Abstract
Objective
This study aims to evaluate differences in the midtrimester cervicovaginal microbiota between women who developed puerperal infections at term and those who did not, and whether obesity modulates this relationship.
Methods
Previously, cervicovaginal swabs were collected at 21 to 25 weeks gestation (stored at −80°C). Samples were identified from Black women with normal vaginal flora (Nugent score: 0–2) delivering term singletons. Patients were in one of four equally sized groups (total n = 120) characterized by absence or presence of puerperal infection and maternal obesity. Samples were thawed, DNA extracted, and polymerase chain reaction with primers targeting the 16S rDNA V4 region was used to prepare an amplicon library sequenced and analyzed using Quantitative Insights into Microbial Ecology (QIIME) suite. Microbiota differences were assessed using permutation-based anodis over three β-diversity measures; Kruskal-Wallis test was used for taxa level analysis.
Results
After quality control measures, 113 samples were analyzed. Overall, there was significant clustering by puerperal infection (p = 0.03), but not by obesity (p > 0.05). Detailed taxa level analysis revealed approximately 66% less Proteobacteria phylum and 400% more BVAB1 genera in the second-trimester microbiota of women who had puerperal infections at term (p < 0.05).
Conclusion
Women who develop puerperal infections at term have a significantly altered midtrimester cervicovaginal microbiome with less Proteobacteria and greater BVAB1. This finding may represent a potential method to identify women at an increased risk of puerperal infection.
Keywords: cervicovaginal microbiome, chorioamnionitis, endometritis, maternal obesity, pregnancy
Puerperal infections (chorioamnionitis and endometritis) are a major source of maternal, neonatal, and long-term morbidity.1 These infections are polymicrobial, most often caused by bacteria that colonize the chorion/amnion or the endomyometrium by ascending from the lower genital tract.1–3 Numerous risk factors for both chorioamnionitis and endometritis have been described, including prolonged rupture of the membranes, prolonged labor, urogenital tract infections, and midtrimester bacterial vaginosis.1,4–6 Obesity has also been noted to be a strong risk factor for puerperal infections.1,7–12
Newer culture-independent molecular techniques using next-generation sequencing of the bacterial 16S rDNA have introduced a novel way to further characterize bacterial communities (microbiome) beyond the scope of traditional culture. Using these techniques, recent studies have not only shown that bacterial communities are relatively stable throughout pregnancy, but certain changes in microbial abundances may be associated with adverse pregnancy outcomes, particularly preterm birth.13–16
As both chorioamnionitis and endometritis are thought to arise from bacteria ascending from the lower genital tract, we hypothesized that (1) there are distinct microbial signatures in the midtrimester cervicovaginal microbiota of pregnant women who later develop puerperal infections at term and (2) maternal obesity affects this relationship. Therefore, we sought to evaluate the second-trimester cervicovaginal microbiota in women, comparing those who did and did not have clinically diagnosed puerperal infections at term. We also sought to evaluate differences in the cervicovaginal microbiota based on maternal obesity and evaluate any interaction between puerperal infection and maternal obesity.
Materials and Methods
Sample Collection
In this institutional review board-approved nested case-control study (N141222001), we utilized stored cervicovaginal samples collected at the University of Alabama at Birmingham during a population-based study of risk factors for preterm birth. Details of the parent study have previously been published.17 Briefly, during a vaginal speculum examination, a Dacron swab sample of the mucus on the ectocervix and in the posterior vaginal fornix was collected from women at a routine prenatal visit between 21 and 25 6/7 weeks gestation. Each swab was left in place for 10 seconds, withdrawn, and placed in 750 μL of fetal fibronectin buffer. These samples were then stored at −80°C. The samples for this study had not been subject to the previous thawing. Concurrently, vaginal smears were also obtained and evaluated by the Nugent score for the presence or absence of bacterial vaginosis.18
Isolation of Microbial DNA and Creation of 16S V4 Amplicon Library
In our current study, microbial genomic DNA was isolated using the fecal DNA isolation kit from Zymo Research (Irvine, CA) (catalog no. D6010) following the manufacturer’s instructions (there are no kits specific for vaginal DNA isolation, and this kit is the standard for isolating DNA that is meant to be analyzed at the University of Alabama at Birmingham Microbiome Resource.). Once the sample DNA was prepared, polymerase chain reaction (PCR) was used with unique barcoded primers to amplify the V4 region of the 16S rDNA to create an “amplicon library” from individual samples.19,20 While previous studies, including those from the Human Microbiome Project, have used V1–V3 or V3–V5 primers, we opted to use V4 primers. Recent literature has shown that for both short and longer read sequences, the V4 region is an appropriate region for capturing microbial diversity.19,21–27 Details of the V4 primers and PCR protocol have been previously published.28
DNA Sequencing and Bioinformatics
The samples were first quantitated using Pico Green (Molecular Probes, Inc., Eugene, OR), adjusted to a concentration of 4 nM, and then used for sequencing.20 The PCR amplicon covered around 255 bases of the V4 region of the 16S rDNA. All the samples were multiplexed using barcodes, and 251 base paired-end reads were sequenced using the Illumina MiSeq platform (San Diego, CA).19,20 FASTQ conversion of the raw data files was performed following demultiplexing. Quality assessment of the FASTQ files was performed using FASTQC, and then quality filtering was done using the FASTX toolkit.29,30 Since the overlap between fragments was approximately 245 bases, both paired reads were merged to generate a single high-quality read using the module “fastq_-mergepairs” of USEARCH (Drive 5). Read pairs with an overlap of less than 200 bases or with too many mismatches (> 20) in the overlapping region were discarded. Chimeric sequences were also filtered using the “identify_chimeric_seqs.py” module of USEARCH.31 The resulting reads with an average base quality Q score of < 20 were discarded. The remainder of the steps (explained below) were performed with the Quantitative Insights into Microbial Ecology (QIIME) suite, version 1.8 and an in-house developed wrapper for QIIME called QWRAP.21,29,32,33 Following the quality control filtering, we excluded any sample containing fewer than 10,000 passing reads. Total reads for the included samples ranged from 40,611 to 227,844 and averaged 115,509.
