Abstract
In plants, the phenylpropanoid pathway is responsible for the synthesis of a diverse array of secondary metabolites that include lignin monomers, flavonoids, and coumarins, many of which are essential for plant structure, biomass recalcitrance, stress defense, and nutritional quality. Our previous studies have demonstrated that Populus trichocarpa PtrEPSP‐TF, an isoform of 5‐enolpyruvylshikimate 3‐phosphate (EPSP) synthase, has transcriptional activity and regulates phenylpropanoid biosynthesis in Populus. In this study, we report the identification of single nucleotide polymorphism (SNP) of PtrEPSP‐TF that defines its functionality. Populus natural variants carrying this SNP were shown to have reduced lignin content. Here, we demonstrated that the SNP‐induced substitution of 142nd amino acid (PtrEPSP‐TFD142E) dramatically impairs the DNA‐binding and transcriptional activity of PtrEPSP‐TF. When introduced to a monocot species rice (Oryza sativa) in which an EPSP synthase isoform with the DNA‐binding helix‐turn‐helix (HTH) motif is absent, the PtrEPSP‐TF, but not PtrEPSP‐TFD142E, activated genes in the phenylpropanoid pathway. More importantly, heterologous expression of PtrEPSP‐TF uncovered five new transcriptional regulators of phenylpropanoid biosynthesis in rice. Collectively, this study identifies the key amino acid required for PtrEPSP‐TF functionality and provides a strategy to uncover new transcriptional regulators in phenylpropanoid biosynthesis.
Keywords: DNA binding, EPSP synthase, phenylpropanoid, rice, transcriptional regulation
1. INTRODUCTION
Rice (Oryza sativa) is currently one of the most important cultivated crops in the world, which feeds about half of the human population. Meanwhile, with the production of approximately 1,140 million tons per year worldwide, the lignocellulosic biomass derived from rice straws has emerged as an attractive renewable feedstock for biofuels (Satlewal, Agrawal, Bhagia, Das, & Ragauskas, 2018). In plants, the phenylpropanoid pathway is responsible for the synthesis of secondary metabolites, including lignin monomers, flavonoids, and coumarins, which play essential roles in determining plant structure, biomass recalcitrance, and stress tolerance (Vogt, 2010). Thus, a comprehensive understanding of the phenylpropanoid pathway and its regulation in rice can lead to more sustainable agriculture and energy.
In plants, the phenylpropanoid pathway consists of a set of enzymes (Xie, Zhang, et al., 2018). Phenylalanine ammonia‐lyase (PAL), cinnamate 4‐hydroxylase (C4H), and 4‐coumarate: CoA ligase (4CL) catalyzes the first three steps of the general phenylpropanoid pathway to convert phenylalanine into p‐coumaroyl‐CoA, which serves as the precursor for phenylpropanoids. Downstream of 4CL, quinate/shikimate p‐hydroxycinnamoyltransferase (HCT), p‐coumaroylshikimate 3′‐hydroxylase (C3′H), caffeoyl shikimate esterase (CSE), caffeic acid O‐methyltransferase (COMT), and caffeoyl‐CoA O‐methyltransferase (CCoAOMT) are involved in the remaining steps of the general phenylpropanoid pathway and the biosynthesis of lignin monomers. In addition, enzymes, including cinnamoyl‐CoA reductase (CCR), ferulate 5‐hydroxylase (F5H), and cinnamyl alcohol dehydrogenase (CAD), specifically catalyze the biosynthesis of lignin monomers.
The phenylpropanoid pathway is tightly regulated by a transcriptional regulatory hierarchy to ensure precise spatial and temporal deposition of phenolics in plant cells. In Arabidopsis and grasses (e.g., rice), several MYB transcription factors directly target phenylpropanoid biosynthetic genes via binding to AC elements, which widely exist in the promoters of major phenylpropanoid pathway genes (e.g., PAL, C4H, 4CL, C3H, HCT, CCoAOMT, CCR, F5H, CAD) (Rogers & Campbell, 2004). MYB46 and its paralog MYB83 activate the expression of phenylpropanoid biosynthetic genes, as well as other MYBs (Kim, Kim, Ko, Kang, & Han, 2014; Zhong et al., 2011; Zhong & Ye, 2012). In rice, the overexpression of OsMYB46 was shown to upregulate the expression of 4CL, MYB58, MYB63, and MYB85 (Zhong et al., 2011). Downstream of MYB46/83, clades of MYBs, such as MYB58/63, MYB42/85, and MYB4/32, are also involved in the regulation of the phenylpropanoid pathway (Rao & Dixon, 2018). In rice, overexpression of OsMYB58/63 or OsMYB42/85 resulted in the upregulation of the expression of one CAD gene and lignin biosynthesis (Hirano, Kondo, et al., 2013). In Arabidopsis, genes in the MYB4/32 clade (e.g., AtMYB4, AtMYB32, and AtMYB7) are proposed to negatively regulate the phenylpropanoid pathway via suppressing the expression of C4H and 4CL genes (Fornalé et al., 2014; Jin et al., 2000; Preston, Wheeler, Heazlewood, Li, & Parish, 2004). However, rice Osmyb4, one member of MYB4 family, was shown to activate the expression of PAL gene and the phenylpropanoid pathway in Arabidopsis, Nicotiana tabacum, and Salvia sclarea (Docimo, Coraggio, Tommasi, & Leone, 2008; Vannini et al., 2004), suggesting that MYB4/32 genes in Arabidopsis and grasses may be involved in different regulatory mechanisms to fine‐tune the phenylpropanoid flux. Upstream of MYB46/83, a group of NAC transcription factors, called secondary wall‐associated NACs (SWNs), function as master switches for the phenylpropanoid pathway and secondary cell wall biosynthesis (Rao & Dixon, 2018). The promoter of OsMYB46 contains secondary wall NAC‐binding elements (SNBEs) and is directly activated by OsSWNs (Zhong et al., 2011). Besides OsMYB46, overexpression of OsSWNs was able to upregulate the expression of SND3, MYB83, MYB85, MYB58, MYB63, etc. (Zhong et al., 2011). Furthermore, the existence of feedforward and feedback regulations, as well as the continual discovery of new transcription factors (e.g., WRKY12, E2Fc, etc.) in the transcriptional regulatory hierarchy (Xie, Zhang, et al., 2018), demonstrates the complexity of the transcriptional regulation of the phenylpropanoid pathway.
5‐enolpyruvylshikimate 3‐phosphate (EPSP) synthase widely exists in plants, bacteria, and fungi. EPSP synthase has carboxyvinyl transferase activity and catalyzes the conversion of phosphoenolpyruvate and shikimate‐3‐phosphate to EPSP in the shikimate pathway, which is responsible for the biosynthesis of aromatic amino acids (Maeda & Dudareva, 2012). Recently, we discovered one isoform of EPSP synthase in Populus (PtrEPSP‐TF) that is involved in the regulation of the phenylpropanoid pathway. Besides the EPSP synthase activity, PtrEPSP‐TF has transcriptional activity and regulates the expression of PtrMYB021 (Xie, Muchero, et al., 2018), one functional homolog of MYB46 in Populus (Wilkins, Nahal, Foong, Provart, & Campbell, 2009; Zhong, McCarthy, Haghighat, & Ye, 2013).
In this study, we identified a SNP critical for PtrEPSP‐TF functionality. The SNP‐induced substitution of the 142nd amino acid dramatically impairs the DNA‐binding and transcriptional activity of PtrEPSP‐TF, which results in the reduced lignin content in Populus natural variants. We also demonstrated the PtrEPSP‐TF‐mediated transcriptional reprogramming of the phenylpropanoid pathway in rice. As a monocot, rice only has an EPSP synthase lacking the DNA‐binding helix‐turn‐helix (HTH) motif, providing a low‐background platform to study the transcriptional effect of PtrEPSP‐TF. We found that the heterologous expression of PtrEPSP‐TF in rice alters the transcription of a bunch of genes involved in the phenylpropanoid pathway. Furthermore, we uncovered five negative regulators of the phenylpropanoid pathway in rice using this heterologous expression system. Our results provide a strategy to better understand the regulation of the phenylpropanoid pathway in rice and Populus, which will complement the strength of the comparative study of Arabidopsis, rice, and Populus transcriptional regulatory elements.
