Abstract
Short-chain enoyl-CoA hydratase (ECHS1) is a mitochondrial beta-oxidation enzyme involved in the metabolism of acyl-CoA fatty acid esters, as well as in valine metabolism. ECHS1 deficiency has multiple manifestations, including Leigh syndrome early at birth or in childhood with poor prognosis, to cutis laxa, exercise-induced dystonia and congenital lactic acidosis.
Here we describe the case of a newborn with mutations in ECHS1 that caught our attention after the incidental finding of 3-hydroxy-butyryl\3-hydroxy-isobutyryl\malonylcarnitine (C4OH\C3DC) and tiglylcarnitine (C5:1) on blood spot in the newborn screening (NBS) program. Diagnosis was suspected based on the analysis of organic acids on dried urine spot. A moderate increase of 2-methyl-2,3-dihydroxybutyric acid, was detected, which is a known marker of this disease. Exome analysis showed c.404A>G (p.Asn135Ser) mutation in homozygosis in the ECHS1 gene. The child was therefore admitted to the hospital. Initial examination showed little response to auditory stimuli and mild hypertonia of the extremities. Clinical deterioration was evident at 4 months of age, including neurological and cardiac involvement, and the patient died at 5 months of age. This case illustrates how an incidental detection in the NBS Program can lead to the diagnosis ECHS1 deficiency. Although it is a severe disease, with no treatment available, early detection would allow adequate genetic counseling avoiding the odyssey that suffered most of these families.
Keywords: ECHS1 deficiency; Mutations in ECHS1; Newborn screening; 2-methyl-2,3-dihydroxybutyric acid; 3-hydroxy-butyrylcarnitine\3-hydoxy-isobutyrylcarnitine; Tiglylcarnitine
Abbreviations: C3DC, malonylcarnitine; C4OH, 3-hydroxy-butyrylcarnitine\3-hydoxy-isobutyrylcarnitine; C5:1, tiglylcarnitine; DBS, dried blood spot; DUS, dried urine spot; ECHS1, short-chain enoyl-CoA hydratase; GC, gas chromatography; HIBCH, 3-hydroxy-isobutyryl-CoA hydrolase; 3MGA, 3-methylglutaconic acid; MRI, magnetic resonance imaging; MS, mass spectrometry; NBS, Newborn Screening; PDH, pyruvate dehydrogenase; TMS, trimethylsilyl
1. Introduction
Short-chain enoyl-CoA hydratase (ECHS1; EC 4.2.1.17) is a mitochondrial beta-oxidation enzyme involved in the metabolism of acyl-CoA fatty acid esters [1]. In addition, it plays a role in the catabolism of isoleucine and valine, converting methacryloyl-CoA to 3-hydroxy-isobutyryl-CoA, and acryloyl-CoA to 3-hydroxypropionyl-CoA [2]. ECHS1 deficiency is an autosomal recessive disease caused by mutations in ECHS1. Although is not fully proven, high concentrations of methacryloyl-CoA and acryloyl-CoA are believed to impair the pyruvate dehydrogenase complex and the mitochondrial respiratory chain [3]. So far, ECHS1 deficiency has been reported in 40 individuals, within 31 families, from different ethnic backgrounds and geographical locations [4]. Biochemically, the disease presents with increased levels of 2-methyl-2,3-dihydroxybutyric acid in urine [2]. Moreover, some of the patients also present high excretion of 3-methylglutaconic acid [5,6], while others present with lactic acidosis [3,7,8]. Additionally, alterations of the acylcarnitine profile have also been reported [9].
ECHS1 deficiency presents with a heterogeneous clinical phenotype, ranging from fatal neonatal onset to adulthood forms [9]. The most common clinical manifestation is Leigh disease (MIM#256000). Other clinical manifestations, such as cardiomyopathy [8,10], cutis laxa [11], but also exercise-induced dystonia at older ages (8 to 17 years of age) [12,13] have also been described for this disease.
Brain MRI findings includes T2 hyperintense signals in the basal ganglia, as well as cerebral atrophy and agenesis of the corpus callosum [3].
