Abstract
Although the physicochemical properties of antimicrobial peptides (AMPs) and anticancer peptides (ACPs) are very similar, it remains unclear which specific parameter(s) of ACPs confer the major anticancer activity. By answering how to construct a short AMP/ACP that could easily be synthesized in the most cost effective way plus conferring a maximum anticancer effect is a very important scientific breakthrough in the development of protein/peptide drugs. In this study, an 18-amino-acids antimicrobial peptide, AcrAP1 (named AP1-Z1), was used as a template. Bioinformatics algorithms were then performed to design its six mutants (AP1-Z3a, AP1-Z3b, AP1-Z5a, AP1-Z5b, AP1-Z7, and AP1-Z9). After a series of in vitro experiments plus intensive computational analysis, the data demonstrated that AP1-Z5a and AP1-Z5b induced both apoptosis and anti-angiogenic effects to achieve the maximum anticancer activity. Specifically, the most effective mutant, AP1-Z5b, exhibited high selectivity for the charged membrane in molecular dynamics simulations. These findings clearly demonstrated that both charge and hydrophobicity play an important role and are necessary to reach an optimum equilibrium for optimizing the anticancer activity of AMPs. Overall, the present study provides a very crucial theoretical basis and important scientific evidence on the key physicochemical parameters of ACP drugs development.
Keywords: antimicrobial peptides, anticancer peptides, molecular dynamics, angiogenesis, apoptosis, drug discovery and development, targeted therapy
Graphical Abstract
Introduction
According to GLOBOCAN 2018, there have been 18.1 million new cancer cases (increased by 28.37% from 14.1 million in 2012) and 9.6 million deaths (increased by 17.07% from 8.2 million in 2012).1,2 The rise in cancer incidence is one of the most concerning issues worldwide, since anticancer drugs, including natural products, hormone agonists/antagonists, and antimetabolites, show insufficient selectivity and non-specific targeting of healthy cells, which increase resistance to drugs.3,4 However, there are still many of the current targeted therapeutic drugs that could only exert their effect on a portion of patients who carried specific gene/protein alternations.5 In view of these problems, it is imperative to find a novel anticancer drug capable of controlling tumor growth and metastasis with minimal toxic side effects, which also could overcome the tumor heterogeneity issue.
Over the last two decades, antimicrobial peptides (AMPs) have received widespread attention due to their ability to kill Gram-negative and -positive bacteria, envelope viruses, fungi, and even transform cancerous cells.6,7 AMPs, also known as host defense peptides (HDPs), are involved in an important part of the innate immune response found in all living species.8 It is noteworthy that approximately 3,000 natural AMPs have been isolated and characterized from bacteria, protozoa, protists, fungi, plants, and animals according to the Antimicrobial Peptide Database (APD, http://aps.unmc.edu/AP/main.php).9 Out of these 3,000 AMPs, 230 of them also possess anticancer activity and classified as anticancer peptides (ACPs).9 The mechanism of action of these peptides has been described as killing at the membrane level, involving the classical “Barrel stave/Carpet/Toroidal” pore-forming mechanism.10 In comparison with specific target drugs, the unique advantage of ACPs is that they also act on intracellular targets to inhibit enzyme activity, proteins, and DNA/RNA synthesis.11,12 Different ACPs can inhibit cancer cell growth through multiple mechanisms.13 Typically, AMPs are relatively smaller (5–50 amino acids) and have a large proportion (generally >30%) of hydrophobic residues and a net positive charge (+2 to +9) due to the presence of multiple arginine and lysine residues.14,15 The APD contains 167 AMPs from Chelicerata, including 43 active peptides from spiders and 84 from scorpions,9 some of which are derived from our laboratory or our collaborating laboratory,16, 17, 18 such as AamAP1, AamAP2, AcrAP1, and AcrAP2. The AMP, AcrAP1, was identified from the venom of the Arabian scorpion (Androctonus crassicauda) using the “shotgun” molecular cloning of their cDNA library.16 AcrAP1 consists of 18 amino acids and is C-terminally amidated; its primary sequence is FLFSLIPHAISGLISAFK. Previous experiments have shown that AcrAP1 possesses antimicrobial activity (minimum inhibitory concentration [MIC], Gram-positive bacterium Staphylococcus aureus 8 μM; Gram-negative bacterium Escherichia coli > 250 μM; and yeast Candida albicans 16 μM).16
Recently, antimicrobial peptides (AMPs) have been classified as a new generation of anticancer drug candidate, which are potentially able to overcome the tumor heterogeneity. Although the physicochemical properties of AMPs and ACPs are very similar, the key specific parameters that confer anticancer activity still remain unclear. Efforts are being made to understand the differences in key physicochemical properties between AMPs and ACPs, which will help to design and modify ACPs with better activity.19 Bioinformatics algorithms are combined with machine learning, where design is automated through selected attributes, taking into account the existing molecular AMP/ACP library, which is considered a future method for rational design.20 These strategies primarily consider improvements in physicochemical properties such as amphipathicity, hydrophilicity, hydrophobicity, and net charge, with the goal of obtaining more active peptide drugs by modification.21 In the present study, the 18-amino-acid antimicrobial peptide, AcrAP1 (named AP1-Z1), was used as a template. Changing the charge (+1 to +9) and hydrophobicity (0.90167−0.38667) was the main approach to study the structure-activity relationship between the physicochemical properties of AcrAP1 and its anticancer activity. Bioinformatics algorithms were used to design 6 mutants (AP1-Z3a, AP1-Z3b, AP1-Z5a, AP1-Z5b, AP1-Z7, and AP1-Z9) of AcrAP1, which were generated by manual and genetic algorithm-based mutation modules.22 The secondary structure changes in aqueous and cell membrane-simulated environments were determined by circular dichroism. The difference in anticancer activity was verified by a series of activity screening methods in vitro. Molecular dynamics (MD) was used to construct the interaction between AP1-Z5b and the cell membrane. The present study provides a theoretical basis for optimizing the physicochemical parameters of ACP anticancer activity.
Results
Construction of the Peptides and Model Prediction
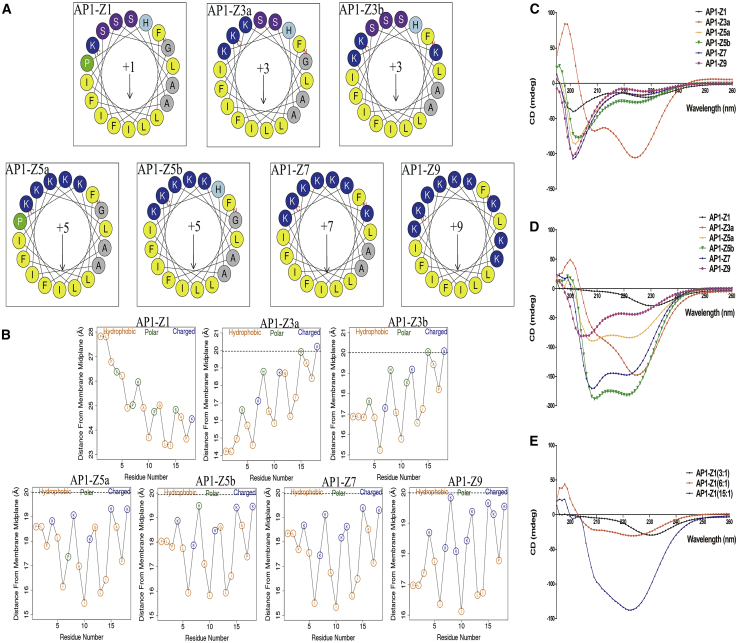
Six mutants of AP1-Z1 were constructed by manual and/or automatic mutation using a genetic algorithm, including two with a net charge (Z) of +3 (AP1-Z3a and AP1-Z3b), two with a net charge of +5 (AP1-Z5a and AP1-Z5b), one with a net charge of +7 (AP1-Z7), and one with a net charge of +9 (AP1-Z9) (Figure 1A; Table S1). The net charge of AP1-Z1 and AP1-Z9 increased from +1 to +9, but the hydrophobicity decreased from 0.90167 to 0.38667 (Table S1). The orientation graph shows the average distance of each residue from the membrane midplane (Figure 1B). Residue types are indicated by circles. The hydrophobic residues (A, F, G, I, L, and V) are yellow, charged residues (K, R, E, and D) are blue, and polar residues are green. The orientation graph indirectly indicates the extent to which the peptide penetrates the cell membrane. According to the primary sequence of the peptide and the position of the phosphate group of the polar lipid head, the position of the peptide is indicated by a horizontal dashed line. Seven peptides were divided into three levels: first level, AP1-Z5a, AP1-Z5b, and AP1-Z7; second level, AP1-Z9, AP1-Z3a, and AP1-Z3b; and third level, AP1-Z1. The first level most easily penetrates the cell membrane and the third level is relatively difficult.
