Abstract
Background
Mesangial collagen synthesis in renal glomeruli contributes to the pathogenesis of diabetic nephropathy (DN) which is one of the most serious complications of diabetes mellitus. However, the underlying mechanism of mesangial collagen synthesis is largely unknown.
Methods
The differential expression of CHOP and TRIM13 which is a well-defined E3 ubiquitin ligase was compared in renal biopsy samples from DN/normal renal tissues, in isolated glomeruli of diabetic/control mice, as well as in high glucose (HG) or TGF-β1-stimulated renal mesangial cells. Then the relationship between TRIM13 and CHOP was explored using the ubiquitination assay.
Findings
We found that the expression of TRIM13 was downregulated in renal biopsies, isolated glomeruli of diabetic mice, and HG/TGF-β1-stimulated renal mesangial cells, while the expression of CHOP was upregulated. An increased level of TRIM13 promoter methylation contributed to the deregulation of TRIM13 in renal glomeruli of DN. The ubiquitination assay confirmed that TRIM13 promoted ubiquitination and degradation of CHOP. Meanwhile, overexpressing TRIM13 attenuated DN-induced collagen synthesis and restored renal function in vitro and in vivo via downregulating CHOP.
Interpretation
Our findings demonstrated that overexpressed TRIM13 suppresses mesangial collagen synthesis in DN by promoting ubiquitination of CHOP, suggesting TRIM13 as a potential therapeutic target in treating DN.
Keywords: Diabetic nephropathy, DNA methylation, TRIM13, CHOP, Ubiquitination
Research in context section.
Evidence before this study
Evidence has demonstrated that the increased mesangial collagen synthesis in renal glomeruli causes mesangial extracellular matrix accumulation, and leads to the pathogenesis of diabetic nephropathy. In vitro experiments indicated that the interference of CHOP in mouse mesangial cells significantly decreased the expression of collagen-associated factors, including Col1a2, TGF-β1, and Col1a4. Therefore, identifying the mechanism of CHOP expression in mesangial cells is important in treating diabetic nephropathy. Ubiquitination is an effective way that affects protein expressions, which is mediated by ubiquitin ligases. As a well-defined ubiquitination ligase, TRIM13 is reported to be involved in complications of diabetes. Based on these findings, we speculated that CHOP expression may be regulated by TRIM13 in mesangial cells.
Added value of this study
We identified that TRIM13 was downregulated in renal biopsies, renal tissues of diabetic mice, and high glucose/TGF-β1-stimulated renal mesangial cells, while the expression of CHOP was upregulated. An increased level of TRIM13 promoter methylation contributed to the deregulation of TRIM13 in renal glomeruli of diabetic nephropathy. TRIM13 promoted ubiquitination and degradation of CHOP. Meanwhile, overexpressing TRIM13 attenuated diabetic nephropathy-induced collagen synthesis and restored renal function via downregulating CHOP.
Implications of all the available evidence
This study demonstrated that overexpressed TRIM13 suppresses mesangial collagen synthesis in diabetic nephropathy by promoting ubiquitination of CHOP, suggesting TRIM13 as a potential therapeutic target in treating diabetic nephropathy. Considering diabetic nephropathy as the most important independent factor in the etiology of chronic renal failure in adults, our findings indicated that the TRIM13 agonist may serve as an ideal renal function protective agent for diabetic patients.
Alt-text: Unlabelled box
1. Introduction
Diabetic nephropathy (DN), a kind of microangiopathy secondary to diabetes mellitus (DM), supervenes as the result of microvascular lesions in the renal glomeruli [1]. The prevalence of DN has reached pandemic proportions around the world and is still increasing. It is reported that in 2017, the prevalence of DN in men and women around the world was 15.48/1000, and 16.50/1000, respectively [2]. Existing research has considered DN as the most important independent factor in the etiology of chronic renal failure in adults [3]. However, the underlying molecular mechanism involved in the pathogenesis of DN is largely unknown. Evidence has demonstrated that the increased mesangial collagen synthesis in renal glomeruli causes mesangial extracellular matrix (ECM) accumulation, and leads to the pathogenesis of DN. With the accumulation of mesangial ECM, renal glomerular basement membranes are thickened and renal fibrosis is finally induced [4, 5]. Mesangial collagen synthesis can be stimulated by multiple cytokines, such as profibrotic connective tissue growth factor (CTGF) and transforming growth factor-β1 (TGF-β1) [6]. Meanwhile, the expression of TGF-β1 can also be enhanced by high glucose (HG) stimulation, further aggravating the severity of DN [7], [8], [9]. Therefore, identifying the mechanism of mesangial collagen synthesis is important in treating DN.
C/EBP homologous protein (CHOP) plays a vital role in DN-associated renal injury [10]. Recent studies have revealed a dramatic increase in CHOP expression in renal tissues of a murine model of DM [11]. CHOP also induced mesangial cell apoptosis in patients with glomerular and renal tubular damage [12]. In vitro experiments indicated that the interference of CHOP in mouse mesangial cells (MMCs) significantly decreased the expression of collagen-associated factors, including Col1a2, TGF-β1, and Col1a4 [13]. These results collectively indicate an involvement of CHOP in the increased collagen synthesis of DN, whereas regulatory mechanisms affecting CHOP expression still need to be elucidated.
Ubiquitination is an effective way that affects protein expressions, which is mediated by ubiquitin ligases. Proteins of the tripartite motif-containing (TRIM) superfamily are members of RING type ubiquitin E3 ligases that consist of three structure sequences: a RING domain in the N-terminal end, followed by a B-Box motif, and a coiled-coil (CC) domain [14]. Among them, the RING domain which is a zinc-binding domain determines the E3 ubiquitin ligase activity of TRIM proteins. Previous studies have demonstrated that proteins of TRIM superfamily are involved in regulating DM complications, including skeletal muscle atrophy [15] and vascular constriction [16]. TRIM13, also known as RFP2, is a member of TRIM superfamily, and is involved in multiple cellular processes, such as apoptosis, survival, and the biogenesis of non-coding RNAs [17]. As a ubiquitin ligase, TRIM13 has been confirmed to involve in physiological and pathological events as a result of the diversity of its substrates. Nevertheless, the role of TRIM13 in mesangial collagen synthesis of DN is currently unknown.
Here, in the current work, we identified the abnormal expression of TRIM13 in renal biopsies, renal tissues of diabetic mice, and HG-stimulated mesangial cells, and investigated the epigenetic-related dysregulation expression of TRIM13 in DN mice and HG-stimulated mesangial cells. Particularly, we elucidated the possible regulatory role of TRIM13 on the ubiquitination and degradation of CHOP in DN pathogenesis.
2. Materials and methods
2.1. Renal biopsy samples
The renal biopsy samples were the residual parts of diagnostic renal biopsies from DN patients (n = 30) or normal renal tissues (n = 30) obtained from the Second Affiliated Hospital of Nanchang University. This study was approved by the ethics committee of the Second Affiliated Hospital of Nanchang University for research (No.20160306018) and the written informed consents were obtained from the patients. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
2.2. Mouse model of DN
All animal studies were approved by the ethics committee of the Second Affiliated Hospital of Nanchang University, and were performed in strict accordance with the Guidelines for Laboratory Animal Care and Use of Nanchang University. Animal experiments were done in the animal laboratory of Nanchang University. Diabetes was induced in C57BL/6 mice with daily intraperitoneal injections (50 mg/kg) of streptozotocin (STZ) (Sigma, St Louis, MO, USA) for 5 consecutive days, as previously described [18]. The kidneys were harvested at 0, 4, 8, and 12 weeks after the establishment of the DN model. The C57BL/6 mice used for controls were administered with the citrate buffer. The 3-week old male db/db mice and db/m mice were purchased from the Model Animal Research Center of Nanchang University. They were acclimated for 1 week before the experiments.
2.3. Glomeruli isolation
The magnetic bead method was used to isolate glomeruli from renal tissues [19]. Briefly, kidneys were perfused with Dynabeads, followed by mincing and digesting with 1 mg/mL collagenase A (Roche, Basel, Switzerland) and 100 U/mL deoxyribonuclease I (Invitrogen, Waltham, MA, USA) in Hanks’ balanced salt solution (HBSS). Then the renal tissues were pressed through 100 µm and 70 µm cell strainers. HBSS was used to wash the 70 µm cell strainer. After that, the glomeruli containing Dynabeads were collected using a magnetic particle concentrator (Thermo Fisher Scientific, Waltham, MA, USA).
2.4. Cell culture and treatment
Primary MMCs were isolated from the kidney tissues of C57BL/6 mice (8–10 weeks old) as described [20], [21], [22]. After the isolation, MMCs were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS with 5% CO2 at 37 °C. MMCs from 6 to 12 passages were used in this study. Human mesangial cell (HMC) line CC-2559 cells were purchased from Lonza Walkersville, Inc. and were cultured as the provider's instructions.
