Abstract
The cellular vesicle is a fluid-filled structure separated from the surrounding environment by a biological membrane. Here, we isolated nanovesicles (NVs) from the juice of clementines using a discontinuous density gradient ultracentrifugation method. To gain information about the protein content of vesicles, mass spectrometry-based organelle proteomics and bioinformatics were applied to the exosome-like vesicle fraction isolated in the 1 mol/L sucrose/D2O cushion. Analysis of 1018 identified proteins revealed a highly complex mixture of different intra, extracellular and artificially-formed vesicle populations. In particular, clathrin-coated vesicles were significantly expressed in this sample. Membrane transporters are significantly represented in clementines nanovesicles. We have found 162 proteins associated with the transport Gene Ontology term (GO: 0006810) which includes; 71 transmembrane transport related, 53 vesicle mediated and 50 intracellular transporters. Platellin-3 like carrier protein containing a Sec14 domain is known to have a role in plant-virus interaction and that is one of the most abundant proteins in our dataset. The presence of transmembrane transporters like ATPases, aquaporins, ATP Binding Cassette (ABC) transporters and tetraspanins, regulators of protein trafficking suggests that nanovesicles of clementines can actively interact with their environment in a controlled way.
Keywords: plant-derived vesicles, tetraspanins, transporters, intercellular communication, ATPases, mass spectrometry, bioinformatics, proteomics, biocargo, Citrus clementina, citrus
1. Introduction
The cellular vesicle is a fluid-filled organelle separated from the surrounding environment by a biological membrane. The different kinds of vesicles, such as vacuoles, lysosomes, transport vesicles and secretory vesicles are important parts of a cell and can be classified based on their size, content such as coat protein, location and function. Plant vacuoles, for example, are large vesicles containing water, inorganic and organic molecules including enzymes, that besides other functions regulate the turgor pressure and the water level of cells [1]. In animal cells, lysosomes are packed with enzymes to digest proteins, polysaccharides, lipids, nucleic acids, damaged organelles, viruses and bacteria. Transport vesicles carry proteins and other molecules within the cell, for example from the endoplasmic reticulum to the Golgi apparatus and vice versa, or from one part of the Golgi to another [2]. Transport vesicles can be distinguished based on their protein coats that polymerize into cages to bend the membrane: clathrin-coated vesicles (CCVs), coat protein complex I (COPI) and coat protein complex II (COPII). The coats also have a function in molecular cargo selection and packing. Secretory vesicles are small membrane-surrounded packages in which nucleic acids, proteins, lipids, and metabolites are transported from organelles to a specific site on the cell membrane [3]. The existence of other small vesicles within cellular organelles like mitochondria, vacuoles, and multivesicular bodies, etc. are also described.
Cells ubiquitously secrete vesicles in all the three domains of life. Vesicles that are released from cells into the extracellular space are called extracellular vesicles (EVs) [4,5]. EVs play important roles in intercellular [5] and interkingdom communications [6,7] as well in immune responses [8]. EVs similar to intracellular vesicles are surrounded by a biological membrane and have a complex biocargo. The biogenesis of EVs is not fully understood, however, it is widely accepted that one type of vesicle called exosomes originates from the multivesicular bodies (MVBs) through the endocytic pathway. Secretion of vesicles in plants might be very similar to that of mammals, however no conclusive evidence has been obtained. Although transmission electron microscopy (TEM) images showed the presence of vesicle-like structures between the plasma membrane and cell wall of fungal hyphae more than 60 years ago [9], it took a long time to purify EV-like vesicles from plants. Regente et al. isolated small EV-like vesicles (20–200 nm in diameter), containing Rab11A from apoplastic washing fluids of sunflower seeds using the ultracentrifugation (UC) isolation method [10]. Recently, isolation methods employed in mammalian EV research have also been used to isolate vesicles from complex plant tissues like root homogenates and fruits. These preparations contain highly heterogeneous populations of both intra- and extracellular vesicles [6,7] and drew attention as alternative therapeutic solutions to mammalian cell-derived EVs [11]. For example, nanovesicles (NVs) isolated from grapes were shown to induce intestinal stem cells and protect against induced colitis in a mouse model. Raimondo et al. demonstrated that Citrus limon-derived nanovesicles inhibit cancer cell proliferation without affecting normal cells by activating TNF-alpha-related-apoptosis-inducing-ligand (TRAIL)-mediated apoptotic cell death [7]. Edible plant-derived vesicles are non-toxic, have tissue-specific targeting properties, and can be easily produced [12] making them promising as vectors for local and systemic delivery of various molecules. Fruit juices are one of the most popular sources to isolate edible plant-derived vesicles, as they are readily available by squeezing. By studying the protein biocargo of citrus fruit-derived vesicles, Pocsfalvi et al. have demonstrated the presence of highly expressed HSP70, HSP80, 14-3-3, G3PD and FBA6, PTL3 and clathirin proteins along with a large number of different enzymes and membrane transporters [13].
In this study, we show that the fruit juice of Citrus clementina contains nanovesicles packed with a complex protein biocargo. We found that the isolated vesicles contain different membrane transporters that may function in the movement of diverse molecular species across the membrane and thus may have an active role in cell–cell and interspecies communication.