Most of the so-far reported variants are missense, but several loss-of-function pathogenic variants have also been described [4]. The aim of this work is to present the case of a newborn with mutations in ECHS1 detected incidentally through the Newborn Screening (NBS) Program of Catalonia.
2. Case report
The child was the fourth son of consanguineous Maghreb parents, who had no family antecedents except for two spontaneous abortions in the first trimester of pregnancy. At birth, the newborn presented dyspeptic stools, with suspicion of allergy to cow milk protein, and continued with exclusive breast feeding. The APGAR score was 8/10/10, and the birth weight was 4080 g. (88 percentile). An increase of 3-hydroxybutyryl\3-hydroxyisobutyryl\malonylcarnitine (C4OH\C3DC) and of tiglylcarnitne (C5:1) on dried blood spots (DBS) in the neonatal screening test was detected (Table 1).
Table 1.
Acylcarnitines on dried blood spots at 3 and 20 days of life.
| 3 days of life |
20 days of life |
|||
|---|---|---|---|---|
| Acylcarnitines (μmol/L) | Results | Cut-off values (p99.5) | Results | Cut-off values (p99.5) |
| C5:1 | 0.04 | <0.02 | 0.02 | <0.02 |
| C4OH\C3DC | 0.73 | <0.5 | 0.12 | <0.29 |
| C2 | 6.5 | 8–60 | 3.31 | 2.9–39.8 |
C2, acetylcarnitine; C4OH\C3DC, 3-hydroxy-isobutyrylcarnitine/3-hydroxy-butyrylcarnitine/malonyl-carnitine; C5:1, tiglylcarnitine.
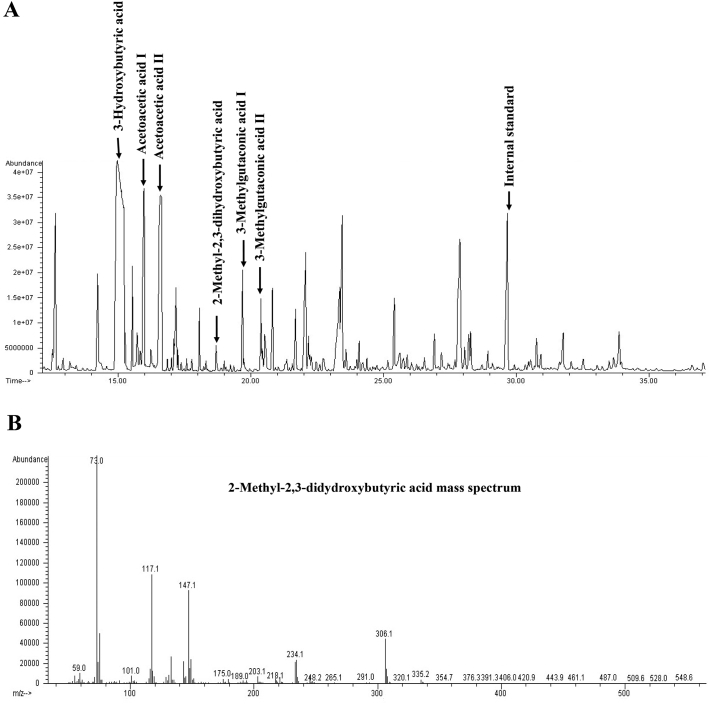
Given these slight elevations, a second DBS and a dried urine spot (DUS) were requested. Results in the second DBS, at 20 days of life, showed normal levels of acylcarnitines (Table 1), but the profile of organic acids on DUS showed a slight increase of 2-methyl-2,3-dihydroxybutyric acid, suggestive of ECHS1 deficiency or 3-hydoxy-isobutyryl-CoA hydrolase (HIBCH) deficiency. Therefore, the newborn was admitted to the hospital for further testing. At admission, physical examination showed a lessened response to auditory stimuli and mild hypertonia of the extremities. Analysis of urine organic acids confirmed the increase of 2-methyl-2,3-dihydroxybutyric acid (13 mmol/mol creatinine, CV <2) as well as the high levels of 3-methylglutaconic acid (3MGA, 82 mmol/mol creatinine, CV <20) (Fig. 1), persistently increased.
Fig. 1.
