Abstract
Retroviruses comprise a large, diverse group that infects a broad range of host organisms. Pathogenicity varies widely; the human immunodeficiency virus is the causative agent of acquired immunodeficiency syndrome, one of the world’s leading infectious causes of death, while many nonhuman retroviruses cause cancer in the host. Retroviruses have been studied intensively, and great strides have been made in understanding aspects of retroviral biology. While the principal functions of the viral structural proteins are well understood, there remain many incompletely characterized domains. One of these is the cytoplasmic tail (CT) of the envelope glycoprotein. Several functions of the CT are highly conserved, whereas other properties are unique to a specific retrovirus. For example, the lentiviruses encode envelope glycoproteins with particularly large cytoplasmic domains. The functions of the long lentiviral envelope CT are still being deciphered. The reported functions of retroviral envelope CTs are discussed in this chapter.
1. INTRODUCTION
Retroviruses are ancient, with the genomes of all vertebrates showing evidence for extensive retroviral activity throughout their evolution, even in species that are not known to harbor active retroviruses at present.1 Retroviruses belong to a larger family of retro-elements that includes long terminal repeat (LTR) retrotransposons, characterized by the presence of the enzymes reverse transcriptase (RT) and integrase (IN), the capsid (CA) protein Gag, and the eponymous LTRs at the 5′ and 3′ ends of the transposon.2 These retrotransposons are transcribed and translated by host cell machinery, then assemble intracellular particles containing the RNA genome, RT and IN, within a core of Gag. They propagate by reverse-transcribing their RNA genome to DNA and re-entering the nucleus where IN inserts the retrotransposon DNA genome back into the host cell genome. By this “copy and paste” mechanism, retrotransposons have accumulated to high numbers in the genomes of many species. The human genome, for example, comprises 42% retrotransposon sequences.3 Retro-elements have therefore contributed significantly to the evolution of species, by continually enlarging and shuffling the genomes in which they reside.
Retroviruses can be viewed as a class of retrotransposon that has acquired both the ability to produce extracellular particles and an envelope (env) gene that encodes a surface-exposed Env glycoprotein to mediate viral entry into new cells.4–6 These env genes have a variety of potential sources; in many cases, they may have been acquired from other viruses by recombination of the retrotransposon RNA with the RNA of the incoming virus. Whatever their origin, retroviral Env glycoproteins share certain general properties (Fig. 1).8 They possess an extracellular domain, which is exposed on the outside of the viral particle. This external domain is responsible for binding to receptor proteins on target cells. Receptor binding triggers conformational changes in the Env glycoprotein to bring a second – fusion – domain into play. The fusion apparatus is principally composed of a hydrophobic fusion peptide and two heptad repeats (HR) that fold into a six-helix bundle during the fusion process.9,10 The concerted action of the fusion peptide and the HRs mediates the fusion of the viral membrane with target cell membrane [typically plasma membrane (PM) or endosomal membrane] during virus entry. The domains that mediate receptor binding and membrane fusion are generally translated as a single polyprotein precursor that is cleaved during trafficking through the Golgi apparatus. The extracellular Env (referred to as the surface, or SU, subunit) and transmembrane (TM) subunit remain covalently or noncovalently associated as heterodimers, which in turn form trimeric Env glycoprotein spikes, in their mature forms. Env is a type I membrane protein, anchored to the membrane by a single transmembrane domain (TMD).11,12 In addition to functioning as an anchor, studies in HIV-1 and SIV indicate that the TMD influences the function of Env. In particular, a conserved arginine residue and a Gly-X-X-X-Gly (where X is any amino acid) motif are required for efficient membrane fusion11,13; the positioning of these features is also important, as the arginine residue is thought to interact with the phosphate head-groups of the phospholipid membrane and the Gly-X-X-X-Gly motif likely mediates interactions between gp41 monomers.14–16 The final major structural and functional region of the Env glycoprotein is the cytoplasmic tail (CT). Retroviral CTs range in size from short peptides of 20–30 amino acids found in most retroviral genera, to the large CT of the lentiviruses, which can be over 200 amino acids in length (Fig. 2). These domains are typically dispensable for receptor binding and fusion activities, but nevertheless play essential roles in viral replication. In particular, the presence of a CT allows communication with the interior of the host cell and, postrelease, the virus particle, and coordination of functions during assembly, release, and infection of a new cell.
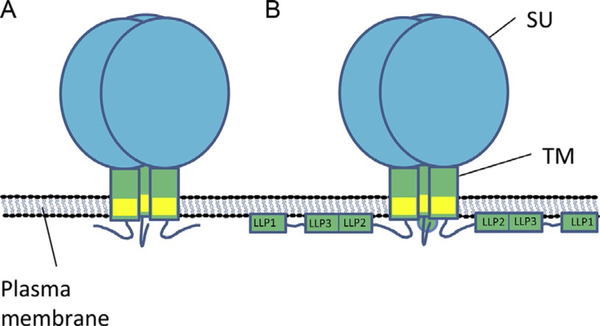
Figure 1.
Schematic representation of retroviral Env glycoproteins. (A) Retroviruses express Env glycoproteins with a large, glycosylated soluble subunit [blue (gray in the print version)], oriented to the outside of the cell/virus, and a smaller transmembrane protein [green (gray in the print version)]. In most retroviral families, the transmembrane domain [yellow (light gray in the print version)] is followed by a short cytoplasmic tail of unknown structure. (B) Lentiviral Env glycoproteins have arrangement of their ectodomains as found in other families, but in most cases possess a larger cytoplasmic tail. Shown here is the cytoplasmic tail for HIV-1 Env, with three alpha-helical lentiviral lytic peptides, although their organization in vivo is unknown. Other lentiviral cytoplasmic tails may contain similar features, but have not been subject to detailed examination. SU, surface unit; TM, transmembrane subunit; LLP, lentiviral lytic peptide. Figure adapted with permission of Cell Press from Ref. 7.
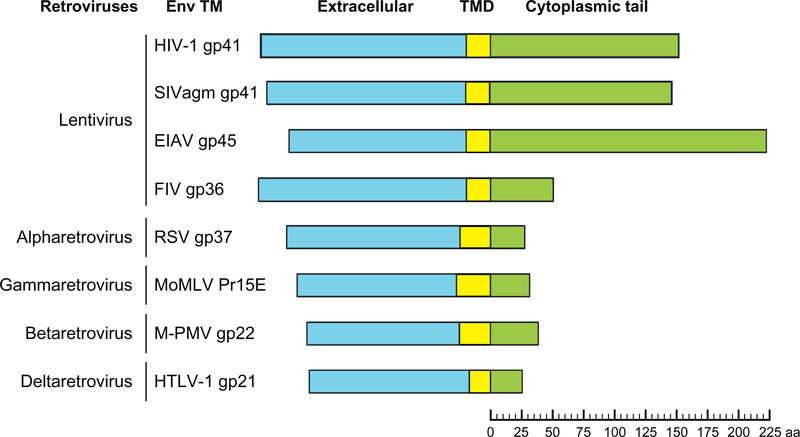
Figure 2.
Schematic representation of retroviral TM subunits, indicating the lengths [in amino acids (aa)] of their respective cytoplasmic tails. As illustrated, lentiviral TM CTs tend to be significantly longer than those of other retroviruses, with the exception of feline immunodeficiency virus (FIV). HIV-1, human immunodeficiency virus 1; SIVagm, simian immunodeficiency virus from African green monkey; EIAV, equine infectious anemia virus; RSV, Rous sarcoma virus; MoMLV, Moloney-MLV; M-PMV, Mason-Pfizer monkey virus; HTLV-1, human T-cell lymphotropic virus type 1. Figure reproduced with permission of Elsevier from Ref. 17.
The acquisition of an Env glycoprotein and consequent addition of a transmission stage to the viral replication cycle dramatically alters the life-style of the virus, as it is no longer confined to the cell in which it was produced or even to the individual organism. The spread of retroviruses can occur through cell-free or cell–cell mechanisms; the latter is often referred to as transmission via a viral synapse.18 In many cases, cell–cell transmission is far more efficient than cell-free transmission, and some viruses, such as human T-lymphotropic virus (HTLV), appear to spread exclusively through this route.19–21 The freedom of the virus to spread from the host favors the development of pathogenic viruses, whose replication takes place at the expense of the host cell or organism. It is not surprising that those viruses that adversely affect the health of their host have been the favored subjects for research, particularly when humans are the host species.