Sequences were grouped into operational taxonomic units (OTUs) using the clustering program UCLUST at a similarity threshold of 97%.31 The Ribosomal Database Program (RDP) classifier was used to make taxonomic assignments (to the genus level) for all OTUs at a confidence threshold of 80% (0.8). The RDP classifier was trained using a version of the Greengenes (v13_8) 16S rRNA database modified to identify bacterial vaginosis-associated bacteria (BVAB).34–36 Details of our BVAB prediction model have been previously described in detail.28 The resulting OTU table included all OTUs, their taxonomic identification, and abundance information. To account for differences in read depth across different samples, the OTUs were rarified at a common sequence depth. OTUs whose average abundance was less than 0.05% were filtered out. OTUs were then grouped together to summarize taxon abundance at different hierarchical levels of classification (e.g., phylum, class, order, family, genus). These taxonomy tables were also used to generate bar charts of taxon abundance. Multiple sequence alignment of OTUs was performed with PyNAST.30 Alpha diversity (within sample diversity) was calculated using a variety of diversity metrics, including Shannon, Chao1, and Simpson, as implemented in QIIME.37 Beta diversity (between sample diversity) among different samples was measured using UniFrac analysis.32 Principal coordinates analysis (PCoA) was performed by QIIME to visualize the dissimilarity matrix (β-diversity) between all the samples, such that samples that are more similar are closer in space than samples that are more divergent.38 Differences between groups of samples at the taxa level were performed by grouping OTUs by phylum, class, order, family and genus, and then testing for differences in frequency by a group using QIIME’s implantation of the Kruskal–Wallis test.
Subject Characteristics and Statistical Analysis
A total of 4,057 samples were available from the parent study. Given the known differences in microbial communities based on race or ethnicity, we chose samples only from Black women.39 In addition, because of the complexity of the microbial communities associated with bacterial vaginosis and the fact that bacterial vaginosis is a risk factor for both chorioamnionitis and endometritis, we included samples with Gram stains scored as normal flora (Nugent score: 0–3).6,18,39 To further eliminate any possible microbial changes associated with preterm birth, only women delivering at term were included.14,16
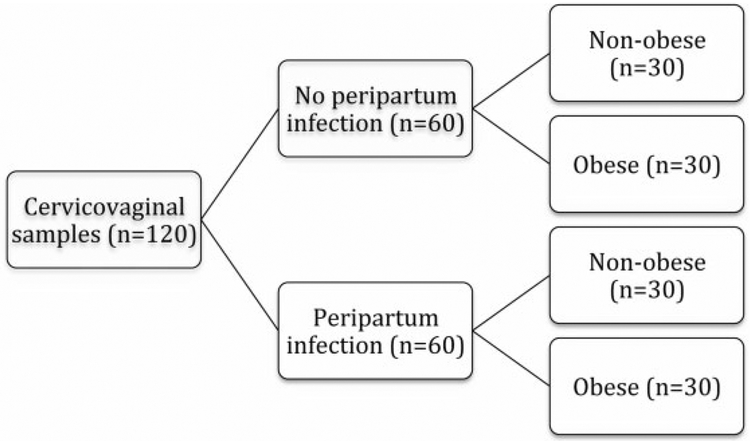
As such, samples from Black women with normal midtrimester flora delivering at term were randomly selected for analysis. These samples were categorized by the presence or absence of clinically diagnosed puerperal infection and secondarily by maternal obesity (body mass index ≥ 30 or < 30 kg/m2)–thus intending to create four equally sized groups (n = 30) with a total sample population of 120 (Fig. 1). Puerperal infection was diagnosed based on standard clinical diagnostic criteria.1,5 No histopathologic correlation was undertaken for the purposes of this study.
Fig. 1.
Flow diagram showing intended sample stratification. These samples were subjected to 16S sequencing methods as described in the text.
