Abstract
Utricularia amethystina Salzm. ex A.St.-Hil. & Girard (Lentibulariaceae) is a highly polymorphic carnivorous plant taxonomically rearranged many times throughout history. Herein, the complete chloroplast genomes (cpDNA) of three U. amethystina morphotypes: purple-, white-, and yellow-flowered, were sequenced, compared, and putative markers for systematic, populations, and evolutionary studies were uncovered. In addition, RNA-Seq and RNA-editing analysis were employed for functional cpDNA evaluation. The cpDNA of three U. amethystina morphotypes exhibits typical quadripartite structure. Fine-grained sequence comparison revealed a high degree of intraspecific genetic variability in all morphotypes, including an exclusive inversion in the psbM and petN genes in U. amethystina yellow. Phylogenetic analyses indicate that U. amethystina morphotypes are monophyletic. Furthermore, in contrast to the terrestrial Utricularia reniformis cpDNA, the U. amethystina morphotypes retain all the plastid NAD(P)H-dehydrogenase (ndh) complex genes. This observation supports the hypothesis that the ndhs in terrestrial Utricularia were independently lost and regained, also suggesting that different habitats (aquatic and terrestrial) are not related to the absence of Utricularia ndhs gene repertoire as previously assumed. Moreover, RNA-Seq analyses recovered similar patterns, including nonsynonymous RNA-editing sites (e.g., rps14 and petB). Collectively, our results bring new insights into the chloroplast genome architecture and evolution of the photosynthesis machinery in the Lentibulariaceae.
Keywords: Utricularia amethystina, Lentibulariaceae, chloroplast phylogenomics, organelle genome, carnivorous plants, polymorphic species, intraspecific variation, ndh genes
1. Introduction
The species of the carnivorous plant family Lentibulariaceae are grouped in three genera: Pinguicula L., Genlisea A.St-Hil., and Utricularia L. [1,2], and are increasingly becoming important plant models mainly due to their alternative nutrient uptake system, their morphological non-orthodox body structure, characterized by Fuzzy Arberian Morphology [3,4], and particular genomic characteristics, such as high mutational levels with nuclear genome shrinkage and expansion in some lineages [5,6].
Utricularia is the biggest genus and most widespread group of carnivorous plants and is very diverse regarding its distribution and habit (e.g., terrestrial, aquatic, lithophytes, epiphytes, and reophytes) [7]. Moreover, several species are polymorphic, which may lead to controversial taxonomic classification. For instance, Utricularia amethystina Salzm. ex A.St.-Hil. & Girard is a terrestrial herb, with petiolate and rosetted leaves. The species is broadly distributed in about 18 different countries of the tropical and subtropical America [7], commonly found in different altitudes (from inselbergs in the Guianas [8] to the coast of Brazil [9]) and habitats such as humid sandy soil of the savannas, swamps, and soil between rocks usually near streams, rivers, and waterfalls.
The species is classified in the Utricularia sect. Foliosa Kamieński, showing common morphological characteristics making it easy to distinguish from other Utricularia sections due to its connate bracts and bracteoles, which is the singular morphology of utricles (carnivorous traps), and the capsule dorsoventrally and bivalvate dehiscence. However, the distinction between the conspecific species is not a trivial task, as U. amethystina shows high intraspecific variation, mainly between reproductive characters, such as the corolla shapes and colors, which can vary from shades of purple (Figure 1A), white (Figure 1B) to yellow (Figure 1C) [7].
Figure 1.
Utricularia amethystina species morphotypes are differentiated mainly by corolla color. (A) U. amethystina purple morphotype; (B) U. amethystina white morphotype; (C) U. amethystina yellow morphotype.
This intraspecific morphological variation resulted in several taxonomic rearrangements since the earliest descriptions at nineteenth century [10,11] and even now there is much controversy about if the species is one or more [12,13]. Taylor (1989) [7] struggled to separate the species based on reproductive characters, such as corolla shape, pedicel sizes, palynological characters, and calyx indumentum, but he couldn’t find traits for enough taxonomical circumscriptions to split the different morphotypes. Indeed, in his Utricularia taxonomic monograph, he synonymized 31 taxa under the binomial “Utricularia amethystina” and he wrote “U. amethystina is a most ‘difficult’ and excessively polymorphic species...” (Taylor, 1989 [7], p. 291). Therefore, he assumed one name for the species, as he was unable to find discontinuities to support taxa separation due to the high degree of polymorphism between populations. However, to date, there is no proper taxonomic treatment to solve this question. In addition, only a few genetic differences have been explored [13], such as the chloroplast regions rps16, trnL-F, trnD-T, and nuclear ITS, but these markers were not able to give enough resolution to distinguish them all. In this context, chloroplast genomes are a valuable resource for phylogenies, and the study of their structure and content can provide clues for improving inter- and intraspecific studies, such as population biology [14], and even the discovery of new species [15].
The chloroplast genomes of most angiosperms have conserved quadripartite structure separated in Large and Small Single Copy regions (LSC and SSC, respectively) and two inverted repetitive regions (IRs) [16]. However, comparative analyses indicate that some plants, such as parasitic [17], mycoheterotrophic (e.g., in [18,19]), and species of carnivorous plants from the order Caryophyllales [20,21], have suffered substantial rearrangement and gene losses throughout plant evolution. For example, across diverse lineages of plants, chloroplast genomes lack NAD(P)H-dehydrogenase (ndh) complex genes, genes that could have been involved in the transition from aquatic to terrestrial habit thought plant evolutionary history [22,23].
Within Lentibulariaceae family, there are published chloroplast genomes (cpDNA) of Pinguicula ehlersiae [24] and seven Genlisea species [24,25]. There are cpDNA genomes available for four Utricularia species: U. gibba [26], U. macrorhiza [24], U. reniformis [27], and U. foliosa [28].
All published Utricularia cpDNAs have the typical quadripartite structure and the same genes as most angiosperms. However, some species exhibit variation, such as two complete copies of ycf1 and ndhF in U. gibba, and U. reniformis, which has reduced chloroplast size due to several losses in all ndhs genes repertoire that could not be integrally found either in the mitochondrial genome [29] nor in the nuclear DNA (unpublished data). Indeed, U. reniformis is a terrestrial species and other assessed Utricularia are aquatic, and based on this observation, Silva et al. (2016) proposed that the loss of ndhs could be related to terrestrial habit [27]. Nonetheless, other studies are still needed for a better understanding of genes especially the evolution of the ndh genes in the genus.
Herein, we present the chloroplast genomes of three Utricularia amethystina morphotypes to assess inter- and intraspecific sequence variability and polymorphic regions that could be used for further phylogenetic studies. In addition, we employed chloroplast transcriptome to assess gene expression and identify the RNA editing sites in each chloroplast. We also compare ndh gene gains and losses across the sequenced Utricularia cpDNA species, to examine the variation of structural changes across the genus.
2. Results
2.1. Structure of Chloroplasts in Utricularia amethystina
A total of 2,873,574 million paired-end reads were generated of all Utricularia amethystina morphotypes. Approximately 7.57%, 9.10%, and 9.56% represents cpDNA-derived reads and were used for the de novo assembly of U. amethystina purple, white, and yellow, respectively. For each morphotype, the assembly using SPAdes resulted in a contig with the entire LSC region, followed by two contigs containing the IR and SSC regions. The cpDNA contigs were joined in a supercontig and circularized using MITOBim iterative read mapping. The three U. amethystina cpDNAs have a consistent quadripartite structure similar to the majority of other angiosperms, and slightly varying in size (Figure 2) (Table 1) (Supplementary Table S1).
