Abstract

Most of the chemotherapeutics and drug-delivery models pose serious health problems and several undesirable side effects due to nonspecificity, lack of proper targeting system, and their large sizes. The rational design and synthesis of target-specific chemotherapeutics are highly important. This research work is focused on the rational design, synthesis, and anticancer studies of fluorescent 1,2,4-triazole–peptide conjugates for the development of target-specific anticancer drugs. Three novel 1,2,4-triazole derivatives: 4-(4-fluorobenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole (4FBAHMT, 2a), 4-(3,4,5-trimethoxybenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole (TMOBAHMT, 2b), and 4-(4-benzyloxy-2-methyloxbenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole (4BO2MOBAHMT, 2c) were synthesized after screening through molecular docking procedures. The docking studies were performed between ligand molecules and αvβ6 integrin protein. Fluorescent carbon nanoparticles (CNPs, 3) were conjugated with 1,2,4-triazole derivatives (2a–c) and l-carnosine (LC) dipeptide to get their corresponding conjugates (4a–c). The title double conjugates were characterized by spectroscopic (UV/vis spectroscopy, fluorescence spectroscopy, and FTIR spectroscopy) and microscopic (scanning electron microscopy, transmission electron microscopy, and atomic force microscopy) techniques. In vitro efficacy of fluorescent 1,2,4-triazole–peptide conjugates was investigated against two pediatric brain tumor cell lines (CHLA-200 & SJGBM2) and human embryonic kidney cell line (HEK293 as a control) by employing cell proliferation assay/MTS assay and fluorescence microscopy. 1,2,4-Triazole derivatives and their conjugates showed potent and selective anticancer activity against CHLA-200 and SJGBM2 cell lines. Cell proliferation assay and fluorescence microscopy results revealed that conjugates were more highly selective and cytotoxic than control drug temozolomide (TM) against both cell lines. CNPs are highly biocompatible and the quantum-sized conjugates were nontoxic for normal embryonic kidney cell line (HEK 293). The experimental results of MTS bioactivity assay and fluorescence microscopy were in close agreement with the theoretical results of molecular docking studies.
1. Introduction
Cancer, a class of diseases recognized by the uncontrolled growth of cells, has gradually become the leading cause of deaths in children and is one of the most dangerous diseases today.1 Currently, three major approaches (chemotherapy, radiation therapy, and surgery) are being used for cancer treatment. Chemotherapy is mostly used in cancer cases where it has spread (metastasized) throughout the body. The efficacy of chemotherapeutics depends upon their ability to target cancerous cells without damaging healthy cells.2 In current chemotherapy, a major challenge is to design novel anticancer drugs that show more selectivity for cancer cells and thus have lesser side effects for normal cells. The most important characteristics of new and potent anticancer drugs should be the high efficacy and selectivity.3
A number of chemotherapeutic agents are now available in the market. Doxorubicin is the most commonly used chemotherapeutic agent, but it has serious problems of cardiotoxicity, liver damage, and myelosuppression.4 Cisplatin is another important anticancer drug which is highly effective against cervical, ovarian, and lung carcinomas.5 Its benefits are limited, however, by serious problems, including inherited and acquired resistance, nephrotoxicity, neurotoxicity, emetogenesis, and oral nonbioavailability.5−7
Various efforts have been made to target these chemotherapeutic drugs to cancer cells by making use of antibodies,8 growth factors,9 folates,10 and nanocomposites.11 The use of pharmaceutical nanocarriers in drug delivery has increased recently.11
Many heterocyclic compounds are found in nature performing different activities crucial for life.12 Nitrogen-containing 5-membered heterocyclic moiety is present in various biologically active natural and synthetic molecules.13 The 1,2,4-triazole molecules show different properties like antimicrobial,14 anti-inflammatory,15 antidepressant,16 antifungal,17 anticonvulsant,18 and antitumor properties.19 Several anticancer drugs (e.g., anastrozole and letrozole) having 1,2,4-triazole scaffold are well known20,21 (Figure 1).
Figure 1.

Commonly used anticancer drugs based on 1,2,4-triazole moiety.
CNPs-based nanocarriers are highly significant due to their excellent fluorescence properties, high solubility in water, excellent chemical inertness, ease of functionalization and modification, high stability against photobleaching, low cytotoxicity, and good biocompatibility.22−25 Consequently, they have received much importance for their potential applications in bioimaging, sensing, and drug delivery.26
Docking procedures are very important in structure-based drug designing because they are important tools to find different poses of ligands within active sites of receptors, binding energies, and types of ligand–receptor interactions.27,28 The αvβ6 integrin is a very important protein which is highly upregulated during embryogenesis and in the carcinoma of breast, lungs, skin, breast, and stomach.29,30 This integrin plays a crucial role in tissue repair, development, and neoplasia.29
Selective targeting approach for cancer cells by using carrier vectors such as peptides can address the issues of selectivity.31 Researchers have made several efforts toward the targeting of anticancer drugs, and peptide-based carriers have become an important component of these targeting approaches.32 The peptides possess outstanding physical and biological properties for tumor targeting.33 LC is a naturally occurring multifunctional dipeptide, which is composed of alanine and histidine amino acids. It is an endogenous peptide which exists widely in healthy muscles, blood, heart, kidney, eyes, and brain tissues in several animal species.34 Recently, LC has been proposed as a potential anticancer therapeutic effective for the inhibition of metastasis and ovarian, gastric, and glioblastoma carcinomas.35−39
The current research work is focused on rational design, synthesis, and anticancer activity assessment of CNPs conjugated with new 1,2,4-triazole derivatives and LC as cancer-targeting peptide for the development of cancer-targeting fluorescent nanoprobes.40 CNPs are tagged with 1,2,4-triazole derivatives and LC for targeting tumor cells and bioimaging.41
2. Experimental Section
2.1. Chemicals and Reagents
Glucose (cat no. G8270), ethanol absolute (>99.5%, cat no. 459844), magnesium sulfate anhydrous (>97%, cat no. 208094), EDC cross-linker (cat no. 7750), and FBS (cat. no. 10437-028) were purchased from Invitrogen. Dialysis membrane (MWCO 3500) was obtained from Thermo Scientific. Two pediatric glioblastoma multiforme (GBM) tumor cell lines (SJGBM2 and CHLA-200) were obtained from Children’s Oncology Group (COG, Lubbock, TX). Cells were cultured in RPMI-1640 (Thermo Fisher Scientific), which was supplemented with 10% heat-inactivated FBS and 1% penicillin–streptomycin. Cell cultures were maintained at 37 °C in a incubator with 5% CO2. Rhodamine 6G mitochondria-specific dye was purchased from Thermo Fisher Scientific. Cell proliferation assay (MTS assay) was performed with CellTiter 96 AQueous One Solution (Promega).
A brief account of the purification procedure of solvents is as follows: (1) Ethanol was refluxed over activated calcium oxide for 6 h followed by distillation and then stored over 4 Å molecular sieves. (2) Methanol was purified by fractional distillation. (3) Chloroform was dried over anhydrous calcium chloride for 24 h followed by distillation and then stored over 4 Å molecular sieves. TLC plates were observed under ultraviolet light (λmax = 254 and 365 nm). N-Hexane/ethyl acetate (2:6) solvent system was used for the development of chromatogram.