Descriptive characteristics of our sample populations were reported. Differences in demographics, labor and intrapartum characteristics, and comorbidities between women with and without puerperal infections were analyzed with chi-square test for categorical variables and Student t-test for continuous variables. For our primary analysis, the overall difference in the microbiome composition between groups defined by puerperal infection was analyzed using permutation-based anodis over all three β-diversity measures (PERMANOVA): Bray–Curtis, weighted UniFrac, and unweighted UniFrac. A planned independent secondary analysis evaluating differences in microbial composition was similarly undertaken to examine the effect of interaction with obesity. Because we anticipated marked predominance of Lactobacillus in all samples, we planned taxa specific analyses with the Kruskal-Wallis test. Differences with false-discovery rate (FDR) corrected p values less than 0.05 were considered significant.
Results
A total of 117 samples were included in the analysis after quality control measures rendered the remaining inadequate for read depth. Analysis of these 117 remaining samples revealed four outliers. These four outliers had a predominant taxon of Gardnerella (96.7 and 24.2%), Mycoplasma (83.1%), or Prevotella (39.4%). These four outliers had no unique demographic characteristics to explain their taxonomic variability. Moreover, given the lack of Lactobacillus (as expected in normal vaginal microbial communities), they were considered false-negative Gram stains for bacterial vaginosis and excluded from further analysis. Demographic characteristics of the analyzed population (n = 113), stratified by puerperal infection, are presented in Table 1. As expected, women who developed puerperal infection were more likely to be nulliparous and have a cesarean for their mode of delivery. Maternal age and maternal morbidities were similar in women who developed a puerperal infection and those who did not. As expected given the study design, body mass index, and rates of maternal obesity were also similar between these two groups.
Table 1.
Baseline characteristics of the sample population: overall and by presence or absence of puerperal infection
| Overall (n = 113) | No infection (n = 58) | Infection (n = 55) | p Valuea | |
|---|---|---|---|---|
| Age (y) | 22.4 ± 4.8 | 22.9 ± 4.8 | 22.2 ± 4.8 | 0.655 |
| Body mass index (mean) | 30.7 ± 8.2 | 30.0 ± 8.4 | 31.4 ± 7.9 | 0.344 |
| Obesity (BMI ≥ 30 kg/m2) | 55 (48.7) | 28 (48.3) | 27 (49.1) | 0.931 |
| Nulliparity | 69 (61.1) | 28 (48.3) | 41 (74.6) | 0.004 |
| Maternal comorbidities | ||||
| Diabetes | 3 (2.7) | 1 (1.7) | 2 (3.6) | 0.612 |
| Preeclampsia | 12 (10.6) | 7 (12.1) | 5 (9.1) | 0.608 |
| Smoking | 19 (16.8) | 11 (19.0) | 8 (14.6) | 0.530 |
| Peripartum infection | ||||
| Chorioamnionitis | 42 (37.2) | 42 (76.4) | ||
| Endometritis | 15 (13.3) | 15 (27.3) | ||
| Gestational age at delivery (wk) | 39.8 ± 1.2 | 39.6 ± 1.2 | 40.0 ± 1.2 | 0.034 |
| Cesarean delivery | 40 (35.4) | 12 (20.7) | 28 (50.9) | 0.0008 |
Abbreviation: BMI, body mass index.
Test of differences between patient with and without puerperal infection.
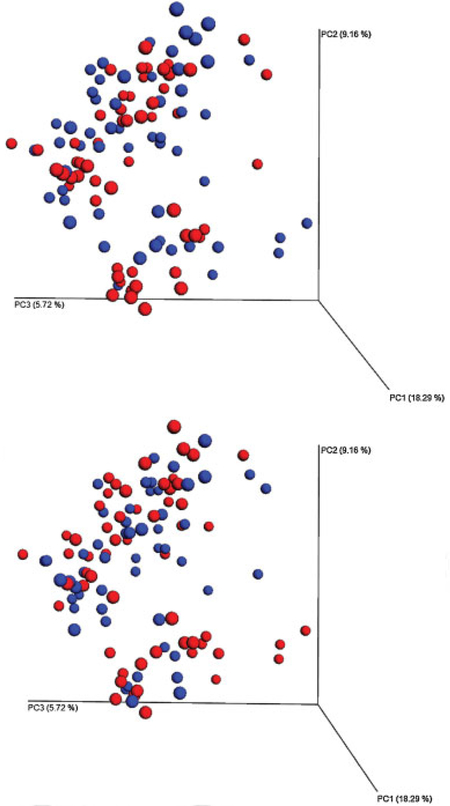
The remaining 113 samples were then analyzed by our primary variable, puerperal infection. When analyzing by PCoA, there were significant differences in β-diversity by unweighted UniFrac clustering (PERMANOVA p = 0.03), but not by weighted UniFrac or Bray–Curtis clustering (PERMANOVA p > 0.05) (Fig. 2). There were no significant differences in β–diversity by the secondary variable of material obesity, using any of the three measures (PERMANOVA p > 0.05) (Fig. 2). For both puerperal infection as well as obesity, subclusters were discernable (Fig. 2). While a small component of these subclusters was attributed to two different Lactobacillus species, no unique demographic characteristics were available to explain the major differences in these unique clusters.