GC–MS of organic acids in urine of ECHS1 patient at 4 months of life.
A) Total ion chromatogram. B) Mass spectra of 2-methyl-2,3-dihydroxybutyric acid.
Pondoestatural and psychomotor development during the first months of life were normal. At the age of 4 months, the child was admitted to the emergency unit due to loss of consciousness, food refusal, and bloody stools. He presented poor general condition (with Glasgow Coma Score of 9), skin pallor, and tachypnea. Routine biochemical tests disclosed metabolic acidosis and a slight lactic acidosis (2.8 mmol/L; Control value: <2.1).
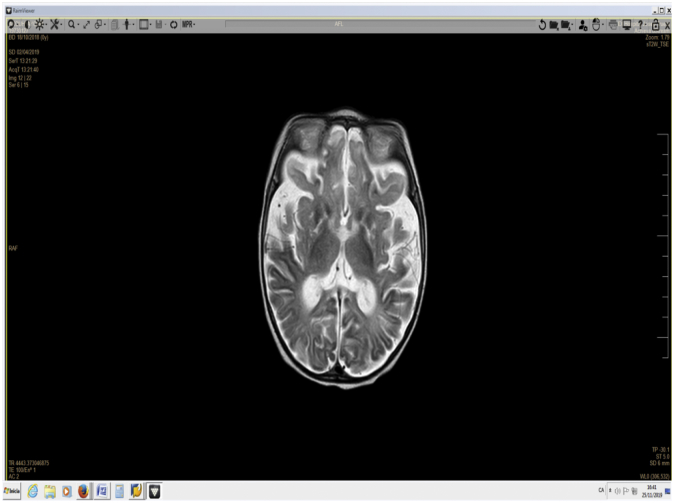
He required orotracheal intubation and mechanical ventilation. Respiratory infection secondary to metapneumovirus was diagnosed. After 48 h, he developed respiratory distress secondary to pneumonia due to bacterial infection; 8 days after admission, acute myocarditis with moderate-severe left heart dysfunction was detected. Magnetic resonance imaging (MRI) (Fig. 2) showed cerebral and cerebellar atrophy, signs of necrosis and atrophy in caudate, pallidum and putamen nucleus. T2 hyperintensity in cerebellar hemispheres with subcortical and inferomedial predominance, were detected. Moreover, a marked alteration of the white matter and of cerebellar hemispheres were evident, suggesting Leigh syndrome. During the following days, the child was irritable, lacked visual tracking, and had hyperreflexia, axial hypotonia, and hypertonia, predominantly in the lower extremities. He was extubated 17 days after admission, but he presented a new respiratory infection with progressive worsening, and died at 5 months of age.
Fig. 2.
MRI image at 4 months of age showing cerebral and cerebellar atrophy.
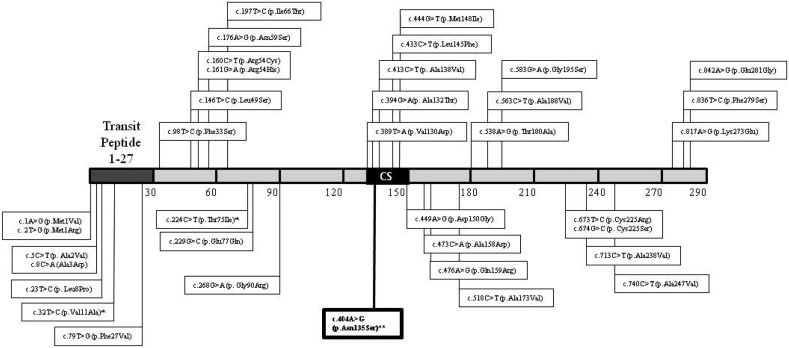
Exome sequencing showed a change in homozygosity in exon 3 of the ECHS1 gene (c.404A>G), which was confirmed by Sanger sequencing. The parents' segregation was also confirmed. This change resulted in the substitution of asparagine for serine at position 135 of the protein (p.Asn135Ser), which was not reported in Human Gene Mutation Database (www.hgmd.cf.ac.uk) [14]. An updated figure (Fig. 3) with the location of the previously reported ECHS1 missense mutations has been added [14].