A great many retroviruses are now known, infecting a wide variety of species (for detailed listings see Ref. 22 and http://www.ncbi.nlm.nih.gov/retroviruses/). The retrovirus family contains two subfamilies, the orthoretrovirinae and the spumavirinae. The spumavirinae contain a single genus, the spumaviruses, while the orthoretrovirinae contain six genera, the alpha-, beta-, gamma-, delta-, and episilon-retroviruses, and the lentiviruses (Table 1). While all share the basic gag, pol, and env genomic arrangement, there is considerable variability in the accessory and regulatory proteins, and in replication strategies and pathogenicity.
Table 1.
Summary of retroviral taxonomy, including the viruses discussed in this review and other commonly studied retroviruses
| Family | Genus | Species |
|---|---|---|
| Orthoretrovirinae | Alpharetrovirus | Avian sarcoma leukosis virus |
| Rous sarcoma virus | ||
| Betaretrovirus | Enzootic nasal tumor virus of goats | |
| Jaagsiekte sheep retrovirus | ||
| Mason-Pfizer monkey virus | ||
| Mouse mammary tumor virus | ||
| Simian retrovirus 4 | ||
| Gammaretrovirus | Feline leukemia virus | |
| Gibbon ape leukemia virus | ||
| Murine leukemia virus | ||
| Spleen focus-forming virus | ||
| Xenotropic murine leukemia virus-related | ||
| virus | ||
| Deltaretrovirus | Bovine leukemia virus | |
| Human T-lymphotropic virus 1, 2, 3, and 4 | ||
| Epsilonretrovirus | Walleye dermal sarcoma virus | |
| Lentivirus | Bovine immunodeficiency virus | |
| Equine infectious anemia virus | ||
| Feline immunodeficiency virus | ||
| Human immunodeficiency virus 1 and 2 | ||
| Simian immunodeficiency virus | ||
| Spumaretrovirinae | Spumavirus | Bovine foamy virus |
| Feline foamy virus | ||
| Simian foamy virus | ||
This review will focus primarily on the functions of the CT of the human immunodeficiency virus (HIV), the causative agent of acquired immunodeficiency syndrome (AIDS), and its close relatives, the simian immunodeficiency viruses (SIVs). Other retroviruses will also be discussed, albeit in less detail, in an attempt to convey the diversity of functions attributed to the CT of retroviral Env proteins.
2. RETROVIRAL ASSEMBLY
Retroviral particles are composed of either an RNA (orthoretroviruses) or DNA (spumaviruses) genome, contained in a protein core.23 This core is surrounded in turn by a lipid membrane, embedded in which are the trimeric Env glycoprotein spikes. The Gag polyprotein is responsible for the formation of the protein core.24 Domain organization varies, but common features are: (a) a membrane-targeting matrix (MA); (b) a multimerizing CA; (c) a nucleic acid-binding nucleocapsid (NC) in orthoretroviruses or an arginine-rich region in foamy viruses25; and (d) one or multiple motifs that recruit the endosomal sorting complex required for transport (ESCRT) proteins to promote virus release. In the pathway exploited by lentiviruses, such as HIV-1, the Gag precursor is translated in the cytoplasm, then forms low-order oligomers through CA–CA interactions. The MA domain mediates Gag binding to the PM.26 In the case of HIV, membrane targeting involves a highly basic patch of residues in MA that bind specifically to phosphatidylinositol-4,5-bisphosphate, and subsequent exposure of an amino-terminal myristic acid moiety that inserts into the lipid bilayer, anchoring Gag to the membrane.27–29 The RNA-binding domain of Gag, typically NC, possesses general nucleic acid-binding properties, but preferentially binds the viral genome.30 In addition to ensuring packaging of the genome within the nascent particle, NC also promotes higher-order multimerization of Gag, which forms an extensive lattice containing several thousand Gag molecules.31 The central feature of the Gag lattice is the hexameric arrangement of CA.32–34 Release of the particle from the host cell is achieved through the recruitment of the ESCRT complex by late domains in Gag.35 Three forms of these domains are known: Pro-[Thr/Ser]-Ala-Pro binds TSG101, a component of ESCRT-I, which is responsible for binding cargo36–39; Tyr-Pro-Xn-Leu (where Xn is a variable number of amino acids) binds to the V-domain of ALIX, which in turn recruits ESCRT-III directly40; Pro-Pro-Pro-Tyr recruits NEDD4-like ubiquitin ligases, ubiquitination is one of the principal signals used by ESCRT to identify cargo.41,42
Following, or concurrent with, release of the virus from the host cell, retroviruses undergo a maturation step wherein the structural proteins of the virus undergo processing by the viral protease (PR), causing the break-down of the immature Gag lattice and the formation of a mature infectious particle.8,43–45 In addition to the dramatic reorganization of Gag (from a shell of the complete Gag protein located immediately beneath the lipid membrane, to a smaller core composed of CA and containing the viral genome), maturation can also influence the function of the Env protein, via interactions with the CT of the TM Env subunit, as discussed below.
The viral particle also contains the viral enzymes PR, RT, and IN.24 Additional accessory proteins may be present, depending on the specific virus. HIV-1 particles, for example, contain Vif and Vpr, and SIV and HIV-2 particles additionally contain Vpx. These accessory proteins often have roles in enhancing the infectivity of the virus by countering host cell defenses; Vif is known to counter the antiviral activity of APOBEC3 proteins,46–48 and Vpx triggers the degradation of the cellular dNTPase SAMHD1.49–51
3. SYNTHESIS AND FUNCTION OF ENV
Retroviral Env glycoproteins are type I membrane proteins, with an extracellular N-terminus and a cytoplasmic C-terminus52; they are synthesized as a precursor at the endoplasmic reticulum (ER), and cotranslationally inserted across the membrane. Following synthesis, the precursor organizes as a trimer and undergoes initial glycosylation steps at the ER. The level of glycosylation is highly variable; for example, mouse mammary tumor virus possesses only 4 putative sites for N-linked glycosylation, while HIV-1 has over 30.8,53,54 Env then traffics through the Golgi apparatus where the mature glycans are added and the precursor is cleaved into the mature forms, the extracellular SU subunit and TM subunit. These mature proteins remain noncovalently associated as heterodimers, which in turn form the homotrimeric Env spikes. Trimerization of Env is driven primarily by motifs in the extracellular portion of the protein; in avian sarcoma leukosis virus (ASLV),55 murine leukemia virus (MLV),56 and HIV,57 truncated, soluble forms of the Env glycoproteins have been shown to form trimers, in the absence of TM-SU cleavage. To obtain structures of a processed, soluble HIV-1 Env trimer, modifications to enhance stability were necessary. Specifically, a disulphide bridge was introduced between residues 501 (in SU) and 605 (in TM), and an Ile to Pro mutation was introduced at residue 559 to improve TM:TM interactions.58,59 All numbering is based on the HXB2 isolate.
