Abstract
The strong dependence of retroviruses, such as human immunodeficiency virus type 1 (HIV-1), on host cell factors is no more apparent than when the endosomal sorting complex required for transport (ESCRT) machinery is purposely disengaged. The resulting potent inhibition of retrovirus release underscores the importance of understanding fundamental structure-function relationships at the ESCRT–HIV-1 interface. Recent studies utilizing advanced imaging technologies have helped clarify these relationships, overcoming hurdles to provide a range of potential models for ESCRT-mediated virus abscission. Here, we discuss these models in the context of prior work detailing ESCRT machinery and the HIV-1 release process. To provide a template for further refinement, we propose a new working model for ESCRT-mediated HIV-1 release that reconciles disparate and seemingly conflicting studies.
Keywords: HIV-1, Gag, ESCRT, superresolution, membranes, abscission
INTRODUCTION
Retroviruses, such as human immunodeficiency virus type 1 (HIV-1), are capable of budding outward from the plasma membrane through the formation of a bud head and stalk. What these viruses cannot do without host cell components is sever their membrane stalk and thereby release from the cell. Viral stalk severance is accomplished by hijacking the host cell’s endosomal sorting complex required for transport (ESCRT) machinery. This machinery drives membrane scission events in numerous cellular contexts, from inward budding of vesicles into multivesicular bodies (MVBs) to cleavage of the cytokinetic furrow (Figure 1a–c). In the context of HIV-1 budding, hijacked HIV-1 ESCRT components redistribute onto newly forming viral buds, resulting in viral particle release from the cell. Here, we discuss the technical and conceptual challenges faced by researchers studying this process, highlighting fundamental structure-function relationships that occur at the ESCRT–HIV-1 interface. Surveying the field, we offer a consensus view of how ESCRTs might function in HIV release.
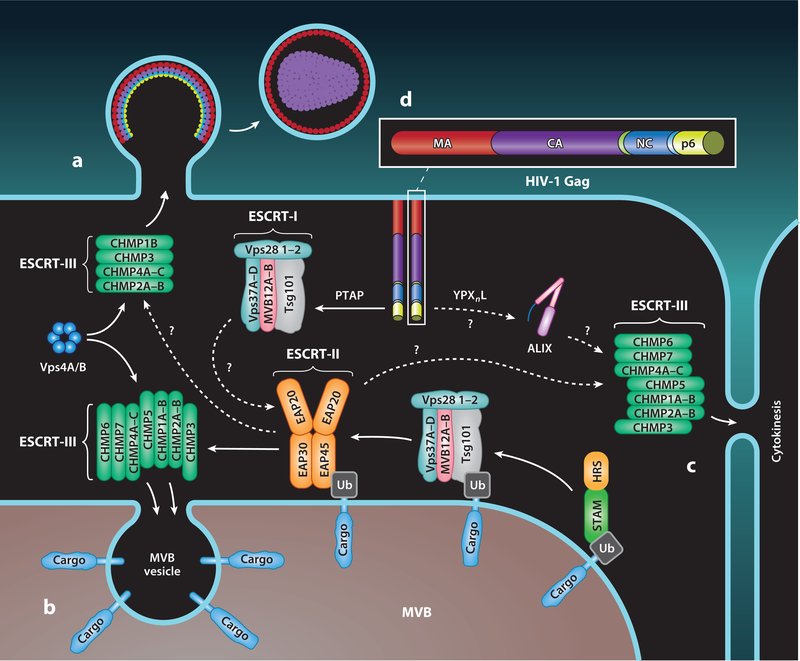
Figure 1.
Interplay between HIV-1 Gag, ESCRT substrates, and the ESCRT machinery. (a–c) Progressive engagement of HIV-1 Gag, ubiquitinated receptor cargo, and the midbody with the ESCRT subcomplexes to mediate (a) virus, (b) intraluminal vesicle, or (c) cytokinetic membrane abscission, respectively. (d) Domain architecture for HIV-1 Gag: The N-terminal MA domain (red) is modified by a myristate group that is required for anchoring Gag to the plasma membrane, the CA domain (purple) is involved in oligomerization and Gag lattice formation, the NC domain (blue) is responsible for viral genome packaging into virus (not shown), and the p6 domain (yellow) is the major determinant for direct interaction with the ESCRT-I subunit Tsg101 and ALIX. Dashed arrows indicate links that are not firmly established to play a dominant role in a specific ESCRT-mediated membrane abscission process. Abbreviations: ALIX, ALG2-interacting protein X; CA, capsid; ESCRT, endosomal sorting complex required for transport; MA, matrix; MVB, multivesicular body; NC, nucleocapsid; PTAP, Pro-Thr/Ser-Ala-Pro; Ub, ubiquitin; YPXnL, Tyr-Pro-Xn-Leu. Figure adapted with permission from Reference 24.
STRUCTURAL FEATURES OF HIV-1 GAG
HIV-1 assembly, like that of other retroviruses, is orchestrated by the Gag polyprotein precursor. HIV-1 Gag is synthesized in the cytosol and is subsequently directed to the plasma membrane, where viral assembly takes place. Assembly involves Gag molecules organizing into a hexagonal lattice that attains an outward curvature (forming a viral bud) through the introduction of gaps in the lattice (1).
The overall organization of orthoretroviral Gag precursor proteins is conserved, with several structural and functional domains—matrix (MA), capsid (CA), and nucleocapsid (NC)—arranged from N to C terminus (Figure 1d). Each of these domains plays distinct and essential roles in the retrovirus replication cycle. The MA domain, often covalently modified with an N-terminal myristate, functions in Gag trafficking to the plasma membrane and in envelope glycoprotein incorporation. The plasma membrane targeting of several different retroviral Gag proteins, including that of HIV-1, is regulated in part by a direct interaction between MA and the phospholipid PI(4,5)P2 (2, 3), which is concentrated in the inner leaflet of the plasma membrane. Assembly takes place in cholesterol-enriched membrane microdomains (4). The CA domain plays a central role in promoting Gag-Gag interactions that drive the assembly process. During particle maturation, when the viral protease (PR) cleaves the Gag precursor to its mature components, the CA protein reassembles to form the virion core that houses the viral RNA genome in complex with the viral enzymes integrase (IN) and reverse transcriptase (RT). The NC domain binds the viral genomic RNA and directs its incorporation into virus particles.