An in vitro model of DN was established by subjecting the MMCs and HMCs to high glucose (HG, 30 mM) medium or recombinant human TGF-β1 (10 ng/ml) (Sigma, St Louis, MO, USA). The cells cultured in mannitol (18.9 mM) /glucose (11.1 mM) were used as the osmotic control for the HG group. A serum depleted (SD) culture with 0.2% FBS was used as the control for the TGF-β1 stimulation group.
2.5. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the kidney tissues and cells using TRIzol reagent (Invitrogen, Waltham, MA, USA). The cDNA was generated from the total RNA (2 μg) by using a cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA). The qRT-PCR was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) on a PRISM 7500 Real-TIME PCR System (Applied Biosystems, Waltham, MA, USA). The 2−ΔΔCt method was used for quantitative analysis. β-actin was used as the internal control. The specific primers used for the amplification were synthesized by Invitrogen (Waltham, MA, USA), with the sequences shown follows: TRIM13, (forward) 5′-CCGGAATTCAACTTCAGCTACTGGAATT-3′ and (reverse) 5′-CGCGGATCCTTATAATAGTTTATATTT-3′; CHOP, (forward) 5′-CTTGGCTG ACTGAGGAGGAG-3′ and (reverse) 5′-TCACCATTCGGTCAATCAGA-3′; Col1a2, (forward) 5′-CCAAAGGTGCTGATGGTTCT-3′ and (reverse) 5′-ACCAGCTTCACCCTTGTCAC-3′; Col4a1, (forward) 5′-GCTCTGGCTGTGGAAAATGT-3′ and (reverse) 5′-CTTGCATCCCGGGA AATC-3′; TGF-β1, (forward) 5′-AGCCCGAAGCGGACTACTAT-3′ and (reverse) 5′-CTGTGTG AGATGTCTTTGGTTTTC-3′; β-actin, (forward) 5′-CCGTGAAAAGATGACCCAGA-3′ and (reverse) 5′-TACGACCAGAGGCATACAG-3′.
2.6. Western blot analysis
Total proteins were obtained from the kidney and cells using protein extraction reagent containing protease and phosphatase inhibitor cocktail (Pierce Chemical Co., Rockford, IL, USA). After quantification of the protein concentration by Bradford method, proteins were separated by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto transfer membranes (Millipore, Billerica, MA, USA), as previously described [23]. The primary antibodies were anti-TRIM13 (1:1000, Abcam, Cambridge, UK), anti-CHOP (1:1000; Abcam), anti-cIAP1 (1:1000; Abcam), and anti-β-actin (1:1000; Cell Signaling Technology, Danvers, MA, USA). After incubation with the horseradish peroxidase-labeled secondary antibody (1:3000; Abcam) for 1 h, an enhanced chemiluminescence kit (Pierce Chemical, Rockford, IL, USA) was used to visualize the protein bands on the transfer membranes.
2.7. Urinary albumin-to-creatinine ratio (UACR) and eGFR
Urine albumin and blood urea nitrogen (BUN) were detected using an ELISA kit (Abnova, Taipei, Taiwan). Urine creatinine was measured using a Creatinine Assay kit (Colorimetric) (Abcam, Cambridge, UK). The UACR of diabetic mice was calculated with the formula: UACR= urinary albumin / urinary creatinine × 100%. Serum creatinine (Scr) was detected using an automatic biochemical analyzer (Hitachi Co., Tokyo, Japan). According to the modified version of the Modification of Diet in Renal Disease (MDRD) equation for the Chinese [24], the eGFR was calculated with the formula: eGFR = 175 × Scr−1.234 × age−0.197 (× 0.79, if female).
2.8. Immunohistochemistry (IHC) and immunofluorescent analysis
For IHC analysis, the paraffin-embedded renal tissues were cut into 4–5 µm slides. After dewaxing and rehydrating, the slides were boiled at 100 °C in 10% citrate buffer for 10 min to unmask the antigens of renal slides. Then the slides were blocked with the blocking solution containing 2% BSA and 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) for 1 h followed by the incubation with primary antibodies against TRIM13 (1:1000) overnight at 4 °C. Then they were incubated with secondary antibodies. The 3,3′-diamino-benzidine (DAB) was used as the chromogen to visualize staining with a microscope.
For double immunofluorescent staining, the slides were treated with 0.25% Triton-100 for 15 min and blocked with the blocking solution for 30 min. Then the slides were incubated with primary antibodies against TRIM13 (1:1000) and CHOP (1:1000) overnight at 4 °C, and further incubated with secondary antibodies. After sealing with the anti-fluorescence quenching sealing agent (Sigma Aldrich, St. Louis, MO, USA), the fluorescence was observed under a Fluorescent Microscope (Nikon, Tokyo, Japan) with excitation 465–495 nm and emission 515–555 nm (green) or excitation 510–560 nm and emission 590 nm (red).
2.9. HE and PAS staining
For hematoxylin-eosin (HE) and periodic acid-Schiff (PAS) staining, the paraffin-embedded renal tissues were sectioned at 3–4 μm thickness. The slides were deparaffinized and stained with H&E and PAS reagent according to the manufacturers’ instructions. The images of each slide were visualized under an optical microscope.
2.10. Morphometric analysis
The morphometric analysis was used to measure the enlargement of glomerular volume (GV) and the mesangial expansion. GV was calculated on periodic acid-Schiff-stained renal sections with a thickness of 4 μm by using the formula of GV=(β/k)(glomerular area)3/2, where β = 1.38 pertains to the sphere and k = 1.10 is the distribution coefficient. The index of the mesangial expansion was defined as ratio of mesangial area/glomerular tuft area.
2.11. Genomic DNA methylation
Genomic DNA was isolated from the kidney tissues of the mouse model using a genomic DNA extraction kit (Takara, Dalian, China). The bisulfite-modified DNAs were obtained using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) with 1 μg genomic DNA, according to the manufacturer's instructions. The bisulfite-modified DNAs were then amplified by PCR with the primers specific for the TRIM13 promoter. The PCR products were cloned with the TOPO Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA), and ten clones of each sample were picked to identify the methylation level of the TRIM13 promoter, as described previously [25]. The primers of TRIM13 promoter were shown as follows: (forward) 5′-AAATAATATGTTGGTGAT GTGATAG-3′ and (reverse) 5′-AGTTTACCTCTGTCTCCTACAAGC-3′.
2.12. Ubiquitination assay
The 293A cells were co-transfected with HA-Ub, His-TRIM13, and pCMV5-myc-CHOP plasmids, with or without MG132 (10 µg/ml) treatment, for 2 h before detection. The ubiquitination of CHOP protein was investigated by immunoprecipitation of the cell lysates with anti-myc antibody. The protein mixtures obtained from immunoprecipitation were then separated by SDS-PAGE and the Ub-CHOP was analyzed by western blotting with anti-HA antibody. The effect of TRIM13 on the ubiquitination of CHOP protein was analyzed by co-transfecting HMCs with HA-Ub and si-TRIM13 or si-Hrd1 or si-gp-78, with or without MG132 (10 µg/ml) treatment, for 2 h before detection. The cell lysates were immunoprecipitated with anti-CHOP antibody, and the ubiquitination levels of CHOP protein in each experimental group were detected by western blotting with anti-HA antibody.
The in vitro ubiquitination assay was performed in a 20 μL reaction system containing ubiquitin conjugation reaction buffer (25 mm Tris-Cl, 5 mm MgCl2, 1 mm dithiothreitol, and 100 mm NaCl), E1 ubiquitin-activating enzymes (UBE1) (100 nM; Boston Biochem, Cambridge, UK), E2 ubiquitin-conjugating enzymes (UbcH5a) (20 μg/mL; Boston Biochem), ubiquitin (50 μM; Boston Biochem), Mg-ATP (5 mM), and complete phosphatase inhibitor mixture and protease inhibitor mixture (Roche, Basel, Schweiz). The reactions were carried out with CHOP protein (0.3 μg) as a substrate and either full-length or truncated TRIM13 recombinant proteins (0.3 μg) at 30 °C for 30 min. The ubiquitination reactions were then stopped by adding LDS sample buffer at 80 °C for 5 min. Western blot analysis was used to analyze the samples. The TRIM13-ΔRING deletion mutant was generated by cutting the TRIM13 construct with PstI and subsequently religating, resulting in the lack of 317 amino acids of the RING domain [26].
2.13. Cell transfection
The pBLAST plasmids containing the full-length human or mouse TRIM13 cDNA were subcloned into the adenoviral (Ad) shuttle plasmids, which were purchased from InvivoGen. The completed TRIM13adv was then transfected into HEK293 cells and purified to obtain Ad-TRIM13 (6 × 109 pfu/mL). The same protocols were used to establish Ad vectors encoding green fluorescent protein (GFP) (Ad-GFP). The Ad-TRIM13 or Ad-GFP vectors were transfected into mesangial cells. The m.o.i of Ad recombinant vectors used for in vitro experiments was 10. HA-Ub, His-TRIM13, and pCMV5-myc-CHOP were transfected into 293A cells using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA). Small interfering RNAs (siRNAs) against TRIM13 (si-TRIM13: 5-CATATCACTTCCGCTAAAT-3) and a scramble control siRNA (5-CTACCTTATTGAGTGGT G-3) (Genechem, Shanghai, China) were transfected into mesangial cells using RNAiMax reagent following the manufacturer's instructions (Invitrogen, Waltham, MA, USA). The working concentration of si-TRIM13 was 30 nM.