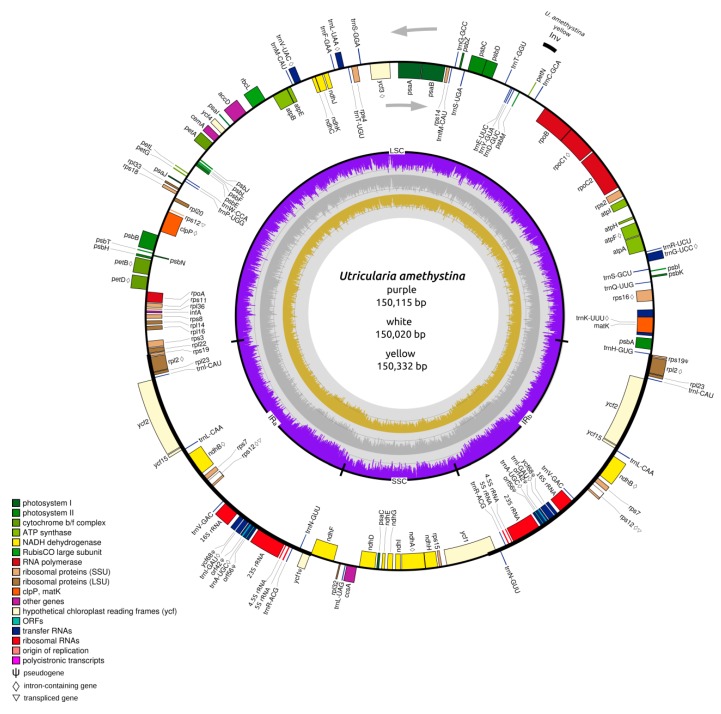
Figure 2.
Chloroplast genome map for Utricularia amethystina purple, white, and yellow. The map represents all three cpDNAs. Gene order and number are the same, except that yellow has an inversion in the petN and psbM genes (see at 2 o’clock in the map). Black thick lines of the outer circle indicate the extension of the inverted repeats. The direction of the arrows denotes the transcription direction. Genes are colored according to their functional groups. The inner graph corresponds to the GC content for each cpDNA region in the chloroplast of each species morphotype. Purple, yellow, and gray bars denote U. amethystina purple, yellow, and white morphotypes, respectively.
Table 1.
The summary of characteristics in Utricularia amethystina chloroplast genomes. Between parentheses the percentage that represents each part in comparison to the cpDNA genome.
| U. amethystina Morphotypes | Purple | White | Yellow |
|---|---|---|---|
| Genbank accession number | MN223721 | MN223722 | MN223720 |
| Genome size (bp) | 150,115 | 150,332 | 150,020 |
| Large single copy (LSC) length (bp) | 82,388 (54.9%) | 82,561 (54.9%) | 82,256 (54.8%) |
| Small single copy (SSC) length (bp) | 16,969 (11.3%) | 17,070 (11.4%) | 16,870 (11.3%) |
| Inverted repeats (IRa+IRb) length (bp) | 25,399 (34.7%) | 25,350 (33.8%) | 25,447 (34.0%) |
| Noncoding regions (bp) | 40,199 | 41,901 | 41,259 |
| Intronic regions(bp) | 18,052 | 19,260 | 20,280 |
| %GC | 37.5 | 37.5 | 37.7 |
| Coverage | 84× | 119× | 123× |
The three cpDNA exhibited in total of 137 annotated genes, including 39 unique protein-coding genes, 30 tRNA, and four rRNA. Eighteen genes (petB, petD, atpF, rpoC1, rps12, rps16, rpl2, ndhA, ndhB (2×), trnK-UUU, trnA-UGC (2×), trnI-GAC (2×), trnG-UCC, trnL-UAA, trnV-UAC) contain one intron, and two genes (clpP, ycf3) have two introns; 18 genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12, trnI-CAU, trnL-CAA, trnV-GAC, 16S rRNA, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, 23S rRNA, 4.5S rRNA, 5S rRNA) have duplicates and five (ycf68, orf42, orf56, ycf1, rps19) partial genes (putative pseudogenes) in the IR regions. All assessed cpDNAs have collinear gene content and arrangement. The main difference among the three morphotypes is the inversion of petN and psbM genes position in U. amethystina yellow in comparison to other Utricularia (Figure 2).
2.2. Repeats and Chloroplast Microsatellites (cpSSR)
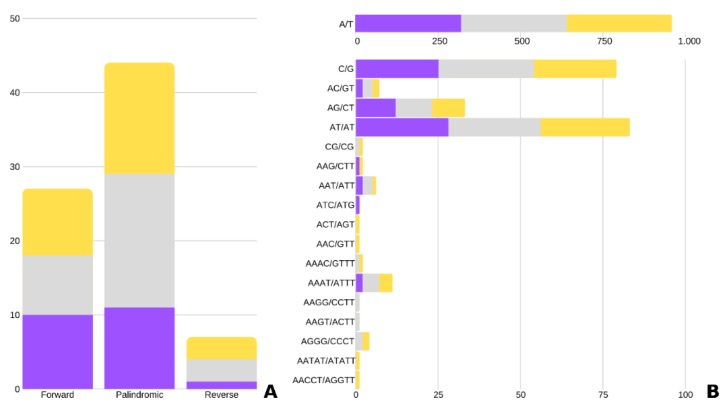
REPuter identified 22, 29, and 27 repeats in U. amethystina purple, white, and yellow, respectively (Figure 3A). Most were characterized as forwarding and palindromic repeats. The repeats were mainly distributed among the ycf2, rpoC1 and trnS-GCU, ycf3, ndhA genes, and rbcL-accD, and rps12-trnV-GAC intergenic region (Supplementary Tables S2–S4). The identified microsatellite (cpSSR), vary from 7 to 369-bp for U. amethystina purple, 7 to 264-bp for U. amethystina yellow, and 7 to 245-bp for U. amethystina white (Figure 3B). The amount of identified cpSSR repeats are similar within all morphotypes, with the mono- (346, 350, 343), and dinucleotide repeats as the most abundant (42, 43, 40), followed by tri- (4, 3, 4) and tetra- repeats (2 for U. amethystina purple and 10 for white and 7 for yellow). Interestingly, penta- nucleotides were only found in U. amethystina yellow, and hexa- repeats were not found in any morphotypes (Supplementary Table S5).
Figure 3.
Quantity of repeats in Utricularia amethystina. (A) Long repeats. (B) Simple sequence repeats (cpSSRs). Purple, yellow, and gray bars denote U. amethystina purple, yellow, and white morphotypes, respectively. (Additional information can be found in Supplementary Table S5).
2.3. Interspecific Comparison
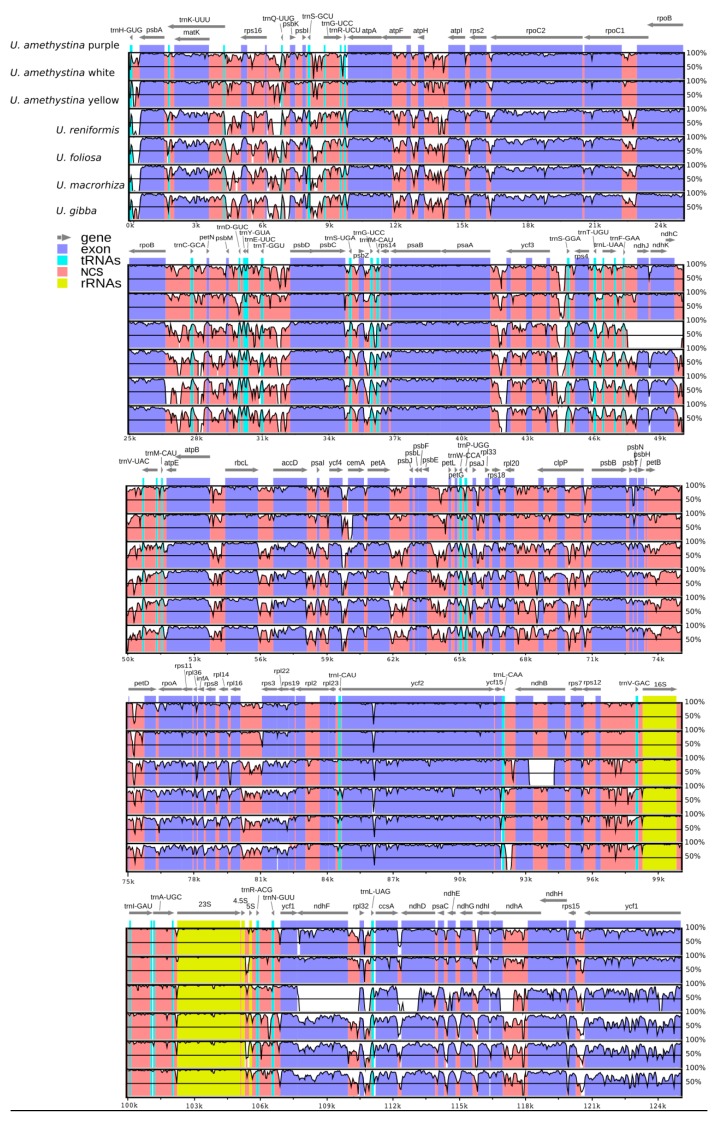
The interspecific chloroplast genome divergence among all available Utricularia cpDNAs, using U. amethystina purple as a reference, showed that U. amethystina specimens have a high degree of synteny; however, for ycf1 and non-coding sequences, they are highly divergent (<50% identity; Figure 4). Furthermore, the coding regions ccsA, matK, rpoC2, rpoC1, rps19, ycf1, ycf2; the introns atpF and rps16; and most intergenic spacers, such as trnK-rps16, rps16-trnQ, psbK-psbI, trnL-trnF, and trnH-psbA, showed high levels of variation that can be used for phylogenetic and DNA-barcoding studies (Figure 4).