2. Results and Discussion
2.1. Isolation of Nanovesicles (NVs) from Clementine Juice
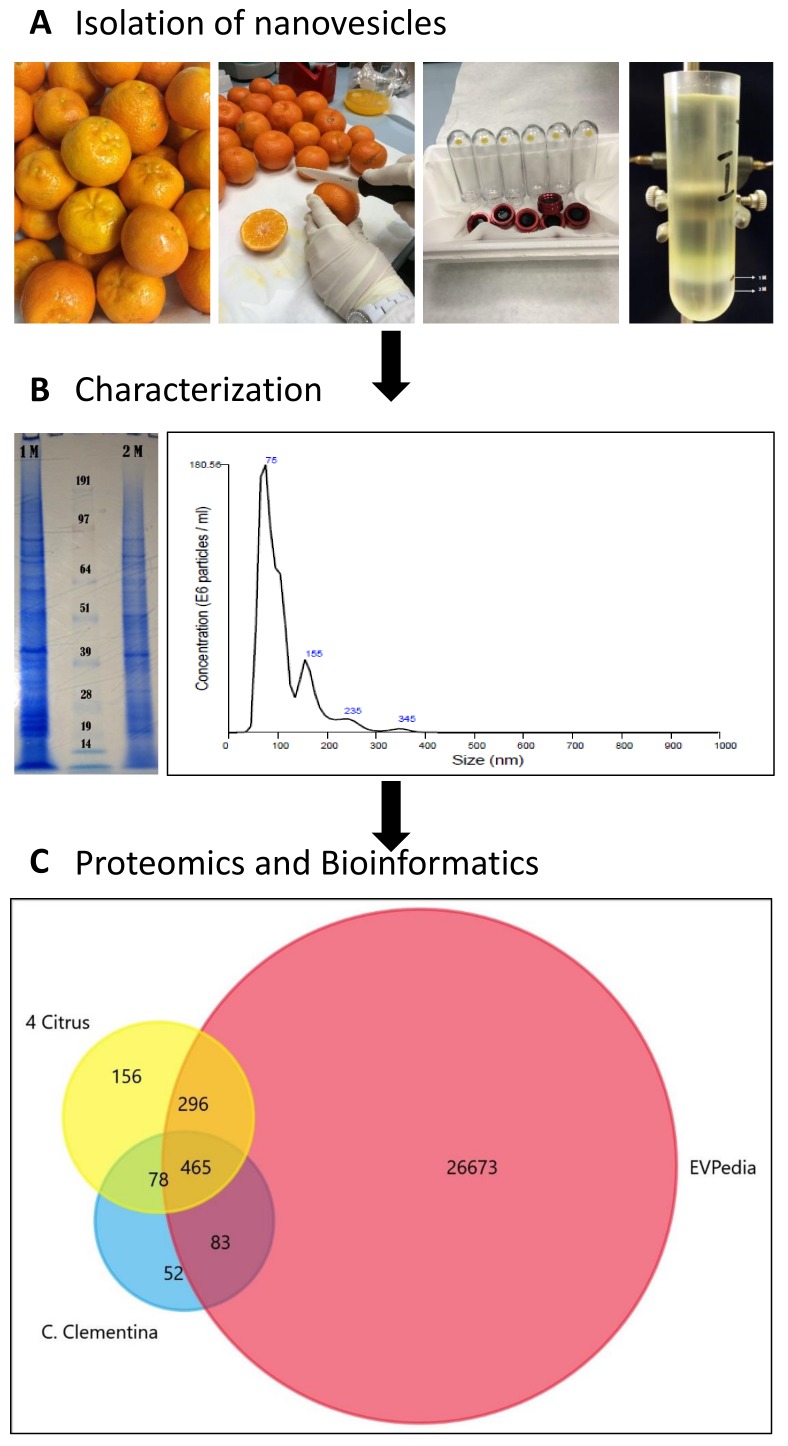
Here, we isolated membrane-bound vesicles from the juice of the clementines using discontinuous density gradient UC. A schematic overview of the experimental workflow is shown in Figure 1 [8,14]. Briefly, juice was subjected to a series of low velocity centrifugation steps to remove sac cells, cellular debris, organelles and medium and large vesicles. Small vesicles containing pellet obtained after differential centrifugation was further purified and separated on 1 mol/L (M) and 2 M sucrose D2O cushions. The layer floating above the 1 M sucrose/D2O cushion (Figure 1A) with density similar to mammalian exosomes (1.15–1.19 g/mL) was collected, washed (refer to material and methods) and used for vesicle characterization and cargo analysis.
Figure 1.
Schematic chart of the experimental work performed to isolate, characterize and analyze C. clementine fruit juice-derived exosome-like nanovesicles. (A) Lower left image shows the pellets obtained after diffferential ultracentrifugation (UC) lower right image shows the separation obtained by sucrose/D2O double cushion UC. The vesicles floating above the 1M sucrose/D2O cushion were found to be similar in density to mammalian extracellular vesicles. (B) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) protein profiles (right) and of vesicle populations in the 1 M and 2 M sucrose/D2O cushions and the particle-size distributions of vesicles isolated in the 1M sucrose/D2O cushion and measured using nanoparticle tracking analysis (NTA). (C). Venn diagram generated by FunRich software [1] shows the numbers of unique and common Orthologous Groups (OGs) of the identified protein. OGs of Citrus clementina (azure) were compared to four citrus species (C. sinensis, C. limon, C. paradise and C. aurantium) (yellow) [13] and EVpedia (red) [14].