2.14. Renal vein injection of adenovirus associated virus (AAV) vectors
AAV vectors encoding TRIM13, CHOP or GFP were established as previously described [27]. Briefly, the AAV9 capsid was used with a chicken β-actin promoter [28]. The TRIM13, CHOP or GFP cDNA was cloned into the AAV9 capsid and the recombinant AAV9 was manufactured by Shanghai GenePharma Co., Ltd. The AAV-TRIM13, AAV-CHOP or AAV-GFP particles were administered to the mice by renal vein injection into the left kidney with a 31 G needle. The dosage of AAV recombinant vectors used for in vivo experiments was 200 μl viruses [1012 genome copy particle (GCP)/ml].
2.15. Statistical analysis
The data are presented as mean ± SD of three independent experiments. All statistical analyses were conducted by using SPSS 17.0 (SPSS, Inc.). Student's t-test and ANOVA with post hoc tests were used to analyze the statistical significance between groups (P<0.05). Correlations between the expression of TRIM13 mRNA in diabetic kidneys and UACR or eGFR were evaluated by Pearson's correlation test. Each experiment was done in triplicate and at least has been repeated three times independently.
3. Results
3.1. TRIM13 is downregulated, while CHOP is upregulated in renal biopsies of DN patients
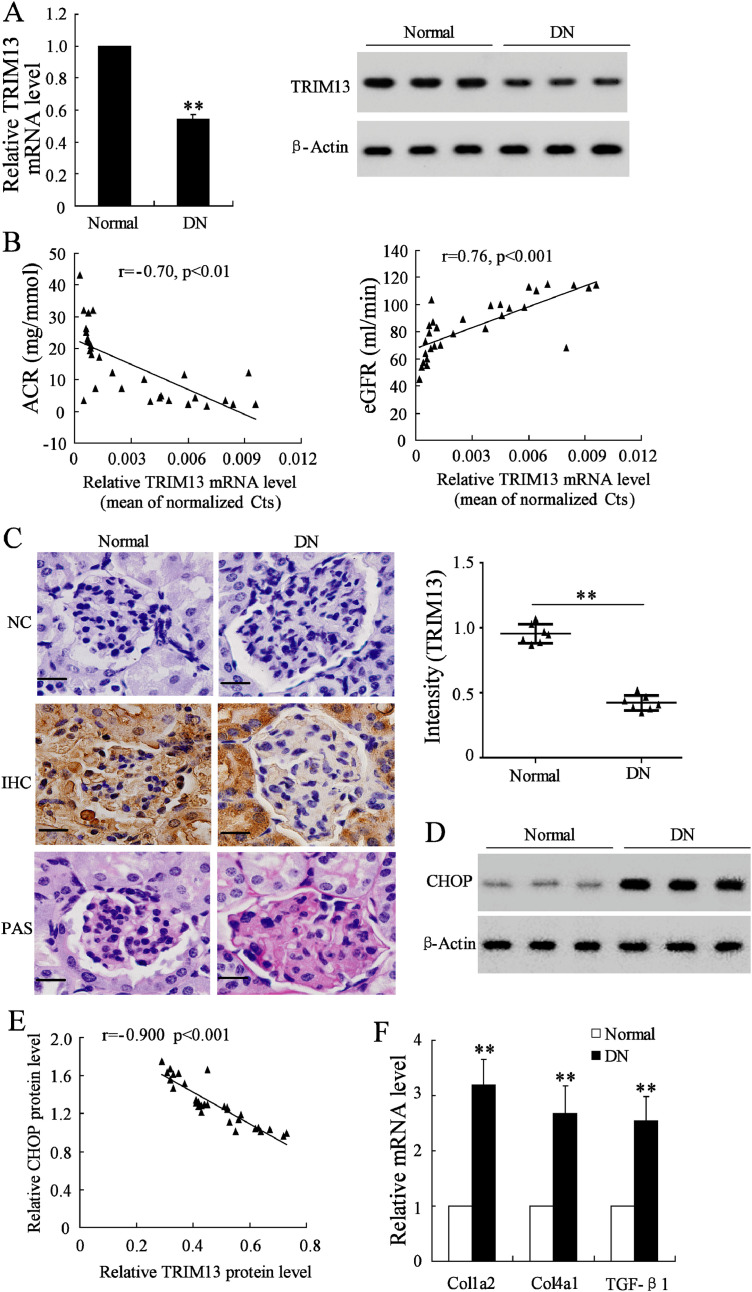
The differential expression of TRIM13 and CHOP in renal biopsies was compared between DN patients and normal controls. We found that both mRNA and protein levels of TRIM13 were markedly reduced in DN patients compared with those in normal controls (Fig. 1A). Then we explored the correlation between TRIM13 and the urinary albumin-to-creatinine ratio (UACR) and estimated glomerular filtration rate (eGFR), the standards by which kidney function is assessed and found that TRIM13 was negatively correlated with UACR, while it was positively correlated with eGFR (Fig. 1B), indicating that the downregulation of TRIM13 is closely correlated to the renal injury of DN patients. The low expression level of TRIM13 in renal biopsies of DN patients was observed in IHC and PAS staining, and the intensity of TRIM13 in IHC staining was significantly reduced compared with normal tissues (Fig. 1C). As previously reported, CHOP is upregulated, and promotes collagen synthesis by increasing the expression of collagen-associated factors, including Col1a2, TGF-β1, and Col1a4 in renal tissues of DN [12, 13]. Such upregulation of CHOP and Col1a2, TGF-β1, and Col1a4 was observed in our experiment (Fig. 1D and 1F). Meanwhile, the CHOP protein level was negatively correlated with the TRIM13 protein level (Fig. 1E).
Fig. 1.
Expression of TRIM13 and CHOP in renal biopsies. (A) The mRNA and protein levels of TRIM13 in renal biopsies from DN patients and normal controls were measured by qRT-PCR and western blot analysis, respectively. (B) The correlation between TRIM13 mRNA and UACR, as well as TRIM13 mRNA and eGFR in renal biopsies of DN patients was analyzed by the Pearson's correlation test. (C) IHC and PAS staining and the IHC intensity analysis of TRIM13 protein in normal and DN biopsies. The scale bar represents 100 μm. Magnification is 400X. (D) The protein level of CHOP. (E) The correlation between TRIM13 protein level and CHOP protein level in renal biopsies from DN patients was analyzed by the Pearson's correlation test. (F) The mRNA levels of collagen-associated factors Col1a2, Col4a1, and TGF-β1. **P<0.01 vs. Normal. Three independent experiments and 10 samples were used in each examination (qRT-PCR, western blot analysis, and IHC/PAS staining). Normal, normal kidney biopsies. DN, kidney biopsies from diabetic nephropathy patients. UACR, urinary albumin-to-creatinine ratio. eGFR, estimated glomerular filtration rate. IHC, immunohistochemistry. NC, negative control.
