and liver injury using adipose-specific Mtor, Raptor, Tsc1, and Atg5 knockout mice.
Materials and Methods
Animals
Male mice (aged 8 to 12 weeks) were used for Gao-binge alcohol model23 or chronic feeding model. For Gao-binge model, Atg5f/f (C57B L/6/129) were generated by Dr. N. Mizushima and were backcrossed with C57BL/6J for another 10 generations, as described previously.24 Adipoq-Cre, Raptorf/f, mTORf/f, and TSC1f/f mice (C57BL/6J background) were all obtained from The Jackson Laboratory (Bar Harbor, ME). All these flox/flox mice were crossed with Adipoq-Cre to generate adipocyte-specific Atg5 knockout (A-Atg5 KO), adipocyte-specific mTOR KO (A-mTOR KO), adipocyte-specific Raptor KO (A-Raptor KO), and adipocyte-specific TSC1 KO (A-TSC1 KO) mice. The Cre- littermates were used as wild-type (WT) control. In brief, mice were acclimated to the Lieber-DeCarli liquid diet (F1259SP; Bio-serv, Flemington, NJ) for 5 days and then given ethanol (Decon, King of Prussia, PA)–containing (5%, v/v) liquid diet (F1258SP; Bio-serv) supplemented with maltose dextrin (Bio-serv) for 10 days. In the morning of the 15th day (ie, day 10 since ethanol feeding started), the mice were orally gavaged with ethanol (5 g/kg) (ethanol group). Mice administered with calorie- and volume-matched control liquid diet and maltose dextrin gavage were used as control (control group). Body weight was monitored every 2 days. Food intake was recorded and matched daily after alcohol feeding started. Eight hours after gavage, mice were euthanized, and blood, epididymal white adipose tissue (eWAT), retroperitoneal WAT, s.c. WAT (sWAT), interscapular brown adipose tissue (iBAT), and liver tissues were collected for the following assays. For chronic feeding model, C57BL/6J wild-type mice were fed with Lieber-DeCarli liquid diet with a gradually increasing ethanol concentration (1% for 2 days, 2% for 2 days, 4% for 1 week, and 5% for 3 weeks). On day 32, mice were euthanized, and blood, adipose tissues, and liver tissue were collected. All procedures were approved by the Institutional Animal Care and Use Committee of The University of Kansas Medical Center (Kansas City, KS).
Protein Extraction and Western Blot Analysis
Fresh tissues were snapped frozen in liquid nitrogen and then homogenized in ice-cold radioimmunoprecipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS), followed by centrifugation at 12,000 × g × 15 minutes at 4°C. The radioimmunoprecipitation assay buffer was supplemented with protease inhibitor cocktail (BioTool, Houston, TX). After centrifugation, the lipid layer was removed, supernatant was collected, and protein concentration was determined by bicinchoninic acid assay (Thermo, Waltham, MA). Samples were mixed with SDS loading buffer containing dithiothreitol and incubated at 95°C for 10 minutes, unless otherwise indicated. For non–heat-denaturing, nonreducing conditions, samples were mixed with SDS loading buffer without dithiothreitol and were not heated. Protein (30 μg) was separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Membranes were incubated in Ponceau S solution (Sigma, St. Louis, MO) for 5 minutes if needed, neutralized with 0.1 mol/L NaOH, and washed in Millipore water (Millipore, Burlington, MA) before blocking. Membranes were blocked in 5% milk in Tris-buffered saline with 0.1% Tween at room temperature for 1 hour, and then incubated with primary antibodies and secondary antibodies prepared in 5% milk in Tris-buffered saline with 0.1% Tween. Signals were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo) and/or Immobilon Western HRP Substrate (Millipore). The following antibodies were used (Table 1). Anti–microtubule-associated protein light chain 3 (LC3) antibody was generated, as previously described.25 Densitometry was analyzed with ImageJ software version 1.8.0 (NIH, Bethesda, MD; https://imagej.nih.gov/ij). Band densities for the proteins of interest were normalized to β-actin unless otherwise indicated.
Table 1.
Summary of Antibody Information
| Antibody | Manufacturer | Catalog no. |
|---|---|---|
| 4E-BP1 | Cell Signaling (Beverly, MA) | 9452 |
| Phosphorylated 4E-BP1 (Ser65) | Cell Signaling | 9451 |
| Adiponectin | Cell Signaling | 2789 |
| AKT | Cell Signaling | 9272 |
| Phosphorylated AKT (Ser473) (D9E) | Cell Signaling | 4060 |
| Atg5 | Santa Cruz Biotechnology (Dallas, TX) | sc-33210 |
| β-Actin | Sigma | A5441 |
| CYP2E1 | Abcam (Cambridge, MA) | ab28146 |
| FGF21 | Abcam | ab64857 |
| FoxO1 | Cell Signaling | 2880S |
| Phosphorylated FoxO1 (Ser256) | Cell Signaling | 9461S |
| GAPDH | Cell Signaling | 2118 |
| GSK-3β | Cell Signaling | 5676 |
| Phosphorylated GSK-3β | Cell Signaling | 5558 |
| MLKL | Cell Signaling | 37705 |
| mTOR | Cell Signaling | 2983 |
| OXPHOS cocktail | Abcam | ab110413 |
| p62/SQSTM1 | Santa Cruz Biotechnology | sc-28359 |
| RIP3 | Cell Signaling | 95702 |
| TSC1 | Cell Signaling | 6935 |
| UCP1 | Abcam | ab10983 |
4E-BP1, 4E-binding protein 1; CYP2E1, cytochrome P450 2E1; FGF21, fibroblast growth factor 21; FOXO1, forkhead box O1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSK-3β, glycogen synthase kinase-3β; MLKL, mixed lineage kinase-like protein; OXPHOS, oxidative phosphorylation; RIP3, receptor-interacting protein kinase 3; SQSTM1, sequestosome 1; TSC1, tuberous sclerosis complex 1; UCP1, uncoupling protein 1.
Protein Oxidation Detection
Protein oxidation/carbonylation was detected with Oxyblot kit (Millipore), following the manufacturer's instructions. The samples were loaded to SDS polyacrylamide gel for electrophoresis. The following steps were similar to common Western blot procedures, as mentioned above, except that the designated primary antibody (specific to the dinitrophenyl moiety) and secondary antibody (goat anti-rabbit IgG) in the kit were used.
RNA Extraction and Real-Time PCR
For liver tissue, RNA was isolated with TRIzol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. For adipose tissue, RNA was isolated with PureLink RNA Mini kit (Invitrogen) with or without TRIzol, following the manufacturer's protocol. RNA concentration and quality were determined by spectrophotometer. RNA (1 to 3 ng) was used for cDNA synthesis with a reverse transcription kit (Fermentas, Vilnius, Lithuania). mRNA levels of target genes were determined on an Applied Biosystems Prism 7900HT real-time PCR instrument (ABI, Foster City, CA) using Maxima SYBR Green/rox qPCR reagents (Fermentas). 18s and 36b4 were used for normalization in liver samples and adipose tissue samples, respectively. The primer sequences are described in Table 2.
Table 2.
Summary of Mouse Primers for Real-Time quantitative PCR
| Gene/primer | Primer sequence |
|---|---|
| 18s | F: 5′-GAGCGAAAGCATTTGCCAAG-3′ |
| R: 5′-GGCATCGTTTATGGTCGGAA-3′ | |
| 36b4 | F: 5′-TAAAGACTGGAGACAAGGTG-3′ |
| R: 5′-GTGTACTCAGTCTCCACAGA-3′ | |
| Adipor1 | F: 5′-CTTGACGATGCTGAGACCAA-3′ |
| R: 5′-GCTGTGGGGAGCAGTAGAAG-3′ | |
| Adipor2 | F: 5′-GCCCAGCTTAGAGACACCTG-3′ |
| R: 5′-GCCTTCCCACACCTTACAAA-3′ | |
| Ccl2 | F: 5′-ACATTCGGCGGTTGCTCTAG-3′ |
| R: 5′-ACATCCTGTATCCACACGGCAG-3′ | |
| Cidea | F: 5′-CTCGGCTGTCTCAATGTCAA-3′ |
| R: 5′-GGGATGGCTGCTCTTCTGTA-3′ | |
| Cox8b | F: 5′-GAACCATGAAGCCAACGACT-3′ |
| R: 5′-GCGAAGTTCACAGTGGTTCC-3′ | |
| Dio2 | F: 5′-CAGTGTGGTGCACGTCTCCAATC-3′ |
| R: 5′-TGAACCAAAGTTGACCACCAG-3′ | |
| F4/80 | F: 5′-CTTTGGCTATGGGCTTCCAGTC-3′ |
| R: 5′-GCAAGGAGGACAGAGTTTATCGTG-3′ | |
| Fgf21 | F: 5′-CTGGGGGTCTACCAAGCATA-3′ |
| R: 5′-CACCCAGGATTTGAATGACC-3′ | |
| Fgfr4 | F: 5′-CGCCAGCCTGTCACTATACAAA-3′ |
| R: 5′-CCAGAGGACCTCGACTCCAA-3′ | |
| Klb | F: 5′-GATGAAGAATTTCCTAAACCAGGTT-3′ |
| R: 5′-AACCAAACACGCGGATTTC-3′ | |
| Il1b | F: 5′-GCCCATCCTCTGTGACTCAT-3′ |
| R: 5′-AGGCCACAGGTATTTTGTCG-3′ | |
| Il6 | F: 5′-ACAAGTCGGAGGCTTAATTACACAT-3′ |
| R: 5′-TTGCCATTGCACAACTCTTTTC-3′ | |
| Ly6g | F: 5′-TGCGTTGCTCTGGAGATAGA-3′ |
| R: 5′-CAGAGTAGTGGGGCAGATGG-3′ | |
| Prdm16 | F: 5′-CAGCACGGTGAAGCCATTC-3′ |
| R: 5′-GCGTGCATCCGCTTGTG-3′ | |
| Tnfa | F: 5′-CGTCAGCCGATTTGCTATCT-3′ |
| R: 5′-CGGACTCCGCAAAGTCTAAG-3′ | |
| Ucp1 | F: 5′-AGGCTTCCAGTACCATTAGGT-3′ |
| R: 5′-CTGAGTGAGGCAAAGCTGATTT-3′ |
F, forward; R, reverse.
Histologic Analysis
Fresh liver and fat tissues were fixed in 10% neutral-buffered formalin overnight, transferred to 70% ethanol for at least 24 hours, embedded in paraffin wax, and cut into sections (5 μm thick). For general morphology, slides were stained with hematoxylin and eosin. For eWAT adipocyte area quantification, at least three fields of ×100 images from each mouse were analyzed in each group using Adipocyte Tools (Montpellier Resources Imagerie, Montpellier, France) with ImageJ version 1.8.0. For BAT lipid droplet (LD) quantification, at least five fields of 200× images from each mouse were analyzed using ImageJ.
Immunohistochemistry Staining
Paraffin-embedded slides were rehydrated, quenched of endogenous peroxidase, and blocked. Then, the tissues were incubated with primary antibodies (1:1000) overnight at 4°C, followed by incubation with biotinylated secondary antibodies. For neutrophil staining, slides were incubated with an anti-Ly6B2 antibody (1:200) overnight at 4°C and then incubated with biotinylated secondary antibody. For quantification, 10 fields of ×20 images were randomly picked for each sample.
Electron Microscopy
Fresh tissues and cells were fixed with 2.5% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer (pH 7.4), followed by 1% OsO4. After dehydration, thin sections were stained with uranyl acetate and lead citrate. Digital images were captured using a JEM 1016CX electron microscope (JEOL, Peabody, MA).