Figure 4.
mVISTA identity plot based in Shuffle-LAGAN alignment for Utricularia amethystina morphotypes and previously reported chloroplasts of other Utricularia species. U. amethystina purple was used as a reference. NCS denotes non-coding sequence.
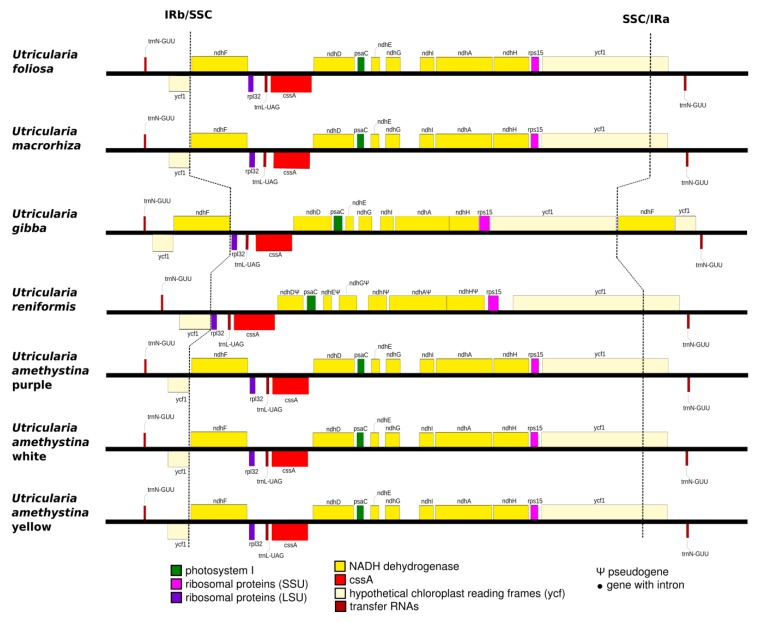
The SSC regions of Utricularia amethystina morphotypes are similar to most Utricularia, except for U. reniformis, due to the deletion of ndhs genes (Figure 5), and U. gibba, which have an extra copy of ycf1 and ndhF.
Figure 5.
Boundaries between SSC regions of all previously published Utricularia species. cpDNA regions color denote different chloroplast gene families.
2.4. Intraspecific Comparison: Species Polymorphism
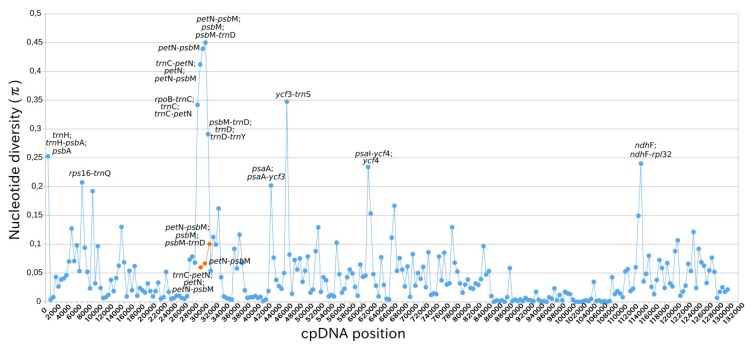
The nucleotide diversity (π) analyses indicated that the IRs exhibited lower variability than LSC and SSC regions (Figure 6). There are twelve spots of chloroplast genome regions that showed remarkably higher π values (>0.02), including nine gene (trnH, psbA, trnC, petN, psbM, trnD, psaA, ycf4 and ndhF) and 12 intergenic regions (trnH-psbA, rps16-trnQ, trnC-petN, rpoB-trnC, petN-psbM, trnC-petN, psbM-trnD, trnD-trnY, psaA-ycf3, ycf3-trnS, psaI-ycf4, ndhF-rpl32). (More details in Supplementary Table S6.)
Figure 6.
Nucleotide diversity (π) for the three Utricularia amethystina morphotypes. Each blue dot represents the nucleotide diversity per 500 bp. Orange dots denote the nucleotide diversity for the un-inverted region in U. amethystina yellow (psbM-petN).
2.5. Chloroplast Expression
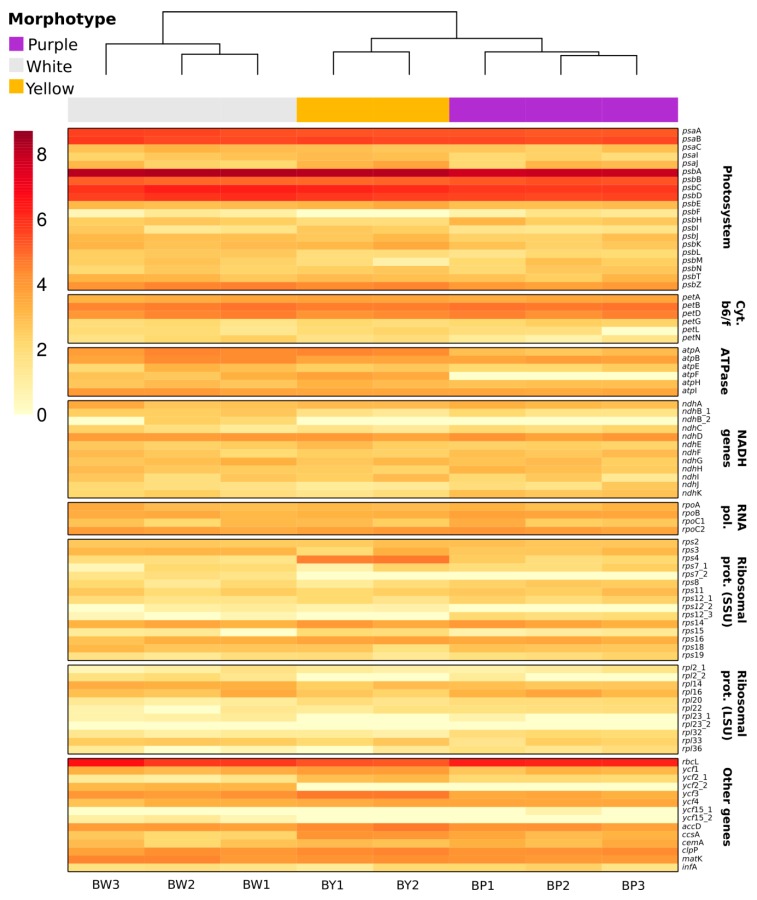
RNAseq clustering analyses indicated distinct expression profiles for each Utricularia amethystina morphotype. The psbA and rbcL genes showed much higher levels of expression, followed by psaA, psaB, psbB, psbC, and psbD, in comparison to other genes in all samples (Figure 7; Supplementary Table S7).
Figure 7.