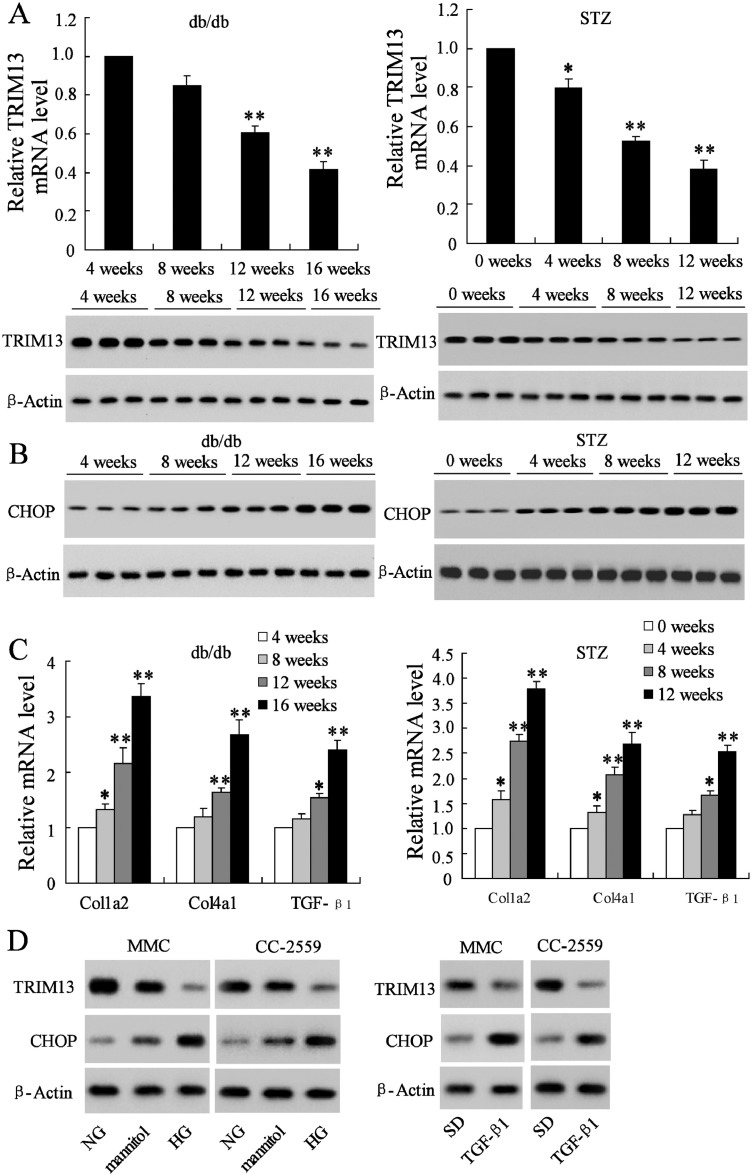
3.2. The expression of TRIM13 and CHOP in glomeruli of diabetic mice and in HG-treated mesangial cells
Given the downregulation of TRIM13 and upregulation of CHOP in renal biopsies of DN patients, we further explore the renal expression level exchange of TRIM13 and CHOP over the duration of the DN model. Renal tissues were harvested and glomeruli was isolated from two types of diabetic mouse model, a db/db mouse model with different ages [from 4 weeks to 16 weeks of db/db mice (n = 8 in each age)], and a streptozotocin- (STZ-) induced mouse model with different time points [from 0 weeks to 12 weeks after STZ treatment (n = 8 in each time point)]. The isolated glomeruli showed highly enriched expression of podocin, a glomerular podocyte marker [29], while cadherin-16, a tubular marker [30], almost was not detected in isolated glomeruli compared with the total kidney lysate (Supplemental Fig. 1A), indicating the high purity of the isolated glomeruli. We observed that, with the age increasing, TRIM13 was gradually decreased, while CHOP was gradually increased in glomeruli as shown in Fig. 2A and 2B. Meanwhile, the expression level of Col1a2, Col4a1, and TGF-β1 was also gradually increased in an age-dependent manner (Fig. 2C). These data suggested that the simultaneous dysregulation of TRIM13 and CHOP was arisen with the development of DN.
Fig. 2.
Expression of TRIM13 and CHOP in glomeruli of diabetic mice and in HG-treated mesangial cells. Renal tissues were collected and glomeruli were isolated from the db/db mice at different ages [from 4 weeks to 16 weeks (n = 8 in each age)] and the streptozotocin- (STZ-) induced mice at different time points [from 0 weeks to 12 weeks after STZ treatment (n = 8 in each time point)]. (A) The mRNA and protein expressions of TRIM13. (B) The protein expression of CHOP. (C) The relative mRNA level of Col1a2, Col4a1, and TGF-β1. In db/db mice, *P<0.05, **P<0.01 vs. 4 weeks; in STZ-induced diabetic C57BL/6 mice, *P<0.05, **P<0.01 vs. 0 weeks. (D) The protein expressions of TRIM13 and CHOP were detected in primary MMCs and HMCs (CC-2559 cells) after the treatment of HG (30 mM) or TGF-β1 (10 ng/ml). Cells treated with NG or mannitol or SD were as the control. *P<0.05 vs. NG; **P<0.01 vs. NG and SD. Three independent experiments. HG, high glucose. STZ, streptozotocin. MMC, mouse mesangial cell. HMC, human mesangial cell. NG, normal glucose. SD, serum depleted.
For the in vitro experiment, mouse mesangial cells (MMCs) and human mesangial cells (HMCs) CC-2559 were stimulated by high glucose (HG, 30 mM), and the protein expression of TRIM13 and CHOP was detected. TGF-β1 (10 ng/ml) was also used for stimulation as the HG can cause the enhancement of TGF-β1, which has been confirmed in DN biopsies and diabetic mice (Fig. 1E and 2C). The result indicated that HG and TGF-β1 stimulation could markedly reduce the protein level of TRIM13 and increase the protein level of CHOP both in MMCs and HMCs (Fig. 2D).
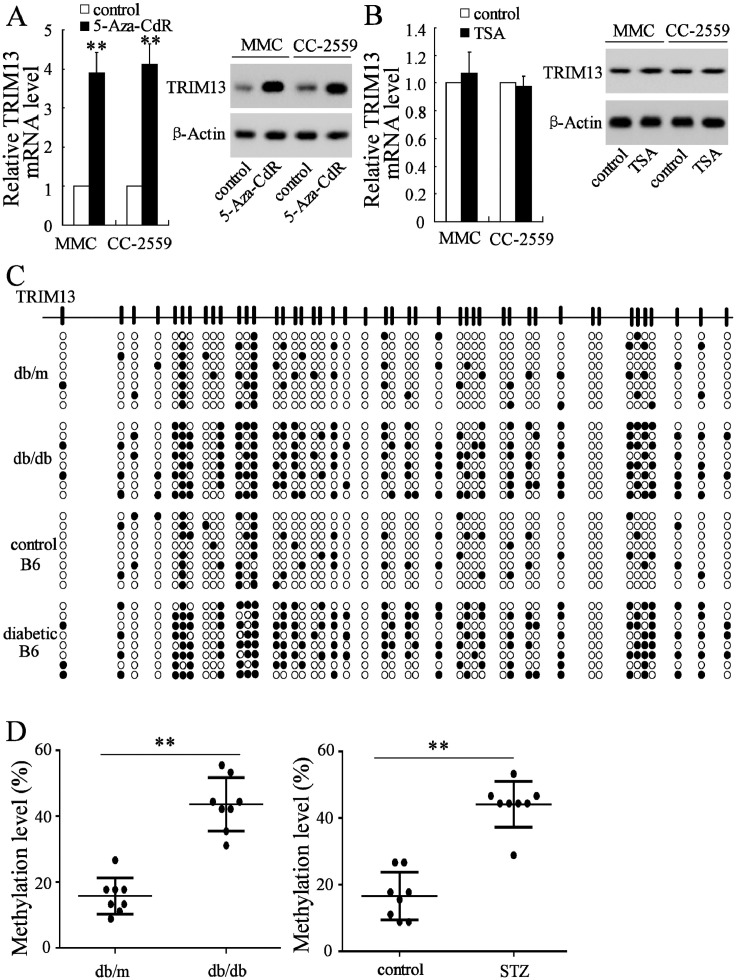
3.3. DNA methylation is associated with the expression of TRIM13
DNA methylation and histone acetylation, belonging to the epigenetic modification, play a critical role in gene expression and cell programing. To verify whether TRIM13 expression is associated with DNA methylation or histone acetylation, MMCs and HMCs were treated with 5-Aza-CdR (5μmol/l), a DNA methyltransferase inhibitor, or TSA (100 nmol/L), a histone deacetylase inhibitor, both of which could inhibit DNA methylation and histone acetylation. We found that the mRNA and protein levels of TRIM13 were dramatically elevated by 5-Aza-CdR treatment (Fig. 3A), whereas they were not significantly affected by TSA treatment (Fig. 3B). Considering that TRIM13 promoter sequences contain multiple CpG islands which could be modified by DNA methylation, we surmised that altered DNA methylation is associated with the expression of TRIM13. Next, we used bisulfite sequencing to measure the methylation levels of the TRIM13 promoter both in renal tissues of db/db mice (n = 8) and STZ-induced mice (n = 8). Results showed that methylation levels of TRIM13 in renal tissues of diabetic mice were higher than those of control mice (Fig. 3C). Consistent with bisulfite sequencing results, the higher methylation levels of TRIM13 in renal tissues of diabetic mice were also verified by qPCR (Fig. 3D). Our data suggested that aberrant methylation was, at least in part, responsible for the downregulated TRIM13 in DN.
Fig. 3.
DNA methylation is associated with the expression of TRIM13. MMCs and HMCs CC-2559 were treated with 5-Aza-CdR (5 μmol/l), a DNA methyltransferase inhibitor, or TSA (100 nmol/l), a histone deacetylase inhibitor, for 48 h. The cell lysate contained TRIM13 mRNA and protein (A) were enhanced in 5-Aza-CdR treated cells, but (B) were with no significant changes in TSA-treated cells. The methylation level of the TRIM13 promoter in renal tissues of db/db mice (20 weeks old, n = 8), db/m mice (n = 8), STZ-induced mice (n = 8) and control mice (n = 8) was detected using bisulfite sequencing (C) and qPCR (D). **P<0.01 vs. control (for 5-Aza-CdR); **P<0.01 vs. db/m or control (for STZ-induced mice). Three independent experiments. MMC, mouse mesangial cells. STZ, streptozotocin.