Fig. 3.
Reported ECHS1 missense mutations. Protein scheme showing the amino acid substitutions associated to ECHS1 deficiency based on the variants annotated in Human Gene Mutation Database [14]. Only missense variants are indicated. Splice site variants, frameshift and nonsense mutations are not represented. CS, enoyl-Coa hydratase conserved site.⁎ Annotated in HGMD as a disease-associated polymorphism.⁎⁎ Variant reported in the present manuscript.
Predictors of pathogenicity (www.varsome.com) [15], based on the guidelines of the American College of Medical Genetics (ACMG), and additional criteria, particularly PM3 and PP4 [16] classified the variant, c.404A > G, as likely pathogenic. In addition, another database, ClinVar (www.ncbi.nlm.nih.gov/clinvar/) [17], also classified it as likely pathogenic.
3. Material and methods
3.1. Acylcarnitine analysis on DBS
Acylcarnitines were analyzed by tandem mass spectrometry (MS/MS) (Xevo TQD, Waters) using NeoBase™ Non-derivatized MSMS kit (Perkin Elmer), as described by the manufacturer.
3.2. Organic acids analysis on DUS
Organic acids on urine or DUS were analyzed as their trimethylsilyl derivatives (TMS) by gas chromatography–mass spectrometry (GS-MS) (Agilent 7890A (GC)/ 5975C MS). The extraction procedure was performed according to Tanaka et al. (1980) [18] with some modifications. Briefly, 4 mL of saturated NaCl was added to DUS, and the mixture was shaken for 30 min. After the addition of 7 μL internal standard (undecanodioic acid) and 275 μL of 4 N HCl, urine was extracted three times with 2 mL ethyl acetate, and the organic phases were mixed and evaporated under nitrogen gas at room temperature. Derivatization with 90 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) at 60 °C for 30 min was performed, and 1 μL was injected into the GC/MS.
3.3. Molecular study
The genetic study was performed using the Nextera® DNA exome kit (Illumina)
that analyzes mutations in the exons and flanking intronic regions of the genome. The bioinformatic analysis was carried out using a self-designed pipeline and the variants were annotated and interpreted using the VariantStudio software version 3.0 (Illumina). The identified variants were prioritized using different databases: 1000 genomes, Exome Aggregation Consortium (ExAC), ClinVar, and Human Gene Mutation Database (HGMD) [14,17,19,20] and predictive models of pathogenicity: Varsome, Sorting Intolerant from Tolerant (SIFT) and Polymorphism Phenotyping v2 (Polyphen-2) [15,21,22]. The variant identified by exome sequencing (c.404A>G) was confirmed by Sanger sequencing. Parents segregation was also confirmed by Sanger sequencing.
3.4. Discussion
Here, we report a newborn with mutations in ECHS1 detected for the first time through the NBS Program. The MS transitions that caught our attention corresponded to C5:1 and C4OH\C3DC, which were included in our panel of acylcarnitines for the detection of other diseases. Currently, the method we use is a non-derivatized assay that is not able to distinguish between isomers. Given this limitation, when these acylcarnitines are elevated, we adopt the strategy of performing organic acid analysis on DUS to establish the differential diagnosis among the corresponding diseases. The increase of 2-methyl-2,3-dihydroxybutyric acid observed in the child's urine (Fig. 1) suggested a possible defect of either a ECHS1 or HIBCH deficiency. The acylcarnitine profile normalized on the second blood spot, but repeated analyses of organic acids in urine showed a persistent elevation of 2-methyl-2,3-dihydroxybutyric acid and of 3MGA. Molecular studies established the diagnosis of ECHS1 deficiency, and although the missense mutation found in this neonate had not been formally proven to be disease-causing, the pathogenic in silico prediction, as well as the clinical phenotype and the organic acid profile, supported this diagnosis. Interestingly, this variant is located at protein position 135, close to several other susbtitutions affecting an enoyl-CoA hydratase conserved site (Fig. 3).