2.2. Characterization of Clementine Nanovesicles
For the characterization of particle size and concentration, nanoparticle tracking analysis (NTA) was used. NVs fraction showed a broad range size distribution from approximately 75 nm to 345 nm with an average size of 103.3 nm with standard deviation 52.3 nm (Figure 1B). Considering the complex character of fruit juice, we expected to purify a mixture of different intracellular and extracellular vesicle populations. In fact, NTA shows vesicle subpopulations at 75, 120, 155 nm (major peaks in the NTA) and at 235 and 345 nm (minor peaks) in the 1M fraction. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the two vesicle fractions purified in the 1 M and 2 M sucrose/D2O cushions (Figure 1B) shows different profiles. The 1 M fraction reveals a highly complex pattern as it is generally observed in EV studies [13]. The yields determined after density cushion UC in the 1 M EV-like fraction are 1.16 × 1012 particles/L of fruit juice and 1.67 mg of NV proteins/L of fruit juice. This is at least 100 times higher than EV yields from mammalian cells and biological fluids.
2.3. The Protein Cargo of Citrus clementina Nanovesicles
Vesicles isolated in the 1 M sucrose/D2O fraction of C. clementina fruit juice were lysed by repeated freeze-thaw cycles in the presence of detergent and protein content was analyzed using mass spectrometry-based organelle proteomics. We identified 1018 proteins against the C. clementina UniProt database (31,274 entries) with log prob >3 values (Table S1). A comparative study was performed to highlight similarities and differences between protein biocargo of clementina vesicles and existing datasets. Orthologous groups (OGs) of each identified protein (678 hits, in Table S2) predicted by EggNOG mapper [15] were searched against (i) the OG accession codes published in EVpedia (27,517 hits, in Table S2) [14] and (ii) four different citrus species (995 unique hits, in Table S2) published recently [13]. The Venn diagram in Figure 1C shows the high overlap percentages found with both EVpedia (548 hits, 85%) and four citrus data sets (543 hit, 84%). The 83 clusters of orthologous groups (COGs) common with EVpedia but not present in the other citrus species studied is a unique feature of C. clementine-derived vesicles.
Most of the identified proteins were “uncharacterized”, therefore, in silico methods were used for their functional elucidation. Table 1 shows the 20 top-ranking proteins together with their functional annotation and identification counts in the EVpedia and 4-citrus data sets [13]. Most of the top-ranking proteins are frequently identified in EV related works as well they have been identified in other citrus species, like clathrin, patellin-3 like, heat shock, actins, ATPase and CDC48 proteins. Especially interesting are the two proteins associated to vesicle formation and transport: clathrin heavy chain and patellin-3 like protein. Clathrin coat protein is a key structural component of membrane budding and vesicle formation and was the first ranking protein in our data set. Patellin-3 like carrier protein is a plasma-membrane protein involved in vesicle and membrane-trafficking associated with cytokinesis and cell plate formation. These two proteins were found to be highly expressed in fruit-derived vesicles from other citrus species, too [13]. Clementina-derived vesicles also contain a variety of enzymes: carboxylase, mutase and ATPase, sucrose synthase, phosphorylase, etc. (Table 1, Tables S1 and S2). Since vesicles transport bioactive cargo within and between cells, enzymes may have a role in the mediation of activities both in local microenvironment and distant inter-organ communications.
Table 1.
List of 20 top-ranking proteins identified in exosome-like nanovesicles containing fraction of Citrus clementina. UniProt accession number indicate the accession number from the UniProt database; UniProt description is the protein identification from the UniProt database; Byonic scores indicate the scores obtained from the byonic software; cluster of orthologous groups (COGs) accession number obtained by EggNOG mapper; EggNOG description was retrieved from the EggNOG database; EVpedia ID count indicates the frequency score published in the EVpedia database; 4 citrus indicates the presence or absence of protein in our previous study [13].