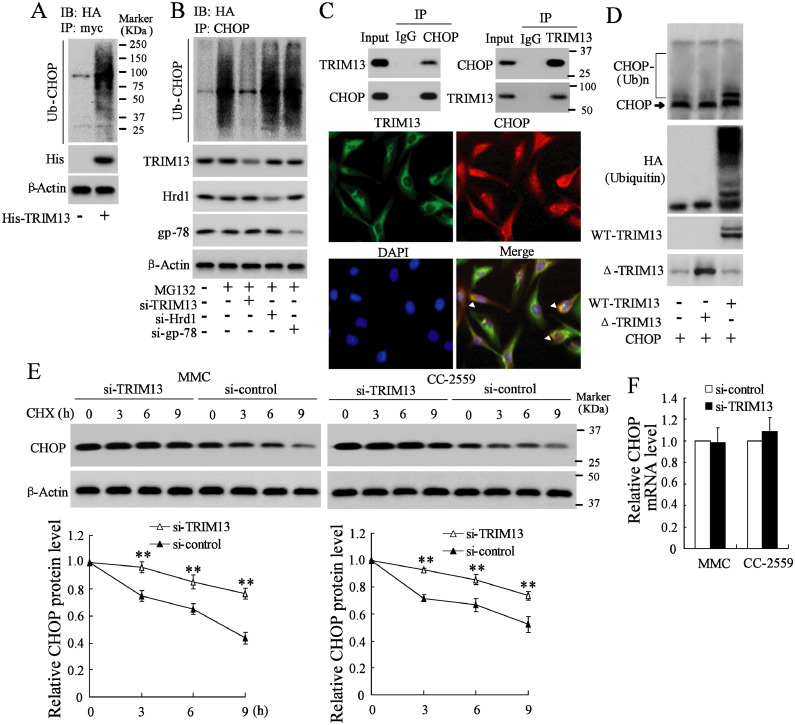
3.4. TRIM13 promotes ubiquitination and degradation of CHOP
As TRIM13 is a well-documented E3 ubiquitin ligase, we next investigated whether CHOP is a substrate of TRIM13. The 293A cells were transiently transfected with Myc-CHOP, and hemagglutinin (HA)-tagged ubiquitin (Ub), and in the presence or absence of His-TRIM13. Cells were harvested at 48 h after transfection. Then we immunoprecipitated Myc-CHOP with anti-Myc, followed by probing the membranes with anti-HA. The high-molecular-weight, polyubiquitinated forms of CHOP which migrated slowly, was observed (Fig. 4A). These data suggested that TRIM13 targeted CHOP for ubiquitination. Next, we transfected CC-2559 cells with HA-Ub, and interfering TRIM13, Hrd1, or gp-78, another two ubiquitin ligases, followed by the addition of MG132, a proteasome inhibitor, to inhibit the degradation of protein via the proteasome pathway. After the immunoprecipitation with anti-CHOP and immunoblotting with anti-HA, we observed polyubiquitinated forms of CHOP after interfering Hrd1 and gp-78, whereas no polyubiquitinated forms of CHOP were shown after interfering TRIM13 (Fig. 4B). These data indicated that CHOP was specifically ubiquitinated by TRIM13. Furthermore, cellular inhibitor of apoptosis protein-1 (cIAP1) is a well-documented CHOP-associated ubiquitin ligase [31]. In the in vivo model, the expression of cIAP1 had no obvious difference in renal tissues of DN mice (n = 8) and the control (n = 8) (Supplemental Fig. 1B). Co-IP assays verified an interaction between TRIM13 and CHOP in CC-2559 cells (Fig. 4C, upper). Immunofluorescent analysis showed that CHOP was predominantly colocalized with TRIM13 in the perinuclear regions (Fig. 4C, lower). These findings indicated that the ubiquitination of CHOP is specifically mediated by TRIM13. To further investigate whether the TRIM13-CHOP is required for CHOP ubiquitination, we used the TRIM13-ΔRING mutant (ΔTRIM13), which lacks a RING domain, a zinc-binding domain determining the E3 ubiquitin ligase activity. The result showed that CHOP was not ubiquitinated by ΔTRIM13 (Fig. 4D), suggesting that the RING domain of TRIM13 is essential for CHOP ubiquitination. We then investigated whether TRIM13-mediated polyubiquitination leads to CHOP degradation. MMCs and CC-2559 cells with or without interfering TRIM13 were treated with cycloheximide (CHX), a protein-synthesis inhibitor, and the expression of CHOP protein was assessed at 3, 6, 9 h after CHX treatment. Treatment with CHX resulted in an accelerated reduction of CHOP in non-interfering TRIM13 cells in comparison with TRIM13-interfering cells (Fig. 4E), which suggested that TRIM13 facilitated degradation of CHOP. However, interfering TRIM13 exerted no significant influence on mRNA expression of CHOP (Fig. 4F).
Fig. 4.
TRIM13 promotes ubiquitination and degradation of CHOP. (A) 293A cells were co-transfected with HA-Ub, pCMV5-Myc-CHOP, and His-TRIM13 for 46 h followed by the treatment of MG132 (10 µg/ml) for 2 h. IP assay with anti-Myc and immunoblotting with anti-HA were performed. (B) CC-2559 cells were co-transfected with HA-Ub and si-TRIM13 or si-Hrd1, or si-gp-78 (another two ubiquitin ligases) for 46 h followed by the treatment of MG132 (10 µg/ml) for 2 h. IP assay with anti-CHOP and immunoblotting with anti-HA were performed. (C) Co-IP assay and the immunofluorescent analysis of TRIM13 and CHOP were performed to investigate the interactions between TRIM13 and CHOP. (D)In vitro ubiquitination assay was performed with recombinant CHOP protein, either recombinant full-length (WT-TRIM13) or ΔTRIM13 that lacks a RING domain. (E) MMCs and CC-2559 cells were transfected with si-TRIM13 or si-control followed by the treatment of CHX (12 μg/ml), a protein-synthesis inhibitor. The level of CHOP protein was detected at 0, 3, 6, and 9 h after CHX treatment. (F) The mRNA level of CHOP after interfering TRIM13 in MMCs and CC-2559 cells. **P<0.01 vs. si-control. Three independent experiments. IP, Immunoprecipitation. CHX, cycloheximide. HA, hemagglutinin. Ub, ubiquitin. WT, wild type. IB, immunoblotting.
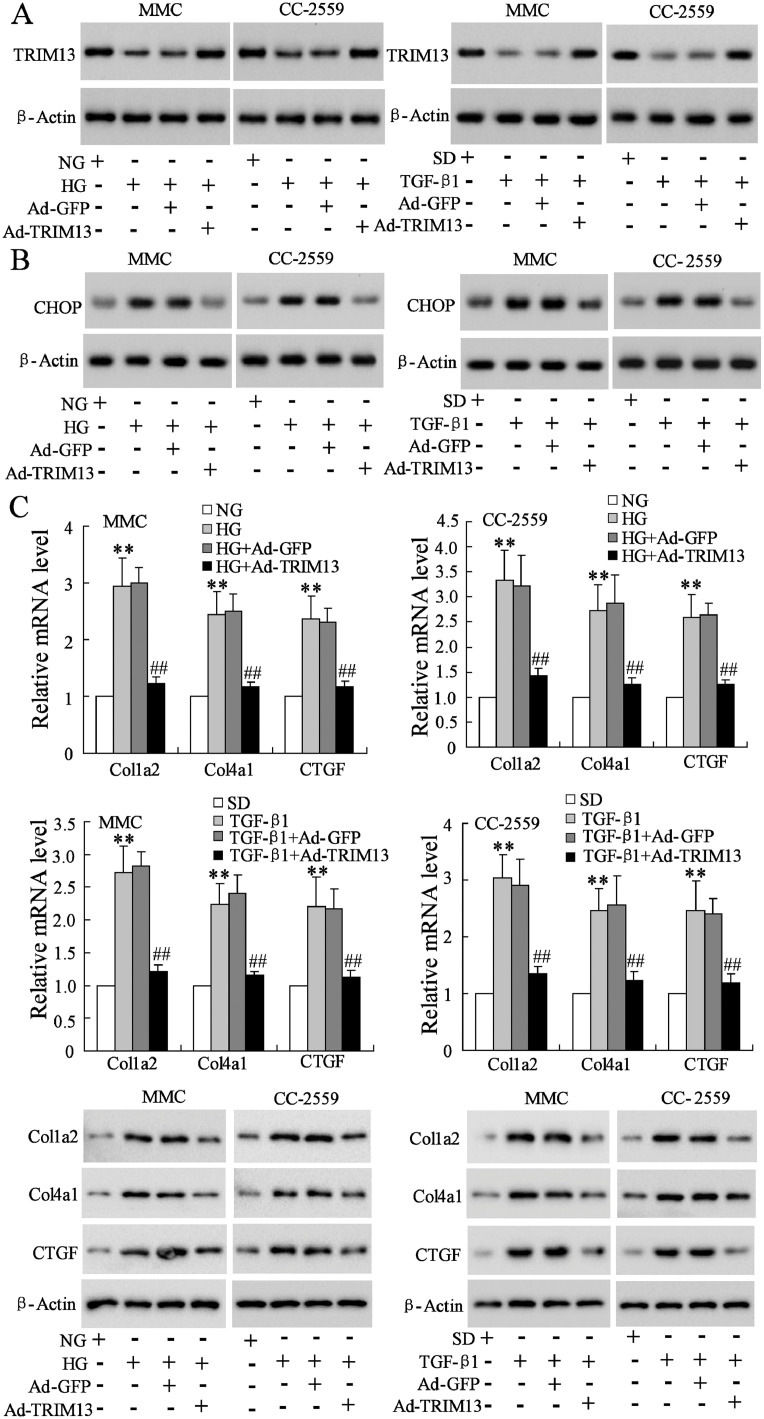
3.5. Overexpressed TRIM13 suppresses HG-induced collagen synthesis via decreasing CHOP
To elucidate whether TRIM13 affected collagen synthesis via regulating CHOP, we transfected MMCs and CC-2559 cells with Ad-TRIM13 to overexpress TRIM13 (Fig. 5A). The result indicated that overexpressing TRIM13 decreased the enhancement of CHOP raised by HG stimulation (Fig. 5B). However, overexpressing TRIM13 did not affect the expression of ATF4 which is an endoplasmic reticulum stress-related factor with a similar function to CHOP (Supplemental Fig. 2). Meanwhile, overexpressing TRIM13 also attenuated collagen synthesis promoted by HG stimulation (Fig. 5C). These data revealed that TRIM13 affects collagen synthesis via specifically regulating CHOP in HG-stimulated mesangial cells. The similar results were observed in TGF-β1-stimulated MMCs and CC-2559 cells as well.