Heat map representation of Utricularia amethystina cpDNA transcript level. The underscore “_1” and “_2” denotes each gene duplicate. The rps12 is a duplicated trans-spliced gene, therefore it was analyzed in three parts.
2.6. RNA Edit
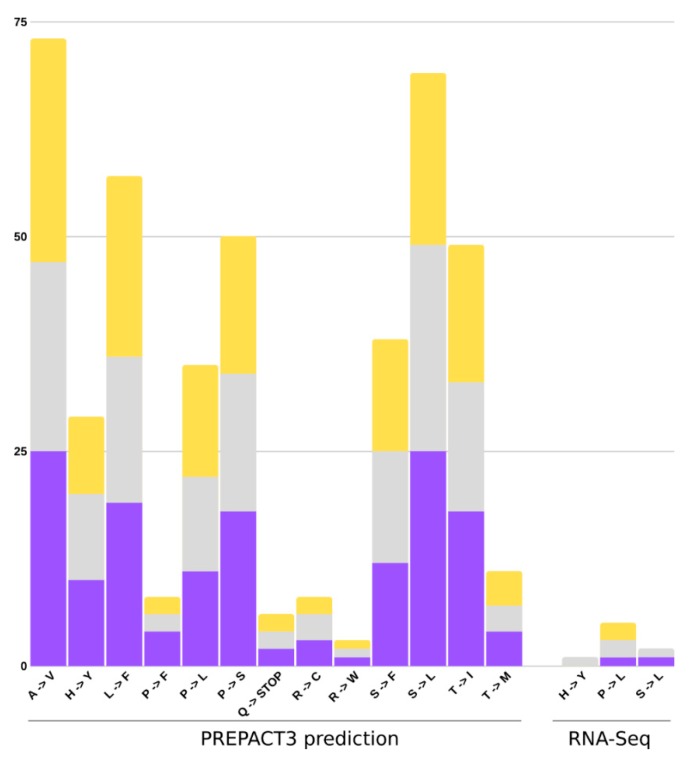
The RNA editing analyses were carried out using the PREPACT3 tool, and in-house script to search for validation of editing sites using RNA-Seq mapped reads. The PREPACT3 predicted 154 sites for U. amethystina purple, 140 for U. amethystina white, and 146 for U. amethystina yellow. Most amino acid changes are shared between populations, and comparison between Utricularia amethystina morphotypes showed 22 genes with the same editing sites and 15 genes with differences in the quantity and amino acid composition changes (for more information see Supplementary Tables S8–S10). According to the results, there are 13 types of amino acid transitions in the three U. amethystina. The most prevalent nonsynonymous substitutional changes occurred between Alanine to Valine and Serine to Leucine, followed by Leucine to Phenylalanine, Proline to Serine, Threonine to Isoleucine, Serine to Phenylalanine, Proline to Leucine, Histidine to Tyrosine, Threonine to Methionine, Proline to Phenylalanine, Arginine to Cysteine, Glutamine to Stop codon, and Arginine to Tryptophan (Figure 8). For RNA-Seq-based results, only three nonsynonymous amino acid substitutions were found (Figure 8; Table 2).
Figure 8.
Quantities of amino acid changes from the prediction and RNA-Seq data of RNA-editing sites for each Utricularia amethystina morphotype. Purple, yellow, and gray bars denote U. amethystina purple, yellow, and white morphotypes, respectively.
Table 2.
RNA editing sites predicted using RNAseq data in Utricularia amethystina cpDNA. The editing level is given in percentage and is showed for each biological replicate. * denotes amino acids with a change in physicochemical composition.
| Morphotype | Gene | Genome (cpDNA) | Codon | Codon Position | Amino Acid | Editing Level | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (Each Biorep) | |||||||||||
| Purple | Position | Strand | from | to | from | to | P1 | P2 | P3 | ||
| rps14 * | 36,640 | - | UCA | UUA | 2 | S | L | 96 | 100 | 100 | |
| petB | 74,797 | + | CCA | CUA | 2 | P | L | 100 | 100 | 97 | |
| White | W1 | W2 | W3 | ||||||||
| ndhB | 138,239 | + | CCA | CUA | 2 | H | Y | 92 | 82 | 0 | |
| ndhD | 113,305 | - | CUA | UUA | 1 | Synonym | 100 | 0 | 100 | ||
| rbcL | 55,651 | + | GCC | GCU | 3 | Synonym | 100 | 100 | 0 | ||
| 55,777 | + | AUC | AUU | 3 | Synonym | 100 | 100 | 0 | |||
| rps14 * | 36,572 | - | UCA | UUA | 2 | S | L | 38 | 14 | 75 | |
| 36,497 | - | CCA | CUA | 2 | P | L | 100 | 0 | 72 | ||
| 36,553 | - | AAC | AAU | 3 | Synonym | 100 | 100 | 0 | |||
| psbB | 72,140 | + | GGC | GGU | 3 | Synonym | 100 | 100 | 0 | ||
| 71,372 | + | UAC | UAU | 3 | Synonym | 100 | 0 | 100 | |||
| psaA | 39,370 | - | AUC | AUU | 3 | Synonym | 100 | 100 | 100 | ||
| petB | 74,816 | + | CCA | CUA | 2 | P | L | 100 | 0 | 97 | |
| Yellow | Y1 | Y2 | - | ||||||||
| rps14 | 36,439 | - | CCA | CUA | 2 | P | L | 69 | 71 | - | |
| ccsA | 111,950 | + | CUA | UUA | 1 | Synonym | 18 | 16 | - | ||
| petB | 74,948 | + | CCA | CUA | 2 | P | L | 100 | 100 | - | |
The results from RNA-Seq data showed eight nonsynonymous and eight synonymous sites for the three U. amethystina morphotypes. Regarding nonsynonymous sites, U. amethystina purple and yellow have rps14 and petB genes edition, and for U. amethystina white, edition sites were two in rps14, petB, and ndhB (Table 2). The same edited sites in the same gene position were found in petB for all U. amethystina samples. In addition, the rps14 gene from purple is the same as rps14 in position 36,572 for white, and rps14 gene from yellow is the same as the rps14 in position 36,497 for white.
2.7. Phylogeny
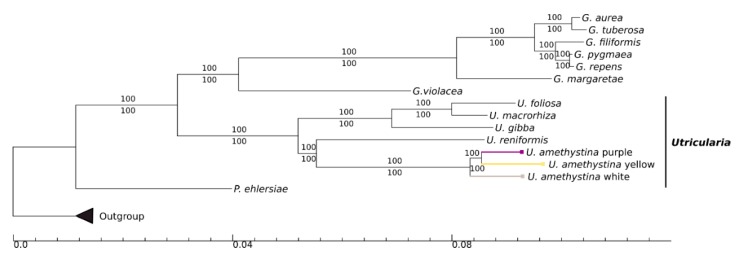
The phylogenetic tree based on available chloroplast genomes of 15 Lentibulariaceae specimens is shown in Figure 9. Both maximum likelihood (ML) and Bayesian inference (BI) trees exhibited identical phylogenetic topologies, and support values (bootstrap for ML and posterior probabilities for BI) are 100% for all clades. The Lentibulariaceae is known to be monophyletic, and Pinguicula is a sister clade to Utricularia-Genlisea. The results show that the Utricularia genus is monophyletic and the three U. amethystina are closely related to U. reniformis. Also, the U. amethystina yellow is sister to purple, with white as having the same common ancestor (Figure 9).
Figure 9.
Bayesian inference tree for available Lentibulariaceae chloroplast genomes and studied Utricularia amethystina. The values above branches denote the posterior probabilities and below maximum likelihood bootstrap. The branch colors correspond to each U. amethystina morphotype flower color. (G. = Genlisea; P. = Pinguicula; U. = Utricularia). The scale represents the expected changes per site.
3. Discussion
Due to the plasticity in corolla shape and color, Utricularia amethystina is one of the most polymorphic species within the Utricularia genus. This polymorphism resulted in a historically taxonomic complicated group with its systematics only partially resolved to date. During the last decades, several efforts attempted to separate the different U. amethystina morphotypes into different species, yet without much success [7,12,13].