Oil Red O Staining
Oil Red O (1 g; Sigma) was dissolved in 200 mL isopropanol to make Oil Red O stock solution. The stock solution and Millipore water (v/v = 6:4) was mixed well and filtered through 125-mm filter paper (Whatman, Maidstone, UK) to make working solution before use. Liver tissues were fixed with 4% paraformaldehyde in phosphate-buffered saline overnight at 4°C, transferred to 20% sucrose overnight at 4°C, and embedded in O.C.T. Compound (Fisher, Hampton, NH). For microscopy, tissues were cut into slides (5 μm thick) using a cryostat. The cryostat sections were dried at 37°C and washed twice with phosphate-buffered saline, incubated in 60% 2-propanol for 1 minute, dried again at 37°C, and incubated with Oil Red O working solution for 10 to 15 minutes at room temperature. Then, the slides were washed with 60% 2-propanol for 2 to 5 minutes one time and with phosphate-buffered saline three times. After counterstaining with hematoxylin for 5 seconds, the slides were washed with reverse osmosis water for at least 3 minutes and mounted with glycerol in phosphate-buffered saline (v/v = 5:1). Images were captured within the same day.
Liver TG and Chol Analysis
Liver triglyceride (TG) and cholesterol (Chol) levels were determined as previously reported.26 Frozen liver tissue (20 to 50 mg) was ground in liquid nitrogen using a mortar and a pestle. Then, the homogenate was incubated with 1 mL of chloroform/methanol (v/v = 2:1) with vigorous shaking for 1 hour at room temperature. Then, 200 μL of Millipore water was added into the mix and centrifuged at 3000 × g for 5 minutes. The lower lipid phase was collected and dried in a fume hood overnight, and the pellet was dissolved in 60 μL of tert-butanol and 40 μL of Triton X-114/methanol (v/v = 2:1) mix. Liver TG and Chol levels were measured with colorimetric assay kits, following the manufacturer's instructions (Pointe Scientific, Canton, MI), and normalized to tissue weight. The tert-butanol and Triton X-114/methanol mixture was used as a blank control.
Serum Biochemical Analysis
Blood samples were collected via cardiac puncture, sat on ice for at least 30 minutes, and centrifuged at 3000 × g at 4°C for 10 minutes; and the supernatant was collected as serum samples. Serum alanine aminotransferase (ALT) and aspartate aminotransferase levels were measured with fluorometric assay kits (Pointe Scientific). Serum TG and Chol levels were measured with colorimetric assay kits (Pointe Scientific). Serum free fatty acid level was measured with a fluorometric assay kit (BioVision, Milpitas, CA). Serum free glycerol level was measured with a colorimetric assay kit (Sigma-Aldrich, St. Louis, MO). Serum β-hydroxybutyrate level was measured with a colorimetric assay kit (BioVision). Millipore water was used as a blank control. All assays were performed following the manufacturer's instructions on a spectrometer, unless otherwise indicated.
Enzyme-Linked Immunosorbent Assay
Serum samples were collected as described above. Serum (2.5 μL) was used to measure the adiponectin level with a commercial kit (Millipore), following the manufacturer's protocol. Serum (10 μL) was used to measure the fibroblast growth factor 21 (FGF21) level with a commercial kit (R&D, Minneapolis, MN), following the manufacturer's protocol.
Ex Vivo Lipolysis Assay
Ex vivo lipolysis assay was performed, as previously described.27 In brief, after mice were euthanized, eWAT and sWAT tissues were collected, cut into smaller pieces, and weighed (10 to 20 mg per piece). Each piece was placed in a 12-well plate (30 to 50 mg each well) and incubated in Dulbecco’s modified Eagle’s medium containing 2% fatty acid–free bovine serum albumin with or without isoproterenol on an orbital shaker for 1 hour. The free glycerol released in medium was measured with free glycerol reagent (Sigma) and normalized to tissue weight.
Protein Oxidation Detection
Protein oxidation/carbonylation was detected with Oxyblot kit, following the manufacturer's instructions. In brief, 5 μL of sample containing 20 μg of protein was denatured by 5 μL of 12% SDS. Samples were incubated with 10 μL of 1× 2-4-dinitrophenyl hydrazine solution at room temperature for 15 minutes, and samples incubated with 1× Derivation-Control Solution were used as a negative control. Later, 7.5 μL of Neutralization Solution was added to stop the derivatization, and 1.5 μL 2-mercaptoethanol was added as a reducing agent. Then, the samples were loaded to SDS polyacrylamide gel for electrophoresis. The following steps were similar to common Western blot procedures, as mentioned above, except that the designated primary antibody (specific to the dinitrophenyl moiety) and secondary antibody (goat anti-rabbit IgG) in the kit were used.
Serum Lipocalin-2 Measurement
A mouse enzyme-linked immunosorbent assay kit (ab199083; Abcam, Cambridge, MA) was used to quantify the serum levels of lipocalin 2, according to the manufacturer's instructions.
Statistical Analysis
Experimental data were subjected to t-test or one-way analysis of variance, where appropriate. Error bars presented SEM. P < 0.05 was considered significant.
Results
Gao-Binge Alcohol Treatment Leads to Adipose Atrophy Associated with Decreased Akt/mTOR Signaling and Increased Autophagy
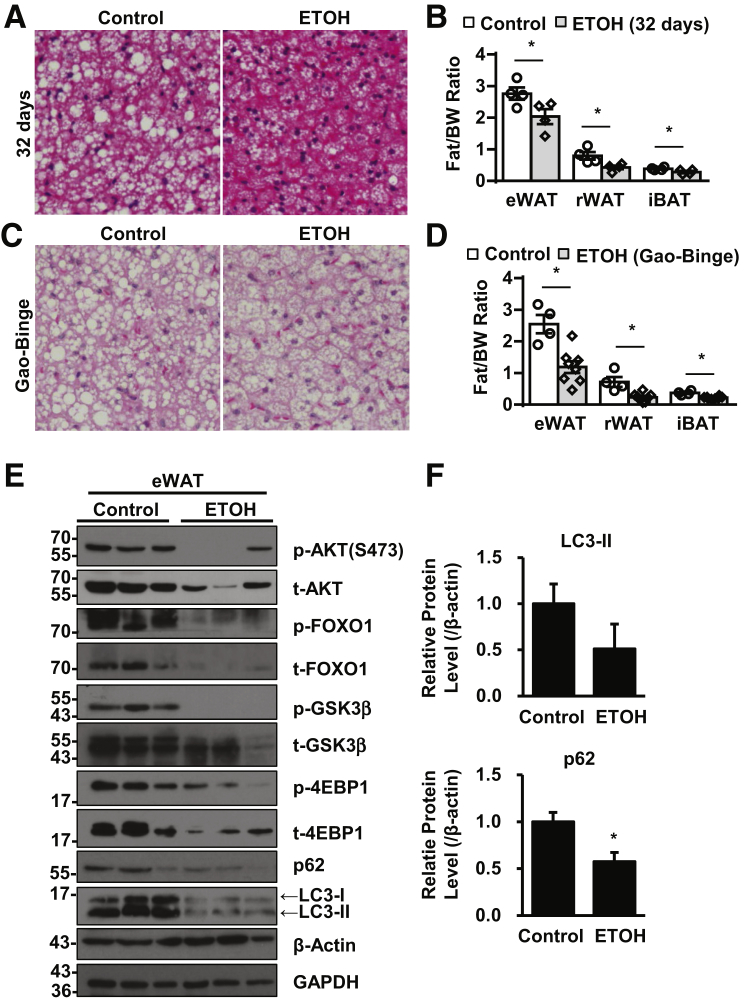
Consistent with previous reports,8,28 the 32-day chronic alcohol feeding significantly decreased the LD size of iBAT (Figure 1A) and weight of eWAT, retroperitoneal WAT, and iBAT (Figure 1B) in C57BL/6J mice. Moreover, Gao-binge alcohol significantly decreased the LD size of iBAT (Figure 1C) and weight of eWAT, retroperitoneal WAT, and iBAT (Figure 1D) in C57BL/6J mice, suggesting that both chronic and Gao-binge alcohol lead to adipose atrophy. Gao-binge alcohol decreased the levels of phosphorylated Akt and total Akt in eWAT. The levels of phosphorylated and total FOXO1 and glycogen synthase kinase-3β, two well-known substrate proteins of Akt, also decreased in eWAT after Gao-binge alcohol (Figure 1E). Moreover, both the levels of total and phosphorylated 4E-binding protein 1, the target substrate of mTOR, decreased after Gao-binge alcohol. These data suggest that Gao-binge alcohol inhibits Akt/mTOR signaling in mouse adipose tissue. mTOR inhibition is a common trigger of autophagy activation. Indeed, the protein levels of both LC3-II and p62 decreased after Gao-binge alcohol (Figure 1, E and F), suggesting a possible increased autophagic flux in mouse adipose tissues after Gao-binge alcohol. Taken together, these data suggest that Gao-binge alcohol may inhibit Akt-mTOR and increase autophagy in mouse adipose tissues.
Figure 1.
Alcohol-induced adipose atrophy associated with mTOR inhibition and increased autophagy in epididymal white adipose tissue (eWAT). WT mice were subjected to either 32-day chronic alcohol feeding or Gao-binge alcohol model. A and C: Representative hematoxylin and eosin staining images of interscapular brown adipose tissue (iBAT). B and D: Quantification of fat tissue weight, including eWAT and retroperitoneal WAT (rWAT) and iBAT. E: Total lysates from eWAT of Gao-binge alcohol–treated mice were subjected to Western blot analysis. F: Densitometry of E. Data are expressed as means ± SEM (B, D, and F). n = 4 to 8 (B and D). n = 5 (F). *P < 0.05 (t-test). Original magnification, ×40 (A and C). 4EBP1, 4E-binding protein 1; BW, body weight; ETOH, ethanol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSK3β, glycogen synthase kinase-3β; p-, phosphorylated; t-, total.