The mature Env subunits have distinct functions, with SU being exposed on the exterior of the virion and possessing the receptor binding functions. The glycosylation of the SU is thought to play roles in avoiding antibody binding,60,61 promoting correct Env folding,62 and receptor binding.63 The binding of receptor (and potentially coreceptor) triggers conformational changes that expose the fusion machinery of TM. The fusion function is conferred by the ecto- and transmembrane-domains of TM, and follows a common mechanism. In HIV-1, −2, and SIV, the critical “fusion peptide” is located at the N-terminus of gp41 (the TM), with two amphipathic HR (HR1 and HR2) C-terminal to the fusion peptide. HR1 and HR2 pack as an antiparallel six-helix bundle within the Env trimer. The structural similarities between gp41 and the influenza fusion protein hemagglutinin 2 suggest a similar spring-loaded mechanism, where the fusion peptide inserts into the PM or endosomal membrane of the target cell, and the energy to promote fusion is derived from the structural changes in the six-helix bundle. Peptides mimicking HR1 can bind to and disrupt the function of the six-helix bundle; this approach yielded the fusion inhibitor enfurvitide. Fusion mediated by retroviral Env glycoproteins often occurs at the PM,64,65 or following endocytosis and low-pH triggering in the endosome.66,67 Some retroviruses, like ASLV, fuse in a pH-dependent fashion at the endosomal membrane after receptor priming at the PM.68
The most C-terminal domain of Env is the CT of TM. This region of the Env is not required for the primary Env functions of receptor binding and membrane fusion, and some retroviruses with deletions of the CT have been shown to replicate in culture69–72; however, the CT has been reported to play a wide variety of regulatory functions in viral biology and in many viruses is required in culture and animal models.69,70,73–75 The CT is the most variable region of Env in terms of its size, presumably reflecting the diverse roles it plays in different viruses. In most retroviruses, the CT is relatively short, 25–35 amino acids, in keeping with the CT of most known viral envelope proteins.17 By contrast, the majority of lentiviral CT domains are large, well over 100 amino acids in length. One exception to this rule is the CT of feline immunodeficiency virus (FIV); at around 50 amino acids it is substantially shorter than those of other lentiviruses, although still longer than most other retroviruses. These larger CT domains have greater potential for function and specific structures. The CT of HIV-1 contains three lentiviral lytic peptides (LLP1–3), highly alpha-helical regions that have been shown to interact with membranes.76–78 The remainder of the CT, approximately 70 amino acids, is of unknown structure. The length of the lentiviral CT also opens the possibility of a more complex topology than the simple cytoplasmic orientation of the short-tailed retroviral Env. A motif in the HIV-1 gp41 CT known as the Kennedy sequence immediately follows the TMD and has been suggested to be exposed outside the membrane, based primarily on detection and neutralization by antibodies.79–81 Such a topology would be in conflict with studies that reported a single membrane-spanning domain.82 Two resolutions of this conflict have been proposed. It is possible that the surface exposure of the Kennedy sequence is context dependent, and these Env molecules are found on infected cells and potentially in fusion intermediates, but are not incorporated into or found on cell-free virions79,80; alternatively, the detection of surface-exposed regions of the CT may be an artifact of the experimental conditions, as regions have been found to be exposed to antibody detection that should be cytoplasmic in both topology models.83 It is noteworthy that viral isolates have been described that have acquired PR cleavage sites in the gp41 CT near the Kennedy sequence84,85 (see below). These cleavage sites would have to be inside the membrane to be accessed by PR, arguing against the three-membrane-pass model for HIV-1 Env topology.
4. FUNCTION OF THE RETROVIRAL ENV CT
4.1. Anterograde trafficking
One function for a cytoplasmic region in any viral Env glycoprotein is to aid in regulating the trafficking of the protein. The vesicular stomatitis virus glycoprotein (VSV-G) has been used extensively as a model protein for ER-to-PM trafficking. The retroviral Env glycoproteins likewise follow an ER-to-PM pathway, and display some specificity of trafficking. Work with several retroviruses has shown that Env traffics preferentially to the basolateral membrane in polarized cells.86–88 Furthermore, HIV-1 Env appears to be able to direct Gag to the site of virus budding in polarized epithelial cells, in a CT- and MA-dependent manner.89,90
The CT also plays a role in determining the site of the PM to which Env localizes. With a few exceptions, such as Rous sarcoma virus (RSV),91 retroviral Env proteins typically demonstrate a tendency to localize at lipid rafts.92–96 In MLV, this function has been shown to require palmitylation of conserved cysteine residues in the CT.93 In contrast, studies of HIV-1 and SIV found that although the CT is palmitylated, this posttranslational modification is not required for raft localization.94,97,98 There are conflicting reports on the role of palmitylation in the HIV-1 Env CT. The existence of HIV-1 isolates that lack cysteine residues in their CT, and efficient replication of palmitylation-deficient mutants, suggest this modification may not be absolutely required98,99; however, it does not rule out a potential contribution to Env function in some isolates.100 The accumulation of HIV-1 Env in lipid rafts may also be influenced by Gag and other motifs of Env, such as a cholesterol recognition amino acid consensus motif between the membrane-proximal external region and the TMD.96,101
In the case of viruses that exploit raft localization of Gag and Env proteins, this colocalization may represent a mechanism to enrich Env at sites of Gag assembly. In HIV-1, a further aspect of this colocalization has been reported; an interaction between the gp41 CT and Rab11-family interacting protein 1c (FIP1c).102 FIP1c appears to mediate an interaction between HIV-1 and the Rab14 trafficking pathway, which targets proteins toward the PM.103,104 It is likely that Env is additionally targeted to specific sites on the PM, as deletion of the CT does not prevent Env from trafficking to the PM, but does prevent it from localizing at sites of viral assembly in physiologically relevant cell types.102,105
4.2. Retrograde trafficking
The CT contains motifs that regulate endocytosis of Env. HIV and SIV are exposed to the host immune system in part through Env on the PM; Env can be recognized by circulating antibodies and triggers antibody-dependent cell-mediated cell killing. To avoid this scenario, the CTs of HIV and SIV Env contain endocytic signals that promote the rapid internalization of Env, limiting its accumulation at the PM.106–109 The primary endocytic signals in the HIV and SIV Env CTs are Tyr-X-X-Φ (Tyr712 in HIV-1 and Tyr721 in SIV, Φ is any hydrophobic amino acid) and the C-terminal dileucine.110–112 Intriguingly, the deletion of Tyr721 and the preceding Gly720 in SIV generates a virus that retains efficient replication in animals, but does not deplete the host CD4 + cells.113 Passage of the virus yielded a compensatory mutation of Ser727 to proline, which restored the ability to deplete host CD4 + cells.114
A Tyr-X-X-Φ motif has also been found to regulate endocytosis of HTLV Env, and its removal was shown to increase both levels of cell-surface Env and Env incorporation into VLPs.115,116 The endocytic motifs recruit subunits of the clathrin adaptor protein complexes AP1–3, with AP1 and AP2 playing the dominant roles in cells.111,112,115,117 A second Tyr-X-X-Φ motif has been identified in the HIV-1 CT, based on Tyr763, and able to interact with AP1 and AP3111; however, this motif appears to be less efficient at recruiting endocytic machinery than the motif based on Tyr712. The rapid endocytosis of HIV-1 Env may only apply to Env molecules outside sites of assembly, as coexpression of Env with high levels of HIV-1 Gag has been reported to reduce the rate of Env internalization.109 The presence of the Gag lattice may prevent the endocytic machinery from interacting with the Env CT at sites of particle assembly. If the Gag lattice indeed blocks AP subunits from accessing the endoctyic signals in the Env CT, then the mechanism could be broadly applicable to other viruses.
The HTLV-1 Env CT contains a second trafficking motif, close to Tyr-X-X-Φ: a PDZ-binding motif that appears to play a role in countering the effects of Env endocytosis.116 Deletion or mutation of the PDZ-binding motif dramatically reduces Env incorporation into VLPs, an effect that can be alleviated by the silencing of AP2 components that interact with the Tyr-X-X-Φ motif. At this time, the identity of the interaction partner(s) for the PDZ-binding motif in the HTLV Env CT is unknown and the precise mechanistic role of the motif is thus unclear. A comparable motif may also exist in the Env CT of RSV.91 It is likely that the interplay of the two trafficking motifs is important in regulating the abundance of Env at the cell surface, with consequences for incorporation into particles, cell–cell fusion and immune recognition.
Deletion of the SIV Env CT leads to enhanced accumulation of Env on the PM; this increase is further associated with elevated incorporation of Env into viral particles.106 When cultured in human cells, SIV isolates will rapidly lose their CT to enhance replication.72 However, the CT is retained during culture in the native host, suggesting that the CT plays a critical role in the natural setting. Indeed, it has been reported that mutation of the Tyr-X-X-Φ motif in SIV did not prevent replication, but did render the virus more susceptible to the host immune response.113
The motifs regulating HIV-1 Env endocytosis may additionally play roles in mediating optimal virus fusion and infectivity.118,119 It has been shown that LLP2 can influence the physical properties of the membrane in the proximity of the CT, which may influence membrane fusion120; however, the specific mechanisms and functional relevance of these findings remain unexplained.
4.3. Env packaging into particles
In addition to the trafficking pathways that direct retroviral Env toward particle assembly sites, there is clear evidence that viral Envs cluster with Gag at sites of assembly. Clustering has been observed by scanning electron microscopy and by super-resolution microscopy.105,121,122 This clustering is seen with a wide variety of Env glycoproteins, including non-retroviral Envs, such as VSV-G and Ebola glycoprotein, suggesting a general property of retroviral assembly sites that is recognized by the Env glycoproteins.122 These data agree with the general observation that many glycoproteins can pseudotype retroviral particles, nevertheless, there is a degree of specificity, as MLV Env will preferentially package into MLV particles over HIV particles when both are assembling in the same cell.74,123 Surprisingly, the specificity of particle selectivity does not appear to be driven by an interaction between the Env CT and MA, as MLV Env continues to select MLV particles containing an HIV-1 MA, over HIV-1 particles with MLV MA123; instead selection appears to be driven by CA and NC domains.