In addition to the MA, CA, and NC domains, individual retroviral genera encode unique, and typically small, peptides at various positions within the Gag precursor. It is these short peptide regions (known as late domains) that recruit cellular ESCRT machinery. Three such motifs have been well characterized; each interacts directly with ESCRT or ESCRT-related factors. The Pro-Thr/Ser-Ala-Pro (PTAP) motif binds the ESCRT-I subunit Tsg101 (5–10); the Tyr-Pro-Xn-Leu (YPXnL, where X is any residue and n = 1–4) motif binds ALG2-interacting protein X (ALIX) (11–14); and the Pro-Pro-Pro-Tyr (PPPY) motif binds to Nedd4-family ubiquitin ligases (15, 16). Retroviral Gag proteins often contain more than one late domain motif; HIV-1 Gag, for example, contains both a PTAP motif and a YPXnL motif (see below). These binding determinants enable establishment of a supercomplex when HIV-1 Gag displays bound ESCRT-I domains to the downstream ESCRT machinery.
ESCRT MACHINERY AND ITS INTERACTIONS WITH GAG
ESCRT proteins were originally described in yeast as playing an essential role in the sorting of cargo proteins into vesicles that bud into late endosomes to form MVBs (17). Genetic screens demonstrated that disruption of components of this machinery led to the formation of swollen, aberrant late endosomes known as class E compartments (18). Subsequent studies revealed that specific components of the ESCRT apparatus are also required for other topologically equivalent membrane scission events in the cell, including the abscission step of cytokinesis, autophagy, membrane wound healing, exosome production, plus-strand virus replication compartment formation, and enveloped virus budding (19). The topology of ESCRT-mediated scission events, in which budding occurs away from the cytosol, is the inverse of the topology of the membrane scission event during endocytosis, in which budding occurs into the cytosol (Figure 1a–c). This reverse-topology scission is the hallmark of most ESCRT functions.
The core ESCRT machinery in mammals comprises ESCRT-I (Tsg101, Vps28, Vps37, and MVB12/UBAP1), ESCRT-II (EAP20, EAP30, and EAP45), and ESCRT-III (CHMP1–7). Up-stream of ESCRT-I is the HRS/STAM complex, often referred to as ESCRT-0. Several ESCRT-associated proteins also play key roles in ESCRT-mediated events: the AAA ATPase Vps4 has been postulated to work in concert with ESCRT-III to drive membrane scission, and it disassembles the ESCRT-III complex after scission. ALIX promotes some ESCRT-mediated activities through direct interactions with ESCRT-I and ESCRT-III. Ubiquitylation of cargo proteins often plays an important role in their sorting into the MVB pathway, and several ESCRT and ESCRT-associated complexes and proteins bear ubiquitin-binding domains (e.g., ESCRT-I, ESCRT-II, and ALIX) (20).
Early studies demonstrated that deletion of p6, or mutation of the PTAP motif within p6, leads to a severe defect in HIV-1 particle production, characterized morphologically by the accumulation of immature budding structures on the plasma membrane of virus-expressing cells (4, 11, 12). Yeast two-hybrid screens identified the ESCRT-I subunit Tsg101 as a direct binding partner for p6, and the p6-Tsg101 interaction was dependent on the PTAP motif in p6 (7–10) and the ubiquitin E2 variant (UEV) domain in Tsg101 (10, 21). RNA interference–mediated depletion of Tsg101, or overexpression of a dominant-negative fragment of Tsg101, recapitulated the late domain phenotype observed earlier for PTAP mutations (8, 10). It was then shown that HIV-1 p6 contains a second, YPXnL-type, late domain that binds directly to ALIX. Although the YPXnL motif in p6 is the primary site for ALIX interaction with HIV-1 Gag, ALIX has also been shown to interact with the NC domain (22–24). The YPXnL motif in p6 binds the central V domain of ALIX (25, 26), whereas the NC domain of Gag interacts with the N-terminal Bro domain of ALIX (22, 23).
Additional studies revealed that different retroviruses seem to preferentially rely on Tsg101 or ALIX for recruitment of ESCRTs to the site of particle budding, and this can be affected by different conditions. For example, the Gag protein of the nonprimate lentivirus equine infectious anemia virus (EIAV) contains a single YPXnL late domain that binds ALIX, and it is well established that ALIX is a dominant player in EIAV release (12, 26, 27). YPXnL mutations in HIV-1 significantly compromise replication in spreading infections (28), but depletion of ALIX (or mutation of the YPXnL ALIX-binding site in p6) has rather mild effects on HIV-1 budding (5, 6, 12, 28–30). A role for ALIX in HIV-1 budding is most pronounced in assays in which overexpression of ALIX can rescue the virus release defect conferred by mutation of the p6 PTAP motif (26, 31). Collectively, these findings underscore the idea that although ALIX can promote HIV-1 release when Tsg101 is absent or when the p6-Tsg101 interaction has been compromised, ALIX generally plays a secondary role in HIV-1 budding.
UNRAVELING A LINK BETWEEN EARLY AND LATE ESCRT MACHINERY IN HIV-1 BUDDING
The links between early cargo sorting and retroviral elements described above established the foundation for understanding the ESCRT components that are hijacked from the host cell machinery and commandeered for virus budding. However, the downstream (late) events and mechanism of ESCRT-III-and Vps4-mediated membrane abscission still remain somewhat enigmatic and controversial.
It is well established for several retroviral Gag proteins that the interaction with one or several of the upstream ESCRT components is necessary for viral membrane abscission. From existing knowledge of the yeast MVB pathway and in vitro biochemical characterization, the logical candidate for recruitment of downstream membrane abscission machinery is ESCRT-II (32, 33). This complex has been shown to act downstream of ESCRT-I to hand off cargo in the MVB pathway and to nucleate the ESCRT-III membrane abscission machinery (Figure 1b) (34). ESCRT-II also has been shown to interact biochemically with ESCRT-I, ubiquitinated membrane cargo, and ESCRT-III subunits (33, 35, 36). It was surprising, therefore, when ESCRT-II was found to be largely dispensable both for ESCRT-mediated HIV-1 release (37) and for the ESCRT-dependent late stages of cytokinesis (38, 39). Moreover, some receptor cargos were found to transit into MVBs via an ESCRT-II-independent pathway (40). These studies questioned the necessity for ESCRT-II in recruiting ESCRT-III for downstream events involved in the virus and cytokinetic abscission pathways. However, recent studies have countered this by suggesting that ESCRT-II bridges link ESCRT-I and ESCRT-III during cytokinetic abscission (41, 42), and in vitro systems have shown that ESCRT-II can be utilized to recruit downstream ESCRT-III onto cargo-containing synthetic membranes (43).