Fig. 5.
Overexpressed TRIM13 suppresses HG-induced collagen synthesis via decreasing CHOP. MMCs and CC-2559 cells were transfected with Ad-GFP or Ad-TRIM13 followed by the treatment of HG or TGF-β1. (A) The protein level of TRIM13. (B) The protein level of CHOP. (C) The mRNA expression and protein level of Col1a2, Col4a1, and CTGF. **P<0.01 vs. NG; ##P<0.01 vs. HG+Ad-GFP; **P<0.01 vs. SD; ##P<0.01 vs. TGF-β1+Ad-GFP. Three independent experiments. MMC, mouse mesangial cell. NG, normal glucose. SD, serum depleted.
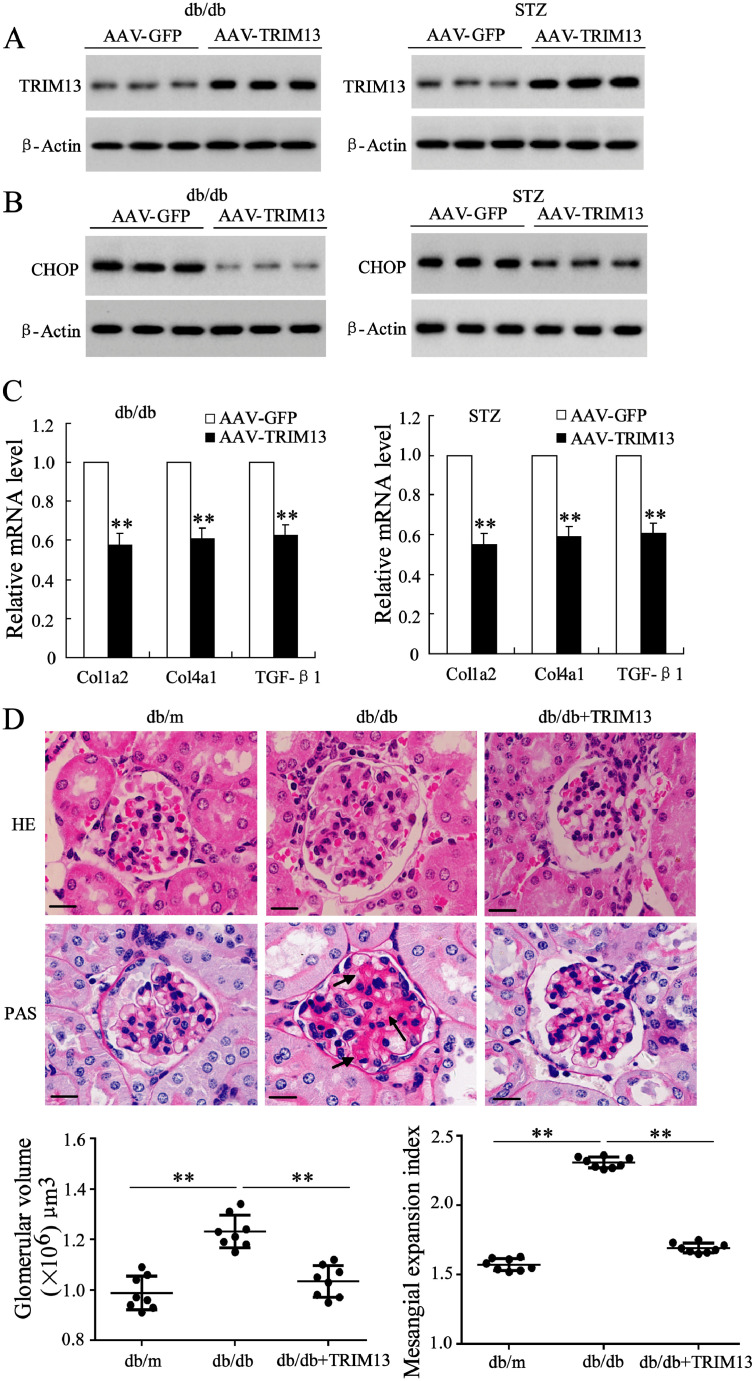
3.6. Overexpressed TRIM13 suppresses collagen synthesis and restores renal function via decreasing CHOP in DN mice
To investigate the physiological function of TRIM13 in the context of DN, we examined the expression of collagen-associated factors after overexpressing TRIM13 in DN mice. TRIM13 overexpression was induced by renal vein injection of adenovirus associated virus (AAV) vectors encoding TRIM13 in db/db mice (n = 8) and STZ-induced mice (n = 8). In isolated glomeruli, the protein expression of TRIM13 was increased (Fig. 6A), while the protein expression of CHOP was decreased by AAV-TRIM13 (Fig. 6B). The overexpressed TRIM13 also markedly suppressed the expression of collagen-associated factors in glomeruli, including Col1a2, Col4a1, and TGF-β1 (Fig. 6C). Morphological images and analysis showed that the glomerular enlargement and the increased mesangial expansion in DN mice (the db/db group) were negated by TRIM13 overexpression (the db/db + TRIM13 group) (Fig. 6D).
Fig. 6.
Overexpressed TRIM13 suppresses collagen synthesis via decreasing CHOP in DN mice. AAV vectors encoding TRIM13 were injected weekly into db/db mice (4 weeks of age) (n = 8) and STZ-induced mice (n = 8) via renal vein for 12 weeks. The expression levels in isolated glomeruli were measured for (A) TRIM13 protein, (B) CHOP protein, and (C) Col1a2, Col4a1, and TGF-β1 mRNA. (D) Representative histology pictures (HE and PAS staining) and morphometric analysis of renal slides of db/m mice, db/db mice and db/db + AAV-TRIM13 mice. Mesangial expansions were shown by black arrows. The scale bars represent 100 μm. Magnification is 400X. **P<0.01 vs. AAV-GFP. Three independent experiments. STZ, streptozotocin. AAV, adenovirus associated virus. DN, diabetic nephropathy.