2. METHODS
2.1. Plant growth conditions
To generate transgenic rice, cDNAs of the archetypic PtrEPSP‐TF, the BESC‐35 PtrEPSP‐TF, and BESC‐876 PtrEPSP‐TF were cloned into binary vector pANIC10A (Mann et al., 2012) and then transformed into Japonica rice Taipei 309, using biolistics and the procedures as described in Mann et al., 2011 (Mann et al., 2011). The empty pANIC10A vector was transformed in parallel as a negative control. Positive transgenic events were selected by measuring PtrEPSP‐TF expression using RT‐PCR in hygromycin‐resistant plants. RNA was isolated as described (Jacobs, Lawler, LaFayette, Vodkin, & Parrott, 2016). Total RNA was treated with RNase‐free DNase I (Thermo Fisher Scientific). Endpoint RT‐PCR was performed with the GoScript Reverse Transcription System (Promega) according to the manufacturer's instructions using primers EPSP‐1219F (5′‐GCTATGACTCTGGCTGTTGTTGC‐3′) and OCS‐R (5′‐CAACGTGCACAACAGAATTGA‐3′). Rice plants were grown in soil under natural sunlight in a closed greenhouse at 22–32°C with controlled water and nutrient supply and a 16‐hr light period.
2.2. RNA‐seq experiments
RNAs were extracted from stem tissues of three‐month‐old rice plants prior to flower emergence (i.e., the vegetative stage). Four biological replicates were collected for each transgenic line. The cDNA libraries were constructed following Illumina standard protocols and sequenced with Illumina HiSeq 2500. After filtering out low‐quality reads, RNA‐Seq reads were aligned to the Oryza sativa genome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Osativa) using TopHat (Kim et al., 2013). Differentially expressed genes (DEGs) were identified using the R package DESeq2 (Love, Huber, & Anders, 2014). Raw p‐values were adjusted for multiple comparison effects using the false discovery rate (FDR). The cutoff for significant DEGs was set as >2 absolute fold change (FC) and FDR ≤ 0.05.
Gene Ontology (GO) enrichment analysis was performed for DEGs using agriGO (Tian et al., 2017). A multiple testing correction was performed using the FDR under dependency (Yekutieli & Benjamini, 1999). GO terms with a corrected p ≤ .05 were considered to be significantly enriched. For transcription factor binding sites (TFBS) prediction, 1‐kb sequences upstream of the translation start site of DEGs in each cluster were analyzed using PlantPAN (Chow et al., 2016).
2.3. Metabolite analysis
Freeze‐dried stems were ground with a micro‐Wiley mill, and ~50 mg DW was subsequently twice extracted with 2.5 ml 80% ethanol overnight and then combined prior to drying a 0.5 ml aliquot in a nitrogen stream. Sorbitol was added before extraction as an internal standard to correct for differences in extraction efficiency, subsequent differences in derivatization efficiency and changes in sample volume during heating. Dried extracts were silylated for 1 hr at 70°C to generate trimethylsilyl (TMS) derivatives, which were analyzed after 2 days with an Agilent Technologies Inc 5975C inert XL gas chromatograph‐mass spectrometer as described elsewhere (Li et al., 2012; Tschaplinski et al., 2012). Metabolite peak extraction, identification, and quantification were described previously (Tschaplinski et al., 2012), and unidentified metabolites were denoted by their retention time as well as key mass‐to‐charge (m/z) ratios. Data were pooled across four independent replicates, and treatment means were tested for statistical significance (p ≤ .05) using Student's t tests.
2.4. Electrophoretic mobility shift assay
The archetypic PtrEPSP‐TF, BESC‐35 PtrEPSP‐TF, and BESC‐876 PtrEPSP‐TF were cloned into the pGEX‐6P‐1 vector (GE Healthcare) by BamHI and EcoRI for GST fusion constructs. The constructs were transformed into E. coli strain BL21(DE3)pLysS (Invitrogen) for protein expression. GST fusion proteins were extracted and purified as previously described using Glutathione Sepharose 4B beads (GE Healthcare) (Xie, Ren, Costa‐Nunes, Pontes, & Yu, 2012). To perform EMSA, proteins were then eluted from beads by incubating with Elution Buffer (50 mM Tris‐HCl pH 8.0, 10 mM reduced glutathione) at 4°C for 30 min. For DNA probes, PtrhAT promoter DNA was PCR amplified, gel purified, and end labeled with biotin as described previously (Xie, Muchero, et al., 2018). The DNA‐binding reaction included 0.25 nM Biotin‐labeled probe, 0.4 µg of purified protein, 10 mM Tris‐HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% Glycerol, 5 mM MgCl2, 1 µg Poly (dI‐dC), 0.05% NP‐40. Reactions were incubated at room temperature for 20 min. The reaction mixtures were then resolved in 6% DNA retardation gel (Novex) by electrophoresis at 100 V for 1–2 hr and electrophoretically transferred to Nylon membrane. Signals of biotin were detected using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) as suggested by the manufacturer.
2.5. Transcriptional activity assay
The protoplast transfection‐based transcriptional activity assay was performed according to the previously described method (Tiwari, Hagen, & Guilfoyle, 2003). Ten µg of effector and reporter plasmids was co‐transfected into 100 µl of Populus protoplasts using PEG‐calcium transfection method and incubated under the darkness for 18–20 hr at room temperature. GUS activity assay was performed as described (Yoo, Cho, & Sheen, 2007). GUS activity was measured using a Fluoroskan microplate reader. To normalize GUS activity, 100 ng of 35S: Luciferase plasmid was co‐transfected for each transfection. Luciferase activity was measured using Promega Luciferase Assay System according to the manufacturer's manual.
2.6. Phloroglucinol‐HCL staining
The S8 segment of rice internode II was sectioned into 60 μm thickness and then stained with 1% phloroglucinol in HCl for 5 min and immediately observed under a light microscope (Ziess). The internal diameters of sclerenchyma cells were measured under the microscope using the manufacturer's software (Ziess).
2.7. Co‐expression analysis
The microarray data of rice were downloaded from the NCBI GEO database under the series accession numbers http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024; 190 samples corresponding to 39 tissues from two indica varieties; Minghui 63 and Zhenshan 97) (Wang et al., 2010) and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51289 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51289; 59 samples corresponding to 20 data points of internodes during secondary cell wall formation and 143 samples corresponding to 48 tissue and organ types) (Hirano, Aya, et al., 2013). For co‐expression analysis, a matrix of DEGs in our RNA‐Seq was constructed by calculating pairwise Pearson correlation coefficients based on the normalized expression data across all samples from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024 and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51289. Cytoscape (Shannon et al., 2003) was used to visualize the resulting network. Only co‐expression relationships with p ≤ .05 were indicated in the network.
3. RESULTS
3.1. DNA binding and transcriptional activities of allelic variants from BESC‐35 and BESC‐876 are impaired
In previous genome‐wide association studies, we identified two Populus natural variants containing PtrEPSP‐TF rare alleles: BESC‐35 and BESC‐876 that showed reduced lignin content compared with the average of the Populus trichocarpa population (Xie, Muchero, et al., 2018). To determine how mutations in these genotypes affect PtrEPSP‐TF protein function, we cloned PtrEPSP‐TF allelic variants from both genotypes. Sequencing and amino acid alignments revealed that both BESC‐35 and BESC‐876 variants carry a point mutation that results in the substitution of the 142nd amino acid from aspartic acid (D) to glutamic acid (E) (D142E, Figure 1a). BESC‐876 has an additional point mutation at position 364 that leads to the substitution of leucine (L) with proline (P) (L364P, Figure 1a).
Figure 1.
