Abstract
Dengue virus (DENV) that caused an outbreak in Dhaka, Bangladesh during 2018 was analysed phylogenetically. DENV samples were classified into type 2-Cosmopolitan genotype (54%) and type 3-genotype I (46%), indicating co-circulation of two DENV types and resurgence of type 3 associated with genotype replacement.
Keywords: Bangladesh, dengue virus, Dhaka, resurgence, type 3
Dengue is a mosquito-borne disease caused by dengue virus (DENV) that is prevalent in urban areas in tropical and subtropical countries. Incidence of dengue has greatly increased in recent decades worldwide, causing an estimated 96 million symptomatic dengue cases per year. DENV, a member of the genus Flavivirus, family Flaviviridae, comprises four types (serotypes) that are further classified into genotypes. Prevalence of DENV types/genotypes differs depending on geographical regions, and changes occasionally. Increased incidence of dengue and severity of the disease have been associated with the change in circulating DENV types as well as virus evolution.
In Bangladesh, dengue has been an emerging public health concern since its resurgence in outbreaks from 2000 to 2002, caused mainly by DENV type 3 (DENV-3) [1,2]. Thereafter, no serotype data were available until the period between 2013 and 2016, when DENV-1 and DENV-2 were reported as dominant serotypes [3]. In 2017, predominance of DENV-2 and re-emergence of DENV-3 were observed [4]. In Dhaka, the capital of Bangladesh, an unprecedented large dengue outbreak has been occurring since August 2018, involving more than 10 000 dengue cases in that year, although between 400 and 6000 annual cases had been reported from 2008 to 2017 [5]. However, for the 2018 outbreak, the DENV type/genotype and its lineage have not been clarified.
During a period between September and November 2018, 90 serum samples were collected from individuals with suspected dengue in four tertiary-care hospitals in Dhaka presenting sudden onset of high fever of unknown origin (≥38.5°C, ≤7 days) with or without rash and joint pain. The presence of DENV protein/gene was tested by three methods: immunochromatography for NS1 antigen (Dengue NS1 Rapid Test Cassette, Acro Biotech Inc., Rancho Cucamonga, CA, USA), reverse transcription-polymerase chain reaction (RT-PCR), and quantitative PCR (qPCR) (RealStar dengue PCR kit 2.0, Altona Diagnostics, Hamburg, Germany). Detection rates of DENV were 71% (64/90), 70% (63/90) and 32% (29/90) by qPCR, immunochromatography and RT-PCR, respectively. Among all the samples tested, 73 were positive for DENV by any of the three methods, whereas 29 samples tested positive by all three methods. By RT-PCR targeting of the C-prM junction [6], 511-bp amplicons were obtained (see Supplementary material, Fig. S1). Among the 29 RT-PCR-positive samples, nucleotide sequences of the partial C-prM gene were determined for 24 samples, and deposited in GenBank under Accession nos MN400313–MN400336. DENV-positive rate was highest in the age group 20–29 years, followed by 30–39 years, and was higher in male than female patients (see Supplementary materials, Tables S1 and S2).