Figure 1.
Helical Wheel Plots, Orientation Graph, and CD Spectra of AP1-Z1 and Its Mutants
(A) Helical wheel plots of AP1-Z1 and its six mutants. The number +1 (+3, +5, +7, and +9) represents the net charge of the peptide. (B) The orientation graph of AcrAP1 and its six mutants: the average distance of each residue from the membrane midplane. The horizontal dotted line marks the location of the phosphate groups of the polar lipid heads. To ensure that the size of all orientation graphs remains consistent, the horizontal dotted line of AP1-Z1 cannot be shown, since the distance of the lipid phosphate groups from the membrane midplane is 20 Å. (C) CD spectra of 0.5 mM peptides AP1-Z1 and its mutants in 1× PBS buffer. (D) 0.5 mM peptides in POPE/POPG (7:3) SUVs suspended in 1× PBS (3:1 lipid:peptide molar ratios). (E) 0.5 mM AP1-Z1 in POPE/POPG (7:3) SUVs suspended in 1× PBS (3:1, 6:1, and 15:1 lipid:peptide molar ratios). *Data analysis by the CDNN software.
Circular Dichroism (CD) Spectroscopy
AP1-Z3a in 1× PBS buffer had a 24.28% higher α helix ratio than the other peptides (Figure 1C); however, the α helix ratio of AP1-Z5b increased from 18.21% in 1× PBS buffer to 48.21% under the conditions of 3:1 lipid:peptide, and that of AP1-Z7 increased from 16.16% to 44.95% (Figures 1C and 1D). CD results were not obtained for AP1-Z3b, since the buffer contained DMSO, which strongly disrupts the CD signal. The α helix ratio of AP1 increased insignificantly from 12.12% to 25.95% with an increase in lipid:peptide ratio from 3:1 to 15:1 (Figure 1E). At higher lipid:peptide molar ratios of 6:1 and 15:1, other peptides induced liposomal disruption to varying degrees.
Anti-proliferative Assay: MTT Assay and Anti-proliferative Curve
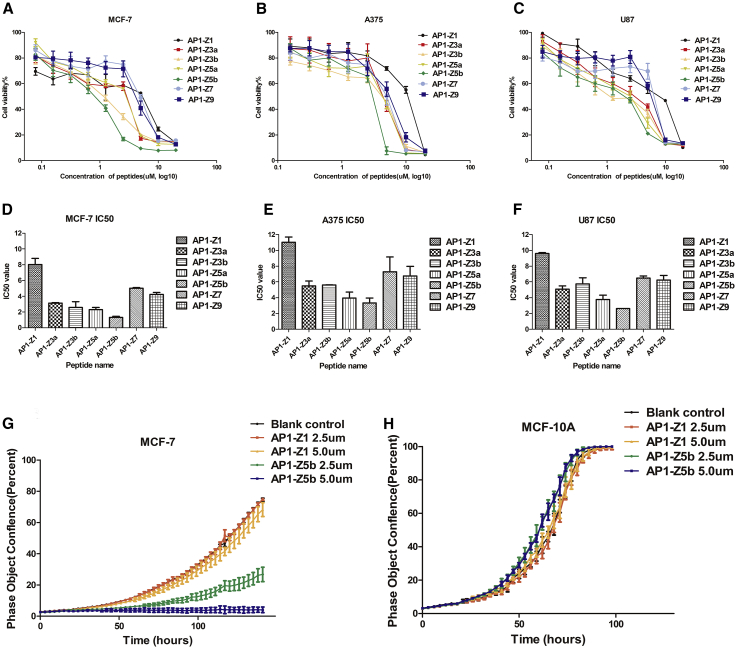
An MTT assay conducted in three cancer cell lines (breast cancer. MCF 7; malignant melanoma. A375; and brain glioma. U87) treated with AP1-Z1 and its six mutants (Figures 2A–2F) showed a similar trend in IC50. The IC50 of mutants AP1-Z5b and AP1-Z5a with a net charge of +5 was relatively lower than that of the other peptides, whereas the IC50 of unmodified AP1-Z1 was relatively higher. Through optimization of the structure, the IC50 of AP1-Z5b was decreased 3- to 6-fold in different cancer cell lines as compared with AP1-Z1: in MCF-7 cell, the IC50 of AP1-Z5b and AP1-Z1 was 1.037 and 7.222 μM, respectively; in A375 cell, the IC50 of AP1-Z5b and AP1-Z1 was 2.607 and 9.478 μM, respectively; and in U87 cell, the IC50 of AP1-Z5b and AP1-Z1 was 3.115 and 10.21 μM, respectively.
Figure 2.
Anti-Proliferative Assay
MTT results of three cancer cell lines treated with AP1-Z1 and its six mutants; proliferation curve of MCF-7 and MCF-10A cells treated with AP1-Z1 and AP1-Z5b. (A and D) Cell viability curve and IC50 of the breast cancer cell line, MCF-7. (B and E) Cell viability curve and IC50 of the malignant melanoma cell line, A375. (C and F) Cell viability curve and IC50 of the brain glioma cell line, U87. (G) Proliferation curve of MCF-7 cells treated with AP1-Z1 and AP1-Z5b (2.5 and 5.0 μM, the blank control was treated with an equal volume of PBS). (H) Proliferation curve of MCF-10A cells treated with AP1-Z1 and AP1-Z5b.
Based on the results of the MTT assay, we selected AP1-Z5b with which to study the anti-proliferative curve of the breast cancer cell line, MCF-7, with the normal breast cell line, MCF-10A, as a control. According to the IC50 of the seven peptides against MCF-7 cells, two concentrations, 2.5 and 5.0 μM, were chosen. In comparison with AP1-Z1, AP1-Z5b significantly inhibited the proliferation of MCF-7 cells in a concentration-dependent manner (Figure 2G). However, AP1-Z1, at a concentration of 2.5 and 5.0 μM, acted the same as AP1-Z5b on MCF-10A cells, which had no significant anti-proliferative effect (Figure 2H).