Fig. 2.
Principle coordinates analysis (unweighted UniFrac) of all operational taxonomic units generated by 16S recombinant RNA sequencing by puerperal infection (top panel: infection blue, no infection = red) and obesity (bottom panel: obese blue, = nonobese = red). Samples closer in space are more similar in microbial composition. When analyzed for all taxa, there was a significant difference between groups for infection, but not obesity.
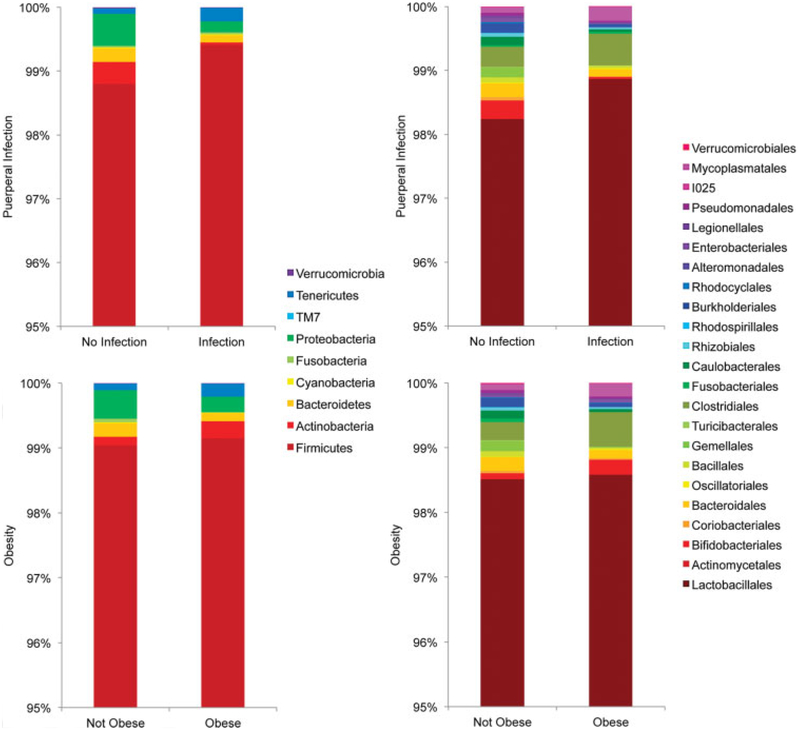
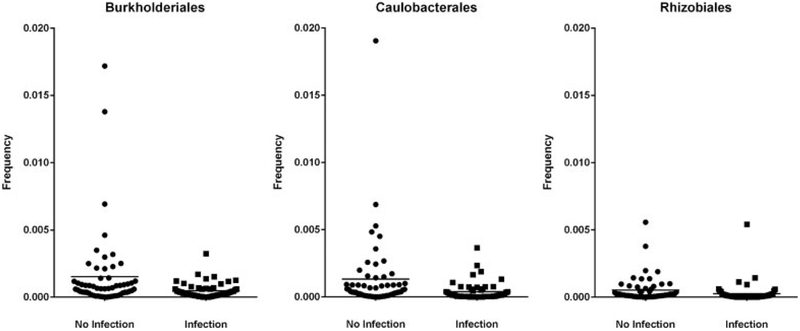
To determine which taxa were driving the differences between the infection and no-infection groups, we also performed taxa level analyses at the phylum, class, order, family, and genus level. We found significant differences at all taxonomic levels regarding puerperal infection, as presented in Table 2. There was decreased relative taxonomic abundance (~66% less) of Proteobacteria phylum seen in the midtrimester cervicovaginal microbiome of women who ultimately developed a puerperal infection at term (FDR-corrected p < 0.01) (Fig. 3). This difference was driven by the reduction of three orders of Proteobacteria (Caulobacterales, Burkholderiales, and Rhizobiales) in the infection group relative to no-infection group (FDR-corrected Kruskal-Wallis p < 0.05) (Table 2 and Fig. 4). Looking at individual samples, these orders were completely absent in patients with puerperal infection as compared with those without puerperal infection (Fig. 3). Several families and genera from these three orders, including a genus from order Pseudomonales, were also significantly reduced in the infection group (FDR-corrected Kruskal-Wallis p < 0.05) (Table 2). In addition to changes in Proteobacteria, there was an approximately 0.6% absolute increase in Firmicutes in the infection group relative to the no-infection group (FDR-corrected Kruskal-Wallis p < 0.05). This mirrors, and may be due to, the reduction in Proteobacteria in the infection group.
Table 2.