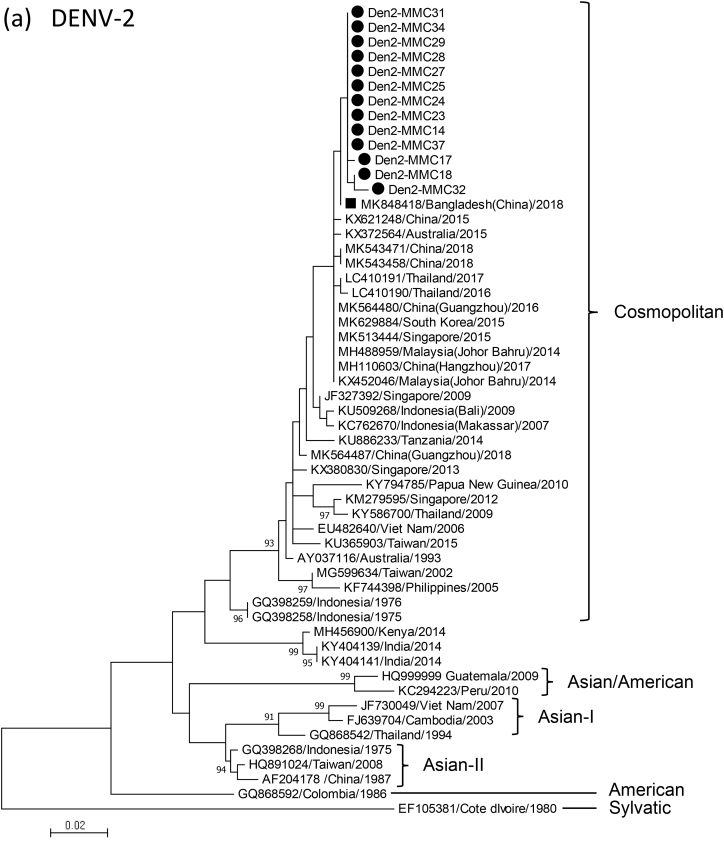
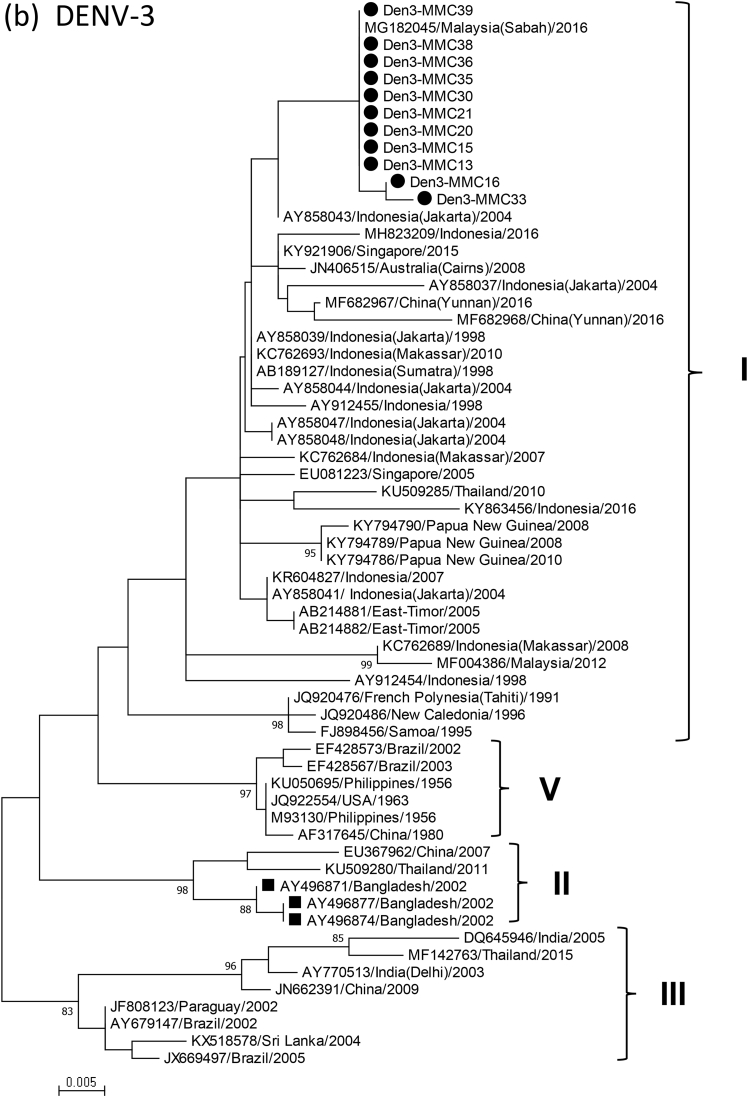
Phylogenetic analysis of the C-prM gene using the MEGA.7 software package revealed that DENV in the 24 samples belonged to type 2 (54%, 13/24) or type 3 (46%, 11/24). All the DENV-2 were assigned to Cosmopolitan genotype and were closely related to strains in China and South-East Asia (Fig.1a). DENV-3 genes in the present study were all grouped into genotype I, and clustered with DENV-3 in Malaysia, Singapore, Indonesia and Australia from the 1990s until recently (Fig.1b). In contrast, DENV-3 detected in Bangladesh in 2002 belonged to genotype II [2]. DENV-3 in 2018 was proved to have a C-prM deduced amino acid sequence (154 amino acids) that was different from those of the 2002 DENV by seven amino acids (see Supplementary material, Fig.S2).
