Abstract
Non-alcoholic fatty liver disease (NAFLD) is an emerging global disease with increasing prevalence. However, the mechanism of NAFLD development is not fully understood. To elucidate the cell-specific role of nuclear factor erythroid-derived 2-like 2 (NRF2) in the pathogenesis of NAFLD, we utilized hepatocyte- and macrophage-specific Nrf2-knockout [Nrf2(L)-KO and Nrf2(Mϕ)-KO] mice to examine the progress of NAFLD induced by high-fat diet (HFD). Compared to Nrf2-LoxP littermates, Nrf2(L)-KO mice showed less liver enlargement, milder inflammation and less hepatic steatosis after HFD feeding. In contrast, Nrf2(Mϕ)-KO mice displayed no significant difference in HFD-induced hepatic steatosis from Nrf2-LoxP control mice. Mechanistic investigations revealed that Nrf2 deficiency in hepatocytes dampens the expression of peroxisome proliferator-activated receptor γ (PPARγ) and its downstream lipogenic genes in the liver and/or primary hepatocytes induced by HFD and palmitate exposure, respectively. While PPARγ agonists augmented PPARγ expression and its transcriptional activity in primary hepatocytes in a NRF2-dependent manner, forced overexpression of PPARγ1 or γ2 distinctively reversed the decreased expression of their downstream genes fatty acid binding protein 4, lipoprotein lipase and/or fatty acid synthase caused by Nrf2 deficiency. We conclude that NRF2-dependent expression of PPARγ in hepatocytes is a critical initiating process in the development of NAFLD, suggesting that inhibition of NRF2 specifically in hepatocytes may be a valuable approach to prevent the disease.
Keywords: Nrf2, NALFD, High-fat diet, Hepatocyte, PPARγ
Graphical abstract
Highlights
-
•
Nrf2(L)-KO, but not Nrf2(Mϕ)-KO, mice showed lessened hepatic steatosis induced by HFD feeding.
-
•
Nrf2 deficiency dampened the expression and transcriptional activity of PPARγ in hepatocytes.
-
•
PPARγ agonists increased PPARγ expression/activity in hepatocytes in a NRF2-dependent manner.
-
•
OE of PPARγ partially reversed the reduction of lipogenic genes in Nrf2(L)-KO hepatocytes.
List of abbreviations
- BODIPY
boron dipyrromethene
- CD
chow diet
- CD36
cluster of differentiation 36
- ChIP
chromatin immunoprecipitation
- CST
cell signaling technology
- FABP4
fatty acid binding protein 4
- FASN
fatty acid synthase
- FFA
free fatty acid
- GCLC
glutamate cysteine ligase catalytic subunit
- HFD
high-fat diet
- H&E
hematoxylin and eosin
- Hrs
hours
- KO
knock out
- min
minutes
- Mlxipl
MLX interacting protein like
- NAFLD
non-alcoholic fatty liver disease
- NAS
NAFLD activity score
- NASH
non-alcoholic steatohepatitis
- NF-κB
nuclear factor kappa-B
- NQO1
NAD(P)H:quinone oxidoreductase 1
- NRF2
nuclear factor erythroid-derived 2-like 2
- Nrf2(L)-KO
hepatocyte-specific Nrf2 knockout
- Nrf2(Mϕ)-KO
macrophage-specific Nrf2 knockout
- OE
overexpression
- PIOG
pioglitazone
- PPAR
peroxisome proliferator-activated receptor
- ROS
reactive oxygen species
- ROSI
rosiglitazone
- SCD1
stearoyl-CoA desaturase-1
- Srebf1
sterol regulatory element-binding transcription factor 1
- TG
triglyceride
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is defined as a metabolic disease related to obesity and type 2 diabetes mellitus, with a diagnostic hallmark of hepatic triglyceride (TG) accumulation above 5% without alcohol consumption [1]. NAFLD is currently a common liver disease with a growing prevalence worldwide. A recent meta-analysis reported that the global prevalence of NAFLD was up to 25% in 2016 [2]. NAFLD can be divided into simple steatosis and steatohepatitis (NASH) [3]. When wide-range inflammation and fibrosis occur, about 10–30% of the cases of simple steatosis develop into NASH [4]. Without effective intervention, 10–29% NASH might progress into cirrhosis in around 10 years [5]. The progression of cirrhosis is dramatic, and 27% of cases, can lead to hepatocellular carcinoma or death without liver transplantation [6]. Therefore, medical intervention of NAFLD should begin as early as possible after diagnosis.
NAFLD is mainly caused by disruption of the homeostasis of lipid metabolism, oxidative stress and subsequent inflammation. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, also well known as NRF2) is a master transcription factor in the regulation of anti-oxidative and anti-inflammatory genes [7]. NRF2 is a member of CNC-bZIP family and binds to the antioxidant response element with small Maf proteins to trigger the transcription of downstream genes [8]. In addition, evidence from a lipopolysaccharide-stimulated Nrf2-knockout (Nrf2-KO) mouse model [9] supports a role of NRF2 as an essential regulator of anti-inflammatory genes, since NRF2-dependent transcription inhibits the activation of nuclear factor kappa-B (NF-κB) and expression of its downstream proinflammatory cytokines, such as inducible nitric oxide synthases, tumor necrosis factor α (TNFα), interleukin 1 beta (IL1β) and interleukin 6 (IL6) [9]. The molecular mechanism by which NRF2 suppresses inflammatory response is demonstrated to be disruption of transcription of proinflammatory cytokines using chromatin immunoprecipitation (ChIP)-seq and ChIP-qPCR analyses in macrophages [10]. In a high-fat diet (HFD)-induced NAFLD model, levels of oxidative stress and inflammation were both increased in the liver of Nrf2-KO mice that were susceptible to NAFLD [11,12]. Moreover, oxidative stress and inflammation appear to be in a vicious cycle, aggravating each other to induce disease progression [13,14].
Aspects of hepatic lipid metabolism associated with NAFLD development includes free fatty acid (FFA) uptake and transport, de novo synthesis, β-oxidation and exportation of TG out of hepatocytes. NRF2 is closely related to lipid homeostasis [7]. Utilizing a transgenic mouse model, many enzymes in these processes were identified and shown to be regulated by NRF2, such as cluster of differentiation 36 (CD36) [15]. Peroxisome proliferator-activated receptor γ (PPARγ) is a classic lipid metabolism regulator in adipocytes and hepatocytes, which is found as an initial factor in the development of NAFLD [16]. Our previous studies indicate that PPARγ is a direct downstream transcriptional target of NRF2 in adipocytes [17]. Hepatic steatosis is mainly attributed to hepatocytes [18], while Kuppfer cells and stellate cells also contribute to the development of NAFLD [19,20]. Kuppfer cells, specialized macrophages located in the liver, might be activated by abnormal lipid accumulation and injury in hepatocytes leading to the development of NASH [21]. Increasing transcription of pro-inflammatory cytokines, such as TNFα, in Kuppfer cells, might activate the stellate cells, leading to collagen synthesis and fibrosis and ultimately cirrhosis. While different types of cells in the liver play distinct roles in the pathogenesis of NAFLD, the initiator of this pathological process is abnormal lipid metabolism within hepatocytes.
In this study, we utilized hepatocyte-specific Nrf2-knockout [Nrf2(L)-KO] and macrophage-specific Nrf2-knockout [Nrf2(Mϕ)-KO] mouse models established by crossing Nrf2-LoxP mice [22] with transgenic mice expressing Cre recombinase under the control of Albumin (Alb-Cre) and Lysozyme 2 (Lyz-Cre), respectively. The mice were then fed with HFD to examine the effect of Nrf2 deficiency in hepatocytes or macrophages on the development of NAFLD. Compared with Nrf2-LoxP littermate controls, Nrf2(L)-KO mice had milder NAFLD, with lower liver weight and hepatic TG levels. However, this effect was not seen in Nrf2(Mϕ)-KO mice. This mitigation of hepatic steatosis was accompanied by decreased PPARγ activity in Nrf2(L)-KO mice. Additional mechanistic investigations in primary hepatocytes indicated that NRF2-dependent expression of PPARγ plays a critical role in the initiation of NAFLD.
2. Materials and methods
2.1. Animals
Nrf2(L)-KO mice and Nrf2(Mϕ)-KO mice on a C57BL/6 background were generated by breeding Nrf2Loxp/LoxP mice [22] with Albumin-Cre mice (Nanjing Biomedical Research Institute of Nanjing University, J003574) and Lysozyme2-Cre mice (Nanjing Biomedical Research Institute of Nanjing University, J004781), respectively. All breeders and littermates were raised at the animal facility of China Medical University. All animal experiments performed were approved by the Institutional Animal Care and Use Committee of China Medical University (Shenyang, China). Mice were housed at 23 ± 1 °C and kept on 12-h light/dark cycles (lights on: 06:00–18:00). Distilled water and standard mouse chow diet (CD, Shukebeita Specific Pathogen Free Mouse Maintenance Diet, Xietong Organism, Jiangsu, China) were provided ad libitum. Tail snips were collected to prepare genomic DNA for genotyping, as described previously [22].