In the case of HIV-1, the clustering of Env at assembly sites requires the Env CT105,121; CT-truncated Env is still observed at the PM, but does not colocalize with Gag. In the 293T cells used for these experiments, CT-truncated Env is still incorporated into particles, despite the lack of colocalization with Gag. However, it is likely that the clustering of Env at sites of assembly (which is also seen in T cells that do require CT for efficient Env packaging) reflects a necessary component of the Env packaging mechanism under physiological conditions. The presence of the CT renders the Env immobile at assembly sites, while the CT-truncated Env retains the ability to diffuse in the membrane, despite the presence of Gag.121 It is not known whether this phenotype is due to a direct interaction between Gag and Env, or a more indirect trapping, where the large CT is unable to diffuse through the densely packed Gag lattice. The clustering of MLV Env appears to occur through a slightly different mechanism, as the truncation of the CT does not prevent clustering of Env at viral assembly sites.74 The MLV Env CT is required, however, for the preferential packaging of MLV Env into MLV particles over HIV-1 particles.74
While trafficking of Env contributes to its incorporation into particles, there are clearly additional factors that influence Env levels in virions, particularly in the long-tailed lentiviruses whose Env glycoproteins do not efficiently package into the particles of other viruses. In the case of HIV-1, point mutations in MA or small deletions in the gp41 CT, which do not affect known trafficking motifs, can inhibit Env incorporation. Furthermore, these Env incorporation defects can be corrected by modifications to the reciprocal domain; that is, a deletion in the CT can be rescued by a point mutation in MA (observed in culture- and in patient-derived samples), and point mutations in MA can be rescued by deletion of the entire gp41 CT.124–127 In a similar vein, it has long been observed that although HIV particles can package the short-CT Env of MLV, MLV particles are unable to package HIV Env, unless the CT is removed.128 A similar phenotype is seen in ASLV, where HIV-1 particles pseudotyped with a CT-truncated Env are far more infectious than those bearing full-length Env.129 In the case of HIV-1, the reciprocal rescue of MA/Env mutations has been interpreted as genetic evidence for an interaction between these protein domains that is required for Env incorporation. Any form of interaction would likely drive coevolution of the Env CT and MA. Accordingly, incorporation of Env from primary isolates into laboratory strain-derived Gag particles can be relatively inefficient relative to incorporation of the cognate Env.130 Also, during infection, coevolution of MA has been observed as compensation for a mutant Env CT.127 However, these phenomena have not been thoroughly explained mechanistically. More recent data additionally point to the importance of the structure of the MA lattice in rescuing or permitting Env incorporation.131 The long-range structure of MA may be important either to allow a MA–Env interaction through avidity effects, compensating for a hypothetically low-affinity interaction between the two proteins; alternatively, the structure of the MA lattice may be an adaptation to the large CT of lentiviruses, potentially accommodating the tail by relieving steric hindrance between MA and the CT.7
A potential role for Env in nucleating assembly, rather than simply packaging into particles has also been reported. In RSV, a correlation was observed between the number of Env spikes on a particle and the completed massembly of the viral core.132 The mechanism by which Env would achieve this nucleation is unknown; an attractive model would involve the Env CT interacting with Gag to promote a region of high Gag concentration, which in turn would promote core formation.132 If such a mechanism is at work in RSV, it would likely be found in other retroviruses, although it would perhaps be less likely in those viruses with low virion Env content, such as HIV-1.133
4.4. Signaling
The CT domain of HIV-1 gp41, in addition to containing motifs involved in regulating its own trafficking, has been shown to impact other cellular functions through signaling motifs. The most persuasive evidence that the CT influences signaling derives from the finding that the CT activates NF-κB through the canonical pathway, via an interaction with the kinase TAK1.134 The same study showed that the conserved CT motif responsible for the interaction in HIV-1 gp41 is also present and functional in SIVmac239 gp41. The critical residues of the signaling motif are also conserved in HIV-2 and a variety of other SIV strains.134 As the HIV-1 5′LTR contains NF-κB responsive elements, this activation of NF-κB may enhance viral gene expression. Activation of NF-κB may also contribute indirectly to enhancing viral gene expression through activation of T cells harboring the provirus.
A potential NF-κB activating domain has also been identified in the Env CT of bovine leukosis virus (BLV), a deltaretrovirus closely related to HTLV.135 This domain contains two motifs of Tyr-X-X-Leu/Ile. BLV Env induces activation of B and T cells, as assayed by induction of IL-2 production.135 The signaling pathway was not investigated in this study, but the similarity between BLV and HIV-1 motifs suggests that the NF-κB pathway may again be involved. In contrast, the CTs of HTLV-1 and simian retrovirus 3 Env are unable to induce activation of NF-κB, suggesting that such signaling is common, but not ubiquitous, in retroviruses.135
The betaretroviruses enzootic nasal tumor virus (ENTV-1) and Jaagsiekte sheep retrovirus (JSRV) disrupt cell signaling through perturbation of phosphatidylinositide 3-kinase (PI3K) signaling, a distinct mechanism from that of BLV or HIV-1. JSRV causes lung cancer in infected animals,136 and induces tumor formation in infected cells.137 In contrast to most transforming retroviruses that carry an activated oncogene or induce tumors via insertional mutagenesis, the JSRV Env glycoprotein is responsible for malignant transformation, with an SH2-binding motif (Tyr-X-X-Met) in the CT of the TM protein playing a central role.138,139 ENTV predominantly infects goats, and shares the Env-mediated ability to transform cells in culture.140 The CTs of both ENTV-1 and JRSV Env contain the Tyr-X-X-Met motif that contributes to the activation of cAkt via PI3K signaling,141,142 and inhibitors of PI3K signaling have been shown to inhibit transformation by ENTV-1 and JSRV.143,144 The specific mechanism by which Env drives transformation remains unclear; in a conventional PI3K docking site, the tyrosine residue would be phosphorylated, however, no evidence for phosphorylation or direct interaction with PI3K has been obtained for JSRV Env.142 In addition, the importance of the CT in relation to other regions of Env varies depending on cell type.142,145–147
4.5. Role of the CT of retroviral TM proteins in regulating Env conformation
A number of studies have demonstrated that the CT of retroviral Env proteins influences the conformation of Env sequences on the exterior of the membrane (for review see Ref. 148). This so-called “inside-out” regulation can affect Env-mediated fusion, reactivity with anti-Env antibodies, and sensitivity to antibody-mediated neutralization. To activate the fusion activity of the MLV Env complex, the MLV PR cleaves the Env CT to remove the C-terminal 16 amino acids from the C-terminus of Env. The removal of the C-terminal so-called “R peptide” produces a glycoprotein with greatly increased fusogenic capacity.149 It should be noted, however, that truncation of the MLV Env CT beyond the R peptide inhibits the fusogenicity of MLV Env; this result was also seen in xenotropic murine leukemia virus-related virus (XMRV).74,150 The truncated MLV Env glycoprotein displays greater reactivity with antibodies to the SU subunit of the Env complex, consistent with an altered conformation.151 Early studies with SIV demonstrated that Env truncation likewise altered antibody reactivity to epitopes in the ectodomain of the TM glycoprotein152 and later work showed that HIV-1 gp41 truncation increased exposure of a CD4-induced epitope in gp120.153,154 Consistent with a connection between the gp41 CT and conformational changes at the gp120 CD4-binding site, truncation of the tail was found to contribute to CD4 independence of an HIV-1 isolate.155,156 Point mutations in LLP2 that did not significantly affect virus infectivity were shown to render mutant virions resistant to neutralizing antibodies157; truncation of the gp41 CT also increased sensitivity to antibody-mediated neutralization in a cell–cell transfer model.158
A role for the Env CT in regulating the conformation and function of the external portions of the glycoprotein complex extends to the betaretrovirus JSRV. Truncation of the JSRV Env CT led to markedly enhanced cell–cell fusion and an increase in the amount of shed SU glycoprotein, suggesting an effect of the truncations on SU/TM interactions.159 This may be related to the observation in HIV-1, that immature particles are more resistant to SU shedding when treated with soluble CD4.160 Both observations are likely reflections of the influence of Gag on Env, mediated via the TM CT.