The CNPs were fabricated using a bottom-up approach (UC-D10 Ultrasonic Bath, BMS, frequency: 35 kHz). The conjugates were characterized by UV–vis spectroscopy (Agilent Technologies Spectrophotometer, Carry Series UV–Vis Spectrophotometer), fluorescence spectroscopy (Horiba Jobin Yvon Fluorolog-3), Fourier transform infrared spectroscopy (PerkinElmer, Spectrum100), scanning electron microscopy (JEOL-Japan, JSM 6490: acceleration voltage, 20 kV), transmission electron microscopy (JEOL, Japan), and atomic force microscopy (Agilent Technologies, 5420 AFM). The samples for UV–vis spectroscopy and fluorescence spectroscopy were prepared in deionized water (20 μg/mL). 1H and 13C NMR spectra were recorded on a Bruker Avance 400 MHz NMR spectrometer using DMSO-d6 and CDCl3 solvents. Mass spectra were recorded using a Bruker TOF mass spectrometer.
2.2. Molecular Dynamic Simulation
The crystal structure of the integrin αvβ6 (PDB code: 4UM9, resolution: 2.50 Å)42 was downloaded from PDB Databank, and globular head of the integrin was used for docking studies. According to the X-ray structure, the bivalent metal cation at MIDAS (MIDAS: metal-ion-dependent adhesion site) was modeled as Mg2+ ion and all other metal bivalent cations were modeled as Ca2+ ions. Water and small ligand molecules were removed from the receptor structure using Sequence Editor of Discovery Studio 2017 R2 Client software. The final optimized shape of integrin αvβ6 is given in Figure S1.
The interactions of three 1,2,4-triazole derivatives and their conjugates with αvβ6 integrin have been investigated by isolating the receptor active site in αvβ6 integrin. A rigid receptor-flexible ligand docking model was used to carry out docking procedures. Insights from the characteristics of active binding site of αvβ6 integrin and analysis of different docking poses of ligands provided the basis for identifying best ligands with high selectivity for αvβ6 integrin.43 The best ligands were identified on the basis of low binding energy values. The MOE software was used to minimize energy of ligand molecules, and final structures were saved in the “moe” file format.44,45 The energy minimization of ligand in gas phase was achieved using MOE with MMFF94x force-field parameter, and London dG was the default scoring method.
2.3. Synthesis of 4-Amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT) (1)
4-Amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT, 1) was synthesized through a simple reaction of thiocarbohydrazide with hydrazine hydrate.46 The thiocarbohydrazide (5 mmol) mixed with 69% hydrazine hydrate (100 mmol) was refluxed for 4 h. The solution was cooled, filtered, and acidified with dil. HCl (pH 6–7). The precipitated product was washed with water and ethanol and recrystallized from boiling water (long lustrous needles, mp 230–232 °C (lit. mp 228 °C)).47
2.4. Synthesis of Derivatives (2a–c) of 4-Amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT, 1)
The derivatives (2a–c) of AHMT (1) were synthesized by the condensation reaction of AHMT (1) with corresponding aldehyde according to a reported method.48 AHMT (10 mmol) was mixed with corresponding aldehyde (10 mmol) in dry ethanol (50 mL) and a few drops of conc. H2SO4 were added to the above solution. The reaction mixture was refluxed at 110 °C for 4 h. The crude product was separated, washed, and recrystallized from ethanol to give the final product in excellent yield.48
2.5. Synthesis of CNPs (3)
CNPs were synthesized by our previously reported method.41 One molar aqueous solution of glucose (50 mL) was mixed with conc. H2SO4 (40 mL, 98%) and ortho-phosphoric acid (10 mL, 98%). The glucose solution was treated ultrasonically for 4 h at 40 °C. The resulting brown solution was kept in an oven at 80 °C for 6 h. Finally, CNPs were purified by centrifugation at 6000 rpm and further dialyzed with 3500 MWCO dialysis membrane for 5 days in deionized water. The final product in powder form was obtained from the dialyzed solution using rotavapor.
2.6. Synthesis of Conjugates (4a–c) of CNPs with 1,2,4-Triazole Derivatives (2a–c) and LC
1,2,4-Triazole derivatives (2a–c) and LC peptide were conjugated with CNPs via standard EDC chemistry41,49 (Scheme 1). CNPs were functionalized through amide bond between carboxylic groups of CNPs and amino groups of 1,2,4-triazole derivatives and LC in a single-step reaction.50 In a typical reaction, 25 mL of aqueous dispersion (1 mg/mL) of CNPs was prepared by sonication for 30 min. Then, 30.00 mg of EDC cross-linker was mixed to activate the CNPs. The whole mixture was vigorously stirred for 30 min. Then, 10.0 mM of the corresponding 1,2,4-triazole derivative in THF and 10.0 mM of LC in water were added to the above activated suspension of CNPs and stirred for 4 h. Excess ethanol was added to the above mixture and kept at 4 °C for overnight. The double conjugates were separated from unreacted CNPs by size exclusion chromatography. The resulting solution containing LC-CNP-1,2,4-triazole conjugates was purified by dialysis (3500 MWCO) for 48 h in deionized water to remove unreacted LC and 1,2,4-triazole derivatives. The final products (4a–c) were obtained by quick freeze/drying (lyophilization) of the above suspension.
Scheme 1. Synthesis of Conjugates of CNPs with 1,2,4-Triazole Derivatives and LC.
2.7. Biological Studies
Two pediatric glioblastoma multiforme (GBM) cell lines (CHLA-200 and SJGBM2 cell lines) were selected for in vitro efficacy and cytotoxic effects. Bioactivity of LC-CNP-1,2,4-triazole conjugates was assessed by cell viability/proliferation (MTS) assay. Microscopy of CHLA-200 cells treated with conjugates was performed using a fluorescence microscope (EVOS Floid Cell Imaging Machine, Life Technologies) at 40× magnification.
2.7.1. Cell Lines and Cell Cultures
Glioblastoma multiforme cell lines (CHLA-200 and SJGBM2) were cultured in RPMI-1640 media with 10% fetal bovine serum (FBS). For MTS assay, CHLA-200 and SJGBM2 cells (second passage) were trypsinized and resuspended in 96-well plates containing corresponding media at a density of 2 × 105 cells/mL.