| Accession UniProt | UniProt Description | MW [kDa] | pI | Scores | COG | EggNOG Description | EVpedia | 4 Citrus |
|---|---|---|---|---|---|---|---|---|
| V4V9V7 | Clathrin heavy chain | 192.8 | 5.3 | 814.0 | ENOG410XPH1 | Clathrin heavy chain | 451 | Y |
| V4T403 | Uncharacterized protein | 64.6 | 4.9 | 906.9 | ENOG410XRSQ | Carrier protein, Patellin-3 like | 44 | Y |
| V4TVM6 | Uncharacterized protein | 101.2 | 5.8 | 740.5 | COG0542 | Heat shock protein | 61 | Y |
| V4VRS8 | Uncharacterized protein | 110.9 | 6.1 | 646.6 | COG2352 | Phosphoenolpyruvate carboxylase | 10 | N |
| V4UFD6 | Uncharacterized protein | 71.0 | 5.1 | 860.1 | COG0443 | Heat shock cognate 70 kDa | 608 | Y |
| V4SV61 | Uncharacterized protein | 61.0 | 5.7 | 880.4 | COG0696 | Phosphoglycerate mutase | 15 | Y |
| V4SQM8 | Uncharacterized protein | 154.8 | 4.7 | 621.9 | n.d. | Unknown | n.d. | N |
| V4WGV2 | Uncharacterized protein | 80.0 | 5.0 | 730.9 | COG0326 | Heat shock protein | 557 | Y |
| V4SQ92 | Uncharacterized protein | 41.7 | 5.3 | 757.5 | COG5277 | Actins | 592 | Y |
| V4ULL2 | Uncharacterized protein | 94.1 | 5.1 | 659.3 | COG0443 | Heat-shock 70 kDa protein | 608 | Y |
| V4SU87 | Plasma membrane ATPase | 105.2 | 6.1 | 810.6 | COG0474 | Plasma membrane ATPase | 378 | Y |
| V4TDB5 | Uncharacterized protein | 68.6 | 5.3 | 710.6 | COG1155 | V-type proton ATPase catalytic subunit | 21 | Y |
| V4U840 | Uncharacterized protein | 93.8 | 5.9 | 759.8 | COG0480 | Elongation factor | 500 | Y |
| V4T1U7 | Uncharacterized protein | 47.1 | 6.0 | 898.2 | COG0446 | Monodehydroascorbate reductase | 112 | Y |
| V4W5U2 | Uncharacterized protein | 63.3 | 5.5 | 696.8 | COG0033 | Phosphoglucomutase | 217 | Y |
| V4RUA3 | Alpha-1,4 glucan phosphorylase | 95.2 | 7.0 | 780.2 | COG0058 | Phosphorylase | 297 | Y |
| V4TVW5 | Sucrose synthase | 92.6 | 6.0 | 660.8 | COG0438 | Sucrose-cleaving enzyme | 154 | Y |
| V4UJC4 | Uncharacterized protein | 90.2 | 6.3 | 649.7 | COG0339 | Oligopeptidase | 142 | Y |
| V4TCA6 | Uncharacterized protein | 252.7 | 6.0 | 601.6 | COG0439 | Acetyl-CoA Carboxylase | 159 | Y |
| V4SL33 | Uncharacterized protein | 89.4 | 5.1 | 575.2 | COG0464 | Cell-division cycle protein 48 | 479 | Y |
2.4. Proteins Involved in Molecular Transport
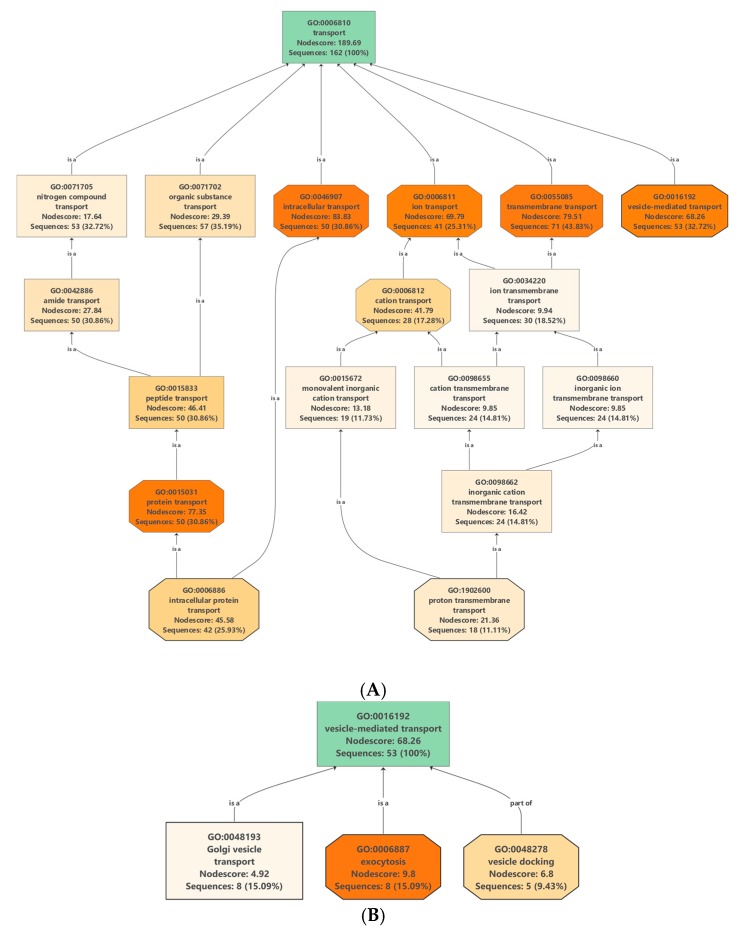
Protein/lipid composition plays a crucial role in the formation of different vesicles [16]. Membrane proteins (MPs), besides being important structural elements of the vesicle membranes, also participate in specific functions such as cargo selection, fusion and interaction with environment. They are transporters, channels, enzymes, receptors, anchorage site and signal transductors. Many integral MPs are transmembrane proteins (TM), which span the lipid bilayer with portions exposed on both sides of the membrane. The presence of tetraspanins [17], and aquaporins [18] MPs has been described in the various vesicle types. In our dataset, we have found 162 proteins associated with transport Gene Ontology (GO) term GO: 0006810 (Table S3) which includes 71 transmembrane transporters, 53 vesicle mediated transporters and 50 intracellular transporters (Figure 2A).