A-mTOR and A-Raptor KO Mice Have Adipose Atrophy Regardless of Alcohol Exposure
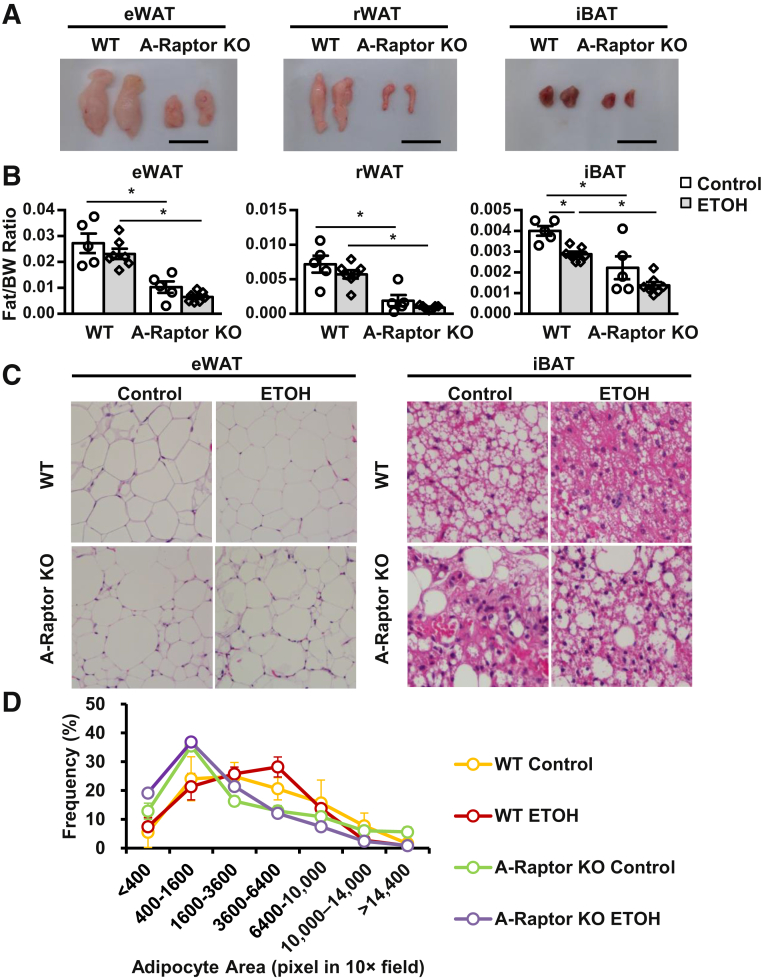
Because Gao-binge alcohol blunted mTOR signaling in adipose tissue and induced adipose atrophy, it was studied whether genetic blunt or activation of mTOR signaling pathways would affect alcohol-induced pathogenesis of adipose and liver tissues. A-mTOR KO, A-Raptor, and A-TSC1 KO mice were generated; and these mice were subjected to Gao-binge alcohol. Both A-mTOR and A-Raptor KO, but not A-TSC1 KO, mice fed control diet had markedly decreased mass of white and brown adipose tissues compared with their matched WT mice, suggesting that ablation of mTOR decreased the basal levels of fat mass. Intriguingly, Gao-binge alcohol decreased the mass of white adipose tissue approximately 30% to 40% in A-Raptor and A-TSC1 KO mice but did not affect the mass of white adipose tissue in A-mTOR KO mice (Figure 2, A and B, and Supplemental Figures S1A and S2A). Western blot analysis revealed successful deletion of Mtor and Tsc1 in both WAT and BAT tissues in A-mTOR and A-TSC1 KO mice (Supplemental Figures S1B and S2B). Notably, the sizes of white adipocytes and brown adipocytes in A-Raptor KO mice were heterogeneous and not ubiquitously smaller (Figure 2, C and D). Despite the decreased fat mass, the body weight of A-Raptor KO mice was similar to WT mice (Supplemental Figure S3A). Moreover, the food intake was similar between WT and A-Raptor KO mice fed control diet or alcohol diet, suggesting that the decreased adipose tissue in A-Raptor KO mice was not due to decreased nutrient intake (Supplemental Figure S3B). Taken together, these data suggest that genetic blunt of Raptor and mTOR in adipose tissue is sufficient to cause adipose atrophy without alcohol feeding, phenocopying Gao-binge alcohol-treated WT mice.
Figure 2.
A-Raptor KO mice have adipose atrophy without alcohol exposure. A-Raptor KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Representative gross images of epididymal white adipose tissue (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT). B: Quantification of fat tissue weight of eWAT, rWAT, and iBAT after Gao-binge treatment. C: Representative hematoxylin and eosin (H&E) staining images of eWAT and iBAT. D: Distribution of eWAT adipocyte area from H&E staining. Data are expressed as means ± SEM (B and D). n = 5 to 7 (B). n = 3 to 6 mice per group (D). *P < 0.05 (one-way analysis of variance). Scale bars = 1 cm (A). Original magnification: ×20 (C, eWAT); ×40 (C, iBAT). ETOH, ethanol.
Adipose-Specific Deletion of Raptor but Not Mtor or Tsc1 Exacerbates Gao-Binge Alcohol-Induced Liver Steatosis
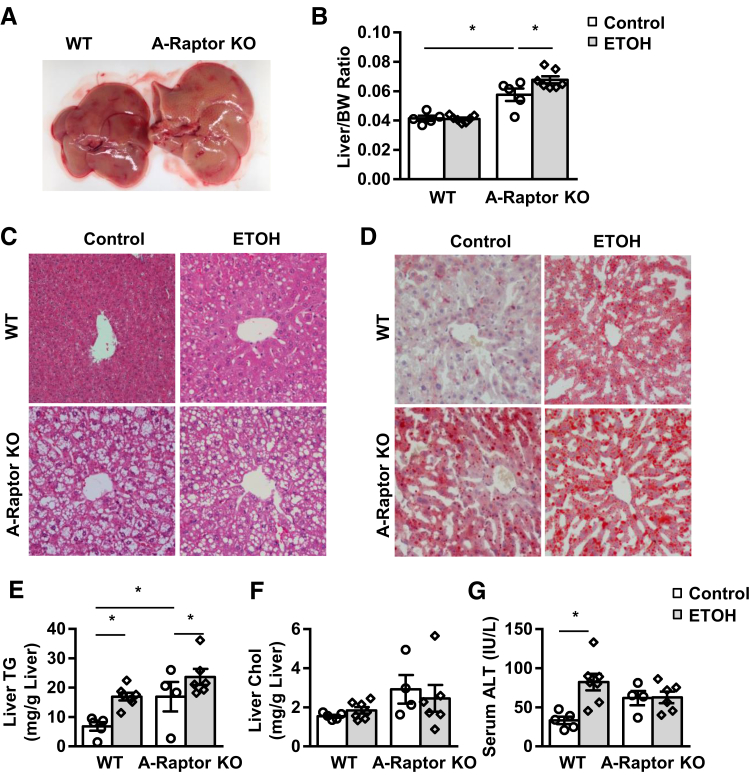
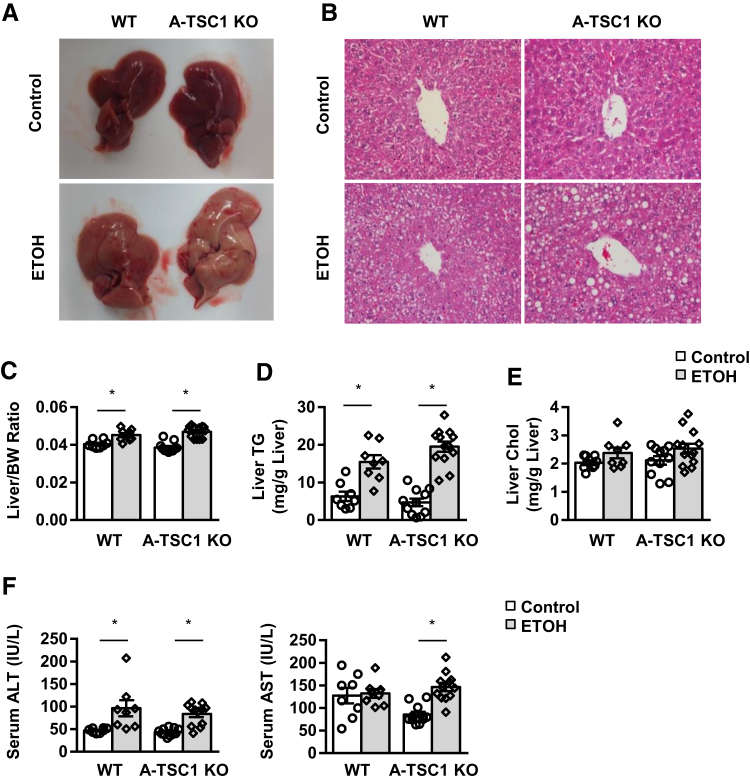
A-Raptor and A-mTOR KO mice fed with control Lieber-DeCarli diet followed by maltose gavage had larger and yellow-colored livers with significantly increased liver mass (Figure 3, A and B, and Supplemental Figure S4A). Compared with matched WT mice, A-Raptor KO and A-mTOR KO mice had increased accumulation of LDs based on hematoxylin and eosin and Oil Red O staining of liver tissues along with increased levels of liver TG (Figure 3, C–E, and Supplemental Figure S4, B and C). Gao-binge alcohol increased LD accumulation, as observed by Oil Red O staining, and increased levels of hepatic TG in WT mice, which were further increased in A-Raptor KO mice (Figure 3, C–E). However, A-Raptor KO (Figure 3F) or A-mTOR KO (data not shown) mice did not have significant differences in liver Chol. Moreover, Gao-binge alcohol did not affect levels of serum ALT in A-Raptor KO and A-mTOR KO mice compared with their respective KO mice fed with the control diet (Figure 3G and Supplemental Figure S4D). In contrast to A-Raptor KO and mTOR KO mice, A-TSC1 KO mice fed with the control Lieber-DeCarli diet had no obvious differences in adipose tissue mass (Supplemental Figure S2A), liver mass, and hepatic steatosis (Figure 4, A–E). Gao-binge alcohol treatment significantly increased serum ALT and aspartate aminotransferase levels in A-TSC1 KO mice compared with the control diet–fed A-TSC1 KO mice. However, the serum ALT and aspartate aminotransferase levels were not different between Gao-binge alcohol–treated A-TSC1 KO mice and their matched WT mice (Figure 4F). Although Gao-binge alcohol did not affect the serum levels of TG and Chol in WT mice, it markedly increased serum levels of TG in A-mTOR KO (13-fold), A-Raptor KO (15-fold), and A-TSC1 KO (2-fold) mice without affecting serum levels of Chol (Supplemental Figure S5). The serum levels of free fatty acid were twofold higher in Gao-binge alcohol–treated A-Raptor KO compared with control A-Raptor KO mice. However, no significant differences were found between WT and A-Raptor KO mice regardless of alcohol treatment. There was no significant difference in serum levels of glycerol between control WT and A-Raptor KO mice, but Gao-binge alcohol significantly increased serum levels of glycerol in A-Raptor KO mice (Supplemental Figure S6). Together, these data indicate that A-Raptor and A-mTOR KO, but not A-TSC1 KO, mice have increased basal hepatic steatosis and liver injury. Gao-binge alcohol further exacerbates steatosis without affecting liver injury in A-Raptor KO mice. Moreover, inhibition of mTOR complex 1 (mTORC1) activity is sufficient to cause adipose atrophy, leading to hepatic steatosis and liver injury.
Figure 3.
A-Raptor KO mice have exacerbated liver steatosis but not liver injury after Gao-binge alcohol. A-Raptor KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Representative gross images of livers from WT and A-Raptor KO mice. B: Liver/body weight (BW) ratio was quantified. C: Representative images of hematoxylin and eosin staining of livers. D: Representative images of liver Oil Red O staining. E–G: Liver triglyceride (TG; E) and cholesterol (Chol; F) and serum alanine aminotransferase (ALT; G) levels were measured. Data are expressed as means ± SEM (B and E–G). n = 5 to 7 (B); n = 4 to 7 (E–G). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (C and D). ETOH, ethanol.
Figure 4.
No difference of liver steatosis and injury between A-TSC1 KO mice and matched WT mice after Gao-binge alcohol. A-TSC1 KO mice and matched WT mice were subjected to Gao-binge alcohol treatment. A: The representative gross liver images of WT and KO mice. B: Representative images of liver hematoxylin and eosin staining. C–E: The liver/body weight (BW) ratio (C), liver triglyceride (TG) levels (D), and cholesterol (Chol) levels (E). F: Serum alanine aminotransferase (ALT) levels and aspartate aminotransferase (AST) levels. Data are expressed as means ± SEM (C–F). n = 8 to 13 (C–F). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (B). ETOH, ethanol.