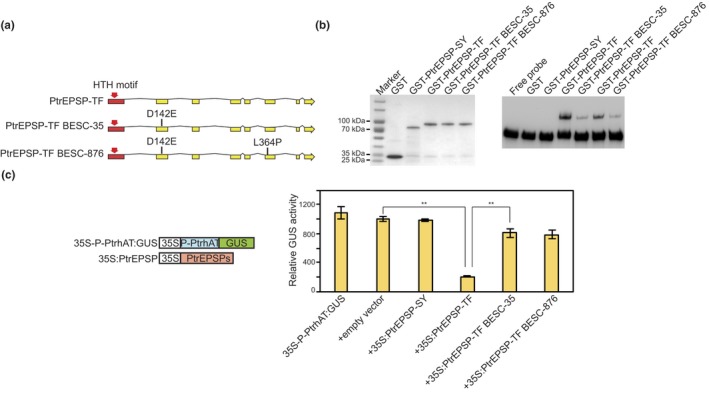
The reduced DNA‐binding and transcriptional activities of BESC‐35 and BESC‐876 variants. (a) A scheme of point mutations in BESC‐35 and BESC‐876 variants. The HTH motif is indicated by a red arrow. Amino acid abbreviation: D, Aspartic acid; E, Glutamic acid; L, Leucine; P, Proline. (b) BESC‐35 and BESC‐876 variants display reduced DNA‐binding affinity to PtrhAT promoter. Left panel: purified GST, GST‐PtrEPSP‐SY (an ESPS homolog of PtrEPSP‐TF that lacks the HTH domain), GST‐PtrEPSP‐TF, GST‐PtrEPSP‐TF from BESC‐35, and GST‐PtrEPSP‐TF from BESC‐876 were resolved in SDS‐PAGE gel and stained with Coomassie Blue. Right panel: EMSA assay of DNA binding to biotin‐labeled PtrhAT promoter. Free probe and GST are negative controls. (c) Transactivation assay of repression activities of archetypic PtrEPSP‐TF, BESC‐35 PtrEPSP‐TF, and BESC‐876 PtrEPSP‐TF on PthAT promoter. The repression activity of the blank vector was analyzed in parallel as a negative control. All transfection assays were performed in triplicate to calculate the mean value and standard deviation (error bar). P‐value comparisons (empty vector and PtrEPSP‐TF, PtrEPSP‐TF and BESC‐35/BESC‐876 PtrESPS‐TF) were calculated using two‐tailed Student t tests (**p ≤ .01)
Given that the HTH motif (amino acids 30–70) is responsible for DNA binding and transcriptional activity of PtrEPSP‐TF (Xie, Muchero, et al., 2018), we speculated that the amino acid substitution adjacent to the HTH motif (i.e., D142E) may affect the transcriptional function of PtrEPSP‐TF. To examine this possibility, we analyzed the binding affinity of BESC‐35 and BESC‐876 variants to the promoter of PtrhAT, which is the direct regulatory target of PtrEPSP‐TF in Populus (Xie, Muchero, et al., 2018). PtrEPSP‐TF and the variant proteins were tagged with GST and expressed in E. coli. After purification, these proteins were subjected to in vitro electrophoretic mobility shift assay (EMSA) to determine their binding affinities to the PtrhAT promoter, which was labeled with biotin for visualization. As shown in Figure 1b, signals of shift bands of both variants were dramatically reduced compared with that of the archetypic PtrEPSP‐TF. These results demonstrate that BESC‐35 and BESC‐876 variants have significantly reduced DNA‐binding activity. Consistent with reduced DNA‐binding activity, both PtrEPSP‐TF variants display impaired transcriptional repression on the PthAT promoter (Figure 1c). In contrast to the archetypic PtrEPSP‐TF, neither BESC‐35 nor BESC‐876 variants can reduce the expression of the GUS gene downstream of the PtrhAT promoter in the transactivation assays using the protoplast transient expression system. Thus, the BESC‐35 and BESC‐876 allelic variants carry mutations impacting the transcriptional repressor activity of PtrEPSP‐TF. Given the promoting effects of the archetypic PtrEPSP‐TF on PtrMYB021 (MYB46 close homolog) (Wilkins et al., 2009) expression and lignin biosynthesis in Populus, the reduced lignin content of BESC‐35 and BESC‐876 Populus genotypes is likely due to the impaired function of PtrEPSP‐TF. As such, BESC‐35 and BESC‐876 variants can be viewed as loss‐of‐function alleles of PtrEPSP‐TF.
3.2. Heterologous expression of PtrEPSP‐TF in transgenic rice alters sclerenchyma thickening and secondary metabolism
The absence of the HTH motif in monocot EPSP synthases (Xie, Muchero, et al., 2018) prompted us to compare functions of archetypic PtrEPSP‐TF with BESC‐35 and BESC‐876 variants via heterologous expression. We generated transgenic rice plants heterogeneously expressing the functional PtrEPSP‐TF, or the BESC‐35 (PtrEPSP‐TFD142E) and BESC‐876 (PtrEPSP‐TFD142E/L364P) alleles. The expression of transgenes is driven by the ZmUbi1 promoter, which allows for high expression levels of transgenes in monocots (Mann et al., 2012). The heterogeneous expression of the functional PtrEPSP‐TF or its loss‐of‐function alleles in rice did not trigger apparent morphological change in leaf and height during the vegetable growth. To visualize secondary cell walls, transverse cross sections of the S8 segment of rice internode II (Lin et al., 2017) were stained with phloroglucinol‐HCl, which turns lignin into a red‐purple color. Under a low‐magnification dissecting microscope, each sclerenchyma cell of the empty‐vector control was clearly observed, and their cell walls were stained red (Figure 2a). Different from the empty‐vector control, the whole sclerenchyma cell region of rice plants expressing the archetypic PtrEPSP‐TF stained red and it was hard to distinguish individual sclerenchyma cells (Figure 2a). On the other side, transgenic rice plants expressing PtrEPSP‐TFD142E or PtrEPSP‐TFD142E/L364P exhibit similar staining results as that of the empty‐vector control (Figure 2a), suggesting the difference of sclerenchyma cell staining is likely induced by difference in functions between PtrEPSP‐TF and PtrEPSP‐TFD142E or PtrEPSP‐TFD142E/L364P. To further determine the cause of the difference of sclerenchyma cell staining, we observed stained stems using a high‐magnification compound microscope. As shown in Figure 2b, expression of the functional PtrEPSP‐TF resulted in thickened secondary cell walls and reduced internal diameter in the sclerenchyma cells that form the cortical layers in bundle sheath fibers surrounding vascular vessels. In contrast, PtrEPSP‐TFD142E and PtrEPSP‐TFD142E/L364P variants resulted in little differences from the empty‐vector control in the thickness of secondary cell walls of the sclerenchyma cells (Figure 2b,c). Unexpectedly, the secondary cell wall thickening induced by the archetypic PtrEPSP‐TF suggested that PtrEPSP‐TF may affect the transcriptional regulation of secondary cell wall biosynthesis in rice as it does in Populus, although the EPSP form found in rice lacks the HTH domain and thus lacking transcriptional control.
Figure 2.
