Abstract
BACKGROUND: High grade glioma molecular profiling is of particular interest in neurooncology. The role of telomerase reverse transcriptase (TERT) varies dependent upon other molecular parameters. We explored the role of TERT in 101 high-grade gliomas. METHODS: A total of 101 patients (pts) with grade III–IV gliomas treated with standard of care and informative tumor genotypes were included in the present study. Of 55 genes targeted with the next-generation sequencing panel, mutations (muts) were found in 37; these were included in the analysis. TERT mut were tested with Sanger sequencing. MGMT promoter methylation status was determined by methylation specific PCR. RESULTS: 270 mut were detected in 92/101 tumors (91.1%). TERT was the most frequently mutated gene (74.3%). IDH1/2 mut were mutually exclusive with mut in the neurofibromin 1 (NF1) gene. Mutated TERT was associated with wild-type (wt) IDH1/2 (p = 0.025). The 12-month overall survival (OS) rate was 74.3% (median OS: 22 months). Pts with TERT and NF1 wt had a median OS of 40.8 months, while among pts with NF1 wt/TERT mutant, the median OS was 18.5 months. NF1 and TERT mut univariately conferred shorter OS (HR = 3.19; p = 0.004 and HR = 2.28; p = 0.002). Upon multivariate analysis, mutated TERT showed marginal unfavorable prognostic significance for OS (p = 0.049), while NF1 lost its unfavorable significance (p = 0.151). CONCLUSIONS: TERT is herein proven to confer poor prognosis in high grade gliomas, independent of IDH and MGMT. NF1 seems to also confer poor prognosis although our small numbers do not allow for firm conclusions.
Introduction
High-grade gliomas are the most common primary central nervous system tumors, and their classification by the World Health Organization has recently undergone a major revision [1,2]. This revision is based on the improved understanding of gliomagenesis and the molecular determinants of glioma behavior. The most clearly defined role is that of IDH mutations (muts), which appear to be the hallmark of lower grade disease and improved prognosis [3]. Oligodendrogliomas are now molecularly defined by the simultaneous presence of IDH muts and codeletion of chromosomal arms 1p and 19q. Furthermore, 1p/19q codeletion has predictive value in terms of response to specific chemotherapy regimens [4], while mutant IDH is becoming the target of new experimental agents [5]. The molecular profiling of gliomas has recently incorporated telomerase reverse transcriptase (TERT) promoter muts as a major determinant of prognosis with opposite effect in low- versus high-grade disease [6].
Neurofibromin 1 (NF1) is a tumor suppressor gene that codes for neurofibromin [7]. Inactivating somatic NF1 muts have also been described to play a significant role in gliomagenesis [8] with preclinical models showing its critical role in combination with p53 [9]. Furthermore, in The Cancer Genome Atlas (TCGA) data, NF1 has been shown to be associated with the mesenchymal subtype of glioblastoma (GBM) [10]. Other biomarkers are being evaluated, whose place in the clinical management of patients (pts) is still not fully understood. Several attempts have been made to include O-6-methylguanine-DNA methyltransferase (MGMT) methylation, TERT promoter, ATRX, and IDH1/2 muts, as well as 1p19q [[11], [12], [13]] in classifications that reflect different glioma prognostic groups and may have therapeutic implications.
Recent technological advances allow a more in-depth study of the molecular profile of glioma with next generation sequencing (NGS) on formalin fixed paraffin-embedded (FFPE) tissue. This has led to a number of studies on the prognostic significance of these parameters with variable results [14,15]. In an attempt to further explore the prognostic role and interrelation of some known biomarkers, we examined the molecular profile of 101 high grade gliomas by NGS and the TERT promoter status by Sanger sequencing. Lastly, the MGMT promoter methylation is a well-known epigenetic event that affects the prognosis of gliomas, is associated with IDH mut and the methylator phenotype and, most importantly, with response to alkylator therapy [12,16]. We had MGMT promoter methylation data on 77 of 101 pts, and we included those in our analysis.
Patients and Methods
One hundred and twenty-six pts with high-grade gliomas and available FFPE tissue samples treated in 2 centers of the Hellenic Cooperative Oncology Group (HeCOG) were registered. Clinical data were retrieved from the HeCOG data office (Athens, Greece). Tumor paraffin blocks were retrieved from the HeCOG biorepository after pt informed consent. We examined 93 blocks from primary tumor and 8 from recurrent tumor. Tumor blocks were stored and centrally processed in the Laboratory of Molecular Oncology (MOL; Hellenic Foundation for Cancer Research/Aristotle University of Thessaloniki) for histology review, including confirmation of tumor tissue on the section; comparison to local typing; Ki67 labeling; histologic grade; areas with necrosis; hemorrhagic infiltrates; microvascular proliferation; assessment of tumor cell content; and selection of tumor areas for macrodissection. The translational protocol was approved by the Bioethics Committee of the Aristotle University of Thessaloniki School of Medicine (No 2/February 4, 2015). MGMT testing was performed at GeneKor Medical SA, Athens, Greece.
Tissue Processing and NGS Genotyping
Tumor dense areas were marked on H&Es and macrodissected manually from 10 μm unstained FFPE sections prior to DNA extraction with the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany), according to standard procedures. DNA concentration was measured with the Qubit fluorometer (Life Technologies, Paisley, UK); 114/126 (90.5%) tumor DNA samples with concentration ≥ 2 ng DNA/μl were processed for NGS.
Tumor genotyping was performed at the MOL with an NGS panel for the most prevalent muts described by TCGA in GBMs [17,18] and with Sanger sequencing for mut hotspots in the TERT promoter [19]. For the panel design, genomic coordinates of targeted regions, based on the GRCh37 assembly, were submitted to the Ampliseq pipeline (Thermo Fisher Scientific/Ion Torrent) to design primers that yield amplicons up to 175 bp in length, adapted for FFPE DNA. Primer specificity was further evaluated with the NCBI's BLAST tool. The applied IAD68363_167 custom Ampliseq panel spanned a total sequence of 31.6 kb and targeted muts in 55 genes (Supplemental Table S1). Following library construction, NGS genotyping was implemented in an Ion Personal Genome Machine (Thermo Fisher/Ion Torrent). Data retrieval, base calling, and the generation of sequence reads were performed on the Torrent Server with Torrent Suite v.4.4.2, followed by adapter sequence trimming, read alignment to the human reference genome, and variant calling. Resulting variants were annotated by Ion Reporter v.4 and accepted for analysis on quality filtering with the following eligibility criteria: p value < 0.0001 (Ion Torrent metric for variant read quality); >100 amplicon reads; position coverage >100; and variant coverage >40 for worse accepted variants (100 reads). Indels involving GC stretches and nonannotated variants were excluded from analysis. With the applied panel, the cutoffs for sample eligibility were set at mean depth 150 and number of variants ≥5. For samples without muts, the cutoff for sample eligibility was set at 300.