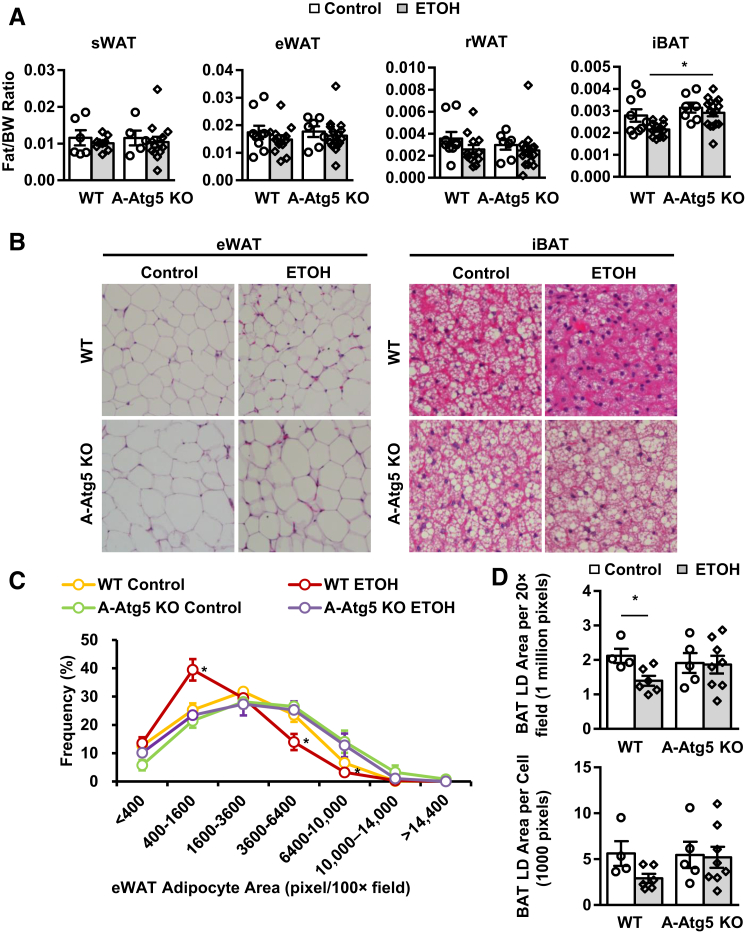
A-Atg5 KO Mice Are Less Sensitive to Gao-Binge Alcohol–Induced Adipose Atrophy
The levels of Atg5-Atg12 conjugate were decreased in eWAT, sWAT, and iBAT; and p62 and LC3-I levels were increased in A-ATG5 KO mice (Supplemental Figure S7A), suggesting successful deletion of Atg5 and inhibition of adipose autophagy in A-Atg5 KO mouse adipose tissues. Notably, despite loading with an equal amount of protein, the basal levels of Atg5-Atg12 were much higher in eWAT and sWAT than muscle and liver, and iBAT had low levels of Atg5-Atg12. The levels of β-actin were higher in eWAT and sWAT than iBAT, but iBAT had much higher levels of glyceraldehyde-3-phosphate dehydrogenase than eWAT and sWAT. These results suggest that autophagy proteins and even internal controls varied among different adipose tissues, perhaps reflecting their respective different physiological functions. Liver and skeletal muscle were also examined because they are crucial in nutrient metabolism, and they are tightly associated with adipose tissues. Atg5-Atg12, p62, and LC3 were barely affected in liver or skeletal muscle of A-Atg5 KO mice, suggesting that the deletion of Atg5 was specific for adipose tissues (Supplemental Figure S7A).
The ratios of various adipose tissues versus body weight in A-Atg5 KO mice were not significantly different compared with matched WT mice (Figure 5A). These results are different from previous reports, which use aP2-Cre, which may delete target genes in both adipose and muscle.12,13 In response to Gao-binge alcohol, WT mice had slightly reduced WAT mass (by 10% to 20%) and significantly reduced iBAT mass (by 40%). However, the mass of iBAT in A-Atg5 KO mice was significantly higher than WT mice after Gao-binge alcohol (Figure 5A). The size of adipocytes in eWAT and iBAT decreased after alcohol treatment in WT mice, but it remained unchanged in A-Atg5 KO mice (Figure 5B). In WT mice, 25% of eWAT adipocytes had an area of 400 to 1600 pixels/×100 field, whereas KO mice had approximately 40% of eWAT adipocytes in the 400 to 1600 range and fewer adipocytes were in the larger area range (Figure 5C). In addition, alcohol decreased LD size per field by approximately 40% per cell and by approximately 50% in iBAT in WT but not in A-Atg5 KO mice (Figure 5D). The body weights on day 1 and the last day of feeding (day 10) were not significantly different between WT and KO mice (Supplemental Figure S7B). In addition, the average food intake from day 1 to day 10 was similar between A-Atg5 KO and the matched WT mice (Supplemental Figure S7C). Taken together, A-Atg5 KO mice are resistant to alcohol-induced adipose atrophy in WAT; and this is not due to increased body weight or nutrient intake.
Figure 5.
A-Atg5 KO mice are partially resistant to Gao-binge alcohol–induced adipocyte atrophy. A-Atg5 KO mice and matched WT mice were subjected to Gao-binge alcohol treatment. A: Quantification of fat tissue weight to body weight (BW) of s.c. white adipose tissue (sWAT), epididymal WAT (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT) after Gao-binge alcohol. B: Representative hematoxylin and eosin (H&E) staining images of eWAT and iBAT. C: Distribution of eWAT adipocyte area in H&E staining. D: Quantification of iBAT lipid droplet (LD) area in H&E staining. Data are expressed as means ± SEM (A, C, and D). n = 5 to 17 (A); n = 3 to 6 mice per group (C); n = 4 to 8 mice per group (D). *P < 0.05 (one-way analysis of variance). Original magnification: ×20 (B, eWAT); ×40 (B, iBAT). ETOH, ethanol.
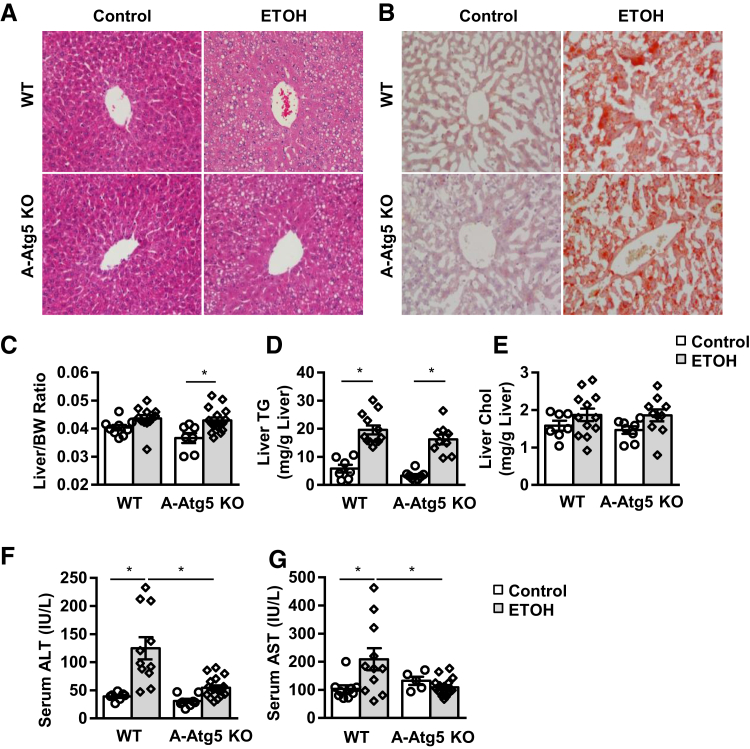
A-Atg5 KO Mice Are Resistant to Gao-Binge Alcohol–Induced Liver Injury but Not Steatosis
Alcohol-induced liver injury and steatosis were examined in A-Atg5 KO mice. Unlike A-mTOR and A-Raptor KO mice, the control A-Atg5 KO mouse livers showed normal liver histology and hepatic lipid levels compared with matched WT mice (Figure 6, A–D). Gao-binge alcohol treatment resulted in liver macrosteatosis and microsteatosis (ie, visualization of big and small LDs in hematoxylin and eosin staining, increased liver Oil Red O staining, and increased liver/body weight ratio and levels of liver TG in both A-Atg5 KO and matched WT mice) (Figure 6, A–D). Liver Chol levels were slightly increased after alcohol, but no difference was found between A-Atg5 KO and WT mice (Figure 6E). These data suggest that A-Atg5 KO mice do not affect alcohol-induced hepatic steatosis.
Figure 6.
A-Atg5 KO mice are resistant to Gao-binge alcohol–induced liver injury but not steatosis. A-Atg5 KO mice and matched WT mice were subjected to Gao-binge alcohol treatment. Representative liver hematoxylin and eosin (A) and Oil Red O (B) staining images. Quantification of liver/body weight (BW) ratio (C), liver triglyceride (TG; D), and cholesterol (Chol; E) levels. Serum alanine aminotransferase (ALT; F) and aspartate aminotransferase (AST; G) levels. Data are expressed as means ± SEM (C–G). n = 5 to 18 (C–G). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (A and B). ETOH, ethanol.
Gao-binge alcohol treatment significantly increased serum levels of free fatty acid in both A-Atg5 KO and matched WT mice compared with their respective control mice (Supplemental Figure S8A), suggesting Gao-binge alcohol may increase lipolysis in both A-Atg5 KO and WT mice. After alcohol treatment, the serum levels of glycerol increased to similar levels in both WT and A-Atg5 KO mice, but no statistical differences were found compared with mice fed with control diet (Supplemental Figure S8B). The levels of serum TG and Chol were not increased by alcohol and were also not different between A-Atg5 KO and matched WT mice (Supplemental Figure S8, C and D). Moreover, the serum levels of β-hydroxybutyrate, a marker to reflect mitochondrial fatty acid β-oxidation, were slightly higher at the basal level in A-Atg5 KO mice (50% increase). The serum levels of β-hydroxybutyrate increased significantly by approximately threefold after Gao-binge alcohol treatment, but no difference was found between A-Atg5 KO and matched WT mice (Supplemental Figure S8E). Collectively, these data suggest that A-Atg5 KO mice have similar serum lipids to WT mice with or without alcohol treatment. However, the serum levels of ALT and aspartate aminotransferase were significantly lower in A-Atg5 KO mice than in WT mice after Gao-binge alcohol (Figure 6, F and G).
The underlying mechanism of liver protection against alcohol in A-Atg5 KO mice was further studied. Previous studies demonstrated that receptor-interacting protein kinase 1–, receptor-interacting protein kinase 3–, and mixed lineage kinase-like protein–mediated necroptosis plays a role in alcohol-induced liver injury.29,30 The levels of receptor-interacting protein kinase 3 and mixed lineage kinase-like protein increased (4.5- and 3.7-fold, respectively) in WT mouse livers in response to alcohol treatment (Supplemental Figure S9). Interestingly, these two proteins increased (2.9- and 2.2-fold, respectively) in A-Atg5 KO livers without alcohol treatment, but they were not further increased in response to alcohol treatment (Supplemental Figure S9). Ethanol metabolism via cytochrome P450 2E1 is largely responsible for ethanol-induced oxidative stress and may contribute to ethanol-induced cell injury.31, 32, 33 Gao-binge alcohol increased hepatic cytochrome P450 2E1 protein to similar levels in A-Atg5 KO and WT mice (Supplemental Figure S10, A and B). Gao-binge alcohol also significantly increased the levels of hepatic protein carbonylation, a biomarker of oxidative stress.34 However, no difference was found between WT and A-Atg5 KO mice (Supplemental Figure S10, C and D).
Gao-binge alcohol slightly increased neutrophil infiltration in the liver in both A-Atg5 KO and WT mice (Supplemental Figure S11, A and B). Consistently, alcohol treatment significantly increased liver Ly6g mRNA levels, which is a marker for neutrophils, monocytes, and granulocytes (Supplemental Figure S11C). In A-Atg5 KO mice, the neutrophil number in each field and the Ly6g mRNA level were similar to WT mice before and after alcohol. There was no significant induction of the proinflammatory cytokines Tnfa, Il1b, or Il6, or macrophage marker F4/80 in liver after alcohol treatment. However, alcohol increased expression of Ccl2, a chemokine regulating recruitment of monocytes and macrophages, to almost 12-fold compared with control WT mice (Supplemental Figure S11C). In A-Atg5 KO mice, the hepatic mRNA levels of Tnfa, Il1b, Il6, or F4/80 were not significantly different than those in WT mice. However, the level of Ccl2 mRNA was much lower in A-Atg5 KO mice than in WT mice after alcohol treatment. The mRNA levels of Tnfa and Il1b in eWAT of A-Atg5 KO and WT mice did not change significantly with or without Gao-binge alcohol treatment (Supplemental Figure S11D). These data suggest that Gao-binge alcohol may lead to mild hepatic inflammation in both WT and A-Atg5 KO mice, and no significant difference was found between these two genotypes of mice. No significant changes of serum levels of lipocalin 2 were found among the WT and A-Atg5 KO mice after alcohol treatment, although the basal levels of serum lipocalin 2 tended to be lower in A-Atg5 KO mice (Supplemental Figure S11E). Taken together, these data suggest that A-Atg5 KO mice are resistant to Gao-binge alcohol–induced liver injury, but not hepatic steatosis. It seems that oxidative stress and hepatic inflammation are not responsible for deceased liver injury in A-Atg5 KO mice after Gao-binge alcohol.