Apoptosis Assay
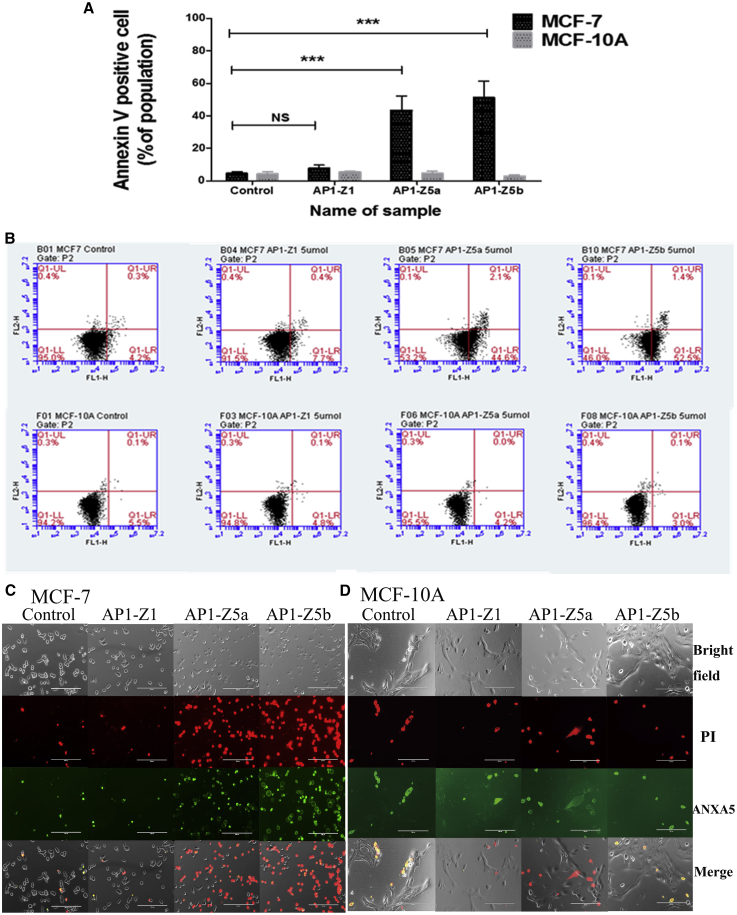
AP1-Z5a and AP1-Z5b were selected to study the apoptosis of the breast cancer cell line, MCF-7, with the normal breast cell line, MCF-10A, as a control. One-way analysis of variance (ANOVA) showed that in comparison with the control group, the mutants AP1-Z5a and AP1-Z5b significantly promoted the apoptosis of the cancer cell line, MCF-7 (AP1-Z5a and AP1-Z5b, ***p < 0.001), but AP1-Z1 had no significant effect. Moreover, in comparison with AP1-Z1, the mutants AP1-Z5a and AP1-Z5b significantly promoted the apoptosis of MCF-7 cells (AP1-Z5a and AP1-Z5b, ***p < 0.001) (Figures 3A and 3B). However, at a concentration of 5 μM, AP1-Z5a and AP1-Z5b did not significantly promote the apoptosis of MCF-10A cells (Figures 3A and 3B).
Figure 3.
Apoptosis Assay
Flow cytometry detection of MCF-7 and MCF-10A cell apoptosis induced by AP1-Z1 and its mutants, AP1-Z5a and AP1-Z5b, at a concentration of 5.0 μM. Fluorescent images of ANXA5/PI in MCF-7 and MCF-10a cells treated with AP1-Z1 and its mutants, AP1-Z5a and AP1-Z5b, at a concentration of 5.0 μM. (A) The mean population (±SD) of ANXA5-positive cells. Statistical significance is marked by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001 between two indicated groups. (B) Apoptotic and necrotic populations of cells double-stained with Alexa Fluor-labeled ANXA5and PI are depicted by flow cytometry. (C) Fluorescent images of MCF-7 cells including bright-field images, PI red-fluorescent images, ANXA5 green-fluorescent images, and merged images. (D) Fluorescent images of MCF-10a cells including bright-field images, PI red-fluorescent images, ANXA5 green-fluorescent images, and merged images.
Annexin V labeled with Alexa Fluor 488 can bind to phosphatidylserine (PS) exposed on the outer leaflet of apoptotic cells, which is transferred from the cytoplasmic membrane to the external cellular environment during apoptosis. In comparison with AP1-Z1, the mutants AP1-Z5a and AP1-Z5b, at a concentration of 5 μM, significantly induced the apoptosis of the breast cancer cell line, MCF-7 (Figure 3C). However, there was no significant change in the apoptosis of MCF-10A cells treated with AP1-Z5a and AP1-Z5b (Figure 3D).
Anti-angiogenesis Assay
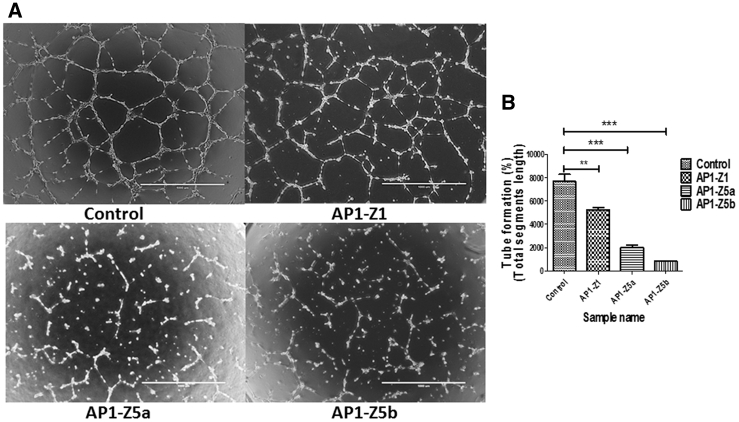
According to the protocol of μ-Slide angiogenesis, human umbilical vein endothelial cells (HUVECs) can form clear blood vessels after 10−12 h at a cell density of 1 × 105 cells/mL. In comparison with the blank control treated with an equal volume of PBS, AP1-Z1 and the mutants AP1-Z5a and AP1-Z5b, at a concentration of 10.0 μM, significantly inhibited angiogenesis, as shown by one-way ANOVA (AP1-Z1, **p < 0.01; AP1-Z5a and AP1-Z5b, ***p < 0.001). Moreover, in comparison with AP1-Z1, the mutants AP1-Z5a and AP1-Z5b significantly inhibited angiogenesis (AP1-Z5a and AP1-Z5b, ***p < 0.001) (Figure 4).
Figure 4.
Anti-Angiogenesis Assay
HUVECs were treated with AP1-Z1 and its mutants, AP1-Z5a and AP1-Z5b, at a concentration of 10.0 μM. (A) The left panel is a representative tube image (bright field) using μ-Slide angiogenesis. (B) The right panel is the quantitation of tube formation using the Angiogenesis Analyzer for ImageJ (*p < 0.05, **p < 0.01, ***p < 0.001 between two indicated groups).
MD Simulations
Structures and Dynamics of Peptide Adsorption
To investigate the selectivity and adsorption behavior of the most effective mutant, AP1-Z5b, on normal and cancer cell membranes, we performed a set of unbiased peptide-membrane simulations. Here, the pure DOPC bilayer represents the normal mammalian cell membrane, while the heterogeneous DOPC/DOPS bilayer represents the anionic cancer cell membrane. For each bilayer type, three 1-μs simulations were run with different starting configurations of the peptide (S1, S2, and S3) (Figure S1); pure membrane simulations were also executed for comparison.