Based on the above, 101/114 (88.6%) FFPE tumors from an equal number of pts were informative for analysis. For these samples, mean/median values for mean read depth were 435/305 (range: 185–3524); mean/median values for the number of eligible variants were 23/22 (range: 5–206). Eligible variants were called muts if these were amino acid or splice-site changing, and if minor allele frequency (MAF) < 0.1% based on dbSNP, 5000 Exomes, and ExAC (http://exac.broadinstitute.org/).
Screening of tumors for TERT promoter muts C228T and C250T, located at positions 1,295,228 and 1,295,250 (GRCh37 assembly) of chromosome 5, was accomplished with nested PCR, using two appropriate primer pairs targeting a 201 bp region within the TERT locus: an outer primer pair (forward 5′-ACCCGTCCTGCCCCTTCACCT-3′ and reverse 5′ CCAGCGGCAGCACCTCGCGGTA-3′) and a nested M13-coupled primer pair (forward 5′-ACCCGTCCTGCCCCTTCACCT-3′ and reverse 5′-CGGGGCCAGGGCTTCCCAC-3′). PCR reactions were performed in a GeneAmp PCR system as follows: 95 °C for 10 min; 18 or 28 cycles (1st or 2nd round PCR, respectively) of denaturation at 95 °C for 30 s; annealing at 68 °C for 30 s; and primer extension at 72 °C for 1 min. For the 2nd PCR, a final extension step at 72 °C for 10 min was added. Sense and antisense sequencing was accomplished with the Big Dye Terminator kit v.1.1 (Applied Biosystems, Foster City, USA), followed by capillary electrophoresis in an ABI3130XL genetic analyzer. Sequence data were visualized and called with the Sequencing Analysis software v5.2.
MGMT methylation detection was also carried out on FFPE tissue. Methylation pattern in the CpG island of MGMT was determined by chemical modification of unmethylated, but not methylated, cytosines to uracil, using the EpiTect Bisulfite Kit (Qiagen). Methylation-specific PCR (MSP) was performed with primers specific for either modified methylated or unmethylated DNA as described previously [20]. MSP reactions were carried out in duplicates. A methylation-positive sample and a negative control were included in each experiment.
Statistical Analysis
Continuous variables are presented as medians with the corresponding range and categorical variables as frequencies with the respective percentages. Chi-square or Fisher's exact test (where appropriate) was used for comparisons of categorical variables, while the nonparametric Wilcoxon rank-sum test was performed to detect differences between categorical and continuous variables.
Overall survival (OS) was measured as the time interval from the date of diagnosis to the date of pt's death or last contact (whichever occurred first). Surviving pts were censored at the date of their last contact. Survival curves were produced using the Kaplan-Meier method and compared across groups with the log-rank test. All parameters were tested for proportionality using time-dependent covariates. Univariate and multivariate Cox regression analyses were performed to assess the association between factors of interest (see below) and mortality rates.
In the entire cohort, a multivariate Cox proportional hazard regression model was applied, including age, TERT mutational status (mutated vs. wild-type [wt]), NF1 mutational status (mutated vs. wt), IDH1/2 mutational status (mutated vs. wt), and MGMT promoter methylation status (methylated vs. unmethylated), parameters that showed statistical significance in the univariate analyses (p < 0.005).
Survival analysis was additionally performed in the subgroup of pts with wt IDH1/2 tumors. Similar to the analyses conducted for the entire cohort, we assessed the impact of TERT muts upon adjustment for all parameters that showed significance or revealed a trend towards significance (p < 0.10) in the univariate analysis (age, NF1 mutational status, TP53 mutational status, number of mutated genes per tumor, and MGMT promoter methylation status).
All tests were two-sided at an alpha 5% level of significance. No adjustment for multiple comparisons was performed. The SAS version 9.3 (SAS Institute) was used for the statistical analysis, and the R studio version 3.5.0 was used to produce survival plots and maps with the mut pattern of genes.
Results
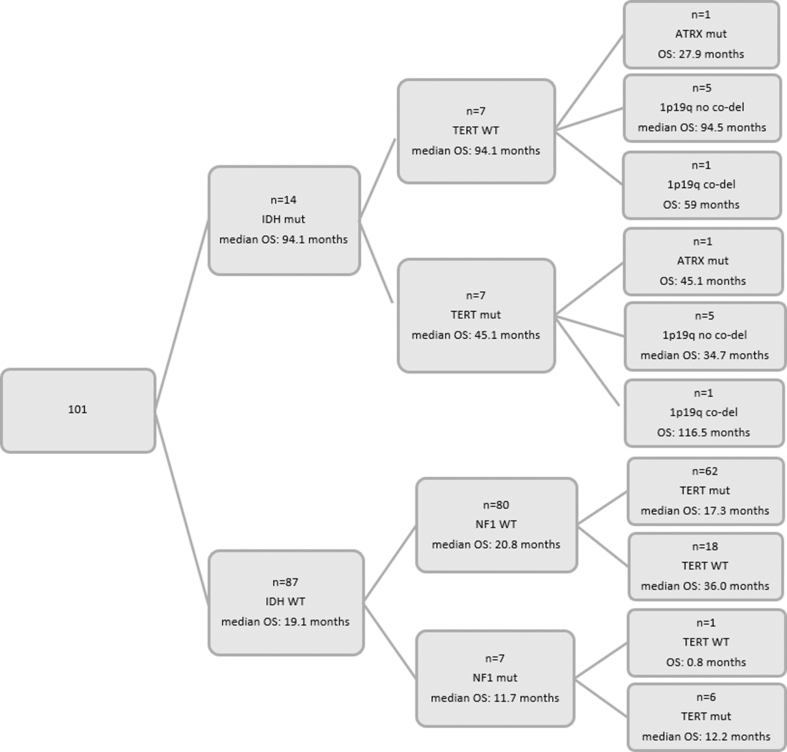
A total of 101 pts with grade III–IV gliomas were included in the current study (Figure 1). Selected pt and tumor characteristics for the entire study population and by grade are presented in Table 1. In total, 85 pts were centrally assessed as grade IV gliomas, while the rest of the pts (15.8%) had grade III tumors. The median age at diagnosis was 54.7 years, with pts with grade IV tumors being diagnosed at an older age compared with those with grade III gliomas (median age: 56.9 [21.3–78.2] vs. 48.3 [27.4–62.9], Wilcoxon rank-sum p = 0.004). Most pts were males (58.4%) and had undergone subtotal tumor excision (59.4%). Four pts had anaplastic oligodendrogliomas (AOs), and one pt had a high grade astroblastoma with areas of astrocytoma as assessed by an experienced pathologist (E.V). Of note, only two out of the four histologically defined AOs were found to carry a codeletion of chromosomal arms 1p and 19q.