2.7.2. Cell Viability/Proliferation Assay (MTS Assay)
After 24 h of seeding, the cells were exposed to 5, 10, 25, and 50 μg/mL concentration of each of the 1,2,4-triazole derivative (2a–c) and corresponding conjugate (4a–c) by adding 100 μL of each concentration in the corresponding well for 72 h. Following treatment for 72 h, cell culture media was removed and a solution of MTS reagent and cell culture media (100 μL, a 1:5 solution of MTS reagent and cell culture media) was added to each well and incubated for 1–4 h. The percent of viable cells was determined by measuring optical density at 490 nm using BoiTek Synergy HT plate reader at each 30 min interval until color of media changed from yellow to purple. To compare the efficacy of title conjugates, cells were also treated with control anticancer drug TM, which is used as first-line treatment for glioblastoma multiforme. The experiments were performed in triplicate, and the viability of cells treated with different conjugates was determined as percent viability compared to nontreated controls.51
2.7.3. Fluorescence Microscopy of Cell Cultures
CHAL-200 cells were exposed to 10, 25, and 50 μg/mL of each of the 1,2,4-triazole derivatives (2a–c) and LC-CNP-1,2,4-triazole conjugates (4a–c) and examined under a fluorescence microscope (EVOS Floid Cell Imaging Machine, Life Technologies) at 40× magnification.52 The cells were seeded on 15 mm coverslips placed in each well of a 24-well plate with RPMI-1640 medium. After 24 h, the cells were treated with various concentrations of 1,2,4-triazole derivatives and their conjugates. Next day, the media was removed gently and the cells were washed with PBS. To fix cells, they were incubated with 4% paraformaldehyde for 20 min. After washing again with PBS, the coverslips treated with conjugates were transferred to glass slides having mounding media without DAPI.
The cells treated with 1,2,4-triazole derivatives were stained with rhodamine 6G dye after fixation with 4% paraformaldehyde following the same procedure mentioned above for conjugates. The samples were prepared for rhodamine 6G staining, as reported by Johnson et al.53 The cells were seeded on 15 mm coverslips placed in a 24-well plate then treated with various concentrations of 1,2,4-triazole derivatives for 24 h. Next day, the cells were stained with rhodamine 6G. Rhodamine 6G (commercial name rhodamine 6GO, Chroma-Gesellschaft, Stuttgart-Unterurkheim) was constituted in deionized water (1 mg/mL stock solution) and stored at 4 °C. The stock solution was diluted in Eagle’s solution (123 mM NaCl, 5.4 mM KCl, 1.4 mM CaCl2, 0.8 mM MgSO4, 1.0 mM NaH2PO4, 13.1 mM NaHCO3, and 5.6 mM glucose, pH 7.3) to 0.5 μg/mL (1.04 μM), then the cells were added, which were incubated at 20 °C for 20 min. These stained cells were rinsed twice with dye-free Eagle’s solution to remove nonspecific fluorescence. The coverslips with the stained cells were then mounted on the glass slides having mounting media.54 The stained cells were examined by fluorescence microscopy. When the rhodamine dye 6G was excited by blue light (λmax = 490 nm), it emitted yellow fluorescence. When excited by green light (λmax = 545 nm), rhodamine 6G emitted red fluorescence.
3. Results and Discussion
3.1. Molecular Docking Studies
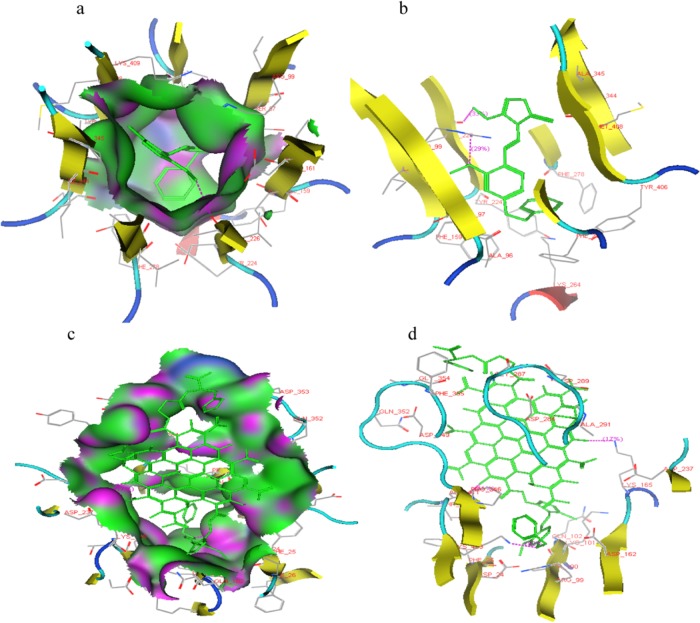
The docking study was performed between ligand molecules and αvβ6 integrin protein. 1,2,4-Triazole-based anticancer drugs showed docking score comparable to that of TM anticancer drug, and the conjugates showed significantly better docking score than 1,2,4-triazole derivatives. The docking score and ligand–receptor interactions are shown in Table 1 and Figures 2a–d and S2a–d. In conjugates, 1,2,4-triazole and CNPs part fits in the active site of αvβ6 integrin and LC stays at the periphery of the active binding site showing its ability as target-specific drug carrier, as displayed by Figure 2c.
Table 1. Docking Score of 1,2,4-Triazole Derivatives (2a–c) and Their Conjugates (4a–c) as a Result of Interactions with αvβ6 Integrin.
| entry | abbreviation (Schiff base) | docking score | entry | abbreviation (conjugate) | docking score |
|---|---|---|---|---|---|
| 2a | 4FBHAMT | –12.50 | 4a | LC-CNP-4FBHAMT | –19.50 |
| 2b | TMOBAHMT | –12.49 | 4b | LC-CNP-TMOBAHMT | –18.06 |
| 2c | 4BO2MOBAHMT | –12.98 | 4c | LC-CNP-4BO2MOBAHMT | –19.74 |
| TM | –11.64 | ||||
| CNPs | –10.34 | ||||
| LC | –10.6 |
Figure 2.
Docking presentation of (a, b) 4BO2MOBAHMT derivative and (c, d) LC-CNP-4BO2MOBAHMT conjugate in the active binding site of αvβ6 integrin.
3.2. Synthesis of 4-Amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT) (1)
Long needles; yield: 42.8%; mp 230–232 °C. IR (ATR, vcm–1): 3261.41, 3194.86 (NH2), 3014.69 (N–H), 2917.02 (N–H), 1637.16 (NH2), 1589.46 (C=N), 1152.94 (C–N), 949.32 (C=S). 1H NMR (400 MHz, DMSO-d6, δ ppm): 12.52 (s, 1H) (NH)ring, 7.13 (s, 1H) (NH)NHNH2, 5.26 (s, 2H) (NH2)NHNH2, 4.09 (s, 2H) (N–NH2). 13C NMR (400 MHz, DMSO-d6, δ ppm): 164.42 (N=C–NH–NH2), 154.67 (C=S). MS-EI: (m/z, relative intensity, %): Calculated [M]+: 146.00, Found [M]+: 146.07 (25%). C2H6N6S: Found: C, 16.33%; H, 4.10%; N, 56.9%. Calculated: C, 16.44%; H, 4.15%; N, 57.53%.