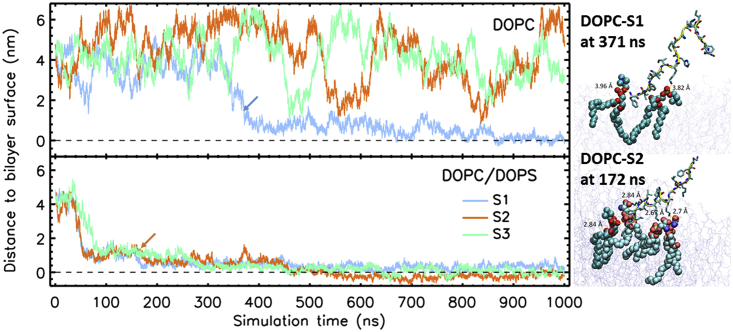
As shown in the summary table of simulation results (Table S2), peptide adsorption was observed in only one of the DOPC simulations, in which the peptide began to associate with the membrane at 371 ns. In contrast, peptide adsorption was observed in all three DOPC/DOPS simulations, with the peptide beginning to associate with the membrane within 50−80 ns, which is much earlier than that observed with the pure DOPC membrane. The formation of uninterrupted hydrogen bonds between the peptide and the membrane was simulated to see Figure S2.
To investigate the process of adsorption of the AP1-Z5b peptide in the two types of membranes, we measured the distance between the peptide and the surface of the membrane as a function of time. Since periodic boundary conditions were applied in the simulations, the peptide can diffuse freely from one leaflet to the other across the water phase. Here, the center of mass (COM) distance of the peptide to the COM of the phosphate group of the closer leaflet was measured. As shown in Figure 5, in the DOPC simulation, the peptide exhibited random diffusion in the water phase at a distance range of 1−6 nm from the membrane. Only in one of the three cases (S1) was the peptide finally stably bound to the membrane. In contrast, in the DOPC/DOPS simulation, the peptide was dragged toward the anionic membrane at a much faster pace, and once arriving at the membrane surface, it bound strongly with the lipids. After a few hundred nanoseconds, the peptide was found embedded at or below the phosphate region of the membrane. In all cases in which adsorption occurred, the peptide inserted into the bilayer via its N terminus.
Figure 5.
Distance (Along the Bilayer) of the Peptide to the Surface of the Membrane
The distance was measured between the center of masses of the peptide and the membrane phosphate atoms of the closest leaflet. Two representative snapshots of the peptide interacting with lipids (<5.0 Å) at initial bilayer insertion. The peptide backbone is outlined as a yellow tube, sidechains are shown as sticks, and proximal lipids are shown as spheres (DOPC in cyan and DOPS in brighter cyan).
By visual inspection, we compared the modes of interaction of the peptide with the two types of membranes. In the DOPC/DOPS simulation, we observed that the AP1-Z5b peptide associated strongly with the lipids. For example, as shown in Figure 5 (molecular images), the peptide in the DOPC/DOPS-S2 simulation bound simultaneously with three DOPS lipids (at their serine or phosphate headgroups) with the sidechains, Lys4, Lys7, and Lys11; all of which maintained a short distance <3.0 Å. Additionally, the N terminus of the peptide was bound to the phosphate group of a DOPC lipid. In contrast, the interaction of the AP1-Z5b peptide with the DOPC membrane was found to be weaker; its initial interaction with the membrane included only two DOPC lipids, with contact distances of almost 4.0 Å.
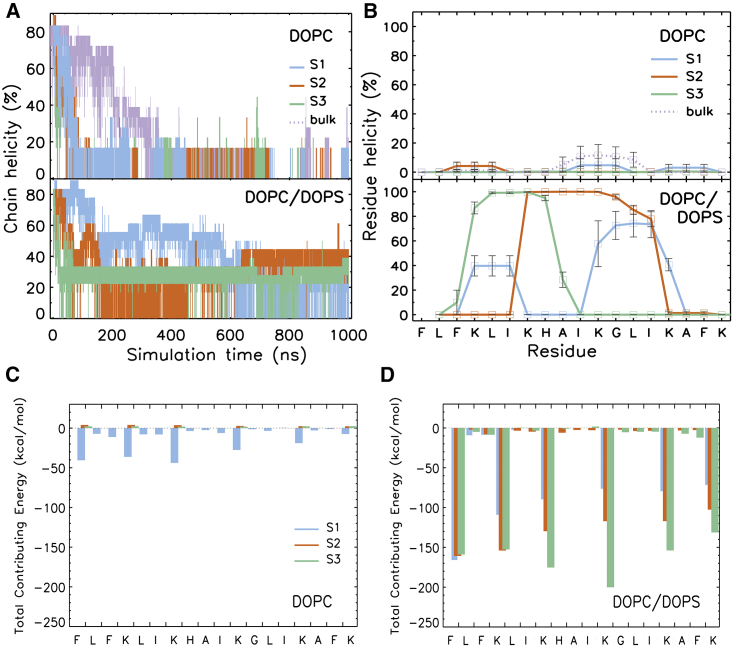
During the simulation, the peptide underwent large structural changes as evidenced by an increase in the root mean square deviation (RMSD) (see Figure S3). The deviation was more pronounced in the DOPC model system (an average of 0.6 nm), with larger fluctuations than in the DOPC/DOPS model system (an average of 0.46 nm). The increased structural stability of the peptide in the latter membrane system is related to its early association with the bilayer and strong binding with the lipids, hence limiting the structural changes to the peptide.
As expected from large RMSDs, the chain helicity of the peptide was reduced over time. As shown in Figures 6A and 6B, at the end of the simulation (800−1,000 ns), the peptide was completely unfolded in the DOPC model system and in the bulk. However, in the DOPC/DOPS simulations, 30−45% of the peptide (equivalent to 5−8 residues) remained helical. The final snapshots of all peptide-membrane simulations are shown in Figure S4.
Figure 6.
The Helicity of AP1-Z5b Observed in the MD Simulations
(A) Total number of residues in the helical structure. (B) Residue-wise helicity from the last 200 ns of simulation. The secondary structure of the peptide was determined using the DSSP program. (C and D) Binding energy contribution of each residue to the peptide-membrane complex.
Energetics of Peptide Adsorption
The strength of the peptide-membrane interaction and the propensity of peptide adsorption in two different membranes can be assessed by analyzing the binding free energy. Table 1 presents the relative binding free energy components of the peptide-membrane complex as calculated using the MM/PBSA method. The results show that electrostatic interaction (ΔGelec) is the major contributor to peptide adsorption in the case of the DOPC/DOPS membrane, whereas polar solvation (ΔGpsolv) is the opposing force. In the DOPC membrane, both polar interactions (ΔGelec and ΔGpsolv) contribute to peptide adsorption but the strength is much weaker. Overall, the AP1-Z5b peptide had a high propensity to adsorb into the anionic DOPC/DOPS membrane, with greater than a 5-fold increase in binding free energy (ΔGbind) as compared with the pure DOPC membrane.
Table 1.
Relative Binding Free Energy Components of the AP1-Z5b-Membrane Complex Averaged from Three Simulations
| Energetic Components | Peptide-DOPC Complex | Peptide-DOPC/DOPS Complex | ΔΔG |
|---|---|---|---|
| ΔGelec | −162.6 | −1754.0 | −1591.4 |
| ΔGpsolv | −116.4 | 110 | 226.4 |
| ΔGvdw | −43.3 | −71.2 | −27.9 |
| ΔGsasa | −5.6 | −10.0 | −4.4 |
| ΔGbind | -327.9 | -1725.2 | -1397.3 |
All energy units are in kcal/mol. The MM/PBSA calculation was performed on 20 snapshots extracted from the last 20 ns of a simulation.