Figure 1.
REMARK diagram.
Table 1.
Selected Patient and Tumor Characteristics by Tumor Grade.
| Total (N = 101) | Grade III (N = 16) | Grade IV (N = 85) | ||||
|---|---|---|---|---|---|---|
| Age at Diagnosis | ||||||
| Median (minimum–maximum) | 54.7 (21.3–78.2) | 48.3 (27.4–62.9) | 56.9 (21.3–78.2) | |||
| Ki67 | ||||||
| N | 100 | 16 | 84 | |||
| Median (minimum–maximum) |
18.0 (0.0–80.0) |
15.0 (0.0–80.0) |
19.0 (0.0–70.0) |
|||
|
N |
% |
N |
% |
N |
% |
|
| Sex | ||||||
| Female | 42 | 41.6 | 4 | 25.0 | 38 | 44.7 |
| Male | 59 | 58.4 | 12 | 75.0 | 47 | 55.3 |
| Type of Surgery | 23 | 22.8 | 4 | 25.0 | 19 | 22.4 |
| Biopsy (<75% of the tumor)/stereotactic biopsy | ||||||
| Subtotal (75–99% of the tumor) | 60 | 59.4 | 9 | 56.3 | 51 | 60.0 |
| Total excision | 13 | 12.9 | 1 | 6.3 | 12 | 14.1 |
| Unknown | 5 | 5.0 | 2 | 12.5 | 3 | 3.5 |
| Necrosis | ||||||
| No | 19 | 18.8 | 14 | 87.5 | 5 | 5.9 |
| Yes | 82 | 81.2 | 2 | 12.5 | 80 | 94.1 |
| Hemorrhage | ||||||
| No | 23 | 22.8 | 12 | 75.0 | 11 | 12.9 |
| Yes | 78 | 77.2 | 4 | 25.0 | 74 | 87.1 |
| Endothelial Hyperplasia | ||||||
| No | 18 | 17.8 | 11 | 68.8 | 7 | 8.2 |
| Yes | 83 | 82.2 | 5 | 31.2 | 78 | 91.8 |
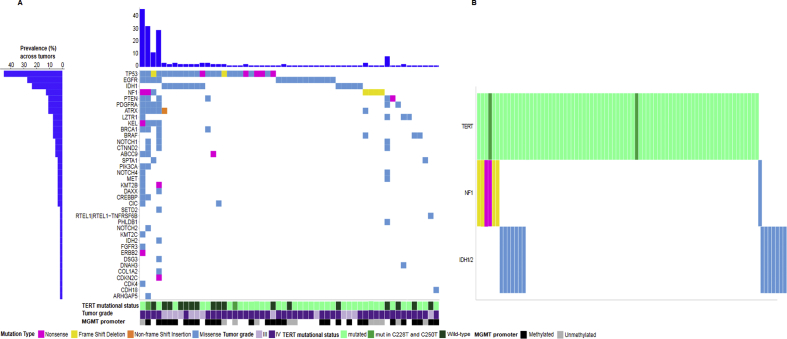
A total of 270 muts were detected in 92 of the 101 tumors (91.1%). Among the 92 tumors with muts, 43 (46.7%) carried only one mut, 35 (38%) carried 2 muts, and 14 (15.2%) carried 3 or more muts. On average, grade III tumors carried 2 muts (range: 1–4), while grade IV tumors carried 1 mut (range 0–45). The distribution of muts per gene per tumor is presented in Figure 2A, while Supplemental Table S2 presents the frequency distribution of mutated genes per grade. TERT, TP53, IDH1/2, EGFR, and NF1 were the most frequently mutated genes. Neither the number of muts nor the number of mutated genes per tumor differed between pts with grade III and grade IV tumors (Wilcoxon rank-sum p = 0.082 and p = 0.055, respectively). Seventy-five pts had tumors with muts in the TERT promoter and 26 pts had wt TERT. Among the 75 pts with TERT muts, 60 (80%) had C228T, 13 had C250T, and 2 both C228T and C250T. TP53 and IDH1/2 muts were identified in 24.8% and 13.9% of the study population, respectively, while 14.9% of pts carried EGFR muts. As described in detail in Figure 2B, NF1 muts were frameshifts and missense muts and occurred at clonal frequencies in the affected tumors (mutated allele frequency > 25% [21]). All NF1 muts were observed in pts with grade IV tumors, while only one tumor with EGFR mut was detected among pts with grade III tumors. All IDH1 muts concerned the same known amino acid change (p.Arg132His). The only IDH2 mut was a not previously reported variant, p.Ala174Val, next to the p.Arg172 hotspot, in a GBM of a 62-year-old female pt. All IDH1/2 muts were present at clonal frequencies in the affected tumors. In contrast to NF1, and as expected, most of the IDH1/2 muts (71.4%) were observed in pts with grade III tumors. The MGMT promoter methylation status was available for 77 of the 101 pts included in the analysis (76.2%). Of note, out of the 14 pts with IDH1/2 muts, those with available MGMT methylation data (10 pts) were all methylated. In contrast, all 22 pts with unmethylated MGMT had wt IDH1/2 tumors. MGMT was informative for 6 of the 7 pts with NF1 muts and 2 of them had methylated MGMT.
Figure 2.
Maps show the distribution of mutations per gene per tumor.A.Mutation profiles for all affected genes.B.Mutual exclusiveness of NF1 and IDH1/2 mutations and their association with TERT mutations. Of the seven tumors with NF1 mutations, 4 (beige bars) had frame shift deletions: p.Tyr2285fs at 57% frequency of the tumor DNA at this position, p.Asp2346fs at 68%, p.Phe1247fs at 80%, and p.Pro678fs at 28%; 2 tumors (purple bar) had nonsense mutations: p.Gln2302Ter at 16% and p.Gln1255Ter at 18% frequency; lastly, one tumor (blue bar) had two missense mutations (p.Gly1133Asp, p.Val1146Ile) at 20% and 30% frequency of the tumor DNA; the latter was the only NF1 mutated tumor without mutated TERT.