3.3. Synthesis of Derivatives (2a–c) of 4-Amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT) (1)
3.3.1. 4-(4-Fluorobenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole (4FBAHMT) (2a)
Yellow crystals; yield: 74.8%; mp 263–264 °C. IR (ATR, vcm–1): 3299.86, 3262.39 (NH2), 3143.37, 3105.90 (N–H), 3024.36 (Ar–H), 2966.93 (N=C–H), 1602.30 (C=N)triazole, 1483.72 (C=N)imine, 1333.15 (C–N), 919.53 (C=S), 832.11 (C–F). 1H NMR (400 MHz, DMSO-d6, δ ppm): 12.94 (s, 1H, NH)endocyclic, 10.66 (s, 1H, N=CH)imine, 8.30 (s, 1H, NH)hydrazine, 7.72–7.61 (m, 2H, Ph), 7.29–7.19 (m, 2H, Ph), 5.50 (s, 2H, NH2)hydrazine.13C NMR (400 MHz, DMSO-d6, δ ppm): 164.45 (C=S), 161.35 (N=CH), (149.60) (C=N), (131.33, 128.46, 128.37, 115.65) (Ar–C). MS-EI: (m/z, relative intensity, %): Calculated [M]+: 252, Found [M]+: 252.06 (20%). C9H9N6FS: Calculated: C, 42.85; H, 3.57; N, 33.33; S, 12.70. Found: C, 42.80; H, 3.53; N, 33.32; S, 12.70.
3.3.2. 4-(3,4,5-Trimethoxybenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole (TMOBAHMT) (2b)
Yellow crystals; yield: 72.52%; mp 242–244 °C. IR (ATR, v cm–1): 3262.39, 3212.43 (NH2), 3093.41 (NH), 2949.42 (Ar–H), 1627.72 (C=N)triazole, 1590.28 (C=N)imine, 1270.70 (C–N), 1019.45 (C=S). 1H NMR (400 MHz, DMSO-d6, δ ppm): 13.17 (s, 1H, NH)endocyclic, 10.13 (s, 1H, N=CH)imine, 8.56 (s, 1H, NH)hydrazine, 7.67 (s, 2H, Ph), 5.45 (s, 2H, NH2)hydrazine, 3.81 (s, 6H, Ph-CH3), 3.72 (s, 3H, Ph-CH3). 13C NMR: (400 MHz, DMSO-d6, δ ppm): 164.67 (C=S), 154.95 (N=CH)imine, 153.05 (N=C)ring, (151.82, 143.64, 140.87, 111.92) (Ph), 57.07 (CH3), 56.71 (CH3). MS-EI: (m/z, relative intensity, %): Calculated [M]+: 325, Found [M]+: 325.09 (100%). C12H16N6O3S: Calculated: C, 44.44; H, 4.94; N, 25.80; S, 9.88. Found: C, 44.40; H, 4.93; N, 25.79; S, 9.86.
3.3.3. 4-(4-Benzyloxy-2-methyloxybenzylidineamino)-3-hydrazino-5-mercapto-1,2,4-triazole (4BO2MOBAHMT) (2c)
White crystalline material; yield: 75.52%; mp162–165 °C. IR (ATR, v cm–1): 3377.13, 3147.75 (NH2), 3022.18 (NH), 2942.62 (Ar–H), 1644.62 (C=N)triazole, 1606.75 (C=N)imine, 1502.94 (C–N), 1138.67 (C–O), 1029.09 (C=S). 1H NMR (400 MHz, DMSO-d6, δ ppm): 13.26 (s, 1H, NH)endocyclic, 9.87 (s, 1H, N=CH)imine, 8.25 (s, 1H, NH)hydrazine, 7.65 (d, J = 1.9 Hz, 1H, Ph), 7.46 (ddt, J = 4.8 Hz, 2H, Ph), 7.41–7.39 (m, 2H, Ph), 7.37–7.34 (m, 1H, Ph), 7.21 (d, J = 8.4 Hz, 1H, Ph), 7.08 (d, J = 2.3 Hz, 1H, Ph), 5.21 (s, 2H, NH2)hydrazine, 3.87 (s, 2H, Ar-O-CH2-Ph), 3.17 (s, 3H, OCH3). 13C NMR: (400 MHz, DMSO-d6, δ ppm): 162.87 (C=S), 159.87 (N=CH)imine, 152.08 (N=C)endocyclic, (149.83, 145.45, 136.99, 128.95, 125.61, 124.80, 121.36, 113.42, 110.57, 108.49) (Ph), 70.37 (CH2), 56.22 (OCH3). MS-EI: (m/z, relative intensity, %): Calculated [M]+: 370, Found [M]+: 370.54 (100%). C17H17N6O2S: Calculated: C, 55.28; H, 4.61; N, 22.76; S, 8.67. Found: C, 55.26; H, 4.60; N, 22.74; S, 8.65.
3.4. Characterization of CNPs and LC-CNP-1,2,4-Triazole Conjugates (4a–c)
3.4.1. CNPs (3)
CNPs fabricated by this approach are highly fluorescent, economical, and have good water solubility and biocompatibility. Acidic conditions impart OH and COOH polar groups to render them highly dispersible in water. CNPs exhibited excellent hydrophilicity due to the presence of OH– and COO– groups. They form a transparent solution under daylight and give excellent blue photoluminescence (PL) when excited under a UV lamp (λmax = 365 nm). The physicochemical properties of CNPs are depicted in Table S1.
The UV/Vis spectra of CNPs showed a strong absorption peak between 255 and 260 nm. The absorption peaks at 260 and 325 nm are due to Π–Π* and n−Π* transitions of extended conjugation of inner core and peripheral −OH functional groups, respectively, as shown in Figure 3a. CNPs displayed strong photoluminescence extended from 300 to 650 nm (Figure 3b) with maximum emission at 450 nm.
Figure 3.
Spectroscopic and microscopic characterization of CNPs: (a) UV/vis spectra, (b) PL spectra (the inset shows the normalized spectra), (c) TEM image, (d) AFM image (the inset shows the height profile of AFM image).
To further explore the fluorescence properties of CNPs, PL spectra were recorded at different excitation wavelengths. The PL emission spectra were obtained with longer wavelengths from 300 to 650 nm. The PL spectra shown in Figure 3b were recorded at different excitation wavelengths (250, 300, 350, 400, 450, 500, 550, and 600 nm), and the corresponding emission spectra were observed at longer wavelengths (300, 350, 450, 500,550, 600, and 650 nm).
The morphological characterization of CNPs was carried out using a scanning electron microscope. The results revealed that CNPs are spherical and uniform in size distribution, as shown in Figure S3a.
Energy-dispersive X-ray spectroscopy (EDS) results demonstrated that carbon (42%) and oxygen (58%) are the major constituents of CNPs due to inner carbon core and peripheral hydroxyl and carboxylic groups, respectively (Figure S4a and Table S2). The TEM and AFM microscopy results showed that the size of CNPs is 2–3 nm. The results of TEM (Figure 3c) were supported by AFM image and the extracted height profile (Figure 3d).
3.4.2. Fluorescent 1,2,4-Triazole–Peptide Conjugates (4a–c)
Double conjugates of CNPs with 1,2,4-triazole derivatives and LC dipeptide were obtained in excellent yield. The physicochemical properties of the LC-CNP-4FBAHMT conjugate are depicted in Table S1.