Most abundant phyla and orders by presence or absence of puerperal infection
| Taxa | Mean taxonomic abundance | FDR-corrected p value | ||
|---|---|---|---|---|
| Infection | No infection | |||
| Phyla | Proteobacteria | 0.001713 | 0.005027 | 0.009 |
| Firmicutes | 0.994103 | 0.988000 | 0.041 | |
| Actinobacteria | 0.000392 | 0.003398 | 0.236 | |
| Tenericutes | 0.002136 | 0.000876 | 0.236 | |
| Bacteroidetes | 0.001121 | 0.002183 | 0.416 | |
| TM7 | 0.000002 | 0.000001 | 0.575 | |
| Verrucomicrobia | 0.000073 | 0.000136 | 0.759 | |
| Fusobacteria | 0.000340 | 0.000269 | 0.759 | |
| Class | Alphaproteobacteria | 0.000668 | 0.001905 | 0.002 |
| Betaproteobacteria | 0.000486 | 0.001671 | 0.022 | |
| Gammaproteobacteria | 0.000559 | 0.001451 | 0.038 | |
| Actinobacteria | 0.000349 | 0.002847 | 0.221 | |
| Bacilli | 0.989100 | 0.984833 | 0.235 | |
| Mollicutes | 0.002136 | 0.000876 | 0.245 | |
| Bacteroidia | 0.001121 | 0.002183 | 0.462 | |
| Coriobacteriia | 0.000043 | 0.000551 | 0.473 | |
| Clostridia | 0.004915 | 0.003138 | 0.536 | |
| TM7–3 | 0.000002 | 0.000001 | 0.536 | |
| Verrucomicrobiae | 0.000073 | 0.000136 | 0.727 | |
| Fusobacteriia | 0.000340 | 0.000269 | 0.727 | |
| Order | Caulobacterales | 0.000388 | 0.001322 | 0.005 |
| Rhizobiales | 0.000261 | 0.000520 | 0.005 | |
| Burkholderiales | 0.000480 | 0.001518 | 0.025 | |
| Enterobacteriales | 0.000115 | 0.000498 | 0.154 | |
| Turicibacterales | 0.000072 | 0.000143 | 0.154 | |
| Pseudomonadales | 0.000283 | 0.000465 | 0.154 | |
| Bacillales | 0.000396 | 0.000758 | 0.164 | |
| Bifidobacteriales | 0.000348 | 0.002846 | 0.164 | |
| Alteromonadales | 0.000032 | 0.000092 | 0.250 | |
| Mycoplasmatales | 0.002136 | 0.000876 | 0.252 | |
| Lactobacillales | 0.988574 | 0.982394 | 0.264 | |
| Legionellales | 0.000128 | 0.000395 | 0.325 | |
| Rhodospirillales | 0.000019 | 0.000064 | 0.325 | |
| Rhodocyclales | 0.000007 | 0.000153 | 0.350 | |
| Bacteroidales | 0.001121 | 0.002183 | 0.370 | |
| Coriobacteriales | 0.000043 | 0.000551 | 0.406 | |
| Clostridiales | 0.004915 | 0.003138 | 0.505 | |
| 1025 | 0.000002 | 0.000001 | 0.511 | |
| Actinomycetales | 0.000002 | 0.000001 | 0.725 | |
| Verrucomicrobiales | 0.000073 | 0.000136 | 0.725 | |
| Fusobacteriales | 0.000340 | 0.000269 | 0.736 | |
| Oscillatoriales | 0.000120 | 0.000111 | 0.803 | |
| Family | Caulobacteraceae | 0.000388 | 0.001322 | 0.009 |
| Hyphomicrobiaceae | 0.000062 | 0.000156 | 0.032 | |
| Oxalobacteraceae | 0.000160 | 0.000485 | 0.045 | |
| Methylocystaceae | 0.000026 | 0.000083 | 0.045 | |
| Rhizobiaceae | 0.000032 | 0.000100 | 0.045 | |
| Methylobacteriaceae | 0.000106 | 0.000106 | 0.063 | |
| Comamonadaceae | 0.000171 | 0.000551 | 0.063 | |
| Moraxellaceae | 0.000245 | 0.000382 | 0.063 | |
| [Tissierellaceae] | 0.000136 | 0.000265 | 0.063 | |
| [Paraprevotellaceae] | 0.000044 | 0.000199 | 0.063 | |
| Streptococcaceae | 0.000517 | 0.001341 | 0.063 | |
| Enterobacteriaceae | 0.000115 | 0.000498 | 0.105 | |
| Turicibacteraceae | 0.000072 | 0.000143 | 0.105 | |
| Clostridiaceae | 0.000141 | 0.000440 | 0.105 | |
| Aurantimonadaceae | 0.000035 | 0.000074 | 0.124 | |
| Paenibacillaceae | 0.000061 | 0.000054 | 0.124 | |
| Bifidobacteriaceae | 0.000348 | 0.002846 | 0.124 | |
| Enterococcaceae | 0.000125 | 0.000258 | 0.124 | |
| Staphylococcaceae | 0.000335 | 0.000704 | 0.147 | |
| Bacteroidaceae | 0.000193 | 0.000530 | 0.147 | |