As shown in Figure 2B, IDH1/2 and NF1 muts were mutually exclusive. In an attempt to explore the relationship between NF1 and TERT, tumors were further classified according to their TERT and NF1 mutational status. Comutation of TERT and NF1 was detected in 6 pts (5.9%), while 25 pts had wt TERT and NF1, 69 pts had wt NF1 and mutated TERT, and in one tumor TERT was wt and NF1 was mutated. It is of note that all 6 comutations of TERT and NF1 were observed in pts with grade IV tumors. In addition, all 6 pts carrying muts in both TERT and NF1, as well as the one pt with mutated NF1 and wt TERT, had wt IDH1/2, while only 7 out of the 25 pts (28%) with wt TERT and NF1 and 7/69 (10.1%) pts with wt NF1 and mutated TERT carried muts in IDH1/2, respectively.
The associations of the mutational status (mutated/wt) with several clinicopathological parameters among the most frequently mutated genes are presented in Table 2. pts carrying muts in IDH1/2 were of younger age compared with those with wt IDH1/2, while pts with TERT muts were older (Wilcoxon rank-sum p = 0.001 and p < 0.001, respectively). No significant differences were detected between Ki67 levels and the mutational status of the examined genes. Tumors with wt IDH1/2 and TP53 were more frequently grade IV.
Table 2.
Associations Between Clinicopathological Parameters and the Mutational Status of Several Genes.
| EGFR |
IDH1/2 |
NF1 |
PTEN |
TERT |
TP53 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | mut | p | wt | Mut | p | wt | mut | p | wt | mut | p | wt | mut | p | wt | mut | p | ||
| Age | Median (minimum–maximum) | 54.6 (21.3–77.9) | 59.0 (40.8–78.2) | 0.209 | 56.2 (21.3–78.2) | 43.2 (25.2–62.9) | 0.001 | 54.4 (21.3–78.2) | 62.7 (45.9–73.8) | 0.064 | 54.5 (21.3–78.2) | 57.1 (53.4–65.7) | 0.423 | 46.5 (21.3–74.1) | 57.7 (33.0–78.2) | <0.001 | 54.6 (23.1–78.2) | 59.0 (21.3–77.9) | 0.805 |
| Ki67 | Median (minimum–maximum) | 17.0 (0–80.0) | 25.0 (0.5–70.0) | 0.538 | 18.0 (0–70.0) | 15.0 (1.5–80.0) | 0.961 | 18.0 (0–80.0) | 16.0 (3.0–20.0) | 0.279 | 18.0 (0–80.0) | 25.0 (8.0–35.0) | 0.675 | 16.5 (0–75.0) | 18 (0–80.0) | 0.802 | 18.0 (0–70.0) | 18.0 (0–80.0) | 0.984 |
| Tumor grade | III | 15 (17.4) | 1 (6.7) | 0.292 | 6 (6.9) | 10 (71.4) | <0.001 | 16 (17.0) | 0 (0.0) | 0.234 | 16 (16.8) | 0 (0.0) | 0.273 | 7(26.9) | 9(12.0) | 0.073 | 8 (10.5) | 8 (32.0) | 0.011 |
| IV | 71 (82.6) | 14 (93.3) | 81 (93.1) | 4 (28.6) | 78 (83.0) | 7 (100.0) | 79 (83.2) | 6 (100.0) | 19(73.1) | 66(88.0) | 68 (89.5) | 17 (68.0) | |||||||
| Sex | Female | 33 (38.4) | 9 (60.0) | 0.117 | 38 (43.7) | 4 (28.6) | 0.287 | 40 (42.6) | 2 (28.6) | 0.469 | 38 (40.0) | 4 (66.7) | 0.199 | 9(34.6) | 33(44.0) | 0.403 | 29 (38.2) | 13 (52.0) | 0.223 |
| Male | 53 (61.6) | 6 (40.0) | 49 (56.3) | 10 (71.4) | 54 (57.4) | 5 (71.4) | 57 (60.0) | 2 (33.3) | 17(65.4) | 42(56.0) | 47 (61.8) | 12 (48.0) | |||||||
| Type of surgery | Biopsy/stereotactic biopsy | 19 (23.2) | 4 (28.6) | 0.892 | 18 (21.7) | 5 (38.5) | 0.368 | 21 (23.6) | 2 (28.6) | 0.553 | 20 (22.2) | 3 (50.0) | 0.256 | 9(36.0) | 14(19.7) | 0.119 | 18 (25.0) | 5 (20.8) | 0.835 |
| Subtotal | 52 (63.4) | 8 (57.1) | 54 (65.1) | 6 (46.2) | 55 (61.8) | 5 (71.4) | 58 (64.4) | 2 (33.3) | 15(60.0) | 45(63.4) | 45 (62.5) | 15 (62.5) | |||||||
| Total excision | 11 (13.4) | 2 (14.3) | 11 (13.3) | 2 (15.4) | 13 (14.6) | 0 (0.0) | 12 (13.3) | 1 (16.7) | 1(4.0) | 12(16.9) | 9 (12.5) | 4 (16.7) | |||||||
| Necrosis | No | 18 (20.9) | 1 (6.7) | 0.192 | 10 (11.5) | 9 (64.3) | <0.001 | 19 (20.2) | 0 (0.0) | 0.187 | 19 (20.0) | 0 (0.0) | 0.224 | 9(34.6) | 10(13.3) | 0.017 | 12 (15.8) | 7 (28.0) | 0.175 |
| Yes | 68 (79.1) | 14 (93.3) | 77 (88.5) | 5 (35.7) | 75 (79.8) | 7 (100.0) | 76 (80.0) | 6 (100.0) | 17(65.4) | 65(86.7) | 64 (84.2) | 18 (72.0) | |||||||
| Hemorrhage | No | 20 (23.3) | 3 (20.0) | 0.781 | 15 (17.2) | 8 (57.1) | 0.001 | 22 (23.4) | 1 (14.3) | 0.579 | 22 (23.2) | 1 (16.7) | 0.713 | 11(42.3) | 12(16.0) | 0.006 | 13 (17.1) | 10 (40.0) | 0.018 |
| Yes | 66 (76.7) | 12 (80.0) | 72 (82.8) | 6 (42.9) | 72 (76.6) | 6 (85.7) | 73 (76.8) | 5 (83.3) | 15(57.7) | 63(84.0) | 63 (82.9) | 15 (60.0) | |||||||
| Endothelial hyperplasia | No | 17 (19.8) | 1 (6.7) | 0.221 | 10 (11.5) | 8 (57.1) | <0.001 | 18 (19.1) | 0 (0.0) | 0.202 | 18 (18.9) | 0 (0.0) | 0.240 | 6(23.1) | 12(16.0) | 0.417 | 13 (17.1) | 5 (20.0) | 0.743 |
| Yes | 69 (80.2) | 14 (93.3) | 77 (88.5) | 6 (42.9) | 76 (80.9) | 7 (100.0) | 77 (81.1) | 6 (100.0) | 20(76.9) | 63(84.0) | 63 (82.9) | 20 (80.0) | |||||||
wt: wild-type; mut: mutated. P-values in bold indicate statistically significant results at the 5% level of significance.