In this study, we analyzed the cpDNAs of three morphologically distinct Utricularia amethystina from different populations: the purple, white, and yellow morphotypes, aiming to detect intra- and interspecific variations and phylogenetic signals and provide new cpDNA regions for evolutionary studies. In addition, we evaluated the transcription and RNA editing sites for U. amethystina populations.
In an attempt to diminish the environmental conditions bias, we have collected the specimens from close populations (~2.8 km between purple and white, 0.2 km between white and yellow, and 2.82 km between purple and yellow. The specimens of U. amethystina cpDNA have a typical quadripartite structure present in most land plants and have a similar organization and GC content to other Utricularia [24,27].
Among the three Utricularia amethystina morphotypes, we found an inversion between the petN and psbM genes in U. amethystina yellow, representing the first known gene inversion in LSC region identified in Lentibulariaceae chloroplast genomes. Indeed, the same inversion was detected in the chloroplast genome of species of Cannabaceae [30], and microstructural short inversions of 10 bp were also found in the petN-psbM region in Solanus species [31]. Some comparative cpDNA studies have also identified structural mutations in monilophyte chloroplast genomes, including as many as six inversions and some gene losses (e.g., in [32,33,34]).
In general, chloroplast deletions/losses are observed among Lentibulariaceae. Indeed, Utricularia reniformis suffered a major SSC region retraction due to the losses of NAD(P)H-dehydrogenase (ndh) complex genes [27]. In contrast, all other sequenced Utricularia cpDNAs have complete ndhs gene complexes. These chlororespiratory genes are ndhA, B, C, D, E, F, G, H, I, J, and K, and encode subunits of the NADH dehydrogenase complex in plant chloroplast genomes that play a role in plant signaling in the photosynthesis reaction [35] and the reduction and oxidation of plastoquinones [36]. As U. reniformis is a terrestrial species, it has been proposed that possibly all terrestrial species of Utricularia may lack members from the ndh genes complex [27,28]. However, U. amethystina is terrestrial, and all three morphotypes retain all plastid ndhs complex genes. Therefore, our results now suggest that the ndhs in terrestrial Utricularia were independently lost and regained, thus refuting the hypothesis (at least for Utricularia) that terrestrial species have experienced the loss of ndhs genes.
Chloroplast repeats are important regions for replication and DNA stability [37]. Microsatellites or SSRs are tandem repeats of 1–6 base pairs units long that can be used as genetic markers [38]. They are most commonly found in plants and due to genetic variation in the number of tandem repeats units. Therefore, as they produce polymorphism detectable with PCR-based methods banding pattern and genotyping, the SSRs are widely used in population genetics and evolutionary studies [39]. Utricularia amethystina has high amounts of mononucleotide repeats in the cpSSR, which is similar to other angiosperms, such as Arabidopsis thaliana [40], and other Lentibulariaceae [24,25,27]. Previous results for Utricularia indicated that most of SSR were found in coding regions for U. gibba, U. macrorhiza, Genlisea margaretae, and Pinguicula ehlersiae. However, for U. reniformis, more cpSSR were found in non-coding regions. In U. amethystina, long repeats have similar quantities between populations, and as seen in other Utricularia, most of them are in coding regions [24,27], an uncommon fact for other angiosperms chloroplast genomes (e.g., see [41]), which could indicate high rates of recombination and rearrangement, as discussed in Silva et al. (2016) [27]. Although long repeats could be the cause for gene rearrangements, we could not find repeats in flanking regions of the genes psbM and petN in U. amethystina yellow. Therefore, this indicates that other evolutionary forces were involved with the observed inversion of these genes in this species.
For some Utricularia, DNA barcoding approaches have been considered a difficult task to perform. For instance, the DNA-barcoding markers, such as ITS, rbcL, and matK, could not discriminate all Utricularia accessions at the species and population level due to their low level of polymorphism (e.g., Utricularia sect. Utricularia in Astuti et al., 2019 [42]). Furthermore, rps16, trnL-F, and trnD-T markers cannot discriminate U. amethystina populations [13]. Therefore, it is important to explore regions with high variability at inter- and intraspecies levels that represent potentially useful markers for future studies. Using mVISTA results for the interspecific divergence analysis, it is noticeable that the LSC and SSC regions are more variable than IR regions, corroborating with the results found for identity analyses with Genlisea species [25] and other angiosperms [41]. The results showed highly variable regions between the different species, mostly represented by intergenic spacers, such as trnH-psbA, trnK-rps16, and rps16-trnQ, which could be used for interspecies identification.
It is previously proposed that populations from closely related environments should be less divergent if they are of the same species. However, we observed high intraspecific chloroplast sequence variability, although geographical sampling covered a restricted area. Among the regions with high nucleotide diversity and intraspecific variations, there is the intergenic spacer, trnH-psbA, which is already being used as DNA barcoding in many studies [43]. This study also revealed spots that can be used for populations and phylogenetic analyses due to high variability, such as the genes trnH, psbA, and intergenic regions, such as trnH-psbA, ycf3-trnS, and rps16-trnQ (see more in the Results section and Supplementary Material S6). Nevertheless, the spots of diversity near the genes petN and psbM should be avoided due to low primer annealing considering that the region could be inverted, as seen in U. amethystina yellow.
The preparation of paired-end libraries was enriched for polyadenylated transcripts which causes the instability of organelle transcripts, therefore there is probably underrepresentation of transcripts [44]. However, we were able to observe that almost all chloroplast protein-coding genes are expressed in all sampled flower tissue of U. amethystina, except for ycf15, both rpl23 duplicated genes in U. amethystina purple and yellow, and the atpF gene in U. amethystina purple (Supplementary Table S7).
The expression profile is similar between samples of the same morphotypes and even expression profile clustering corresponds to the phylogenetic hypothesis proposed in this research. The rbcL gene was one of the most highly expressed genes and encodes for one of the most abundant enzymes in nature, the large subunit of ribulose-1-5-biphosphate carboxylase [45]. This protein is involved in fixing CO2 and photorespiration [46]. Moreover, high levels of gene expression were found in Photosystem I (PSI) and II genes (PSII), such as psaA and psaB, and psbA, psbB, psbC, and psbD, these proteins are involved in photosynthesis [47]. Studies of barley leaf activities showed that dark-grown plants were deficient in PSI and PSII proteins [48]. Moreover, Klein et al. (1988) showed that the elongation of translation in psaA, psaB, psbA, and rbcL are regulated by light [49,50]. Therefore, considering that the corollas were collected during the day, our results are congruent with the hypothesis of a protein exhibiting light-induced translation.
Interestingly, the petN and psbM genes are expressed in all Utricularia amethystina biological samples, indicating that, the inversion observed in U. amethystina yellow did not affect the expression of these genes.
RNA editing sites are common features of a plant chloroplast. These mutations usually occur from C-to-U in mRNA molecules, and thus have an important role in the differential amino acid generation that can lead to different proteins originated from the same gene [51]. RNA-Seq-based results showed that there is a sum of eight editing sites for all U. amethystina morphotypes (Table 2).
The PREPACT3′s prediction showed that most nonsynonymous substitutions were characterized as Alanine to Valine and Serine to Leucine. Both lead to protein variations (Table 2, Tables S8–S10), whereas amino acid changes from Alanine to Valine, Histidine to Tyrosine, Leucine to Phenylalanine, Proline to Phenylalanine, Proline to Leucine, Proline to Serine, Arginine to Tryptophan, Threonine to Isoleucine, and Threonine to Methionine result in no physicochemical properties changes in protein. In addition, the Arginine to Cysteine, Serine to Leucine, and Serine to Phenylalanine mutations can modify protein formation due to hydrophilic (Serine and Arginine) to hydrophobic (Leucine, Phenylalanine, and Cysteine) molecule changes [52,53]. Moreover, PREPACT3 has predicted that the genes rps2 and rpl32 can be edited from Glutamine into a Stop codon, and despite they could be polycistronic genes as in other plants [54], these genes are still transcribed according to RNA-Seq data.