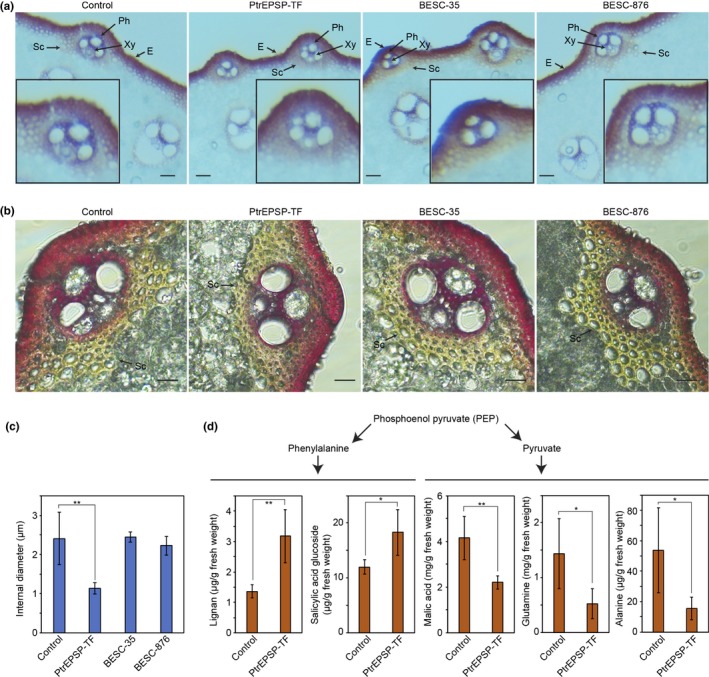
Transgenic rice plants expressing PtrEPSP‐TF display thickened sclerenchyma and altered secondary metabolism. (a) Dissecting microscopy images of phloroglucinol‐HCl stained rice stems from empty‐vector control, archetypic PtrEPSP‐TF, BESC‐35 PtrEPSP‐TF, and BESC‐876 PtrEPSP‐TF. E, Epidermis; Ph, phloem; Sc, sclerenchyma cells; Xy, xylem. Scale bar: 200 µm. (b) Compound microscopy images of phloroglucinol‐HCl stained rice stems empty‐vector control, archetypic PtrEPSP‐TF, BESC‐35 PtrEPSP‐TF, and BESC‐876 PtrEPSP‐TF. Scale bar: 10 µm. Sc, sclerenchyma cells. (c) The Sclerenchyma cells of PtrEPSP‐TF have smaller internal diameter. The internal diameters of ten sclerenchyma cells were measured under a microscope to calculate mean values and standard errors (error bars). P‐value comparison was calculated using two‐tailed Student t tests (**p ≤ .01). (d) Rice plants expressing functional archetypic PtrEPSP‐TF have increased accumulation of phenylpropanoid metabolites, but reduced accumulation of pyruvate metabolites. Four independent replicates were measured to calculate mean values and standard errors (error bars). P‐value comparison was calculated using two‐tailed Student t tests (**p ≤ .01, *p ≤ .05)
Besides sclerenchyma thickening, the lignin content of rice expressing archetypic PtrEPSP‐TF (31.98 ± 0.24 [% of dry weight]) was significantly higher (p ≤ .01) than that of the empty‐vector control (29.11 ± 0.00 [% of dry weight]). Gas chromatography‐mass spectrometry (GC‐MS) analysis revealed that secondary metabolism of transgenic rice was altered by archetypic PtrEPSP‐TF, where metabolites in the phenylalanine branch (Figure 2d), including salicylic acid glucoside exhibited 52.3% (p = .028) and lignan exhibited 132.5% (p = .007) increases, respectively, relative to the empty‐vector controls (Figure 2d). This same response occurred in Populus overexpressing PtrEPSP‐TF (Xie, Muchero, et al., 2018). Phenylalanine and pyruvate are synthesized using the same precursor phosphoenol pyruvate (PEP) (Davies, 1979), and phenylalanine is the precursor of the phenylpropanoid pathway (Fraser & Chapple, 2011). Opposite to the pattern of the phenylalanine metabolites, metabolites in the pyruvate branch (Figure 2d), including malic acid, alanine, and glutamine, exhibited significant reductions, 46.8% (p = .008), 63.8% (p = .039), and 71.3% (p = .037) respectively, in rice expressing archetypic PtrEPSP‐TF (Figure 2d).
3.3. Heterologous expression of PtrEPSP‐TF in rice alters transcriptional regulation of the phenylpropanoid pathway
To explore the molecular effects of the archetypic PtrEPSP‐TF in transgenic rice, we extracted total RNAs from stems of three‐month‐old transgenic rice plants and performed RNA‐seq analysis. For each sample, four biological replicates were analyzed, and highly consistent results were obtained (Pearson correlation r ≥ .93). Differential expression analysis identified a large collection of mRNAs whose abundance was significantly affected (Fold change ≥ 2, p ≤ .01, false discovery rate [FDR] ≤ 0.05) by the archetypic PtrEPSP‐TF. Compared with transcriptomes of empty‐vector control, a total of 3,953 upregulated genes and 1,471 downregulated genes were identified (Figure 3a and Dataset S1). Consistent with results that the PtrEPSP‐TFD142E and PtrEPSP‐TFD142E/L364P variants have impaired DNA‐binding and transcriptional repressor activity, differentially expressed genes (DEGs) in BESC‐35 and BESC‐876 alleles expression lines were far fewer compared to the functional archetypic PtrEPSP‐TF expression line (Figure 3a). The BESC‐35 variant led to 729 upregulated genes and 1,091 downregulated genes, whereas the BESC‐876 variant resulted in 353 upregulated and 172 downregulated genes (Figure 3a, Dataset S1). In addition, among the 3,953 upregulated genes by the archetypic PtrEPSP‐TF, only 134 of them (3.39%) were also upregulated by BESC‐35 and BESC‐876 variants (Figure S1). Similarly, only 1.63% (24 out of 1,471) downregulated genes by the archetypic PtrEPSP‐TF were downregulated by BESC‐35 and BESC‐876 variants (Figure S1).
Figure 3.
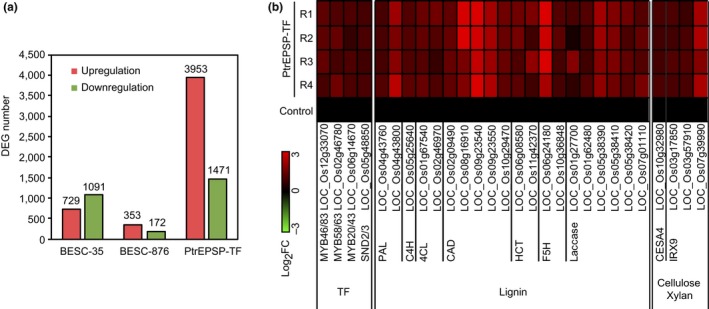
The heterologous expression of the archetypic PtrEPSP‐TF in rice induces the upregulation of genes involved in lignin and secondary cell wall biosynthesis. (a) Rice plants expressing archetypic PtrEPSP‐TF have more DEGs than rice plants expressing BESC35 PtrEPSP‐TF or BESC876 PtrEPSP‐TF. Red bar: upregulated genes; Green bar: downregulated genes. (b) Heat map showing expression patterns of PAL, C4H, 4CL, CAD, F5H, LACCASE, CESA4, IRX9, MYB46/83, MYB58/63, MYB20/43, and SND2/3 in four independent replicates (R1 to R4) of the archetypic PtrEPSP‐TF expression rice. Log2FC of mean expression levels of the four replicates of empty‐vector control was set as 0. The color scale represents the value of Log2FC
We then performed Gene Ontology (GO) enrichment analysis on DEGs of rice expressing the functional archetypic PtrEPSP‐TF to determine the global effect of PtrEPSP‐TF. Among upregulated ones, genes associated with cell walls, lipids, secondary metabolism, and light reactions were significantly enriched (Figure S2a). Further GO enrichment analysis showed that genes involved in phenylpropanoid, flavonoid, and lignin metabolism were overrepresented among upregulated genes associated with secondary metabolism (Figure S2b). In contrast, downregulated genes did not show any enrichment in metabolic pathways (Figure S2c and d).
To determine whether the differential sclerenchyma cell staining between rice plants expressing the archetypic PtrEPSP‐TF and BESC‐35 or BESC‐876 alleles was related to transcriptional reprogramming of secondary cell wall biosynthesis, we looked for genes related to secondary cell wall biosynthesis from DEGs of transgenic rice. A total of twenty lignin, one cellulose, and three xylan biosynthetic genes were found. Interestingly, four master regulators of secondary cell wall biosynthesis were also found (Figure 3b). All of these were upregulated by the functional PtrEPSP‐TF (Figure 3b). In contrast, expression of these genes only showed modest upregulation or even downregulation in the BESC‐35 and BESC‐876 lines (Figure S3), suggesting that the archetypic PtrEPSP‐TF, but not BESC‐35 or BESC‐876 variants, can trigger the transcriptional reprogramming of secondary cell wall biosynthesis in rice.
Among these 28 upregulated genes involved in secondary cell wall biosynthesis, four genes encode transcription factors (Figure 3b), and their Arabidopsis homologs were found to be master regulators of secondary cell wall/lignin biosynthesis, including LOC_Os12g33070 (MYB46/83)(Zhong et al., 2011), LOC_Os02g46780 (MYB58/63)(Zhou, Lee, Zhong, & Ye, 2009), LOC_Os06g14670 (MYB20/43)(Zhong, Lee, Zhou, McCarthy, & Ye, 2008), and LOC_Os05g48850 (SND2/3)(Hussey et al., 2011). Downstream of these transcription factors, PAL, C4H, and 4CL catalyze the first three steps of the phenylpropanoid pathway and are essential for the biosynthesis of monolignols (Fraser & Chapple, 2011). We observed that two PAL genes (LOC_Os04g43760 and LOC_Os04g43800), one C4H gene (LOC_Os05g25640), and two 4CL genes (LOC_Os01g67540 and LOC_Os02g46970) were upregulated by the archetypic PtrEPSP‐TF (Figure 3b). In addition, genes critical for monolignol biosynthesis were also upregulated by the archetypic PtrEPSP‐TF, such as five CADs (LOC_Os02g09490, LOC_Os08g16910, LOC_Os09g23540, LOC_Os09g23550, and LOC_Os10g29470), two HCTs (LOC_Os06g08580 and LOC_Os11g42370), and two F5Hs (LOC_Os06g24180 and LOC_Os10g36848) (Figure 3b). Additionally, six Laccase genes were upregulated by PtrEPSP‐TF: LOC_Os01g27700, LOC_Os01g62480, LOC_Os05g38390, LOC_Os05g38410, LOC_Os05g38420, and LOC_Os07g01110 (Figure 3b). The four cellulose and xylan biosynthetic genes upregulated by PtrEPSP‐TF were CESA4 (LOC_Os10g32980) and IRX9 (LOC_Os03g17850, LOC_Os03g57910, and LOC_Os07g39990) (Figure 3b).