| Lachnospiraceae | 0.000171 | 0.000120 | 0.147 | |
| Lactobacillaceae | 0.987615 | 0.980379 | 0.147 | |
| [Chromatiaceae] | 0.000032 | 0.000092 | 0.168 | |
| Mycoplasmataceae | 0.002136 | 0.000876 | 0.175 | |
| Pseudomonadaceae | 0.000038 | 0.000083 | 0.175 | |
| Rikenellaceae | 0.000099 | 0.000083 | 0.193 | |
| Coxiellaceae | 0.000128 | 0.000395 | 0.260 | |
| Rhodospirillaceae | 0.000019 | 0.000064 | 0.261 | |
| Rhodocyclaceae | 0.000007 | 0.000153 | 0.293 | |
| Aerococcaceae | 0.000317 | 0.000416 | 0.347 | |
| Coriobacteriaceae | 0.000043 | 0.000551 | 0.353 | |
| Rs-045 | 0.000002 | 0.000001 | 0.485 | |
| Prevotellaceae | 0.000553 | 0.001164 | 0.488 | |
| Veillonellaceae | 0.004402 | 0.002238 | 0.504 | |
| Actinomycetaceae | 0.000002 | 0.000001 | 0.699 | |
| Verrucomicrobiaceae | 0.000073 | 0.000136 | 0.699 | |
| Leptotrichiaceae | 0.000340 | 0.000269 | 0.726 | |
| Phormidiaceae | 0.000120 | 0.000111 | 0.788 | |
| S24–7 | 0.000232 | 0.000208 | 0.788 | |
| Genera | Bvab1 | 0.000124 | 0.000028 | 0.046 |
| Pleomorphomonas | 0.000026 | 0.000083 | 0.046 | |
| Acinetobacter | 0.000152 | 0.000324 | 0.046 | |
| Agrobacterium | 0.000032 | 0.000100 | 0.046 | |
| Methylobacterium | 0.000106 | 0.000106 | 0.087 | |
| Limnohabitans | 0.000091 | 0.000283 | 0.092 | |
| Lactococcus | 0.000183 | 0.000242 | 0.106 | |
| Ureaplasma | 0.000314 | 0.000379 | 0.125 | |
| SMB53 | 0.000103 | 0.000356 | 0.138 | |
| Turicibacter | 0.000072 | 0.000143 | 0.138 | |
| Paenibacillus | 0.000061 | 0.000054 | 0.169 | |
| Comamonas | 0.000030 | 0.000076 | 0.169 | |
| Enterococcus | 0.000125 | 0.000258 | 0.170 | |
| [Prevotella] | 0.000024 | 0.000101 | 0.179 | |
| Staphylococcus | 0.000335 | 0.000704 | 0.179 | |
| Bacteroides | 0.000193 | 0.000530 | 0.179 | |
| Finegoldia | 0.000066 | 0.000064 | 0.179 | |
| Lactobacillus | 0.987615 | 0.980379 | 0.179 | |
| Parvimonas | 0.000003 | 0.000112 | 0.187 | |
| Rheinheimera | 0.000032 | 0.000092 | 0.187 | |
| Gardnerella | 0.000311 | 0.002765 | 0.187 | |
| Escherichia | 0.000053 | 0.000351 | 0.187 | |
| Pseudomonas | 0.000038 | 0.000083 | 0.202 | |
| Paraprevotella | 0.000019 | 0.000097 | 0.217 | |
| Peptoniphilus | 0.000066 | 0.000089 | 0.235 | |
| Streptococcus | 0.000334 | 0.001099 | 0.251 | |
| Coxiella | 0.000128 | 0.000395 | 0.278 | |
| Hydrogenophilus | 0.000007 | 0.000153 | 0.317 | |
| Atopobium | 0.000042 | 0.000550 | 0.342 | |
| Aerococcus | 0.000317 | 0.000416 | 0.372 | |
| Bifidobacterium | 0.000037 | 0.000080 | 0.407 | |
| Veillonella | 0.000105 | 0.000155 | 0.447 | |
| Prevotella | 0.000553 | 0.001164 | 0.521 | |
| Dialister | 0.000042 | 0.000192 | 0.523 | |
| Megasphaera | 0.004255 | 0.001891 | 0.549 | |
| Blautia | 0.000047 | 0.000092 | 0.592 | |
| Enhydrobacter | 0.000094 | 0.000059 | 0.617 | |
| Actinomyces | 0.000002 | 0.000001 | 0.706 | |
| Akkermansia | 0.000073 | 0.000136 | 0.706 | |
| Sneathia | 0.000340 | 0.000269 | 0.738 | |
| Oscillatoria | 0.000120 | 0.000111 | 0.811 | |
| Mycoplasma | 0.000156 | 0.000471 | 0.824 | |
| Gemella | 0.000058 | 0.001539 | 0.830 | |
Abbreviation: FDR, false-discovery rate.
Note: Bold text signifies FDR-approved p value < 0.05.
Fig. 3.
Phylum-level (left) and order-level (right) relative abundance (y-axis) by puerperal infection (top panel) and obesity (bottom panel). Stacked bar plots indicate the dominant phyla (left) or orders (right) for each group.
Fig. 4.