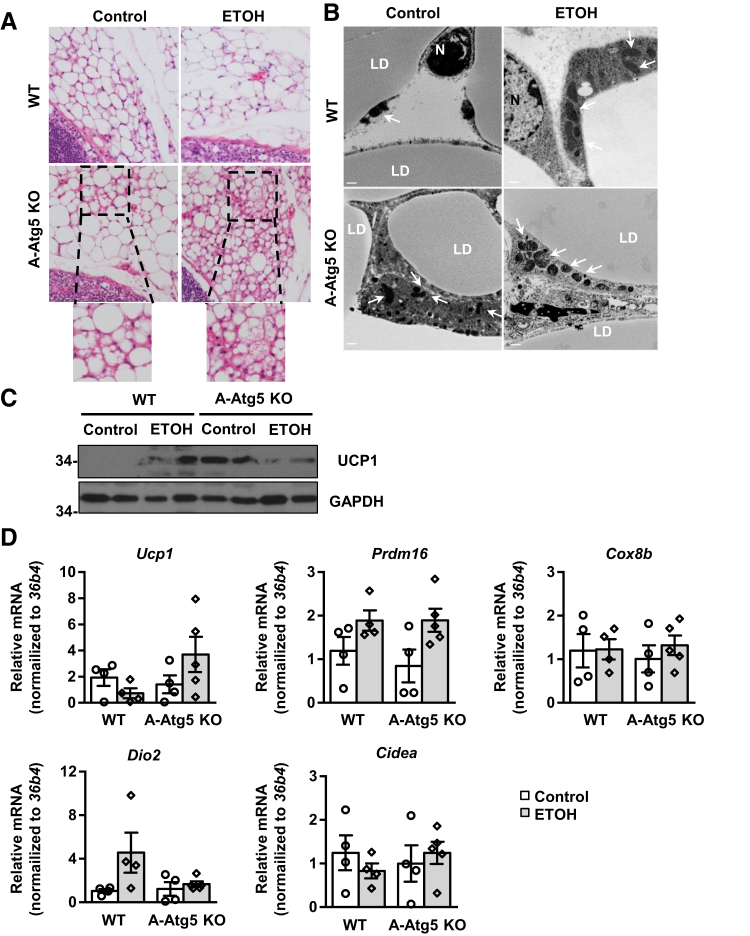
A-Atg5 KO Mice Have Increased Features of sWAT Browning
Autophagy-mediated degradation of mitochondria is a hallmark of the transition of browning adipocytes to white adipocytes.35 Therefore, lack of autophagic removal of mitochondria may favor the browning of white adipocytes. Multilocular LDs were rarely seen in sWAT of WT mice. However, depots of adipocytes with multilocular LDs around lymph nodes were readily detected in sWAT of A-Atg5 KO mice and were increased by Gao-binge alcohol treatment (Figure 7A). Electron microscopy studies revealed that there were increased mitochondria numbers in sWAT adipocytes of control A-Atg5 KO mice compared with control WT mice. Gao-binge alcohol increased the numbers of mitochondria in both WT and A-Atg5 KO mice, but increased mitochondria seemed more evident in A-Atg5 KO mice (Figure 7B). Gao-binge alcohol increased the levels of uncoupling protein 1 (UCP1) in WT mice. The basal levels of UCP1 markedly increased in sWAT of A-Atg5 KO mice compared with matched WT mice, which slightly decreased after Gao-binge alcohol (Figure 7C). However, the mRNA levels of genes specifically expressed in brown/beige adipocytes, including Prdm16, Cox8b, Dio2, and Cidea, were not different between WT and A-Atg5 KO mice. However, UCP1 levels increased twofold in Gao-binge alcohol–treated A-Atg5 KO mice (Figure 7D). Overall, these data suggest that Gao-binge alcohol and deletion of Atg5 in mouse adipocytes may individually lead to mild white adipose browning.
Figure 7.
A-Atg5 KO mice have increased features of s.c. white adipose tissue (sWAT) browning. A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Representative hematoxylin and eosin staining images of sWAT. The bottom panels are enlarged images from the above boxed areas. B: Representative electron microscopy images of sWAT are shown. Arrows indicate mitochondria. C: Total lysates of sWAT were subjected to Western blot analysis. D: sWAT samples were subjected to RNA extraction and real-time quantitative PCR. Data are expressed as means ± SEM (D). n = 4 to 5 (D). Scale bars = 500 nm (B). Original magnification, ×40 (A). ETOH, ethanol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LD, lipid droplet; N, nucleus; UCP1, uncoupling protein 1.
No Difference in Isoproterenol-Induced Lipolysis in ex Vivo WT and A-Atg5 KO Adipocytes
eWAT and sWAT were next isolated from WT and A-Atg5 KO mice, and their response to isoproterenol, a β-adrenergic agonist that induces lipolysis, was tested. In WT mice, isoproterenol incubation increased free glycerol release by 1.4-fold in eWAT (Supplemental Figure S12A) and by 2-fold in sWAT (Supplemental Figure S12B). The average free glycerol released in response to isoproterenol was slightly lower in A-Atg5 KO mice, but the induction fold was not significantly different from WT mice. This suggests that constitutional Atg5 deficiency in adipose tissue may not affect isoproterenol-induced lipolysis ex vivo.
A-Atg5 KO Mice Have Increased Basal Levels of Fgf21 and Adiponectin
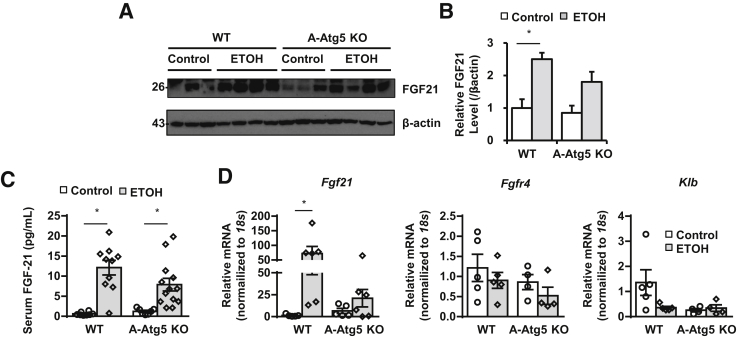
FGF21 is an antiobesity, antidiabetes hormone secreted by liver.36,37 Gao-binge alcohol increased the expression of liver Fgf21 both at the mRNA level (72-fold increase) and protein level (2.5-fold increase) as well as increased circulating Fgf21 levels in response to alcohol (Figure 8). Interestingly, A-Atg5 KO mice had higher basal levels of liver Fgf21 mRNA (fivefold) and circulating Fgf21 levels (twofold) compared with WT mice. Liver Fgf21 mRNA and protein levels as well as serum Fgf21 levels were also increased after alcohol, but by a smaller extent (20-, 1.8-, and 4-fold, respectively) in A-Atg5 KO mice (Figure 8). FGF21 binds to isoforms of FGF receptors facilitated by transmembrane protein β-Klotho (KLB). In liver, the mRNA level of Klb decreased after Gao-binge alcohol in WT mice. The mRNA level of Klb was already lower in control A-Atg5 KO mice, which was not altered after Gao-binge alcohol (Figure 8). There were no significant changes in mRNA levels of Fgfr4 in A-Atg5 KO mice or WT mice regardless of alcohol treatment (Figure 8). These results suggest that, although A-Atg5 KO mice have higher basal circulating Fgf21 than WT mice, the decreased liver injury in A-Atg5 KO mice was likely not directly due to circulating Fgf21 after alcohol.
Figure 8.
Gao-binge alcohol increases hepatic and circulating fibroblast growth factor 21 (Fgf21) in both WT and A-Atg5 KO mice. A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Total liver lysates were subjected to Western blot analysis. B: Densitometry of A. C: Serum Fgf21 levels were measured by enzyme-linked immunosorbent assay. D: Livers were subjected to RNA extraction and real-time quantitative PCR. Data are expressed as means ± SEM (B–D). n = 3 to 4 (B); n = 8 to 13 (C); n = 4 to 5 (D). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
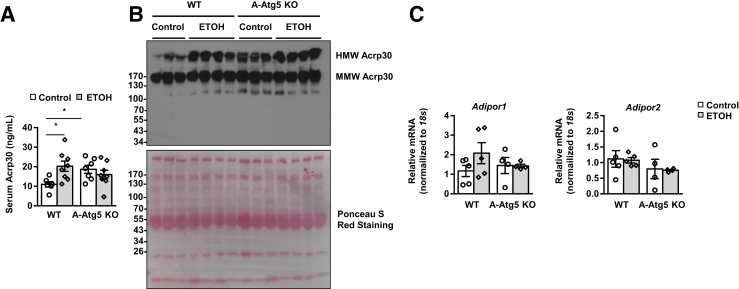
Adiponectin regulates lipid and glucose metabolism and plays a protective role in various metabolic scenarios, but its response to alcohol exposure is still debatable.38, 39, 40 The levels of circulating adiponectin increased from 9.97 to 16.36 ng/mL in response to Gao-binge alcohol (Figure 9A). Interestingly, the serum levels of adiponectin were almost twofold higher in control A-Atg5 KO mice than control WT mice, which slightly decreased after alcohol. Increased serum levels of high-molecular-weight (HMW) adiponectin were found in WT mice after alcohol. A-Atg5 KO mice already had increased basal serum levels of HMW adiponectin, which were not further changed after alcohol treatment (Figure 9B). Adiponectin is secreted by adipose tissue and targets liver via receptors AdipoR1 and AdipoR2. The mRNA levels of liver Adipor1 and Adipor2 remained unchanged in response to Gao-binge alcohol in both WT and A-Atg5 KO mice (Figure 9C). These results suggest that A-Atg5 KO mice or Gao-binge alcohol treatment increased adipose adiponectin secretion, which possibly serves as an adaptive protective mechanism. Therefore, it is possible that the higher basal levels of circulating adiponectin and Fgf21 could prime the A-Atg5 KO mice to be less sensitive to alcohol-induced liver injury.
Figure 9.
Increased circulating adiponectin (Acrp) in mice with either Gao-binge alcohol treatment or adipose-specific deletion of Atg5. A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Serum adiponectin levels were measured by enzyme-linked immunosorbent assay. B: Equal amounts of serum samples were subjected to Western blot in nonheated, nondenatured conditions. Ponceau S Red staining was performed as a loading control. C: Livers were subjected to RNA extraction and real-time quantitative PCR. Data are expressed as means ± SEM (A and C). n = 6 to 8 (A). n = 4 to 5 (C). *P < 0.05 (t-test). ETOH, ethanol; HMW, high molecular weight; MMW, medium molecular weight.