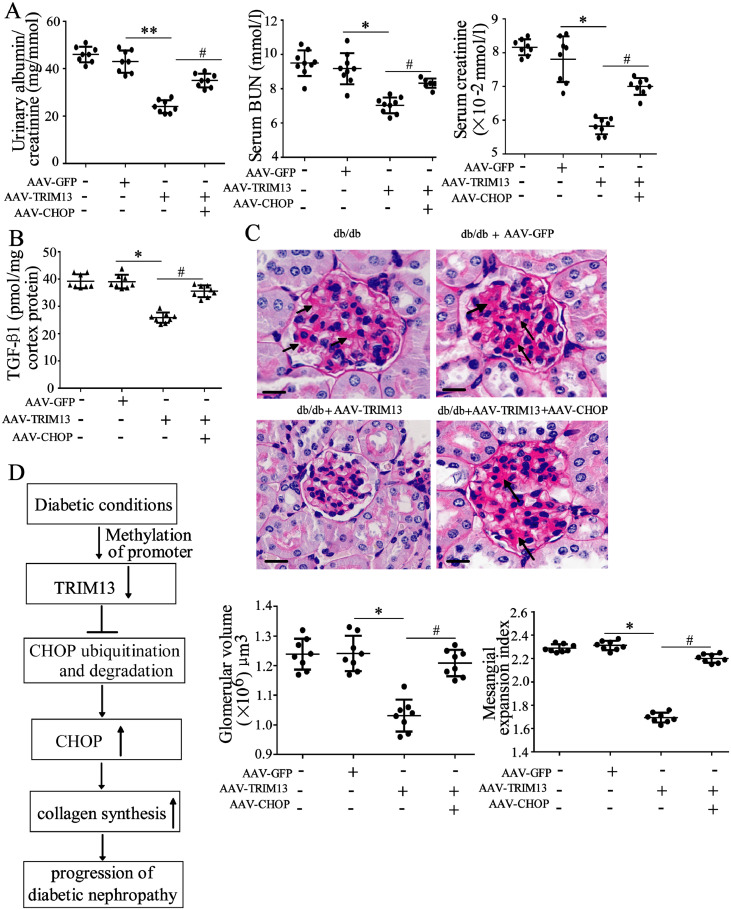
Next, we performed rescue experiments in vivo to determine whether CHOP was a main downstream molecule for TRIM13 in DN. The db/db mice received AAV-TRIM13 injection alone or combined with the injection of AAV-CHOP. Results showed that TRIM13 overexpression substantially reduced the level of UACR, blood urea nitrogen (BUN), and creatinine, whereas the combined injection abrogated this response (Fig. 7A). These findings were also confirmed in STZ-induced mice and control mice. Compared with the control mice [C57BL/6 mice (Supplemental Fig. 3A) and db/m mice (Supplemental Fig. 3B) administered with the citrate buffer], the combined injection restored the decrease of UACR, BUN, and creatinine levels, which was caused by AAV-TRIM13 injection alone (Supplemental Fig. 3C). The expression of TGF-β1 in tissues was significantly reduced after AAV-TRIM13 injection, while such response was negated by the combined injection with AAV-CHOP (Fig. 7B). As shown in Fig. 7C, PAS staining and morphometric analysis showed that AAV-TRIM13 reduced the glomerular size and mesangial expansion (black arrows), whereas combined injection of AAV-TRIM13 and AAV-CHOP partly negated this response. Taken together, our findings indicated that overexpressed TRIM13 could suppress collagen synthesis and restore renal function in DN mice.
Fig. 7.
Overexpressed TRIM13 restores renal function in DN mice. The db/db mice were randomly divided into 4 groups: Control, AAV-GFP, AAV-TRIM13, AAV-TRIM13+AAV-CHOP. AAV-TRIM13 and/or AAV-CHOP vectors were injected weekly via the renal vein for 12 weeks. (A) The UACR, BUN, and creatinine levels. (B) The TGF-β1 level in tissues was detected using ELISA. (C) PAS staining and morphometric analysis of renal glomeruli. Mesangial expansions were shown by black arrows. The scale bars represent 100 μm. Magnification is 400X. (D) Schema depicting the mechanisms of the involvement of TRIM13/CHOP axis in DN. *P<0.05, **P<0.01 vs. AAV-GFP; #P<0.05 vs. AAV-TRIM13. Three independent experiments. UACR, urinary albumin-to-creatinine ratio. AAV, adenovirus associated virus. DN, diabetic nephropathy. BUN, blood urea nitrogen.
4. Discussion
DN, a consequence of sustained hyperglycemia, often progresses to end-stage renal disease which is responsible for considerable individual and socioeconomic costs. Mesangial ECM accumulation reflects increased protein synthesis such as collagen, fibronectin, and laminin, and is the leading cause of DN [32, 33]. As the major component of glomeruli, renal mesangial cells play an important role in collagen synthesis and mesangial ECM accumulation. Recently, accumulating studies have identified several agents that alleviate mesangial cell dysfunction in DN, such as perilla frutescent [34], ramipril [35], and daphnetin [36]. TRIM13, as a member of the TRIM superfamily, exerts E3 ubiquitin ligase activity and is involved in multiple cellular processes, including cell apoptosis and survival, differentiation, and tumorigenesis [17]. However, its role in the mesangial collagen synthesis are rarely reported. In our study, TRIM13 was markedly downregulated in renal biopsies and tissues of DN, and was gradually decreased in an age-dependent manner of diabetic mice. In addition, overexpression of TRIM13 significantly suppressed collagen-associated gene expression in vitro and in vivo, indicating TRIM13 as a potential therapeutic agent in alleviating mesangial cell dysfunction.
The post-genomic era provided insight into the complexity of the ubiquitin system, and more than 1000 proteins regulate ubiquitination in human cells. Proteins of TRIM superfamily exert their E3 ubiquitin ligase activity by the RING domain in the N-terminal end of the typical structure [14]. TRIM13 is a member of TRIM superfamily and mediate various numbers of physiological and pathological events as a result of the diversity of its substrates. To the best of our knowledge, the role of TRIM13 in DN pathogenesis has been rarely reported. Notably, previous studies have demonstrated that CHOP shows a close relationship with the development of DN, and CHOP protein ubiquitination and degradation could be induced by cIAP1, which has E3 ubiquitin ligase activity [31, 37]. In the current study, TRIM13 was predominantly localized in the perinuclear regions (large amount) of mesangial cells using immunofluorescent analysis, as well as in the nuclear (small amount). Although the definite localization of TRIM13 in mesangial cells remains unclear, some studies indicated a perinuclear localization [38] and nuclear localization (small amount [26] or large amount [39]). The large amount of CHOP localized in nuclear was also observed in our study, which consistent with the previous study [40]. We also used IHC and PAS staining to evaluate the TRIM13 expression in glomeruli and tubuli (Fig. 1C). To our surprise, the tubuli level of TRIM13 was not reduced, even a little elevated, which is differ from the glomeruli level of TRIM13. We supposed that the effect of TRIM13 in tubuli may be different from that in glomeruli, which deserve more investigation in our future studies.
Our findings revealed that TRIM13 participates in the course of DN by mediating CHOP ubiquitination and degradation. This finding identified a novel substrate of TRIM13 and provided a new insight in the regulatory mechanism of TRIM13 in DN pathogenesis. Recently, Kato and his colleagues demonstrated that the increased splicing of transcription factor Xbp1 and downregulation of the transcriptional repressor Atf3 co-operate to upregulate CHOP in mesangial cells in response to high levels of glucose or TGF-β [13]. Their study identified the transcriptional regulation of CHOP in DN, while in our study, the mRNA expression of CHOP was also detected in mesangial cells transfected with Ad-GFP or Ad-TRIM13, followed by treatment with high glucose or TGF-β. The result indicated that the mRNA level of CHOP was not affected by TRIM13 (Supplemental Fig. 4), which supported the post-transcriptional regulation between TRIM13 and CHOP. Additionally, CHOP has been defined as a critical mediator of endoplasmic reticulum (ER) stress which is also an initiation factor for the progression of DN [41]. Recent experimental findings have demonstrated that TRIM13, localized in ER, participates in the program of ER-associated protein degradation (ERAD) for evasion of ER stress [26]. We assumed that TRIM13/CHOP axis may affect the process of ER stress in DN, which deserves further investigations.
As a stable system of metabolic information, chromatin modification can be observed at regulatory and coding elements of many genes implicated in metabolism. DNA methylation is one of the major forms of chromatin modification. DNA methylation which is frequently described as a “silencing” epigenetic mark often occurs at 5′-cytosines of CpG dinucleotide, and is reported to be changed by hyperglycemia [42]. Some genes have different methylation between DM patients with or without DN, including UNC13B, PPAPR3, and TRPS1, suggesting altered DNA methylation is relevant to the initiation and pathogenesis of DN [43]. In the current study, we detected the level of DNA methylation and histone acetylation in murine and human mesangial cells, and found that the mRNA and protein levels of TRIM13 were dramatically elevated by DNA methyltransferase inhibitor, whereas they were not significantly affected by histone deacetylase inhibitor. Meanwhile, the methylation level of TRIM13 promoter was increased in renal tissues of diabetic mice. Our findings indicated that the downregulation of TRIM13 in DN is partly related to the high DNA methylation of its promoter under the pathological condition of DM. Consistent with our results, protein expressions of TRIM superfamily members, such as TRIM58 and TRIM28, were regulated by DNA methylation of promoters [44, 45]. Taken together, our data highlight the role of DNA methylation in the pathogenesis of DN, and suggest that suppressing DNA methylation of TRIM13 promoter thus increasing TRIM13 expression may provide a potential therapeutic strategy in treating DN.
In summary, this study demonstrated that DNA methylation-mediated downregulation of TRIM13 is observed in glomeruli and mesangial cells of DN, and overexpression of TRIM13 suppressed mesangial collagen synthesis and restored renal function via promoting CHOP ubiquitination and degradation (Fig. 7D). To the best of our knowledge, it is the first to identify the important role of TRIM13 and its possible mechanism in the context of DN. Our findings add to current knowledge of the pathogenesis of DN and provide new insights for DN treatment.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81970583 and No. 81560132), the Supporting Project for the Foregoers of Main Disciplines of Jiangxi Province (No. 20162BCB22023), and the “5511” Innovative Drivers for Talent Teams of Jiangxi Province (No. 20165BCB18018).