The 2 × 2 associations of the mutational status between EGFR, IDH1/2, NF1, PTEN, TERT, and TP53 are presented in Table 3. pts with TERT muts as compared with those with wt TERT had more frequently wt TP53 and IDH1/2(chi-square p = 0.016 and p = 0.025, respectively). In addition, wt TP53 was more frequent in pts with wt IDH1/2 (p < 0.001).
Table 3.
Associations of the Mutational Status Between EGFR, IDH1/2, NF1, PTEN, TERT, and TP53.
| EGFR |
IDH1/2 |
NF1 |
PTEN |
TERT |
TP53 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mut | wt | p | mut | wt | p | mut | Wt | p | mut | wt | p | mut | wt | p | mut | wt | p | ||
| EGFR | wt | 0 (0.0) | 86 (100.0) | ||||||||||||||||
| mut | 15 (100.0) | 0 (0.0) | |||||||||||||||||
| IDH1/2 | wt | 14 (93.3) | 73 (84.9) | 0.382 | 0 (0.0) | 87 (100.0) | |||||||||||||
| mut | 1 (6.7) | 13 (15.1) | 14 (100.0) | 0 (0.0) | |||||||||||||||
| NF1 | wt | 12 (80.0) | 82 (95.3) | 0.031 | 14 (100.0) | 80 (92.0) | 0.271 | 0 (0.0) | 94 (100.0) | ||||||||||
| mut | 3 (20.0) | 4 (4.7) | 0 (0.0) | 7 (8.0) | 7 (100.0) | 0 (0.0) | |||||||||||||
| PTEN | wt | 12 (80.0) | 83 (96.5) | 0.013 | 13 (92.9) | 82 (94.3) | 0.838 | 5 (71.4) | 90 (95.7) | 0.009 | 0 (0.0) | 95 (100.0) | |||||||
| mut | 3 (20.0) | 3 (3.5) | 1 (7.1) | 5 (5.7) | 2 (28.6) | 4 (4.3) | 6 (100.0) | 0 (0.0) | |||||||||||
| TERT | wt | 2 (13.3) | 24 (27.9) | 0.234 | 7 (50.0) | 19 (21.8) | 0.025 | 1 (14.3) | 25 (26.6) | 0.472 | 1 (16.7) | 25 (26.3) | 0.600 | 0 (0.0) | 26 (100.0) | ||||
| mut | 13 (86.7) | 62 (72.1) | 7 (50.0) | 68 (78.2) | 6 (85.7) | 69 (73.4) | 5 (83.3) | 70 (73.7) | 75 (100.0) | 0 (0.0) | |||||||||
| TP53 | wt | 11 (73.3) | 65 (75.6) | 0.852 | 5 (35.7) | 71 (81.6) | <0.001 | 4 (57.1) | 72 (76.6) | 0.250 | 2 (33.3) | 74 (77.9) | 0.014 | 61 (81.3) | 15 (57.7) | 0.016 | 0 (0.0) | 76 (100.0) | |
| mut | 4 (26.7) | 21 (24.4) | 9 (64.3) | 16 (18.4) | 3 (42.9) | 22 (23.4) | 4 (66.7) | 21 (22.1) | 14 (18.7) | 11 (42.3) | 25 (100.0) | 0 (0.0) | |||||||
wt: wild-type; mut: mutated. P-values in bold indicate statistically significant results at the 5% level of significance.
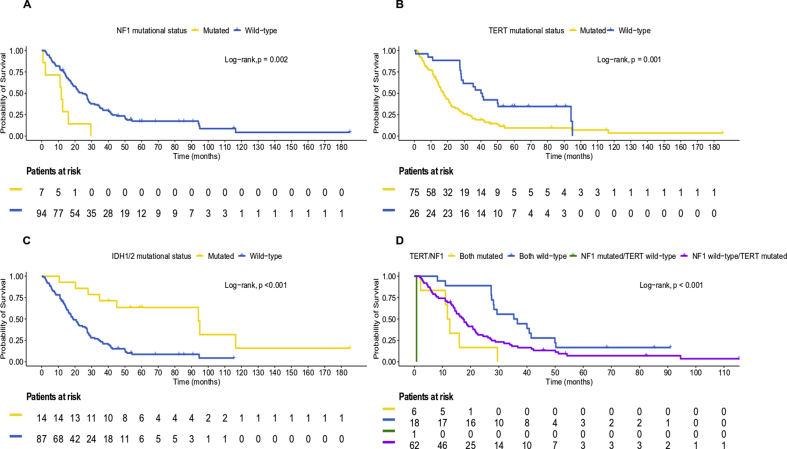
Pts were followed up for a median of 82.3 months (95% CI 59.0–185.1), and throughout this period 87 deaths were reported. The median OS for all pts included in the study was 22.0 months (95% CI 17.3–27.9), while 74.3% survived one year since diagnosis. The 6-month and the 9-month survival rates were 87.1% and 81.2%, respectively. The median OS for pts with grade IV tumors was 20.8 months (95% CI 15.8–27.0) and was significantly shorter compared with that of pts with grade III tumors whose median OS was 64.6 months (95% CI 20.3–116.5) (log-rank p = 0.007).
Pts with wt TERT had significantly longer survival compared with those with TERT muts (median OS: 40.4 months [95% CI 28.1–94.1] vs. 17.7 months [95% CI 14.4–21.5], log-rank p = 0.001). Among pts with wt NF1, those with wt TERT had median OS of 40.8 months (95% CI 28.3–94.1), while those with mutated TERT had median OS of 18.5 months (95% CI 14.4–21.7). All six pts with comutation of TERT and NF1 had died at the final follow-up (median OS: 12.2 months [95% CI 2.2–29.5]), while the pt with wt TERT and mutated NF1 died within 24 days after the diagnosis of grade IV glioma. Figure 3(A–C) presents the Kaplan–Meier plots based on the mutational status of NF1, TERT, and IDH1/2 with respect to OS in the entire cohort.