One possibility for explaining these discrepancies in the involvement of ESCRT-II in recruiting downstream membrane fission machinery (e.g., ESCRT-III) is if an unidentified ESCRT-II adapter acts as surrogate for ESCRT-II when it is silenced during viral and cytokinetic membrane abscission. This hypothetical factor would be absent or not utilized in the yeast MVB pathway and thus not identified in the early genetic screens for the MVB ESCRT pathway. Another possibility is that only minute quantities of ESCRT-II are required to initiate linkage among retroviral cargo, ESCRT-I, and the ESCRT-III membrane abscission machinery. Because the studies suggesting ESCRT-II is not required for cytokinesis and retroviral membrane abscission rely on small interfering RNA (siRNA) knockdown of ESCRT-II components, it cannot be completely ruled out that very low levels of this protein complex survive the knockdown to link ESCRT-I and ESCRT-III. However, if linkage between ESCRT-I and ESCRT-III in the HIV-1 abscission pathway is solely dependent on ESCRT-II, then complete ESCRT-II knockout should yield as potent a reduction in virus release as the knockdown of ESCRT-I. In recent studies, this was not observed: CRISPR-Cas9 knockout of the mammalian ESCRT-II component EAP45 showed only a modest effect on HIV-1 release when compared with loss of the dominant ESCRT-I complex (44). It thus seems likely that ALIX or other unidentified factors can serve as surrogates for ESCRT-II to link ESCRT-I and ESCRT-III activities during HIV-1 budding.
A close spatial link between the HIV-1 Gag lattice and the ESCRT-III membrane abscission machinery has been observed by Cashikar et al. (45) through the use of scanning electron microscopy (SEM) of unroofed cells. They found what appear to be intimate contacts between HIV-1 Gag assemblies and ESCRT-III filaments, with the latter confirmed by immunogold labeling. This result was captured by depletion of both human isoforms of Vps4A/B in order to trap virus budding structures associated with ESCRT-III filaments. In striking images, ESCRT-III filaments were observed encircling HIV-1 Gag lattices. The lattices represented a very early stage of viral budding because they were seen from the inner leaflet of the plasma membrane and had incomplete shell morphology. Despite a lack of conclusive evidence for a specific intermediary linking HIV-1 Gag/ESCRT-I and ESCRT-III, these results suggest that once the HIV-1 Gag lattice begins to form a nascent bud, ESCRT-III filaments become associated with the bud.
SEM analysis of unroofed cells provides only superficial topological information on the inner leaflet of the plasma membrane, however, not revealing the interior of a fully budded virus particle. Whether ESCRT-III filaments associate there cannot be determined by this technique (Figure 2a). To address this question, recent work used interferometric photoactivated localization microscopy (iPALM), which achieves ~20-nm lateral and ~10-nm axial resolution (46), to examine ESCRT-I and ESCRT-III in cells producing budding HIV-1 particles (47). The results showed significant colocalization of ESCRT-I and ESCRT-III in the head of fully formed viral buds still attached to the plasma membrane (Figure 2c). Minimal labeling was seen at the base of the bud stalk, with maximal density of ESCRT probes residing within the radius of the HIV-1 Gag shell. These results suggested that as a bud head and stalk form on the cell surface, ESCRT-III filament assembly is initiated and sustained in the head of the virus bud, where Gag and ESCRT-I components now exclusively reside.
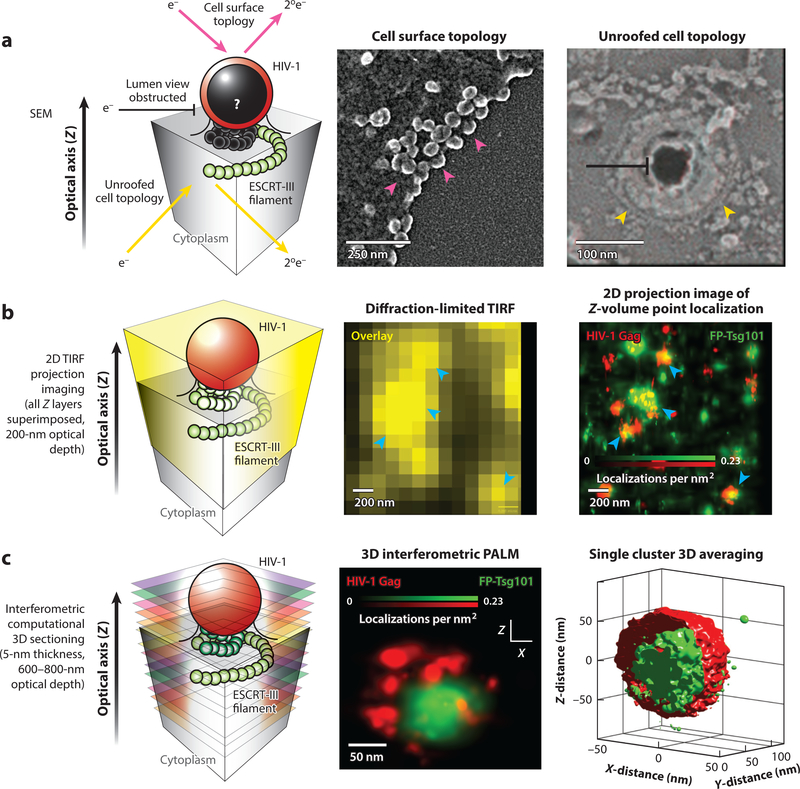
Figure 2.