Discussion
In this study, we characterized the effects of Gao-binge alcohol on mTOR signaling and autophagy in mouse adipose tissues and the adipose-liver axis in alcohol-induced liver injury. Gao-binge alcohol induced white and brown adipocyte atrophy, increased adipose tissue lipolysis and release of free fatty acid, and increased circulating adiponectin and FGF21. Mechanistically, Gao-binge alcohol inactivated Akt/mTOR signaling and increased autophagic flux in eWAT. Genetic deletion of mTOR or Raptor in adipose tissues significantly decreased adipose mass and increased liver injury and hepatic steatosis. Deletion of Atg5 in adipose tissue did not significantly affect adipose mass but partially protected against alcohol-induced decrease in adipocyte size and liver injury.
In the present study, Gao-binge alcohol induced WAT and BAT adipose tissue atrophy, which is consistent with previous reports in chronic ethanol-fed mice and rats.7,27 It has been suggested that adipose atrophy induced by chronic ethanol exposure is the consequence of increased adipose tissue lipolysis, resulting from the impaired insulin-mediated suppression of lipolysis, but not from the β-adrenergic receptor activation.7 In addition, a recent study reported that FGF21-mediated eWAT lipolysis can also contribute to adipose atrophy in a modified Gao-binge mouse model (chronic alcohol feeding for 12 days plus acute binge).37 Consistent with this report, increased circulating Fgf21 levels were also found in Gao-binge alcohol–treated mice. In addition to lipolysis, it has been reported that genetic ablation of mTOR or Raptor in mouse adipose tissue leads to decreased adipose tissue mass.20,41 In line with these previous findings, mice with genetic ablation of adipose Raptor or Mtor, but not Tsc1, showed lipodystrophy in both WAT and BAT. mTOR has two complexes, which are called mTORC1 and mTORC2, and Raptor is one of the main components for mTORC1. Gao-binge alcohol further lowered adipose mass in A-Raptor KO mice but not in A-mTOR KO mice, suggesting that loss of mTORC2 functions may also affect alcohol-induced adipose mass changes. Interestingly, Gao-binge alcohol decreased AKT-mTOR signaling in WAT, suggesting that Gao-binge alcohol–induced adipose atrophy may also be partially mediated by mTOR inhibition.
mTOR is an intracellular nutrient sensor and negatively regulates autophagy. Indeed, the levels of p62 and LC3-II proteins decreased in mouse eWAT after Gao-binge alcohol, suggesting an overall increased catabolic status, which is likely due to increased autophagic degradation in mouse adipose tissues. Notably, studies using Adipoq-Cre to delete Raptor/Mtor or Tsc1 exhibited reduced adipose tissue mass and body weight in mice at early ages of 4 to 8 weeks.20,41,42 Because the mTOR signaling pathway determines many biological processes, including protein translation and lipid metabolism,43 it is likely that impaired adipogenesis in mice with genetic deletion of mTOR or its regulators is mediated by both autophagy-dependent and autophagy-independent mechanisms. For instance, genetic deletion of Tsc1 results in the persistent activation of mTOR, which generally leads to decreased autophagy. However, changes in adipose tissue and liver injury were different in A-Atg5 and A-TSC1 KO mice, further supporting the notion that mTOR may have multiple functions in addition to autophagy regulation.
Although previous studies showed that autophagy inhibition genetically or pharmacologically impairs adipogenesis in vitro and tends to decrease adipose mass in mice,11,13,44 no significant difference was found in body weight or fat mass in A-Atg5 KO mice. Several possibilities exist to explain these differences. First, Adipoq-Cre was used to generate adipocyte-specific Atg5 KO mice, which is more specific for adipose tissue than the aP2-Cre used in previous studies.45 Second, although the partial Atg5 deletion and autophagy inhibition were confirmed in WAT and BAT, there were still Atg5 and LC3-II proteins in WAT and BAT, suggesting the recombination was likely not 100%, and the remaining Atg5 might be enough for some normal adipogenesis. Third, mice with deletion of Atg12 in brown and beige adipocytes using Ucp1-Cre did not have reduced adipose mass or impaired adipogenesis, although Atg12 KO mice had less adiposity expansion after 8-week high-fat diet challenge.35 Although these mice may have different response on obesogenic diet, diabetic diet, or other metabolic stress, it is a possibility that inhibiting autophagy using Adipoq-Cre or Ucpl-Cre is not potent enough to impair adipogenesis. It will be intriguing to see whether A-Atg5 KO mice are less resistant to high-fat diet–induced obesity or glucose challenge in the future. One notable limitation of this study is using constitutional Adipoq-Cre, which may mix the impact of autophagy deficiency on adipogenesis at the early adipose development stage and in mature adipocytes. Cai et al46 used the inducible Cre system based on a tamoxifen-dependent Cre recombinase and found that deleting Atg3 or Atg16L, two other important components in the autophagosome maturation process, increased adipose inflammation, induced adipose and systematic insulin resistance, and increased lipid peroxide-induced nuclear factor erythroid 2–related factor 2 activation in adipose tissue and liver. This supports the post-developmental role of adipose autophagy in the adipose-liver axis and the whole body metabolic system. It will be interesting to investigate the role of alcohol in the pathogenesis of adipose and liver using these inducible mouse models in the future.
How do changes in adipose tissue impact alcohol-induced liver injury? It is well known that both adipose tissues and liver can actively secrete proteins and other mediators into the circulation, such as adipokines and hepatokines, to regulate the function of other tissues/organs in a paracrine manner. Among them, adiponectin, the most abundant gene product in adipose tissue, protects against alcohol-induced liver injury by coordinating multiple signaling pathways in the liver, leading to enhanced fat oxidation, reduced lipid synthesis, and prevention of hepatic steatosis.47 The effect of alcohol on adiponectin levels has been controversial in human studies48,49 and in rodent models.38,40,50, 51, 52 It seems that adiponectin is induced by alcohol as an adaptive response, and the decrease in adiponectin occurs during chronic exposure at a later time point. Gao-binge alcohol increased serum total adiponectin levels in WT mice, and A-Atg5 KO mice had higher adiponectin at the basal level, although adiponectin levels were not further increased after alcohol. Circulating adiponectin contains three major oligomeric forms: a low-molecular-weight trimer, a medium-molecular-weight hexamer, and an HMW multimer. It has been suggested that different oligomers of adiponectin may have different physiological functions.53 The levels of HMW adiponectin correlated with total adiponectin levels, and medium-molecular-weight adiponectin levels did not differ much between treatments or genotypes after Gao-binge alcohol. Notably, A-Atg5 KO mice had increased total serum adiponectin and HMW adiponectin at basal levels.
Another potential mediator of the adipose-liver axis is FGF21. Consistent with previous studies,36,37 Gao-binge alcohol markedly increased liver and serum levels of Fgf21. Interestingly, the basal levels of liver Fgf21 mRNA and serum FGF21 were already increased significantly, although the levels of Fgf21 only increased mildly after exposure to alcohol in A-Atg5 KO mice compared with WT mice. In A-Atg5 KO mice, liver Fgf21 protein levels were not paralleled with Fgf21 mRNA, and it is possible that part of the Fgf21 protein synthesized in liver was secreted to circulation. Alcohol binge alone could induce serum FGF21 elevation, peaking at 6 hours and gradually restoring to normal in human and mice.54 Therefore, the effect of Gao-binge alcohol on serum FGF21 is likely a mixture of chronic exposure and acute binge. Our results suggest that it is possible that the increased basal levels of adiponectin and Fgf21 in A-Atg5 KO mice may prime these mice to be more resistant to alcohol-induced liver injury. Future studies are needed to further dissect how alcohol induces adiponectin and FGF21 and how increased basal levels of adiponectin and FGF21 affect the liver response to alcohol.
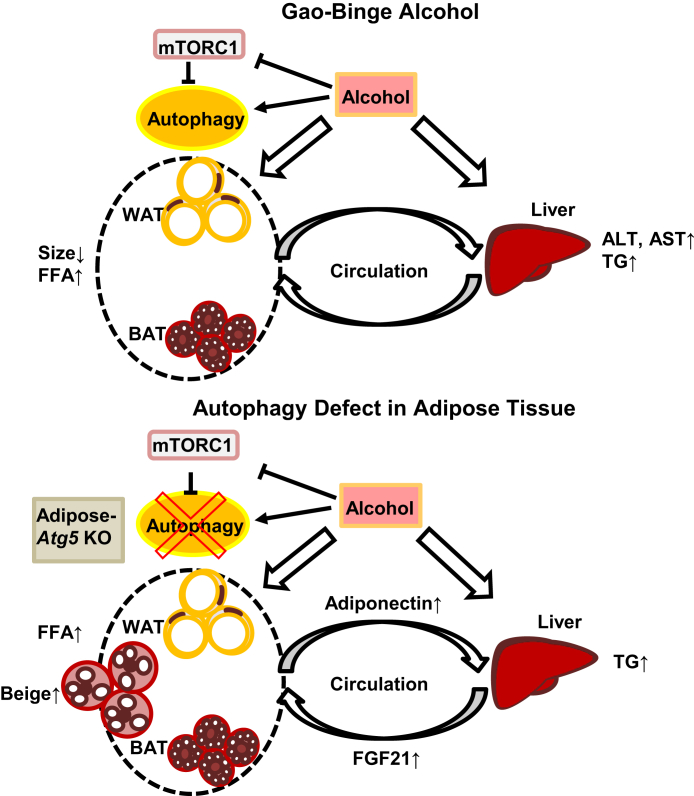
In conclusion, Gao-binge alcohol leads to decreased adipocyte/LD size in mouse adipose tissues, which is associated with decreased Akt/mTOR signaling and enhanced autophagy in adipose tissues. A-Atg5 KO mice are resistant to Gao-binge alcohol–induced adipose atrophy and liver injury, but not hepatic steatosis. Increased levels of circulating adiponectin and FGF21 may enable these mice to be more resistant to alcohol-induced liver injury. The molecular signaling events in adipose, liver, and blood circulating after alcohol exposure are summarized in Figure 10. In the future, it may be beneficial to preventing/treating alcohol-induced adipose atrophy and liver injury by developing some lipophilic autophagy/lysosomal inhibitors to specifically inhibit autophagy in adipose tissues.
Figure 10.
Scheme of the role of autophagy in Gao-binge alcohol–induced adipose atrophy and liver injury. Alcohol suppresses mTOR and increases autophagy in adipocytes. Alcohol also increases adipocyte atrophy, lipolysis, and circulation levels of serum adiponectin and fibroblast growth factor 21 (FGF21). In A-Atg5 KO mice, adipocyte autophagy is inhibited, which is associated with increased beige adipocyte appearance, and alleviates alcohol-induced adipose atrophy and liver injury. The increased circulating FGF21 and adiponectin likely play a protective role against alcohol-induced liver injury. ALT, alanine aminotransferase; AST, aspartate aminotransferase; BAT, brown adipose tissue; FFA, free fatty acid; mTORC1, mTOR complex 1; TG, triglyceride; WAT, white adipose tissue.
Acknowledgments
We thank Barbara Fegley (University of Kansas Medical Center Electron Microscopy Research Laboratory) for assistance with the electron microscopy studies; Dr. Noboru Mizushima (University of Tokyo) for providing Atg5f/f mice; and Tara McKeen for critically reading the manuscript.
Author Contributions
Y.L. designed the study, acquired and analyzed the data, performed statistical analysis, and wrote the manuscript; X.C., S.W., J.A.W., and H.-M.N. acquired and analyzed the data; W.-X.D. conceived and designed the study, revised the manuscript, and supervised the study; all authors had access to the study data and have reviewed and approved the final manuscript.
Footnotes
Supported in part by NIH grants R01 AA020518, U01 AA024733, R21 AA027250, P20 GM 103549, and P30 GM 118247 (all to W.-X.D.); and a pilot project from the University of Kansas Cancer Center Support grant P30CA168524. Y.L. is a recipient of the Biomedical Research Training Program, funded by University of Kansas Medical Center.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2019.09.023.