Collectively, we demonstrated that the heterologous expression of the archetypic PtrEPSP‐TF in rice resulted in the thickening of secondary cell wall and increased lignin content, which may be through transcriptional reprogramming of genes critical for lignin/secondary cell wall biosynthesis in rice. This observation is consistent with the analysis of transgenic Populus overexpressing PtrEPSP‐TF (Xie, Muchero, et al., 2018), suggesting that besides Populus, PtrEPSP‐TF is capable of affecting the lignin/secondary cell wall biosynthesis when introduced to rice in which the native rice EPSP synthase lacks the HTH domain.
3.4. PtrEPSP‐TF mediates a transcriptional regulatory network in rice
In Populus, the archetypic PtrEPSP‐TF positively regulates PtrMYB021 expression by directly repressing the expression of PtrhAT, a SLEEPER‐like gene that directly represses PtrMYB021 expression (Xie, Muchero, et al., 2018). As a close homolog of Arabidopsis MYB46, a master regulator of secondary cell wall biosynthesis, PtrMYB021 functions as a transcriptional activator and activates the biosynthesis pathways for lignin, cellulose, and xylan (Zhong et al., 2013), the major components of secondary cell walls. Similarly, in the present study, we demonstrate that the archetypic PtrEPSP‐TF is able to upregulate rice MYB46 (LOC_Os12g33070) and phenylpropanoid/lignin biosynthetic genes (PAL, 4CL, C4H, F5H, CAD, CCR, and LACCASE) when introduced to rice. Therefore, we hypothesize that an analogous PtrEPSP‐TF‐mediated transcriptional mechanism, in which PtrEPSP‐TF represses negative regulators to activate lignin/secondary cell wall biosynthesis, may exist in rice.
In support of this hypothesis, we found three tandemly duplicated hAT SLEEPER‐like genes, LOC_Os09g21410 (−7.3, p = .0049), LOC_Os09g21420 (−3.1, p = 2.7E‐11), and LOC_Os09g21430 (−1.7, p = 1.1E‐06), that were significantly downregulated in the archetypic PtrEPSP‐TF expression lines (Dataset S1). In contrast, the expression of LOC_Os09g21410 and LOC_Os09g21420 only had moderate reduction or even no significant change in the BESC‐35 and BESC‐876 lines (Dataset S1). The expression of LOC_Os09g21430 exhibited a higher reduction in the BESC‐35 line than that in the line expressing the archetypic PtrEPSP‐TF, but had no significant change in the BESC‐876 line (Dataset S1). Besides hAT SLEEPER‐like genes, we found an additional 82 transcription factors that were significantly downregulated in the same line (Dataset S2). Among them, we found twelve NAC and MYB transcription factors (Dataset S2). Among the twelve NAC and MYB transcription factors, XND1 (LOC_Os02g34970) is a homolog of Arabidopsis AT5G64530, which has been reported to negatively regulate lignin biosynthesis (Zhao, Avci, Grant, Haigler, & Beers, 2008). MYB48 (LOC_Os01g74410) has a close homolog MYB12 in Arabidopsis, which was found to activate the biosynthesis of flavonoids, which, interestingly, competes for precursors with the biosynthesis of monolignols (Mehrtens, Kranz, Bednarek, & Weisshaar, 2005). Also, MYB3R1 (LOC_Os12g13570 and AT4G32730 in Arabidopsis) and MYB3R3 (LOC_Os05g38460 and AT3G09370 in Arabidopsis) are putative transcriptional repressors (Kobayashi et al., 2015) and NAC047 (LOC_Os11g03300 and AT3G04070 in Arabidopsis) is associated with cell wall re‐organization (Rauf et al., 2013). To determine whether these five transcription factors (XND1, MYB48, MYB3R1, MYB3R3, and NAC047) are indeed targets of PtrEPSP‐TF in rice, we tested the repression of their promoter activity by PtrEPSP‐TF using the Populus mesophyll protoplast transient expression system. A 500‐bp fragment of the promoter region of each putative target gene (−500 to −1 from start codon) was inserted between the 35S promoter and the GUS gene in reporter constructs. Then, each reporter construct was co‐transfected with the effector construct expressing PtrEPSP‐TF into protoplasts. GUS activity was measured to represent the activity of the tested promoters. As illustrated by the reduced GUS activity, PtrEPSP‐TF repressed the activity of all five tested promoters (Figure 4a). In contrast, PtrEPSP‐TF did not repress the promoter of LOC_Os03g01870 (NAC1) (Figure 4a), a NAC transcription factor downregulated in all the three transgenic lines (PtrEPSP‐TF, BESC‐35, and BESC‐876) (Dataset S1), suggesting that the altered expression of LOC_Os03g01870 (NAC1) may be from background signals (e.g. sequencing noises). The combined RNA‐seq and transactivation assay results demonstrate that expression of the five rice NAC and MYB transcription factors was negatively regulated by PtrEPSP‐TF.
Figure 4.
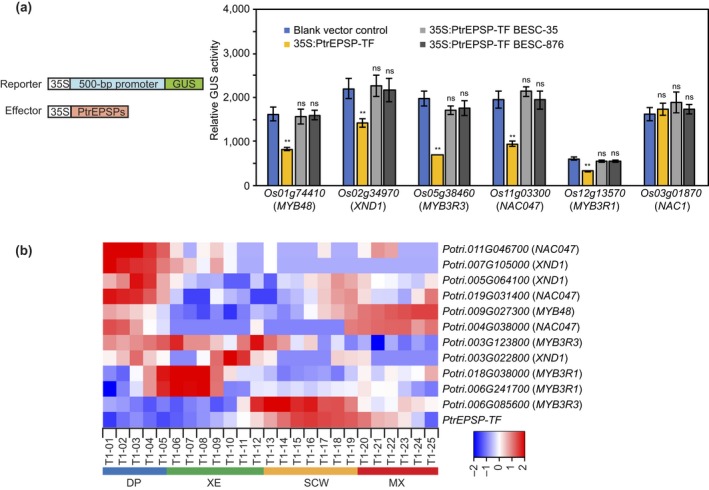
PtrEPSP‐TF represses the expression of five genes. (a) Left panel: a scheme of reporter and effector constructs used in transactivation assays. Right panel: transactivation assays determining the transcriptional repression of PtrEPSP‐TF on LOC_Os02g34970 (XND1), LOC_Os01g74410 (MYB48), LOC_Os12g13570 (MYB3R1), LOC_Os05g38460 (MYB3R3), and LOC_Os11g03300 (NAC047). All transfection assays were performed in triplicate to calculate the mean value and standard deviation (error bar). P‐value comparison with blank vector control was calculated using two‐tailed Student t tests (**p ≤ .01, *p ≤ .05, ns p > .05). (b) Heatmap of expression levels of PtrEPSP‐TF, Potri.003G022800 (XND1), Potri.005G064100 (XND1), Potri.007G105000 (XND1), Potri.009G027300 (MYB48), Potri.018G038000 (MYB3R1), Potri.006G241700 (MYB3R1), Potri.003G123800 (MYB3R3), Potri.006G085600 (MYB3R3), Potri.011G046700 (NAC047), Potri.004G038000 (NAC047), and Potri.019G031400 (NAC047) in wood‐forming zones (http://aspwood.popgenie.org). DP, differentiating phloem zone; XE, xylem expansion zone; SCW, secondary cell wall deposition zone; MX, mature xylem zone. Color bar at the bottom depicts the relative expression levels
To further investigate the analogous PtrEPSP‐TF‐mediated transcriptional mechanism in Populus and rice, we analyzed the expression patterns of Populus homologs of XND1, MYB48, MYB3R1, MYB3R3, and NAC047 using the AspWood database (http://aspwood.popgenie.org/). During Populus wood formation, the phenylpropanoid pathway is activated to generate lignified secondary cell wall. AspWood provides high‐spatial‐resolution gene expression profiles spanning the wood‐forming zone of Populus (Sundell et al., 2017). As shown in Figure 4b, PtrEPSP‐TF gene has a peak expression in the secondary cell wall deposition zone, which is similar to expression patterns of positive regulators of the phenylpropanoid and lignin biosynthesis (e.g., SND1, SND2, VND7; Sundell et al., 2017). In contrast, Populus homologs of XND1, MYB48, MYB3R1, MYB3R3, and NAC047 (except Potri.006G085600, one homolog of MYB3R3) have low expression levels in the secondary cell wall deposition zone, but have peak expression levels in the differentiating phloem zone, xylem expansion zone, or the mature xylem zone (Figure 4b). These opposite expression patterns suggest that PtrEPSP‐TF may also have repression effect on Populus XND1, MYB48, MYB3R1, MYB3R3, and NAC047 genes.