To determine the critical residues for adsorption, we performed per-residue free energy decomposition analysis. Figures 6C and 6D indicate that in the case of the DOPC/DOPS membrane, all lysine residues (Lys 4, 7, 11, 15, and 18) contributed significantly to peptide adsorption. Additionally, the N-terminal residue, which also carries a positive charge, made an equally important contribution as the other charged residues. In the case of the neutral DOPC membrane in which adsorption occurred (DOPC-S1), all charged residues showed contributions, albeit small as compared with those in the anionic DOPC/DOPS simulations.
Effect of Peptide Adsorption on Membrane Structure
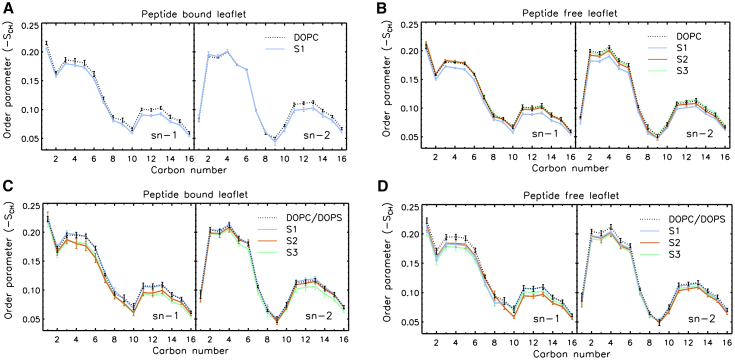
To understand changes in the membrane structure due to peptide adsorption, we compared the properties of the two different membranes, DOPC and DOPC/DOPS, on the bilayer thickness, lateral headgroup orientation, and lipid tail order parameters. As shown in Table S2, the bilayer thickness was reduced upon peptide adsorption. For peptide-adsorbed DOPC (DOPC-S1), the bilayer was thinned by 8 Å as compared with the pure DOPC bilayer; similarly, for peptide-adsorbed DOPC/DOPS (an average of three simulations, S1−S3), the bilayer was thinned by 5.7 Å. Consistently, the headgroups of the peptide-adsorbed bilayers were seen to increase tilting, presumably due to the charge-charge interactions induced by the membrane-bound peptide (by 1.8° in DOPC and 0.7° and 1.8° for DOPC and DOPS in DOPC/DOPS). The adsorbed peptide affected not only the headgroup region of the membrane but also the structure of the hydrophobic core. As shown in Figure 7, when the peptide bound to one leaflet of the bilayer, the SCD values for both leaflets were reduced, suggesting a decrease in the order of the hydrophobic tails.
Figure 7.
The AP1-Z5b (S1, S1 & S3) Adsorption on Membrane Structure in the MD Simulations
(A–D) Deuterium order parameters of the acyl chains in the pure DOPC (A and B) and anionic DOPC/DOPS (C and D) membranes. Mean values were computed from the last 200-ns trajectory with SEs from block averaging displayed as error bars. Pure membrane simulations are displayed as dotted lines.
Discussion
The International Agency for Research on Cancer (IARC) has identified seven major infectious agents that are carcinogenic: the Gram-negative bacterium Helicobacter pylori (H. pylori), hepatitis B virus (HBV), hepatitis C virus (HCV), certain strains of the human papillomavirus (HPV), Epstein-Barr virus (EBV), Human immunodeficiency virus type-1 (HIV-1), and human T cell lymphotropic virus type-1 (HTLV-1).23 The incidence of infection-related cancers is still relatively high, especially in underdeveloped and developing countries.1 Approximately two million new cancer cases are due to infectious pathogens such as bacteria and viruses.24 AMPs are considered the most promising alternative to conventional anti-infection molecules, some of which have been shown to possess dual antimicrobial and anticancer activities. ACPs have an innate advantage over targeted antibody drugs, since ACPs against cancer cells are not affected by tumor heterogeneity.25 In the inner leaflet of the plasma membrane of healthy cells, a negatively charged phospholipid, PS, is present. This asymmetry between the inner and outer membrane leaflets is lost in cancer cells, resulting in the presence of PS in the outer leaflet.26 The presence of PS, heparin sulfate, sialylated gangliosides, and O-glycosylated mucin, combined with increased transmembrane potential, surface area, and membrane fluidity, promotes the specific activity of ACPs toward cancer cells without being affected by tumor heterogeneity.25 The present study selected three kinds of tumor cells: the breast cancer cell line, MCF-7; the malignant melanoma cell line, A375; and the brain glioma cell line, U87, which were treated with the ACPs, AP1-Z1, and its six mutants. A similar trend in the anti-proliferative effects on the different tumor cells was achieved, as shown by the MTT results. At the same time, it was found that the net charge is an important physicochemical parameter that plays a key role in improving anticancer activity and reducing IC50. However, did the anticancer activity increase as the net charge of the peptide increased? The answer is no. AP1-Z5a and AP1-Z5b had a better anticancer activity than did AP1-Z9, which had a net charge of +9.
Although AMPs and ACPs share many characteristics, it remains unclear which physicochemical parameters of ACPs confer anticancer activity. It is generally accepted that most AMPs and ACPs do not have an exact secondary structure when free in solution, but attain an α-helical or β sheet structure when electrostatically interacting with the membrane.27 As observed in the present study, with the exception of AP1-Z3a, the peptides were predominantly in a random coil structure in PBS solution; however, the percentage of α-helical structure was increased in the environment of SUVs simulating the plasma membrane, especially with AP1-Z5b and AP1- Z7.
In our experiments, the AP1 mutants were clearly selective for cancer cells in which the outer leaflet of the membrane contains a higher ratio of negatively charged lipids. To gain molecular insight into the selectivity and membrane binding interaction of the anticancer peptides, we simulated AP1-Z5b in the two membrane types, DOPC (represents the normal mammalian cell membrane) and DOPC/DOPS (represents the anionic cancer cell membrane), and in the bulk. In all the DOPC/DOPS simulations, the peptide quickly associated with the membrane and progressively penetrated the phosphate region of the bilayer. In contrast, the adsorption of the peptide into DOPC was a less frequent event, since it was observed in only one of the three cases. Moreover, it occurred at a much later time in the simulation, indicating that the membrane was not highly attractive to the peptide. Regarding the structure of the peptide, the results of our simulations suggest that AP1-Z5b was mostly a random coil in the bulk solution; however, once it was adsorbed into the bilayer, the peptide remained partially helical. In the DOPC/DOPS bilayer, the middle chain segment stayed as an α helix from residue 4 to 15, and both N- and C-terminal residues adopted a coil structure. In all simulations in which peptide adsorption occurred, the peptide entered the membrane bilayer via its N terminus.