Imaging modalities for interrogating the structural organization of the interface between HIV-1 Gag and the endosomal sorting complex required for transport (ESCRT) machinery. (a) Scanning electron microscopy (SEM) can interrogate exposed surface topologies of the HIV-1 budding process, probing either the surface of intact cells (magenta arrows; left micrograph) or the inner leaflet of the plasma membrane from mechanically unroofed cells (yellow arrows; right micrograph). This technique has very high spatial resolution (typically 1–5 nm) and easily resolves individual HIV-1 Gag particles (magenta arrowheads) and putative ESCRT machinery (yellow arrowheads). However, it does not enable direct visualization of the luminal contents of unperturbed HIV-1 Gag lattices as the budding membrane neck constricts (black). Right micrograph reproduced from Reference 45 with permission from Dr. Phyllis Hanson. (b) Total internal reflection fluorescence (TIRF) microscopy is an imaging technique that provides high contrast for structures of interest close to the plasma membrane (within ~200 nm). This technique, as with all fluorescence microscopy techniques, provides inherent molecular specificity due to direct fluorescent labeling of proteins of interest. TIRF microscopy is inherently diffraction limited and precludes resolution of objects much closer than 200–250 nm apart, resulting in the blurring of images between single HIV-1 assembly sites when the membrane is densely populated (blue arrowheads in left micrograph). 2D point localization microscopy techniques, such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), bypass the diffraction limit and allow for nearly 10-fold improvements in resolution between HIV-1 assembly sites (blue arrowheads in right micrograph), as shown with HIV-1 Gag (red) and fluorescent protein (FP)-tagged Tsg101 (green; overlay yellow). TIRF and 2D point localization microscopy alone provide only projection images of all the fluorescent probes found within a ∼200-nm volume, which can complicate interpretation of 3D membrane remodeling processes such as HIV-1 budding. (c) Interferometric PALM (iPALM) utilizes light wave interference to extract the 3D position of a single fluorescent molecule, which enables computational sectioning of individual budding HIV-1 Gag lattices (red) and precise positioning of associated cellular components such as Tsg101 (green). As with all fluorescent techniques, any unlabeled structures, such as membranes, remain transparent, which enables luminal views into the HIV-1 Gag lattice. This technique also enables 3D single HIV-1 Gag cluster averaging of registered fluorescent channels for statistical assessment of ESCRT spatial distributions; at the right is an isosurfaced probability density image with n = 450 particles.
In other studies using 2D projection superresolution imaging (47, 48), offsets between the HIV-1 Gag lattice and ESCRT-I and ESCRT-III probes were observed during the budding process. Although the authors interpreted this as evidence for ESCRT subunits localizing within the stalk of viral buds, this must be viewed cautiously, as interpretation of virus budding structures from 2D projections presents a challenge for quantitatively describing any 3D membrane remodeling process (see Figure 2b,c).
Results from dynamic total internal reflection fluorescence (TIRF) microscopy of HIV-1 assembly and ESCRT recruitment are consistent with major ESCRT-III assembly in the head of the virus (47, 49, 50). These data revealed that despite the encircling and relatively low copy number of ESCRT-III filaments at the plasma membrane observed by SEM during early bud formation (45), there is a late and much more extensive recruitment of ESCRT-III and Vps4 to the Gag lattice in late-stage buds (47, 49, 50). Transient spikes in the fluorescent signal from ESCRT-III and Vps4 probes occurred when HIV-1 Gag lattice formation was completed. Assuming the transient spikes in ESCRT-III and Vps4 signals reflect their accumulation in the bud head and neck, the data fit well with the iPALM data (51), which also investigated late-stage viral buds. Many open questions remain, however, as to the nature of the decline in ESCRT-III and Vps4 levels after the rapid polymerization of ESCRT-III subsides, as observed in the TIRF study (49). It is not clear, for example, whether the decline in ESCRT-III levels at the bud site results from filament depolymerization and whether there is diffusional exchange between ESCRT-III pools in the viral head and cytoplasm prior to membrane fission.
METHODOLOGICAL LIMITATIONS IN STUDYING ESCRT FUNCTION
Before discussing below the various proposed mechanisms for how ESCRTs sever HIV-1 buds, it is worth surveying the limitations of the various methods used for studying ESCRT structure and function, which include biochemical, genetic, and imaging approaches. Biochemical purification of native and mutant ESCRT components has permitted in vitro reconstitution for analysis of the shape and behavior of ESCRT filaments on membranes. Genetic tools have allowed ESCRT function to be tested in different conditions in live cells, pinpointing the role of different components. Finally, imaging approaches using antibodies and fluorescent protein tags have enabled ESCRTs to be localized in time and space, revealing that assembly of ESCRTs is a highly ordered and choreographed process during HIV-1 release. Because each of these approaches has different strengths and limitations, understanding how ESCRTs drive HIV-1 budding has been challenging.
For example, as mentioned above in the context of ESCRT-II, an unfortunate consequence of siRNA depletion experiments used in studying HIV-1 budding is that undetectable levels of knocked-down ESCRT-II subunits may be able to mediate virus abscission, making genetic knockout technologies necessary for resolving any discrepancies. The redundancy of ESCRT-III subunits may make it necessary to knock out a significant repertoire to achieve significant defects in HIV-1 abscission. Such knockouts would likely be accompanied by significant toxicities, and ambiguity as to the role of specific ESCRT-III subunits could still exist due to perturbation of unrelated ESCRT-dependent processes that indirectly affect HIV-1 biogenesis.
The dearth of suitable tools for probing the spatiotemporal organization of ESCRT proteins in cellulo has hindered our understanding of the mechanism of ESCRT-mediated membrane abscission. For example, commercial antibodies against ESCRT-III components can often be unreliable for microscopy and even western blotting. The lack of antigenicity of some ESCRT-III subunits could be due to the conformational flexibility of these subunits (52, 53). The fact that the antibodies require fixation and permeabilization of intact cells limits their use in understanding the temporal dynamics of ESCRT assembly.
As an alternative to direct labeling of endogenous ESCRTs, researchers commonly employ epitope-tagged ESCRT probes, which can be used in live or fixed cells and tissues. Unfortunately, modern fluorescent protein tagging approaches are still regarded in the ESCRT field as highly disruptive to ESCRT function. The concerns about fluorescently tagged ESCRT components could be due in part to early reports that expression of such probes has dominant-negative effects on HIV-1 budding, cytokinesis, and MVB biogenesis (14, 54, 55). Fluorescent protein–tagged ESCRT subunits have been used in many recent studies, however, and it has been demonstrated that, if expressed at low levels, they do not perturb HIV-1 budding (47, 49, 51) or cytokinesis and associated processes (56–58). Given these results, and specific studies examining this issue in HIV-1 budding (59), fluorescent protein tags themselves likely are not perturbing. Rather, problems likely arise when the ESCRT protein of interest is overexpressed, which could lead to titration of essential complex components and cause dominant-negative effects. Indeed, an early study demonstrated that overexpression of untagged Tsg101 was highly disruptive to the ESCRT machinery (59). It is therefore important to ensure that ESCRT probes are expressed at low levels, regardless of the epitope or fluorescent tag, and that proper controls are performed to demonstrate normal HIV-1 egress, cytokinesis, and MVB biogenesis in the presence of these probes.