Both female and male 12–16 weeks old Nrf2(L)-KO mice and littermate control Nrf2-LoxP mice were randomly divided into 2 groups, respectively. The mice in Nrf2-LoxP, CD and Nrf2(L)-KO, CD groups were provided with standard CD. In contrast, the mice in Nrf2-LoxP, HFD and Nrf2(L)-KO, HFD groups were fed with a HFD (60% kcal fat; Research diets #D12492) for 12 weeks. Distilled water was provided ad libitum to all mice. Food and water consumption, body weight and blood glucose were monitored regularly. At necropsy, the liver was weighed and a portion was excised and fixed in 4% paraformaldehyde buffer for histopathology. Blood and various tissue samples were collected and frozen at −80 °C.
2.2. Intraperitoneal glucose tolerance test (IPGTT)
The mice fed with HFD were given an intraperitoneal injection of 1.0 g/kg BW of D-(+)-glucose (G8769; Sigma, St. Louis, MO) following overnight fasting, and blood glucose levels were measured at 0, 15, 30, 60 and 120 min after the glucose injection using the FreeStyle Blood Glucose Monitoring System (TheraSense, Alameda, CA). Blood samples were collected from the tail bleeds and analyzed as described previously [22].
2.3. Measurement of body composition
The fat mass of mice was measured by a Minispec LF-50 NMR body composition analyzer with the Minispec NF software (Bruker BioSpin GmbH, Rheinstetten, Germany) according to the manufacture's protocol [23].
2.4. Analysis of TG, glycerol and free fatty acids (FFA)
Heparin-treated fresh blood samples were centrifuged at 5,000 g for 5 min at 4 °C. The liver samples or the primary hepatocytes were homogenized in cold PBS (or chloroform:methanol = 2:1) and then centrifuged at 12,000 g for 5 min at 4 °C. The resultant supernatants were used for measurements. The levels of glycerol and total TG in the plasma samples and liver supernatants were assessed using specific kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China) adhering by the manufacturer's instructions. FFA levels were measured using a standard protocol of a mouse enzyme linked immunosorbent assay (ELISA) kit (Jiangsu Meimian Industrial Co., Ltd, Jiangsu, China).
2.5. Measurement of malondialdehyde (MDA) levels
The liver tissues were homogenized in the lysis buffer (#P0013, Beyotime Institute of Biotechnology, Shanghai, China). Total protein content was determined by using the Enhanced BCA Protein Assay Kit (#P0009, Beyotime). MDA levels were measured with Lipid Peroxidation MDA assay kit according to the instructions (#S0131, Beyotime).
2.6. Liver histological and immunohistochemical analysis
Liver samples were fixed in 4% paraformaldehyde buffer overnight after harvesting and then transferred to 70% ethanol. After dehydrating with graded ethanol solutions, tissues were then embedded in paraffin, sectioned at 5 μm thickness and stained with hematoxylin-eosin stain (H&E). For Oil Red O staining, 20 μm thickness frozen section was used. The stained sections were assessed visually by light microscopy.
For immunohistochemistry, the macrophage marker F4/80 (sc-377009, 1:1000, Santa Cruz Biotechnology, Santa Cruz, CA) were assessed following IHC-DAB kit instructions (Beijing Zhongshan Jinqiao Biological Technology Co., Beijing, China) as described previously [24].
2.7. Primary hepatocytes isolation and treatments
Primary hepatocytes were isolated from Nrf2-LoxP and Nrf2(L)-KO mice fed with CD. A modified collagenase method for hepatocyte isolation described previously [25] was utilized. In brief, mice were first anaesthetized with pentobarbital sodium (50 mg/kg BW). Then the liver was perfused with Hank's balanced salt solution buffer (37 °C) via the vena cava, followed by a collagenase buffer (1 mg/ml, C-5138, Sigma) for cell dissociation. Digested livers were centrifuged at 30 g for 3 min to concentrate pure hepatocytes at the bottom of tubes. Isolated primary hepatocytes were cultured in M199 medium with 100 units/mL penicillin, 100 μg/mL streptomycin and 2 mM l-glutamine. After culture for 6 h, primary hepatocytes were treated with rosiglitazone (ROSI, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China; 0, 1, 10 μM) and pioglitazone (PIOG, Beijing Solarbio; 0, 0.25, 0.5 μM) for 24 h, respectively.
Sodium palmitate (P9767, Sigma) was dissolved to 0.15 M in 0.1 mM NaOH solution via heating at 90 °C and oscillating for 5 min. 10% BSA (abs49001012, Absin, Shanghai, China) solution was immediately added to make 15 mM and filtered with 0.22 μm filter as stock (pH ≈ 7.65). Before application to isolated primary hepatocytes, stock solution was incubated in a water bath at 37 °C for conjugation, and then diluted to 0.5 mM into M199 medium.
2.8. Measurement of fatty acid uptake
Boron dipyrromethene (BODIPY, Sigma, #790389) was used to measure fatty acid uptake as described previously [26]. Isolated primary hepatocytes were seeded in 96-well plates at 25,000 cells per well. After 6 h culture (without FBS), the medium was replaced with 10 μg/L BODIPY in 200 μl PBS. The fluorescence values were measured immediately utilizing fluorescence microplate reader (Molecular Devices FlexStation 3) with kinetic mode for 45 min (once every 3 min, totally 16 reads). The excitation and emission wavelengths of BODIPY were set to 493 nm and 520 nm, respectively.
2.9. Measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in primary hepatocytes
The Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience, Agilent, Billerica, MA, USA) was used to measure OCR and ECAR as previously described [27]. Isolated primary hepatocytes from Nrf2(L)-KO and Nrf2-LoxP mice were seeded in XF24-well microplates at 5.0 × 103 cells per well in 200 μl medium for 2 h. Then the medium was replaced with regular culture medium without FBS overnight. Before the measurement, the primary hepatocytes was balanced in XF base medium with 2.5 mM glucose, 2 mM l-glutamine and 1 mM sodium pyruvate for 1 h in no-CO2 incubator. ECAR and OCR were calculated under basal condition and after the additions of mitochondrial inhibitors, oligomycin (5 μM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 2 μM), and a mixture of rotenone (1 μM) and antimycin-A (1 μM), in succession. Three independent experiments were performed, and representative results were showed in figures.
2.10. Overexpression of PPARγ in hepatocytes
Transient transfection was performed in primary hepatocytes isolated from Nrf2(L)-KO and Nrf2-LoxP mice fed with CD with Lipofectamine 3000 (Life Technologies, Grand Island, NY) following standard procedures [25]. Briefly, 2.5 μg plasmids in 7.5 μl Lipofectamine 3000 were administrated to primary hepatocytes of both genotypes for 48 h. The PPARγ1 plasmid was purchased from Addgene (plasmid #8886; Cambridge, MA) [28] and the PPARγ2 with HA tag was gifted from Dr. Wen Xie (University of Pittsburgh) [29].
2.11. Western blotting
Liver tissues and harvested cells were prepared and lysed according to standard protocols as previously described [22,30]. Antibodies for PPARγ (#2435; 1:1000; Cell Signaling Technology (CST), Danvers, MA), pNF-κB (#3033; 1:1000; CST), NF-κB (#8242; 1:1000; CST), PPARα (ab8934; 1:1000; Abcam, Cambridge, United Kingdom), NRF2 (sc-13032; 1:800; Santa Cruz Biotechnology, CA), α-TUBULIN (sc-5286; 1:1000, Santa Cruz) and β-ACTIN (sc-47778; 1:1000; Santa Cruz) were used. Blotting membranes were incubated with the primary antibody at 4 °C overnight and the secondary IgG-horse radish peroxidase (#31460; 1:10000; Thermo Scientific) at room temperature for 1 h. The resulting bands were visualized using Tanon 5500 (Tanon, Shanghai, China) as previously described [30]. Quantification of the results was accomplished using ImageJ software (National Institute of Mental Health, USA).