The observed repression of SLEEPER‐like genes in addition to known negative regulators of cell wall biosynthesis, such as XND1, is consistent with the proposed transcriptional regulatory model in Populus (Xie, Muchero, et al., 2018), that established that the phenylpropanoid pathway is induced upon overexpression of PtrEPSP‐TF, which suppresses the expression of a SLEEPER‐like repressor of the pathway. To confirm this hypothesis, we looked for transcriptional regulators related to cell wall biosynthesis among a total of 221 predicted transcriptional regulators. A total of 43 NAC and MYB transcription factors were upregulated by PtrEPSP‐TF (Dataset S3). Among these transcription factors, four master regulators of phenylpropanoid biosynthesis were found, including NAC and MYB transcription factors LOC_Os12g33070 (MYB46/83), LOC_Os02g46780 (MYB58/63), LOC_Os06g14670 (MYB20) (Zhong et al., 2008), and LOC_Os05g48850 (SND2/3). These four transcription factors and/or their Arabidopsis orthologs activate lignin and secondary cell wall biosynthesis (Nakano, Yamaguchi, Endo, Rejab, & Ohtani, 2015). For the remaining 39 NAC and MYB transcription factors, we searched for their predicted cis‐element‐binding sites in promoter regions of 3,423 genes specifically upregulated by the archetypic PtrEPSP‐TF (Figure S1). Five transcription factors are presented to highlight using this approach (Figure S4 and Dataset S4): LOC_Os11g08210 (NAC081), LOC_Os03g42630 (NAC058), LOC_Os05g35500 (MYB4), LOC_Os08g43550 (MYB4), LOC_Os12g07640 (MYB4).
Cis‐elements recognized by the five transcription factors were enriched in promoters of these genes. Specifically, the cis‐element (TFmatrixID_0382) targeted by LOC_Os11g08210 (NAC081) was found in 1,952 of the 3,423 (57.03%; Figure S4). LOC_Os03g42630 (NAC058) targets the cis‐element TFmatrixID_0391, which was found in promoters of 984 out of the 3,423 (28.75%; Figure S4). The cis‐element TFmatrixID_0336 targeted by the four MYB4 paralogs LOC_Os05g35500, LOC_Os08g43550, and LOC_Os12g07640 was found in promoters of 963 out of the 3,423 genes (28.13%; Figure S4). Overall, we propose that these five transcription factors in addition to LOC_Os12g33070 (MYB46/83), LOC_Os02g46780 (MYB58/63), LOC_Os06g14670 (MYB20), and LOC_Os05g48850 (SND2/3) may function downstream of the MYB48, XND1, MYB3R3, MYB3R1, and NAC047, which are individually repressed by the archetypic PtrEPSP‐TF (Figures 4 and 5).
Figure 5.
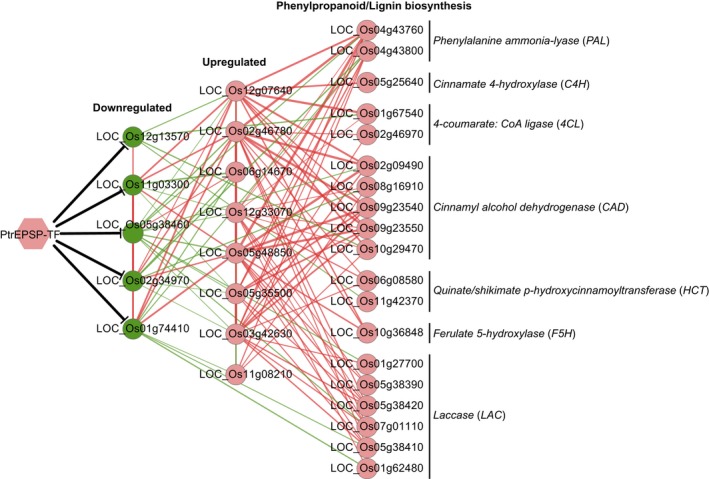
Co‐expression analysis of members of PtrEPSP‐TF‐mediated transcriptional network in rice. For co‐expression relationships, only correlation coefficiency (r) >0.1 or <−0.1 and p ≤ .05 are shown. Black edges indicate experimentally validated relationships. Red edges indicate positive co‐relationship. Green edges indicate negative co‐relationship. The strength of co‐relationship (r) is represented by the thickness of edges
To validate this proposed PtrEPSP‐TF‐mediated transcriptional network, we conducted co‐expression analysis using existing developmental transcriptome datasets of rice (GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51289 (Hirano, Aya, et al., 2013) and GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024 (Wang et al., 2010); Dataset S5). In the co‐expression analysis, LOC_Os08g43550 (MYB4) and LOC_Os06g24180 (F5H) were not present in the two microarray datasets and were not included in the proposed network (Figure 5). Co‐expression analyses of the rest of genes except LOC_Os12g33070 (MYB46/83) were performed using dataset GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51289 because LOC_Os12g33070 (MYB46/83) is not included in the dataset. We used dataset GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024 to analyze the co‐expression of LOC_Os12g33070 (MYB46/83) (Hirano, Aya, et al., 2013) with other members in the PtrEPSP‐TF‐mediated network. Expressions of most Layer one genes (downregulated by PtrEPSP‐TF) were negatively co‐related with expressions of Layer two genes (upregulated by PtrEPSP‐TF) and even some lignin/phenylpropanoid biosynthetic genes (green lines, Figure 5). As predicted, expressions of Layer two genes displayed significant positive co‐expression with expression of PALs (LOC_Os04g43760, LOC_Os04g43800), C4H (LOC_Os05g25640), 4CLs (LOC_Os01g67540, LOC_Os02g46970), CADs (LOC_Os02g09490, LOC_Os08g16910, LOC_Os09g23540, LOC_Os09g23550, LOC_Os10g29470), HCTs (LOC_Os06g08580, LOC_Os11g42370), F5H (LOC_Os10g36848), and Laccases (LOC_Os01g27700, LOC_Os01g62480, LOC_Os05g38390, LOC_Os05g38410, LOC_Os05g38420, LOC_Os07g01110) (Figure 5).
Collectively, our transcriptomics analysis and transactivation assay identified five repression targets of PtrEPSP‐TF, which may negatively regulate lignin and phenylpropanoid biosynthesis.
4. DISCUSSION
Populus plants containing rare mutations at PtrEPSP‐TF locus show reduced lignin content (Xie, Muchero, et al., 2018). However, the underlying molecular mechanism had not been established. Here, by functionally analyzing two natural variants of PtrEPSP‐TF, we unveiled that the reduced lignin content and increased sugar release (Lu, Hao, et al., 2019) are likely caused by the reduced DNA binding and subsequently reduced transcriptional activities of the variant PtrEPSP‐TF. An EMSA assay demonstrated that the two PtrEPSP‐TF variants occurring in genotypes BESC‐35 and BESC‐876 have reduced DNA‐binding affinity to the PtrhAT promoter, which is the direct target of PtrEPSP‐TF. Moreover, the transcriptional repression effects of the BESC‐35 and BESC‐876 variants on PtrhAT promoter displayed significant reduction in transactivation assays. The HTH motif of PtrEPSP‐TF was shown to be responsible for its DNA‐binding activity (Xie, Muchero, et al., 2018). Given BESC‐35 and BESC‐876 variants share the same point mutation close to the HTH motif leading to the D142E amino acid substitution, the 142nd amino acid may play an important role in assisting the proper function of the HTH motif (e.g., protein conformation, target recognition). Although PtrEPSP‐TF BESC‐876 has another point mutation leading to the L364P amino acid substitution, it did not display additional reduction of DNA‐binding activity, suggesting that this point mutation may not affect the DNA binding of PtrEPSP‐TF. In fact, the L364P substitution is further away from the HTH domain than is the D142E substitution.