Figure 3.
Kaplan–Meier plots with respect to overall survival based on the mutational status of (A) NF1, (B) TERT, and (C) IDH1/2 genes in the entire cohort and (D) TERT/NF1 among patients with IDH1/2 wild-type tumors.
As expected, among IDH1/2 negative pts, those with wt TERT had median OS of 35.3 months (95% CI 27.3–41.5), while those with mutated TERT had median OS of 16.1 months (95% CI 13.8–20.8). The 7 pts with both TERT and IDH1/2 muts had a median OS of 45.1 months, while those with IDH1/2 muts had a median OS of only 94.1 months (95% CI 27.9–95.0). In addition, pts with wt NF1 and TERT presented with longer survival compared with those with wt NF1 and mutated TERT (median OS: 36.0 months [95% CI 27.4–41.5] vs. 17.3 months [95% CI 14.0–20.8]) among pts with IDH1/2 wt tumors (Figure 3D).
As depicted in Supplemental Table S3, pts with muts in NF1 and TERT were at significantly higher risk of death compared with those without (hazard ratio [HR] = 3.19, 95% CI 1.44–7.06, Wald's p = 0.004 and HR = 2.28, 95% CI 1.36–3.83, p = 0.002, respectively). In contrast, pts with muts in IDH1/2 presented with longer survival compared with pts with wt IDH1/2 (HR = 0.24, 95% CI 0.11–0.52, p < 0.001), while pts with methylated MGMT promoter were at significantly lower risk of death compared with those with unmethylated MGMT (HR = 0.47, 95% CI 0.28–0.80, p = 0.006). Age at the time of diagnosis was also a significant prognostic factor for OS with the expected hazard ratio of death increasing with each year increase in age (HR = 1.05, 95% CI 1.03–1.07, p < 0.001).
On multivariate analysis, adjusting for age, NF1, IDH1/2 mutational status, and MGMT promoter methylator status, TERT muts were found to be marginally associated with shorter OS (HR = 1.88, 95% CI 1.00–3.51, p = 0.049), whereas mutated NF1 lost its unfavorable prognostic significance for OS (HR = 1.92, 95% CI 0.79–4.70, p = 0.151). Age, MGMT promoter status, and IDH1/2 mutational status remained significant factors in the multivariate analysis, underlining their independent prognostic significance for OS in the entire cohort (Table 4).
Table 4.
Results of the Cox Multivariate Regression Analyses with Respect to OS (A) in the Entire Cohort and (B) in Patients with Wild-type IDH1/2 Tumors.
| Parameter | N Patients | N Events | HR (95% CI) | p |
|---|---|---|---|---|
| (A) Entire Cohort (77 Patients; 65 Events) | ||||
| Age∗ | 1.04 (1.01–1.07) | 0.016 | ||
| TERT Mutational Status | ||||
| Mutated vs. wild-type | 55 vs. 22 | 50 vs. 15 | 1.88 (1.00–3.51) | 0.049 |
| NF1 Mutational Status | ||||
| Mutated vs. wild-type | 6 vs. 71 | 6 vs. 59 | 1.92 (0.79–4.70) | 0.151 |
| IDH1/2 Mutational Status | ||||
| Mutated vs. wild-type | 10 vs. 67 | 5 vs. 60 | 0.30 (0.10–0.86) | 0.026 |
| MGMT Promoter Status | ||||
| Methylated vs. unmethylated | 55 vs. 22 | 44 vs. 21 | 0.49 (0.28–0.87) | 0.014 |
| (B) IDH1/2 Wild-type Tumors (67 Patients; 60 Events) | ||||
| Age∗ | 1.03 (1.00–1.06) | 0.054 | ||
| TERT Mutational Status | ||||
| Mutated vs. Wild-type | 51 vs. 16 | 47 vs. 13 | 2.26 (1.09–4.66) | 0.028 |
| NF1 Mutational Status | ||||
| Mutated versus wild-type | 6 vs. 61 | 6 vs. 54 | 1.82 (0.59–5.64) | 0.301 |
| N Mutated Genes Per Tumor∗ | 1.00 (0.89–1.20) | 0.935 | ||
| TP53 Mutational Status | ||||
| Mutated vs. wild-type | 11 vs. 56 | 11 vs. 49 | 1.62 (0.68–3.83) | 0.275 |
| MGMT Promoter Status | ||||
| Methylated vs. unmethylated | 45 vs. 22 | 39 vs. 21 | 0.49 (0.28–0.86) | 0.013 |
N, number; HR, hazard ratio; CI, confidence interval. P-values in bold indicate statistically significant results at the 5% level of significance.
Continuous variable.
Similar results were observed in the subgroup of pts with wt IDH1/2 tumors for NF1 and TERT (HR = 2.70, 95% CI 1.22–5.97, p = 0.014 and HR = 1.99, 95% CI 1.14–3.47, p = 0.015, respectively), while a trend towards longer survival was observed for pts with methylated MGMT promoter (HR = 0.62, 95% CI 0.36–1.05, p = 0.076). Mutated TP53 univariately showed a trend for worse survival (HR = 1.63, 95% CI 0.94–2.85, p = 0.085), whereas increase in pt's age and in the number of mutated genes per tumor was associated with higher risk of death (HR = 1.05, 95% CI 1.02–1.07, p < 0.001 and HR = 1.09, 95% CI 1.02–1.16, p = 0.009, respectively) (Supplemental Table S4).
In multivariate analysis, in pts with IDH1/2 wt tumors, adjusting for age, the number of mutated genes per tumor, TP53, NF1 and MGMT promoter methylator status, mutated TERT was associated with shorter OS (HR = 2.26, 95% CI 1.09–4.66, p = 0.028) (Table 4).
Discussion
We presented herein a series of high-grade glioma pts treated in two institutions in Greece, whose tumors were examined by NGS for a number of muts previously described in gliomas. Subsequently, we evaluated their association with one another and with pt outcome. NF1 was found to be among the most frequently mutated genes, and all such muts were found in GBMs, none of which carried an IDH mut. TERT was also tested in all pts. As expected, IDH mutant tumors were associated with younger age and, as our cohort included only high-grade tumors, TERT muts were associated with poorer outcome.