2.12. RT-qPCR
Liver tissues were lysed using TissueLyser II (#85300, Qiagen, Germany). Total RNA was isolated from liver lysates and primary hepatocytes with RNAiso Plus kits (Code No. 9109, Takara, Dalian, China). cDNA was generated using Prime Script RT reagent Kits (Takara, Dalian, China). Real-time PCR reactions were performed using the SYBR Premix EX Taq Kit (Takara, Dalian, China) and the QuantStudio 6 Flex real-time PCR system (Applied Biosystems, Waltham, Massachusetts, USA) [22,24]. cDNA was amplified using the following PCR conditions: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Data were analyzed using the 2−ΔΔCT method as previously described [31]. Primers for Pparg1/2, fatty acid binding protein 4 (Fabp4), Cd36, sterol regulatory element-binding transcription factor 1 (Srebf1), MLX interacting protein like (Mlxipl), fatty acid synthase (Fasn), stearoyl-CoA desaturase-1 (Scd1), Actin and other genes are listed in supplementary Table S1. Primers were synthesized by Shanghai Sangon Biotech Co., Ltd. (China). All mRNA levels were presented relative to Actb of Nrf2-LoxP mice fed with CD or control untreated hepatocytes.
2.13. Statistical analysis
All data were expressed as mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 6 software (Graphpad Software Inc., San Diego, CA) with p < 0.05 considered as significant. Statistical differences were determined by an unpaired Student's t-test or by two-way analysis of variance followed by a Dunnett's multiple comparison test. Analysis for Table 1 was performed using Statistical Products and Services software (v.16.0, SPSS Institute Cary, Chicago, IL, USA) with a Kruskal-Wallis test for comparing all groups and a Mann-Whitney test for the comparison of two specific groups.
Table 1.
Liver steatosis analyzed based on NAS.
| NAS Steatosis levels | CD |
HFD |
||
|---|---|---|---|---|
| Nrf2-LoxP | Nrf2(L)-KO | Nrf2-LoxP | Nrf2(L)-KO | |
| 0: <5% | 10 | 11 | 0 | 0 |
| 1: 5%–33% | 0 | 0 | 2 | 5 |
| 2: 33%–67% | 0 | 0 | 4 | 2 |
| 3: >67% | 0 | 0 | 2 | 0 |
CD: chow diet; HFD: high-fat diet; NAS: NAFLD Activity Score.
Note: 16 wks old Nrf2-LoxP and Nrf2(L)-KO female mice were fed with CD and HFD for 12 wks and the steatosis of livers were evaluated based on H&E staining. Based on NAS, the primary histologic features were classified into four grades of steatosis severity (% cells involved in lesion; evaluation of parenchymal involvement by steatosis) [32]. According to the Kruskal-Wallis test, p = 0.058 in HFD fed Nrf2(L)-KO vs Nrf2-LoxP mice.
3. Results
3.1. Nrf2(L)-KO mice fed with HFD show smaller livers, milder hepatic steatosis and less hepatic TG accumulation
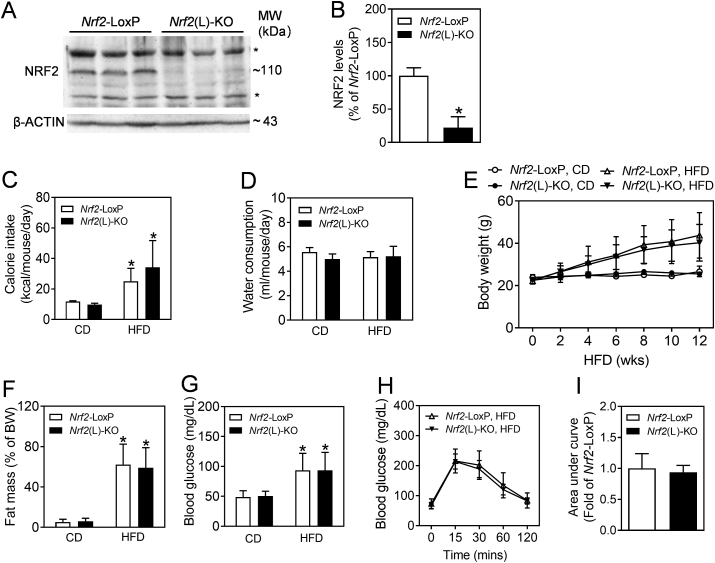
To confirm the knockout efficiency of Nrf2(L)-KO mice, an immunoblotting assay against NRF2 was performed using the whole tissue lysates of liver. As shown in Fig. 1A and B, the protein levels of NRF2 were dramatically decreased in the liver of Nrf2(L)-KO mice relative to Nrf2-LoxP littermate controls. To determine the potential effect of Nrf2 ablation in hepatocytes on general energy metabolism, the body weight, food intake, water consumption, fat mass and blood glucose levels were monitored over 12 weeks for all groups. As shown in Fig. 1C, caloric intake was calculated based on food consumption and the energy content of the diets (CD, 3.616 kcal/g; HFD, 5.243 kcal/g). While the energy intake of HFD-fed mice were significantly higher than those of CD groups, there were no significance between genotypes, either under CD or HFD. In addition, the water consumption also showed minimal fluctuation among the 4 groups (Fig. 1D). Furthermore, Nrf2(L)-KO mice fed with either CD or HFD showed no significant differences in body weight gain (Fig. 1E), relative fat mass measured by a body composition analyzer (Fig. 1F) and fasting blood glucose levels (Fig. 1G) from the corresponding littermate controls, whereas the mice responded well to HFD exposure, showing elevated body weight gain, relative fat mass and blood glucose levels compared to CD controls. In agreement with the measurements above, IPGTT performed in the HFD groups following HFD exposure also displayed no significant difference between the genotypes (Fig. 1H and I). The same tendency was also seen in the measurements of male Nrf2(L)-KO mice (supplementary Fig. S1).
Fig. 1.
Impact of consumption of HFD on food intake, body weight gain, fat mass and blood glucose levels in Nrf2-LoxP and Nrf2(L)-KO female mice. (A) Representative image of immunoblots of NRF2 using the liver tissues of Nrf2-LoxP and Nrf2(L)-KO mice. *, non-specific bands. (B) Relative quantitative protein expression of NRF2. n = 3. *p < 0.05 vs. Nrf2-LoxP mice. (C and D) Calorie intake (C) and water consumption (D) during the HFD exposure period. n = 10 for Nrf2-LoxP, CD; n = 11 for Nrf2(L)-KO, CD; n = 8 for Nrf2-LoxP, HFD; n = 7 for Nrf2(L)-KO, HFD. *p < 0.05 vs. CD of the same genotype. (E) Body weight of age-matched Nrf2-LoxP and Nrf2(L)-KO mice fed with CD or HFD diet for 12 wks from 16 wks of age. (F) Fat mass as percentages of body weight measured by a Body Composition Analyzer. (G) Fasting blood glucose levels. n = 7–11 females. *p < 0.05 vs. CD of the same genotype. (H) Intraperitoneal glucose tolerance test. Nrf2(L)-KO and Nrf2-LoxP control mice fed with HFD for 10 wks were challenged with 1.0 mg of glucose per gram of BW. n = 7–8 females. (I) Areas under the curves of (H).
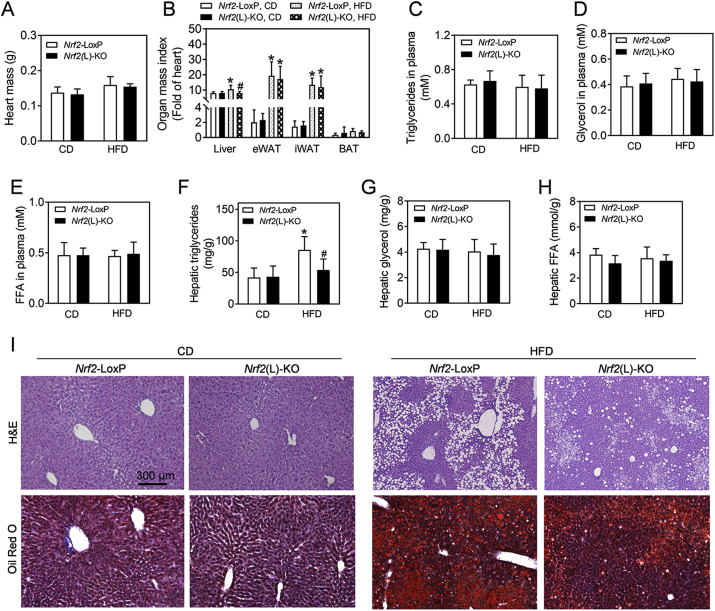
To further ascertain the effect of Nrf2 deletion on body composition, the organ mass indexes were determined in Nrf2(L)-KO mice and their Nrf2-LoxP controls following HFD exposure. Because the heart weight showed no significant difference between the genotypes, either treated with CD or HFD (Fig. 2A), we used the heart weight instead of body weight to calculate relative organ weight in order to eliminate the possible bias of altered amounts of fat tissues (Fig. 2B). Compared to CD, consumption of HFD by control mice resulted in enlargement of the liver. Nrf2(L)-KO mice fed with HFD showed significantly less increase in liver size than control mice (Fig. 2B and S1F). In contrast, there were no significant differences in the mass of adipose tissues between the genotypes under either CD- or HFD-fed conditions. In addition, there were no significant differences in the levels of TG, glycerol and FFA in plasma among various situations (Fig. 2C–E). In contrast, hepatic TG, but not glycerol and FFA, in Nrf2-LoxP mice exposed to HFD were significantly higher than those in control mice treated with CD (Fig. 2F–H). Interestingly, hepatic TG in the Nrf2(L)-KO mice fed with HFD were significantly lower than those in the Nrf2-LoxP mice on the same diet (Fig. 2F). In addition, the livers of mice fed with HFD generally had more fat deposition evaluated by histological analysis than those fed with CD, whereas Nrf2(L)-KO mice fed with HFD had less hepatic lipid deposition than their Nrf2-LoxP littermates with the same diet (Fig. 2I and S2). A severity analysis of liver steatosis calculated by a NAFLD Activity Score as described previously [32], also displayed a relative attenuated steatosis in HFD-fed Nrf2(L)-KO mice compared to Nrf2-LoxP controls (Table 1). Taken together, the analyses above clearly elucidated that Nrf2(L)-KO mice are resistant to HFD-induced hepatic steatosis compared to Nrf2-LoxP control mice.