The presented evolutionary history, based on whole chloroplast DNA genomes, and reconstructed by ML and BI approaches supported the same relationship within the Lentibulariaceae when compared with one or few loci–loci analyses (Figure 8) [2,55]. These analyses and many other studies indicated that Utricularia amethystina can be paraphyletic [13]. However, in this study, despite the differences in the specimen, they are still a monophyletic taxon. This indicated that U. amethystina morphotypes have a common ancestry, but the sampling of other species from sect. Foliosa (U. tricolor and U. tridentata) and species from the close phylogenetically related sect. Psyllosperma would be necessary to shine this issue.
Our results support that the sampling based on three different morphotypes proved to be insufficient to allow firm conclusions on the U. amethystina species separation, considering we sampled one individual per morphotype. However, the scenario presented here based on chloroplast genomes suggests that U. amethystina morphotypes may be different species as previous studies based on morphometric approach [12] and phylogeny with few loci [13], but with more populations, have suggested.
Moreover, the comparative and functional analyses provided by this study bring new insights into the Utricularia chloroplast genome architecture, in particular, the evolutionary history of ndh complex genes and other important photosynthesis-related genes. Taken together, these results prove that we are just in the beginning for the understanding of the evolution of chloroplast photosynthesis machinery in the Lentibulariaceae.
4. Materials and Methods
4.1. Sampling and DNA Extraction
The three Utricularia amethystina morphotypes were collected from natural populations geographically close to each other (~2.8 km between purple and white; 0.2 km between white and yellow; and 2.82 km between purple and yellow). The samples were preserved in silica gel and stored at room temperature. Vouchers were deposited at Universidade Estadual Paulista (UNESP) in the Herbarium JABU (Table S1). The total genomic DNA was extracted from approximately 0.1 µg of flowers with Qiagen DNeasy Plant Mini extraction kit (Qiagen, Hilden, Germany) following manufacturer’s protocol. The quality and quantity of DNA were estimated with Nanodrop 2000 (Thermo Scientific, MA, USA) and Qubit fluorometer (Life Technologies, CA, USA), respectively.
4.2. Organellar Genome Sequencing, Assembly and Annotation
Sequence libraries were quantified using Bioanalyzer 2100 (Agilent, CA, USA). The paired-end libraries were prepared using Illumina library preparation manufacturer’s protocol, and genomic DNA of 2 × 100 bp and insert size of ~200 bp was sequenced using Illumina MiSeq platform (Illumina, San Diego, CA, USA)
The produced paired-end reads filtered for adapters, low-quality bases (Phred score Q > 24) and size (length cutoff for 50 bp), and possible contaminants using Trimmomatic v. 0.38 [56]. The resulting paired-end reads were mapped in the search for discarding mitochondria and nuclear reads with Bowtie2 v.2.2.3 [57] using very sensitive local and -N 1 parameters and Utricularia spp. (NC_021449, KY025562, and KT336489) chloroplasts as reference genomes. The resulting reads were assembled using SPAdes v. 3.7.1 [58] and regions with assembly uncertainties were extended using iterative read mapping performed using MITObim v.1.8 [59].
The organelles genomes were primarily annotated using DOGMA [60], cross checked with GeSeq [61], and start and stop codons were adjusted manually for annotation refinements. The tRNAs were annotated using tRNA-scan [62], implemented in DOGMA and Aragorn [63]. The rRNAs were annotated using RNAmmer and BLASTn searches with available Utricularia cpDNA genomes. The cp genome map was constructed using the Organellar Genome Draw program [64].
4.3. Repeats and SSR Analyses
To avoid redundant results, only one IR of each Utricularia amethystina cpDNA was used and direct, forward, reverse, and palindromic repeats were identified using REPuter [65] with a minimal size of 30 pb and Hamming distance of 3. Simple sequence repeats (SSR) were detected using MISA-web [66] by setting the minimum number of repeats to 7, 4, 4, 3, 3, and 3, for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively.
4.4. Phylogenetic Reconstruction
Sequence alignment for all published and available Lentibulariaceae cpDNA genomes (Species and accession number, respectively: Utricularia foliosa, KY025562; U. gibba, KC997777; U. reniformis, KT336489; U. macrorhiza, HG803177; Pinguicula ehlersiae, HG803178, Genlisea aurea, MF593121; G. violacea, MF593126; G. tuberosa, NC_037082; G. filiformis, MF593122; G. pygmaea, MF593123; G. repens, MF593124; G. margaretae, HG530134) were performed with online MAFFT v.7 [67] with default parameters. The phylogenetic tree reconstruction was performed using the Bayesian inference (BI) and maximum likelihood (ML) approaches under the best-of-fit model GTR+G+I, in accordance with Akaike Information Criterion (AIC), assessed by Mr.ModelTest v.2.4 [68]. For BI, set for the substitution model accordingly, we employed Mr.Bayes v.3.2.6 software with 5 × 107 generations sampled for each 1000 generations using two runs and four chains, until the average standard deviation of split frequencies became less than 0.01, beginning with random trees. The initial trees were discarded after reaching stationary (~25%). For ML we used the software RAxML v.8.2.10 [69] with 1000 bootstrap pseudoreplicates. The analyses were performed using implementations within CIPRES Science Gateway v.3.3 (https://www.phylo.org/) and cladograms were edited using TreeGraph v.2.15 beta [70]. The species Tectona grandis (NC020098), Sesamum indicum (NC016433), and Tanaecium tetragonolobum (NC027955) were used as outgroup.
4.5. Intraspecific Polymorphism Analyses
For polymorphism analyses, the Utricularia amethystina chloroplasts were aligned using MAFFT v.7, with default parameters. Based on the cpDNA multiple alignments, polymorphism analysis was conducted for coding genes, introns, and intergenic spacers. The nucleotide diversity was calculated using Tassel v.5.2.54 [71] with a sliding window of 500 bp length.
4.6. Interspecific Comparison
Using the Utricularia amethystina purple as a reference and previously published Utricularia (Utricularia foliosa, KY025562; U. gibba, KC997777; U. reniformis, KT336489; U. macrorhiza, HG80317), the identity of cpDNA was assessed using mVISTA online software (http://genome.lbl.gov/vista/mvista/submit.shtml) with Shuffle-LAGAN Mode.
4.7. RNA Extraction, Sequencing and RNA Editing Site Analyses
The corollas of Utricularia amethystina were stored in RNAlater® (Thermo Fisher Scientific, MA, USA) from each analyzed population and were used as plant tissues for RNA-Seq. The corollas (~5 per specimen) were pooled in three replicates for U. amethystina white and purple and two for U. amethystina yellow, and total RNA was extracted using PureLink RNA MiniKit (Thermo Fisher Scientific, MA, USA), according to manufacturer’s protocol. The extracted RNA was analyzed with Agilent 2100 Bioanalyzer and Qubit 2.0 Fluorometer for quality and quantity assessment, and only samples with RNA Integrity Number (RIN) > 7.0 were used for the sequencing.
The eight libraries (3 libraries for each U. amethystina purple and white and 2 for U. amethystina yellow) were constructed following the TruSeq Stranded mRNA LS Protocol sample preparation protocol. The paired-end (2 × 100 pb) sequencing was performed in one lane on an Illumina platform (HiSeq 2500) following supplier-provided protocols (Illumina, San Diego, CA, USA).
The raw sequencing data, was preprocessing with high stringency using the following steps. (1) For the 3′ end, the adapter and low-quality reads were removed using Scythe (https://github.com/vsbuffalo/scythe; default parameters, except for -n 5 and -M 15); (2) for the 5′ end, the removal of adapter and low quality reads were performed with Cutadapt [72]; default parameters, except for –overlap 5; –minimum-length = 15; --times = 2); (3) to filter reads with more than 30% of unknown base (Ns), polyA/T tails we used the software Prinseq [73].