Although dicots and monocots have numerous morphological differences in the cell wall, the transcriptional regulatory mechanism appears to be relatively conserved, especially surrounding the critical role of NAC and MYB transcription factors (Nakano et al., 2015; Rao & Dixon, 2018). The ortholog of PtrEPSP‐TF in rice lacks the DNA‐binding HTH motif, thus lacking the transcriptional activity. By heterogeneously expressing PtrEPSP‐TF in rice, we observed altered thickness of sclerenchyma cell wall, lignin level, secondary metabolism, and transcript levels of genes associated with lignin/phenylpropanoid biosynthesis, which are similar to Populus deltoides plants overexpressing PtrEPSP‐TF (Xie, Muchero, et al., 2018). As the major mechanical tissues in rice stem, sclerenchyma fibers have heavily thickened secondary cell walls and are the major contributor of straw biomass. As such, increasing sclerenchyma cell wall thickness is a viable option to enhance the production of straw biomass.
We also observed that lignin/phenylpropanoid biosynthetic genes including PAL, 4CL, C4H, F5H, CAD, CCR, and LACCASE were significantly upregulated by PtrEPSP‐TF in rice. Moreover, master regulators of lignin/ phenylpropanoid biosynthesis, such as MYB46/83, MYB58/63, MYB20/43, and SND2/3 also displayed increased expression levels in transgenic rice expressing PtrEPSP‐TF. Collectively, we conclude that PtrEPSP‐TF may have a relatively conserved effect on the lignin/phenylpropanoid biosynthesis in Populus and rice. The fact that transgenic rice expressing PtrEPSP‐TF does not exhibit as many upregulated cellulose and xylan biosynthetic genes as Populus deltoides overexpressing PtrEPSP‐TF (Xie, Muchero, et al., 2018) further demonstrates the divergence of secondary cell wall biosynthesis in dicots and monocots. Other regulatory mechanisms of secondary cell wall biosynthesis contain both conserved and divergent parts between dicots and monocots (Rao & Dixon, 2018).
The current understanding of the regulation of rice secondary cell wall biosynthesis mainly derives from comparative studies of the main transcriptional regulatory mechanism defined in Arabidopsis. Based on molecular and genetic analyses, we were able to identify five repression targets of PtrEPSP‐TF in rice: MYB48 (LOC_Os01g74410), XND1 (LOC_Os02g34970), MYB3R3 (LOC_Os05g38460), NAC047 (LOC_Os11g03300), and MYB3R1 (LOC_Os12g13570). Studies on Arabidopsis homologs of these repression targets of PtrEPSP‐TF suggest that these five transcription factors may negatively affect lignin biosynthesis (Kobayashi et al., 2015; Mehrtens et al., 2005; Rauf et al., 2013; Zhao et al., 2008). However, their detailed regulatory roles in lignin and secondary cell wall biosynthesis remain undefined. Based on RNA‐seq and co‐expression analyses, we defined their negative regulatory relationships with positive regulators (i.e., MYB46/83, MYB58/63, MYB20/43, SND2/3, NAC081, NAC058, and MYB4) and biosynthetic genes (i.e., PAL, 4CL, C4H, F5H, CAD, CCR, and LACCASE) of lignin and secondary cell wall, which demonstrates the link of the five repression targets of PtrEPSP‐TF and the regulation of lignin and secondary cell wall biosynthesis.
With the production of approximately 1,140 million tons biomass per year worldwide, rice straw has been considered as an attractive biomass feedstock for producing second‐generation biofuels and bioproducts (Satlewal et al., 2018). However, to date, the transcriptional regulatory network of lignin and secondary cell wall biosynthesis in rice remains poorly studied. The discovery of the five new regulators extends our understanding of the regulatory mechanism of lignin and secondary cell wall biosynthesis in rice and will potentially be bioengineered to improve rice biomass for the production of biofuels and bioproducts. It remains unknown though, which native transcription factors in rice (equivalent to the function of PtrEPSP‐TF) regulate the transcription of these five new regulators.
Collectively, our studies of the heterologous expression of PtrEPSP‐TF and its two alleles defined one critical amino acid for PtrEPSP‐TF transcriptional function and uncovered five negative regulators of secondary cell wall biosynthesis in rice, which enrich the toolbox of the bioengineering for an improved biofuel feedstock. Additionally, the integration of heterologous expression and systems biological analyses in this study demonstrates a strategy complementing the approach of referring the established transcriptional network in Arabidopsis to understand the regulation of the phenylpropanoid pathway in rice.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
W.M., J.‐G.C., G.A.T., and M.X. designed experiments. M.X., W.M., and J.‐G.C. wrote the manuscript. G.A.T., T.J.T., and F.C. edited the manuscript. M.X. and S.S.J performed molecular experiments. J.Z. and M.X. performed co‐expression analysis. M.J.M., P.R.L., and W.P. generated transgenic plants. N.E. and T.J.T. performed metabolomic analysis. C.D and M.F.D analyzed lignin levels. J.Z., V.R.S., E.L., and J.S. analyzed the data.
Supporting information
ACKNOWLEDGMENTS
This research was funded by The BioEnergy Science Center and The Center for Bioenergy Innovation from 2007 to 2017 and from 2017 to 2019, respectively. Both are the U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science. This research used resources of the Compute and Data Environment for Science (CADES) at the Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT‐Battelle, LLC for the U.S. Department of Energy under Contract No. DE‐AC05‐00OR22725. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE‐AC02‐05CH11231.
Xie M, Zhang J, Singan VR, et al. Identification of functional single nucleotide polymorphism of Populus trichocarpa PtrEPSP‐TF and determination of its transcriptional effect. Plant Direct. 2020;4:1–13. 10.1002/pld3.178
This manuscript has been authored by UT‐Battelle, LLC under Contract No. DE‐AC05‐00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non‐exclusive, paid‐up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for the United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe‐public‐access‐plan).
Contributor Information
Jin‐Gui Chen, Email: chenj@ornl.gov.
Wellington Muchero, Email: mucherow@ornl.gov.