Fig. 2.
Nrf2(L)-KO female mice show reduced liver enlargement, milder hepatic steatosis and lower hepatic TG accumulation than Nrf2-LoxP mice fed with HFD. (A) Heart mass and (B) organ mass index expressed as a fold of heart. n = 10 for Nrf2-LoxP, CD; n = 11 for Nrf2(L)-KO, CD; n = 8 for Nrf2-LoxP, HFD; n = 7 for Nrf2(L)-KO, HFD. Values are mean ± SD. *p < 0.05 vs. CD of the same genotype; #p < 0.05 vs. Nrf2-LoxP mice with the same diet. (C–E) Levels of TG, glycerol and FFA in plasma. n = 5 for Nrf2-LoxP, CD; n = 5 for Nrf2(L)-KO, CD; n = 8 for Nrf2-LoxP, HFD; n = 7 for Nrf2(L)-KO, HFD. (F–H) Levels of hepatic TG (F), glycerol (G) and FFA (H). n = 7–11 for TG contents; n = 5–8 for glycerol and FFA measurements. (I) Representative histological images of H&E and Oil Red O staining in the liver. Magnification = 100 × ; bar scale = 300 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Nrf2(L)-KO mice have attenuated expression of PPARγ2 induced by HFD exposure in the liver
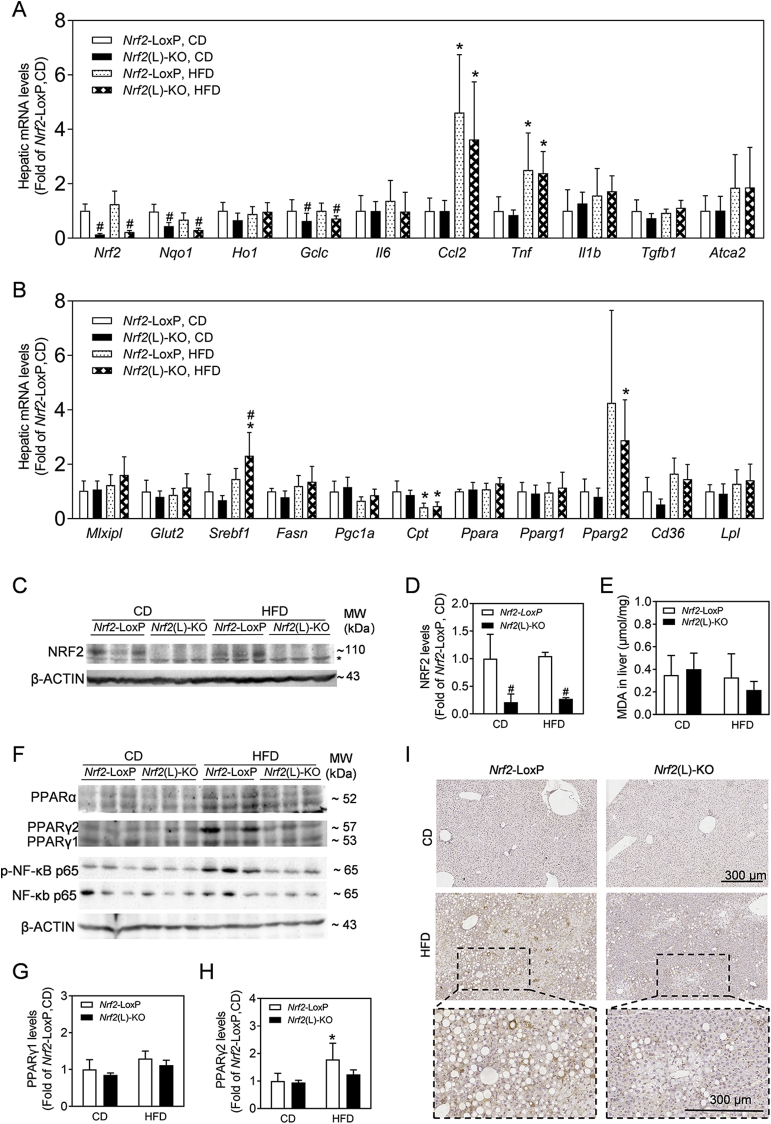
To understand the mechanism that Nrf2(L)-KO mice become resistant to HFD-induced hepatic steatosis, the expression of mRNA related to oxidative stress, glucose and lipid metabolism, inflammation and fibrosis was measured by RT-qPCR (Fig. 3A and B). Consistent with the deficiency of Nrf2 in hepatocytes, the mRNA levels of Nrf2 and its downstream genes, such as NAD(P)H:quinone oxidoreductase 1 (Nqo1), glutamate-cysteine ligase catalytic subunit (Gclc) and hemeoxygenase 1 (Ho1), were all decreased in the liver of Nrf2(L)-KO mice compared to Nrf2-LoxP under CD or HFD exposure, while HFD had no significant effect on these mRNA expression. The expression of the rate-limiting enzyme of β-oxidation, carnitine palmitoyl transferase (Cpt), was reduced to nearly 50% by HFD exposure in both Nrf2-LoxP and Nrf2(L)-KO genotypes, which is consistent with TG accumulation in hepatocytes (Fig. 3A). In contrast, the mRNA levels of pro-inflammatory cytokines, including Ccl2 and Tnf, were significantly induced by HFD exposure in both Nrf2(L)-KO and control mice, but there were no differences between the genotypes either under CD or HFD (Fig. 3A). Hepatic mRNA level of a key lipid synthesis gene, Srebf1, was increased in HFD-fed Nrf2(L)-KO mice compared to control mice with the same diet, which is not consistent with the reduced liver steatosis seen in HFD-fed Nrf2(L)-KO mice (Fig. 3B). This finding suggests that other mechanism(s) might play more dominant role in the regulation of lipid metabolism in Nrf2(L)-KO hepatocytes. Importantly, the levels of Pparg2 mRNA were found to be induced by HFD exposure in both genotypes, whereas the expression of Pparg2 showed a trend to be lower in Nrf2(L)-KO mice compared to control mice (Fig. 3B). No significant differences between the Nrf2(L)-KO and their littermate controls were noted for any other measured genes (Fig. 3A and B). In agreement with the mRNA expression, the protein levels of NRF2 in the liver showed a dramatic reduction in Nrf2(L)-KO mice compared to controls (Fig. 3C and D). In contrast, the MDA levels in the liver showed no significant difference between the two genotype mice with CD or HFD (Fig. 3E). Consistent with the mRNA expression of Pparg2, the protein levels of PPARγ2 in the liver of Nrf2(L)-KO mice fed with HFD were also lower than those in Nrf2-LoxP group (Fig. 3F–H). In addition, that the levels of phosphorylated NF-κB p65, but not total NF-κB p65, were substantially induced in the liver of Nrf2-LoxP mice by HFD exposure, whereas Nrf2(L)-KO mice showed a lower expression compared to control mice with the same diet (Fig. 3F). Furthermore, immunohistochemistry analysis against F4/80, a macrophage marker, showed that HFD exposure induced macrophage infiltration in the livers of Nrf2-LoxP control mice (Fig. 3I). Compared to control mice, Nrf2(L)-KO mice have less hepatic macrophage infiltration following HFD exposure (Fig. 3I).
Fig. 3.