In live cells, the process of viral budding occurs within minutes after the initiation of assembly and is subdiffractive (residing below the resolution limit of the light microscope, ~250 nm), requiring microscopes with rapid imaging speeds and superresolution capabilities. Imaging HIV-1 budding has been possible using TIRF microscopy, which has provided valuable information regarding the timing of recruitment of different ESCRT components to single viral buds (47, 49, 60). However, because the images were diffraction limited, conclusions regarding the 3D spatial organization of different budding components could not be made (Figure 2b).
To analyze the fine distribution of ESCRT components in viral buds, researchers have turned to superresolution imaging and electron microscopy approaches. These techniques have the ability to decipher protein distributions and fine architecture, respectively. Even so, these approaches have limitations. These techniques are performed on fixed cells, so the temporal dynamics of HIV-1 budding are missing. Conventional point-localization superresolution approaches, such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) (61, 62), can examine ESCRT protein distributions within the virus with remarkable lateral resolution (~20–30 nm) (47, 48), but they are severely compromised in axial resolution (~200 nm), making interpretations of 3D protein organization within the viral bud difficult (Figure 2b).
Three-dimensional imaging approaches, combined with PALM/STORM techniques, overcome the problem of poor axial resolution (46, 63–65). For example, iPALM achieves ∼10-nm axial resolution with photoconvertible probes tagged to proteins of interest (46), offering a powerful approach for deciphering the spatial organization of ESCRT proteins in viral buds (Figure 2c). That said, careful titration of endogenously expressed, tagged proteins is crucial to avoid any overexpression artifacts. A study using low expression of ESCRT subunits combined with iPALM examined the 3D organization of Gag, ESCRT-I, ESCRT-III, and Vps4A in viral buds. This study revealed a pool of ESCRT-I, ESCRT-III, and Vps4A residing within the head of the virus particle, suggesting a model for HIV-1 Gag scaffolding of growing ESCRT-III filament(s) (see Figure 2c) (51).
SEM has even better lateral resolution than PALM/STORM techniques (typically 1–5 nm). Its use in examining the organization of ESCRT-III in HIV-1 buds on the plasma membrane of unroofed cells showed ropelike ESCRT-III filaments surrounding HIV-1 Gag lattices during early stages of viral budding (45). These early stages of viral budding are ideal for imaging by SEM, because early in the budding process HIV-1 Gag has not yet fully deformed the flat plasma membrane and the newly forming stalk (>100 nm in diameter) is fully visible (Figure 2a). In contrast, late stages of viral budding are difficult to visualize using surface topology methods such as SEM. At a later stage in the budding process, viral buds have stalks with much smaller diameters (<50 nm) and heads further away from the cell surface, preventing optical access to these interior structures by SEM (see diagram in Figure 2a).
Despite their limitations, different imaging modalities have begun to shape our understanding of the in cellulo organization and dynamics of ESCRT components. This is significant given the multitude of shapes and sizes observed for ESCRT-III filaments produced in vitro. The in cellulo imaging approaches are beginning to allow researchers to determine which of the in vitro structures are relevant in the context of virus abscission.
THE ROLES OF ESCRT-III AND VPS4 IN HIV-1 BUDDING
Our knowledge of how ESCRT-III and Vps4 function in HIV-1 budding has been greatly influenced by the various methods described above. We now know that upon recruitment to the nascent Gag lattice, ESCRT-III subunits nucleate from early ESCRT scaffolds and membranes into filamentous structures of variable morphology, composition, and stoichiometry. These assemblies can bend and shape membranes and are thought to be the main drivers of membrane remodeling of the bud stalk leading to membrane severing and HIV-1 release. The hexameric AAA ATPase Vps4 triggers ESCRT-III disassembly and filament remodeling, then helps orchestrate the final ESCRT-III-dependent scission process (66–70). In the absence of activating and recruiting signals, however, ESCRT-III subunits are soluble and reside in the cytoplasm (71).
Evolution has likely specified particular ESCRT-III subunits for cooperative and effective membrane abscission at the stalk of the HIV-1 bud. In support of this hypothesis, viruses such as HIV-1 and EIAV have been shown to utilize a very limited subset of the ESCRT-III repertoire found in mammals. Comprehensive siRNA knockdown studies have demonstrated that only one of the CHMP4 homologs and one of the CHMP2 homologs are heavily relied upon for HIV-1 and EIAV release (27, 39). These studies revealed relatively minor virus release defects with the loss of CHMP3 and CHMP1B compared with CHMP4 and CHMP2 isoforms. This result in some ways contradicts in vitro studies by Lata et al. (72), which demonstrated a strict requirement for CHMP3 in the formation of highly ordered tubes composed of a copolymer between CHMP2 and CHMP3; however, CHMP2B has been shown to form structurally similar tubes in cells (73) and could potentially compensate for loss of CHMP3 during HIV-1 abscission. Although HIV-1 and EIAV release did not show strong requirements for CHMP1B, this ESCRT-III subunit has been localized to EIAV assembly sites (49).
In detailing the ESCRT-III subunit requirement for HIV-1 budding, Morita et al. (39) demonstrated that HIV-1 budding was unperturbed by knockdown of CHMP6. This result is striking because ESCRT-II is believed to interface with ESCRT-III via the CHMP6 subunit. This finding, combined with the above-mentioned data pertaining to knockout of ESCRT-II (44), strongly suggests that the HIV-1 Gag–ESCRT-I supercomplex does not rely solely on ESCRT-II and CHMP6 to nucleate CHMP4/CHMP2 filaments. One interpretation of these results is that, when present, nonessential ESCRT-III subunits will participate in virus abscission, but when those subunits are absent from an infected cell, other subunits may compensate, with other ESCRT-II-like and ESCRT-III isoforms playing a dominant role in mediating virus membrane remodeling in the absence of ESCRT-II or CHMP6.
Adding to the complexities in understanding the role of specific ESCRT-III subunits in viral budding, many ESCRT-III proteins change their conformation markedly over the course of their functions (52, 53). This may help explain why, in vitro, different polymerized ESCRT-III subunits can exhibit many forms and structures, including spirals, coils, cones, and tubes. Different ESCRT-III filaments also have a preferred radius of curvature (52, 53), raising the possibility that different ESCRT-III subunits play distinct roles at specific points in the abscission process. For example, when purified ESCRT-III subunits are incubated with lipids, they can form tubular ESCRT assemblies that extend to various lengths and have closures consisting of cones and domes (72). These properties, together with microscopic observations of ESCRT-III dynamics in vitro (74, 75), have led to the view that ESCRT-III filaments act as spiral springs that grow outward from the cargo nucleation site, with the energy of deformation greatest in the cargo-distal coil (Figure 3a,b). As the filament polymerizes, these distal or outer coils narrow the membrane tube to increasingly approach their preferred diameter (Figure 3c). A dome-like end cap is then formed that drives the membrane around the cap, narrowing it sufficiently to drive membrane fission (~3 nm) (Figure 3d,e) (76).