Supplemental Data
A-mTOR KO mice have adipose atrophy without alcohol exposure. A-mTOR KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Quantification of fat tissue weight of s.c. white adipose tissue (sWAT), epididymal WAT (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT) after Gao-binge treatment. B: Total lysates from eWAT, sWAT, and iBAT from A-mTOR KO mice and matched Cre mice were subjected to Western blot analysis. Data are expressed as means ± SEM (A). n = 5 to 8 (A). *P < 0.05 (one-way analysis of variance). BW, body weight; ETOH, ethanol.
A-TSC1 KO mice do not alter the adipose mass compared with their matched WT mice. A-TSC1 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Quantification of fat tissue weight of s.c. white adipose tissue (sWAT), epididymal WAT (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT) after Gao-binge treatment. B: Total lysates from eWAT, sWAT, and iBAT from A-TSC1 KO mice and matched Cre- mice were subjected to Western blot analysis. Data are expressed as means ± SEM (A). n = 8 to 13 (A). *P < 0.05 (one-way analysis of variance). BW, body weight; ETOH, ethanol.
No difference of body weight and food intake between A-Raptor KO mice and their matched WT mice. A-Raptor KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: The average body weight of A-Raptor KO mice and WT mice on day 1 (since alcohol feeding started) and day 10. B: The average food intake of A-Raptor KO mice and WT mice. Data are expressed as means ± SEM (A and B). n = 5 to 7 (A and B). ETOH, ethanol.
A-mTOR KO mice have hepatomegaly and increased liver steatosis regardless of alcohol exposure. A-mTOR KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: The liver weight was recorded and normalized to body weight (BW). B: Representative liver hematoxylin and eosin staining images are shown. C and D: Liver triglyceride (TG) levels (C) and serum alanine aminotransferase (ALT) levels (D) were measured. Data are expressed as means ± SEM (A, C, and D). n = 5 to 8 (A, C, and D). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (B). ETOH, ethanol.
Serum lipid changes in A-Raptor KO, A-mTOR KN, and A-TSC1 KO mice after Gao-binge alcohol. A-Raptor KO, A-mTOR KO, and A-TSC1 KO mice and their matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum triglyceride (TG; A) and cholesterol (Chol; B) levels were measured. Data are expressed as means ± SEM (A and B). n = 4 to 13 (A and B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Increased serum free fatty acid (FFA) and glycerol in A-Raptor KO mice after Gao-binge alcohol. A-Raptor KO and the matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum free FFA (A) and glycerol (B) levels were measured. Data are expressed as means ± SEM (A and B). n = 4 to 7 (A and B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Generation and validation of A-Atg5 KO mice. A: Total lysates from skeletal muscle, liver, epididymal white adipose tissue (eWAT), s.c. WAT (sWAT), and interscapular brown adipose tissue (iBAT) of A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Western blot analysis. B: The average body weight on day 1 (since alcohol feeding started) and day 10. C: The average food intake. Data are expressed as means ± SEM (B and C). n = 8 to 12 (B and C). ETOH, ethanol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
No significant changes of serum lipids between A-Atg5 KO mice and the matched WT mice after Gao-binge. A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum free fatty acid (FFA; A), free glycerol (B), triglyceride (TG; C), cholesterol (Chol; D), and β-hydroxybutyrate (β-HB; E) levels were measured. Data are expressed as means ± SEM (A–E). n = 7 to 12 (A–E). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Gao-binge alcohol does not increase hepatic levels of receptor-interacting protein kinase 3 (RIP3) and mixed lineage kinase-like protein (MLKL) in A-Atg5 KO mice. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Total liver lysates were subjected to Western blot analysis. B: Densitometry of A. Data are expressed as means ± SEM (B). n = 3 to 4 (B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Gao-binge alcohol induces similar levels of hepatic cytochrome P450 2E1 (CYP2E1) and protein carbonylation in A-Atg5 KO and matched WT mice. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Total liver lysates were subjected to Western blot analysis. B: Densitometry of A. C: Representative Western blot analysis, detecting hepatic carbonylation levels. D: Densitometry of C. Data are expressed as means ± SEM (B and D). n = 3 to 4 (B and D). *P < 0.05 (one-way analysis of variance). ETOH, ethanol; NC, negative control.
There is no significant difference of hepatic neutrophil infiltration between A-Atg5 KO and matched WT mice after Gao-binge alcohol. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Representative images of liver Ly6B2 immunohistochemistry staining. Arrows denote neutrophils. B: Quantification of A. C and D: Livers (C) and epididymal white adipose tissues (D) were subjected to RNA extraction and real-time quantitative PCR. E: Serum lipocalin 2 was measured using the enzyme-linked immunosorbent assay kit. Data are expressed as means ± SEM (B–E). n = 4 to 6 (B and C); n = 3 (D); n = 8 to 13 (E). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (A). ETOH, ethanol.
There is no significant difference of lipolysis between WT and Atg5 KO adipocytes ex vivo. Epididymal white adipose tissue (eWAT; A) and s.c. WAT (sWAT; B) samples were collected from A-Atg5 KO mice and matched Cre- (WT) mice and subjected to ex vivo lipolysis assay. Pieces of adipose tissues were incubated with or without 10 μmol/L isoproterenol [ISO or non-treated (NT)]. The free glycerol released in the medium in 1 hour was normalized to tissue weight and compared with nonchallenged tissues. Data are expressed as means ± SEM (A and B). n = 9 (A and B). *P < 0.05 (one-way analysis of variance).
References
- 1.Gao B., Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams J.A., Manley S., Ding W.X. New advances in molecular mechanisms and emerging therapeutic targets in alcoholic liver diseases. World J Gastroenterol. 2014;20:12908–12933. doi: 10.3748/wjg.v20.i36.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy L.E., Ding W.X., Cresci G., Saikia P., Shah V.H. Linking pathogenic mechanisms of alcoholic liver disease with clinical phenotypes. Gastroenterology. 2016;150:1756–1768. doi: 10.1053/j.gastro.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Wang S., Ni H.M., Huang H., Ding W.X. Autophagy in alcohol-induced multiorgan injury: mechanisms and potential therapeutic targets. BioMed Res Int. 2014;2014:498491. doi: 10.1155/2014/498491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Ding W.X. Adipose tissue autophagy and homeostasis in alcohol-induced liver injury. Liver Res. 2017;1:54–62. doi: 10.1016/j.livres.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Sebastian B.M., Tang H., McMullen M.M., Axhemi A., Jacobsen D.W., Nagy L.E. Taurine supplementation prevents ethanol-induced decrease in serum adiponectin and reduces hepatic steatosis in rats. Hepatology. 2009;49:1554–1562. doi: 10.1002/hep.22811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang L., Chen X., Sebastian B.M., Pratt B.T., Bederman I.R., Alexander J.C., Previs S.F., Nagy L.E. Chronic ethanol and triglyceride turnover in white adipose tissue in rats: inhibition of the anti-lipolytic action of insulin after chronic ethanol contributes to increased triglyceride degradation. J Biol Chem. 2007;282:28465–28473. doi: 10.1074/jbc.M705503200. [DOI] [PubMed] [Google Scholar]
- 8.Zhong W., Zhao Y., Tang Y., Wei X., Shi X., Sun W., Sun X., Yin X., Sun X., Kim S., McClain C.J., Zhang X., Zhou Z. Chronic alcohol exposure stimulates adipose tissue lipolysis in mice: role of reverse triglyceride transport in the pathogenesis of alcoholic steatosis. Am J Pathol. 2012;180:998–1007. doi: 10.1016/j.ajpath.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebastian B.M., Roychowdhury S., Tang H., Hillian A.D., Feldstein A.E., Stahl G.L., Takahashi K., Nagy L.E. Identification of a cytochrome P4502E1/Bid/C1q-dependent axis mediating inflammation in adipose tissue after chronic ethanol feeding to mice. J Biol Chem. 2011;286:35989–35997. doi: 10.1074/jbc.M111.254201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky D.J. The molecular machinery of autophagy and its role in physiology and disease. Semin Cell Dev Biol. 2010;21:663. doi: 10.1016/j.semcdb.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Baerga R., Zhang Y., Chen P.H., Goldman S., Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K., Tang Y., Pessin J.E., Schwartz G.J., Czaja M.J. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovsan J., Bluher M., Tarnovscki T., Kloting N., Kirshtein B., Madar L., Shai I., Golan R., Harman-Boehm I., Schon M.R., Greenberg A.S., Elazar Z., Bashan N., Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 15.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J., Dibble C.C., Matsuzaki M., Manning B.D. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.E., Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 19.Polak P., Cybulski N., Feige J.N., Auwerx J., Ruegg M.A., Hall M.N. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Shan T., Zhang P., Jiang Q., Xiong Y., Wang Y., Kuang S. Adipocyte-specific deletion of mTOR inhibits adipose tissue development and causes insulin resistance in mice. Diabetologia. 2016;59:1995–2004. doi: 10.1007/s00125-016-4006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti P., English T., Shi J., Smas C.M., Kandror K.V. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowell K.T., Steiner J.L., Coleman C.S., Lang C.H. Decreased whole-body fat mass produced by chronic alcohol consumption is associated with activation of S6K1-mediated protein synthesis and increased autophagy in epididymal white adipose tissue. Alcohol Clin Exp Res. 2016;40:1832–1845. doi: 10.1111/acer.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni H.M., Woolbright B.L., Williams J., Copple B., Cui W., Luyendyk J.P., Jaeschke H., Ding W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., Lu B., Stolz D.B., Clemens D.L., Yin X.M. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buettner R., Newgard C.B., Rhodes C.J., O'Doherty R.M. Correction of diet-induced hyperglycemia, hyperinsulinemia, and skeletal muscle insulin resistance by moderate hyperleptinemia. Am J Physiol Endocrinol Metab. 2000;278:E563–E569. doi: 10.1152/ajpendo.2000.278.3.E563. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W., Zhong W., Sun X., Sun Q., Tan X., Li Q., Sun X., Zhou Z. Visceral white adipose tissue is susceptible to alcohol-induced lipodystrophy in rats: role of acetaldehyde. Alcohol Clin Exp Res. 2015;39:416–423. doi: 10.1111/acer.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X., Tang Y., Tan X., Li Q., Zhong W., Sun X., Jia W., McClain C.J., Zhou Z. Activation of peroxisome proliferator-activated receptor-gamma by rosiglitazone improves lipid homeostasis at the adipose tissue-liver axis in ethanol-fed mice. Am J Physiol Gastrointest Liver Physiol. 2012;302:G548–G557. doi: 10.1152/ajpgi.00342.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roychowdhury S., McMullen M.R., Pisano S.G., Liu X., Nagy L.E. Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology. 2013;57:1773–1783. doi: 10.1002/hep.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Ni H.M., Dorko K., Kumer S.C., Schmitt T.M., Nawabi A., Komatsu M., Huang H., Ding W.X. Increased hepatic receptor interacting protein kinase 3 expression due to impaired proteasomal functions contributes to alcohol-induced steatosis and liver injury. Oncotarget. 2016;7:17681–17698. doi: 10.18632/oncotarget.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford B.U., Kono H., Isayama F., Kosyk O., Wheeler M.D., Akiyama T.E., Bleye L., Krausz K.W., Gonzalez F.J., Koop D.R., Rusyn I. Cytochrome P450 CYP2E1, but not nicotinamide adenine dinucleotide phosphate oxidase, is required for ethanol-induced oxidative DNA damage in rodent liver. Hepatology. 2005;41:336–344. doi: 10.1002/hep.20532. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y., Cederbaum A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardag-Gorce F., Yuan Q.X., Li J., French B.A., Fang C., Ingelman-Sundberg M., French S.W. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279:23–29. doi: 10.1006/bbrc.2000.3889. [DOI] [PubMed] [Google Scholar]