Vertical scatter plot demonstrating relative abundance frequencies of phylum: Proteobacteria, orders: Burkholderiales, Caulobacterales, Rhizobialis by the presence of puerperal infection. The image demonstrates a decreased relative taxonomic abundance of all orders in second-trimester samples from women who developed a puerperal infection at term (false-discovery rate corrected p < 0.05).
Interestingly, we detected a bacterial vaginosis associated genus known as BVAB1 in both infection and no-infection patients.35,36 Although it was present at low frequencies (< 0.02% on average in both groups), there was four times the abundance of BVAB1 in the infection group relative to the no-infection group (FDR-corrected Kruskal-Wallis p < 0.05) (Table 2).
Although there were no significant differences in clustering by obesity, we still tested for differences in taxa frequency. There were no taxa with significant differences in frequency between the obese and nonobese groups (Kruskal-Wallis p > 0.05 for all taxa).
Comment
We have demonstrated significant clustering of the midtrimester cervicovaginal microbiota between women who do and do not develop puerperal infections at term. In addition, we have shown that women who develop puerperal infections have less abundance of three orders of Proteobacteria and increased abundance of BVAB1 genera in their midtrimester cervicovaginal microbiome. Based on this, it appears that assessment of the midtrimester microbiome may be able to identify unique microbial signatures for women at an increased risk for puerperal infection months later, even when limited to lower risk women without bacterial vaginosis. Obesity, in this cohort of patients without bacterial vaginosis, did not affect the midtrimester cervicovaginal microbiome and there was no interaction between obesity and microbiome with relation to puerperal infection.
Prior studies have reported that deficiencies in Lactobacillus are associated with preterm premature rupture of the membranes, histological chorioamnionitis, and intra-amniotic infection.14,16,40–42 Our findings did not demonstrate deficiencies in Lactobacillus among women who developed puerperal infections. In fact, we noted a small increase in Lactobacillus that mirrored a lower abundance of three orders of Proteobacteria in the midtrimester cervicovaginal microbiome of women delivering at term who developed puerperal infections. Possible explanations for the differences seen in our analysis include analysis of a different clinical endpoint (puerperal infection) as well as analysis of only term births (as opposed to preterm births included in the above-mentioned studies). We did, however, show increased relative abundance of BVAB1 genera. While the abundance was not significantly increased to result in a clinical diagnosis of bacterial vaginosis, it does support the argument that microbes involved in bacterial vaginosis are a risk factor for chorioamnionitis and endometritis.6,18,39 These microbial changes should thus be further investigated prospectively to discern if they can be used to predict women (term or preterm) that may be at an increased risk of puerperal infection.
The strengths of our study include the use of samples from a well-described prospective cohort of patients. Data regarding maternal height, weight, and pregnancy outcomes were collected as part of the parent study. In addition, use of a standard definition of puerperal infections applied prospectively (chorioamnionitis and endometritis) at one institution allows a greater precision in the analysis of this outcome despite the fact that both diagnoses are made based on clinical criteria. Furthermore, the use of samples from women of a single race and exclusion of women with bacterial vaginosis avoids the complexities of microbial differences by ethnicity and a microbial process that is a risk factor for puerperal infections.
However, this is a retrospective analysis of a subset of a previously collected cohort of patients. We recognize that the relatively small sample size necessitated by limited resources could be viewed as a limitation, but we did demonstrate small, but statistically significant differences in bacterial composition by puerperal infection. Another possible perceived limitation is the use of the V4 region for our 16S analysis. However, our group has published utilizing these methods, and justification for V4 usage is described in the Methods section above.19,21–27 Some region of 16S must be chosen unless whole genome sequencing is undertaken, and that latter approach is cost prohibitive for a large number of samples. Another potential limitation is the use of midtrimester cervicovaginal samples. While the stability of the microbiome in pregnancy has been described, it is possible that the midtrimester microbiome is not reflective of the cervicovaginal microbiome at term.13–16 In addition, the population studied in our analysis was Black women with normal flora delivering term gestations. We acknowledge that this is a select, low-risk population–arguing to generalizability. However, the ability to identify these microbial differences in a low-risk population could result in an even greater ability to identify microbial aberrations in high-risk populations (including women with BV). Thus, our findings, if replicated in a variety of populations, may represent a potential biomarker to detect patients at risk of puerperal infections with delivery. Rapid and cheaper testing for these biomarkers in the midtrimester could be accomplished through rapid PCR-based tests in routine clinical-care settings. This may provide a way to screen and identify, patients who are at risk for developing puerperal infections. Furthermore, identification of such a biomarker could provide a target for treatment such as probiotic therapy.
In summary, while obesity does not appear to affect the cervicovaginal microbiota in the midtrimester of pregnancy, there are differences in the cervicovaginal microbiome between women who proceed to develop puerperal infections at term and those who do not. The relative reduced abundance of Proteobacteria and relative increase in abundance of BVAB1 in women who develop puerperal infections that were observed in this study should be further explored prospectively to assess the presence of bacteria from this phylum as a potential biomarker of protection against puerperal infections.