Nrf2(L)-KO female mice show reduced expression of PPARγ2 in the liver. (A and B) Relative mRNA levels in the liver. n = 5 for Nrf2-LoxP, CD; n = 5 for Nrf2(L)-KO, CD; n = 8 for Nrf2-LoxP, HFD; n = 7 for Nrf2(L)-KO, HFD. Values are mean ± SD. *p < 0.05 vs. CD of the same genotype; #p < 0.05 vs. Nrf2-LoxP mice with the same diet. (C) Representative images of immunoblots of NRF2. n = 3. (D) Quantification of NRF2 protein levels. (E) MDA levels in liver tissues. n = 10 for Nrf2-LoxP, CD; n = 11 for Nrf2(L)-KO, CD; n = 8 for Nrf2-LoxP, HFD; n = 7 for Nrf2(L)-KO, HFD. (F) Representative images of immunoblots of PPARα, PPARγ, phosphorylated NF-κB p65 and NF-κB p65. n = 3. β-ACTIN is a loading control. (G–H) Relative quantitative expression of PPARγ1 and PPARγ2 calculated according to (F). (I) Immunohistochemical analysis of F4/80 in the liver. Magnification = 80 × ; bar scale = 300 μm.
3.3. Nrf2(Mϕ)-KO mice display no significant difference in hepatic steatosis from Nrf2-LoxP control mice
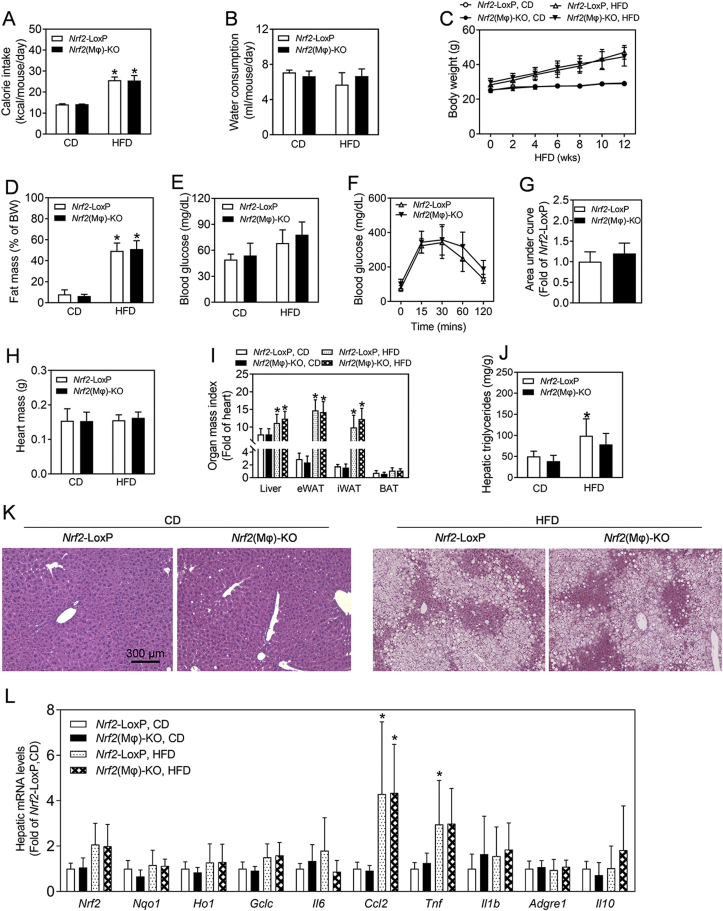
To further clarify that NRF2 in hepatocytes play a critical role in the development of hepatic steatosis, a parallel investigation was conducted in Nrf2(Mϕ)-KO mice and their Nrf2-LoxP controls. As shown in Fig. 4, there were no significant differences in calorie intake, water consumption, body weight, relative fat mass, blood glucose level, glucose tolerance test, major organ mass index, and hepatic lipid accumulation between the genotypes under either CD or HFD exposure conditions, whereas HFD exposure significantly elevated most of those parameters in Nrf2(Mϕ)-KO and Nrf2-LoxP control mice (Fig. 4A–K).
Fig. 4.
Measurements in Nrf2(Mϕ)-KO mice and Nrf2-LoxP littermates. (A–C) Calorie intake (A), water consumption (B) and body weight (C) during the HFD exposure period. Age-matched Nrf2-LoxP and Nrf2(Mϕ)-KO mice were fed with CD or HFD diet for 12 wks from 16 wks of age. *p < 0.05 vs. CD of the same genotype. (D) Fat mass as percentages of body weight measured by a Body Composition Analyzer. (E) Fasting blood glucose levels. n = 5 for Nrf2-LoxP, CD; n = 9 for Nrf2(Mϕ)-KO, CD; n = 9 for Nrf2-LoxP, HFD; n = 13 for Nrf2(Mϕ)-KO, HFD. (F) Intraperitoneal glucose tolerance test following HFD exposure. Nrf2(L)-KO and control mice fed with HFD 12 wks were challenged with 1.0 mg of glucose per gram of BW. n = 9–13. (G) Areas under the curves of (F). (H–I) Heart mass (H) and organ mass index measured as a fold of heart (I). n = 5–13. (J) Levels of hepatic TG. n = 5 for Nrf2-LoxP, CD; n = 3 for Nrf2(Mϕ)-KO, CD; n = 6 for Nrf2-LoxP, HFD; n = 5 for Nrf2(Mϕ)-KO, HFD. (K) Representative histological images of the H&E staining in the liver. Magnification = 100 × ; bar scale = 300 μm. (L) Hepatic mRNA levels in Nrf2(Mϕ)-KO and Nrf2-LoxP control mice.
To further validate the phenotype of Nrf2(Mϕ)-KO mice, we measured the mRNA levels of oxidative stress and inflammation-related genes in the liver. As shown in Fig. 4L, the mRNA expression of Ccl2 and Tnf in the liver was significantly elevated by HFD exposure in both genotypes of mice. However, there were no significant difference in the mRNA expression between the genotypes either under CD or HFD exposure. Other mRNAs measured showed no significance among the four groups.
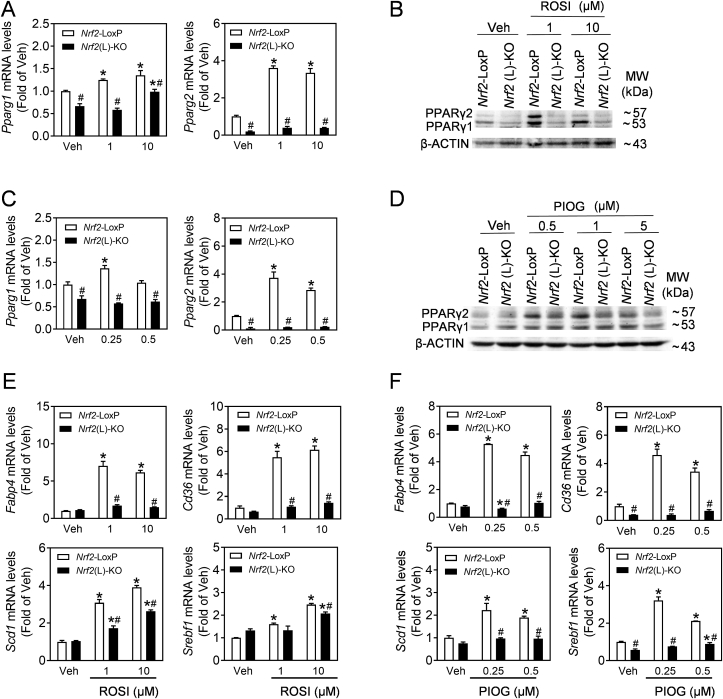
3.4. PPARγ agonists increase the expression of PPARγ and its downstream target genes in primary hepatocytes in a NRF2-dependent manner
To investigate the role of PPARγ in lipid deposition in Nrf2-deficient hepatocytes, the PPARγ agonists, ROSI and PIOG, were used in the primary hepatocytes isolated from Nrf2-LoxP and Nrf2(L)-KO mice. As shown in Fig. 5, the mRNA and protein expression of PPARγ1 and PPARγ2 were significantly induced by ROSI (Fig. 5A and B) or PIOG (Fig. 5C and D) in Nrf2-LoxP hepatocytes. Nrf2(L)-KO hepatocytes showed dramatically attenuated mRNA and protein expression of PPARγ1 and PPARγ2 under vehicle or agonist-challenged conditions. In addition, the mRNA expression of Fabp4 and Cd36, which are known PPARγ target genes, Scd1 and Srebf1 displayed a similar pattern as PPARγ (Fig. 5E and F), suggesting that NRF2 is critical in PPARγ activation in hepatocytes.
Fig. 5.