Secondary deficiencies of pyruvate dehydrogenase (PDH) and mitochondrial respiratory chain activities have been reported in this deficiency [5,10,[23], [24], [25]]. We could not measure these activities in our patient, as no tissues were available, but the slight increase of plasma lactate in addition to the elevated 3MGA in urine (Fig. 1) is in agreement with a generalized mitochondrial dysfunction and supports the observations made by others [3,5,6,9,26] suggesting that ECHS1 deficiency is a novel disease to be included in the differential diagnosis of 3MGA aciduria [5].
As observed for our patient, the age of clinical onset is early at birth, followed by a fatal prognosis in most of the described cases [2,3,5,7,10,24,26,27], and although it has been reported that low valine diet and administration of antioxidants were of benefit in some patients [13,28], these therapies were not considered in our patient, because by the time we reached the diagnosis the clinical deterioration was very severe.
It is worth considering that HIBCH deficiency shares the clinical phenotype and some of the metabolic features with ECHS1 deficiency. HIBCH deficiency is also characterized by Leigh syndrome and is likewise involved in valine pathway metabolism. HIBCH catalyzes the conversion of 3-hydroxy-isobutyryl-CoA to 3-hydroxy-isobutyrate. Consequently, a rise of plasma 3-hydroxy-isobutyryl-carnitine is detected in this disease, while the levels in ECHS1 deficiency are normal [[29], [30], [31]], making this metabolite appropriate for the differential diagnosis between both diseases. Our case was complicated by the fact that 3-hydroxy-isobutyryl-carnitine cannot be formally distinguished from its isomer (3-hydroxy-butyryl-carnitine) or from the isobaric compound (malonyl-carnitine). However, exome sequencing followed by filtering of the genes of interest easily solved the diagnosis. Consequently, after the diagnosis of ECHS1 was established, the only possibility for the elevation of C4OH was 3-hydroxy-butyryl-carnitine, which is not a rare finding since a hug peak of 3-hydroxybutyric acid was detected in urine (Fig. 1). Interestingly, C4, one of the acylcarnitines most commonly elevated in this disease [3,4,7,27] was normal in our patient, while C5:1 (tiglylglycine), one of the metabolites that caught our attention in the initial sample, might be elevated as a consequence of a block in the catabolism of tiglyl-CoA in the Isoleucine pathway.
4. Conclusions
In sum, an infant child with mutations in ECHS1 was detected for the first time through the NBS program. This is a severe disease with no available treatment. The incidental elevation of C5:1 and C4OH\C3DC and the strategy of analyzing organic acids on DUS led us to the suspicion of ECHS1 or HIBCH deficiencies, and exome sequencing established the differential diagnosis of a disease that otherwise could have been missed. Based on an accurate diagnosis, the child's family has now access to genetic counseling.
Funding
This research was supported by the Instituto de Salud Carlos III (PI16/01048) and the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), an initiative of the Instituto de Salud Carlos III (Ministerio de Ciencia e Innovación, Spain). This study was also supported by the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) and the CERCA Programme/Generalitat de Catalunya.
Financial disclosure
The authors have no financial relationships relevant to this article to disclose.
Declaration of Competing Interest
The authors have no conflicts of interest to disclose.
Acknowledgments
We thank to C. Martínez, E. Ramón, M. Fernández, C. Zaragoza and C. Cutillas their excellent technical assistance in the acylcarnitine and organic acid analysis.