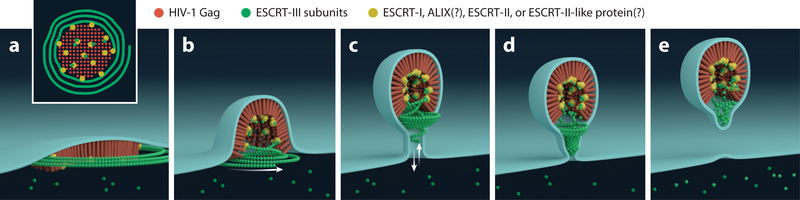
Figure 3.
Hypothetical mechanistic model for endosomal complex required for transport (ESCRT)-mediated HIV-1 abscission. (a) ESCRT-I and associated early factors coassemble with HIV-1 Gag (red) oligomerization. Upon packing of the HIV-1 Gag lattice and initial membrane buckling, ESCRT-III filaments (green) nucleate and encircle the lattice. ESCRT-III filament nucleation is dependent upon ESCRT-I binding to HIV-1 Gag and should require ESCRT-III-linking proteins such as ESCRT-II, ALG2-interacting protein X (ALIX), or unidentified ESCRT-II-like complexes (yellow linkers). (b) Further lattice packing leads to HIV-1 Gag–dependent membrane budding. (c) Prolonged association of the early ESCRT-III filaments with the HIV-1 Gag–ESCRT-I supercomplex leads to incorporation of the filaments into the head of the budding virus. This stage of budding establishes a bud stalk, which allows for ESCRT-III filament–mediated narrowing. During this stage, the stalk aperture allows for free diffusion of ESCRT-III subunits between the compartments (white arrows). (d) Further polymerization of ESCRT-III filaments leads to dome cap formation and results in narrowing of the bud stalk. This stage of budding is accompanied by Vps4-mediated decoupling of ESCRT-III from the HIV-1 Gag lattice and enables sliding of the ESCRT-III dome cap structure to the abscission site. (e) Narrowing of the bud stalk to ~3 nm enables spontaneous membrane fission of the viral bud (81). The remaining pool of head-localized ESCRT-III would be trapped within the released particle, but the pool of ESCRT-III that was disassembled prior to stalk aperture closure and virus abscission would be recycled back into the cytoplasm.
Vps4 interacts with ESCRT-III filaments and is an essential cofactor for all the known cellular and pathogenic processes requiring the ESCRT machinery. This is evident in the potent arrest of ESCRT-dependent viruses such as HIV-1 when Vps4 is absent or a dominant-negative form of Vps4 is expressed. The role Vps4 plays is essential and conserved, as evidenced by the use of Vps4 by highly divergent lineages of viruses infecting archaeal species (77). Despite the solid evidence base for the dependence on Vps4, there is still an open question in the field as to the exact role that Vps4 plays in mediating ESCRT-dependent membrane abscission. While convincing in vitro evidence has demonstrated global unfolding and disassembly of polymerized ESCRT-III subunits (72, 78), it is possible Vps4 plays a much more active role in abscission than simple disassembly of ESCRT-III filaments. It is interesting to speculate that specific ESCRT filaments may be selectively remodeled by the ATPase to bring about global rearrangements in the filaments themselves. This could be accomplished by selective removal of specific ESCRT-III subunits due to the biochemical strengths of their interacting domains (66–68). The destabilization of specific filament-cargo or filament-filament interactions could promote constriction and abscission of membrane cargo. Indeed, as proposed by Cashikar et al. (45), Vps4 could displace the putative linkage between the ESCRT-III filament and the HIV-1 Gag lattice, and this could be the critical catalytic step to trigger spontaneous filament coils and release of the virus. Further studies will be required to establish the precise role of Vps4 in ESCRT-mediated HIV-1 abscission.
TOWARD A CONSENSUS MODEL FOR ESCRT-MEDIATED HIV-1 ABSCISSION
A broadly accepted model for ESCRT-mediated virus abscission is still lacking, but consensus on several aspects of this pathway is emerging. Core requirements that are widely acknowledged include (a) direct linkages between HIV-1 Gag, early ESCRT components, and associated factors to nucleate ESCRT-III filaments on the viral bud stalk and head; (b) ESCRT filaments acting as spiral springs to constrict the stalk connecting the virus head and plasma membrane; and (c) further narrowing of the spiral springs into a dome-like cap structure that brings the stalk membranes into such close proximity that severing of the virus from the plasma membrane occurs (Figure 3).
The established direct binding between the ESCRT-I component Tsg101 and HIV-1 Gag, as well as the dominant role ESCRT-I plays in mediating HIV-1 release, sets a focal point for orchestrating subsequent downstream recruitment of later-acting ESCRT components. Models depicting the association of ESCRT-I only with the bud neck are inconsistent with the measurement of Tsg101 residing in the head of the HIV-1 Gag lattice (51). Indeed, HIV-1 Gag possesses one Tsg101 binding site (PTAP) for every Gag molecule comprising the budding virion. Further evidence from iPALM data has documented a population of ESCRT-III probes residing in the head or lumen of the HIV-1 Gag lattice (51). Therefore, a direct interface between HIV-1 Gag, ESCRT-I (and, perhaps to a lesser extent, ALIX), and an as-yet-unknown connector between ESCRT-I and ESCRT-III (potentially ESCRT-II) is likely required to form a supercomplex capable of directly, and thus proximally, nucleating ESCRT-III filaments for membrane neck constriction and virus abscission (Figure 3a,b). Supporting this, a topology proximal to the deformed membrane bud is seen by polymerizing ESCRT-III subunits in vitro (74), which mirrors that observed for the localization of ESCRT-III subunits within the head of budding HIV-1 (51). Indeed, superresolution imaging of individual HIV-1 assembly sites and single-cluster averaging of the corresponding regions of interest for CHMP4B probes determined that recruitment of this ESCRT-III subunit to virus assembly sites was dependent on the presence of an intact ESCRT-I-binding PTAP motif. These results collectively suggest that to initiate polymerization of ESCRT-III at HIV-1 assembly sites, it is necessary to first recruit ESCRT-I to the HIV-1 Gag lattice.