Nrf2(L)-KO hepatocytes exhibit decreased response to PPARγ agonists. (A–D) Primary hepatocytes isolated from Nrf2-LoxP and Nrf2(L)-KO mice fed with CD were cultured for 6 h in normal media and then exposed to rosiglitazone (ROSI, Veh, 1, 10 μM) or pioglitazone (PIOG, Veh, 0.25, 0.5 μM) for 24 h. (A–D) The mRNA expression (A and C) and representative images of immunoblots of PPARγ (B and D) in the cells treated with ROSI (A and B) or PIGO (C and D). *p < 0.05 vs. Veh of the same genotype; #p < 0.05 vs. Nrf2-LoxP with the same treatment. (E and F) mRNA expression of Fabp4, Cd36, Scd1 and Srebf1 induced by ROSI (E) and PIOG (F).
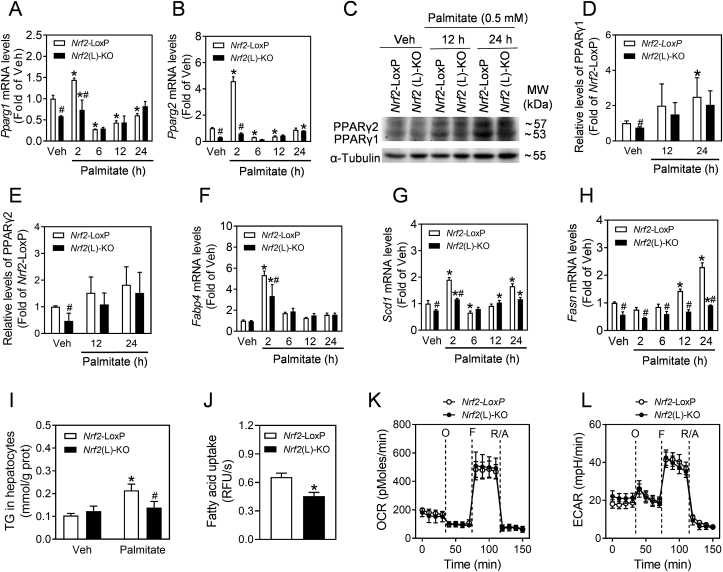
3.5. Nrf2 deficiency down-regulates PPARγ activation and lipogenesis induced by palmitate treatment in primary hepatocytes
Primary hepatocytes isolated from Nrf2(L)-KO and Nrf2-LoxP control mice were treated with palmitate (0.5 mM) to simulate PPARγ and subsequent lipogenesis. The mRNA levels of Pparg1 and Pparg2 increased and peaked at 2 h after treatment (Fig. 6A and B), and decreased thereafter (6, 12 and 24 h). Compared to Nrf2-LoxP cells, Nrf2(L)-KO hepatocytes were less responsive in their Pparg1 and Pparg2 mRNA induction. In agreement with the mRNA expression, the protein levels of PPARγ1 and PPARγ2 also displayed a trend of increase in both genotypes, while the Nrf2 deletion weakened the tendency (Fig. 6C–E). In addition, the mRNA expression of Fabp4, Scd1 and Fasn, the major downstream lipigenic genes of PPARγ, also showed significant increases in response to palmitate treatment in Nrf2-LoxP cells, whereas Nrf2(L)-KO hepatocytes had reduced induction in their expression (Fig. 6F–H). Following a 24-hrs palmitate treatment, the TG levels in Nrf2(L)-KO hepatocytes were much lower than those in Nrf2-LoxP cells (Fig. 6I and Fig. S4).
Fig. 6.
Nrf2(L)-KO hepatocytes show reduced activation of PPARγ in response to palmitate challenge. (A–B) The mRNA levels of Pparg1 and Pparg2. Isolated primary hepatocytes from Nrf2-LoxP and Nrf2(L)-KO mice were challenged with 0.5 mM palmitate for indicated time. *p < 0.05 vs. Veh of the same genotype; #p < 0.05 vs. Nrf2-LoxP with the same treatment. (C) Representative image of immunoblots of PPARγ1 and PPARγ2 in hepatocytes. (E and F) Relative quantitative expression of PPARγ1 and PPARγ2 in (C). (F–H) mRNA levels of Fabp4, Scd1 and Fasn. (I) TG content in hepatocytes. The cells were treated with 0.5 mM palmitate for 24 h. (J) Fatty acid uptake rate in primary hepatocytes. RFU, relative fluorescence unit. (K) OCR and (L) ECAR measured by a Seahorse XF24 Extracellular Flux Analyzer in primary hepatocytes.
To evaluate the effect of Nrf2 deficiency on fatty acid uptake in hepatocytes, a BODIPY uptake assay was performed. As shown in Fig. 6J, the rate of BODIPY uptake in Nrf2(L)-KO hepatocytes was slower than control cells. In contrast, the mitochondrial function in hepatocytes, as measured by OCR and ECAR, showed no significant difference between the two types of cells (Fig. 6K and L).
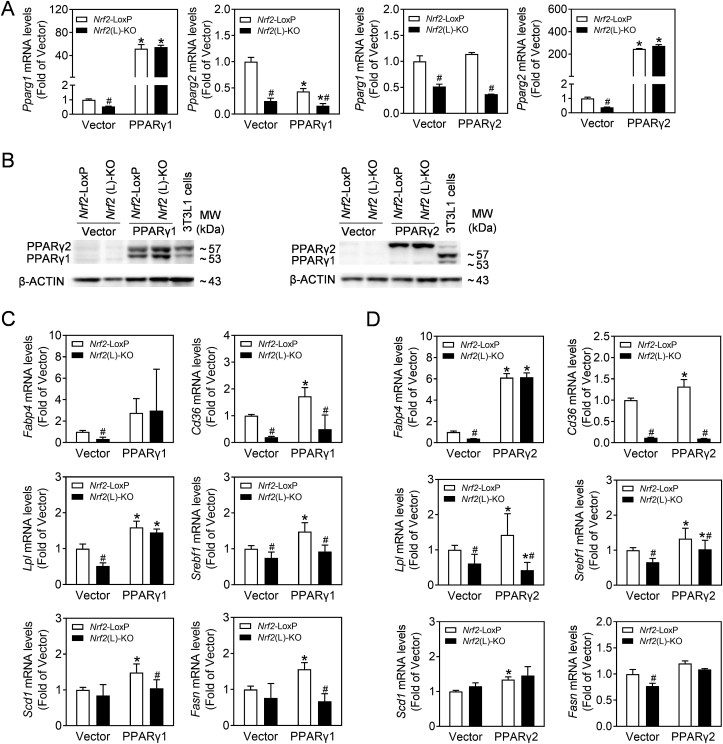
3.6. Overexpression of PPARγ1 or γ2 distinctively reverses the reduced expression of lipogenic genes caused by Nrf2 deficiency in primary hepatocytes
To validate the involvement of PPARγ1 or 2 in the alteration of lipogenic genes in Nrf2(L)-KO hepatocytes, we overexpressed Pparg1 and Pparg2 in primary hepatocytes from Nrf2(L)-KO and Nrf2-LoxP mice. As shown in Fig. 7A and B, both mRNA and protein levels were measured to confirm the efficacy of overexpression. Accordingly, the direct downstream gene of PPARγ, Fabp4, increased in both Nrf2-deleted and control hepatocytes to the same extent (Fig. 7C and D), which indicate that PPARγ1 or PPARγ2 overexpression reverses NRF2-dependent reduction of PPARγ activity. However, the reversal was not observed in the mRNA levels of Cd36, a direct target gene of NRF2 [33,34], suggesting that the relationship between CD36 and NRF2 was more prominent than CD36 and PPARγ in the process of hepatic steatosis. Consistent with this notion, the mRNA levels of Lpl and Scd1 and Fasn were also reversed by overexpression of Pparg1 (Fig. 7C) and Pparg2 (Fig. 7D), respectively. While other PPARγ target genes reported in adipose tissues were also induced by the overexpression, their expression were not fully reversed in Nrf2(L)-KO cells, compared to Nrf2-LoxP hepatocytes (Fig. 7C and D).
Fig. 7.
The reduced expression of PPARγ-related genes in Nrf2(L)-KO hepatocytes is partially reversed by PPARγ overexpression. Hepatocytes isolated from Nrf2-LoxP and Nrf2(L)-KO mice were transfected with Pparg1 and Pparg2 plasmids. (A) mRNA levels of Pparg1 and Pparg2 in the cells. *p < 0.05 vs. Veh of the same genotype; #p < 0.05 vs. Nrf2-LoxP with the same treatment. (B) Representative images of immunoblots of PPARγ. (C) mRNA expression of PPARγ downstream gene Fabp4 and other lipid metabolism-related genes.