Upon insertion, the peptide orientated parallel to the membrane plane, with its N terminus pointing slightly toward the hydrophobic core and its C terminus pointing toward the bulk. According to the MM/PBSA calculation, the most critical residues in the peptide for binding with membrane lipids were the five lysine residues and the phenylalanine residue at the N terminus. The strength of binding was dependent on the composition of the membrane. In our simulations, the peptide bound to the anionic 3:1 DOPC/DOPS membrane with a 5-fold increased binding free energy as compared with the pure DOPC membrane. The surface-bound peptide exhibited a disruptive effect on the structure of the bilayer; it decreased the order parameters of the acyl chains of the lipid tails, increased the tiling angle of the headgroups, and decreased the thickness of the membrane. These changes in membrane structure are in accordance with those observed in previous electroporation experiments in which an external electric field was applied along the bilayer. Under a strong electric field, water channels were formed.28
In our peptide-membrane MD, although each of the simulations was run up to 1 μs simulation time (with a total of 7 μs simulation time), neither penetration of the peptide across the membrane nor formation of water channels was observed. Previous studies of other cationic AMPs over shorter or longer timescales using MD simulations have also been unable to observe membrane penetration.29, 30, 31 This may be due to the longer transition time required for the penetration event to happen, or more likely that the penetration event requires a number of peptides to act together, for example to form aggregates and transmembrane pores. Studying the complexation of dimers, trimers, or multimers is a challenging task with fully atomic simulations, which will require sampling over many multi-μs timescales or the use of enhanced sampling techniques.29
Natural ACPs with higher anticancer activity usually have a sequence of more than 30 amino acids, which greatly increases the cost of synthetic production.11 Currently, only a small number of ACPs enter clinical trials, such as ITK-1 (Green Peptide), ACG-1005 (AngioChem), and MBI-226 (Cadence Pharmaceuticals), since the synthesis cost is higher than that of organic small molecule drugs. At present, automatic peptide synthesizers can synthesize peptides less than 30 amino acids in length; however, those of more than 30 amino acids require greater human input and material resources. Therefore, the relatively low-cost, short peptide ACPs have an effective anticancer activity, which is an urgent problem to be solved. The present study selected the 18-amino-acid AP1-Z1 as a template to explore the physicochemical parameters that confer anticancer activity. The combination of bioinformatics algorithms and machine learning is considered one of the most important methods for the future rational design of peptides and protein drugs. These strategies mainly consider improvements in the physical and chemical properties such as amphiphilicity, hydrophobicity, and overall positive charge, with the purpose of efficiently designing more active drugs. Our peptides were designed using this method with a view to understanding the differences in physicochemical properties between AMPs and ACPs.
Conclusions
From the current literature, ACPs possess the following anticancer mechanisms: recruitment of immune cells, such as dendritic cells, to kill tumor cells, induction of necrosis or apoptosis of cancer cells, inhibition of angiogenesis to eliminate tumor nutrition and prevent metastasis, and inhibition or activation of certain regulatory functional proteins to interfere with gene transcription and translation.32 Analysis of the results shows that AP1-Z5a and AP1-Z5b induced apoptosis and anti-angiogenesis dual effects to achieve maximum anticancer activity than the other mutants. Both charge and hydrophobicity play an important role in optimizing the anticancer activity of ACPs. To improve the anticancer activity, the net charge and hydrophobicity do not need to be as high as possible; instead, they mutually restrict and influence to reach an equilibrium that achieves better anticancer activity. Thus, ACPs have great potential to become a next-generation anticancer drug with low drug resistance and side effects.
Materials and Methods
Cell Lines
The breast cancer cell line (MCF 7), malignant melanoma cell line (A375), brain glioma cell line (U87), human mammary epithelial cell line (MCF 10A), and HUVECs were obtained from ATCC. The A375 and U87 cancer cell lines were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin. HUVECs were maintained in Medium 200 with the addition of low serum growth supplement (LSGS). The MCF 7 cell line was maintained in DMEM supplemented with 5% (v/v) FBS. The MCF 10A cell line was maintained in DMEM/F12 supplemented with 5% horse serum, 1% penicillin/streptomycin, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 20 ng/mL recombinant human EGF, and 100 ng/mL cholera toxin. The MCF 10A cell line was employed in the screening assays as a control, i.e., a normal human cell line. HUVECs were used in the angiogenesis assay. All cells were seeded onto 75 cm2 culture flasks (Falcon, Fisher Scientific) and cultured at 5% CO2, 37°C. All laboratory reagents were of analytical grade or better.
Methods
Construction of the Peptides and Model Prediction
The mutants of the antimicrobial peptide AP1-Z1 were designed by HeliQuest.22 HeliQuest screens the SwissProt database to identify protein fragments of similar characteristics based on the physicochemical properties and amino acid composition of the input sequence. The server is divided into a sequence analysis module and a screening module, wherein the sequence analysis module has a mutation submodule that allows the user to mutate helices manually or automatically using a genetic algorithm to produce a mutant with specific properties. To increase the net charge and hydrophobicity of the AP1-Z1 target sequence, we mutated the polar, uncharged residues, His, Ser, and Gly, etc., to the charged residue, Lys. All peptides were synthesized by GenScript Nanjing. To improve chemical stability, we amidated all peptides at the N terminus.
The Monte Carlo simulation model was used to simulate the peptide-membrane interactions with the MCPep web server.33 The parameters were as follows: membrane hydrophobic thickness, 30 Å; fraction of charged lipids, 30%; concentration of monovalent salts, 0.15 M; number of independent MC runs, 3; number of MC cycles in each independent run, 500,000; and root mean square deviation (RMSD) cut-off for clustering of resultant conformations, 3 Å.
CD Spectroscopy
Small unilamellar vesicles (SUVs) were prepared according to the following process. The lipid mixture (palmitoyloleoyl phosphatidylethanolamine, POPE; palmitoyloleoyl phosphatidyl-glycerol, POPG) POPE/POPG (7:3) was dissolved in chloroform and dried under N2 overnight in a glass tube. The dried lipid powder was resuspended in 1× PBS buffer (pH 7.4) at room temperature. The resulting multi-lamellar vesicle suspension was press-filtered through a liquid extruder (Northern Lipids) equipped with a 0.1-μm pore size Whatman Nuclepore hydrophilic membrane to produce homogeneous liposomes (SUVs), which were used within 6 h of production.
CD spectra were recorded on a Chirascan Circular Dichroism Spectrometer. The AP1-Z1 and its six mutants (AP1-Z3a, AP1-Z3b, AP1-Z5a, AP1-Z5b, AP1-Z7, and AP1-Z9) were dissolved in 1× PBS, pH 7.4 at a concentration of 0.5 mM, and the negative control was treated with equal volumes of PBS. Spectra were run at 25°C from 250 to 190 nm using a quartz cell that was 0.1 mm in length. Data were collected at 1-nm intervals at a scan rate of 60 nm/min. All CD spectra were the average of three scans, and the final spectrum was corrected by subtracting the corresponding baseline spectrum obtained under identical conditions. The secondary structure content was estimated by CDNN 2.1 standard analysis.
Anti-Proliferative Assay: MTT Assay and Anti-Proliferative Curve
Cells were plated into 96-well microplates at a density of 5,000 cells/well and incubated for 24 h. The peptide stock solutions were prepared at a concentration of 10 mM and further diluted in the corresponding medium to obtain a final concentration. The growth medium containing peptides of different concentrations was added to the cells, and the negative control was treated with equal volumes of PBS. Each sample was prepared in triplicate. Following a 24-h incubation, 10 μL 5 mg/mL MTT solution (in 0.01 M sterile PBS, pH 7.4) was added to each well, and the plate was incubated for an additional 4 h at 37°C, 5% CO2. After removal of the medium, 100 μL/well DMSO was added to each well and the plate was shaken for 10 min to dissolve the formazan product. The OD of each well was measured at 550 nm using a PerkinElmer VICTOR X3 multimode plate reader.
Cells were plated onto 96-well microplates at a density of 3,000 cells/well and incubated for 18−24 h. The IncuCyte ZOOM Continuous Live-Cell Imaging & Analysis System was employed to record cell growth. The instrument can automatically generate cell growth curves, which are calculated from the cell area attached to the bottom of each well according to images taken by the instrument every 3 h. The peptide stock solutions were prepared at a concentration of 10 mM and further diluted in the corresponding medium to obtain a final concentration. The growth medium containing peptides of different concentrations was added to the cells, and the negative control was treated with equal volumes of PBS. Each sample was prepared in triplicate.