The striking dome cap formed by the heterofilament between mammalian CHMP2A and CHMP3 has been observed in the presence of synthetic phospholipid bilayers (72). Although direct cellular visualization of the dome cap ESCRT-III filament has remained elusive, the dome cap explains how ESCRT-III can act to constrict and taper the membrane to a narrow aperture to enable spontaneous membrane fission (76). Observations from iPALM (51) and SEM (45) studies have shown intimate association of ESCRT-III filaments proximal to, and within, the HIV-1 Gag lattice, suggesting the filaments begin polymerization in cis relative to the Gag lattice and constrict to form the tapering dome cap as it approaches the plane of the plasma membrane and aperture of the bud neck (Figure 3b,c).
This view posits that the direct upstream interaction between Gag and ESCRT-I leads to local nucleation of ESCRT-III filament(s) proximal to the Gag lattice by an unknown mediator (potentially low levels of ESCRT-II or an ESCRT-II-like protein). Multiple filaments could nucleate from this source due to the potential for stoichiometric Gag–ESCRT-I supercomplexes and the observed multistart helices observed both in vitro and during cytokinesis. These potential nucleation sites would, in turn, lead to polymerization and growth of ESCRT-III filaments. Ingression and constriction into a dome cap structure would then result in a corresponding membrane ingression to the point of spontaneous fission of the membrane and ultimate release of the virus (Figure 3d,e). HIV-1 budding thus would occur by ESCRT-III filaments growing from the viral head inward toward the plasma membrane, by an outside-in mechanism.
Countering this view of HIV-1 budding has been the reverse proposal of an inside-out ESCRT-dependent pinching mechanism, in which ESCRT-III filaments inside the plasma membrane grow outward into the viral stalk and head. This model is based primarily on reports that overexpression of the ESCRT-III subunit CHMP4B and inhibition of ESCRT-III disassembly by coexpression with a dominant-negative Vps4B construct both lead to flat, ropelike coils of CHMP4B appearing on the inner leaflet of the plasma membrane (79). Tubelike appendages emanated from the membrane surface in these experiments. Drawing from the MVB field, the idea emerged that the ropelike coils of ESCRT-III encircling Gag cargo served to push the viral head outward (away from the plasma membrane), creating a stalk, which later would be severed as it narrowed by constriction of the coils. The observations from SEM showing ESCRT-III-positive filaments on the plasma membrane encircling nascent HIV-1 Gag lattices were interpreted as supporting this inside-out pinching model (45).
Several conceptual problems, however, make the inside-out pinching model unlikely. First, HIV-1 Gag molecules have the capacity to oligomerize into a 2D lattice and to differentiate a head and stalk domain independently of ESCRT action, as evidenced by the exaggerated teardrop-shaped buds that lack the ability to recruit ESCRT-I (7). This suggests ESCRTs serve not to form the stalk, but primarily to further narrow and abscise it. Second, it is unclear how ESCRT-III filaments at the base of the stalk in the inside-out pinching model could nucleate to drive abscission once the HIV-1 Gag lattice has disappeared from the flat cell surface and redistributed into the bud head. As mentioned above, ESCRT-III has been shown to be recruited to membranes only by early ESCRT components, which directly link it to the Gag lattice. As long as the Gag lattice is proximal to the flat cell surface, ESCRT-III should be seen on the plasma membrane as encircling filaments [consistent with the SEM data of early buds (45)]. However, superresolution iPALM images showed very little ESCRT-III signal on the adjacent plasma membrane of late-stage and nonreleased virus particles (51). Significant ESCRT-III signal was present only in the viral head and stalk, supporting the idea that ESCRT-III filaments seen in nonreleased, late-stage buds grow only from the viral head domain (where Gag lattices are localized), functioning by an outside-in pinching mechanism (Figure 3b,c).
Notably, the proposed consensus model for ESCRT-mediated HIV-1 fission parallels the current model of abscission of the midbody during cytokinesis, which is also mediated by ESCRTs (57, 80, 81). During this process, nucleation of ESCRT-III filaments is observed in cis relative to the site of cargo at the midbody (i.e., CEP55). This requires binding of ESCRT-I and ALIX to mediate ESCRT-III filament nucleation at the midbody (42). Once recruited to the midbody, ESCRT-III polymerizes into filaments that move outward from the midbody toward the plasma membrane of the daughter cell (57). At some distance from the midbody, membrane neck ingression and abscission occur, with the tip of the dome cap structure predicted to face away from the midbody (81), as is proposed in our consensus model for HIV-1 abscission (Figure 3d).
The Gag scaffolding model for HIV-1 budding makes several predictions for the terminal stage of the process. Prior to neck closure, significant remodeling of ESCRT-III structures is likely to occur (Figure 3c). This could potentially allow for exchange of liberated ESCRT-III subunits from the head of the virus to the cytoplasm, and lead to priming of the ESCRT-III dome cap (which is enriched in CHMP2A and CHMP3). Indeed, biochemical studies have shown an apparent reduction in the levels of CHMP2A subunits relative to CHMP4B subunits in released particles (51). This mechanism could also serve to disengage the ESCRT-III nucleation sites at the spoke-like linkage sites connecting to the HIV-1 Gag lattice as observed by SEM (45). The ESCRT-III remodeling would be Vps4 mediated and could enable further constriction or sliding of the narrow dome cap to the abscission site, as proposed for ESCRT-mediated abscission during cytokinesis (57, 81).
During Vps4-mediated remodeling of the ESCRT-III filament, ESCRT-III subunits should become solubilized from the filament, but a fraction of the pool could be trapped in cis relative to the abscised particle (Figure 3e). The locations of Vps4 have not been modeled explicitly, but both biochemical and imaging-based approaches have detected Vps4 in released particles and within the head of budding HIV-1 particles (14, 51, 82). Future studies will need to assess the spatial relationships between Vps4, ESCRT-III, and the site of membrane abscission to test the model that Vps4 plays an active role in abscission.