Acknowledgments
The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical and Translational Science (UL1TR000165), and Heflin Center.
Footnotes
The authors report no conflict of interest.
This study was presented in part at the 36th Annual Meeting of the Society for Maternal-Fetal Medicine; February 1–6, 2016; Atlanta, GA.
References
- 1.Tita AT, Andrews WW. Diagnosis and management of clinical chorioamnionitis. Clin Perinatol 2010;37(2):339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahey JO. Clinical management of intra-amniotic infection and chorioamnionitis: a review of the literature. J Midwifery Womens Health 2008;53(3):227–235 [DOI] [PubMed] [Google Scholar]
- 3.Hastings-Tolsma M, Bernard R, Brody MG, Hensley J, Koschoreck K, Patterson E. Chorioamnionitis: prevention and management. MCN Am J Matern Child Nurs 2013;38(4):206–212, quiz 213–214 [DOI] [PubMed] [Google Scholar]
- 4.Curtin WM, Katzman PJ, Florescue H, Metlay LA. Accuracy of signs of clinical chorioamnionitis in the term parturient. J Perinatol 2013;33(6):422–428 [DOI] [PubMed] [Google Scholar]
- 5.Soper DE, Mayhall CG, Froggatt JW. Characterization and control of intraamniotic infection in an urban teaching hospital. Am J Obstet Gynecol 1996;175(2):304–309, discussion 309–310 [DOI] [PubMed] [Google Scholar]
- 6.Clark P, Kurtzer T, Duff P. Role of bacterial vaginosis in peripartum infections. Infect Dis Obstet Gynecol 1994;2(4):179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabiru W, Raynor BD. Obstetric outcomes associated with increase in BMI category during pregnancy. Am J Obstet Gynecol 2004; 191(3):928–932 [DOI] [PubMed] [Google Scholar]
- 8.Menon R, Taylor RN, Fortunato SJ. Chorioamnionitis–a complex pathophysiologic syndrome. Placenta 2010;31(2):113–120 [DOI] [PubMed] [Google Scholar]
- 9.Myles TD, Gooch J, Santolaya J. Obesity as an independent risk factor for infectious morbidity in patients who undergo cesarean delivery. Obstet Gynecol 2002;100(5 Pt 1):959–964 [DOI] [PubMed] [Google Scholar]
- 10.Cottam DR, Mattar SG, Barinas-Mitchell E, et al. The chronic inflammatory hypothesis for the morbidity associated with morbid obesity: implications and effects of weight loss. Obes Surg 2004;14(5):589–600 [DOI] [PubMed] [Google Scholar]
- 11.Ramsay JE, Ferrell WR, Crawford L, Wallace AM, Greer IA, Sattar N. Maternal obesity is associated with dysregulation of metabolic, vascular, and inflammatory pathways. J Clin Endocrinol Metab 2002;87(9):4231–4237 [DOI] [PubMed] [Google Scholar]
- 12.Friis CM, Paasche Roland MC, Godang K, et al. Adiposity-related inflammation: effects of pregnancy. Obesity (Silver Spring) 2013; 21(1):E124–E130 [DOI] [PubMed] [Google Scholar]
- 13.Romero R, Hassan SS, Gajer P, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014;2(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyman RW, Fukushima M, Jiang H, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci 2014;21(1): 32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A 2015;112(35):11060–11065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero R, Hassan SS, Gajer P, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2014;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neggers Y, Goldenberg R, Cliver S, Hauth J. Effects of domestic violence on preterm birth and low birth weight. Acta Obstet Gynecol Scand 2004;83(5):455–460 [DOI] [PubMed] [Google Scholar]
- 18.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol 1991;29(2):297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79(17):5112–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar R, Eipers P, Little RB, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet 2014;82:1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 2011;108(1, Suppl 1):4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 2007;35(18):e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6(8):1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuczynski J, Lauber CL, Walters WA, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 2012;13(1):47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 2008;36(18):e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS ONE 2013;8(1):e53608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE 2014;9(4):e94249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramaniam A, Kumar R, Cliver SP, et al. Vaginal microbiota in pregnancy: evaluation based on vaginal flora, birth outcome, and race. Am J Perinatol 2016;33(4):401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 2013;531:371–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010;26(2):266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26(19):2460–2461 [DOI] [PubMed] [Google Scholar]
- 32.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 2006;7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73(5):1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6(3): 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005; 353(18):1899–1911 [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan S, Hoffman NG, Morgan MT, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE 2012;7(6):e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2013;2(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108(1, Suppl 1):4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol 2015;212(5):653.e1–653.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldwin EA, Walther-Antonio M, MacLean AM, et al. Persistent microbial dysbiosis in preterm premature rupture of membranes from onset until delivery. PeerJ 2015;3:e1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kacerovsky M, Vrbacky F, Kutova R, et al. Cervical microbiota in women with preterm prelabor rupture of membranes. PLoS ONE 2015;10(5):e0126884. [DOI] [PMC free article] [PubMed] [Google Scholar]