The NF1 gene is a large tumor suppressor gene, spread on the 17q11.2 locus [22]. Its product neurofibromin is expressed at high levels in the CNS and in association with tubulin. As its role is the regulation of the RAS/MAPK pathway, its absence leads to prolonged RAS/RAF/MARK activation, as well as decreased PI3K/AKT-regulated apoptosis. In fact, the mTOR pathway is constitutively activated in NF-deficient cells [8]. NF1 germline mut is inherited in an autosomal dominant pattern leading to neurofibromatosis 1, a disease most commonly associated with cutaneous neurofibromas, cafe au lait macules, and optic gliomas as well as peripheral nerve sheath tumors [23]. Interestingly, one study showed that the few GBMs that occur in pts with germline NF1 muts have a more benign course than sporadic GBMs [24].
NF1 may undergo proteasomal and genetic inactivation in gliomas [25]. Somatic NF1 muts have been reported in 11% [22] or up to 23% of GBMs [8] and at a lower rate in lower grade gliomas. Here, we identified NF1 muts in GBMs only, they were single base pair alterations and were considered pathogenic, as previously described [8]. NF1 mutations are reported as drivers in low-grade gliomas [26] where they seem to confer adverse prognosis [22]. Their role as drivers in GBMs is less clear though: NF1 muts with PTEN mut were considered the hallmark of the mesenchymal subtype in the TCGA data, as they were found in 53% of such tumors [10]. Of note, in an extended series, only 13% of mesenchymal tumors carried muts in this gene [17], and in another study, NF1 muts were not associated with GBM pt outcome [22]. Our pts with NF1 mutated tumors had a poor outcome, as 6/7 died within the first 16 months and only one, who also carried a TERT mut, survived up to 30 months. In fact, 6/7 pts with NF1 muts also carried muts in TERT, while only one pt had wt TERT and died within 24 days.
Whether the poor outcomes were a result of the mut of NF1 alone or of its comutation with other genes such as TP53, cannot be concluded safely [25]. Of note, NF1 muts were mutually exclusive with IDH1/2 muts in our series, as in previous reports [17,26], thus underlining their association with poor prognosis. Unfortunately, the examined numbers of NF1 mutated GBMs, both here [7] and in the aforementioned study [22,30], are too small for definitive conclusions.
Our study adds more evidence in favor of a critical prognostic role of TERT in high-grade gliomas, which seems to lose its significance in the presence of other muts (MGMT and IDH) [6,27]. Given the variable function of TERT in gliomas of different grade, one wonders whether it is an independent molecular marker or whether its significance is surrogate to other somatic muts encountered in these tumors. This is further supported by published data on TERT and IDH muts in gliomas [11].
TERT promoter muts attract transcriptional factors that should not normally act at this site and thus contribute to the activation of TERT expression [26]. TERT muts have originally been associated with tumors of low self-renewal [19] including GBMs; in the latter, TERT muts increase the capacity of self-renewal and thus survival and growth [28], despite the fact that they do not contribute to increased size of the telomeres [26]. Previously, TERT promoter muts were shown to interact with IDH muts conferring a dual prognostic impact [11,29]. We showed that TERT/IDH comutation conferred worse outcome than IDH-only in this cohort of high-grade gliomas, while TERT/NF1 comutation was a worse prognosticator compared with TERT-only. In larger pt series, the same will probably be proven for comutation of the TERT promoter with other driver genes in gliomas.
Our findings regarding TERT could be explained in the context of epistasis, where the effect of a given mut depends on the mutational context [30]. In addition, reactivation of telomerase in cancer has been described as a double-edged sword [31], in the sense that it promotes the perpetuation of already established genomic alterations, which drive malignancy. Obviously, because of the small size of our cohort, the presented data should be regarded as hypothesis generating needing validation in larger series of pts with high-grade glioma.
As expected, the promoter of MGMT in IDH mutant tumors was methylated, and all unmethylated tumors were IDH wt. The interdependence of TERT and MGMT has been shown in two significant papers [32,33]. In the first one, the significance of TERT muts was further defined when MGMT was included in the prognostic analysis of molecular characteristics of gliomas [32], while in the second, conversely, the effect of MGMT promoter methylation was enhanced by TERT muts [33], thus reinforcing the theory of a potential codependent role of TERT. However, we must underline that in our cohort, its effect was independently significant in multivariate analysis, albeit with a marginal p-value, in the entire group.
Of note, in our group, PTEN muts were more frequent in NF1 mutant cases, as were EGFR muts. In addition, we could not confirm the data showing BRAF muts to be mutually exclusive with NF1. However, the number of pts with NF1 and BRAF muts was too small to allow for the drawing of robust conclusions.
The small numbers of NF1 mutated pts also prevent the drawing of safe conclusions regarding the role of MGMT in the setting of NF1. Overall, MGMT was significantly associated with OS as expected. In the search of a more “constitutional” mut in gliomas, MGMT promoter methylation represents a significant prognosis altering epigenetic event, but whether it can change the prognosis of NF1 mut tumors, would need a larger number of pts to assess. Furthermore, the stability of this alteration in the course of a GBM is uncertain [34].
Overall, we studied several genes of interest in high-grade gliomas, and, like other investigators, as well as like in other tumors, we did not identify many new significant muts beyond those already known in the literature. Our sample appears to be representative of the known molecular biology of high-grade gliomas. Ideally, one would want to study these tumors prospectively using a uniformly grade III or grade IV group of pts, who are all treated in the same way.
Barring these reservations, this study adds to the growing body of evidence on the molecular complexity of high-grade gliomas. The role of NF1 in the sensitivity and resistance to specific therapeutic approaches and its ability to help us identify pts with a particularly poor prognosis needs to be further clarified. As the experience with TERT teaches us, these data are developing gradually and are open to revision, if further analysis of the mutational landscape of GBMs reveals new or more potent muts. Being that as it may, TERT remains of interest, and its significance in IDH wt gliomas is reinforced by our data.
Funding
This work was supported by a research grant from F. Hoffmann-La Roche and by an internal HeCOG research grant. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
ER: Advisory Board: Novartis, Honoraria: Novartis, Travel: Genesis Pharmaceuticals, Pfizer, Roche, Bristol-Myers Squibb, Genekor.