Figure 2.
Enrichment and distribution of (A) transport-related Gene Ontology (GO): 0006810 and (B) vesicle mediated transport-related GO: 16192 terms in the protein data set of nanovesicles isolated from the juice of C. clementine.
2.4.1. Transmembrane Transporters in Clementine Vesicles
Transmembrane transporters are involved in the movement of ions, small molecules, macromolecules and cellular components across the lipid bilayer. Most of the identified transmembrane transporters in our dataset are ATPases (25 ATPases and 12 ABC subunits, Table S3) and aquaporins (6 proteins, Table S3). ATPases operate within the biological membranes moving water, sugars, amino acids, proton, cations and anions across the membranes. There are four major types of ATPases, namely P, V, F and ABC types. Our study revealed the presence of 10 vacuolar H+-ATPase (V-ATPase) subunits. V-ATPase is directly involved in the establishment and maintenance of the acidic pH of cells, endocytic and secretory organelles [19]. Besides their canonical functions, the roles of V-ATPases in membrane fusion, endocytosis and vesicular trafficking have also been reported [19]. In plant, electrochemical H+-gradient generated by V-ATPases energizes secondary transport of metabolites across the vacuolar membrane (tonoplast) and other endomembrane. In fact, tonoplast has been shown to be highly enriched in V-ATPases. Proteomics revealed the presence of prominent V-type ATPase subunits in tonoplast vesicle preparations [20]. One reasonable explanation of the high expression of V-ATPases in our C. clementina juice sac cells-derived vesicle sample, therefore, could be the presence of tonoplast vesicles. During the production of the juice, low-density small vesicles can easily have formed from the rupture and re-vesiculation of tonoplast and been purified by density gradient ultracentrifugation. The protein dataset revealed four P-type ATPases in C. clementina-derived vesicles. The P-type ATPases by translocating cations, heavy metals and lipids maintain the electrochemical gradients across cellular membranes. In fruit juice cells of Citrus species, the vacuole can be very acidic (pH 2). Recent work shows that for this hyper acidification a vacuolar proton-pumping P-ATPase complex could be responsible [19].
Plants have many more ATP Binding Cassette (ABC) transporters than mammalian cells [21]. ABC transporters are grouped into 8 subfamilies. In our samples, we have found 11 ABC transporter subunits from 6 different subfamilies A, B, C, E, F and G. While some ABC transporters have specific endogenous secondary metabolite substrates including lipids, carbohydrates, phytohormones, etc., others may transport many different chemically unrelated substances. Besides ATPases, the presence of aquaporin channel proteins were also characteristic of clementine vesicles.
Aquaporins by regulating the flux of water and other small solute molecules across membranes are not only important for plant physiology but also for the interaction between plants and the environment. Vesicles containing aquaporin have recently come into the focus of EV research [18]. Here, the first time we demonstrate that vesicles of clementina contain tip (tonoplast intrinsic protein) and pip (plasma membrane intrinsic proteins) types of aquaporins.
2.4.2. Vesicle-Mediated Transport: Clathrin-Coated Vesicle (CCV), Coat Protein Complex I (COPI) and Coat Protein Complex II (COPII) Vesicles
Putative vesicle related transport proteins of clementina were identified by analyzing the proteins that are associated with transport GO terms (GO: 0006810) (Table S3). Out of 162 proteins in our experimental data set associated with transport 53 were related to “vesicle-mediated transport” (GO: 0016192) (Figure 2B). Analysis of the dataset shows the presence of proteins related to all the three main types of intracellular vesicles: CCVs, COPI and COPII vesicles. CCV related proteins were clathrin light- and heavy-chain coat proteins as well different subunits of AP-1, AP-2 and AP-4 complexes. Several proteins of COPI transport vesicles were also identified, including 7 coatomer complex subunits (alpha-1, beta-1, delta, epsilon-1, zeta-1, gamma-1 and beta-2). COPII vesicle-related proteins including 11 sec proteins and two GTP binding proteins SARA1 were also present in clementina vesicle. Proteomic data confirms our previous observations [13] that the clementina similarly to bitter orange, grapefruit, lemon and sweet orange-derived vesicles contains vesicular transport-related proteins characteristic to CCVs, COPI and COPII trafficking vesicles and EVs.
2.4.3. Regulators of Trafficking: The Tetraspanins
Tetraspanins are the main structural elements of the membrane microdomains (referred to as tetraspanin-enriched microdomains) and regulators of protein trafficking. Numerous physiological and pathological roles have been associated to tetraspanins. Recently, the role of tetraspanins and tetraspanin-associated proteins in the biogenesis of EVs has been proposed. Tetraspanins CD9, CD63, and CD81 are frequently present in EV samples and are often used as EV biomarkers. Plant tetraspanins have been overlooked for a long time but research is emerging thanks to their newly discovered roles in intercellular and interspecies communication [22]. Seventeen tetraspanin genes have been described in the model plant Arabidopsis. Here we were interested in whether tetraspanins are also a structural part of the plant derived vesicles. Blast sequence similarity searching was applied to identify Arabidopsis tetraspanin “homologues” (20 proteins) in the proteome of C. clementine (Table S3). The 13 homologues identified were then searched for in our vesicle related protein data set. In clementine-derived nanovesicles (1 M fraction) two tetraspanins, tetraspanin-3-like and tetraspanin-8-like were identified. Both tetraspanins are known to be present in Arabidopsis female reproductive organ (pistil or carpel). Moreover, proteomic analysis of plasmodesmata, the channels that link adjacent cells in plant tissues isolated from populus cell suspension, has also shown enrichment of the same tetraspanin-3 and 8 [23]. Based on this data, we can speculate that the vesicle population expressing these tetraspanins in our sample can be those that formed from the membrane of the plasmodesmata.