3.4.2.1. LC-CNP-4FBAHMT Conjugate (4a)
The UV/vis spectra of LC, 4FBAHMT derivative (2a), and 4FBAHMT conjugate (4a) are shown in Figure 4a. The LC molecule is UV/vis-inactive, and the 4FBAHMT derivative showed two absorption peaks around 260 and 325 nm due to Π–Π* and n−Π* transitions, respectively. The 4FBAHMT conjugate showed a red shift due to the extended conjugation resulting from amide bond between carboxylic groups of CNPs and amino groups of 1,2,4-triazole derivative and LC. In this conjugate, two peaks are observed at 270 and 340 nm due to this red shift. The absorption peaks of CNPs at 260 and 325 nm resulting from Π–Π* and n−Π* transitions are shifted to longer wavelengths in the conjugate. The LC-CNP-4FBAHMT conjugate maintained absorbance properties after functionalization and conjugation process.
Figure 4.
Spectroscopic and microscopic characterization of LC-CNP-4FBAHMT conjugate: (a) UV–vis spectra, (b) PL spectra (the inset shows the normalized spectra), (c) FT-IR spectra, (d) TEM image of conjugate, and (e) AFM image of conjugate (the inset shows the extracted height profile of AFM image).
The 4FBAHMT conjugate showed strong photoluminescence ranging from 300 to 650 nm with maximum emission at 500 nm (Figure 4b). The functional group modifications and attachment of LC and 1,2,4-triazole molecules do not change the PL properties of CNPs.
PL spectra of 4FBAHMT conjugate showed excellent emission from 300 to 650 nm when excited at different wavelengths (250–600 nm).
The PL spectra shown in Figure 4b were recorded at different excitation wavelengths (250, 300, 350, 400, 450, 500, 550, and 600 nm). The PL emission spectra were observed at progressively longer wavelengths (300, 350, 400, 450, 500, 550, 600, and 650 nm).
The comparison of the FTIR spectra of CNPs, LC, 4FBAHMT derivative, and LC-CNP-4FBAHMT conjugate is shown in Figure 4c. The absorption bands at 1700 and 1620 cm–1 can be attributed to C=O and C=C stretching vibrational modes, respectively. Very small bands around 2150 and 3000 cm–1 are characteristic peaks of C=C and C–H stretching. The bands in the range 1300–1000 cm–1 are due to C–OH stretching and O–H bending vibrations. A very broad and high intensity peak around 3400 cm–1 is a feature peak of O–H stretching. Low-intensity vibrations around 570 cm–1 are characteristic peaks of CNPs.
The vibrational stretching bands between 3300 and 3010 cm–1 are characteristic for NH2 and NH groups of LC and 4FBAHMT derivative. Strong vibrational bands around 1645 (C=O str.), 1610 (N–H bend), 1400 (C–N stretch), and 1200 cm–1 (C–O stretch) suggest the presence of LC. The presence of two stretching bands around 1620 (N=CH)triazole ring and 1580 cm–1 (N=CH)imine is characteristics of 4FBAHMT derivative.
The FTIR spectra of the LC-CNP-4FBAHMT conjugate (Figure 4c) displayed the resemblance of some feature bands with CNPs, LC, and 4FBAHMT derivative. The appearance of a broad peak between 3260 and 3250 cm–1 and shifting of carboxylic group peak to lower wavelength (∼1680 cm–1) confirmed the formation of amide bond in LC-CNP-4FBAHMT conjugate. This broad band is due to the presence of OH and NH2 groups of CNPs and LC, respectively. The presence of −OH and N–H bands and the relatively low value of C=O stretching band suggest the formation of an amide bond in the LC-CNP-4FBAHMT conjugate.
SEM results showed that LC-CNP-4FBAHMT conjugates are also spherical and monodispersed, as shown in Figure S3b. The conjugates are larger than CNPs. The EDS profile of the LC-CNP-4FBAHMT conjugates showed the presence of sulfur coming from 1,2,4-triazole derivative (Figure S4b). The TEM and AFM results showed that the average size of the LC-CNP-4FBAHMT conjugates is 8–10 nm (Figure 4d,e). TEM image was supported by AFM image and the extracted height profile (Figure 4e).
The LC-CNP-TMOBAHMT and LC-CNP-4BO2MOBAHMT conjugates displayed similar features, and their characterization is discussed in supplementary data (Figures S5 and S6).
3.5. Kinetic Studies (in Vitro Drug Loading and pH-Triggered Drug Release)
Drug loading and pH-triggered drug release of 4FBAHMT derivative from LC-CNP-1,2,4-triazole conjugate were performed using UV/vis spectroscopy, taking advantage of the absorption properties of 4FBAHMT derivative and CNPs. The LC-CNP-4FBAHMT conjugate displayed a feature peak at 340 nm, which corresponds to 320 nm of free 4FBAHMT derivative. The results showed that 4FBAHMT derivative was loaded onto the CNPs. The amount of loaded 4FBAHMT derivative was estimated using a calibration curve (Figure S7). The amount of 4FBAHMT derivative loaded on the conjugate was 300 μg/mg of LC-CNP-4FBAHMT conjugate.
LC-CNP-4FBAHMT conjugates were dialyzed in acetate buffer (pH 5.0) and phosphate-buffered saline (PBS, pH 7.4) for 100 h for drug release studies. The results indicated that there is a continuous release of drug with time at both pH values. However, at physiological pH (7.4), the drug release is very slow and only 20% free drug is detected after 80 h. On the other hand, under acidic conditions (pH 5.0), the release significantly increases with time (60% after 80 h; Figure 5).
Figure 5.

Kinetics of 4FBAHMT derivative release from LC-CNP-4BAHMT conjugate at different pH values.
3.6. Biological Studies
3.6.1. Cell Viability/Proliferation/MTS Assay
Cell viability/proliferation/MTS assay results are presented in Figure 6a–f. Our in vitro results revealed that CNPs are highly biocompatible and LC-CNP-1,2,4-triazole conjugates (4a–c) have significantly high cytotoxicity against both cell lines with varying drug concentration.
Figure 6.
Cell viability/proliferation/MTS assay of (a, d) 4FBAHMT conjugate, (b, e) TMOBAHMT conjugate, (c, f) 4BO2MOBAHMT conjugate against CHLA-200 and SJGBM2 cell lines. The cells were exposed to different concentrations (5, 10, 25, and 50 μg/mL) of CNPs, TM, LC, 1,2,4-triazole derivatives, and LC-CNP-1,2,4-triazole conjugates.
The viability assay also indicated that LC-CNP-1,2,4-triazole conjugates (4a–c) have significantly higher efficacies in terms of selectivity and cytotoxic effects than 1,2,4-triazole derivatives (2a–c), and TM against both cell lines and maximum cytotoxicity is observed at 50 μg/mL. The three 1,2,4-triazole derivatives, i.e., 4FBAHMT, TMOBAHMT, and 4BO2MOBAHMT, and their conjugates showed better cytotoxicity against CHLA-200 cells than SJGBM2 cell line. All of the conjugates demonstrated more than 50% cytotoxicity against both cell lines at 50 μg/mL, whereas CNPs and LC are nontoxic to both cell lines up to 50 μg/mL. Among three conjugates, the LC-CNP-4BO2MOBAHMT conjugate showed better cytotoxicity against both cell lines.
The title compounds were nontoxic to embryonic kidney cell line (HEK293 as a control). Furthermore, the cell viability/proliferation assay results also indicated that these drug conjugates are more effective than control anticancer drug TM (one of the anticancer drugs used for brain tumor). Thus, fluorescent nanoconjugates based on 1,2,4-triazole derivatives are potent and more selective toward two pediatric brain tumor cell lines (CHLA-200 and SJGBM2) compared to nontreated controls and can prove to be future drug candidates for targeting brain tumor.
3.6.2. Fluorescence Imaging of CHLA-200 Cells Treated with Conjugates
CHLA-200 cells treated with various concentrations (10, 25, 50 μg/mL) of fluorescent conjugates were observed directly under a fluorescence microscope. The cells treated with 1,2,4-triazole derivatives were stained with rhodamine 6G dye (mitochondrial stain) before visualization.
The microscopic examination results of CHLA-200 cells treated with various concentrations of 1,2,4-triazole derivatives (2a–c) and their conjugates (4a–c) are presented in Figures 7–9. Microscopic images of CHLA-200 cells showed that conjugates have pronounced cytotoxic effects as the viability of drug-treated cells decreases significantly compared to that of nontreated controls with increasing drug concentration. Furthermore, cytotoxic effects of conjugates on CHLA-200 cell cultures were more promising than cytotoxic effects of 1,2,4-triazole derivative (Figures 7–9b) within the concentration range of 10–50 μg/mL. These results are consistent with MTS assay results.
Figure 7.
Microscopic images of CHLA-200 cells treated with various concentrations (10, 25, 50 μg/mL) of (a) LC-CNP-4FBAHMT conjugate and (b) 4FBAHMT derivative. Cells treated with 1,2,4-triazole derivative were stained with rhodamine 6G dye.
Figure 9.
Microscopic images of CHLA-200 cells treated with various concentrations (10, 25, 50 μg/mL) of (a) LC-CNP-4BO2MOBAHMT conjugate and (b) 4BO2MOBAHMT derivative. Cells treated with 1,2,4-triazole derivative were stained with rhodamine 6G dye.
Figure 8.
Microscopic images of CHLA-200 cells treated with various concentrations (10, 25, 50 μg/mL) of (a) LC-CNP-TMOBAHMT conjugate and (b) TMOBAHMT derivative. Cells treated with 1,2,4-triazole derivative were stained with rhodamine 6G dye.
4. Conclusions
CNPs fabricated by bottom-up approach are highly fluorescent, with good water solubility and biocompatibility. This method is very simple, economical, and produces CNPs on a large scale with tunable surface functionalities and high quantum yield. CNPs can be functionalized easily for advanced drug-delivery system. Three new 1,2,4-triazole derivatives (2a–c) were synthesized after screening through molecular docking procedures with αvβ6 integrin protein using Molecular Operating Environment (MOE) docking software. 1,2,4-Triazole derivatives (2a–c) showed docking scores comparable to anticancer drug TM, whereas the LC-CNP-1,2,4-triazole conjugates (4a–c) displayed significantly high docking scores for αvβ6 integrin protein. The double conjugates of CNPs were synthesized with 1,2,4-triazole derivatives and LC dipeptide through amide bond using EDC coupling chemistry. CNPs and their conjugates exhibited excellent fluorescence properties. Electron microscopic examination showed that CNPs are spherical and quantum-sized (2–3 nm) and their conjugates are nanodrug systems having size around 8 nm. Fluorescent LC-CNP-1,2,4-triazole conjugates (4a–c) showed potent and selective anticancer activity against two pediatric brain tumor cell lines (CHLA-200 & SJGBM2) compared to control drug TM. These conjugates were more cytotoxic than the corresponding 1,2,4-triazole derivatives against both cell lines. The title conjugates were nontoxic for normal embryonic kidney cell line (HEK 293 cell line). CNPs were highly biocompatible for both types of cell lines. The experimental results of MTS bioactivity assay were in close agreement with the theoretical results. This research work offered a lead to the design of novel anticancer therapeutics. However, further studies are needed to explore their binding mechanism with αvβ6 integrin and effects of this binding on biological activities. In vivo biological activity, biophysical assays, and mechanistic studies are the future prospective of this research work. These CNPs-based nanomaterials along with the potential anticancer activity of 1,2,4-triazole derivatives and tailored targeting ability of LC can prove to be excellent alternatives for conventional drug-delivery models in vivo.
Acknowledgments
The authors acknowledge the Higher Education Commission of Pakistan and the University of Miami, FL, USA, for providing research facility and support for this research work.
Glossary
Abbreviations
- AHMT
4-amino-3-hydrazino-5-mercapto-1,2,4-triazole
- 4BO2MOBAHMT
4-(4-benzyloxy-2-methyloxbenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole
- CNPs
carbon nanoparticles
- DAPI
4′,6-diamidino-2-phenylindole
- EDC
1-ethyl-3-[3-dimethylaminopropyl]carbodiimide HCl
- EDS
energy-dispersive X-ray spectroscopy
- 4FBAHMT
4-(4-fluorobenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole
- FBS
fetal bovine serum
- L-Car, LC
l-carnosine
- LC-CNP-4BO2MOBAHMT
l-carnosine-carbon nanoparticles-4-(4-benzyloxy-2-methyloxybenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole conjugate
- LC-CNP-4FBAHMT
l-carnosine-carbon nanoparticles-4-(4-fluorobenzylidenamino)-3-aminomethyl-5-mercapto-1,2,4-triazole conjugate
- LC-CNP-TMOBAHMT
l-carnosine-carbon nanoparticles-4-(3,4,5-trimethoxybenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole conjugate
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium, inner salt
- PL
photoluminescence
- RPMI-1640
Roswell Park Memorial medium
- TMOBAHMT
4-(3,4,5-trimethoxybenzylidenamino)-3-hydrazino-5-mercapto-1,2,4-triazole
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b01903.
Optimized crystal structure of αvβ6 integrin (Figure S1); docking presentation of 4a and 4b conjugates in the active binding site of αvβ6 integrin (Figure S2); physiochemical properties of CNPs and 4a conjugate (Table S1); electron microscopy of CNPs and 4a conjugate (Figures S3, S4 and Table S2); characterization of 4a and 4b conjugates (Figures S5 and S6); and calibration curve used for the determination of drug loading (Figure S7) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Siegel R. L.; Miller K. D.; Jemal A. Cancer statistics. CA Cancer J. Clin. 2016, 66, 7–30. 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Dianzani C. H.; Zara G. P.; Maina G.; Pettazzoni P.; Pizzimenti S.; Rossi F.; Gigliotti C. L.; Ciamporcero E. S.; Daga M.; Barrera G. Drug delivery nanoparticles in skin cancers. BioMed Res. Int. 2014, 2014, 1–13. 10.1155/2014/895986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R.; Madanmohan S.; Kesavan A.; Baskar G.; Krishnamoorthy Y. R.; Santosham R.; Ponraju D.; Rayala S. K.; Venkatraman G. Nanomedicine: Towards development of patient-friendly drug-delivery systems for oncological applications. Int. J. Nanomed. 2012, 7, 1043–1060. 10.2147/IJN.S25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S.; Lee K. H.; Oh J. E.; Park T. G. In vitro and in vivo anti-tumor activities of nanoparticles based on doxorubicin-PLGA conjugates. J. Controlled Release 2000, 68, 419–431. 10.1016/S0168-3659(00)00280-7. [DOI] [PubMed] [Google Scholar]
- Zhang C. X.; Lippard S. J. New metal complexes as potential therapeutics. Curr. Opin. Chem. Biol. 2003, 7, 481–489. 10.1016/S1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- Kelland L. R. Preclinical perspectives on platinum resistance. Drugs 2000, 59, 1–8. 10.2165/00003495-200059004-00001. [DOI] [PubMed] [Google Scholar]
- Rosenberg B. Noble metal complexes in cancer chemotherapy. Adv. Exp. Med. Biol. 1978, 91, 129–150. 10.1007/978-1-4684-0796-9_10. [DOI] [PubMed] [Google Scholar]
- Hebert C.; Norris K.; Sauk J. J. Targeting of human squamous carcinomas by SPA470-doxorubicin immunoconjugates. J. Drug Targeting 2003, 11, 101–107. 10.1080/1061186031000121478. [DOI] [PubMed] [Google Scholar]
- Arteaga C. Targeting HER1/EGFR: A molecular approach to cancer therapy. Semin. Oncol. 2003, 30, 3–14. 10.1016/S0093-7754(03)70010-4. [DOI] [PubMed] [Google Scholar]
- Liu J.; Kolar C.; Lawson T. A.; Gmeiner W. H. Targeted drug delivery to chemoresistant cells: Folic acid derivatization of FdUMP [10] enhances cytotoxicity toward 5-FU-resistant human colorectal tumor cells. J. Org. Chem. 2001, 66, 5655–5663. 10.1021/jo005757n. [DOI] [PubMed] [Google Scholar]
- Panyam J.; Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv. Drug Delivery Rev. 2003, 55, 329–347. 10.1016/S0169-409X(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Kalluraya B.; Lingappa B.; Rai N. S. Synthesis and Biological study of some novel 4-[5-(4,6-disubstituted-2-thiomethylpyrimidyl)-4′-amino-1,2,4-triazolo-3-yl]thioacetyl-3- arylsydnones. Phosphorus, Sulfur Silicon Relat. Elem. 2007, 182, 1393–1401. 10.1080/10426500601161049. [DOI] [Google Scholar]
- Lednicer D.; Mitscher L. A.. Organic Chemistry of Drug Synthesis; Wiley Interscience: New York, 1997; Vol. 1, p 226. [Google Scholar]
- Meenakshi V. A.; Sabir H. B. Synthesis and anti-inflammatory evaluation of 1,2,4-triazole derivatives. RRJC 2014, 3, 24–26. [Google Scholar]
- Rakesh K.; Shahar M. Y.; Birendra S. Synthesis, characterization and biological evaluation of novel 1,2,4-triazole derivatives as potent anti-bacterial and anti-inflammatory agents. Pharma Chem. 2014, 6, 137–143. [Google Scholar]
- Chelamalla R.; Venkatesham A.; Sarangapani M. Synthesis and anti-depressant activity of some novel 1,2,4-triazole derivatives. J. Pharm. Res. 2012, 5, 4739. [Google Scholar]
- Ram J. S.; Dharmendra K. S. Syntheses of some 3,5-diaryl-4H-1,2,4-triazole derivatives and their anti-fungal activity. Eur. J. Chem. 2009, 6, 219–224. [Google Scholar]
- Plech T.; Kapron B.; Luszczki J. J.; Paneth A.; Siwek A.; Kolaczkowski M.; Zolnierek M.; Nowak G. Studies on the anti-convulsant activity of 4-alkyl-1,2,4-triazole-3-thiones and their effect on GABAergic system. Eur. J. Med. Chem. 2014, 86, 690–699. 10.1016/j.ejmech.2014.09.034. [DOI] [PubMed] [Google Scholar]
- Li X.; Li X. Q.; Liu H. M.; Zhou X. Z.; Shao Z. H. Synthesis and evaluation of anti-tumor activities of novel chiral 1,2,4-triazole Schiff bases bearing γ-butenolide moiety. Org. Med. Chem. Lett. 2012, 2, 26. 10.1186/2191-2858-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler J. Differences between the non-steroidal aromatase inhibitors anastrozole and letrozole – of clinical importance?. Br. J. Cancer 2011, 104, 1059–1066. 10.1038/bjc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharb R.; Sharma P. C.; Yar M. S. Pharmacological significance of triazole scaffold. J. Enzyme Inhib. Med. Chem. 2011, 26, 1–21. 10.3109/14756360903524304. [DOI] [PubMed] [Google Scholar]
- Yu S.; Kang M. W.; Chang H. C.; Chen K. M.; Yu Y. C. Bright fluorescent nanodiamonds: No photobleaching and low cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. 10.1021/ja0567081. [DOI] [PubMed] [Google Scholar]
- Bourlinos A. B.; Stassinopoulos A.; Anglos D.; Zboril R.; Georgakilas V.; Giannelis E. P. Photoluminescent carbogenic dots. Chem. Mater. 2008, 20, 4539–4541. 10.1021/cm800506r. [DOI] [Google Scholar]
- Neugart F.; Zappe A.; Jelezko F.; Tietz C.; Boudou J. P.; Krueger A.; Wrachtrup J. Dynamics of diamond nanoparticles in solution and cells. Nano Lett. 2007, 7, 3588–3591. 10.1021/nl0716303. [DOI] [PubMed] [Google Scholar]
- Baker S. N.; Baker G. A. Luminescent carbon nanodots: Emergent nanolights. Angew. Chem., Int. Ed. 2010, 49, 6726–6744. 10.1002/anie.200906623. [DOI] [PubMed] [Google Scholar]
- da Silva J. C. G. E.; Goncalves H. M. R. Analytical and bioanalytical applications of carbon dots. TrAC, Trends Anal. Chem. 2011, 30, 1327–1336. 10.1016/j.trac.2011.04.009. [DOI] [Google Scholar]
- Meng X. Y.; Zhang H. X.; Mezei M.; Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetényi C.; Spoel D. V. D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002, 11, 1729–1737. 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuss J. M.; Gallo J.; DeLisser H. M.; Klimanskaya I. V.; Folkesson H. G.; Pittet J. F.; Nishimura S. L.; Aldape K.; Landers D. V.; Carpenter W.; Gillett N.; Sheppard D.; Matthay M. A.; Albelda S. M.; Kramer R. H.; Pytela R. Expression of the beta 6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J. Cell Sci. 1995, 108, 2241–2251. [DOI] [PubMed] [Google Scholar]
- Breuss J. M.; Gillett N.; Lu L.; Sheppard D.; Pytela R. Restricted distribution of integrin beta 6 mRNA in primate epithelial tissues. J. Histochem. Cytochem. 1993, 41, 1521–1527. 10.1177/41.10.8245410. [DOI] [PubMed] [Google Scholar]
- Schally A. V.; Nagy A. Chemotherapy targeted to cancers through tumoral hormone receptors. Trends Endocrinol. Metab. 2004, 15, 300–310. 10.1016/j.tem.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Nakase I.; Konishi Y.; Ueda M.; Saji H.; Futaki S. Accumulation of arginine-rich CPPs in tumors and the potential for anti-cancer drug delivery in vivo. J. Controlled Release 2012, 159, 181–188. 10.1016/j.jconrel.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Moorthi C.; Manavalan R.; Kathiresan K. Nanotherapeutics to overcome conventional cancer chemotherapy limitations. J. Pharm. Pharm. Sci. 2011, 14, 67–77. 10.18433/J30C7D. [DOI] [PubMed] [Google Scholar]
- Iovine B.; Oliviero G.; Garofalo M.; Orefice M.; Nocella F.; Borbone N.; Piccialli V.; Centore R.; Mazzone M.; Piccialli G.; Bevilacqua M. A. The anti-proliferative effect of L-carnosine correlates with a decreased expression of hypoxia inducible factor 1 alpha in human colon cancer cells. PLoS One 2014, 9, e96755 10.1371/journal.pone.0096755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid I.; van Reyk D. M.; Davies M. J. Carnosine and its constituents inhibit glycation of low-density lipoproteins that promotes foam cell formation in vitro. FEBS Lett. 2007, 581, 1067–1070. 10.1016/j.febslet.2007.01.082. [DOI] [PubMed] [Google Scholar]
- Gaunitz F.; Hipkiss A. R. Carnosine and cancer: A perspective. Amino Acids 2012, 43, 135–142. 10.1007/s00726-012-1271-5. [DOI] [PubMed] [Google Scholar]
- Pietrasik J. M.; Książek K. L-carnosine prevents the pro-cancerogenic activity of senescent peritoneal mesothelium towards ovarian cancer cells. Anticancer Res. 2016, 36, 665–672. [PubMed] [Google Scholar]
- Rybakova Y. S.; Kalen A. L.; Eckers J. C.; Fedorova T. N.; Goswami P. C.; Sarsour E. H. Increased manganese superoxide dismutase and cyclin B1 expression in carnosine induced inhibition of glioblastoma cell proliferation. Biomed. Khim. 2015, 61, 510–518. 10.18097/PBMC20156104510. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Yang J.; Li J.; Shi X.; Ouyang L.; Tian Y.; Lu J. Carnosine inhibits the proliferation of human gastric cancer SGC-7901 cells through both of the mitochondrial respiration and glycolysis pathways. PLoS One 2014, 9, e104632 10.1371/journal.pone.0104632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiyants V.; Perekhoda L.; Saidov N.; Kadamov I. Docking studies and <tep-common:author-query>AQ7: Please provide a DOI number for ref 40 or indicate if one doesn&#x2019;t exist.</tep-common:author-query>biological evaluation of anti-cancer activity of new 1,2,4-triazole (4H) derivatives. Scr. Sci. Pharm. 2014, 2, 46–53. 10.14748/ssp.v1i2.778. [DOI] [Google Scholar]
- Ajmal M.; Yunus U.; Matin A.; Haq N. U. Synthesis, characterization and in vitro evaluation of methotrexate conjugated fluorescent carbon nanoparticles as drug delivery system for human lung cancer targeting. J. Photochem. Photobiol., B 2015, 153, 111–120. 10.1016/j.jphotobiol.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Xianchi D.; Nathan E.; Hudson C. L.; Timothy A. S. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat. Struct. Mol. Biol. 2014, 21, 1091–1096. 10.1038/nsmb.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civera M.; Arosio D.; Bonato F.; Manzoni L.; Pignataro L.; Zanella S.; Gennari C.; Piarulli U.; Belvisi L. Investigating the interaction of cyclic RGD peptidomimetics with αvβ6 integrin by biochemical and <tep-common:author-query>AQ8: Please provide a DOI number for ref 43 or indicate if one doesn&#x2019;t exist.</tep-common:author-query>molecular docking studies. Cancers 2017, 9, 1–13. 10.3390/cancers9100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba Y.; Kumagai T.; Yamamoto A.; Yoshitsu H.; Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 2006, 281, 8981–8990. 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- Croney J. C.; Jameson D. M.; Learmonth R. P. Fluorescence spectroscopy in biochemistry: teaching basic principles with visual demonstrations. Biochem. Mol. Biol. Educ. 2001, 29, 60–65. 10.1016/S1470-8175(01)00019-4. [DOI] [Google Scholar]
- Dickinson R. G.; Jacobsen N. W. Detection of thioureido groups in open chain and heterocyclic cornpounds by hydrazinolysis. Anal. Chem. 1969, 41, 1324–1326. 10.1021/ac60279a045. [DOI] [Google Scholar]
- Stollé R.; Bowles P. E. Über Thiocarbohydrazid. Ber. Dtsch. Chem. Ges. 1908, 41, 1099–1110. 10.1002/cber.190804101217. [DOI] [Google Scholar]
- Kalyaanamoorthy S.; Chen Y. P. Structure-based drug design to augment hit discovery. Drug Discovery Today 2011, 16, 831–839. 10.1016/j.drudis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Shanti K. A.; Radhakumary C.; Molly A.; Sreenivasan K. Functionalized carbon dots enable simultaneous bone crack detection and drug deposition. J. Mater. Chem. B 2014, 2, 8626–8632. 10.1039/C4TB00918E. [DOI] [PubMed] [Google Scholar]
- Ray S. C.; Saha A.; Jana N. R.; Sarkar R. Fluorescent carbon nanoparticles: synthesis, characterization, and bioimaging application. J. Phys. Chem. C 2009, 113, 18546–18551. 10.1021/jp905912n. [DOI] [Google Scholar]
- Shanghao L.; Daniel A.; Zhili P.; Steven V.; Scott R.; Guillermo D. A.; Abdelhameed M. O.; Regina M. G.; Roger M. L. Transferrin conjugated nontoxic carbon dots for doxorubicin delivery to target pediatric brain tumor cells. Nanoscale 2016, 8, 16662–16669. 10.1039/C6NR05055G. [DOI] [PubMed] [Google Scholar]
- Kaufmann R.; Müller P.; Hildenbrand G.; Hausmann M.; Cremer C. Analysis of Her2/Neu membrane protein clusters in different types of breast cancer cells using localization microscopy. J. Microsc. 2011, 242, 46–54. 10.1111/j.1365-2818.2010.03436.x. [DOI] [PubMed] [Google Scholar]
- Johnson L. V.; Walsh M. L.; Chen L. B. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. U.S.A. 1980, 77, 990–994. 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi T.; Tohru Y.; Yosinobu K.; Keiji Y. Fluorescent dye monitoring of mitochondrial changes associated with malignant cell transformation. Cell Struct. Funct. 1987, 12, 525–537. 10.1247/csf.12.525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.