REFERENCES
- Chow, C. N. , Zheng, H. Q. , Wu, N. Y. , Chien, C. H. , Huang, H. D. , Lee, T. Y. , … Chang, W. C. (2016). PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research, 44, D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D. (1979). The central role of phosphoenolpyruvate in plant metabolism. Annual Review of Plant Physiology, 30, 131–158. 10.1146/annurev.pp.30.060179.001023 [DOI] [Google Scholar]
- Docimo, T. , Coraggio, I. , De Tommasi, N. , & Leone, A. (2008). Enhancing phenylpropanoid secondary metabolites in Nicotiana tabacum and Salvia sclarea by overexpression of a rice Myb4 transcription factor. Planta Medica, 74, PG87 10.1055/s-0028-1084839 [DOI] [Google Scholar]
- Fornalé, S. , Lopez, E. , Salazar‐Henao, J. E. , Fernández‐Nohales, P. , Rigau, J. , & Caparros‐Ruiz, D. (2014). AtMYB7, a new player in the regulation of UV‐sunscreens in Arabidopsis thaliana. Plant and Cell Physiology, 55, 507–516. 10.1093/pcp/pct187 [DOI] [PubMed] [Google Scholar]
- Fraser, C. M. , & Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book, 9, e0152 10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, K. , Aya, K. , Morinaka, Y. , Nagamatsu, S. , Sato, Y. , Antonio, B. A. , … Matsuoka, M. (2013). Survey of genes involved in rice secondary cell wall formation through a co‐expression network. Plant and Cell Physiology, 54, 1803–1821. 10.1093/pcp/pct121 [DOI] [PubMed] [Google Scholar]
- Hirano, K. , Kondo, M. , Aya, K. , Miyao, A. , Sato, Y. , Antonio, B. A. , … Matsuoka, M. (2013). Identification of transcription factors involved in rice secondary cell wall formation. Plant and Cell Physiology, 54, 1791–1802. 10.1093/pcp/pct122 [DOI] [PubMed] [Google Scholar]
- Hussey, S. G. , Mizrachi, E. , Spokevicius, A. V. , Bossinger, G. , Berger, D. K. , & Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biology, 11, 173 10.1186/1471-2229-11-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T. B. , Lawler, N. J. , LaFayette, P. R. , Vodkin, L. O. , & Parrott, W. A. (2016). Simple gene silencing using the trans‐acting si RNA pathway. Plant Biotechnology Journal, 14, 117–127. 10.1111/pbi.12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Cominelli, E. , Bailey, P. , Parr, A. , Mehrtens, F. , Jones, J. , … Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV‐protecting sunscreens in Arabidopsis. The EMBO Journal, 19, 6150–6161. 10.1093/emboj/19.22.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , & Salzberg, S. L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W.‐C. , Kim, J.‐Y. , Ko, J.‐H. , Kang, H. , & Han, K.‐H. (2014). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Molecular Biology, 85, 589–599. 10.1007/s11103-014-0205-x [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. , Suzuki, T. , Iwata, E. , Nakamichi, N. , Suzuki, T. , Chen, P. , … Ito, M. (2015). Transcriptional repression by MYB3R proteins regulates plant organ growth. The EMBO Journal, 34, 1992–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Tschaplinski, T. J. , Engle, N. L. , Hamilton, C. Y. , Rodriguez, M. Jr , Liao, J. C. , … Graham, D. E. (2012). Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnology for Biofuels, 5, 2 10.1186/1754-6834-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. , Williams, B. J. , Thangella, P. A. V. , Ladak, A. , Schepmoes, A. A. , Olivos, H. J. , … Bartley, L. E. (2017). Proteomics coupled with metabolite and cell wall profiling reveal metabolic processes of a developing rice stem internode. Frontiers in Plant Science, 8, 1134 10.3389/fpls.2017.01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K. , Hao, N. , Meng, X. , Luo, Z. , Tuskan, G. A. , & Ragauskas, A. J. (2019). Investigating the correlation of biomass recalcitrance with pyrolysis oil using poplar as the feedstock. Bioresource Technology, 289, 121589 10.1016/j.biortech.2019.121589 [DOI] [PubMed] [Google Scholar]
- Maeda, H. , & Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology, 63, 73–105. 10.1146/annurev-arplant-042811-105439 [DOI] [PubMed] [Google Scholar]
- Mann, D. G. , King, Z. R. , Liu, W. , Joyce, B. L. , Percifield, R. J. , Hawkins, J. S. , … Mazarei, M. (2011). Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnology, 11, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, D. G. J. , LaFayette, P. R. , Abercrombie, L. L. , King, Z. R. , Mazarei, M. , Halter, M. C. , … Neal Stewart Jr, C. (2012). Gateway‐compatible vectors for high‐throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnology Journal, 10, 226–236. 10.1111/j.1467-7652.2011.00658.x [DOI] [PubMed] [Google Scholar]
- Mehrtens, F. , Kranz, H. , Bednarek, P. , & Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol‐specific regulator of phenylpropanoid biosynthesis. Plant Physiology, 138, 1083–1096. 10.1104/pp.104.058032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, Y. , Yamaguchi, M. , Endo, H. , Rejab, N. A. , & Ohtani, M. (2015). NAC‐MYB‐based transcriptional regulation of secondary cell wall biosynthesis in land plants. Frontiers in Plant Science, 6, 288 10.3389/fpls.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, J. , Wheeler, J. , Heazlewood, J. , Li, S. F. , & Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal, 40, 979–995. 10.1111/j.1365-313X.2004.02280.x [DOI] [PubMed] [Google Scholar]
- Rao, X. , & Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front Plant Sci, 9, 399 10.3389/fpls.2018.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf, M. , Arif, M. , Fisahn, J. , Xue, G. P. , Balazadeh, S. , & Mueller‐Roeber, B. (2013). NAC transcription factor speedy hyponastic growth regulates flooding‐induced leaf movement in Arabidopsis. The Plant Cell, 25, 4941–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, L. A. , & Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytologist, 164, 17–30. 10.1111/j.1469-8137.2004.01143.x [DOI] [PubMed] [Google Scholar]
- Satlewal, A. , Agrawal, R. , Bhagia, S. , Das, P. , & Ragauskas, A. J. (2018). Rice straw as a feedstock for biofuels: Availability, recalcitrance, and chemical properties. Biofuels, Bioproducts and Biorefining, 12, 83–107. 10.1002/bbb.1818 [DOI] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell, D. , Street, N. R. , Kumar, M. , Mellerowicz, E. J. , Kucukoglu, M. , Johnsson, C. , … Nilsson, O. (2017). AspWood: High‐spatial‐resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. The Plant Cell, 29, 1585–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, T. , Liu, Y. , Yan, H. , You, Q. , Yi, X. , Du, Z. , … Su, Z. (2017). agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research, 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S. B. , Hagen, G. , & Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin‐responsive transcription. The Plant Cell, 15, 533–543. 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaplinski, T. J. , Standaert, R. F. , Engle, N. L. , Martin, M. Z. , Sangha, A. K. , Parks, J. M. , … Mielenz, J. R. (2012). Down‐regulation of the caffeic acid O‐methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnology for Biofuels, 5, 71 10.1186/1754-6834-5-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini, C. , Locatelli, F. , Bracale, M. , Magnani, E. , Marsoni, M. , Osnato, M. , … Coraggio, I. (2004). Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal, 37, 115–127. [DOI] [PubMed] [Google Scholar]
- Vogt, T. (2010). Phenylpropanoid biosynthesis. Molecular Plant, 3, 2–20. 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Wang, L. , Xie, W. , Chen, Y. , Tang, W. , Yang, J. , Ye, R. , … Zhang, Q. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. The Plant Journal, 61, 752–766. 10.1111/j.1365-313X.2009.04100.x [DOI] [PubMed] [Google Scholar]
- Wilkins, O. , Nahal, H. , Foong, J. , Provart, N. J. , & Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3‐MYB family of transcription factors. Plant Physiology, 149, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, M. , Muchero, W. , Bryan, A. C. , Yee, K. L. , Guo, H.‐B. , Zhang, J. , … Payyavula, R. S. (2018). A 5‐enolpyruvylshikimate 3‐phosphate synthase functions as a transcriptional repressor in Populus. The Plant Cell, 30, 1645–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, M. , Ren, G. , Costa‐Nunes, P. , Pontes, O. , & Yu, B. (2012). A subgroup of SGS3‐like proteins act redundantly in RNA‐directed DNA methylation. Nucleic Acids Research, 40, 4422–4431. 10.1093/nar/gks034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, M. , Zhang, J. , Tschaplinski, T. J. , Tuskan, G. A. , Chen, J.‐G. , & Muchero, W. (2018). Regulation of lignin biosynthesis and its role in growth‐defense tradeoffs. Frontiers in Plant Science, 9, 1427 10.3389/fpls.2018.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekutieli, D. , & Benjamini, Y. (1999). Resampling‐based false discovery rate controlling multiple test procedures for correlated test statistics. Journal of Statistical Planning and Inference, 82, 171–196. 10.1016/S0378-3758(99)00041-5 [DOI] [Google Scholar]
- Yoo, S. D. , Cho, Y. H. , & Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols, 2, 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Avci, U. , Grant, E. H. , Haigler, C. H. , & Beers, E. P. (2008). XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. The Plant Journal, 53, 425–436. 10.1111/j.1365-313X.2007.03350.x [DOI] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , McCarthy, R. L. , Reeves, C. K. , Jones, E. G. , & Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant and Cell Physiology, 52, 1856–1871. 10.1093/pcp/pcr123 [DOI] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , Zhou, J. , McCarthy, R. L. , & Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell, 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , McCarthy, R. L. , Haghighat, M. , & Ye, Z. H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS ONE, 8, e69219 10.1371/journal.pone.0069219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , & Ye, Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant and Cell Physiology, 53, 368–380. 10.1093/pcp/pcr185 [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Lee, C. , Zhong, R. , & Ye, Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. The Plant Cell, 21, 248–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