GF: Advisory Board: Pfizer, Sanofi and Roche, Honoraria: Astra-Zeneca.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.10.016.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Referencesd
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Wesseling P., Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 3.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Bent M.J., Brandes A.A., Taphoorn M.J., Kros J.M., Kouwenhoven M.C., Delattre J.Y., Bernsen H.J., Frenay M., Tijssen C.C., Grisold W. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 5.Burris H., Mellinghoff I., Maher E., Wen P., Beeram M., Touat M., Faris J., Azad N., Cloughesy T., Gore L. Abstract PL04-05: The first reported results of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a Phase I study of patients with advanced IDH1-mutant solid tumors, including gliomas. Mol Cancer Ther. 2015;14(12 Supplement 2) PL04-5-PL-5. [Google Scholar]
- 6.Yang P., Cai J., Yan W., Zhang W., Wang Y., Chen B., Li G., Li S., Wu C., Yao K. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neuro Oncol. 2016;18(8):1099–1108. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffzek K., Welti S. Neurofibromin: Protein domains and functional characteristics. In: Upadhyaya M., Cooper D., editors. Neurofibromatosis Type 1. Springer; Berlin, Heidelberg: 2012. pp. 305–3226. [Google Scholar]
- 8.Philpott C., Tovell H., Frayling I.M., Cooper D.N., Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Human Genom. 2017;11(1):13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y., Guignard F., Zhao D., Liu L., Burns D.K., Mason R.P., Messing A., Parada L.F. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer cell. 2005;8(2):119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H., Pekmezci M., Rice T., Kosel M.L., Smirnov I.V. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegi M.E., Diserens A.C., Gorlia T., Hamou M.F., de Tribolet N., Weller M., Kros J.M., Hainfellner J.A., Mason W., Mariani L. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 13.Wiestler B., Capper D., Holland-Letz T., Korshunov A., von Deimling A., Pfister S.M., Platten M., Weller M., Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 14.Zacher A., Kaulich K., Stepanow S., Wolter M., Kohrer K., Felsberg J., Malzkorn B., Reifenberger G. Molecular Diagnostics of Gliomas Using Next Generation Sequencing of a Glioma-Tailored Gene Panel. Brain Pathol (Zurich, Switzerland) 2017;27(2):146–159. doi: 10.1111/bpa.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H.S., Kwon M.J., Song J.H., Kim E.S., Kim H.Y., Min K.W. Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol Res Pract. 2018;214(6):881–888. doi: 10.1016/j.prp.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Binabaj M.M., Bahrami A., ShahidSales S., Joodi M., Joudi Mashhad M., Hassanian S.M., Anvari K., Avan A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J Cell Physiol. 2018;233(1):378–386. doi: 10.1002/jcp.25896. [DOI] [PubMed] [Google Scholar]
- 17.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frattini V., Trifonov V., Chan J.M., Castano A., Lia M., Abate F., Keir S.T., Ji A.X., Zoppoli P., Niola F. The integrated landscape of driver genomic alterations in glioblastoma. Nature Genet. 2013;45(10):1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr., Friedman A.H., Friedman H., Gallia G.L., Giovanella B.C. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteller M., Garcia-Foncillas J., Andion E., Goodman S.N., Hidalgo O.F., Vanaclocha V., Baylin S.B., Herman J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 21.McGranahan N., Favero F., de Bruin E.C., Birkbak N.J., Szallasi Z., Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283):283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vizcaíno M.A., Shah S., Eberhart C.G., Rodriguez F.J. Clinicopathologic implications of NF1 gene alterations in diffuse gliomas. Human Pathol. 2015;46(9):1323–1330. doi: 10.1016/j.humpath.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cawthon R.M., Weiss R., Xu G.F., Viskochil D., Culver M., Stevens J., Robertson M., Dunn D., Gesteland R., O'Connell P. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62(1):193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 24.Shibahara I., Sonoda Y., Suzuki H., Mayama A., Kanamori M., Saito R., Suzuki Y., Mashiyama S., Uenohara H., Watanabe M. Glioblastoma in neurofibromatosis 1 patients without IDH1, BRAF V600E, and TERT promoter mutations. Brain Tumor Pathol. 2018;35(1):10–18. doi: 10.1007/s10014-017-0302-z. [DOI] [PubMed] [Google Scholar]
- 25.McGillicuddy L.T., Fromm J.A., Hollstein P.E., Kubek S., Beroukhim R., De Raedt T., Johnson B.W., Williams S.M.G., Nghiemphu P., Liau L.M. Proteasomal and Genetic Inactivation of the NF1 Tumor Suppressor in Gliomagenesis. Cancer Cell. 2009;16(1):44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceccarelli M., Barthel Floris P., Malta Tathiane M., Sabedot Thais S., Salama Sofie R., Murray Bradley A., Morozova O., Newton Y., Radenbaugh A., Pagnotta Stefano M. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y., Koh J., Kim S.-I., Won J.K., Park C.-K., Choi S.H., Park S.H. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol Commun. 2017;5:62. doi: 10.1186/s40478-017-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong D.E., Woo S.R., Nam H., Nam D.-H., Lee J.-H., Joo K.M. Preclinical and clinical implications of TERT promoter mutation in glioblastoma multiforme. Oncol Lett. 2017;14(6):8213–8219. doi: 10.3892/ol.2017.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuong H.G., Altibi A.M.A., Duong U.N.P., Ngo H.T.T., Pham T.Q., Chan A.K., Park C.K., Fung K.M., Hassell L. TERT promoter mutation and its interaction with IDH mutations in glioma: Combined TERT promoter and IDH mutations stratifies lower-grade glioma into distinct survival subgroups-A meta-analysis of aggregate data. Crit Rev Oncol Hematol. 2017;120:1–9. doi: 10.1016/j.critrevonc.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Chandler C.H., Chari S., Dworkin I. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 2013;29(6):358–366. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiyama K. Springer Science & Business Media; 2009. Telomeres and Telomerase in Cancer. [Google Scholar]
- 32.Arita H., Yamasaki K., Matsushita Y., Nakamura T., Shimokawa A., Takami H., Tanaka S., Mukasa A., Shirahata M., Shimizu S. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen H.N., Lie A., Li T., Chowdhury R., Liu F., Ozer B., Wei B., Green R.M., Ellingson B.M., Wang H.J. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017;19(3):394–404. doi: 10.1093/neuonc/now189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park C.K., Kim J.E., Kim J.Y., Song S.W., Kim J.W., Choi S.H., Kim T.M., Lee S.H., Kim I.H., Park S.H. The Changes in MGMT Promoter Methylation Status in Initial and Recurrent Glioblastomas. Transl Oncol. 2012;5(5):393–397. doi: 10.1593/tlo.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.