4. Discussion
Previous studies utilizing global Nrf2-KO or constitutive NRF2-activated mice showed that NRF2 plays a critical role in the development of NAFLD, in which NRF2 is involved in the regulation of lipid metabolism, inflammation and antioxidant response [7]. However, the conclusions from those models on whether, how and where NRF2 participates in the development of NAFLD are not quite clear, which might be partially due to the modifications of Nrf2 gene in those models occurred throughout the body, but not any cell type-specific. Here, we used hepatocyte- and macrophage-specific Nrf2-knockout mouse models to induce NAFLD with a prolonged HFD exposure. Nrf2 deletion in hepatocytes, but not in macrophages, resulted in attenuated hepatic steatosis. Mechanistic investigations indicate that the attenuation from HFD-induced NAFLD in Nrf2(L)-KO mice might be attributable to the decreased expression of PPARγ, PPARγ2 in particular. These findings demonstrate that NRF2-dependent expression of PPARγ plays a critical role in hepatocytes for the initiation of NAFLD induced by HFD exposure.
Consistent with our previous findings in adipocytes [17], the results in primary hepatocytes confirmed that PPARγ2 is down-regulated with Nrf2 deletion, in particular under the PPARγ agonist-challenged conditions. In addition, PPARγ1 is also expressed in the liver and exhibits the same tendency as PPARγ2 in Nrf2(L)-KO hepatocytes and liver tissues. In a NAFLD model using hepatocyte-specific Pparg-knockout mice, a set of enzymes related to de novo lipogenesis were down-regulated, while PPARγ activation by an agonist might reverse the phenotype [35], indicating that PPARγ is a crucial factor for the initiation of NAFLD. The results in the present study indicate that PPARγ activation induced by PPARγ agonists (ROSI and PIOG) or palmitate treatment is dependent, at least in part, on the presence of NRF2 in hepatocytes. Forced expression of either PPARγ1 or PPARγ2 was able to restore the alterations in the expression of lipogenic genes, such as Fabp4 and Lpl, in Nrf2(L)-KO hepatocytes. Therefore, NRF2-mediated activation of PPARγ and subsequent de novo lipogenesis are crucial for hepatic lipid accumulation. Since Cd36 is a downstream target gene of NRF2, the mechanism on Nrf2(L)-KO hepatocytes had decreased fatty acid uptake rate in response to FFA challenge might be a result of lowered expression of CD36 in the cells. These findings indicate that PPARγ is one of the major factors mediating NRF2-regulated lipogenesis in hepatocytes. In addition, NRF2 might also play a distinct role in hepatocytes to regulate lipid homeostasis in a PPARγ-independent way.
SREBP1 is a master regulator of de novo lipogenesis [36]. In the current study, Srebf1 was found to be the only gene elevated in HFD-fed Nrf2(L)-KO mice compared to control group with the same diet. This is consistent with the results of many other studies using Nrf2 transgenic mouse models [11,37,38]. However, our Nrf2(L)-KO mice fed with HFD showed lower liver TG accumulation, which highlights the important role of PPARγ in lipogenesis in hepatocytes. Keap1-knockdown mice, which makes NRF2 constitutively active, fed with HFD for 24 weeks exhibited body weight gain, increased inflammation and lipogenic gene expression [39], indicating that elevated NRF2 aggravates the NAFLD process. To some extent, these results were in accordance with our present study showing that NRF2 appears to play a pathogenic role in the initiation of NAFLD. Other studies utilizing genetic Nrf2 models also drew similar conclusions [15,40,41].
Mice fed with HFD developed hepatic inflammation in both Nrf2-LoxP and Nrf2(L)-KO genotypes. The inflammation in Nrf2(L)-KO mice fed with HFD was generally lower than the Nrf2-LoxP HFD group, as indicated by the subdued phosphorylation of NF-κB, the mRNA levels of proinflammatory cytokines Ccl2 and Tnf and reduced macrophage infiltration. Although NRF2 can repress inflammation [10], we believe that the difference of inflammation between genotypes fed with HFD is a consequence of reduced hepatic steatosis in Nrf2(L)-KO group in this study. Consistent with this conclusion, there was no significant difference of NAFLD determination in the Nrf2(Mϕ)-KO mouse model, which indicates that Nrf2 deficiency in macrophage is not the key in the initiation of NAFLD.
Previous studies using mice with global changes in Nrf2 expression displayed unexpected phenotypes with inconsistency in body and liver weights, as well as expression of lipid metabolism-related genes [42]. This may be due to the fact that Nrf2 expression in different cell types or organs is well controlled and varies due to diverse status or diseases. In vivo, there may be compensation for the loss of global Nrf2 balance by subtle changes in expression that alter phenotype and conceal the effect of NRF2 on a specific disease [43]. In agreement with this explanation, we could not find significant increases in hepatic MDA levels, NRF2 protein levels, the mRNA expression of Nrf2 and the target genes after HFD feeding, which is consistent with the findings reported by Zhang et al. [44]. A more recent study used adipocyte- and hepatocyte-specific Nrf2 deletion mouse models to explore the role of NRF2 in obesity [45]. After 170 days of HFD feeding, the mice with hepatocyte Nrf2 deletion showed lower plasma insulin levels and improved insulin sensitivity without changing hepatic TG accumulation [45], which is not in line with our current findings in Nrf2(L)-KO mice with 12 weeks of HFD challenge. It suggests that NRF2 in hepatocytes possibly plays different roles in the initiation or progression of various NAFLD stages in different HFD exposure situations.
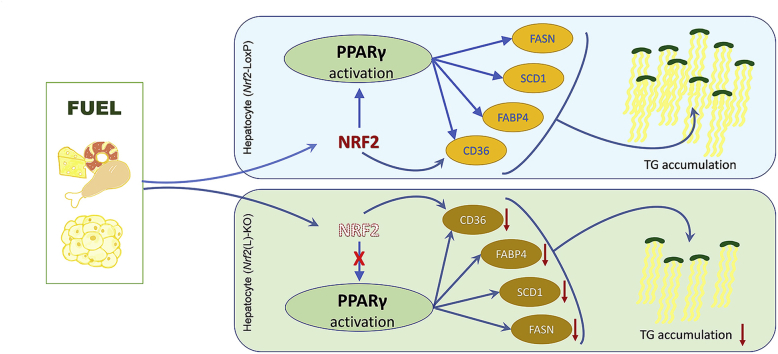
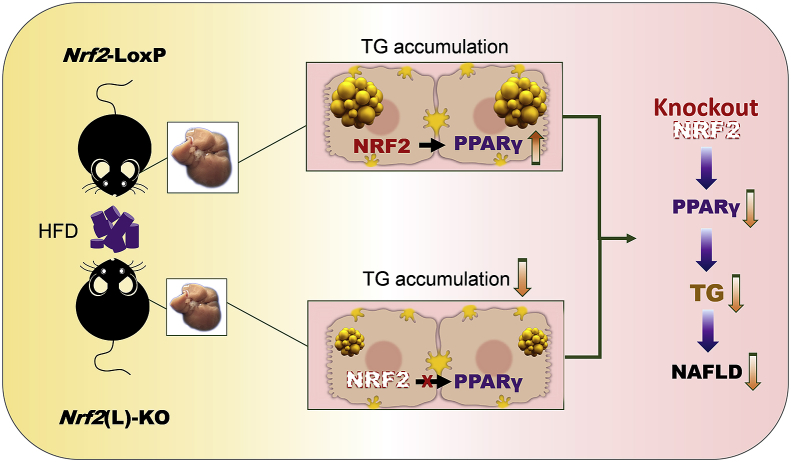
In conclusion, Nrf2(L)-KO mice showed many signs of diminished NAFLD after HFD feeding, including lower accumulation of TG in the liver and minimal liver weight gain. A key aspect of this appears to be the ability of NRF2 to regulate PPARγ expression and influence lipid metabolism in hepatocytes (Fig. 8). Thus, NRF2 in hepatocytes is a critical factor in the initiation of NAFLD and a potential intervention molecular target. Further study is needed to elucidate the specific stages of NAFLD with NRF2 modulation as an effective prevention or therapeutic strategy.
Fig. 8.
Schematic illustration of the proposed role of NRF2 in hepatocytes in HFD-induced NAFLD. Following a prolonged HFD exposure, the two genotypes of mice displayed different liver mass and TG accumulation in the liver. A possible mechanism is that NRF2 in hepatocyte is responsible for the expression of PPARγ to positively regulate lipid accumulation. Ablation of Nrf2 in hepatocytes results in down-regulation of PPARγ expression and subsequent reduction of lipid accumulation in the cells. The key findings in the present study demonstrate an essential role of NRF2 in the initiation of NAFLD via PPARγ expression.
Author contributions
Designed, supervised the experiments and finalized the manuscript: Jingbo Pi, Yuanyuan Xu, Jingqi Fu.
Acquisition of data: Lu Li, Dan Liu, Jing Sun, Jingqi Fu, Yongyong Hou, Chengjie Chen, Junbo Shao, Xin Wang.