Author contributions
YL conceived and designed the study and drafted the manuscript. DR and YS collected the data and contributed to the statistical analysis. XZ interpreted the data. GX puts forward the concept of the study and reviewed the manuscript. All authors read and approved the final manuscript.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files, or are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.043.
Appendix. Supplementary materials
References
- 1.Sharma D. Diabetic nephropathy: new insights into established therapeutic paradigms and novel molecular targets. Diabetes Res Clin Pract. 2017;128:91–108. doi: 10.1016/j.diabres.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Thomas B. The global burden of diabetic kidney disease: time trends and gender gaps. Curr Diab Rep. 2019;19(4):18. doi: 10.1007/s11892-019-1133-6. [DOI] [PubMed] [Google Scholar]
- 3.Forbes J.M., Coughlan M.T., Cooper M.E. Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes. 2008;57(6):1446–1454. doi: 10.2337/db08-0057. [DOI] [PubMed] [Google Scholar]
- 4.Qian Y. From fibrosis to sclerosis: mechanisms of glomerulosclerosis in diabetic nephropathy. Diabetes. 2008;57(6):1439–1445. doi: 10.2337/db08-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dugbartey G.J., nephropathy Diabetic. A potential savior with 'rotten-egg' smell. Pharmacol Rep. 2017;69(2):331–339. doi: 10.1016/j.pharep.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Sharma K., Ziyadeh F.N. Biochemical events and cytokine interactions linking glucose metabolism to the development of diabetic nephropathy. Semin Nephrol. 1997;17(2):80–92. [PubMed] [Google Scholar]
- 7.Solini A. Angiotensin-II and rosuvastatin influence matrix remodeling in human mesangial cells via metalloproteinase modulation. J Hypertens. 2011;29(10):1930–1939. doi: 10.1097/HJH.0b013e32834abceb. [DOI] [PubMed] [Google Scholar]
- 8.Ziyadeh F.N. Stimulation of collagen gene expression and protein synthesis in murine mesangial cells by high glucose is mediated by autocrine activation of transforming growth factor-beta. J Clin Invest. 1994;93(2):536–542. doi: 10.1172/JCI117004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman B.B. Transcriptional activation of transforming growth factor-beta1 in mesangial cell culture by high glucose concentration. Kidney Int. 1998;54(4):1107–1116. doi: 10.1046/j.1523-1755.1998.00119.x. [DOI] [PubMed] [Google Scholar]
- 10.Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 11.Morse E. TRB3 is stimulated in diabetic kidneys, regulated by the er stress marker CHOP, and is a suppressor of podocyte MCP-1. Am J Physiol Renal Physiol. 2010;299(5):F965–F972. doi: 10.1152/ajprenal.00236.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao D. CHOP mediates XBP1S-induced renal mesangial cell necrosis following high glucose treatment. Eur J Pharmacol. 2015;758:89–96. doi: 10.1016/j.ejphar.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 13.Kato M. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayan K. TRIM13 is a negative regulator of MDA5-mediated type i interferon production. J Virol. 2014;88(18):10748–10757. doi: 10.1128/JVI.02593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D. Resveratrol improves muscle atrophy by modulating mitochondrial quality control in STZ-Induced diabetic mice. Mol Nutr Food Res. 2018;62(9) doi: 10.1002/mnfr.201700941. -e1700941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y. Regulation of vascular large-conductance calcium-activated potassium channels by NRF2 signalling. Diabet Vascular Disease Res. 2017;14(4):353–362. doi: 10.1177/1479164117703903. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama S., Family Proteins T.R.I.M. Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Furman B.L. Streptozotocin-Induced diabetic models in mice and rats. Curr Protoc Pharmacol. 2015;70 doi: 10.1002/0471141755.ph0547s70. 5.47.1-20. [DOI] [PubMed] [Google Scholar]
- 19.Takemoto M. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161(3):799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.S. Differential behavior of mesangial cells derived from 12/15-lipoxygenase knockout mice relative to control mice. Kidney Int. 2003;64(5):1702–1714. doi: 10.1046/j.1523-1755.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 21.Kang S.W. 12-lipoxygenase is increased in glucose-stimulated mesangial cells and in experimental diabetic nephropathy. Kidney Int. 2001;59(4):1354–1362. doi: 10.1046/j.1523-1755.2001.0590041354.x. [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi K. Inhibition of SRC kinase blocks high glucose-induced EGFR transactivation and collagen synthesis in mesangial cells and prevents diabetic nephropathy in mice. Diabetes. 2013;62(11):3874–3886. doi: 10.2337/db12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M.Z. Epidermal growth factor receptor inhibition slows progression of diabetic nephropathy in association with a decrease in endoplasmic reticulum stress and an increase in autophagy. Diabetes. 2014;63(6):2063–2072. doi: 10.2337/db13-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y.C. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol. 2006;17(10):2937–2944. doi: 10.1681/ASN.2006040368. [DOI] [PubMed] [Google Scholar]
- 25.Croes L. DFNA5 promoter methylation a marker for breast tumorigenesis. Oncotarget. 2017;8(19):31948–31958. doi: 10.18632/oncotarget.16654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner M. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18(5):1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dashkoff J. Tailored transgene expression to specific cell types in the central nervous system after peripheral injection with AAV9. Mol Ther Methods Clin Dev. 2016;3:16081. doi: 10.1038/mtm.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocca C.J. rAAV9 combined with renal vein injection is optimal for kidney-targeted gene delivery: conclusion of a comparative study. Gene Ther. 2014;21(6):618–628. doi: 10.1038/gt.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleem M.A. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 30.Thedieck C. Expression of KSP-cadherin during kidney development and in renal cell carcinoma. Br J Cancer. 2005;92(11):2010–2017. doi: 10.1038/sj.bjc.6602597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y., Xia P. Cellular inhibitor of apoptosis protein-1 (cIAP1) plays a critical role in beta-cell survival under endoplasmic reticulum stress: promoting ubiquitination and degradation of C/EBP homologous protein (CHOP) J Biol Chem. 2012;287(38):32236–32245. doi: 10.1074/jbc.M112.362160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanwar Y.S. Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med. 2008;233(1):4–11. doi: 10.3181/0705-MR-134. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q.-l. and W. wang, lncrna-h19 induces retinal müller cell apoptosis via mir-29b/foxo4 axis in diabetic retinopathy. Clinical Surgery Res Commun. 2018;2(4):11–18. [Google Scholar]
- 34.Kim H.-.R., Kim S.-.Y. Perilla frutescens sprout extract protect renal mesangial cell dysfunction against high glucose by modulating AMPK and NADPH oxidase signaling. Nutrients. 2019;11(2):356. doi: 10.3390/nu11020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren W. Ramipril can alleviate the accumulation of renal mesangial matrix in rats with diabetic nephropathy by inhibiting insulin-like growth factor-1. J Acta Cirurgica Brasileira. 2019;34(1) doi: 10.1590/s0102-865020190010000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K. Daphnetin inhibits high glucose-induced extracellular matrix accumulation, oxidative stress and inflammation in human glomerular mesangial cells. J Pharmacol Sci. 2019;139(2):91–97. doi: 10.1016/j.jphs.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y.B. CHOP/ORP150 ratio in endoplasmic reticulum stress: a new mechanism for diabetic peripheral neuropathy. Cell Physiol Biochem. 2013;32(2):367–379. doi: 10.1159/000354444. [DOI] [PubMed] [Google Scholar]
- 38.Tomar D. TRIM13 regulates er stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta. 2012;1823(2):316–326. doi: 10.1016/j.bbamcr.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Joo H.M. Ret finger protein 2 enhances ionizing radiation-induced apoptosis via degradation of AKT and MDM2. Eur J Cell Biol. 2011;90(5):420–431. doi: 10.1016/j.ejcb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 40.He D. C/EBP homologous protein induces mesangial cell apoptosis under hyperglycemia. Mol Med Rep. 2013;7(2):445–448. doi: 10.3892/mmr.2012.1234. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang A., Forbes J.M. Stress in the kidney is the road to pERdition: is endoplasmic reticulum stress a pathogenic mediator of diabetic nephropathy? J Endocrinol. 2014;222(3):R97–111. doi: 10.1530/JOE-13-0517. [DOI] [PubMed] [Google Scholar]
- 42.Marumo T. Diabetes induces aberrant DNA methylation in the proximal tubules of the kidney. J Am Soc Nephrol JASN. 2015;26(10):2388–2397. doi: 10.1681/ASN.2014070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell C.G. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-3-33. -33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W. TRIM58/cg26157385 methylation is associated with eight prognostic genes in lung squamous cell carcinoma. Oncol Rep. 2018;40(1):206–216. doi: 10.3892/or.2018.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbez-Masson L. The hush complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28(6):836–845. doi: 10.1101/gr.228171.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files, or are available from the corresponding author upon reasonable request.