3. Materials and Methods
3.1. Fruit Material and Vesicle Isolation
Fruits of Citrus clementina, a seedless Italian cultivar produced around Naples collected in Naples, Italy on 24 February 2018. 20 pieces of fresh fruits were gently squeezed with the help of a glass lemon/orange squeezer and filtered using filter paper to obtain about 800 mL of fruit juice. The isolation procedure was performed in duplicates at room temperature. For the second isolation, 400 mL of fruit juice was used. Protease inhibitor cocktail containing 0.5 mL, 1 mg/mL Leupeptine, 2.5 mL 100 mM Phenylmethylsulfonyl fluoride (PMSF) and 1.6 mL 1 M sodium azide were added to every L of sample. Vesicles were isolated by differential UC following the procedure described by Stanly et al. [24] Briefly, low-velocity centrifugation steps were performed at 400× g and 800× g for 20 min at 22 °C to eliminate cells and cell debris, and at 15,000× g for 20 min at 22 °C to collect the fraction enriched in microvesicles. The supernatant was ultracentrifuged at 150,000× g for 60 min at 4 °C using a Type 70 Ti Beckman rotor. The obtained pellet was resuspended in 28 mL of 20 mM Tris-HCl (pH 8.5), under-layered by two cushions composed of 1 and 2 mol/L sucrose prepared in Tris-HCl/D2O, and centrifuged at 110,000× g for 180 min at 4 °C using SW 32 Ti rotor. The resulting vesicle layers were collected separately, washed twice in 20 mM Tris-HCl and collected by centrifugation at 110,000× g for 60 min at 4 °C using a SW 32 Ti rotor. The pellet was resuspended in 100–150 µL 20 mM Tris-HCl buffer and micro bicinchoninic acid (BCA) assay was performed to determine the protein concentration.
3.2. Physiochemical Characterization of Different Vesicle Populations
3.2.1. Nanoparticle Tracking Analysis
Particle size distribution was determined by NTA using a NanoSight NS300 system (Malvern Technologies, Malvern, UK) configured with a 488 nm laser and a high sensitivity scientific complementary metal-oxide semiconductor (CMOS) camera. Sample was diluted 1:200 in particle-free PBS. Samples were analyzed under constant flow conditions (flow rate = 50) at 25 °C. Data were analyzed using NTA 3.1.54 software with a detection threshold of 5.
3.2.2. Proteomics and Data analysis
SDS-PAGE Analysis
The quality of the vesicle samples and the reproducibility of the isolation was checked using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Samples (20 μg protein measured by micro BCA assay) were used as described in paragraph “Lysis of the Vesicles and Shotgun Proteomics Analysis” and electrophoretically separated under reducing conditions on a precast Novex Bolt 4%–12% Bis-Tris Plus gel using Bolt (3-(N-morpholino)propanesulfonic acid) MOPS SDS running buffer (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions and stained with colloidal Coomassie blue.
Lysis of the Vesicles and Shotgun Proteomics Analysis
For in-solution proteomics analysis, samples (100 μg protein measured by micro Bicinchoninic acid assay) were resuspended in 0.2% RapiGest SF (Waters Corp., Milford, MA, USA) and vesicles were lysed using five freeze-thaw cycles consisting of alternative freezing the samples for 5 min in a dry ice–ethanol slurry followed by thawing them in a bath of water under sonication at room temperature for 5 min. Proteins were reduced using 5 mM DL-dithiothreitol (Sigma-Aldrich, Saint Louis, MO, USA,), alkylated using 15 mM iodoacetamide (Sigma-Aldrich, Saint Louis, MO, USA), and digested using MS grade trypsin (Pierce, Thermo Sci. Rockford, IL, USA) as described previously [24]. Samples were vacuum dried and solubilized in 5% acetonitrile and 0.5% formic acid (liquid chromatography–mass spectrometry (LC–MS) grade, Thermo Sci. Rockford, IL, USA) prior to nano high-performance liquid chromatography–tandem mass spectrometry (nano-HPLC-MS/MS) analysis.