The take-home message from these studies is that mutations (both deletions and point mutations) in the CT of retroviral Env proteins can induce changes in the conformation and activity of the ectodomain of TM and in SU; the mechanism by which this occurs remains to be clarified, but could in some cases relate to altered interaction of the CT with Gag or with the inner leaflet of the cellular/viral membrane.
4.6. Role of Gag and virion maturation in regulating Env function
In multiple retroviral systems, the fusogenicity of the viral Env glycoprotein complex is linked to the state of virion maturation. This regulation, which is presumably in place to limit premature membrane fusion during particle production, is achieved via two main mechanisms: (1) cleavage of the Env CT by the viral PR concomitant with particle release and maturation, and (2) suppression of Env fusogenicity by unprocessed Gag in the immature virion. Cleavage of the CT by PR has been observed with a gammaretrovirus [MLV73,161,162]; a betaretrovirus [Mason-Pfizer monkey virus (M-PMV)163]; and a lentivirus [equine infectious anemia virus (EIAV)164]. This strategy allows the C-terminal end of the CT to be present during assembly to facilitate Env incorporation into virions, and then be removed to activate fusion. The mechanism by which Env CT truncation enhances fusion is not fully understood, but, as mentioned above, a number of studies have demonstrated that such truncations alter the conformation of the ectodomain of the TM protein (e.g., Refs. 151–154).
In the case of HIV-1, which does not normally undergo PR-mediated processing of the gp41 CT, two initial reports observed that the fusogenic activity of the Env complex was markedly lower in immature virions (containing uncleaved Gag) than in mature virions.165,166 In studies of cell–cell transfer, it was similarly observed that although immature particles can transfer into target cells, they do not fuse until maturation has occurred.167
This suppression of Env fusion activity could be overcome by deletion of the gp41 CT, suggesting that an interaction between the long CT and uncleaved Gag was responsible for the fusion defect, perhaps by limiting the mobility of the Env complex in the viral membrane or by constraining the ability of Env to undergo conformational changes necessary to catalyze membrane fusion. Indeed, super-resolution microscopy analysis revealed that Env clustered on the surface of HIV-1 virions in a manner that depended upon the gp41 CT.168 Env on immature and mature virions was found to react differentially with several antibodies specific for the gp41 ectodomain169; this effect was partially alleviated by gp41 CT truncation.
As mentioned above, the HIV-1 Env CT does not typically undergo PR-mediated processing. However, propagation of HIV-1 in the presence of a cholesterol-binding entry inhibitor [amphotericin B methyl ester (AME)] led to the selection of resistant viruses that had acquired mutations in the gp41 CT that resulted in PR-mediated cleavage at the site of the mutations.84,85 These HIV-1 isolates thus behaved like MLV, M-PMV, and EIAV in that PR cleaved their CTs within the virus particle. This strategy for resistance allowed the CT to be present during assembly to promote Env incorporation into virions, and then be removed to activate fusion activity and confer AME resistance. When selection for AME resistance was performed with SIVmac, which tolerates gp41 CT truncations when virus is propagated in human cells, the virus acquired a stop codon in the gp41 CT rather than a site for PR-mediate CT cleavage.84 There is also evidence that mutations in the HIV-1 gp41 CT can contribute to PR-inhibitor resistance; however, the mechanism for this resistance is not clear.170 Because interactions between uncleaved Gag and the gp41 CT suppress Env fusion activity,165,166 PR inhibitors act at both the level of virus maturation and Env function during entry.170 One could therefore speculate that mutations in the CT that modulate interactions with Gag could allow escape from the Env-dependent component of PR-inhibitor activity.
The hypothesis that uncleaved Gag regulates Env mobility in the membrane gains support from several recent studies that used super-resolution microscopy to measure the localization of Env at budding sites and on virions. Env was found to cluster at sites of Gag assembly; deletion of the gp41 CT, or mutations in MA that block Env incorporation, resulted in a loss of Env/Gag coclustering.105 Similarly, the mobility of Env on the cell surface was constrained by Gag in a gp41 CT-dependent manner.121 The hypothesis that the gp41 CT interacts directly with uncleaved Gag in immature virions is supported by the following observations: removal of the viral membrane of mature virions by detergent treatment results in the loss of Env from the detergent-stripped virions; however, when the same experiment is performed with immature virions (composed of unprocessed Gag) Env is retained following removal of membrane. This retention of Env in the detergent-stripped particles is lost if the gp41 CT is truncated.171
4.7. HIV-1 versus SIV gp41 CT
Although, there have been occasional reports of naturally occurring isolates of HIV-1 that bear truncated gp41s,172,173 the loss of the gp41 CT is most commonly observed with SIV propagated in human cells.72,174,175 Intriguingly, such SIV isolates lacking their gp41 CTs reacquire full-length CTs when passaged in simian cells in vitro72,174 and in monkeys.75 These differences may reflect use of alternative trafficking pathways by HIV-1 and SIV Env, and the interaction motifs for these putative anterograde trafficking cofactors may reside in different parts of the CT; however, the basis for this species-specific preference for long versus short gp41 CT in simian versus human cells, respectively, has not been elucidated.
4.8. Role of Env in counteracting BST-2
BST-2 (also known as CD317, HM1.24, and tetherin) is an interferon-induced viral restriction factor, with activity against a wide range of enveloped viruses.176–179 It is expressed on the PM and causes the accumulation of viral particles, which are trapped at the cell surface after completion of the budding process.180 The primary mechanism of viral restriction by BST-2 in vivo is unclear, but may include direct inhibition of release through tethering, enhanced exposure of virions to the immune system, and signaling through NF-κB.178,179,181–185 Whatever the mechanism, the potency of BST-2 restriction in vivo is illustrated by the fact that a number of viruses have evolved strategies to counteract BST-2.186 HIV-1 Vpu and SIV Nef proteins have been shown to counter BST-2187; however, in the case of HIV-2 and FIV, this function is associated with the Env protein. The ability of HIV-2 Env to enhance virus release in HeLa and T-cell lines was identified before the antiviral role of BST-2 was discovered188,189; that activity was mapped to the CT of the HIV-2 Env.190,191 HIV-2 Env is able to interact with and down-regulate BST-2 from the PM, sequestering it in the trans-Golgi network.192,193 Two specific regions of HIV-2 Env are involved in counteracting BST-2. The membrane-proximal endocytic signaling motif, Gly-Tyr-X-X-Φ, is required for the down-regulation of surface BST-2; however, neither this motif nor any other part of the CT confers specificity for human BST-2.193 Interestingly, when nef-deficient SIV was passaged in a tetherin-expressing cell line, it acquired the ability to counteract BST-2 through mutations in the gp41 CT, mimicking the activity of HIV-2 gp41.194
The BST-2-countering function of FIV is less well understood than that of the primate lentiviruses, although like HIV-2, the Env protein confers this activity.195,196 Unlike HIV-2 Env, FIV Env must be packaged into particles to counter restriction by BST-2.195 It is not clear which domains of FIV Env are required to counteract BST-2, although it appears that the CT is dispensable.195 EIAV is also restricted by equine BST-2, when replicating in physiologically relevant cells, and the EIAV Env protein has been shown to interact with equine BST-2 and redistribute it from the PM to intracellular sites.197 As is the case for FIV, the specific domain of Env responsible for this activity is unknown; however, the mechanism by which EIAV counteracts BST-2 more closely resembles that of HIV-2 than FIV. Like HIV-2 Env, EIAV Env can counter BST-2 without being incorporated into particles, and does not cause BST-2 degradation.197 It is, therefore, probable that the EIAV Env CT is involved in counteracting BST-2-mediated restriction.
4.9. Env CT as a therapeutic target?
As detailed above, the CT of many retroviruses performs a variety of functions critical to the virus replication cycle; these functions include the “classical” functions of Env (packaging into viral particles and mediating fusion with target cells) as well as functions in regulating the cellular environment through signaling or interaction with cellular factors. The Env CT could then be amenable to targeting for antiviral therapy in conditions such as HIV-AIDS. Unfortunately, at the present time, there is a lack of information regarding the structure of any retroviral CT, particularly in the context of their interactions with the membrane and other proteins. As more information becomes available, it may become possible to target the Env CT to inhibit viral replication and pathogenicity, providing a novel approach to alleviating the problems of viral drug resistance.
5. CONCLUSIONS
All retroviral Env glycoproteins share a common structural arrangement, with the CT being the most variable region. It seems likely that all CTs play a role in the trafficking and incorporation of Env into particles. In addition, for many retroviruses, the Env CT functions in the regulation of Env fusogenicity. In some cases, the CT influences signaling within the cell, typically stimulating pathways that contribute to activating cells that may otherwise remain in a resting, inactive state unfavorable for viral gene expression. In the case of lentiviruses, the large CT poses particular questions. Most viruses, including most retroviruses, have only a small CT; organisms as genetically efficient as viruses do not encode large non-functional domains. Additional functions can be attributed to the large lentiviral CT that are not shared by their smaller-CT relatives, such as the countering of BST-2 by the HIV-2 Env CT. In many cases, however, the activities ascribed to the large lentiviral CT are functions that in theory could also be performed by a small CT. It is unclear whether the additional size is required for specific functions or represents a domain with a greater number of functions. The reduction in replication efficiency by CT-deleted retroviruses in their authentic hosts clearly demonstrates that this region of the virus plays important roles, including some that no doubt have yet to be described.
ACKNOWLEDGMENTS
We thank the members of the Freed laboratory for productive discussions. Work in the Freed laboratory is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, and by the Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134(1–2):221–234. 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Lerat E, Capy P. Retrotransposons and retroviruses: analysis of the envelope gene. Mol Biol Evol. 1999;16(9):1198–1207. [DOI] [PubMed] [Google Scholar]
- 5.McClure MA. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991;8(6):835–856. [DOI] [PubMed] [Google Scholar]
- 6.Kim FJ, Battini J-L, Manel N, Sitbon M. Emergence of vertebrate retroviruses and envelope capture. Virology. 2004;318(1):183–191. 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 7.Tedbury PR, Freed EO. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol. 2014;22(7):372–378. 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt VM. Virion proteins In: Coffin JM, Hughes SH, Varmus HE, eds. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 9.Hunter E Membrane fusion In: Coffin JM, Hughes SH, Varmus HE, eds. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 10.Blumenthal R, Durell S, Viard M. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem. 2012;287(49):40841–40849. 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64(12):6314–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabuzda D, Olshevsky U, Bertani P, Haseltine WA, Sodroski J. Identification of membrane anchorage domains of the HIV-1 gp160 envelope glycoprotein precursor. J Acquir Immune Defic Syndr. 1991;4(1):34–40. [PubMed] [Google Scholar]
- 13.Shang L, Yue L, Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell–cell fusion and virus infection. J Virol. 2008;82(11):5417–5428. 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Yue L, Shang L, Hunter E. Truncation of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein defines elements required for fusion, incorporation, and infectivity. J Virol. 2009;83(22):11588–11598. 10.1128/JVI.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyauchi K, Curran AR, Long Y, et al. The membrane-spanning domain of gp41 plays a critical role in intracellular trafficking of the HIV envelope protein. Retrovirology. 2010;7:95 10.1186/1742-4690-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410(4):582–608. 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolly C, Sattentau QJ. Retroviral spread by induction of virological synapses. Traffic. 2004;5(9):643–650. 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 19.Bangham CRM. The immune control and cell-to-cell spread of human T-lymphotropic virus type 1. J Gen Virol. 2003;84(Pt 12):3177–3189. [DOI] [PubMed] [Google Scholar]
- 20.Majorovits E, Nejmeddine M, Tanaka Y, Taylor GP, Fuller SD, Bangham CRM. Human T-lymphotropic virus-1 visualized at the virological synapse by electron tomography. PLoS One. 2008;3(5):e2251. 10.1371/journal.pone.0002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto N, Okada M, Koyanagi Y, Kannagi M, Hinuma Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science. 1982;217(4561):737–739. 10.1126/science.6980467. [DOI] [PubMed] [Google Scholar]
- 22.Petropoulos C Retroviral taxonomy, protein structures, sequences, and genetic maps In: Coffin JM, Hughes SH, Varmus HE, eds. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 23.Freed EO, Martin MA. HIVs and their replication In: Knipe DM, Howley PM, eds. Field’s Virology. 6th ed Philadelphia, PA: Lippincott, Williams, and Wilkins; 2013:1502–1560. [Google Scholar]
- 24.Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2(7):a006924. 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müllers E, Uhlig T, Stirnnagel K, Fiebig U, Zentgraf H, Lindemann D. Novel functions of prototype foamy virus Gag glycine- arginine-rich boxes in reverse transcription and particle morphogenesis. J Virol. 2011;85(4):1452–1463. 10.1128/JVI.01731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono A, Orenstein JM, Freed EO. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol. 2000;74(6):2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101(41):14889–14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006;103(30):11364–11369. 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saad JS, Loeliger E, Luncsford P, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366(2):574–585. 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3(8):643–655. 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 31.Rein A, Datta SAK, Jones CP, Musier-Forsyth K. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem Sci. 2011;36(7):373–380. 10.1016/j.tibs.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huseby D, Barklis RL, Alfadhli A, Barklis E. Assembly of human immunodeficiency virus precursor gag proteins. J Biol Chem. 2005;280(18):17664–17670. 10.1074/jbc.M412325200. [DOI] [PubMed] [Google Scholar]
- 33.Briggs JAG, Kräusslich H-G. The molecular architecture of HIV. J Mol Biol.2011;410(4):491–500. 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 34.Pornillos O, Ganser-Pornillos BK, Banumathi S, Hua Y, Yeager M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J Mol Biol. 2010;401(5):985–995. 10.1016/j.jmb.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14(3):232–241. 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrus JE, von Schwedler UK, Pornillos OW, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med.2001;7(12):1313–1319. 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 38.VerPlank L, Bouamr F, LaGrassa TJ, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc Natl Acad Sci U S A. 2001;98(14):7724–7729. 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99(2):955–960. 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack B, Calistri A, Craig S, Popova E, Go€ttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. [DOI] [PubMed] [Google Scholar]
- 41.Kikonyogo A, Bouamr F, Vana ML, et al. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A. 2001;98(20): 11199–11204. 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A. 2000;97(24): 13063–13068. 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell NM, Lever AML. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 2013;21(3):136–144. http://dx.doi.org/101016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Müllers E. The foamy virus Gag proteins: what makes them different? Viruses. 2013;5(4):1023–1041. 10.3390/v5041023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller PW, Huang RK, England MR, et al. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol. 2013;87(24):13655–13664. 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9(11):1398–1403. 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 47.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 48.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5(1):51 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laguette N, Sobhian B, Casartelli N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353): 654–657. 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hrecka K, Hao C, Gierszewska M, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaller T, Bauby H, Hué S, Malim MH, Goujon C. New insights into an X-traordinary viral protein. Front Microbiol. 2014;5:126 10.3389/fmicb.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haffar OK, Dowbenko DJ, Berman PW. Topogenic analysis of the human immune-deficiency virus type 1 envelope glycoprotein, gp160, in microsomal membranes. J Cell Biol. 1988;107(5):1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Felkner RH, Roth MJ. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol. 1992;66(7): 4258–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dedera DA, Gu RL, Ratner L. Role of asparagine-linked glycosylation in human immunodeficiency virus type 1 transmembrane envelope function. Virology. 1992;187(1):377–382. [DOI] [PubMed] [Google Scholar]
- 55.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc Natl Acad Sci U S A. 1988;85(22):8688–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tucker SP, Srinivas RV, Compans RW. Molecular domains involved in oligomerization of the Friend murine leukemia virus envelope glycoprotein. Virology. 1991;185(2):710–720. [DOI] [PubMed] [Google Scholar]
- 57.Earl PL, Moss B. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res Hum Retroviruses. 1993;9(7):589–594. [DOI] [PubMed] [Google Scholar]
- 58.Julien J-P, Cupo A, Sok D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyumkis D, Julien J-P, de Val N, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montefiori DC, Robinson WE, Mitchell WM. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1988;85(23):9248–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raska M, Takahashi K, Czernekova L, et al. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem. 2010;285(27):20860–20869. 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Chien PC, Tuen M, et al. Identification of an N-linked glycosylation in the C4 region of HIV-1 envelope gp120 that is critical for recognition of neighboring CD4 T cell epitopes. J Immunol. 2008;180(6):4011–4021. [DOI] [PubMed] [Google Scholar]
- 63.Raska M, Novak J. Involvement of envelope-glycoprotein glycans in HIV-1 biology and infection. Arch Immunol Ther Exp (Warsz). 2010;58(3):191–208. 10.1007/s00005-010-0072-3. [DOI] [PubMed] [Google Scholar]
- 64.Felts RL, Narayan K, Estes JD, et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci U S A. 2010; 107(30):13336–13341. 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68(2):1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3): 433–444. 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sloan RD, Kuhl BD, Mespléde T, Münch J, Donahue DA, Wainberg MA. Productive entry of HIV-1 during cell-to-cell transmission via dynamin-dependent endocytosis. J Virol. 2013;87(14):8110–8123. 10.1128/JVI.00815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103(4):679–689. [DOI] [PubMed] [Google Scholar]
- 69.Murakami T, Freed EO. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci U S A. 2000;97(1):343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74(10):4891–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez LG, Davis GL, Hunter E. Mutants of the Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J Virol. 1987;61(10):2981–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirsch VM, Edmondson P, Murphey-Corb M, Arbeille B, Johnson PR, Mullins JI. SIV adaptation to human cells. Nature. 1989;341(6243):573–574. 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 73.Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68(3):1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucas TM, Lyddon TD, Grosse SA, Johnson MC. Two distinct mechanisms regulate recruitment of murine leukemia virus envelope protein to retroviral assembly sites. Virology. 2010;405(2):548–555. 10.1016/j.virol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shacklett BL, Weber CJ, Shaw KE, et al. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J Virol. 2000;74(13):5836–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viard M, Ablan SD, Zhou M, et al. Photoinduced reactivity of the HIV-1 envelope glycoprotein with a membrane-embedded probe reveals insertion of portions of the HIV-1 Gp41 cytoplasmic tail into the viral membrane. Biochemistry. 2008;47(7):1977–1983. 10.1021/bi701920f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisenberg D, Wesson M. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers. 1990;29(1):171–177. 10.1002/bip.360290122. [DOI] [PubMed] [Google Scholar]
- 78.Srinivas S, Srinivas R, Anantharamaiah G, Segrest J, Compans R. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992; 267(10):7121–7127. [PubMed] [Google Scholar]
- 79.Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes. PLoS One. 2010;5(12):e15261. 10.1371/journal.pone.0015261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. Detailed topology mapping reveals substantial exposure of the “cytoplasmic” C-terminal tail (CTT) sequences in HIV-1 Env proteins at the cell surface. PLoS One. 2013;8(5):e65220. 10.1371/journal.pone.0065220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy RC, Henkel RD, Pauletti D, et al. Antiserum to a synthetic peptide recognizes the HTLV-III envelope glycoprotein. Science. 1986;231(4745):1556–1559. [DOI] [PubMed] [Google Scholar]
- 82.Liu S, Kondo N, Long Y, Xiao D, Iwamoto A, Matsuda Z. Membrane topology analysis of HIV-1 envelope glycoprotein gp41. Retrovirology. 2010;7:100 10.1186/1742-4690-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Postler TS, Martinez-Navio JM, Yuste E, Desrosiers RC. Evidence against extracellular exposure of a highly immunogenic region in the C-terminal domain of the SIVmac gp41 transmembrane protein. J Virol. 2012;86(2):1145–1157. 10.1128/JVI.06463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Waheed AA, Ablan SD, Roser JD, et al. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc Natl Acad Sci U S A. 2007;104(20):8467–8471. http://dx 10.1073/pnas.0701443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waheed AA, Ablan SD, Sowder RC, et al. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J Virol. 2010;84(6):3121–3126. 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ball JM, Mulligan MJ, Compans RW. Basolateral sorting of the HIV type 2 and SIV envelope glycoproteins in polarized epithelial cells: role of the cytoplasmic domain. AIDS Res Hum Retroviruses. 1997;13(8):665–675. [DOI] [PubMed] [Google Scholar]
- 87.Lodge R, Delamarre L, Lalonde JP, et al. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J Virol. 1997;71(7):5696–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Owens RJ, Compans RW. Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J Virol.1989;63(2):978–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Owens RJ, Dubay JW, Hunter E, Compans RW. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci U S A. 1991;88(9):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lodge R, Göttlinger H, Gabuzda D, Cohen EA, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68(8):4857–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ochsenbauer C, Dubay SR, Hunter E. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol Cell Biol. 2000;20(1):249–260. 10.1128/MCB.20.1.249-260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beer C, Pedersen L, Wirth M. Amphotropic murine leukaemia virus envelope protein is associated with cholesterol-rich microdomains. Virol J. 2005;2:36 10.1186/1743-422X-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li M, Yang C, Tong S, Weidmann A, Compans RW. Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J Virol. 2002;76(23):11845–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vzorov AN, Weidmann A, Kozyr NL, Khaoustov V, Yoffe B, Compans RW. Role of the long cytoplasmic domain of the SIV Env glycoprotein in early and late stages of infection. Retrovirology. 2007;4:94 10.1186/1742-4690-4-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74(7): 3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schwarzer R, Levental I, Gramatica A, et al. The cholesterol-binding motif of the HIV-1 glycoprotein gp41 regulates lateral sorting and oligomerization. Cell Microbiol. 2014;16(10):1565–1581. 10.1111/cmi.12314. [DOI] [PubMed] [Google Scholar]
- 97.Yang P, Ai L-S, Huang S-C, et al. The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants. J Virol. 2010;84(1):59–75. 10.1128/JVI.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan W-E, Lin H-H, Chen SS-L. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J Virol. 2005;79(13):8374–8387. 10.1128/JVI.79.13.8374-8387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J Virol. 2004;78(10):5500–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rousso I, Mixon MB, Chen BK, Kim PS. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc Natl Acad Sci U S A. 2000;97(25):13523–13525. 10.1073/pnas.240459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J Virol. 2006;80(11):5292–5300. 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qi M, Williams JA, Chu H, et al. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 2013;9(4):e1003278. 10.1371/journal.ppat.1003278, Swanstrom R, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Linford A, Yoshimura S, Nunes Bastos R, et al. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22(5):952–966. 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Junutula JR, De Maziére AM, Peden AA, et al. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15(5):2218–2229. 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muranyi W, Malkusch S, Müller B, Heilemann M, Kräusslich H-G. Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 2013;9(2):e1003198. 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.LaBranche CC, Sauter MM, Haggarty BS, et al. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69(9):5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Polydefkis M, Koenig S, Flexner C, et al. Anchor sequence-dependent endogenous processing of human immunodeficiency virus 1 envelope glycoprotein gp160 for CD4 + T cell recognition. J Exp Med. 1990;171(3):875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Callahan KM, Rowell JF, Soloski MJ, Machamer CE, Siliciano RF. HIV-1 envelope protein is expressed on the surface of infected cells before its processing and presentation to class II-restricted T lymphocytes. J Immunol. 1993;151(6):2928–2942. [PubMed] [Google Scholar]
- 109.Egan MA, Carruth LM, Rowell JF, Yu X, Siliciano RF. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70(10):6547–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rowell JF, Stanhope PE, Siliciano RF. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155(1):473–488. [PubMed] [Google Scholar]
- 111.Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238(2):305–315. 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 112.Wyss S, Berlioz-Torrent C, Boge M, et al. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter]. J Virol. 2001;75(6):2982–2992. 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Breed MW, Jordan APO, Aye PP, et al. Loss of a tyrosine-dependent trafficking motif in the simian immunodeficiency virus envelope cytoplasmic tail spares mucosal CD4 cells but does not prevent disease progression. J Virol. 2013;87(3):1528–1543. 10.1128/JVI.01928-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Breed MW, Jordan APO, Aye PP, et al. A single amino acid mutation in the envelope cytoplasmic tail restores the ability of an attenuated simian immunodeficiency virus mutant to deplete mucosal CD4 + T cells. J Virol. 2013;87(23):13048–13052. 10.1128/JVI.02126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berlioz-Torrent C, Shacklett BL, Erdtmann L, et al. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73(2):1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ilinskaya A, Heidecker G, Derse D. Opposing effects of a tyrosine-based sorting motif and a PDZ-binding motif regulate human T-lymphotropic virus type 1 envelope trafficking. J Virol. 2010;84(14):6995–7004. 10.1128/JVI.01853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Byland R, Vance PJ, Hoxie JA, Marsh M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell. 2007;18(2):414–425. 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Day JR, Münk C, Guatelli JC. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J Virol. 2004;78(3):1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bhakta SJ, Shang L, Prince JL, Claiborne DT, Hunter E. Mutagenesis of tyrosine and dileucine motifs in the HIV-1 envelope cytoplasmic domain results in a loss of Env-mediated fusion and infectivity. Retrovirology. 2011;8(1):37 10.1186/1742-4690-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boscia AL, Akabori K, Benamram Z, et al. Membrane structure correlates to function of LLP2 on the cytoplasmic tail of HIV-1 gp41 protein. Biophys J. 2013;105(3):657–666. 10.1016/j.bpj.2013.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roy NH, Chan J, Lambele M, Thali M. Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell–cell fusion. J Virol. 2013;87(13):7516–7525. 10.1128/JVI.00790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jorgenson RL, Vogt VM, Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol. 2009;83(9):4060–4067. 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gregory DA, Lyddon TD, Johnson MC. Multiple Gag domains contribute to selective MLV Env recruitment to MLV virions. J Virol. 2013;87(3). 10.1128/JVI.02604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74(8):3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Freed EO, Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69(3):1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mammano F, Kondo E, Sodroski J, Bukovsky A, Go€ttlinger HG. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69(6):3824–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beaumont E, Vendrame D, Verrier B, et al. Matrix and envelope coevolution revealed in a patient monitored since primary infection with human immunodeficiency virus type 1. J Virol. 2009;83(19):9875–9889. 10.1128/JVI.01213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nack U, Schnierle BS . Replacement of the murine leukemia virus (MLV) envelope gene with a truncated HIV envelope gene in MLV generates a virus with impaired replication capacity. Virology. 2003;315(1):209–216. 10.1016/S0042-6822(03)00519-1. [DOI] [PubMed] [Google Scholar]
- 129.Lewis BC, Chinnasamy N, Morgan RA, Varmus HE. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J Virol. 2001;75(19):9339–9344. 10.1128/JVI.75.19.9339-9344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lambeleé M, Labrosse B, Roch E, et al. Impact of natural polymorphism within the gp41 cytoplasmic tail of human immunodeficiency virus type 1 on the intracellular distribution of envelope glycoproteins and viral assembly. J Virol. 2007;81(1):125–140. 10.1128/JVI.01659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tedbury PR, Ablan SD, Freed EO. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog. 2013;9(11): e1003739. 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content:implications that nucleation controls morphogenesis. J Mol Biol. 2008;376(4): 1168–1181. 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu P, Chertova E, Bess J, et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100(26):15812–15817. 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Postler TS, Desrosiers RC. The cytoplasmic domain of the HIV-1 glycoprotein gp41 induces NF-κB activation through TGF-β-activated kinase 1. Cell Host Microbe. 2012;11(2):181–193. 10.1016/j.chom.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Beaufils P, Choquet D, Mamoun RZ, Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12(13):5105–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hofacre A, Fan H. Jaagsiekte sheep retrovirus biology and oncogenesis. Viruses. 2010;2(12):2618–2648. 10.3390/v2122618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maeda N, Palmarini M, Murgia C, Fan H. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc Natl Acad Sci U S A. 2001;98(8):4449–4454. 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu S-L, Miller AD. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene. 2007;26(6):789–801. 10.1038/sj.onc.1209850. [DOI] [PubMed] [Google Scholar]
- 139.Cousens C, Maeda N, Murgia C, Dagleish MP, Palmarini M, Fan H. In vivo tumor-igenesis by Jaagsiekte sheep retrovirus (JSRV) requires Y590 in Env TM, but not full-length orfX open reading frame. Virology. 2007;367(2):413–421. 10.1016/j.virol.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alberti A, Murgia C, Liu S-L, et al. Envelope-induced cell transformation by ovine betaretroviruses. J Virol. 2002;76(11):5387–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Palmarini M, Maeda N, Murgia C, De-Fraja C, Hofacre A, Fan H. A phosphatidylinositol 3-kinase docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J Virol. 2001;75(22):11002–11009. 10.1128/JVI.75.22.11002-11009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu S-L, Lerman MI, Miller AD. Putative phosphatidylinositol 3-kinase (PI3K) binding motifs in ovine betaretrovirus Env proteins are not essential for rodent fibroblast transformation and PI3K/Akt activation. J Virol. 2003;77(14):7924–7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maeda N, Fu W, Ortin A, de las Heras M, Fan H. Roles of the Ras-MEK-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt-mTOR pathways in Jaagsiekte sheep retrovirus-induced transformation of rodent fibroblast and epithelial cell lines. J Virol. 2005;79(7):4440–4450. 10.1128/JVI.79.7.4440-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maeda N, Fan H. Signal transduction pathways utilized by enzootic nasal tumor virus (ENTV-1) envelope protein in transformation of rat epithelial cells resemble those used by jaagsiekte sheep retrovirus. Virus Genes. 2008;36(1):147–155. 10.1007/s11262-007-0193-x. [DOI] [PubMed] [Google Scholar]
- 145.Allen TE, Sherrill KJ, Crispell SM, Perrott MR, Carlson JO, DeMartini JC. The jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J Gen Virol. 2002;83(Pt 11):2733–2742. [DOI] [PubMed] [Google Scholar]
- 146.Hofacre A, Fan H. Multiple domains of the Jaagsiekte sheep retrovirus envelope protein are required for transformation of rodent fibroblasts. J Virol. 2004;78(19):10479–10489. 10.1128/JVI.78.19.10479-10489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hull S, Lim J, Hamil A, Nitta T, Fan H. Analysis of jaagsiekte sheep retrovirus (JSRV) envelope protein domains in transformation. Virus Genes. 2012;45(3):508–517. 10.1007/s11262-012-0793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Steckbeck JD, Kuhlmann A-S, Montelaro RC. Structural and functional comparisons of retroviral envelope protein C-terminal domains: still much to learn. Viruses. 2014;6(1):284–300. 10.3390/v6010284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao Y, Zhu L, Benedict CA, Chen D, Anderson WF, Cannon PM. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72(7):5392–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Côté M, Zheng Y-M, Liu S-L. Membrane fusion and cell entry of XMRV are pH-independent and modulated by the envelope glycoprotein’s cytoplasmic tail. PLoS One. 2012;7(3):e33734. 10.1371/journal.pone.0033734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aguilar HC, Anderson WF, Cannon PM. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J Virol. 2003;77(2):1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Spies CP, Ritter GD, Mulligan MJ, Compans RW. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68(2):585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Edwards TG, Wyss S, Reeves JD, et al. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol. 2002;76(6):2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wyss S, Dimitrov AS, Baribaud F, Edwards TG, Blumenthal R, Hoxie JA. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J Virol. 2005;79(19):12231–12241. 10.1128/JVI.79.19.12231-12241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.LaBranche CC, Hoffman TL, Romano J, et al. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73(12):10310–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Edwards TG, Hoffman TL, Baribaud F, et al. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol. 2001;75(11):5230–5239. 10.1128/JVI.75.11.5230-5239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kalia V, Sarkar S, Gupta P, Montelaro RC. Antibody neutralization escape mediated by point mutations in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41. J Virol. 2005;79(4):2097–2107. 10.1128/JVI.79.4.2097-2107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Durham ND, Yewdall AW, Chen P, et al. Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol. 2012;86(14):7484–7495. 10.1128/JVI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Côté M, Zheng Y-M, Albritton LM, Liu S-L. Fusogenicity of Jaagsiekte sheep retrovirus envelope protein is dependent on low pH and is enhanced by cytoplasmic tail truncations. J Virol. 2008;82(5):2543–2554. 10.1128/JVI.01852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hammonds J, Chen X, Ding L, et al. Gp120 stability on HIV-1 virions and Gag-Env pseudovirions is enhanced by an uncleaved Gag core. Virology. 2003;314(2):636–649. 10.1016/S0042-6822(03)00467-7. [DOI] [PubMed] [Google Scholar]
- 161.Green N, Shinnick TM, Witte O, Ponticelli A, Sutcliffe JG, Lerner RA. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci U S A. 1981;78(10):6023–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Henderson LE, Sowder R, Copeland TD, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52(2):492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brody BA, Rhee SS, Sommerfelt MA, Hunter E. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci U S A. 1992;89(8):3443–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rice NR, Henderson LE, Sowder RC, Copeland TD, Oroszlan S, Edwards JF. Syn thesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64(8):3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Murakami T, Ablan S, Freed E, Tanaka Y. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J Virol. 2004;78(2):1026–1031. 10.1128/JVI.78.2.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wyma DJ, Jiang J, Shi J, et al. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol. 2004;78(7):3429–3435. 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dale BM, McNerney GP, Thompson DL, et al. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe. 2011;10(6):551–562. 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chojnacki J, Staudt T, Glass B, et al. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338(6106):524–528. 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 169.Joyner AS, Willis JR, Crowe JE, