Abstract
Next-generation sequencing (NGS) methods for cancer testing have been rapidly adopted by clinical laboratories. To establish analytical validation best practice guidelines for NGS gene panel testing of somatic variants, a working group was convened by the Association of Molecular Pathology with liaison representation from the College of American Pathologists. These joint consensus recommendations address NGS test development, optimization, and validation, including recommendations on panel content selection and rationale for optimization and familiarization phase conducted before test validation; utilization of reference cell lines and reference materials for evaluation of assay performance; determining of positive percentage agreement and positive predictive value for each variant type; and requirements for minimal depth of coverage and minimum number of samples that should be used to establish test performance characteristics. The recommendations emphasize the role of laboratory director in using an error-based approach that identifies potential sources of errors that may occur throughout the analytical process and addressing these potential errors through test design, method validation, or quality controls so that no harm comes to the patient. The recommendations contained herein are intended to assist clinical laboratories with the validation and ongoing monitoring of NGS testing for detection of somatic variants and to ensure high quality of sequencing results.
Next-generation sequencing (NGS) for the detection of somatic variants is being used in a variety of molecular oncology applications and scenarios, ranging from sequencing entire tumor genomes and transcriptomes for clinical research to targeted clinical diagnostic gene panels. This guideline will focus on targeted gene panels and their diagnostic use in solid tumors and hematological malignancies. The expanding knowledge base of molecular alterations that initiate and drive tumor growth and metastasis has resulted in the development and clinical laboratory implementation of a diversity of targeted gene panels. Individual gene panels may focus on solid tumors, or hematological malignancies, or may be technically designed to interrogate both, with interpretation focused on the tumor phenotype. The information generated by targeted gene panels can inform diagnostic classification, guide therapeutic decisions, and/or provide prognostic insights for a particular tumor. The numbers of genes included in panels can differ substantially between laboratories. Some laboratories include only core genes for which substantial literature exists with regard to their diagnostic, therapeutic, or prognostic relevance. Other panels include a larger gene set that includes the aforementioned core set of genes and additional genes that are being investigated in clinical trials and/or for which evidence is still accruing. The analysis of genes in a panel may be restricted to mutational hotspots relevant to a therapeutic agent or it may be broader and include flanking regions or the entire gene sequence. When planning the development of a targeted gene panel, the laboratory needs to define its intended use, including what types of samples will be tested (eg, testing only primary tumor samples or also used to monitor residual disease post-therapy) and what types of diagnostic information will be evaluated and reported. These considerations, among others, will influence the design, validation, and quality control of the test. The Association of Molecular Pathology (AMP) convened a working group of subject matter experts with liaison representation from the College of American Pathologists to address the many issues pertaining to the analytical validation and ongoing quality monitoring of NGS testing for detection of somatic variants and for ensuring high quality of sequencing results. These developed professional recommendations will be described in detail in the following sections.
Overview of Targeted NGS for Oncology Specimens
General Considerations
NGS offers multiple approaches for investigation of human genome, including sequencing of whole genome, exome, and transcriptome. However, targeted panels are often practical in the clinical setting for detection of clinically informative genetic alterations. They are currently the most frequently used type of NGS analysis for molecular diagnostic somatic testing for solid tumors and hematological malignancies. Before introducing clinical NGS testing, several issues need to be considered. The choice of a commercially available targeted NGS panel, or whether to design one’s own, is dependent on clinical indication of the test and the genes to be tested. Germline applications may necessitate different genes/panels than sporadic cancer applications. Solid tumor applications may necessitate different choices than hematological malignancies. Available pan-cancer panels are attractive in that they permit batching of samples across multiple indications with resultant savings on cost, human labor, and turnaround time.
Targeted NGS panels can be designed to detect single-nucleotide variants (SNVs; alias point mutations), small insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs), or gene fusions. Within a single panel, target sequences can be designed to cover hotspot regions of a single gene (eg, exons 9 and 20 of PIK3CA, exon 15 of BRAF, exons 18 to 21 of EGFR, or exons 12 and 14 of JAK2) or to cover the entirety of the coding and noncoding sequences relevant to a given gene (eg, KRAS, NRAS, or TP53) or SV. This design is important, as it relates ultimately to the potential capability of the panel to be used for detection of CNAs versus SNVs and small indels. SNVs are the most common mutation type in solid tumors and hematological malignancies [eg, KRAS p.Gly12 variants (eg, p.Gly12Asp), PIK3CA p.His1047Arg, EGFR p.Leu858Arg, and JAK2 p.Val617Phe]. Indels include nucleotide insertions, deletions, or both insertion and deletion events within close proximity. This can include the loss of one wild-type allele accompanied by duplication of a mutation-bearing allele, resulting in maintenance of overall copy number. Indels range in size from 1 to <1 kb in length, although most indels are only several base pairs (bp) to several dozen bp in length. These changes may be in-frame, resulting in the loss and/or gain of amino acids from the protein sequence (eg, EGFR exon 19 deletions), or frameshift, resulting in a change to the protein’s amino acid sequence downstream of the indel (eg, NPM1 p.Trp288 frameshift variants).
Another consideration in choosing or designing a gene panel is whether gene copy number will be assessed as part of the analysis. CNAs are structural changes resulting in gain or loss of genomic DNA in a chromosomal region, common in solid tumors and affecting both tumor suppressor genes and oncogenes. One example is TP53, one of the most frequently mutated genes in cancer; these mutations are often accompanied by loss of the remaining wild-type allele. Other examples include PTEN, CDKN2A, and RB1, losses of which may have clinical implications. Increased copy number can also be important, as in the case of ERBB2 (HER 2) in breast and gastric cancers. Similarly, copy number gains in MET, RICTOR, MDM2, and other genes are of clinical interest. Algorithms for assessing copy number have been established for sequencing data derived from both hybridization-capture and amplicon-based libraries. Regardless of the method, CNA assessment is influenced by the number of probes or amplicons covering the gene of interest. Copy number estimates from a single hotspot region in a gene are not as accurate as measurements averaged from probes or amplicons covering all exonic regions. The limit of detection (particularly for gene losses) is heavily dependent on the fraction of tumor cells present in the tested sample. SVs include translocations and other chromosomal rearrangements. SVs have been identified in many types of human malignancies and serve as important markers for cancer diagnosis, patient prognostication, and for selection of targeted therapies (eg, RET/PTC fusions are used for diagnosis of papillary thyroid carcinoma; TMPRSS2/ERG to predict favorable outcome in prostate cancer; EML4/ALK fusion for selection of targeted therapies in lung adenocarcinomas). There are two major approaches used for detection of gene fusions in targeted oncology NGS panels, either to sequence DNA using hybridization capture method or to sequence RNA (cDNA) by amplification-based methods.1,2 Most of the breakpoints occur in the introns of genes. Therefore, if DNA is used as a starting material, hybrid capture probes have to be designed either to span the whole gene, including intron regions, or to capture those exons/introns that are most frequently involved in the fusion of interest. Another practical approach is to use RNA, reverse transcribe it to cDNA, and to amplify it with fusion-specific primers or using other approaches (eg, hybrid capture). Each of these methods is currently used in the clinical setting. It is important to select and appropriately validate the bioinformatics pipeline for fusion detection.
Targeted NGS Method Overview
Overall, targeted NGS methods include four major components: sample preparation, library preparation, sequencing, and data analysis.3
Sample Preparation
The first step in clinical NGS analysis of a tumor is to assess the submitted sample. In the case of hematological specimens, tumor cell content may be inferred from separate analyses. For example, a white blood cell count differential from a peripheral blood sample or flow cytometric data from a bone marrow aspirate may establish the approximate fraction of tumor cells in the material used for nucleic acid extraction. Solid tumor samples, however, require microscopic review by an appropriately trained and certified pathologist before being accepted for NGS testing. This review ensures that the expected tumor type has been received and that there is sufficient, nonnecrotic tumor for NGS analysis. Microscopic review can be used to mark areas for macrodissection or microdissection (eg, through use of a dissecting microscope), thereby enriching the tumor fraction and increasing sensitivity for gene alterations. Estimation of tumor cell fraction, which is critical information when interpreting mutant allele frequencies and CNAs, should be performed. However, the estimation of tumor percentages based purely on review of hematoxylin and eosin–stained slides can be affected by many factors and experience significant interobserver variability.4 Non-neoplastic cells, such as inflammatory infiltrates and endothelial cells for example, which are often smaller than the neoplastic cells and intimately associated with the tumor, may remain inconspicuous and lead to gross underestimation of tumor proportion. In cases with more abundant inflammation and necrosis, it is important to remain conservative in the estimations and further correlate with the sequencing results. Review of mutant allele fractions (including silent mutations) in these cases would allow for more precise estimates of tumor purity and would allow for more accurate results and confident recommendations for further testing if needed.
Library Preparation
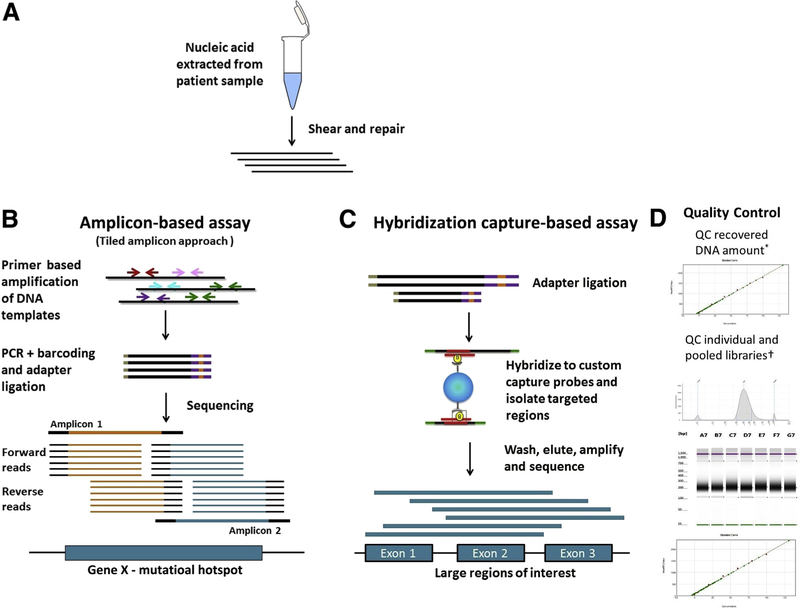
Library preparation is the process of generating DNA or cDNA fragments of specific size range. Two major approaches are used for targeted NGS analysis of oncology specimens: hybrid capture–based and amplification-based approaches (Figure 1).
Figure 1.
High-level comparison of target enrichment workflow for amplicon and capture hybridization NGS assays. A: Nucleic acid is extracted and quantified. The DNA is sheared and repaired to generate fragments of uniform size distribution, and the fragment size can be monitored by gel electrophoresis or Agilent Bioanalyzer. B: Amplification-based assays: Target enrichment in amplification-based assays consists of PCR amplification of the desired region using primers. A tiled amplicon approach is depicted in which primers are designed to generate multiple overlapping amplicons of the same region to avoid allele dropout. The sequencing reads generated will have the same start and stop coordinates dictated by the primer design. C: Hybridization capture–based assays: Target enrichment in hybridization capture–based assays uses long biotinylated oligonucleotide probes complementary to a region of interest. Probes hybridize to target regions contained within larger fragments of DNA. As a result, regions flanking the target will also be isolated and sequenced. Targeted fragments are isolated using streptavidin magnetic beads, followed by washing, elution, amplification, and sequencing. The sequencing reads from these molecules will have unique start and stop coordinates when aligned to a reference, allowing identification and removal of PCR duplicates. D: Quality control: The size distribution pattern of the individual and pooled libraries are quality controlled using Agilent TapeStation and quantified using a Spectramax microplate reader. Example images as visualized using an Agilent Bioanalyzer and Spectramax microplate reader are shown. QC, quality control.
Hybrid capture NGS
Hybrid capture–based enrichment methods use sequence-specific capture probes that are complementary to specific regions of interest in the genome. The probes are solution-based, biotinylated oligonucleotide sequences that are designed to hybridize and capture the regions intended in the design. Capture probes are significantly longer than PCR primers and therefore can tolerate the presence of several mismatches in the probe binding site without interfering with hybridization to the target region. This circumvents issues of allele dropout, which can be observed in amplification-based assays. Because probes generally hybridize to target regions contained within much larger fragments of DNA, the regions flanking the target are also isolated and sequenced. Compared to amplicon-based assays, hybrid capture–based assays enable the interrogation of neighboring regions that may not be easily captured with specific probes. However, hybrid capture–based assays can also isolate neighboring regions that are not of interest, thereby reducing overall coverage in the regions of interest if the off-target sequencing is not appropriately balanced. Also, in cases with rearrangements, isolated neighboring regions may also be from genomic areas far from the intended or predicted targets. Fragment sizes obtained by shearing and other fragmentation approaches will have a large influence over the outcome of the assays. Shorter fragments will be captured with higher specificity than longer fragments as they will contain a lower proportion of off-target sequences. On the other hand, longer reads would be expected to map to the reference sequence with less ambiguity than shorter reads.
Examples of hybridization capture technology that are currently commercially available include Agilent SureSelect (Agilent Technologies, Santa Clara, CA), Nimblegen(F. Hoffmann-La Roche Ltd, Basel, Switzerland), and Illumina TruSeq (Illumina, San Diego, CA). Custom panels can be developed to interrogate large regions of the genome (typically 50 to several thousand genes). Once regions of interest are determined, the size of the target regions will determine the number of probes required to capture each specific region. Probe densities can be increased for regions that prove difficult to enrich. General sample preparation steps for hybridization capture enrichment include initial DNA shearing, followed by several enzymatic steps encompassing end repair, A-base addition and ligation of sequence adaptors followed by PCR amplification, and clean-up. Next, the NGS library is hybridized to the custom biotinylated oligonucleotide capture probes. Because of the random nature of shearing, the size and nucleotide content of the individual captured fragments will differ. The resulting sequencing reads from the captured fragments will contain unique start and stop coordinates once they are aligned to the reference sequence. This enables the identification and removal of PCR duplicates from the data set, allowing a more accurate determination of depth of coverage and variant frequencies.
A modified approach to the hybridization capture options described above is used by the HaloPlex target enrichment system (Agilent Technologies). The HaloPlex technique is based on restriction enzyme digestion of genomic DNA followed by hybridization of biotinylated DNA probes, which are designed with homology only to the 5′ and 3′ ends of the regions of interest. This promotes the circularization of the regions of interest and increases capture specificity. The probe/fragment circular hybrids are captured using streptavidin-coated beads, which are then ligated, purified, and amplified.5,6
Hybridization capture is sensitive to sample base composition. Sequences that are adenine-thymine rich can be lost through poor annealing, whereas regions with high guanine-cytosine content can be lost through formation of secondary structures.7
Amplification-based NGS
Amplification-based library preparation methods rely on a multiplex PCR amplification step to enrich for target sequences. Target sequences are tagged with sample-specific indexes and sequencing adaptors used to anchor the amplicons to complimentary oligonucleotides embedded in the platform’s sequencing substrate before initiation of the sequencing process. Depending on target sequence primer and kit design, the amplification step in library preparation can be either a one-stage or a two-stage PCR approach. Amplification-based library preparation methods are versatile and scalable, and can be used to construct libraries of a range of sizes (eg, Illumina’s 26-gene TruSight Tumor panel and ThermoFisher’s 409-gene AmpliSeq Comprehensive Cancer Panel (ThermoFisher Scientific, Waltham, MA). The hands-on time in the laboratory setting for amplification-based library preparation is typically shorter than for hybridization capture methods.
Amplification-based library preparation methods are vulnerable to chemistry issues associated with PCR primer design. For example, allele dropout may occur if there is a single-nucleotide polymorphism or short indel in the primer region of the sequence, as the primer will be mismatched and not bind. This will result in lower-than-expected coverage for the amplicon and potential for incorrect assessment of variant allele frequencies for any variants in that amplicon. In addition, amplification-based library preparation is less likely to work effectively for genes with high guanine-cytosine content (eg, CEBPA8) or regions with highly repetitive sequences. Furthermore, amplification-based library preparation may not enable detection of indels if the indel removes the primer region of the sequence, or the indel sufficiently alters the size of the amplicon. Finally, sequencing quality diminishes at the ends of amplicons, leading to potential miscalling of variants in poor quality regions. This last issue can be ameliorated by tiling amplicons to ensure overlap if a critical hotspot region lies at the end of an amplicon.
Sequencing
Currently available sequencing platforms have different chemistries for sequencing that include sequencing by synthesis (Illumina NGS platforms) and ion semiconductor–based sequencing (ThermoFisher’s Ion systems), as well as different detection methods. Given the market share and popularity of both platforms, and the engineering differences between them, head-to-head comparison of the two sequencing technologies across multiple applications has been frequently undertaken. A number of studies have examined the performance of both NGS platforms across a diverse set of applications and found that the Illumina and Ion sequencers produce comparable results.9–12 Illumina and Ion sequencers have been evaluated for potential uses in clinical microbiology,9 germline variant detection,11 and prenatal testing.12,13 Recently, an evaluation of the Illumina MiSeq and ThermoFisher Ion Proton systems for detection of somatic variants in oncology determined that both platforms showed equal performance in detection of somatic variants in DNA derived from formalin-fixed, paraffin-embedded (FFPE) tumor samples using amplicon-based commercial panels.14 These comparisons are routinely reported with a caveat associated with the Ion sequencer’s ability to accurately detect homopolymer tracts, which is because of limitations in the linear range of detection of voltage changes associated with the addition of multiple identical nucleotides during sequencing. Despite the similarities in technical performance, differences between platforms exist, notably different DNA input requirements, different cost of reagents, run time, read length, and cost per sample. These differences have implications to the instruments’ capacity to handle low-quality samples, capability of detecting insertions/deletions, and sample throughput.
It is recognized by the Working Group that technological improvements in NGS will outpace published method and/or platform-specific clinical practice recommendations for the foreseeable future. The authors anticipate that detailed discussions of newer methods will be incorporated into a revised and updated version of this article in the near future.
Data Analysis
The data analysis pipeline (alias the bioinformatics pipeline) of NGS can be divided into four primary operations: base calling, read alignment, variant identification, and variant annotation.15,16 A wide number of commercial, open source, and laboratory-developed resources are available for each of these steps. Although detailed information on the requirements of the data analysis pipeline is available, two general points need emphasis. First, it is well established that the four main classes of sequence variants (SNVs, indels, CNAs, and SVs) each require a different computational approach for sensitive and specific identification. Second, the range of software tools, and the type of validation required, depends on assay design. For SNV detection, many popular NGS analysis programs are designed for constitutional genome analysis with algorithms that may ignore SNVs with variant allele frequencies (VAFs) falling outside the expected range for homozygous and heterozygous variants. Published comparisons of various bioinformatics tools for SNV detection may be helpful.17,18
Alignment of indel-containing sequence reads is technically challenging, and algorithms specifically designed for the task are required. One such specialized approach is called local realignment, which essentially tweaks the local alignment of bases within each mapped read so as to minimize the number of base mismatches.19 Probabilistic modeling based on mapped sequence reads can be used to identify indels that are up to 20 bp, but these methods do not provide an acceptable sensitivity for detection of larger indels, such as FLT3 internal tandem duplications that may exceed 300 bp in length.20 Split-read analysis approaches to indel detection use algorithms that can appropriately map the two ends of a read that is interrupted (or split) by insertion or deletion. These algorithms can also manage reads that have been trimmed (soft-clipped) because of misalignments caused by indels.20,21
Although less common than SNVs, CNAs account for the majority of nucleotide differences between any two genomes because of the large size of individual CNAs.22 Detection of CNAs is conceptually different from identification of SNVs or indels because the individual sequence reads arising from CNAs often do not have sequence changes at the bp level but instead are simply underrepresented or overrepresented. Assuming deep enough sequencing coverage, the relative change in DNA content will be reflected in the number of reads mapping within the region of the CNA after normalization to the average read depth across the same sample.23–25 Analysis of allele frequency at commonly occurring SNVs can be a useful indicator of CNAs or loss of heterozygosity in NGS data.26
Finally, detection of SVs also presents some challenges. The breakpoints for interchromosomal and intrachromosomal rearrangements are usually located in noncoding DNA sequences, introns of genes, often in highly repetitive regions, and therefore are difficult to both capture and to map to the reference genome. In addition, SV breakpoints often contain superimposed sequence variation ranging from small indels to fragments from several chromosomes.27,28 Discordant mate-pair methods (with analysis of associated soft-clipped reads) and split-read methods can be used to identify SVs,29–32 and often provide single base accuracy for the localization of the breakpoint, which is a significant advantage in that such precise localization of the breakpoint facilitates orthogonal validation by PCR. Multiple tools should be evaluated to determine which has optimal performance characteristics for the particular assay under consideration, because, depending on the design of capture probes and specific sequence of the target regions, different SV detection tools have large differences in sensitivity or specificity.
Detection of SVs using RNA (cDNA) as starting material uses different bioinformatics approaches, especially when it is performed using amplification-based sequencing. In this case, fused transcripts are aligned to a gene reference of targeted chimeric fusion transcripts.
Considerations for Test Development, Optimization, and Familiarization
Designing Panel Content
Targeted NGS panels can range from hotspot panels focused on individual codons to more comprehensive panels that include the coding regions of hundreds of genes. When designing the NGS panel content, it is important to understand the panel’s intended use. Is it going to be used to search for therapeutic targets and enrolling a patient in a specific clinical trial? Such panels are usually designed as pan-cancer panels and contain a large number of genes, including many genes with scientific evidence of therapeutic response. Panels designed for diagnosis and patient prognostication are usually tumor specific, tend to be smaller in size, and include only those genes that are directly implicated in the oncobiology of the tumor. Overall, selection of specific genes and determining the number of genes in the NGS panel has to be thoroughly considered by laboratories during test development. The scientific evidence for including specific genes in a panel needs to be documented in the validation protocol. The size of the panel may affect sequencing reagent cost, depth of sequencing, laboratory productivity, and complexity of analytical and clinical interpretation. It is recommended to include only those genes that have sufficient scientific evidence for the disease diagnosis, prognostication, or treatment [eg, professional practice guidelines, published scientific literature, test registries (eg, National Center for Biotechnology Information Genetic Testing Registry, http://www.ncbi.nlm.nih.gov/gtr and Eurogen Tests, http://www.eurogentest.org/index.php?id=160, both last accessed January 8, 2016)].33
We recommend that the laboratory should determine gene content based on available scientific evidence and clinical validity and utility of the NGS assay. The scientific evidence used to support NGS panel design should be documented in the validation protocol.
Choosing Sequencing Platform and Sequencing Method
When deciding on a clinical sequencing platform and method, there are numerous considerations that must be taken into account. Important components in the decision-making process include required turnaround time, samples to be tested, required sensitivity, expected volume of testing, type and complexity of the genetic variants to be assessed, degree of bioinformatics support, infrastructure, and resources available in the laboratory (particularly computational resources), and expenses associated with the instrument and test validation. Other considerations include, but are not limited to, the ability to achieve a simple and reproducible workflow in the clinical laboratory, regulatory issues, and reimbursement. Choice of sequencing method will also depend highly on the number of genes required for the panel and the specific needs for the regions of interest.
At the time of this article’s preparation, the most commonly used NGS platforms in a clinical laboratory include the Illumina series and ThermoFisher’s Ion Torrent series. Each platform has pros and cons; therefore, good knowledge of the limitations and advantages of each is important. Illumina platforms provide high versatility and scalability to perform a wide spectrum of assays from small and targeted panels to highly comprehensive. However, they require higher DNA and RNA input and have longer sequencing time. Illumina instruments also require more comprehensive bioinformatics support and are associated with higher cost of instruments. Ion Torrent series, on the other hand, has much shorter sequencing time and may be the platform of choice for many institutions to run small gene panels (<50 genes) and on samples with limited amount of DNA or RNA (ie, biopsy specimens). In addition, this platform is less expensive and comes with sufficient build-in bioinformatics pipelines. However, Ion Torrent series have increased error rate in homopolymer regions and have low scalability.
We recommend that the laboratory directors should consider the following during clinical NGS platform selection: size of the panel (number of genes and the extent of gene coverage); expected testing volume; required test turnaround time; availability of bioinformatics support; provider’s degree of technological innovation, platform flexibility, and scalability; and laboratory resources, technical expertise, and manufacturer’s level of technical support.
Assessing Potential Sources of Error during the NGS Assay Development Process
Careful evaluation of the intended use of an assay will determine potential sources of error that must be addressed. This error-based approach is explained in the Clinical and Laboratory Standards Institute guidance document EP23, which states: “the laboratory should systematically identify the potential failure modes … and estimate the likelihood that harm would come to a patient.”34,pp16 That is to say, the likelihood, detectability, and severity of harm are determined at each step throughout a process. Each source of error can then be addressed at three different levels–assay design, method validation, and/or quality control.
With a complex process such as NGS, this error-based approach to design and optimization is exceedingly important. A thorough understanding of the probability of potential failure points helps determine what level of validation and quality control is needed for particular steps in the process. It will also assist in troubleshooting errors that may arise as well as validating modifications to parts of the test system.
There are potential errors associated with the detection of somatic variants in tumor tissue by NGS that bear specific consideration. Table 1 summarizes a number of preanalytical and analytical factors that can negatively affect NGS assay performance. During the process of nucleic acid extraction, it is critical to avoid cross-contamination between samples, by changing scalpel blades between tissue dissections, wiping work surfaces frequently with bleach, and ensuring that samples are handled only one at a time. Nucleic acid yield can be a problem when working with small samples, particularly FFPE samples; therefore, optimization of the entire extraction procedure is often necessary to minimize transfers and loss of material through multiple steps.35–37 DNA obtained from older FFPE blocks (eg, >3 years) often shows evidence of deamination, which can significantly increase background noise in the final NGS reads, depending on the sequencing method used.35 Treatment with uracil N-glycolase can be helpful with such samples,37 but this may require increasing input DNA into the library step and should be validated thoroughly before being adopted routinely. Stochastic bias is also a concern when working with small samples, as the number of genome equivalents present in the sample may be insufficient to consistently detect variants with low allele burden. In addition, during library preparation, it is important to keep in mind the possible impact of amplification errors and content bias related to the library method used. Because potential sources of error can be addressed through assay design (in addition to method validation and quality controls), these should be considered early in the design phase of test development.
Table 1.
Potential Sources of Error Affecting Next-Generation Sequencing Assays Designed for Formalin-Fixed, Paraffin-Embedded Tissue
| Step | Assay design considerations | Quality assessment during Validation |
|---|---|---|
| DNA yield | Optimize extraction | Measure yield |
| DNA purity and integrity | Optimize DNA library preparation | Monitor DNA library preparation |
| Deamination or depurination | Ung treatment, duplex reads | Confirm all positives with orthogonal method |
| Contamination | Change blades during tissue dissection | No template control |
| Stochastic bias | Increase input, multiple displacement amplification, single-molecule barcoding | Sensitivity control |
| Amplification errors | High-fidelity polymerase, duplex reads | Confirm all positives with orthogonal method |
| Capture bias | Optimize enrichment, long-range PCR | Define minimum coverage, back-fill with orthogonal method |
| Primer bias and allele dropout | Assess causes of false negatives, design overlapping regions | Bioinformatically flag homozygosity of rare variants |
This is not a comprehensive list.
We recommend that the likelihood, detectability, and severity of harm of potential errors should be determined at each step. Anticipated potential errors specific to the detection of somatic variants in tumor tissue by NGS should be addressed. Potential errors should be addressed through assay design, method validation, and/or quality controls.
Optimization and Familiarization Process
Before the formal process of assay validation can begin, a phase of assay development generally referred to as optimization and familiarization (O&F) is required. O&F is the process by which physical samples, supplemented by model data sets (eg, well-curated data sets available in the public domain as well as so-called in silico mutagenized data sets), are subjected to the NGS test to systematically evaluate whether the test meets design expectations. O&F invariably uncovers unanticipated assay design and bioinformatics problems. In addition, by providing laboratory technologists with the opportunity to become familiar with the testing procedures, O&F often uncovers logistical issues. The O&F phase should address library complexity, required depth of sequence, and preliminary performance specifications using well-characterized reference materials.
Preliminary Performance Specifications
The O&F process includes all aspects of NGS test, from sample and library preparation to sequencing and variant calling. Because O&F is performed to identify unanticipated problems with an NGS test, and to make necessary test changes, by definition O&F involves running samples before the formal assay validation process begins. Therefore, it is recommended that the O&F process would involve well-characterized normal cell lines (eg, the HapMap cell line NA12878; Coriell Institute for Medical Research, Camden, NJ), tumor cell lines with well-characterized alterations, as well as patient specimens of different clinical specimen types (eg, fresh, FFPE tissue, cytology specimens as dictated by the assay’s intended use), different technologists performing the testing on different days, and so on, as part of a systematic process to seek and correct unanticipated quality issues associated with the so-called wet bench portion of the test.
Likewise, the O&F phase should include a systematic evaluation of the bioinformatics component to ensure that the pipeline performs as expected based on the sequencing depth of coverage achieved in actual testing, variant class, and VAF. The O&F phase for the bioinformatics pipeline should be performed on sequence files from physical samples, as well as on model data sets designed to challenge particular aspects of the pipeline.
Although some bioinformatics tools make it possible to detect VAFs of 1% (or even lower), validation of assays with such low levels of detection must take into account two confounding factors. First, the current intrinsic error rates of NGS library preparation approaches, sequencing chemistries, and platforms complicate reliable discovery of variants at low VAFs <2% without compromising specificity, although recent advances in NGS methods that employ unique molecular identifiers increase sequencing accuracy and permit reliable detection of low-frequency variants.38–40 Second, the presence of contaminants in clinical NGS data sets can interfere with reliable detection of low-frequency variants.41,42
In the setting of inherited disease testing, the minimum VAF (which is essentially the minimum allelic ratio for those diseases not characterized by CNAs) indicates the lowest level of mosaicism that can be detected. For testing of oncology specimens, because of the intrinsic genetic instability that is a feature of many tumor types, the minimum VAF for detection of a sequence variant is not highly correlated with the percentage tumor cellularity of the specimen or the percentage of tumor cells that harbor the sequence change.
In the setting of cancer, two different features of tumor samples affect the metrics of the limit of variant detection (namely, tissue heterogeneity in that no tumor specimen is composed of 100% neoplastic cells and tumor cell heterogeneity in that malignant neoplasms often contain multiple clones).43–45 Interpretation of test results when NGS is performed on actual tumor samples must take this heterogeneity into account because it affects the lower limit of minor variant allele detection (eg, an assay with a validated lower limit of detection of 10% VAF will fail to detect a heterozygous mutation present in 50% of tumor cells if the percentage tumor cellularity is <40%).
Whether the sample is fresh or fixed also affects the limit of detection. It is well established that formaldehyde reacts with DNA and proteins to form covalent crosslinks, engenders oxidation and deamination reactions, and leads to the formation of cyclic base derivatives,46–49 and these chemical changes can lead to errors in low coverage NGS data sets50,51 or assays designed to detect variants at low VAFs.52
Although highly optimized bioinformatic tools can be used for extremely sensitive detection of specific classes of variants in NGS data, in practice the lower limit of detection is usually defined by the intended use. As examples, among laboratories that perform NGS of tumor samples to guide targeted therapy, the lower limit of VAF that has clear clinical utility is generally in the range of 5%, but in the setting of minimal residual disease testing, accurate detection of variants at frequencies substantially <1% may be required.53
Laboratories should use reference materials composed of well-characterized normal cell lines (eg, the HapMap cell line NA12878) and allogenic or isogenic cell line mixtures to estimate performance specifications given defined quality metrics and thresholds for each type of genetic alteration intended to detect. Initially, the laboratory needs to sequence a normal reference cell line (eg, HapMap cell line NA12878) and compare the sequencing results against the reference sequence provided in an external database (eg, The International Genome Sample Resource, http://www.1000genomes.org, last accessed June 30, 2016). For each reported variant class (ie, SNVs, indels, CNAs, SVs), the performance as positive percentage agreement (PPA) and positive predictive value (PPV) has to be established and documented (Table 2).54 If this experiment does not allow documentation of multiple genomic alterations using the reference cell line because of small size of targeted regions of the NGS panel and absence of a specific variant class, a well-characterized reference material can be used [eg, National Institute of Standards and Technology Reference Material 8398 (https://www.nist.gov/programs-projects/genome-bottle, last accessed January 5, 2017), other commercially available reference materials]. Next, a mixing experiment of reference cell lines (eg, HapMap cell lines NA12878 and NA12877) needs to be performed. If a mix of HapMap cell lines does not allow testing for all types of genetic alterations intended to be detected by the panel, a mixture of tumor cell lines with well-characterized alterations can be used (eg, SW620 cell line; ATCC, Manassas, VA). These reference materials can be used to estimate the performance for different variant types and to evaluate the limits of detection (LODs). The Working Group has developed a series of templates and resources to assist the laboratory in documenting both mixing studies and performance specifications (AMP Validation Resources, http://www.amp.org/committees/clinical_practice/ValidationResources.cfm, last accessed August 22, 2016).
Table 2.
Grid to Assist Laboratories in Calculation of Positive Percentage Agreement and Positive Predictive Value
| Next-generation sequencing testing result | Orthogonal method positive | Orthogonal method negative | Total |
|---|---|---|---|
| Positive | A | B | A + B |
| Negative | C | D | C + D |
| Total | A + C | B + D | A + B + C + D |
Positive percentage agreement (PPA; PPA = [A/(A + C)]).
Positive predictive value (PPV; PPV = [A/(A + B)]).
Reproduced and modified from G. A. Barnard.54 Reproduced with the permission of the Council of the Royal Society from The Philosophical Transactions © Published by Oxford University Press.
We recommend that the O&F phase must be performed before NGS test validation, the wet bench protocol and the bioinformatics pipeline should be established for all clinically relevant variant types (eg, SNVs, indels, CNAs, SVs), reference cell lines or reference materials should be used for initial evaluation of panel performance, mixing studies should be performed to estimate assay performance for each variant type intended for clinical use, and whenever possible, all specimen types and preparations intended for clinical use should be tested during O&F phase to evaluate the sequencing process in an end-to-end manner.
Library Complexity
At a specific depth of sequence coverage, the number of independent DNA template molecules (sometimes referred to as genome equivalents) sequenced by the assay has an independent impact on variant detection. For example, the information content of 1000 sequence reads derived from only 10 genome equivalents of a heterogeneous tumor sample (via a higher number of amplification cycles) is less than the information content of 1000 sequence reads derived from 100 different genome equivalents (via a lower number of amplification cycles). The number of unique genome equivalents sequenced by the assay is often referred to as the library complexity. The library complexity affects objective measures of assay performance in ways that are analogous to the impact of depth of sequence coverage.
It is well recognized that quantitation of input nucleic acid by simple measurement of the mass of DNA in the sample often provides an unreliable estimate of library complexity because the efficiency with which a given mass of DNA can be sequenced is variable. This is not surprising given the wide range of preanalytic variables that affect DNA quality (eg, the presence or absence of fixation; the type of fixative; the length of fixation). Measurement of library complexity is straightforward in a hybrid capture–based assay because the sequence reads have different 5′ and 3′ termini reflecting the population of DNA fragments captured during the hybridization step. However, measurement of library complexity in a DNA library produced by an amplification-based method is more difficult because all amplicons have identical 5′ and 3′ termini regardless of the size of the population of DNA fragments from which they originated. Dilution experiments using various amounts of DNA input during assay O&F phase can provide data on library complexity.
Traditionally, methods for accurate measurement of amplifiable input nucleic acid involve a quantitative PCR–based approach to measure the cycle threshold of amplification of the test sample. The quantitative PCR approach is cumbersome in routine practice, and indirect. Methods that involve unique molecule identifiers or single molecule tags (eg, single-molecule molecular inversion probes38,40,55; HaloPlex target enrichment system5,56) make it possible to directly measure library complexity. The method selected for evaluation of library complexity should be at the discretion of the laboratory director.
We recommend that the library complexity should be evaluated by dilution experiments using various amounts of DNA input during assay O&F phase or by other methods involving unique molecular identifiers as per discretion of the laboratory director.
Establishing Criteria for Depth of Sequencing
Depth of sequencing, or depth of coverage, is defined as the number of aligned reads that contain a given nucleotide position, and bioinformatics tools are extremely dependent on adequate depth of coverage for sensitive and specific detection of sequence variants. The relationship between depth of coverage and the reproducibility of variant detection from a given sample is straightforward in that a higher number of high-quality sequence reads lends confidence to the base called at a particular location, whether the base call from the sequenced sample is the same as the reference base (no variant identified) or is a nonreference base (variant identified).17,57–59 However, many factors influence the required depth of coverage, including the sequencing platform,9 the sequence complexity of the target region (regions with homology to multiple regions of the genome, the presence of repetitive sequence elements or pseudogenes, and increased guanine-cytosine content).57,58 In addition, the library preparation used for target enrichment and the types of variant being evaluated are important considerations. Thus, the coverage model for every NGS test must be systematically evaluated during assay development and validation.
In general, a lower depth of coverage is acceptable for constitutional testing where germline alterations are more easily identified because they are in either a heterozygous or homozygous state. However, in the setting of constitutional testing, the presence of mosaicism may complicate the interpretation of the presence (or absence) of a variant, which is not a trivial issue because it is clear that a large number of diseases are characterized by mosaicism [eg, neurofibromatosis type 1 (NF1)60; McCune-Albright syndrome61; PIK3CA-related segmental overgrowth62]. A minimum of 30× coverage with balanced reads (forward and reverse reads equally represented) is usually sufficient for germline testing.63,64 In contrast, much higher read depths are necessary to confidently identify somatic variants in tumor specimens because of tissue heterogeneity (malignant cells, as well as supporting stromal cells, inflammatory cells, and uninvolved tissue), intratumoral heterogeneity (tumor subclones), and tumor viability. An average coverage of at least 1000× may be required to identify heterogeneous variants in tissue specimens of low tumor cellularity. For NGS of mitochondrial DNA, an average coverage of at least 5000× is required to reliably detect heteroplasmic variants.65,66
The required depth of coverage can be estimated based on the required lower limit of detection, the quality of the reads, and tolerance for false-positive or false-negative results. Base calls at a specified genomic coordinate are fundamentally different from many quantitative properties that involve measurement of continuous variables, such as serum sodium concentration. Instead, each base call in a DNA sequence is a so-called nominal property in that it is drawn from a limited set of discontinuous values. This has implications for the statistical calculation of assay metrics.19,67–70
These performance parameters can and should be estimated during the development phase to help define acceptance criteria for validation. For example, for a given proportion of mutant alleles, the probability of detecting a minimum number of alleles can be determined using the binomial distribution equation:
| (1) |
Where P(x) is the probability of x variant reads, x is the number of variant reads, n is the number of total reads, and p is the probability of detecting a variant allele (ie, the proportion of mutant alleles in the sample). Excel allows one to calculate the binomial probability directly using the following formula: =BINOM.DIST(number_s, trials, probability_s, cumulative), where number s is the number of successes (x in the binomial equation), trials is the number of reads (n in the binomial equation), probability_s is the probability of success (p in the binomial equation), and cumulative refers to whether the determination should be the exact probability for a given x and n (FALSE) or the cumulative probability (TRUE).
By calculating the binomial probability for a given number of trials and probability of successes, one can define the binomial distribution (Figure 2). For example, for a given mutant allele frequency of 5% and 250 reads, the probability of detecting four or fewer mutations would be 0.457%. Therefore, the probability of detecting of five or more mutations is 1 minus 0.457% (or 99.543%). Thus, if the threshold for a variant call were set at five or more reads, the probability of a false negative would be <0.5% provided a minimum of 250 reads were obtained. For clinical NGS panels, a minimal depth of coverage of 250 reads per tested amplicon or target is strongly recommended.
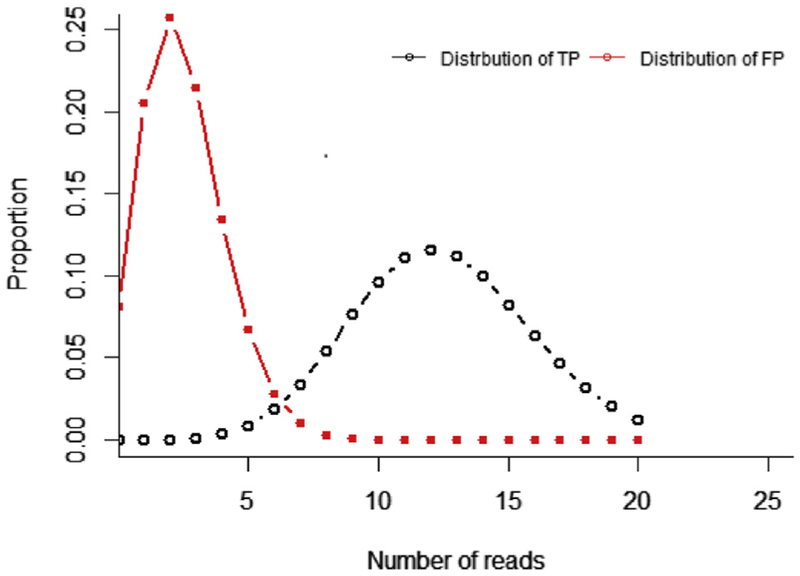
Figure 2.
Determining depth of sequence. Given an allele burden of 5% and 250 read depth, the binomial distribution of true positives (TPs) can be calculated. Also, given a sequence error rate of 1%, the binomial distribution of false-positive (FP) results can also be calculated and shown to overlap the true positive distribution. The overlap of true-positive and false-positive distributions should be considered when determining minimum depth of sequence needed to reliably detect a given allele burden.
The binomial probability distribution could also be used to calculate the probability of a false positive for a given error rate and threshold for variant calling (Figure 2). In this case, the probability of success would depend on the error rate. For example, if the test system has an error rate of 1% (ie, a sequence quality equivalent to a Phred Q score of 20), the probability of getting five or more errors at a particular base would be 10.78%. However, if the threshold was set at five or more reads, that rate of false positives would not be realized assuming the nucleotide errors would be random. For example, the probability that five or more random errors would all have the same nucleotide change is 0.01%. Of course, raising the threshold or reducing the error rate could reduce the probability of false positives yet further.
However, not all errors are random and platform-specific systemic errors do occur. Therefore, estimation of needed depth of coverage using the binomial distribution is only an estimate and determination of false-positive and false-negative rates for a given depth and threshold must be validated.
We recommend a minimal depth of coverage >250 reads per tested amplicon or target for somatic variant detection. In certain limited circumstances, minimal depth <250 reads may be acceptable but the appropriateness should be justified based on intended limit of detection, the quality of the reads, and tolerance for false-positive or false-negative results.
NGS Test Validation
After determining initial assay conditions and establishing bioinformatics pipeline configurations during O&F phase, the NGS test needs to be validated. Regulatory requirements under Clinical Laboratory Improvement Amendments call for all non–Federal Drug Administration–approved/cleared tests (alias laboratory-developed procedures or tests) to address accuracy, precision (repeatability), reportable range, reference range (normal range), analytical sensitivity (limits of detection or quantification), analytical specificity (interfering substances), and any other parameter that may be relevant (eg, carryover). Various guidance documents provide definitions for these performance characteristics, but the definition of accuracy can be refined to better meet the needs for NGS somatic analysis.3,71,72 Therefore, it is recommended that accuracy should be stated in terms of PPA and PPV. NGS panel validation should include the outlined validation protocol with defined types and number of samples, established PPA and PPV, reproducibility and repeatability of variant detection, reportable range, reference range, limits of detection, interfering substances, clinical sensitivity and specificity, if appropriate, validation of bioinformatics pipelines, and other parameters as described below.
Validation Protocol
The validation protocol should be completed before accumulating validation data. That is to say, data collected during development, optimization, and familiarization are not part of the validation. However, those data can be used to estimate test performance and thereby set performance criteria for acceptance as well as determine the number and types of samples as discussed below.
The validation protocol should start with an explicit statement of the intended use, which will determine the types of samples and the performance characteristics that need to be addressed. For example, a test that is intended to detect known hotspot mutations, including large insertions or deletions in formalin-fixed tissue, will need to include formalin-fixed samples with these types of mutations. The lower limit of detection that is clinically indicated should also be defined.
Careful design of the validation protocol is necessary to ensure that all relevant parameters are addressed as efficiently as possible. It may be helpful to include a validation matrix of the planned validation (Table 3). The validation protocol needs to be approved by the laboratory director before validation begins. Ideally, the standard operating procedures for generating sequence data and bioinformatics analysis, as well as the validation samples, should be given to technologists that were not involved in the development or optimization of the test so that they can acquire the validation data in a blinded manner. However, it is recognized that not all laboratories have sufficient staffing to support this approach. The Working Group has developed a template to assist the laboratory in documenting and describing studies performed in the validation phase (AMP Validation Resources, http://www.amp.org/committees/clinical_practice/ValidationResources.cfm, last accessed August 22, 2016).
Table 3.
Sample Next-Generation Sequencing Validation Matrix
| Next-generation sequencing run | Technologist | Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| 1 | A | PS1 | LOD1 | PS2 | PS3 | PS4 | LOD1 | PS5 | PS6 | PS7 | PS8 | PS9 | PS10 |
| 2 | B | PS11 | PS12 | LOD2 | PS13 | PS14 | PS15 | LOD2 | PS16 | PS17 | PS18 | PS19 | PS20 |
| 3 | A | PS21 | PS22 | PS23 | LOD3 | PS24 | PS25 | PS26 | LOD3 | PS27 | PS28 | PS29 | PS30 |
| 4 | B | PS31 | PS32 | PS33 | PS34 | PS35 | PS36 | PS37 | PS38 | LOD1 | PS39 | LOD1 | PS40 |
| 5 | A | PS41 | PS42 | PS43 | PS44 | PS45 | PS46 | PS47 | PS48 | PS47 | LOD2 | PS50 | LOD2 |
| 6 | B | LOD3 | PS51 | PS52 | PS53 | LOD3 | PS54 | PS55 | PS56 | PS57 | PS58 | PS59 | PS60 |
Technologist A and B represent two individual technologists performing the validation testing. A validation matrix can help ensure that all performance parameters are addressed as efficiently as possible. Testing personnel should be blinded to the identity of samples and controls.
LOD, limit of detection sample; PS, previously tested patient sample.
Types and Number of Samples Required for Test Validation
Assay validation should be performed using samples of the type intended for the assay so that test performance is representative of the larger population. However, massively parallel sequencing of multiple genes cannot be validated as if it were a single-analyte test. There is far too much variation in the types of samples, types of variants, allele burden, and targeted exons or regions. Therefore, an error-based approach to validation must be used.
To use an error-based approach, the question that must be addressed is, to what extent can the performance of the test for a given sample type, variant type, genomic region, or allele burden be extrapolated to other sample types, variant types, genomic regions, and allele burdens? Performance is certainly expected to vary considerably for different sample types, variant types, and allele burden, and therefore it is essential to establish performance characteristics by these factors. A range of well-characterized samples should be selected that maximizes the variation by these factors, as indicated given the stated intended use of the test. The number of each sample type tested during validation should be in proportion to the anticipated sample types to be tested within the clinical service. However, if a sample type is known to be problematic (eg, FFPE tissue), additional validation samples are recommended to determine the impact of the sample quality or quantity on the test results regardless of the number that are anticipated in typical patient samples.
It is recognized that for most panels it is not practical to obtain well-characterized samples representing all of the pathogenic variants that might be detected. However, laboratories should strive to include samples with hotspot mutations relevant to the test’s intended use (eg, mutations involving KRAS codons 12, 13, and 61 for a colon cancer panel). Although sourcing these samples is not trivial, it is critical that laboratories make a substantial effort to show that their assay can actually detect common and clinically relevant mutations that they state they can detect. In silico data sets can augment, but not supplant, real samples. So a mix of real and in silico samples can be envisioned.
Test performance is less likely to vary by genomic region provided that quality metrics are met (eg, read quality, read length, strand bias, read depth). However, systematic errors do occur based on regional variation (eg, repetitive sequence, pseudogenes). Therefore, it is important to include at least two well-characterized samples that have known sequence for all targeted regions. Some commercially available cell lines have been well characterized and can serve such purpose (eg, HapMap cell line NA12878). In some cases, such reference cell lines have been subjected to formalin fixation and paraffin embedding, resulting in lower-quality DNA that may more closely mimic the material intended for use with a new assay. The intent is to detect potential systematic errors that are likely to be evident because of their recurrent nature. Such errors would be seen in many samples, including those for which known sequence was not available. The cell lines with known sequence of all regions could then be used to ascertain the cause of systematic errors in these regions.
A perennial question is how many samples need to be tested. Although performance is often stated in terms of CIs, the CI of the mean only gives an estimate of the population mean with a stated level of confidence. It does not define the distribution of the underlying population and does not give an indication of the performance of any given sample.
To estimate the distribution of the underlying population and the performance of individual samples, the tolerance intervals should be used. For a normally distributed population, the lower tolerance interval could be determined: , Where is the sample mean, s is the sample SD, and k is a correction factor for a two-sided tolerance interval, and defines the number of sample SDs required to cover the desired proportion of the population. The two-sided k value for 95% confidence and n = 20 is 2.75, which is significantly higher than the 1.96 corresponding to the z-score of a normal population distribution because the tolerance interval is based on the sample size and the error-prone estimates of the underlying population mean and population SD. As the number of samples increases, the k value approaches the z-score.
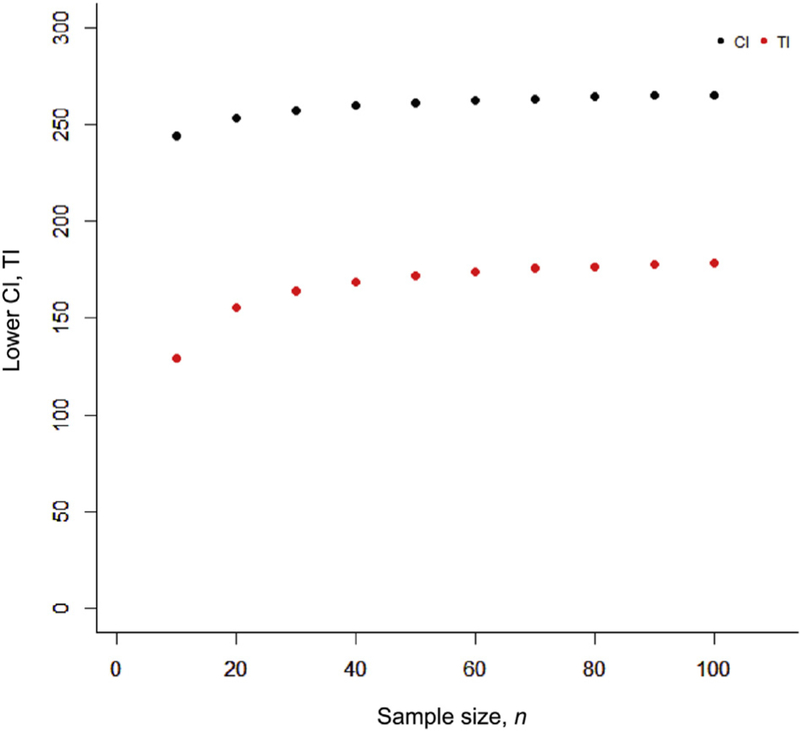
For example, perhaps we want to determine the probability of getting a minimum of 250 reads for a given region. If a validation set of samples shows a mean depth of coverage 275 reads and an SD of 50 reads, after running 100 samples the 95% lower confidence limit would indicate that we would be confident that our average depth of coverage would be >266 (Figure 3). However, our 95% lower tolerance interval indicates that for any given sample we could only be confident of reliably getting a read depth of 179 or greater (Figure 3).
Figure 3.
Determining the minimum number of reads: CI versus tolerance interval (TI). Determination of the CI and tolerance interval for minimum read depth (average of 275 reads with an SD of 50). The lower CI determines with 95% confidence the lower level of the average across the population. As the sample size increases, this estimate improves. The tolerance interval determines with 95% confidence the minimum number of reads above which 95% of the population will fall. The CI can be used to predict the average performance of a population, and tolerance interval can be used to predict the performance of a given sample.
The above estimate of the tolerance interval would only be applicable to a population that is normally distributed. However, the distribution of the underlying population is often not normal [eg, when there is a natural boundary that the data cannot exceed (ie, 0% or 100%)]. Therefore, it is helpful to define the tolerance intervals using nonparametric methods to estimate the performance parameters regardless of the distribution of the underlying population. The one-sided nonparametric tolerance interval can be determined by finding the value for k that satisfies the cumulative binomial equation.73
| (2) |
where
| (3) |
when k is an integer between 0 and n, 0 ≤ k ≤ n and CL is the confidence level (eg, 0.95). By setting k = 0 (ie, 0 failures), the formula can be simplified to: pn = 1 − CL
| (4) |
This equation is often used to determine the number of samples needed to verify a predetermined reliability and CI. For example, a one-sided tolerance interval with 95% confidence and 95% reliability could be determined by the performance on a set of 59 or more samples regardless whether that metric was parametric or nonparametric.73 This, of course, assumes that the performance of each sample is independent of others and the samples are representative of the population from which they are drawn. For example, a laboratory director may want to assess the maximum false-positive rate for his or her test (ie, false-positive variants to total number of variants per sample). After performing the test on 59 representative samples, the highest false-positive rate is 1.9%. Therefore, he or she could be 95% confident that 95% or more of his her samples will have a false-positive rate ≤ 1.9%.
By testing a minimum of 59 samples during validation, conclusions can be drawn as to the tolerance intervals of essentially any performance characteristic whether it is parametric or nonparametric in nature. It is expected that laboratories would be able to acquire quality metric data (eg, read depth, read length, bias, and quality scores) for 59 samples that contain SNVs. Ideally, these 59 samples would also have other variants such as indels. It is acknowledged that ascertainment of samples containing indels is more challenging and laboratories are encouraged to source as many samples with indels as possible to adequately determine assay performance. Variants that are more complex may be difficult to source so assay design approaches or quality controls may be needed to confidently detect these, as discussed below.
We recommend that the validation samples include previously characterized clinical samples of the specimen type intended for the assay (FFPE, blood, bone marrow); previously characterized clinical samples with each type of pathogenic alteration that the assay is intended to detect (eg, SNVs, indels, CNAs, SVs); samples with most common mutations relevant to the intended clinical use of the panel; two or more samples for which a consensus sequence has been previously established (eg, National Institute of Standards and Technology reference material) for all regions covered by the panel; and a minimum of 59 samples to assess quality metrics and performance characteristics.
PPA and PPV
PPA is the proportion of known variants that were detected by the test system. It requires that all true variants must be known. The true presence of genetic variants can be determined using reference samples or reference methods (eg, Sanger sequencing). When using reference methods, it is possible to use a combined reference method (eg, Sanger sequencing coupled with targeted mutation analysis when the allele burden is expected to be low). However, the combined reference method must be determined before collecting validation data and cannot be used for discrepancy resolution because this will bias the data.74 Because the performance will likely vary by mutation type, the PPA should be determined for each (eg, SNVs, small indels, larger indels, CNAs, SVs) (Table 2). Sourcing sufficient samples with known SNVs and perhaps small indels should not be a problem. It is not necessary that these be pathogenic variants because the goal is to demonstrate the analytical performance of the test. Nevertheless, common pathogenic variants should be included whenever possible. It may be difficult or impossible to find sufficient numbers of samples with larger indels or SVs. In such cases, the laboratory may choose to supplement the NGS test with another validated test (eg, FLT3-ITD fragment analysis, RT-PCR) until sufficient number of cases is reached, or include appropriate controls, or clearly state the test limitations within the report. When analyzing the validation data, the rate of detection of known positives for each sample should be determined and documented (ie, mean, SD, CIs, and tolerance intervals, or reliability). For example, a sample with 100,000 bp of known, targeted sequence may have 90 true SNVs or a set of 59 samples may have 62 true known variants. Assuming coverage and quality indicators met quality thresholds, the PPA would be the proportion of 90 or 62 variants that were detected, respectively. The discrepancy resolution should not be performed because it will bias the data.74
PPV is the proportion of detected variants that are true positives. Again, it requires that all true variants must be known and the true presence of genetic variants can be determined using reference samples or reference methods (eg, Sanger sequencing) (Table 2).
Similarly to PPA, the PPV should be determined for each mutation type (eg, SNVs, small indels, and larger indels, CNAs, SVs). When analyzing the validation data, the proportion of variants that are true positives should be determined. For example, a sample with 100,000 bp of known, targeted sequence may have 90 true SNVs. Assuming coverage and quality indicators for this 100,000 bp met quality thresholds, if all of these were detected and an additional 10 false-positive variants were detected, the PPV would be 90/100 or 90%. Again, discrepancy resolution should not be performed because it will bias the data74 and the overall performance for the validation set should also be determined.
We recommend that PPA and PPV should be documented for each variant type (eg, SNV, small indels, large indels, CNAs, SVs). For variant types for which 59 validation samples are not available, the laboratory should supplement the NGS test with another validated test until the number of samples reached, or to include appropriate controls, or clearly state the limitations in the report.
Repeatability/Reproducibility
Complex, multistep processes can introduce random error (or imprecision) at every step because of variation in instrumentation, reagents, and technique. To minimize variation, the instruments, reagents, and personnel must be qualified for the intended purpose. Nevertheless, variation can occur and this should be quantified through the method validation. Given the number of possible sources of variation, it is not practical to exhaustively assess all sources of variation independently. Rather, it is recommended to assess a minimum of three samples across all steps and over an extended period to include all instruments, testing personnel, and multiple lots of reagent. Replicate (within run) and repeat (between run) testing should be performed. Of course, acceptance criteria need to be set before the acquisition of validation data. For example, SNV allele frequency or CNA has to be within a specified range of variation from run to run. If acceptance criteria are not met, additional precision studies may be required to assess sources of variation. Given the extensive quality controls and quality metrics that are included in most steps, sources of variation should be identifiable and quantified.
We recommend that a minimum of three samples should be tested across all NGS testing steps to include all instruments, testing personnel, and multiple lots of reagents. Variance should be quantified at each NGS testing step for which data are available.
Reportable Range and Reference Range
The reportable range is the span of all test results that are considered valid. This should include the targeted regions that meet the minimum quality requirements, the variant types that have been validated, and the limits of detection for these. The reportable range should be included in the report, perhaps together with the methods and limitations so that it is clearly understood by the ordering provider what regions, variants, and allele burdens would not be detected.
The reference range is the range of normal values. It is not simply the reference Human Genome, which is a compilation of multiple genomes from healthy individuals, but rather the variants that are considered benign or nonpathogenic. For genetic variants, this could be difficult to define and may vary by intended use of the test. Because our understanding of genotype-phenotype correlations is far from complete, some laboratories may opt to report all detected variants, whereas others may opt to report only those that are considered clinically informative. Regardless, the reference range should be included in the report so that it is clearly understood by the ordering provider what types of variants would or would not be reported.
We recommend that the appropriate reportable range and reference range will depend on the intended use of the test and should be determined as part of the validation process. Reportable range and reference range should be included in the patient report.
Limits of Detection
The LOD for each type of genetic alteration is recommended to estimate during O&F phase using cell line mixing experiments, as described above and shown in a series of templates (AMP Validation Resources, http://www.amp.org/committees/clinical_practice/ValidationResources.cfm, last accessed August 22, 2016).
The lower LOD (LLOD) could be defined as the minor allele fraction at which 95% of samples would reliably be detected. Often, a laboratory director may choose to run 20 validation samples to demonstrate the LLOD assuming that 19 or 20 correct results would indicate a reliability ≥95%. However, by testing just 20 samples, the director could not be confident the test would reliably detect 95% or more samples at that lower limit of detection. If the true reliability were 90%, what would be the probability of getting 20 of 20 correct? That could easily be calculated as (0.90)20 and would show that the probability of getting 20 of 20 correct when the reliability is 90% would be 12%. In other words, it would not be particularly unlikely and 20 samples therefore would not give confidence that the reliability is at least 95%. If we had run 100 samples and all were correct, we could show that (0.90)100 would equal 0.003%, which would be unlikely. We would therefore feel confident that our reliability must be >90%.
The number of samples that would be required could therefore be calculated by defining the reliability and confidence that we would like to demonstrate: rn = α, where r is the reliability, n is the number of samples, and α is the confidence level (ie, probability of a type I error). By solving for n, it can be shown:
| (5) |
If we want to be 95% confident (α = 0.05) of at least 95% reliability (r = 0.95), the minimum number of samples could be calculated to be 59. If we wanted more reliability or confidence, of course that number of samples would be more and could be calculated. This minimum number of samples assumes that all results are correct. If a proportion is incorrect, the reliability together with CIs could be calculated using one of several complex statistical methods.73
Interestingly, the natural log of 0.05 is −3.00. Therefore, for a 95% confidence level, the rule of three can be applied.74,75 For example, for fluorescent in situ hybridization test, a director may choose to count 100 cells and seeing no translocation, claim that <1% of cells have the translocation. However, to be 95% confident that <1% of cells have the translocation, 300 cells would have to be counted without a single positive. Likewise, if a director wants to claim a reliability ≥95% with 95% confidence, he or she would need to test 3 × 20 or 60 samples. The rule of three is convenient and an accurate estimate of the binomial CI for sample sizes of 30 or more.
Different mutation types would likely have different lower limits of detection and therefore LLOD should be determined for each variant type. Of course, it may prove difficult to source 59 or more validation samples with the targeted mutations and VAF needed to validate the LLOD. Therefore, sensitivity controls would be needed to ensure detection of targeted mutations at the LLOD.
Plasmid controls and other artificial constructs may be used during validation and clinical testing to demonstrate accurate detection of certain variants. However, plasmids are far less complex than typical clinical samples, and it has been shown that they are more readily detected at a given allele burden.75 More recently, it has been shown that linearized plasmid controls performed with similar efficiency as formalin-fixed cell line genomic DNA and could be used to assess or monitor LLOD provided that the genomic DNA and plasmids were fragmented to comparable size.76 Therefore, plasmid controls and other artificial constructs potentially could be used for the validation of the LLOD and the monitoring of assay performance.77
We recommend that the LLOD for each variant type should be determined. A minimum of 59 samples should be used to establish the LLOD. If sufficient samples cannot be sourced, sensitivity controls should be used.
Interfering Substances and Carryover
Interfering substances that are known to affect molecular testing, particularly amplification-based methods, should be addressed during validation (eg, heavy metal fixation, melanin, hemoglobin). Nucleic acid extraction and purification steps normally eliminate possible contaminants. However, consideration must be given to the types of samples that are used in the validation to be sure all intended types can be amplified and sequenced. For example, melanomas need to be included in the validation if the intended use is to detect BRAF mutations. In addition, consideration should be given to interference from repetitive sequence and pseudogenes. For highly fragmented DNA, short reads can be misaligned if derived from pseudogenes and yield false-positive results. Alternatively, highly repetitive sequences may reduce on-target reads by depleting capture probes.
Carryover is a recognized problem with NGS sequencing tests that are designed to detect variants with low allele burden. Carryover should be addressed through test design and validation as well as through the inclusion of no template controls (NTCs). Bioinformatics approaches have been described that can be used to detect human-human sample contamination, and should also be used to monitor carry-over.42,78 During test design, procedures should be in place to avoid carryover from one sample to another (eg, changing scalpel blades between samples). In addition, during validation and every clinical run thereafter, an NTC should be included in every run to verify no carryover from neighboring wells during amplification steps. It is not necessary to take the NTC all of the way through sequencing as a quality check on amplified product may suffice.
We recommend that the possible sources of test interference should be specifically identified, with the impact of each systematically evaluated during validation. The risk of carryover should be evaluated at each step of the assay. A no template control should be included in every run but need not be evaluated all of the way through sequencing.
Clinical Validation and Clinical Utility
The quantitative analytical performance of a laboratory test, in this case an NGS test, does not necessarily predict performance at a clinical level. The intrinsic biological variability of disease has the greatest impact on the clinical sensitivity and specificity of NGS testing. The intrinsic biological variability results in a scenario in which, generally, only a subset of cases of a specific disease or tumor type harbors a characteristic mutation; more than one characteristic genetic abnormality is associated with a specific disease or tumor type; more than one disease or tumor type can share the same mutation and the clinical significance of the same mutation will be different for different tumor types (eg, theranostic significance of epidermal growth factor receptor mutation will be different when detected in lung adenocarcinoma versus glioma); a mutation that is characteristic of a tumor type or disease can be identified in a subset of healthy individuals; and so on.33 Thus, even an NGS assay with perfect analytical performance (ie, 100% analytical sensitivity and 100% analytical specificity) may have a lower clinical sensitivity and specificity and the differences between analytical performance and clinical performance have to be recognized. What is the intended use of the panel? Is it going to be used for tumor diagnosis, prognostication, or making therapeutic decisions? Some NGS assays will sequence a more limited target region (eg, a hotspot test), which might be sufficient if intended use of the panel is limited to specific genetic alterations (ie, KRAS/NRAS mutations in colorectal cancer for guiding treatment with cetuximab), but this panel might have a lower clinical sensitivity than a more comprehensive test if intended use of this panel is tumor diagnosis and prognostication because other variants outside the hotspot region will not be detected. Similarly, a test that detects a limited range of variant types (eg, only SNVs and small indels) might have a lower clinical sensitivity (but higher clinical specificity) than a test that also detects larger indels, CNAs, and SVs. Therefore, the clinical validity (ie, clinical sensitivity and specificity) and clinical utility of the NGS test needs to be determined during the assay design and evaluated during the validation process. The validation protocol has to include the NGS panel content, the intended use of the test, potential clinical validity and utility, and references.
In addition, if the intended use of the NGS panel is to establish diagnosis of cancer based on combination of genetic alterations or this is a multianalyte NGS test with a prediction algorithm, a full scale of clinical validation using defined patient cohort is required and has to be performed to ensure the diagnostic accuracy of the panel. For such NGS tests, the same guidelines and calculations as outlined for analytical validation (listed above) should be followed.
We recommend that clinical validity (ie, clinical sensitivity and specificity) and clinical utility of the NGS assays needs to be defined during design of the test and need to be evaluated during the validation process. The validation protocol has to include the NGS panel content, the intended use of the test, potential clinical validity and utility, and references. Full scale of clinical validation is required for multianalyte NGS tests with prediction algorithms and should be performed using the guidelines and calculations as defined for an analytical validation.
Validation of Bioinformatics Pipelines
The current discussion is limited to those general aspects of NGS bioinformatics that affect assay design and clinical use, and that therefore must be considered as part of overall assay validation. Detailed discussion of the validation requirements for clinical bioinformatics pipelines for variant detection will be reported in a separate companion article currently under development by an AMP Clinical Practice Committee Working Group.
Given the number of genes and the range of mutations for which testing is performed by NGS, it is impractical (if not impossible) to follow an analyte-specific validation approach. For this reason, methods-based paradigms have been developed, which are centered on the method of analysis rather than the specific analyte being tested,79,80 and several different reagents can be used to validate the bioinformatics (BI) pipeline via a methods-based paradigm.
Because cell lines that are genetically well characterized are an inexhaustible reagent, they are a particularly useful source of reference material for assay validation, especially for characterization of assay sensitivity and limit of detection. The Centers for Disease Control and Prevention’s genetic reference material coordination program (https://wwwn.cdc.gov/clia/Resources/GETRM/default.aspx, last accessed January 5, 2017) has evaluated several well-characterized cell lines for various variants specific to many genetic conditions, as have the National Institute of Standards and Technology and several commercial vendors (AMP Validation Resources, http://www.amp.org/committees/clinical_practice/ValidationResources.cfm, last accessed August 22, 2016). Likewise, DNA oligonucleotides or plasmids can be engineered to incorporate specific sequence variants, at known ratios, at known positions, and in known allelic ratios, to simultaneously evaluate many aspects of BI analysis.76,81 A novel type of approach that can be used for BI validation parallels so-called in silico proficiency testing.79,82 By this approach, the actual sequence files from NGS of a well-characterized specimen are manipulated by computerized algorithms that introduce relevant sequence variants into the reference sequence files83,84; the resulting simulated files are an ideal reagent for BI validation because they challenge every step in the BI pipeline from alignment through variant detection, annotation, and interpretation; can be designed for all four major classes of variants, either alone or in combination, at any VAF; and can be developed for any genetic locus, either alone or in combination to generate complex mixtures of variants that mimic clinical samples.
We recommend that available reference materials that are appropriate for the type of variants, and their anticipated ranges, should be used to validate the bioinformatics pipeline.
Validating a Modified Component of a Test or Platform
NGS is an evolving technology with frequent improvements in techniques and technology. After completion of test validation, the laboratory may face a necessity to modify or improve one or more components of the test (eg, add additional genetic locus, use new version of reagents or bioinformatics pipeline, or move validated assay to the new model of the sequencing instrument). If any component of the test is to be changed, consideration must be given to the potential errors that may be introduced. For some changes, the potential errors may be minimal or readily detected using quality control procedures should they occur. For example, changing an extraction protocol may pose little risk if quality metrics are routinely used to verify the purity, concentration, and integrity of every sample. On the other hand, some changes, like a change in platform, may require a complete validation to be performed. It is the responsibility of the laboratory director to determine the potential error of any change in protocol and provide documented evidence that potential errors are unlikely to occur or should be readily detected so patients are not harmed. Regardless of the significance of the change, it is recommended that verification of continued performance include testing of at least several known samples in an end-to-end manner to help detect unanticipated errors.
When new genes or genomic regions are added to the existing test, the laboratory should perform a supplemental validation analyzing a number of samples and establishing that the performance of the panel was not altered by the newly added genes and the performance characteristics of newly added genes are as expected. However, it does not require a full validation process as for the original test. It is the responsibility of the laboratory director to determine the number of samples that are required to demonstrate that panel’s performance is not affected by the addition of new sequencing targets and to provide documented evidence.
We recommend that any additions or changes to a validated NGS test should have a supplemental validation, including a validation protocol and summary. Potential sources of error that may result from the changes should be assessed during the supplemental validation. The supplemental validation should include a number of known samples in an end-to-end manner. Samples selected for this process should specifically address potential errors that are introduced by the modifications.
Implementation and Quality Control Metrics
The implementation of NGS technology in the clinical diagnostic environment is complex and requires significant changes in the infrastructure of the laboratory. Because of the marked differences compared to older technology, the technical aspects of quality management for test and system validation, quality control, quality assurance, and the measurement of quality characteristics is more challenging and with degrees of complexity that vary depending on the platform and comprehensiveness of the assays. Quality control measures must be established to the specific assay and these quality control measures must be applied to every run.
Specimen Requirements
Specimen requirements for NGS can be highly variable depending on the disease setting, individual practices of acquisition, tissue handling, processing protocols, and testing method. During NGS assay validation, laboratories must validate all potential specimen types (ie, FFPE, fresh tissue, blood/bone marrow, cell-free DNA) and establish criteria for specimen acceptability. Nonvalidated specimens should be considered as inappropriate for NGS testing and rejected. Laboratories must also establish the individual requirements of minimum tumor content to match the sensitivity of the assay and qualification criteria for quality and quantity of DNA based on the assay validation to ensure maximal performance and accurate results. Samples with tumor content below the established cutoff should be considered as unacceptable for testing. Before testing, samples should be reviewed by an appropriately trained and board-certified pathologist for specimen suitability, including specimen type, tumor quality and quantity, and selection of areas for macrodissection/microdissection. Minor deviations from the established validation criteria may be warranted in rare cases; the acceptability and reporting of these cases are at the discretion of the laboratory director or designee.
We recommend that the laboratory should monitor specimen acceptability. Specimens that are not validated for NGS assay should be rejected. Before testing, tumor tissue samples should be reviewed by an appropriately trained and board-certified pathologist for specimen suitability and selection of areas for macrodissection/microdissection. Enrichment for neoplastic cells should be considered (eg, macrodissection/microdissection). For liquid samples, flow cytometry or other methods should be used to evaluate the sample’s percentage of neoplastic cells. Laboratories should archive either a representative slide or image of the tissue tested.
DNA Requirements
DNA quantification is a critical component in NGS, which not only affects the sensitivity of the assay but also the number and the accuracy of the variants reported. As DNA quality and quantity can vary widely based on the source and extraction method, and requirements may differ depending on the NGS method, each laboratory must establish a standardized and cost-effective workflow for nucleic acid quantitation to guarantee accurate and reproducible results. Quality control methods must be well developed to clearly define the preanalytical sample quality, accurately assess the minimum amount of DNA required to detect a variant depending on the sensitivity of the assay, and must establish a way to adjust DNA inputs.
There are multiple methods of DNA and RNA quantification. Spectrophotometric methods, although the most commonly used across laboratories, should be avoided. These measure total nucleic acids (including double-stranded DNA, single-stranded DNA, oligo, and free nucleotides) as well as impurities that markedly overestimate amplifiable DNA or RNA content. Double-stranded DNA–specific fluorometric quantitation methods are recommended to inform DNA input. Fluorescence-based DNA measurements are far lower than those quantified by spectrophotometry, but results are more accurate and precise, particularly at lower concentration ranges.
We recommend that the laboratory should monitor quality and quantity of nucleic acids (DNA, RNA) using fluorometric quantitation methods.
Library Qualification and Quantification Requirements
Accurate library qualification and quantification are pivotal to obtaining optimal NGS data. Underloading or using libraries with inappropriate fragment size ranges can lead to reduced coverage and read depth. Conversely, overloading would cause saturation of the flow cell or beads and lead to read problems.
Library qualification should be performed to detect potential problems such as high percentage of short DNA fragments or adapter dimers. Electrophoretic methods are generally used to assess the overall range of DNA fragment sizes constituting the library. The laboratory must standardize protocols to obtain fragment sizes of the expected molecular weight narrow range. Specific measures should be taken to minimize primer dimers, adapter dimers, and broader bands of higher molecular weight. Primer dimers that are usually minimized by the use of magnetic beads do not constitute a significant problem unless they dominate the reaction. Adapter dimers, however, can be problematic as they sequence much more efficiently, leading to much higher percentage in final data files.
Library quantification is recommended, and the laboratory should decide on the method depending on what is suitable for their needs. Real-time quantitative PCR assays that use primers specific for the adapter sequence are a common method of quantification, as only sequences that have adapters and thus will be sequenced and included in the measurement. Digital PCR assays also targeting the adapter sequence provide an alternate method with increased accuracy by providing absolute quantification.
We recommend that library qualification should be performed. An appropriate method for quantitation should be defined during the O&F and validation phases.
Core Metrics of Analytical Performance Quality Control
The performance requirements for the assay must be established during the validation procedure, and the same specifications must be used to monitor the performance of the assay each time a sample is processed. Given the inherent differences among platforms, specific applications and informatics tools, specific recommendations for ranges and thresholds cannot be offered and each laboratory must define the criteria and means to monitor all quality metrics to ensure optimal analytical performance. Quality metrics that require ongoing review are summarized in Table 4.
Table 4.
Core Quality Metrics for Validation and Ongoing Quality Control
| Core quality metric | Validation parameters | Ongoing quality control |
|---|---|---|
| Nucleic acid quality and quantity | Requirements of DNA quality and quantity differ depending on the tissue source, extraction method, and next-generation sequencing method. Each laboratory must establish:
|
A plan for ongoing monitoring must be established. Any changes on extraction protocols should be followed by close monitoring of all downstream processes to ensure adequate performance of the assay. |
| Library qualification and quantification | The laboratory must standardize protocols for library qualification and quantification. Library quantification is recommended—each laboratory must validate a method that is suitable for its needs. |
The fragment sizes of the library must be measured to ensure they fall within the expected molecular weight narrow range. Ongoing measures should be taken to minimize primer dimers, adapter dimers, and broader bands of higher molecular weight. |
| Depth of coverage | Requirements vary depending on the platform used and the application. Coverage must be defined to achieve adequate sensitivity and specificity in the regions of interest. Each laboratory must establish the minimum criteria for depth of coverage characteristic of a particular region under standard assay conditions (coverage threshold). |
Ongoing measures should be taken to monitor the overall coverage and region coverage in each run. If coverage thresholds are outside the validated range, the samples should be subjected to reanalysis. If only local regions are affected, testing of that region by an alternate method may be performed. |
| Uniformity of coverage | The required level of coverage across the targeted regions must be defined during the vaLidation stage. | The uniformity of coverage must be monitored and compared to the levels established during the validation. If the coverage uniformity profile falls outside of the expected profile as established by the validation, this may be indicative of errors during the testing process. |
| GC bias | GC content affects sequencing efficiency and the uniformity of coverage of the targeted regions. The extent of GC bias in all parts of the genome included in the assay should be determined during validation. | GC bias should be monitored with every run to detect changes in test performance or sample quality issues. |
| Cluster density and alignment rate | The laboratory must define the right balance between overclustering and underclustering and outline the steps to prevent and resolve both instances. As a general guideline, the percentage of clusters passing filter should be >80% and alignment rate should be >95%. | Cluster density and alignment rate should be monitored in every run. |
| Transition/transversion ratio | Important parameter for whole-exome or whole-genome sequencing. Not required for targeted panels. The ratio of transitions/transversions should be comparable to published values. | The transition/transversion ratio should be monitored with every sample to assess test performance. Ratios lower or higher than expected may indicate that the quality of base calls was Low. |
| Base call quality scores | The Laboratory must establish acceptable raw base call quality score thresholds for the assay during validation. Preprocessing methods to remove low-quality base calls should be established to reduce the false-positive rate. |
Quality scores and quality of signal/noise ratio should be monitored in every run. Low-quality scores can lead to increased false-positive variant calls; thus, result must be interpreted with caution and repeat testing may be indicated. |
| Mapping quality | Parameters for mapping quality must be established during validation and should demonstrate that the test only analyzes reads that map to the regions targeted by the assay. Steps should be established to filter reads that map to nontargeted regions. |
The proportion of reads that do not map to target regions must be monitored during each run. Poor mapping quality may be a result of non-specific amplification, capture of off-target DNA, or contamination. |
| Duplication rate | Acceptable parameters for maximum duplication rate should be established for each assay. Filtering of duplicate reads by the analysis pipeline should be established to increase the number of usable sequencing data and prevent skewing of allelic fractions. |
The duplication rate should be monitored in every run and for each sample independently to monitor library diversity. |
| Strand bias | Strand bias occurs when the genotype inferred from information presented by the forward strand and the reverse strand disagrees. Each laboratory must define the tolerance level for strand bias and outline specific criteria for when alternate testing should be instituted. |
The degree of strand bias must be monitored in all samples. |
This is not a comprehensive list.
GC, guanine-cytosine content.
Controls
Control samples generally fall into three categories. The first category is reference cell lines; the most widely used are HapMap cell lines (eg, NA12878, NA19240, NA18507, NA19129; Coriell, Camden, NJ); they can be used either alone or as mixtures to model different variant allele frequencies, the lower limit of detection, and so on, in library preparation and bioinformatics analysis. The public availability of the reference sequence for the HapMap cell lines enhances their utility (National Center for Biotechnology Information FTP site, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp, last accessed October 23, 2016). The second type is synthetic DNA fragments, which have particular advantages because they can be designed to incorporate specific sequence variants at known positions. They, likewise, can be mixed in known allelic ratios, to simultaneously evaluate many aspects of platform performance, library preparation, and bioinformatics analysis.79 The third type is genetically characterized cell lines. Because cell lines are an inexhaustible reagent, and because FFPE cell blocks can easily be produced from cell lines, they are a particularly useful source of reference material.
Control samples can be used to readily detect sources of error and avoid potential harm to the patient. These should be used to monitor steps of the assay when validation data are insufficient to ensure that potential errors are exceedingly unlikely to occur. Of course, it is possible to design controls for each step within the process, but it is also possible to have a single control monitor multiple steps. In the latter case, the challenge would be troubleshooting where the error occurred. As mentioned in Interfering Substances and Carryover, an NTC should be included in every run to verify no carryover from neighboring wells during amplification steps. It is not necessary to take the NTC all of the way through sequencing as a quality check on amplified product may suffice. Also, each run should include sensitivity controls to ensure detection of targeted mutations at the LLOD, where the validation data are insufficient to ensure >95% reliability and confidence. Additional control samples may also be needed when validation data are lacking or residual risk of error remains unacceptably high. Clinical laboratory professionals typically derive the set of controls needed to suit the specifics of the methods used with respect to their intended clinical use of the test.
We recommend that for targeted NGS, a no template control should be included into library preparation to verify that there is no contamination of reagents. Sensitivity controls should be included when necessary to ensure the detection of variants at the LLOD. Cell line mixtures (with different variants and different VAFs) are a convenient source of positive controls and sensitivity controls in the same sample.
Confirmatory Testing
It is improbable that validation material would be available that has all mutation types in all genomic regions or exons. However, as discussed in Types and Number of Samples Required for Test Validation, random errors are not likely to vary by genomic region provided that quality metrics are met (eg, read quality, read length, strand bias, read depth), although systematic errors may occur because of repetitive sequence or pseudogenes. Such errors would be seen in many samples, including those for which known sequence was not available. Therefore, during validation, it is essential to confirm variants in targeted regions that are recurrent. For samples with sequence that is unknown, this would require testing with an orthogonal method (eg, PCR, interphase fluorescence in situ hybridization, microarray analysis). After validation, such confirmation may also prove useful to confirm unexpected or perplexing results that arise in routine clinical use.
Although each orthogonal validation method has advantages and disadvantages, several issues are common to all.79,81,82,84 First, although the lower limit of sensitivity of optimized conventional approaches is similar to that of routine NGS tests for SNVs, enhanced NGS bioinformatics analysis methods enable detection of variants present at a frequency of <1%, a level of sensitivity significantly better than can be achieved by conventional techniques. Second, some discrepancies between SNVs detected by NGS assays and an orthogonal validation method may actually represent tissue heterogeneity and/or intratumoral heterogeneity rather than errors. Third, orthogonal validation used as confirmatory testing of positive results but not of negative results can raise the issue of discrepant analysis (alias discordant analysis or review bias) that may poorly estimate test performance.85–87 This last issue is especially problematic because some current guidelines recommend the use of confirmatory testing for positive results,3,72,88 without associated testing of negative results (ie, wild-type results).
Conventional orthogonal validation approaches that have been used to confirm SNVs and small indels in NGS test results include Sanger sequencing, restriction fragment length polymorphism analysis, allele-specific PCR, and single-nucleotide polymorphism arrays. Common technologies used for orthogonal CNA validation are real-time quantitative PCR, interphase fluorescence in situ hybridization, and array-based comparative genomic hybridization. Classic cytogenetics, metaphase fluorescence in situ hybridization, and interphase fluorescence in situ hybridization are commonly used to confirm the presence of SVs.
We recommend that confirmation by conventional orthogonal method should be considered for unexpected or perplexing results. The choice of the method for variant confirmation is at the discretion of the laboratory director.
Proficiency Testing
Proficiency testing determines the performance of individual laboratories for specific tests or measurements and is used to monitor laboratories continuing performance. In addition, an important element of proficiency testing is interlaboratory comparison. For laboratories in the United States, participation in proficiency testing is a requirement for accreditation by the Centers for Medicare and Medicaid Services and for certification under the Clinical Laboratory Improvement Amendments. Most proficiency testing is conducted through the mechanism of laboratories receiving blinded samples from an external agency that has received deemed status via Clinical Laboratory Improvement Amendments for conducting external quality assessment (EQA) of laboratory performance. When there are no EQA surveys available for a given analyte, laboratories are required to conduct alternative EQA. Examples of alternative EQA include splitting samples and repeat testing within an individual laboratory, or splitting samples and performing an interlaboratory comparison.
The advent of NGS has challenged the ability of external agencies to develop and implement proficiency testing surveys that match the analytical complexity of multigene panels for somatic variants. Recognizing that it is not possible to source samples for proficiency testing for every potential sequence variant that may be encountered during clinical testing, the concept of methods-based proficiency testing(PT) was earlier proposed for molecular genetic testing, including NGS-based testing.79,80 Methods-based PT for molecular genetic testing is an EQA strategy that diverges from traditional analyte-specific proficiency testing (eg, for a single gene or sequence variant) to encompass the ability of the test to detect a representative spectrum of the types of sequence variants that the test is designed to analyze (eg, single-nucleotide variants or indels). Methods-based PT for molecular-based testing has been approved as a PT approach by the Centers for Medicare and Medicaid Services. To augment DNA-based PT approaches, in silico–based PT for somatic variants has been developed and its feasibility demonstrated.89 At the time of publication, multiple EQA programs have been initiated for NGS testing of germline and somatic variants, respectively (eg, College of American Pathologists, UK National External Quality Assessment).
We recommend that the laboratory should participate in an appropriate proficiency testing program.
Validation Documentation and Summary
Documentation of the validation of a laboratory test serves several important functions. A thoroughly documented validation provides a key reference for the laboratory and its personnel and is a required element to demonstrate when seeking laboratory certification and accreditation (Table 5). Extracts from the validation document also typically serve as the core information for the generation of a standard operating procedure followed by laboratory personnel.
Table 5.
Recommended Components of a Next-Generation Sequencing Validation Document
| Component | Description |
|---|---|
| Purpose of the clinical test | Detailed description of how the test will be implemented in clinical practice |
| Acceptable clinical sample types | Include all pertinent information, to include sample preservation types, rejection criteria, and other aspects that can affect test performance |
| Rationale for the inclusion of specific genes | Description based on expert review of the scientific literature |
| It is recognized that the evidentiary strength of the gene-disease associations for a given test can vary from case reports to extensive clinical trials and expert consensus guidelines | |
| Methodological approach | Detailed documentation of testing method with supporting references as needed |
| Types and sources of reagents and test instrumentation | Detailed listing of all testing components |
| Bioinformatics pipeline used for data processing and analysis | Comprehensive description to include all algorithms, software, scripts, reference sequences, and databases, whether in-house developed, or open source or vendor supplied |
| Detailed step-by-step testing procedure | Should include sufficient detail to process a patient sample from the point of entry into the assay (eg, extraction of DNA from a paraffin-embedded tissue block or blood) and subsequent steps through next-generation sequencing library preparation, sequencing, and data processing and analysis. |
| The level of detail in the validation document must be sufficient to allow the method to be reproduced | |
| Validation sample description | Comprehensive description of the source of the samples used for validation and accompanying controls |
| Optimization and familiarization results | Full and complete description of the results obtained, to include changes to protocols and retesting as needed |
| Validation results | Full and complete description of the validation results obtained During validation, unexpected and/or discordant results may be generated. Full and complete description of the validation results obtained as compared to the reference method(s). These additional studies must be described and summarized in the validation |
| Performance characteristics | Documentation of the performance characteristics (eg, sensitivity, reproducibility) of the assay that are determined during validation; can be presented in various formats with tables commonly used |
| Assay acceptance and rejection criteria | Should include any determined contingency or corrective actions that can be implemented when an analytical process fails to meet acceptance criteria |
| Assay limitations | As determined by the validation studies |
| Quality control/assurance metrics | Description of how test quality will be monitored over time after clinical implementation |
| Other information | Any additional information pertinent to the validation and performance of the clinical test |
This is not a comprehensive list, but represents the panel’s best practices recommendation of minimal components that should be included in the validation document for clinical next-generation sequencing testing.
Summary
A targeted NGS method brings a unique advantage for detection of multiple somatic alterations using a single platform and is successfully used in oncology specimens for prediction of response to targeted therapies, disease diagnosis, and patient prognostication. It has become a method of choice for detection of somatic variants and was rapidly adopted by clinical laboratories. However, this new method is challenging and requires thorough analytical validation to ensure the high quality of sequencing results. This first version of the Guidelines for Validation of NGS-Based Oncology Panels provides consensus recommendations on validation and ongoing monitoring of targeted NGS panels in the clinical setting and covers a broad spectrum of topics, including NGS platform overview, test design, potential sources of error during NGS assay development process, optimal number of samples for validation, establishing minimal depth of sequencing, implementation and quality control metrics, and others. This document summarizes a current knowledge about targeted NGS in the field of molecular diagnostics, exposes challenges of this technology, and provides guidance on how to ensure high-quality sequencing when it is used for patient care.
Acknowledgments
We thank many colleagues from the Association for Molecular Pathology and the College of American Pathologists communities for their valuable comments and suggestions and Mrudula Pullambhatla for her outstanding administrative support to the project.
Supported by the Association for Molecular Pathology.
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The Association for Molecular Pathology (AMP) Clinical Practice Guidelines and Reports are developed to be of assistance to laboratory and other health care professionals by providing guidance and recommendations for particular areas of practice. The Guidelines or Report should not be considered inclusive of all proper approaches or methods, or exclusive of others. The Guidelines or Report cannot guarantee any specific outcome, nor do they establish a standard of care. The Guidelines or Report are not intended to dictate the treatment of a particular patient. Treatment decisions must be made based on the independent judgment of health care providers and each patient’s individual circumstances. AMP makes no warranty, express or implied, regarding the Guidelines or Report and specifically excludes any warranties of merchantability and fitness for a particular use or purpose. AMP shall not be liable for direct, indirect, special, incidental, or consequential damages related to the use of the information contained herein.
Disclosures: K.V.V. and C.C. received consulting fees from PierianDx and ThermoFisher Scientific, respectively.
The Next-Generation Sequencing Analytical Validation Working Group of the Clinical Practice Committee is a working group of the Association for Molecular Pathology Clinical Practice Committee with liaison representation from the College of American Pathologists (K.V.V.). The AMP 2016 Clinical Practice Committee consisted of Marina N. Nikiforova (Chair), Monica J. Basehore, Jennifer Crow, Linda Cook, Birgit Funke, Meera R. Hameed, Lawrence J. Jennings, Arivarasan Karunamurthy, Benjamin Pinsky, Somak Roy, Mark J. Routbort, Ryan Schmidt, and David S. Viswanatha.
Standard of practice is not defined by this article and there may be alternatives. See Disclaimer for further details.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control or the US Agency for Toxic Substances and Disease Registry, the Food and Drug Administration, the National Institute of Standards and Technology, or the NIH.
References
- 1.Beadling C, Wald A, Warrick A, Neff T, Zhong S, Nikiforov Y, Corless C, Nikiforova M: A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing. J Mol Diagn 2016, 18:165–175 [DOI] [PubMed] [Google Scholar]
- 2.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Berger MF: Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015, 17:251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. Nucleic Acid Sequencing Methods in Diagnostic Laboratory Medicine; Approved Guideline CLSI document MM09-A2. ed 2. Wayne, PA: Clinical and Laboratory Standards Institute, 2014 [Google Scholar]
- 4.Smits AJJ, Kummer JA, de Bruin PC, Bol M, van den Tweel JG, Seldenrijk KA, Willems SM, Offerhaus GJA, de Weger RA, van Diest PJ, Vink A: The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014, 27: 168–174 [DOI] [PubMed] [Google Scholar]
- 5.Coonrod EM, Durtschi JD, VanSant Webb C, Voelkerding KV, Kumnovics A: Next-generation sequencing of custom amplicons to improve coverage of HaloPlex multigene panels. Biotechniques 2014, 57:204–207 [DOI] [PubMed] [Google Scholar]
- 6.Samorodnitsky E, Jewell BM, Hagopian R, Miya J, Wing MR, Lyon E, Damodaran S, Bhatt D, Reeser JW, Datta J, Roychowdhury S: Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat 2015, 36:903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ: Target-enrichment strategies for next-generation sequencing. Nat Methods 2010, 7: 111–118 [DOI] [PubMed] [Google Scholar]
- 8.Yan B, Hu Y, Ng C, Ban KH, Tan TW, Huan PT, Lee PL, Chiu L, Seah E, Ng CH, Koay ES, Chng WJ: Coverage analysis in a targeted amplicon-based next-generation sequencing panel for myeloid neoplasms. J Clin Pathol 2016, 69:801–804 [DOI] [PubMed] [Google Scholar]
- 9.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ: Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012, 30: 434–439 [DOI] [PubMed] [Google Scholar]
- 10.Quail M, Smith ME, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y: A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genomics 2012, 13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boland JF, Chung CC, Roberson D, Mitchell J, Zhang X, Im KM, He J, Chanock SJ, Yeager M, Dean M: The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet 2013, 132: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Li S, Xie W, Li X, Zhang C, Jiang HH, Zheng J, Pan X, Zheng H, Liu JS, Deng Y, Chen F, Jiang HH: Performance comparison between rapid sequencing platforms for ultra-low coverage sequencing strategy. PLoS One 2014, 9:e92192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon YJ, Zhou Y, Li Y, Guo Q, Chen J, Quan S, Zhang A, Zheng H, Zhu X, Lin J, Xu H, Wu A, Park SG, Kim BC, Joo HJ, Chen H, Bhak J: The feasibility study of non-invasive fetal trisomy 18 and 21 detection with semiconductor sequencing platform. PLoS One 2014, 9:e110240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misyura M, Zhang T, Sukhai MA, Thomas M, Garg S, Kamel-Reid SST: Comparison of next generation sequencing panels and platforms for detection and verification of somatic tumor variants for clinical diagnostics. J Mol Diagn 2016, 18:842–850 [DOI] [PubMed] [Google Scholar]
- 15.Gargis AS, Kalman L, Bick DP, da Silva C, Dimmock DP, Funke BH, Gowrisankar S, Hegde MR, Kulkarni S, Mason CE, Nagarajan R, Voelkerding KV, Worthey EA, Aziz N, Barnes J, Bennett SF, Bisht H, Church DM, Dimitrova Z, Gargis SR, Hafez N, Hambuch T, Hyland FC, Lubin IM: Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol 2015, 33:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crockett DK, Voelkerding KV: Bioinformatics tools in clinical genomics Genomic Applications in Pathology. Edited by Netto GJ, Schrijver I. New York, NY: Springer, 2015. pp. 177–196 [Google Scholar]
- 17.Spencer DH, Tyagi M, Vallania F, Bredemeyer AJ, Pfeifer JD, Mitra RD, Duncavage EJ: Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J Mol Diagn 2014, 16:75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallania FLM, Druley TE, Ramos E, Wang J, Borecki I, Province M, Mitra RD: High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res 2010, 20:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011, 43:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer DH, Abel HJ, Lockwood CM, Payton JE, Szankasi P, Kelley TW, Kulkarni S, Pfeifer JD, Duncavage EJ: Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn 2013, 15:81–93 [DOI] [PubMed] [Google Scholar]
- 21.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK: VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012, 22:568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AWC, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME: Origins and functional impact of copy number variation in the human genome. Nature 2010, 464:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE: Copy number variation detection and genotyping from exome sequence data. Genome Res 2012, 22:1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Speed TP: Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 2012, 40: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amarasinghe KC, Li J, Halgamuge SK: CoNVEX: copy number variation estimation in exome sequencing data using HMM. BMC Bioinformatics 2013, 14 Suppl 2:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D: Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008, 40: 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nambiar M, Raghavan SC: How does DNA break during chromosomal translocations? Nucleic Acids Res 2011, 39:5813–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roukos V, Misteli T: The biogenesis of chromosome translocations. Nat Cell Biol 2014, 16:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R: Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, Holmfeldt L, Payne-Turner D, Fan X, Wei L, Zhao D, Obenauer JC, Naeve C, Mardis ER, Wilson RK, Downing JR, Zhang J: CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods 2011, 8:652–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki S, Yasuda T, Shiraishi Y, Miyano S, Nagasaki M: ClipCrop: a tool for detecting structural variations with single-base resolution using soft-clipping information. BMC Bioinformatics 2011, 12:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER: BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009, 6:677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph L, Cankovic M, Caughron S, Chandra P, Emmadi R, Hagenkord J, Hallam S, Jewell KE, Klein RD, Pratt VM, Rothberg PG, Temple-Smolkin RL, Lyon E: The spectrum of clinical utilities in molecular pathology testing procedures for inherited conditions and cancer. J Mol Diagn 2016, 18:605–619 [DOI] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. Laboratory Quality Control Based on Risk Management: Approved Guideline CLSI document E23-A: Wayne, PA: Clinical and Laboratory Standards Institute, 2011 [Google Scholar]
- 35.Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR: Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther 2014, 18:587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Park C, Ji Y, Kim DG, Bae H, van Vrancken M, Kim D-H, Kim K-M: Deamination effects in formalin-fixed, paraffin-embedded tissue samples in the era of precision medicine. J Mol Diagn 2016, 19: 137–146 [DOI] [PubMed] [Google Scholar]
- 37.Serizawa M, Yokota T, Hosokawa A, Kusafuka K, Sugiyama T, Tsubosa Y, Yasui H, Nakajima T, Koh Y: The efficacy of uracil DNA glycosylase pretreatment in amplicon-based massively parallel sequencing with DNA extracted from archived formalin-fixed paraffin-embedded esophageal cancer tissues. Cancer Genet 2015, 208:415–427 [DOI] [PubMed] [Google Scholar]
- 38.Hiatt JB, Pritchard CC, Salipante SJ, O’Roak BJ, Shendure J: Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res 2013, 23:843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA: Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA 2012, 109:14508–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B: Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA 2011, 108:9530–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, Boehnke M, Kang HM: Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet 2012, 91:839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sehn JK, Spencer DH, Pfeifer JD, Bredemeyer AJ, Cottrell CE, Abel HJ, Duncavage EJ: Occult specimen contamination in routine clinical next-generation sequencing testing. Am J Clin Pathol 2015, 144:667–674 [DOI] [PubMed] [Google Scholar]
- 43.Renovanz M, Kim EL: Intratumoral heterogeneity, its contribution to therapy resistance and methodological caveats to assessment. Front Oncol 2014, 4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C: Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012, 366:883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA: Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467: 1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auerbach C, Moutschen-Dahmen M, Moutschen J: Genetic and cytogenetical effects of formaldehyde and related compounds. Mutat Res 1977, 39:317–361 [DOI] [PubMed] [Google Scholar]
- 47.Bresters D, Schipper ME, Reesink HW, Boeser-Nunnink BD, Cuypers HT: The duration of fixation influences the yield of HCV cDNA-PCR products from formalin-fixed, paraffin-embedded liver tissue. J Virol Methods 1994, 48:267–272 [DOI] [PubMed] [Google Scholar]
- 48.Feldman MY: Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol 1973, 13:1–49 [DOI] [PubMed] [Google Scholar]
- 49.Karlsen F, Kalantari M, Chitemerere M, Johansson B, Hagmar B: Modifications of human and viral deoxyribonucleic acid by formaldehyde fixation. Lab Invest 1994, 71:604–611 [PubMed] [Google Scholar]
- 50.Loudig O, Brandwein-Gensler M, Kim RS, Lin J, Isayeva T, Liu C, Segall JE, Kenny PA, Prystowsky MB: Illumina whole-genome complementary DNA-mediated annealing, selection, extension and ligation platform: assessing its performance in formalin-fixed, paraffin-embedded samples and identifying invasion pattern-related genes in oral squamous cell carcinoma. Hum Pathol 2011, 42: 1911–1922 [DOI] [PubMed] [Google Scholar]
- 51.Kerick M, Isau M, Timmermann B, Sültmann H, Herwig R, Krobitsch S, Schaefer G, Verdorfer I, Bartsch G, Klocker H, Lehrach H, Schweiger MR: Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics 2011, 4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spencer DH, Sehn JK, Abel HJ, Watson MA, Pfeifer JD, Duncavage EJ: Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn 2013, 15:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ben Lassoued A, Nivaggioni V, Gabert J: Minimal residual disease testing in hematologic malignancies and solid cancer. Expert Rev Mol Diagn 2014, 14:699–712 [DOI] [PubMed] [Google Scholar]
- 54.Barnard G: Studies in the history of probability and statistics, IX: Thomas Bayes’s an essay towards solving a problem in the doctrine of chances. Biometrika 1958, 45:296–315 [Google Scholar]
- 55.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R: Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci USA 2011, 108:20166–20171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoddard JL, Niemela JE, Fleisher TA, Rosenzweig SD: Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Front Immunol 2014, 5:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M: Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 2011, 29: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP: Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 2014, 15:121–132 [DOI] [PubMed] [Google Scholar]
- 59.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Getz G, Golub TR: Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014, 25:91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kehrer-Sawatzki H, Cooper DN: Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J Med Genet 2008, 45:622–631 [DOI] [PubMed] [Google Scholar]
- 61.Narumi S, Matsuo K, Ishii T, Tanahashi Y, Hasegawa T: Quantitative and sensitive detection of GNAS mutations causing McCune-Albright syndrome with next generation sequencing. PLoS One 2013, 8:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurek KC, Luks VL, Ayturk UM, Alomari AI, Fishman SJ, Spencer SA, Mulliken JB, Bowen ME, Yamamoto GL, Kozakewich HPW, Warman ML: Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet 2012, 90:1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asan, Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, Wang J, Wu M, Liu X, Tian G, Wang J, Wang J, Yang H, Zhang X: Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol 2011, 12:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parla JS, Iossifov I, Grabill I, Spector MS, Kramer M, McCombie WR: A comparative analysis of exome capture. Genome Biol 2011, 12:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cui H, Li F, Chen D, Wang G, Truong CK, Enns GM, Graham B, Milone M, Landsverk ML, Wang J, Zhang W, Wong L-JC: Comprehensive next-generation sequence analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med 2013, 15:388–394 [DOI] [PubMed] [Google Scholar]
- 66.Dames S, Chou L-S, Xiao Y, Wayman T, Stocks J, Singleton M, Eilbeck K, Mao R: The development of next-generation sequencing assays for the mitochondrial genome and 108 nuclear genes associated with mitochondrial disorders. J Mol Diagn 2013, 15:526–534 [DOI] [PubMed] [Google Scholar]
- 67.Pyzdek T: What Every Engineer Should Know about Quality Control, ed 1 New York, NY: Mardel Dekker, Inc., 1988 [Google Scholar]
- 68.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA: The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010, 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R: The variant call format and VCFtools. Bioinformatics 2011, 27:2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ajay SS, Parker SCJ, Abaan HO, Fuentes Fajardo KV, Margulies EH: Accurate and comprehensive sequencing of personal genomes. Genome Res 2011, 21:1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA: Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012, 30:1033–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015, 17:405–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balakrishnan N: Methods and Applications of Statistics in Engineering, Quality Control, and the Physical Sciences, ed 1 Hoboken, NJ: John Wiley & Sons, Inc., 2011 [Google Scholar]
- 74.Alonzo TA, Pepe M: Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat Med 1999, 18:1987–3003 [DOI] [PubMed] [Google Scholar]
- 75.Whale AS, Cowen S, Foy CA, Huggett JF: Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS One 2013, 8:e58177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sims DJ, Harrington RD, Polley EC, Forbes TD, Mehaffey MG, McGregor PM, Camalier CE, Harper KN, Bouk CH, Das B, Conley BA, Doroshow JH, Williams PM, Lih CJ: Plasmid-based materials as multiplex quality controls and calibrators for clinical next-generation sequencing assays. J Mol Diagn 2016, 18:336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duncavage EJ, Abel HJ, Pfeifer JD: In silico proficiency testing for clinical next-generation sequencing. J Mol Diagn 2017, 19:35–42 [DOI] [PubMed] [Google Scholar]
- 78.Mathias PC, Turner EH, Scroggins SM, Salipante SJ, Hoffman NG, Pritchard CC, Shirts BH: Applying ancestry and sex computation as a quality control tool in targeted next generation sequencing. Am J Clin Pathol 2016, 145:308–315 [DOI] [PubMed] [Google Scholar]
- 79.Schrijver I, Aziz N, Jennings LJ, Richards CS, Voelkerding KV, Weck KE: Methods-based proficiency testing in molecular genetic pathology. J Mol Diagn 2014, 16:283–287 [DOI] [PubMed] [Google Scholar]
- 80.Richards CS, Palomaki GE, Lacbawan FL, Lyon E, Feldman GL: Three-year experience of a CAP/ACMG methods-based external proficiency testing program for laboratories offering DNA sequencing for rare inherited disorders. Genet Med 2014, 16:25–32 [DOI] [PubMed] [Google Scholar]
- 81.Zook JM, Samarov D, McDaniel J, Sen SK, Salit M: Synthetic spike-in standards improve run-specific systematic error analysis for DNA and RNA sequencing. PLoS One 2012, 7:e41356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kalman LV, Lubin IM, Barker S, Du Sart D, Elles R, Grody WW, Pazzagli M, Richards S, Schrijver I, Zehnbauer B: Current landscape and new paradigms of proficiency testing and external quality assessment for molecular genetics. Arch Pathol Lab Med 2013, 137: 983–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duncavage EJ, Advani RH, Agosti S, Foulis P, Gibson C, Kang L, Khoury JD, Medeiros LJ, Ohgami RS, O’Malley DP, Patel KP, Rosenbaum JN, Wilson C; Members of the Cancer Biomarker Reporting Committee of CAP: Template for reporting results of biomarker testing of specimens from patients with chronic lymphocytic leukemia/small lymphocytic lymphoma. Arch Pathol Lab Med 2016, [Epub ahead of print] doi: 10.5858/arpa.2016-0045-CP [DOI] [PubMed] [Google Scholar]
- 84.Frampton M, Houlston R: Generation of artificial FASTQ files to evaluate the performance of next-generation sequencing pipelines. PLoS One 2012, 7:e49110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lipman HB, Astles JR: Quantifying the bias associated with use of discrepant analysis. Clin Chem 1998, 44:108–115 [PubMed] [Google Scholar]
- 86.Hadgu A: Discrepant analysis is an inappropriate and unscientific method. J Clin Microbiol 2000, 38:4301–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller WC: Bias in discrepant analysis: when two wrongs don’t make a right. J Clin Epidemiol 1998, 51:219–231 [DOI] [PubMed] [Google Scholar]
- 88.Clinical and Laboratory Standards Institute. Molecular Methods for Clinical Genetics and Oncology Testing. CLSI guideline MM01-A3. ed 3 Wayne, PA: Clinical and Laboratory Standards Institute, 2012 [Google Scholar]
- 89.Duncavage EJ, Abel H, Merker J, Bodner J, Zhao Q, Voelkerding KV, Pfeifer JD: A model study of in silico proficiency testing for clinical next generation sequencing. Arch Pathol Lab Med 2016, 140:1085–1091 [DOI] [PubMed] [Google Scholar]