Nano Liquid Chromatography-Electrospray Ionization–Tandem Mass Spectrometry (NanoLC-ESI-MS/MS)
Shotgun proteomics analysis was carried out on 5 μg of tryptic digest using a Dionex Ultimate 3000 nanoRSLC (Dionex, Sunnyvale, CA, USA) coupled to a Bruker Maxis II mass spectrometer (Bruker Daltonics GmbH, Bremen, Germany) via CaptiveSpray nanobooster ionsource. Tryptic digest samples were desalted on an Acclaim PepMap100 C-18 trap column (100 μm × 20 mm, Thermo Scientific, Sunnyvale, CA, USA) using 0.1% TFA for 8 min at a flow rate of 5 μL/min and separated on the ACQUITY UPLC M-Class Peptide BEH C18 column (130 Å, 1.7 μm, 75 μm × 250 mm, Waters, Milford, MA, USA) at 300 nL/min flow rate, 48 °C column temperature. Solvent A was 0.1% formic acid, solvent B was acetonitrile, 0.1% formic acid and a linear gradient from 4% B to 50% B in 90 min was used. Mass spectrometer was operated in the data dependent mode using a fix cycle time of 2.5 s. MS spectra was acquired at 3 Hz, while MS/MS spectra at 4 or 16 Hz depending on the intensity of the precursor ion. Singly charged species were excluded from the anaylsis.
Protein Identification, Quantification and Statistical Analysis
Raw data files were processed using the Compass Data Analysis software (Bruker, Bremen, Germany). Proteins were identified using the Byonicv.3.4.0 software against the Citrus clementina UniProt database (31,274 entries). The search criteria were the following: 20 ppm precursor and fragment mass tolerance. Two missed cleavages were allowed, and the following modifications were set: carbamidomethylation on cysteine as fixed modification, while methionine oxidation and asparagine and glutamin deamidation as variable modifications. Protein identifications were validated by the Percolator algorithm [25] false discovery rate was <1%. Protein data is available at EV-Track under ID: EV190071.
Bioinformatics
Tag distributions of OmicsBox, version 1.1.74: 1018 sequences were blasted using QBlast service against the National Center for Biotechnology Information (NCBI) public databases using taxonomy filter green plants (taxa: 33,090, Viridiplantae), number of blast hits 20 and expectation value 1.0 × 10−3. The InterPro domain searches were performed using the public European Molecular Biology Lab-European Bioiformatics Institute (EMBL-EBI) InterPro web services to seen sequences against Interpro’s signatures (CDD, HAMAP, HMMpfam, HMMPIR, Fprintscan, BlastproDom). All signatures generated interpro results. All together 949 sequences were annotated and 939 were mapped against exclusively created GO-annotated proteins to obtain functional labels of GO-associated and Uniprot’s ID mapping. Orthology assignment and COGs annotation were performed by EggNOg Mapper version 5.4.1 [15]. EggNOG OGs were used to compare protein datasets between different taxa and EVpedia [14,26].
4. Conclusions
In this study, exosome-like nano-sized vesicles have been isolated with a high yield (1.6 mg/L) from the fruit juice of C. clementine. NTA and proteomic analysis show a heterogeneous mixture of different vesicle subpopulations. Proteomic and bioinformatic studies revealed that all the three major classes of coated vesicles, i.e., CCV, COPI and COPII, distinct in their protein compositions as well as tonoplast, apoplast and plasmodesmata-derived nanovesicles, present in this preparation. Amongst the top-ranking proteins are clathrin heavy chains associated with vesicle formation and patellin-3 like carrier proteins associated with cytokinesis-related cell division. Besides the natural intra- and extracellular vesicles, we propose that plasma and vacuole membranes as well as the limiting membrane of the plasmodesmata can give rise to the formation of artificial vesicles during the preparation of the fruit juice and isolation of the vesicles. Vesicle samples isolated from edible-plants foods such as whole organs, tissues, fruit juice, etc. therefore inherently contain a complex mixture of natural and artificial vesicles. The presence of different membrane transporters like aquaporin, ATPases and ABC transporters as well as tetraspanin regulators implies their possible participation in translocation and transportation of various substances between the vesicle interior and environment. However, to gain a better insight, the results of this study need to be integrated with biochemical, physiological and functional assays to establish that the identified transporters are indeed functional. Understanding the molecular components of the plant food-derived vesicles, including enzymes and transporters, is expected to widen our knowledge on how vesicles can be used as vectors for local and systemic molecular delivery. More generally, this information can be of value in the exploitation of edible plant-derived vesicles in biotechnology and biomedicine.
Acknowledgments
The authors thank Alfatest srl, Italy for the NTA measurements on the Nanosight instrument.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/24/6205/s1. Table S1. Statistical analysis, protein and peptide data related to the mass spectrometry-based proteomics analysis. Tab (A) Experimental details and statistical data, Tab (B) identified proteins in the vesicle fraction isolated by gradient ultracentrifugation in 1 M sucrose/D2O gradient of C. clementina fruit juice with log prob >3 values, (C) identified peptide data. Table S2. Orthology assignment and clusters for orthologous groups (COG) annotation Tab (A) of proteins identified in the nanovesicles fraction of C. clementina (Table S1B), Tab (B) of four different citrus species [13] performed by EggNOg Mapper [15], and Tab (C) taken from EVPedia [16,26]. Table S3. List of proteins isolated from fruit juice of C. clementina and associated with transport Gene Ontology (GO) term GO: 0006810. Highlighted in separated columns are transmembrane transport related proteins (71 proteins), vesicle mediated transport (53 proteins) and intracellular transport (50 proteins) and tetraspanin-like proteins (2 proteins).