References
- 1.Stern J.R., del Campillo A. Enzymatic reaction of crotonyl coenzyme A. J. Am. Chem. Soc. 1953;75:2277–2278. [Google Scholar]
- 2.Peters H., Buck N., Wanders R., Ruiter J., Waterham H.R., Koster J., Pitt J.J. ECHS1 mutations in Leigh disease: a new inborn error of metabolism affecting valine metabolism. Brain. 2014;137:2903–2908. doi: 10.1093/brain/awu216. [DOI] [PubMed] [Google Scholar]
- 3.Ferdinandusse S., Friederich M.W., Burlina A., Ruiter J.P., Coughlin C.R., Dishop M.K., Gallagher R.C., Bedoyan J.K., Vaz F.M., Waterham H.R. Clinical and biochemical characterization of four patients with mutations in ECHS1. Orphanet J.Rare Dis. 2015;18(10):79. doi: 10.1186/s13023-015-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Ganetzky, C. Stojinski, Mitochondrial short-chain enoyl-CoA hydratase 1 deficiency, in: Adam MP, Ardinger HH, Pagon RA, et al., (Eds.), GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2019, Available from: https://www.ncbi.nlm.nih.gov/books/NBK542806/.2019 Jun 20. [PubMed]
- 5.Fitzsimons P.E., Alston C.L., Bonnen P.E., Hughes J., Crushell E., Geraghty M.T., Tetreault M., O'Reilly P., Twomey E., Sheikh Y. Clinical, biochemical, and genetic features of four patients with short-chain enoyl-CoA hydratase(ECHS1) deficiency. Am. J. Med. Genet. A. 2018;176:1115–1127. doi: 10.1002/ajmg.a.38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffnagel I.C., Redeker E.J.W., Reneman L., Vaz F.M., Ferdinandusse S., Poll-The B.T. Mitochondrial encephalopathy and transient 3-methylglutaconic aciduria in ECHS1 deficiency: long-term follow-up. JIMD Rep. 2018;39:83–87. doi: 10.1007/8904_2017_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Mutairi F., Shamseldin H.E., Alfadhel M., Rodenburg R.J., Alkuraya F.S. A lethal neonatal phenotype of mitochondrial short-chain enoyl-CoA hydratase-1 deficiency. Clin. Genet. 2017;91:629–633. doi: 10.1111/cge.12891. [DOI] [PubMed] [Google Scholar]
- 8.Ganetzky R.D., Bloom K., Ahrens-Nicklas R., Edmondson A., Deardorff M.A., Bennett M.J., Ficicioglu C. ECHS1 deficiency as a cause of severe neonatal lactic acidosis. JIMD Rep. 2016;30:33–37. doi: 10.1007/8904_2016_538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharpe A.J., McKenzie M. Mitochondrial fatty acid oxidation disorders associated with short-chain enoyl-CoA hydratase (ECHS1) deficiency. Cells. 2018;7 doi: 10.3390/cells7060046. (pii: E46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haack T.B., Jackson C.B., Murayama K., Kremer L.S., Schaller A., Kotzaeridou U., de Vries M.C., Schottmann G., Santra S., Büchner B. Deficiency of ECHS1 causes mitochondrial encephalopathy with cardiac involvement. Ann. Clin. Transl. Neurol. 2015;2:492–509. doi: 10.1002/acn3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramaniam S., Riley L.G., Bratkovic D., Ketteridge D., Manton N., Cowley M.J., Gayevskiy V., Roscioli T., Mohamed M., Gardeitchik T., Morava E., Christodoulou J. Unique presentation of cutis laxa with Leigh-like syndrome due to ECHS1 deficiency. J. Inherit. Metab. Dis. 2017;40:745–747. doi: 10.1007/s10545-017-0036-4. [DOI] [PubMed] [Google Scholar]
- 12.Olgiati S., Skorvanek M., Quadri M., Minneboo M., Graafland J., Breedveld G.J., Bonte R., Ozgur Z., van den Hout M.C., Schoonderwoerd K. Mov. Disord. 2016;31:1041–1048. doi: 10.1002/mds.26610. [DOI] [PubMed] [Google Scholar]
- 13.Mahajan A., Constantinou J., Sidiropoulos C. ECHS1 deficiency-associated paroxysmal exercise-induced dyskinesias: case presentation and initial benefit of intervention. J. Neurol. 2017;264:185–187. doi: 10.1007/s00415-016-8381-z. [DOI] [PubMed] [Google Scholar]
- 14.Human Gene Mutation Database, www.hgmd.cf.ac.uk, Accessed November 2019.
- 15.Varsome, www.varsome.com, Accessed July 2019.
- 16.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ClinVar, www.ncbi.nlm.nih.gov/clinvar/, Accessed July 2019.
- 18.Tanaka K., West-Dull A., Hine D.G., Lynn T.B., Lowe T. Gas-chromatographic method of analysis for urinary organic acids. II. description of the procedure, and its application to diagnosis of patients with organic acidurias. Clin. Chem. 1980;26:1847–1853. [PubMed] [Google Scholar]
- 19.1000 genomes, https://www.internationalgenome.org/about/, Accessed July 2019.