Apoptosis Assay
Flow Cytometry: Fluorescence-Activated Cell Sorting (FACS) Assay
An Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit (cat. no. V13245, Invitrogen) was used to study apoptosis following treatment with the peptides. A volume of 2.5 mL cell suspension/well was seeded onto a 6-well plate at a density of 2 × 105 cells/mL. Following culture for 24 h, an optimum concentration of peptides was added to the cells, and the negative control was treated with equal volumes of PBS. Cells were harvested after 1−3 h, washed twice in cold 1× PBS, pH 7.4, centrifuged, and resuspended in 1× Annexin-binding buffer. Cell density was determined and diluted to 1 × 106 cells/mL in 1× Annexin-binding buffer to prepare a sufficient volume for 100 μL per well. Volumes of 5 μL Alexa Fluor 488 Annexin V and 1 μL 100 μg/mL PI working solution were added to each 100 μL cell suspension. The cells were incubated for 10 min at room temperature, after which 400 μL 1× Annexin-binding buffer was added, gently mixed, and the samples were stored on ice. The stained cells were analyzed using a BD Accuri C6 flow cytometer, and the fluorescence emission at 530 nm and 575 nm was measured following excitation at 488 nm.
PI and Annexin V Staining
A volume of 250 μL cell suspension/well was seeded onto a 48-well plate at a density of 2 × 104 cells/well. Following culture for 24 h, an optimum concentration of peptides was added to the cells, and the negative control was treated with equal volumes of PBS. After 1−3 h, Alexa Fluor 488 Annexin V (ANXA5) and PI were added to each well at a ratio of 1:250 and 1:1,000 (V/V), respectively, and the cells were incubated for 30 min at 37°C. The stained cells were analyzed using EVOS FL Auto Cell Imaging System.
Anti-angiogenesis Assay
Matrigel was thawed slowly overnight in an icebox at 4°C. A volume of 10 μL gel was added to each well of the μ-Slide, which was placed in a Petri dish containing paper towel soaked in water for additional humidity, and the entire assembly was then placed in an incubator for polymerization for 30−60 min. During this time, the cell suspension was prepared at a concentration of 1 × 10 5 cells/mL and 50 μL was added to each well. The assembly was incubated at 37°C, 5% CO2 for 10−12 h and subsequently analyzed using an EVOS FL Auto Cell Imaging System.
MD Simulations
System Construction and Simulation Conditions
Fully atomistic MD simulations were performed to gain further insight into the behavior of the most effective AP1 mutant, AP1-Z5b, in cell membranes. Two membrane models were constructed to simulate the different physiochemical properties of normal mammalian and cancer cell membranes. The former was represented by a model of 102 pure zwitterionic DOPC lipids and the latter by a 3:1 mixture of zwitterionic DOPC and anionic DOPS lipids. NaCl ions were added to neutralize the systems and to reach physiological conditions of 150 mM. All membrane models were constructed using the Membrane Builder module of the CHARMM-GUI server to obtain the correct topology and force field files for the systems.34 The systems were energy minimized and equilibrated with and without restraints following the procedures suggested by CHARMM-GUI.
The initial configuration of the peptide (FLFKLIKHAIKGLIKAFK) was generated using the PEP-FOLD server (version 3.5).35 Under experimental conditions at pH 7, the side chains of K4, K7, K11, K15, and K18 were protonated (charge +1), and that of H8 was neutral. The N terminus of the peptide was capped with a positively charged NH3+ group, and the C terminus was amidated such that it was neutral. The CHARMM-GUI generated peptide system was first equilibrated in solvent for 200 ns, then the trajectory was subjected to clustering analysis to select the three most representative peptide conformations for subsequent peptide-membrane simulations (see Figure S1). To minimize biasing the simulation results by initial peptide conformations, we inserted the peptide into the middle of the water phase with a distance of at least 3 nm to the membrane surface. All peptide-membrane systems were first energy minimized using the steepest descent algorithm, and equilibrated for 50 ps using the NVT ensemble followed by 300 ps using the NPT ensemble with position restraints on the protein atoms. Finally, for production runs, all restraints were removed, and trajectory data were collected every 2 ps.
MD simulations were performed using the GROMACS 5.0.7 package,36 with the CHARMM36m force field37 and the TIP3P water model.38 All production simulations were run with the following parameters. The leapfrog algorithm was used to integrate the Newton’s equations of motion with a time step of 2 fs. Periodic boundary conditions were applied in all directions. Long-range electrostatic interactions were computed using particle-mesh Ewald (PME),39 and a distance cut-off of 1.2 nm was used for the short-range electrostatic and van der Waals interactions. Hydrogen atoms were restrained using the LINCS algorithm.40,41 The temperature was maintained at 310 K by a Nose-Hoover thermostat and the pressure at 1 atm by a Parrinello-Rahman barostat with semi-isotropic coupling for membrane systems and isotropic coupling for peptide-only systems.
The list of membrane and peptide-membrane simulation systems is presented in Table S3. The systems of three different initial conformations of the peptide are denoted as S1, S2, and S3. The six peptide-membrane systems were run for 1 μs each, and the two membrane-only systems were run for 200 ns each. Additionally, the peptide-only system was run for 1 μs. Together, these yielded a total simulation time of 7.4 μs.
Binding Free Energy Calculation
To investigate the binding energy of AP1-Z5b in the two types of membranes, we used the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method via the g_mmpbsa program,42 which is a widely used program to estimate the binding free energy of protein-ligand and protein-protein complexes. Here, the binding free energy of a peptide-membrane complex was estimated using the following equation:
where , , and are the total free energy if the peptide-bilayer complex, peptide, and bilayer, respectively. The total free energy of each component was in turn calculated using the following equation:
where and are the electrostatic energy, based on the Coulomb potential, and van der Waals energy, based on the Lennard-Jones potential, respectively. Both energies were computed using the CHARMM36m force field. The polar solvation free energy, , was estimated by solving the nonlinear Poisson-Boltzmann equation using dielectric constants of 1 for the vacuum, 7 for the membrane, and 80 for the solvent, with an ionic strength of 0.15 M. The nonpolar solvation free energy, , was estimated using the solvent-accessible surface area (SASA) model, with a surface tension constant of 0.0226778 kJ/mol/Å2, probe radius of 1.4 Å, and offset of 3.84928 kJ/mol. For each peptide-membrane production simulation, 100 snapshots were extracted from the last 20 ns trajectory to compute the binding free energy.