Author Contributions
C.S.; sample collection, isolation and physiochemical characterization of nanovesicles, writing-original draft preparation. M.M.; sample collection and carried out experiments, I.F.; sample preparation for proteomic analysis, L.T.; Proteomic analysis, G.P.; conceptualization, supervision, bioinformatics analysis, writing-review and editing.
Funding
This research was funded by the European Union’s Horizon 2020 Research and innovation programme under grant agreement No 801338. G. Pocsfalvi and M. Moubarak thank the grant N. 1525025487 for foreign students 2018–2019 awarded by the Italian Government to M Moubarak.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Echeverría E. Vesicle-mediated solute transport between the vacuole and the plasma membrane. Plant Physiol. 2000;123:1217–1226. doi: 10.1104/pp.123.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang I., Robinson D.G. Transport vesicle formation in plant cells. Curr. Opin. Plant Biol. 2009;12:660–669. doi: 10.1016/j.pbi.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Van de Meene A.M.L., Doblin M.S., Bacic A. The plant secretory pathway seen through the lens of the cell wall. Protoplasma. 2017;254:75–94. doi: 10.1007/s00709-016-0952-4. [DOI] [PubMed] [Google Scholar]
- 4.Regente M., Pinedo M., Elizalde M., de la Canal L. Apoplastic exosome-like vesicles: A new way of protein secretion in plants? Plant Signal. Behav. 2012;7:544–546. doi: 10.4161/psb.19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krämer-Albers E.-M., Hill A.F. Extracellular vesicles: Interneural shuttles of complex messages. Curr. Opin. Neurobiol. 2016;39:101–107. doi: 10.1016/j.conb.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Mu J., Zhuang X., Wang Q., Jiang H., Deng Z.-B., Wang B., Zhang L., Kakar S., Jun Y., Miller D., et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014;58:1561–1573. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raimondo S., Naselli F., Fontana S., Monteleone F., Lo Dico A., Saieva L., Zito G., Flugy A., Manno M., Di Bella M.A., et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget. 2015;6:19514–19527. doi: 10.18632/oncotarget.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P., et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girbardt M. Über die Substruktur von Polystictus versicolor L. Arch. Für Mikrobiol. 1957;28:255–269. doi: 10.1007/BF00411497. [DOI] [PubMed] [Google Scholar]
- 10.Regente M., Corti-Monzón G., Maldonado A.M., Pinedo M., Jorrín J., de la Canal L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009;583:3363–3366. doi: 10.1016/j.febslet.2009.09.041. [DOI] [PubMed] [Google Scholar]
- 11.Rome S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019;10:529–538. doi: 10.1039/C8FO02295J. [DOI] [PubMed] [Google Scholar]
- 12.Yang C., Zhang M., Merlin D. Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. J. Mater. Chem. B. 2018;6:1312–1321. doi: 10.1039/C7TB03207B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pocsfalvi G., Turiák L., Ambrosone A., del Gaudio P., Puska G., Fiume I., Silvestre T., Vékey K. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J. Plant Physiol. 2018;229:111–121. doi: 10.1016/j.jplph.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Kim D.-K., Lee J., Kim S.R., Choi D.-S., Yoon Y.J., Kim J.H., Go G., Nhung D., Hong K., Jang S.C., et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M., et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill S., Catchpole R., Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2018;43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreu Z., Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Ballesta M.C., Garcia-Ibañez P., Yepes-Molina L., Rios J.J., Carvajal M. The expanding role of vesicles containing aquaporins. Cells. 2018;7:179. doi: 10.3390/cells7100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxson M.E., Grinstein S. The vacuolar-type H+-ATPase at a glance—more than a proton pump. J. Cell Sci. 2014;127:4987–4993. doi: 10.1242/jcs.158550. [DOI] [PubMed] [Google Scholar]
- 20.Trentmann O., Haferkamp I. Current progress in tonoplast proteomics reveals insights into the function of the large central vacuole. Front. Plant Sci. 2013;4:34. doi: 10.3389/fpls.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., Martinoia E. Plant ABC transporters. Arabidopsis Book. 2011;9:e0153. doi: 10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez-Jimenez S., Hashimoto K., Santana O., Aguirre J., Kuchitsu K., Cárdenas L. Emerging roles of tetraspanins in plant inter-cellular and inter-kingdom communication. Plant Signal. Behav. 2019;14:e1581559. doi: 10.1080/15592324.2019.1581559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leijon F., Melzer M., Zhou Q., Srivastava V., Bulone V. Proteomic analysis of plasmodesmata from populus cell suspension cultures in relation with callose biosynthesis. Front. Plant Sci. 2018;9:1681. doi: 10.3389/fpls.2018.01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanly C., Fiume I., Capasso G., Pocsfalvi G. Isolation of exosome-like vesicles from plants by ultracentrifugation on sucrose/Deuterium Oxide (D2O) density cushions. In: Pompa A., De Marchis F., editors. Unconventional Protein Secretion: Methods and Protocols. Springer; New York, NY, USA: 2016. pp. 259–269. [Google Scholar]
- 25.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 26.Pathan M., Keerthikumar S., Ang C.-S., Gangoda L., Quek C.Y.J., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.