- 34.Fedorova M., Bollineni R.C., Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev. 2014;33:79–97. doi: 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- 35.Altshuler-Keylin S., Shinoda K., Hasegawa Y., Ikeda K., Hong H., Kang Q., Yang Y., Perera R.M., Debnath J., Kajimura S. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016;24:402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Zhao C., Xiao J., Liu L., Zhang M., Wang C., Wu G., Zheng M.-H., Xu L.-M., Chen Y.-P., Mohammadi M., Chen S.-Y., Cave M., McClain C., Li X., Feng W. Fibroblast growth factor 21 deficiency exacerbates chronic alcohol-induced hepatic steatosis and injury. Sci Rep. 2016;6:31026. doi: 10.1038/srep31026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C., Liu Y., Xiao J., Liu L., Chen S., Mohammadi M., McClain C.J., Li X., Feng W. FGF21 mediates alcohol-induced adipose tissue lipolysis by activation of systemic release of catecholamine in mice. J Lipid Res. 2015;56:1481–1491. doi: 10.1194/jlr.M058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Z., Zhou Z., Deaciuc I., Chen T., McClain C.J. Inhibition of adiponectin production by homocysteine: a potential mechanism for alcoholic liver disease. Hepatology. 2008;47:867–879. doi: 10.1002/hep.22074. [DOI] [PubMed] [Google Scholar]
- 39.Shen Z., Liang X., Rogers C.Q., Rideout D., You M. Involvement of adiponectin-SIRT1-AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2010;298:G364–G374. doi: 10.1152/ajpgi.00456.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Sebastian B.M., Nagy L.E. Chronic ethanol feeding to rats decreases adiponectin secretion by subcutaneous adipocytes. Am J Physiol Endocrinol Metab. 2007;292:E621–E628. doi: 10.1152/ajpendo.00387.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee P.L., Tang Y., Li H., Guertin D.A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol Metab. 2016;5:422–432. doi: 10.1016/j.molmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magdalon J., Chimin P., Belchior T., Neves R.X., Vieira-Lara M.A., Andrade M.L., Farias T.S., Bolsoni-Lopes A., Paschoal V.A., Yamashita A.S., Kowaltowski A.J., Festuccia W.T. Constitutive adipocyte mTORC1 activation enhances mitochondrial activity and reduces visceral adiposity in mice. Biochim Biophys Acta. 2016;1861:430–438. doi: 10.1016/j.bbalip.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Mao Z., Zhang W. Role of mTOR in glucose and lipid metabolism. Int J Mol Sci. 2018;19:2043. doi: 10.3390/ijms19072043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C., He Y., Okutsu M., Ong L.C., Jin Y., Zheng L., Chow P., Yu S., Zhang M., Yan Z. Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARγ2 degradation. Am J Physiol Endocrinol Metab. 2013;305:E530–E539. doi: 10.1152/ajpendo.00640.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeffery E., Berry R., Church C.D., Yu S., Shook B.A., Horsley V., Rosen E.D., Rodeheffer M.S. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai J., Pires K.M., Ferhat M., Chaurasia B., Buffolo M.A., Smalling R., Sargsyan A., Atkinson D.L., Summers S.A., Graham T.E., Boudina S. Autophagy ablation in adipocytes induces insulin resistance and reveals roles for lipid peroxide and Nrf2 signaling in adipose-liver crosstalk. Cell Rep. 2018;25:1708–1717.e5. doi: 10.1016/j.celrep.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogers C.Q., Ajmo J.M., You M. Adiponectin and alcoholic fatty liver disease. IUBMB Life. 2008;60:790–797. doi: 10.1002/iub.124. [DOI] [PubMed] [Google Scholar]
- 48.Beulens J.W.J., van Loon L.J.C., Kok F.J., Pelsers M., Bobbert T., Spranger J., Helander A., Hendriks H.F.J. The effect of moderate alcohol consumption on adiponectin oligomers and muscle oxidative capacity: a human intervention study. Diabetologia. 2007;50:1388–1392. doi: 10.1007/s00125-007-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sierksma A., Patel H., Ouchi N., Kihara S., Funahashi T., Heine R.J., Grobbee D.E., Kluft C., Hendriks H.F. Effect of moderate alcohol consumption on adiponectin, tumor necrosis factor-alpha, and insulin sensitivity. Diabetes Care. 2004;27:184–189. doi: 10.2337/diacare.27.1.184. [DOI] [PubMed] [Google Scholar]
- 50.Fulham M.A., Mandrekar P. Sexual dimorphism in alcohol induced adipose inflammation relates to liver injury. PLoS One. 2016;11:e0164225. doi: 10.1371/journal.pone.0164225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu A., Wang Y., Keshaw H., Xu L.Y., Lam K.S., Cooper G.J. The fat-derived hormone adiponectin alleviates alcoholic and nonalcoholic fatty liver diseases in mice. J Clin Invest. 2003;112:91–100. doi: 10.1172/JCI17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pravdova E., Macho L., Fickova M. Alcohol intake modifies leptin, adiponectin and resistin serum levels and their mRNA expressions in adipose tissue of rats. Endocr Regul. 2009;43:117–125. [PubMed] [Google Scholar]
- 53.Tsao T.S., Tomas E., Murrey H.E., Hug C., Lee D.H., Ruderman N.B., Heuser J.E., Lodish H.F. Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity: different oligomers activate different signal transduction pathways. J Biol Chem. 2003;278:50810–50817. doi: 10.1074/jbc.M309469200. [DOI] [PubMed] [Google Scholar]
- 54.Desai B.N., Singhal G., Watanabe M., Stevanovic D., Lundasen T., Fisher F.M., Mather M.L., Vardeh H.G., Douris N., Adams A.C., Nasser I.A., FitzGerald G.A., Flier J.S., Skarke C., Maratos-Flier E. Fibroblast growth factor 21 (FGF21) is robustly induced by ethanol and has a protective role in ethanol associated liver injury. Mol Metab. 2017;6:1395–1406. doi: 10.1016/j.molmet.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-mTOR KO mice have adipose atrophy without alcohol exposure. A-mTOR KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Quantification of fat tissue weight of s.c. white adipose tissue (sWAT), epididymal WAT (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT) after Gao-binge treatment. B: Total lysates from eWAT, sWAT, and iBAT from A-mTOR KO mice and matched Cre mice were subjected to Western blot analysis. Data are expressed as means ± SEM (A). n = 5 to 8 (A). *P < 0.05 (one-way analysis of variance). BW, body weight; ETOH, ethanol.
A-TSC1 KO mice do not alter the adipose mass compared with their matched WT mice. A-TSC1 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: Quantification of fat tissue weight of s.c. white adipose tissue (sWAT), epididymal WAT (eWAT), retroperitoneal WAT (rWAT), and interscapular brown adipose tissue (iBAT) after Gao-binge treatment. B: Total lysates from eWAT, sWAT, and iBAT from A-TSC1 KO mice and matched Cre- mice were subjected to Western blot analysis. Data are expressed as means ± SEM (A). n = 8 to 13 (A). *P < 0.05 (one-way analysis of variance). BW, body weight; ETOH, ethanol.
No difference of body weight and food intake between A-Raptor KO mice and their matched WT mice. A-Raptor KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: The average body weight of A-Raptor KO mice and WT mice on day 1 (since alcohol feeding started) and day 10. B: The average food intake of A-Raptor KO mice and WT mice. Data are expressed as means ± SEM (A and B). n = 5 to 7 (A and B). ETOH, ethanol.
A-mTOR KO mice have hepatomegaly and increased liver steatosis regardless of alcohol exposure. A-mTOR KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. A: The liver weight was recorded and normalized to body weight (BW). B: Representative liver hematoxylin and eosin staining images are shown. C and D: Liver triglyceride (TG) levels (C) and serum alanine aminotransferase (ALT) levels (D) were measured. Data are expressed as means ± SEM (A, C, and D). n = 5 to 8 (A, C, and D). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (B). ETOH, ethanol.
Serum lipid changes in A-Raptor KO, A-mTOR KN, and A-TSC1 KO mice after Gao-binge alcohol. A-Raptor KO, A-mTOR KO, and A-TSC1 KO mice and their matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum triglyceride (TG; A) and cholesterol (Chol; B) levels were measured. Data are expressed as means ± SEM (A and B). n = 4 to 13 (A and B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Increased serum free fatty acid (FFA) and glycerol in A-Raptor KO mice after Gao-binge alcohol. A-Raptor KO and the matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum free FFA (A) and glycerol (B) levels were measured. Data are expressed as means ± SEM (A and B). n = 4 to 7 (A and B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Generation and validation of A-Atg5 KO mice. A: Total lysates from skeletal muscle, liver, epididymal white adipose tissue (eWAT), s.c. WAT (sWAT), and interscapular brown adipose tissue (iBAT) of A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Western blot analysis. B: The average body weight on day 1 (since alcohol feeding started) and day 10. C: The average food intake. Data are expressed as means ± SEM (B and C). n = 8 to 12 (B and C). ETOH, ethanol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
No significant changes of serum lipids between A-Atg5 KO mice and the matched WT mice after Gao-binge. A-Atg5 KO mice and matched Cre− (WT) mice were subjected to Gao-binge alcohol treatment. Serum free fatty acid (FFA; A), free glycerol (B), triglyceride (TG; C), cholesterol (Chol; D), and β-hydroxybutyrate (β-HB; E) levels were measured. Data are expressed as means ± SEM (A–E). n = 7 to 12 (A–E). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Gao-binge alcohol does not increase hepatic levels of receptor-interacting protein kinase 3 (RIP3) and mixed lineage kinase-like protein (MLKL) in A-Atg5 KO mice. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Total liver lysates were subjected to Western blot analysis. B: Densitometry of A. Data are expressed as means ± SEM (B). n = 3 to 4 (B). *P < 0.05 (one-way analysis of variance). ETOH, ethanol.
Gao-binge alcohol induces similar levels of hepatic cytochrome P450 2E1 (CYP2E1) and protein carbonylation in A-Atg5 KO and matched WT mice. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Total liver lysates were subjected to Western blot analysis. B: Densitometry of A. C: Representative Western blot analysis, detecting hepatic carbonylation levels. D: Densitometry of C. Data are expressed as means ± SEM (B and D). n = 3 to 4 (B and D). *P < 0.05 (one-way analysis of variance). ETOH, ethanol; NC, negative control.
There is no significant difference of hepatic neutrophil infiltration between A-Atg5 KO and matched WT mice after Gao-binge alcohol. A-Atg5 KO mice and matched Cre- (WT) mice were subjected to Gao-binge alcohol treatment. A: Representative images of liver Ly6B2 immunohistochemistry staining. Arrows denote neutrophils. B: Quantification of A. C and D: Livers (C) and epididymal white adipose tissues (D) were subjected to RNA extraction and real-time quantitative PCR. E: Serum lipocalin 2 was measured using the enzyme-linked immunosorbent assay kit. Data are expressed as means ± SEM (B–E). n = 4 to 6 (B and C); n = 3 (D); n = 8 to 13 (E). *P < 0.05 (one-way analysis of variance). Original magnification, ×20 (A). ETOH, ethanol.
There is no significant difference of lipolysis between WT and Atg5 KO adipocytes ex vivo. Epididymal white adipose tissue (eWAT; A) and s.c. WAT (sWAT; B) samples were collected from A-Atg5 KO mice and matched Cre- (WT) mice and subjected to ex vivo lipolysis assay. Pieces of adipose tissues were incubated with or without 10 μmol/L isoproterenol [ISO or non-treated (NT)]. The free glycerol released in the medium in 1 hour was normalized to tissue weight and compared with nonchallenged tissues. Data are expressed as means ± SEM (A and B). n = 9 (A and B). *P < 0.05 (one-way analysis of variance).