Read of pathology picture: Linlin Wang, Rui Zhao, Lu Li.
Interpreted the data and wrote the manuscript: Lu Li, Jingqi Fu, Jingbo Pi.
Critical revision of the manuscript for important intellectual content: Huihui Wang, Yuanyuan Xu, Melvin E. Andersen, Qiang Zhang.
All authors approved the final manuscript.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by National Natural Science Foundation of China 81830099 (J.P.), 81573106 (J.P.), 81573187 (Y.X.) and 81402635 (J.F.), Shenyang Municipal Bureau of Science and Technology Support Program for Young Innovation Scholar (RC180207, J.F.), Liaoning Province Natural Science Foundation (20180530011, J.F.), the Startup Funding of China Medical University (J.P.), Liaoning Pandeng Scholar Program (J.P.), China Medical University Training Program for National Natural Science Fund for Excellent Young Scholars (YQ20170001, J.F.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101412.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.redox.2019.101412.
Contributor Information
Yuanyuan Xu, Email: yyxu@cmu.edu.cn.
Jingbo Pi, Email: jbpi@cmu.edu.cn, jingbopi@163.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Multimedia component 2
Transparency document
References
- 1.Stefan N., Haring H.U., Cusi K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diab. Endocrinol. 2019;7(4):313–324. doi: 10.1016/S2213-8587(18)30154-2. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69(6):2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 3.Ioannou G.N. The role of cholesterol in the pathogenesis of NASH. Trends Endocrinol. Metab. 2016;27(2):84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Williams C.D. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Argo C.K. Systematic review of risk factors for fibrosis progression in non-alcoholic steatohepatitis. J. Hepatol. 2009;51(2):371–379. doi: 10.1016/j.jhep.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Starley B.Q., Calcagno C.J., Harrison S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51(5):1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 7.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98(3):1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin S. Nrf2 is essential for the anti-inflammatory effect of carbon monoxide in LPS-induced inflammation. Inflamm. Res. 2015;64(7):537–548. doi: 10.1007/s00011-015-0834-9. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi E.H. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meakin P.J. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol. Cell. Biol. 2014;34(17):3305–3320. doi: 10.1128/MCB.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhry S. Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic. Biol. Med. 2010;48(2):357–371. doi: 10.1016/j.freeradbiomed.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Lugrin J. The role of oxidative stress during inflammatory processes. Biol. Chem. 2014;395(2):203–230. doi: 10.1515/hsz-2013-0241. [DOI] [PubMed] [Google Scholar]
- 14.Dandekar A., Mendez R., Zhang K. Cross talk between ER stress, oxidative stress, and inflammation in Health and disease. In: Oslowski C.M., editor. Stress Responses: Methods and Protocols. Springer; New York: New York, NY: 2015. pp. 205–214. [DOI] [PubMed] [Google Scholar]
- 15.Huang J. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299(6):G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souza-Mello V. Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J. Hepatol. 2015;7(8):1012–1019. doi: 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pi J. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285(12):9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki A., Diehl A.M. Nonalcoholic steatohepatitis. Annu. Rev. Med. 2017;68:85–98. doi: 10.1146/annurev-med-051215-031109. [DOI] [PubMed] [Google Scholar]
- 19.Baffy G. Kupffer cells in non-alcoholic fatty liver disease: the emerging view. J. Hepatol. 2009;51(1):212–223. doi: 10.1016/j.jhep.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nati M. The role of immune cells in metabolism-related liver inflammation and development of non-alcoholic steatohepatitis (NASH) Rev. Endocr. Metab. Disord. 2016;17(1):29–39. doi: 10.1007/s11154-016-9339-2. [DOI] [PubMed] [Google Scholar]
- 21.Byrne C.D., Targher G. NAFLD: a multisystem disease. J. Hepatol. 2015;62(1 Suppl):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Xue P. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes. 2013;62(3):845–854. doi: 10.2337/db12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou Y. Adipocyte-specific deficiency of Nfe2l1 disrupts plasticity of white adipose tissues and metabolic homeostasis in mice. Biochem. Biophys. Res. Commun. 2018;503(1):264–270. doi: 10.1016/j.bbrc.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Sun J. NRF2 mitigates acute alcohol-induced hepatic and pancreatic injury in mice. Food Chem. Toxicol. 2018;121:495–503. doi: 10.1016/j.fct.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 25.Fu J., Akhmedov D., Berdeaux R. The short isoform of the ubiquitin ligase NEDD4L is a CREB target gene in hepatocytes. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalifeh-Soltani A. Mfge8 promotes obesity by mediating the uptake of dietary fats and serum fatty acids. Nat. Med. 2014;20(2):175–183. doi: 10.1038/nm.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng H. CNC-bZIP protein Nrf1-dependent regulation of glucose-stimulated insulin secretion. Antioxidants Redox Signal. 2015;22(10):819–831. doi: 10.1089/ars.2014.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tontonoz P. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 29.Gao J. CAR suppresses hepatic gluconeogenesis by facilitating the ubiquitination and degradation of PGC1α. Mol. Endocrinol. (Baltimore, Md.) 2015;29(11):1558–1570. doi: 10.1210/me.2015-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui Q. Deficiency of long isoforms of Nfe2l1 sensitizes MIN6 pancreatic beta cells to arsenite-induced cytotoxicity. Toxicol. Appl. Pharmacol. 2017;329:67–74. doi: 10.1016/j.taap.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z. Nrf2 deficiency aggravates the increase in osteoclastogenesis and bone loss induced by inorganic arsenic. Toxicol. Appl. Pharmacol. 2019;367:62–70. doi: 10.1016/j.taap.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Brown G.T., Kleiner D.E. Histopathology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Metabolism. 2016;65(8):1080–1086. doi: 10.1016/j.metabol.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olagnier D. Nrf2, a PPARgamma alternative pathway to promote CD36 expression on inflammatory macrophages: implication for malaria. PLoS Pathog. 2011;7(9) doi: 10.1371/journal.ppat.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubouy A. Nrf2-driven CD36 and HO-1 gene expression in circulating monocytes correlates with favourable clinical outcome in pregnancy-associated malaria. Malar. J. 2015;14:358. doi: 10.1186/s12936-015-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran-Salvador E. Role for PPARgamma in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 2011;25(8):2538–2550. doi: 10.1096/fj.10-173716. [DOI] [PubMed] [Google Scholar]
- 36.Bertolio R. Sterol regulatory element binding protein 1 couples mechanical cues and lipid metabolism. Nat. Commun. 2019;10(1) doi: 10.1038/s41467-019-09152-7. 1326-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka Y. Dysregulated expression of fatty acid oxidation enzymes and iron-regulatory genes in livers of Nrf2-null mice. J. Gastroenterol. Hepatol. 2012;27(11):1711–1717. doi: 10.1111/j.1440-1746.2012.07180.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramadori P. Hepatocyte-specific Keap1 deletion reduces liver steatosis but not inflammation during non-alcoholic steatohepatitis development. Free Radic. Biol. Med. 2016;91:114–126. doi: 10.1016/j.freeradbiomed.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 39.More V.R. Keap1 knockdown increases markers of metabolic syndrome after long-term high fat diet feeding. Free Radic. Biol. Med. 2013;61:85–94. doi: 10.1016/j.freeradbiomed.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider K. Increased energy expenditure, Ucp1 expression, and resistance to diet-induced obesity in mice lacking nuclear factor-erythroid-2-related transcription factor-2 (Nrf2) J. Biol. Chem. 2016;291(14):7754–7766. doi: 10.1074/jbc.M115.673756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rui W. Nuclear factor erythroid 2-related factor 2 deficiency results in amplification of the liver fat-lowering effect of estrogen. J. Pharmacol. Exp. Ther. 2016;358(1):14–21. doi: 10.1124/jpet.115.231316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L. Is Nrf2-ARE a potential target in NAFLD mitigation? Curr. Opin. Toxicol. 2019;13:35–44. [Google Scholar]
- 43.Sauer V. Induced pluripotent stem cells as a source of hepatocytes. Curr. Pathobiol. Rep. 2014;2(1):11–20. doi: 10.1007/s40139-013-0039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y.K. Nrf2 deficiency improves glucose tolerance in mice fed a high-fat diet. Toxicol. Appl. Pharmacol. 2012;264(3):305–314. doi: 10.1016/j.taap.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chartoumpekis D.V. Nrf2 deletion from adipocytes, but not hepatocytes, potentiates systemic metabolic dysfunction after long-term high-fat diet-induced obesity in mice. Am. J. Physiol. Endocrinol. Metab. 2018;315(2):E180–e195. doi: 10.1152/ajpendo.00311.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 2