In conclusion, the working consensus model proposed here involving an outside-in pinching mechanism satisfies the structural linkage of early and late ESCRT components with the HIV-1 Gag lattice. Additionally, it is consistent with the observations of ESCRT-III filaments associating with spoke-like structures presumed to be early ESCRT components, and mirrors the role of ESCRTs in midbody abscission during cytokinesis. Although the model begs for refinement through elaboration, subtraction, and transformation as new studies test various aspects and intricacies, it should assist future studies attempting to disentangle the mechanistic details of the complicated and elusive ESCRT machine usurped by HIV-1. Finally, further studies aimed at determining the precise mechanism(s) used by HIV-1 to hijack the ESCRT machinery may help uncover new therapeutic avenues for targeting a range of pathogenic ESCRT-dependent viruses.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Phyllis Hanson for kindly allowing reproduction of the SEM image in Figure 2a.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. PNAS 106:11090–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. PNAS 101:14889–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. PNAS 103:11364–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono A, Waheed AA, Freed EO. 2007. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology 360:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol 69:6810–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. PNAS 88:3195–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. PNAS 98:7724–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 9.Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med 7:1313–19 [DOI] [PubMed] [Google Scholar]
- 10.Demirov DG, Ono A, Orenstein JM, Freed EO. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. PNAS 99:955–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puffer BA, Parent LJ, Wills JW, Montelaro RC. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol 71:6541–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. PNAS 100:12414–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–99 [DOI] [PubMed] [Google Scholar]
- 14.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, et al. 2003. The protein network of HIV budding. Cell 114:701–13 [DOI] [PubMed] [Google Scholar]
- 15.Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol 68:6605–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol 168:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough J, Colf LA, Sundquist WI. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem 82:663–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzmann DJ, Odorizzi G, Emr SD. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol 3:893–905 [DOI] [PubMed] [Google Scholar]
- 19.Odorizzi G. 2015. Membrane manipulations by the ESCRT machinery. F1000Research 4:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt O, Teis D. 2012. The ESCRT machinery. Curr. Biol 22:R116–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popov S, Popova E, Inoue M, Gottlinger HG. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol 82:1389–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, et al. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLOS Pathog. 5:e1000339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii K, Hurley JH, Freed EO. 2007. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat. Rev. Microbiol 5:912–16 [DOI] [PubMed] [Google Scholar]
- 25.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. 2007. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol 14:194–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–52 [DOI] [PubMed] [Google Scholar]
- 27.Sandrin V, Sundquist WI. 2013. ESCRT requirements for EIAV budding. Retrovirology 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, et al. 2009. Functional role of Alix in HIV-1 replication. Virology 391:284–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demirov DG, Orenstein JM, Freed EO. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol 76:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem 282:3847–55 [DOI] [PubMed] [Google Scholar]
- 31.Usami Y, Popov S, Gottlinger HG. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol 81:6614–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283–89 [DOI] [PubMed] [Google Scholar]
- 33.Teo H, Perisic O, Gonzalez B, Williams RL. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 7:559–69 [DOI] [PubMed] [Google Scholar]
- 34.Tang S, Buchkovich NJ, Henne WM, Banjade S, Kim YJ, Emr SD. 2016. ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. eLife 5:e15507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hierro A, Sun J, Rusnak AS, Kim J, Prag G, et al. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature 431:221–25 [DOI] [PubMed] [Google Scholar]
- 36.Im YJ, Wollert T, Boura E, Hurley JH. 2009. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell 17:234–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, et al. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol 80:9465–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, et al. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, et al. 2006. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem 281:5094–105 [DOI] [PubMed] [Google Scholar]
- 41.Goliand I, Nachmias D, Gershony O, Elia N. 2014. Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol. Biol. Cell 25:3740–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christ L, Wenzel EM, Liestol K, Raiborg C, Campsteijn C, Stenmark H. 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol 212:499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlson LA, Hurley JH. 2012. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. PNAS 109:16928–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng B, Ip NC, Prestwood LJ, Abbink TE, Lever AM. 2015. Evidence that the endosomal sorting complex required for transport-II (ESCRT-II) is required for efficient human immunodeficiency virus-1 (HIV-1) production. Retrovirology 12:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cashikar AG, Shim S, Roth R, Maldazys MR, Heuser JE, Hanson PI. 2014. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3:e02184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, et al. 2009. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. PNAS 106:3125–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleck M, Itano MS, Johnson DS, Thomas VK, North AJ, et al. 2014. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. PNAS 111:12211–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prescher J, Baumgartel V, Ivanchenko S, Torrano AA, Brauchle C, et al. 2015. Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLOS Pathog. 11:e1004677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol 13:394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, et al. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol 13:469–74 [DOI] [PubMed] [Google Scholar]
- 51.Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, et al. 2014. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science 343:653–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, et al. 2015. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife 4:e12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, et al. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350:1548–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. PNAS 103:19140–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howard TL, Stauffer DR, Degnin CR, Hollenberg SM. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci 114:2395–404 [DOI] [PubMed] [Google Scholar]
- 56.Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336:220–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. PNAS 108:4846–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. 2015. ESCRT-III controls nuclear envelope reformation. Nature 522:236–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol 77:6507–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–45 [DOI] [PubMed] [Google Scholar]
- 62.Rust MJ, Bates M, Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3:793–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang B, Wang W, Bates M, Zhuang X. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319:810–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, et al. 2008. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5:527–29 [DOI] [PubMed] [Google Scholar]
- 65.Pavani SR, Thompson MA, Biteen JS, Lord SJ, Liu N, et al. 2009. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. PNAS 106:2995–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, et al. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449:735–39 [DOI] [PubMed] [Google Scholar]
- 67.Merrill SA, Hanson PI. 2010. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem 285:35428–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–44 [DOI] [PubMed] [Google Scholar]
- 69.Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babst M, Sato TK, Banta LM, Emr SD. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, et al. 2009. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol 16:754–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lata S, Schoehn G, Jain A, Pires R, Piehler J, et al. 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science 321:1354–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, et al. 2011. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J. Biol. Chem 286:40276–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee IH, Kai H, Carlson LA, Groves JT, Hurley JH. 2015. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. PNAS 112:15892–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, et al. 2015. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163:866–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. 2009. Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comput. Biol 5:e1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Snyder JC, Samson RY, Brumfield SK, Bell SD, Young MJ. 2013. Functional interplay between a virus and the ESCRT machinery in archaea. PNAS 110:10783–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. 2015. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol 22:492–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanson PI, Roth R, Lin Y, Heuser JE. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol 180:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carlton JG, Martin-Serrano J. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908–12 [DOI] [PubMed] [Google Scholar]
- 81.Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. 2012. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J 102:2309–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ladinsky MS, Kieffer C, Olson G, Deruaz M, Vrbanac V, et al. 2014. Electron tomography of HIV-1 infection in gut-associated lymphoid tissue. PLOS Pathog. 10:e1003899 [DOI] [PMC free article] [PubMed] [Google Scholar]