Filtered RNA-seq reads were mapped against the assembled chloroplast genome using STAR version 2.7.2a [74], using default parameters except for adjusted parameters to perform an end-to-end mapping, diminish multiple mapping of the same reads, minimum and maximum size of introns and the number of allowed mismatches (--outFilterMultiMapMax = 3; --outFilterMismatchNmax = 2; --outFilterMismatchNoverLmax = 0.1; --outSJfilterReads = Unique; --alignEndsType = EndtoEnd; --alignIntronMin = 70; --alignIntronMax = 2500). To estimate differential transcripts abundance between biological replicates, normalized count data was obtained using relative log expression (RLE) method in DESeq2 version 3.9 [75] and results were showed following with log2(norm. counts+1). The rps12 is a duplicated trans-spliced gene, therefore it was analyzed in three parts and “_2” and “_3” represent the duplicated regions. The putative RNA edit sites were predicted following PREPACT3 software [76] with BlastX searches (using default parameters) against the Nicotiana tabacum, as reference. The prediction results were compared with the results obtained with an in-house script that counts the number of editing sites according to the previous STAR mapped reads, except for the number of mapped reads, which was set to 1 (only uniquely mapped reads). All of the sites were inspected for C to U nucleotide substitutions by a custom Perl script, with the use of the following parameters; presence in at least two of the biological replicates, editing set with a minimum coverage of 10×.
Acknowledgments
We thank Cristine Gobbo Menezes and all colleagues from the Laboratory of Plant Systematics (Unesp/FCAV) for collaboration in searching for Utricularia natural populations and fruitful discussions. We also thank Tiago Santana Balbuena (Unesp/FCAV) for providing the opportunity to use the server to support the analyses.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/24/6130/s1.
Author Contributions
Conceptualization, S.R.S., V.F.O.M., and A.M.V.; methodology, S.R.S., A.M.V., and D.G.P., V.F.O.M., T.P.M., H.A.P., and E.J.M.; software, S.R.S., A.M.V., V.F.O.M., and D.G.P.; validation, S.R.S., D.G.P., V.F.O.M., and A.M.V.; formal analysis, S.R.S., A.M.V., V.F.O.M., and D.G.P.; investigation, S.R.S., A.M.V., V.F.O.M., and D.G.P.; resources, S.R.S., V.F.O.M., T.P.M., E.J.M., and A.M.V.; data curation, S.R.S., V.F.O.M. and A.M.V.; writing—original draft preparation, S.R.S., A.M.V., and V.F.O.M.; writing—review and editing, S.R.S., A.M.V., D.G.P., T.P.M., H.A.P., B.J.P., and E.J.M.; visualization, S.R.S., V.F.O.M., A.M.V., and T.P.M.; supervision, V.F.O.M. and A.M.V.; project administration, S.R.S., V.F.O.M., and A.M.V.; funding acquisition, V.F.O.M., A.M.V., and T.P.M.
Funding
This research was funded by FAPESP—Fundação de Amparo à Pesquisa do Estado de São Paulo (Proc. #2013/05144-0, #2013/25164-6, #2018/02285-6) and in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES), Finance Code 001 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the fellowships (Bolsa de Produtividade—Grant # 312908/2018-0 for VFOM and #302599/2016-9 for AMV).
Conflicts of Interest
The authors declare no conflict of interest.
Ethical Statements
The Utricularia amethystina samples were collected with the SisBio authorization #48516-1, by the Ministério do Meio Ambiente (MMA) and Instituto Chico Mendes de Conservação da Biodiversidade (IGMBio).
References
- 1.Müller K.F., Borsch T., Legendre L., Porembski S., Barthlott W. Recent progress in understanding the evolution of carnivorous Lentibulariaceae (Lamiales) Plant Biol. (Stuttg) 2006;8:748–757. doi: 10.1055/s-2006-924706. [DOI] [PubMed] [Google Scholar]
- 2.Silva S.R., Gibson R., Adamec L., Domínguez Y., Miranda V.F.O. Molecular phylogeny of bladderworts: A wide approach of Utricularia (Lentibulariaceae) species relationships based on six plastidial and nuclear DNA sequences. Mol. Phylogenet. Evol. 2018;118:244–264. doi: 10.1016/j.ympev.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Rutishauser R., Isler B. Developmental genetics and morphological evolution of flowering plants, especially bladderworts (Utricularia): Fuzzy Arberian Morphology complements Classical Morphology. Ann. Bot. 2001;88:1173–1202. doi: 10.1006/anbo.2001.1498. [DOI] [Google Scholar]
- 4.Reut M.S., Płachno B.J. Unusual developmental morphology and anatomy of vegetative organs in Utricularia dichotoma – leaf, shoot and root dynamics. Protoplasma. 2019 doi: 10.1007/s00709-019-01443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert V.A., Jobson R.W., Michael T.P., Taylor D.J. The carnivorous bladderwort (Utricularia, Lentibulariaceae): A system inflates. J. Exp. Bot. 2010;61:5–9. doi: 10.1093/jxb/erp349. [DOI] [PubMed] [Google Scholar]
- 6.Veleba A., Bureš P., Adamec L., Šmarda P., Lipnerová I., Horová L. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol. 2014;203:22–28. doi: 10.1111/nph.12790. [DOI] [PubMed] [Google Scholar]
- 7.Taylor P. The Genus Utricularia—A Taxonomic Monograph. The Royal Botanic Gardens, Kew; London, UK: 1989. [Google Scholar]
- 8.Seine R., Porembski S., Barthlott W. A neglected habitat of carnivorous plants: Inselbergs. Feddes Repert. 1996;106:555–562. doi: 10.1002/fedr.19961060525. [DOI] [Google Scholar]
- 9.BFG Brazilian Flora 2020: Innovation and collaboration to meet Target 1 of the Global Strategy for Plant Conservation (GSPC) Rodriguésia. 2018;69:1513–1527. doi: 10.1590/2175-7860201869402. [DOI] [Google Scholar]
- 10.Vellozo J.M.C. Florae Fluminensis, Seu, Descriptionum Plantarum Praefectura Fluminensi Sponte Mascentium Liber Primus ad Systema Sexuale Concinnatus. Typographia National; Rio de Janeiro, Brazil: 1825. 19. Utricularia; p. 18. [Google Scholar]
- 11.Saint-Hilaire A.F.C.P., Girard F. Comptes Rendus Hebdomadaires des Séances de L’académie des Sciences. Volume 7. French Academy of Sciences; Paris, France: 1838. [Google Scholar]
- 12.Baleeiro P.C., Jobson R.W., Sano P.T. Morphometric approach to address taxonomic problems: The case of Utricularia sect. Foliosa (Lentibulariaceae). J. Syst. Evol. 2016;54:175–186. doi: 10.1111/jse.12186. [DOI] [Google Scholar]
- 13.Baleeiro P.C., Sano P.T., Jobson R.W. Molecular Phylogeny of the Utricularia amethystina Complex (Utricularia sect. Foliosa) Assessed Using Plastid and Nuclear Sequence Data. Syst. Bot. 2019;44:398–404. [Google Scholar]
- 14.Zhang Y., Iaffaldano B.J., Zhuang X., Cardina J., Cornish K. Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Plant Biol. 2017;17:1–14. doi: 10.1186/s12870-016-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S.H., Suh H.J., Park J., Kim Y., Kim S. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA Part B Resour. 2019;4:626–627. doi: 10.1080/23802359.2018.1564390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen R.K., Ruhlman T.A. Plastid Genomes of Seed Plants. In: Bock R., Knoop V., editors. Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes) Springer; Dordrecht, The Netherlands: 2012. [Google Scholar]
- 17.Westwood J.H., Yoder J.I., Timko M.P., DePamphilis C.W. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Delannoy E., Fujii S., Colas Des Francs-Small C., Brundrett M., Small I. Rampant Gene loss in the underground orchid Rhizanthella Gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braukmann T., Stefanović S. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 2012;79:5–20. doi: 10.1007/s11103-012-9884-3. [DOI] [PubMed] [Google Scholar]
- 20.Gruzdev E.V., Kadnikov V.V., Beletsky A.V., Kochieva E.Z., Mardanov A.V., Skryabin K.G., Ravin N.V. Plastid Genomes of Carnivorous Plants Drosera rotundifolia and Nepenthes × ventrata Reveal Evolutionary Patterns Resembling Those Observed in Parasitic Plants. Int. J. Mol. Sci. 2019;20:4107. doi: 10.3390/ijms20174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevill P.G., Howell K.A., Cross A.T., Williams A.V., Zhong X., Tonti-Filippini J., Boykin L.M., Dixon K.W., Small I. Plastome-wide rearrangements and gene losses in carnivorous droseraceae. Genome Biol. Evol. 2019;11:472–485. doi: 10.1093/gbe/evz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martín M., Sabater B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010;48:636–645. doi: 10.1016/j.plaphy.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Ruhlman T.A., Chang W.-J., Chen J.J.W., Huang Y.-T., Chan M.-T., Zhang J., Liao D.-C., Blazier J.C., Jin X., Shih M.-C., et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 2015;15:100. doi: 10.1186/s12870-015-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicke S., Schäferhoff B., Depamphilis C.W., Müller K.F. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol. Biol. Evol. 2014;31:529–545. doi: 10.1093/molbev/mst261. [DOI] [PubMed] [Google Scholar]