Fig. 1.
Phylogenetic dendrogram of partial C-prM gene of dengue virus type 2 (DENV-2) (a) and DENV-3 (b) causing the 2018 outbreak in Dhaka, Bangladesh (closed circle), and those from other diverse geographical locations, constructed by maximum likelihood method using the MEGA.7 software package. The tree was statistically supported by bootstrapping with 1000 replicates, and genetic distances were calculated using the Kimura two-parameter model. The variation scale is described at the bottom. Percentage bootstrap support is indicated by the values at each node (values <80 are omitted). Black square indicates a DENV-2 strain that was imported from Bangladesh to China (a) and DENV-3 strains detected in Bangladesh in 2002 (b). Genotypes of DENV-2 and DENV-3 are shown at right.
A recent study in 2017 (until February 2018) in Dhaka, described DENV-2 as the dominant type, but also detected some DENV-1 and DENV-3 [4]. In the present study, despite a small sample size, DENV-3 accounted for 46% of the samples in September and October 2018, suggesting its increase during the outbreak starting from August 2018. As explained previously, serotype shift is considered to be a result of failure in heterotypic immunity by antibody induced by the first predominantly circulating serotype in the population [7]. In Bangladesh, it is suggested that DENV-3 might have been recently introduced and spread, in a population that had immunity mainly to the predominant DENV-2. The replacement of DENV serotype/genotype has been reported to be associated with increases in incidence and severity of dengue [8]. Accordingly, it is important to survey serotypes and genotypes of circulating DENV to prepare for potential outbreaks and occurrence of severe cases.
Conflict of interest
None to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2019.100629.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Podder G., Breiman R.F., Azim T., Thu H.M., Velathanthiri N., Mai le Q. Origin of dengue type 3 viruses associated with the dengue outbreak in Dhaka, Bangladesh, in 2000 and 2001. Am J Trop Med Hyg. 2006;74:263–265. [PubMed] [Google Scholar]
- 2.Islam M.A., Ahmed M.U., Begum N., Chowdhury N.A., Khan A.H., Parquet Mdel C. Molecular characterization and clinical evaluation of dengue outbreak in 2002 in Bangladesh. Jpn J Infect Dis. 2006;59:85–91. [PubMed] [Google Scholar]
- 3.Muraduzzaman A.K.M., Alam A.N., Sultana S., Siddiqua M., Khan M.H., Akram A. Circulating dengue virus serotypes in Bangladesh from 2013 to 2016. Virusdisease. 2018;29:303–307. doi: 10.1007/s13337-018-0469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K., Phadungsombat J., Nakayama E.E., Saito A., Egawa A., Sato T. Genotype replacement of dengue virus type 3 and clade replacement of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017. Infect Genet Evol. 2019;75:103977. doi: 10.1016/j.meegid.2019.103977. [DOI] [PubMed] [Google Scholar]
- 5.Institute of Epidemiology, Disease Control and Research Dengue data analysis from reporting hospitals of Dhaka City Date 27.07.2019. https://iedcr.gov.bd/index.php/dengue/219 Dengue Current Situation Available at:
- 6.Lanciotti R.S., Calisher C.H., Gubler D.J., Chang G.J., Vorndam A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotaki T., Yamanaka A., Mulyatno K.C., Churrotin S., Labiqah A., Sucipto T.H. Continuous dengue type 1 virus genotype shifts followed by co-circulation, clade shifts and subsequent disappearance in Surabaya, Indonesia, 2008–2013. Infect Genet Evol. 2014;28:48–54. doi: 10.1016/j.meegid.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Nunes P.C., Sampaio S.A., da Costa N.R., de Mendonça M.C., Lima Mda R., Araujo S.E. Dengue severity associated with age and a new lineage of dengue virus-type 2 during an outbreak in Rio De Janeiro, Brazil. J Med Virol. 2016;88:1130–1136. doi: 10.1002/jmv.24464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.