- 20.ExAC (Exome Aggregation Consortium), http://exac.broadinstitute.org/, Accessed July 2019.
- 21.SIFT Sorting Intolerant from Tolerant, https://sift.bii.a-star.edu.sg/, Accessed July 2019.
- 22.Polyphen-2 (Polymorphism Phenotyping v2), http://genetics.bwh.harvard.edu/pph2/, Accessed July 2019.
- 23.Tetreault M., Fahiminiya S., Antonicka H., Mitchell G.A., Geraghty M.T., Lines M., Boycott K.M., Shoubridge E.A., Mitchell J.J., Michaud J.L. Whole-exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum. Genet. 2015;134:981–991. doi: 10.1007/s00439-015-1577-y. [DOI] [PubMed] [Google Scholar]
- 24.Sakai C., Yamaguchi S., Sasaki M., Miyamoto Y., Matsushima Y., Goto Y. ECHS1 mutations cause combined respiratory chain deficiency resulting in Leigh syndrome. Hum. Mutat. 2015;36:232–239. doi: 10.1002/humu.22730. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa E., Shimura M., Fushimi T., Tajika M., Ichimoto K., Matsunaga A., Tsuruoka T., Ishige M., Fuchigami T., Yamazaki T. Clinical validity of biochemical and molecular analysis in diagnosing leigh syndrome: a study of 106 japanese patients. J. Inherit. Metab. Dis. 2017;40:685–693. doi: 10.1007/s10545-017-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedoyan J.K., Yang S.P., Ferdinandusse S., Jack R.M., Miron A., Grahame G., DeBrosse S.D., Hoppel C.L., Kerr D.S., Wanders R.J.A. Lethal neonatal case and review of primary short-chain enoyl-CoA hydratase (SCEH) deficiency associated with secondary lymphocyte pyruvate dehydrogenase complex (PDC) deficiency. Mol. Genet. Metab. 2017;120:342–349. doi: 10.1016/j.ymgme.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair P., Hamzeh A.R., Mohamed M., Malik E.M., Al-Ali M.T., Bastaki F. Novel ECHS1 mutation in an Emirati neonate with severe metabolic acidosis. Metab. Brain Dis. 2016;31:1189–1192. doi: 10.1007/s11011-016-9842-x. [DOI] [PubMed] [Google Scholar]
- 28.Shayota B.J., Soler-Alfonso C., Bekheirnia M.R., Mizerik E., Boyer S.W., Xiao R., Yang Y., Elsea S.H., Scaglia F. Case report and novel treatment of an autosomal recessive leigh syndrome caused by short-chain enoyl-CoA hydratase deficiency. Am. J. Hum. Genet. 2019;179:803–807. doi: 10.1002/ajmg.a.61074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferdinandusse S., Waterham H.R., Heales S.J., Brown G.K., Hargreaves I.P., Taanman J.W., Gunny R., Abulhoul L., Wanders R.J., Clayton P.T. HIBCH mutations can cause leigh-like disease with combined deficiency of multiple mitochondrial respiratory chain enzymes and pyruvate dehydrogenase. Orphanet J. Rare Dis. 2013;4:188. doi: 10.1186/1750-1172-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuter M.S., Sass J.O., Leis T., Köhler J., Mayr J.A., Feichtinger R.G., Rauh M., Schanze I., Bähr L., Trollmann R. HIBCH deficiency in a patient with phenotypic characteristics of mitochondrial disorders. Am. J. Med. Genet. A. 2014;164:3162–3169. doi: 10.1002/ajmg.a.36766. [DOI] [PubMed] [Google Scholar]
- 31.Soler-Alfonso C., Enns G.M., Koenig M.K., Saavedra H., Bonfante-Mejia E., Northrup H. Identification of HIBCH gene mutations causing autosomal recessive leigh syndrome: a gene involved in valine metabolism. Pediatr. Neurol. 2015;52:361–365. doi: 10.1016/j.pediatrneurol.2014.10.023. [DOI] [PubMed] [Google Scholar]