- 25.Silva S.R., Michael T.P., Meer E.J., Pinheiro D.G., Varani A.M., Miranda V.F.O. Comparative genomic analysis of Genlisea (corkscrew plants—Lentibulariaceae) chloroplast genomes reveals an increasing loss of the ndh genes. PLoS ONE. 2018;13:e0190321. doi: 10.1371/journal.pone.0190321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibarra-Laclette E., Lyons E., Hernández-Guzmán G., Pérez-Torres C.A., Carretero-Paulet L., Chang T.H., Lan T., Welch A.J., Juárez M.J.A., Simpson J., et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva S.R., Diaz Y.C.A., Penha H.A., Pinheiro D.G., Fernandes C.C., Miranda V.F.O., Michael T.P., Varani A.M. The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS ONE. 2016;11:e0165176. doi: 10.1371/journal.pone.0165176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva S.R., Pinheiro D.G., Meer E.J., Michael T.P., Varani A.M., Miranda V.F.O. The complete chloroplast genome sequence of the leafy bladderwort, Utricularia foliosa L. (Lentibulariaceae) Conserv. Genet. Resour. 2017;9:213–216. doi: 10.1007/s12686-016-0653-5. [DOI] [Google Scholar]
- 29.Silva S.R., Alvarenga D.O., Aranguren Y., Penha H.A., Fernandes C.C., Pinheiro D.G., Oliveira M.T., Michael T.P., Miranda V.F.O., Varani A.M. The mitochondrial genome of the terrestrial carnivorous plant Utricularia reniformis (Lentibulariaceae): Structure, comparative analysis and evolutionary landmarks. PLoS ONE. 2017;12:e0180484. doi: 10.1371/journal.pone.0180484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Jin J., Moore M.J., Yi T., Li D. Plastome characteristics of Cannabaceae. Plant Divers. 2018;40:127–137. doi: 10.1016/j.pld.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gargano D., Scotti N., Vezzi A., Bilardi A., Valle G., Grillo S., Cozzolino S., Cardi T. Genome-wide analysis of plastome sequence variation and development of plastidial CAPS markers in common potato and related Solanum species. Genet. Resour. Crop Evol. 2012;59:419–430. doi: 10.1007/s10722-011-9692-7. [DOI] [Google Scholar]
- 32.Gao L., Zhou Y., Wang Z.W., Su Y.J., Wang T. Evolution of the rpoB-psbZ region in fern plastid genomes: Notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011;11:1–13. doi: 10.1186/1471-2229-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L., Yi X., Yang Y.X., Su Y.J., Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evol. Biol. 2009;9:1–14. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf P., Roper J., Duffy A. The evolution of chloroplast genome structure in ferns. Genome. 2010;53:731–738. doi: 10.1139/G10-061. [DOI] [PubMed] [Google Scholar]
- 35.Shikanai T. The NAD(P)H Dehydrogenase Complex in Photosynthetic Organisms: Subunit Composition and Physiological Function. Funct. Plant Sci. Biotechnol. 2007;1:129–137. [Google Scholar]
- 36.Sazanov L.A., Burrows P.A., Nixon P.J. The plastid ndh genes code for an NADH-specific dehydrogenase: Isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. USA. 2002;95:1319–1324. doi: 10.1073/pnas.95.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson C.E., Zorbas H., Price G.B., Zannis-Hadjopoulos M. Inverted repeats, stem-loops, and cruciforms: Significance for initiation of DNA replication. J. Cell. Biochem. 1996;63:1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Gupta P.K., Balyan H.S., Sharma P.C., Ramesh B. Microsatellites in plants: A new class of molecular markers. Curr. Sci. 1996;70:45–54. [Google Scholar]
- 39.Gao C., Ren X., Mason A.S., Li J., Wang W., Xiao M., Fu D. Revisiting an important component of plant genomes: Microsatellites. Funct. Plant Biol. 2013;40:645–661. doi: 10.1071/FP12325. [DOI] [PubMed] [Google Scholar]
- 40.Lawson M.J., Zhang L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol. 2006;7:R14. doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao X., Tang P., Li Z., Li D., Liu Y., Huang H. The first complete chloroplast genome sequences in Actinidiaceae: Genome structure and comparative analysis. PLoS ONE. 2015;10:e0129347. doi: 10.1371/journal.pone.0129347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Astuti G., Petroni G., Adamec L., Miranda V.F.O., Peruzzi L. DNA barcoding approach fails to discriminate Central European bladderworts (Utricularia, Lentibulariaceae), but provides insights concerning their evolution. Plant Biosyst. 2019 doi: 10.1080/11263504.2019.1610112. [DOI] [Google Scholar]
- 43.Kress W.J., Wurdack K.J., Zimmer E.A., Weigt L.A., Janzen D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rorbach J., Bobrowicz A., Pearce S., Minczuk M. Polyadenylation in bacteria and organelles. In: Rorbach J., Bobrowicz A., editors. Methods in Molecular Biology. Volume 1125. Humana Press; Totowa, NJ, USA: 2014. pp. 211–227. [DOI] [PubMed] [Google Scholar]
- 45.Saschenbrecker S., Bracher A., Rao K.V., Rao B.V., Hartl F.U., Hayer-Hartl M. Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco. Cell. 2007;129:1189–1200. doi: 10.1016/j.cell.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 46.Andrews T.J., Lorimer G.H. Rubisco: Structure, Mechanisms, and Prospects for Improvement. In: Hatch M.D., Boardman N.K., editors. Photosynthesis. Academic Press; Cambridge, MA, USA: 1987. pp. 131–218. [Google Scholar]
- 47.Nelson N., Yocum C.F. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 48.Plesnicar M., Bendall D.S. The photochemical activities and electron carriers of developing barley leaves. Biochem. J. 1973;136:803–812. doi: 10.1042/bj1360803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein R.R., Mason H.S., Mullet J.E. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoschke R., Bock R. Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell. 2018;30:745–770. doi: 10.1105/tpc.18.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf P.G., Rowe C.A., Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: Analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 52.Betts M.J., Russell R.B. Amino-Acid Properties and Consequences of Substitutions. In: Barnes M.R., Gray I.C., editors. Bioinformatics for Geneticists: A Bioinformatics Primer for the Analysis of Genetic Data. 2nd ed. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2007. [Google Scholar]
- 53.Wang M., Liu H., Ge L., Xing G., Wang M., Weining S., Nie X. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes (Basel) 2017;8:13. doi: 10.3390/genes8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugita M., Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 55.Müller K., Borsch T. Phylogenetics of Utricularia (Lentibulariaceae) and molecular evolution of the trnK intron in a lineage with high substitutional rates. Plant Syst. Evol. 2005;250:39–67. doi: 10.1007/s00606-004-0224-1. [DOI] [Google Scholar]
- 56.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn C., Bachmann L., Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 61.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurtz S., Choudhuri J.V., Ohlebusch E., Scheleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katoh K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nylander J.A.A. MrModeltest v2.3. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 69.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stöver B.C., Müller K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010;11:1–9. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 72.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 73.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lenz H., Hein A., Knoop V. Plant organelle RNA editing and its specificity factors: Enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinform. 2018;19:255. doi: 10.1186/s12859-018-2244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.