Structure Analysis of Peptide and Membranes
Standard analysis, such as RMSD, box size, and peptide-membrane distance, were performed using the tools provided in the GROMACS package. The DSSP program43 was used for peptide secondary structure analysis (hence, the peptide helicity). Membrainy44 was employed for membrane property analysis, including membrane thickness, headgroup orientation, and lipid order parameters. The bilayer thickness was measured as a distance between the COMes of the phosphorous atoms in the two leaflets; the headgroup orientation was computed as an angle between the headgroup vector (P-N) and the membrane normal; and the lipid tail order parameters were computed from the C-H bond vectors and the membrane normal. Plots were created using IDL 8.4.1, and molecular images were created using VMD 1.9.2.45
Author Contributions
R.M. and S.W.W. carried out the experiments, while R.M., L.G., and H.F.K. designed the experiments; S.W.I.S. and R.M. analyzed and constructed the calculation model; R.M., S.W.I.S., C.S., and H.F.K. drafted and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was funded by the Science and Technology Development Fund, Macau SAR (file number 019/2017/A1). R.M. was in receipt of a PhD studentship from the Science and Technology Development Fund (FDCT) and the Faculty of Health Science (FHS), University of Macau. All simulations were performed at the High-Performance Computing Cluster (HPCC) provided by the Information and Communication Technology Office (ICTO) of the University of Macau. We would like to thank Dr. Xiaohui Hu for his guidance and advice on the circular dichroism experiment.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.12.001.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Smith L.L., Brown K., Carthew P., Lim C.K., Martin E.A., Styles J., White I.N. Chemoprevention of breast cancer by tamoxifen: risks and opportunities. Crit. Rev. Toxicol. 2000;30:571–594. doi: 10.1080/10408440008951120. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y.S., Li D., Shi H.S., Wen Y.J., Yang L., Xu N., Chen X.C., Chen X., Chen P., Li J. Intratumoral expression of mature human neutrophil peptide-1 mediates antitumor immunity in mice. Clin. Cancer Res. 2009;15:6901–6911. doi: 10.1158/1078-0432.CCR-09-0484. [DOI] [PubMed] [Google Scholar]
- 5.Verma S., Miles D., Gianni L., Krop I.E., Welslau M., Baselga J., Pegram M., Oh D.Y., Diéras V., Guardino E., EMILIA Study Group Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy K.V., Yedery R.D., Aranha C. Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Hancock R.E.W., Haney E.F., Gill E.E. The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 2016;16:321–334. doi: 10.1038/nri.2016.29. [DOI] [PubMed] [Google Scholar]
- 8.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals (Basel) 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee T.H., Hall K.N., Aguilar M.I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016;16:25–39. doi: 10.2174/1568026615666150703121700. [DOI] [PubMed] [Google Scholar]
- 11.Felício M.R., Silva O.N., Gonçalves S., Santos N.C., Franco O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front Chem. 2017;5:5. doi: 10.3389/fchem.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 13.Xu H., Chen C.X., Hu J., Zhou P., Zeng P., Cao C.H., Lu J.R. Dual modes of antitumor action of an amphiphilic peptide A(9)K. Biomaterials. 2013;34:2731–2737. doi: 10.1016/j.biomaterials.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 14.Hancock R.E.W., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 15.Ma R., Mahadevappa R., Kwok H.F. Venom-based peptide therapy: insights into anti-cancer mechanism. Oncotarget. 2017;8:100908–100930. doi: 10.18632/oncotarget.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Q., Hou X., Ge L., Li R., Zhou M., Wang H., Wang L., Wei M., Chen T., Shaw C. Cationicity-enhanced analogues of the antimicrobial peptides, AcrAP1 and AcrAP2, from the venom of the scorpion, Androctonus crassicauda, display potent growth modulation effects on human cancer cell lines. Int. J. Biol. Sci. 2014;10:1097–1107. doi: 10.7150/ijbs.9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B., Lyu P., Xi X., Ge L., Mahadevappa R., Shaw C., Kwok H.F. Triggering of cancer cell cycle arrest by a novel scorpion venom-derived peptide-Gonearrestide. J. Cell. Mol. Med. 2018;22:4460–4473. doi: 10.1111/jcmm.13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almaaytah A., Zhou M., Wang L., Chen T., Walker B., Shaw C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides. 2012;35:291–299. doi: 10.1016/j.peptides.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Dennison S.R., Whittaker M., Harris F., Phoenix D.A. Anticancer α-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes. Curr. Protein Pept. Sci. 2006;7:487–499. doi: 10.2174/138920306779025611. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y.C., Lim Y.F., Russo E., Schneider P., Bolliger L., Edenharter A., Altmann K.H., Halin C., Hiss J.A., Schneider G. Multidimensional Design of Anticancer Peptides. Angew. Chem. Int. Ed. Engl. 2015;54:10370–10374. doi: 10.1002/anie.201504018. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y.B., Wang X.F., Wang H.Y., Liu Y., Chen Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol. Cancer Ther. 2011;10:416–426. doi: 10.1158/1535-7163.MCT-10-0811. [DOI] [PubMed] [Google Scholar]
- 22.Gautier R., Douguet D., Antonny B., Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 23.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 24.Vedham V., Divi R.L., Starks V.L., Verma M. Multiple infections and cancer: implications in epidemiology. Technol. Cancer Res. Treat. 2014;13:177–194. doi: 10.7785/tcrt.2012.500366. [DOI] [PubMed] [Google Scholar]
- 25.Kelly G.J., Kia A.F.A., Hassan F., O’Grady S., Morgan M.P., Creaven B.S., McClean S., Harmey J.H., Devocelle M. Polymeric prodrug combination to exploit the therapeutic potential of antimicrobial peptides against cancer cells. Org. Biomol. Chem. 2016;14:9278–9286. doi: 10.1039/c6ob01815g. [DOI] [PubMed] [Google Scholar]
- 26.Leite N.B., Aufderhorst-Roberts A., Palma M.S., Connell S.D., Ruggiero Neto J., Beales P.A. PE and PS Lipids Synergistically Enhance Membrane Poration by a Peptide with Anticancer Properties. Biophys. J. 2015;109:936–947. doi: 10.1016/j.bpj.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoskin D.W., Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siu S.W.I., Böckmann R.A. Electric field effects on membranes: gramicidin A as a test ground. J. Struct. Biol. 2007;157:545–556. doi: 10.1016/j.jsb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Zhao T., Wei D., Strandberg E., Ulrich A.S., Ulmschneider J.P. How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim. Biophys. Acta. 2014;1838:2280–2288. doi: 10.1016/j.bbamem.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Lee J., Jung S.W., Cho A.E. Molecular Insights into the Adsorption Mechanism of Human β-Defensin-3 on Bacterial Membranes. Langmuir. 2016;32:1782–1790. doi: 10.1021/acs.langmuir.5b04113. [DOI] [PubMed] [Google Scholar]
- 31.Sahoo B.R., Fujiwara T. Membrane Mediated Antimicrobial and Antitumor Activity of Cathelicidin 6: Structural Insights from Molecular Dynamics Simulation on Multi-Microsecond Scale. PLoS ONE. 2016;11:e0158702. doi: 10.1371/journal.pone.0158702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D., Gao Y., Qi Y., Chen L., Ma Y., Li Y. Peptide-based cancer therapy: opportunity and challenge. Cancer Lett. 2014;351:13–22. doi: 10.1016/j.canlet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Gofman Y., Haliloglu T., Ben-Tal N. Monte Carlo simulations of peptide-membrane interactions with the MCPep web server. Nucleic Acids Res. 2012;40:W358–W363. doi: 10.1093/nar/gks577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 35.Maupetit J., Derreumaux P., Tufféry P. A fast method for large-scale de novo peptide and miniprotein structure prediction. J. Comput. Chem. 2010;31:726–738. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- 36.Mark J.A., Teemu M., Roland S., Szilárd P., Jeremy C.S., Berk H. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 37.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 39.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 40.Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 41.Hess B., Bekker H., Berendsen H.J.C., Fraaije J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 42.Kumari R., Kumar R., Lynn A., Open Source Drug Discovery, C.; Open Source Drug Discovery Consortium g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 44.Carr M., MacPhee C.E. Membrainy: a ‘smart’, unified membrane analysis tool. Source Code Biol. Med. 2015;10:3. doi: 10.1186/s13029-015-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14 doi: 10.1016/0263-7855(96)00018-5. 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.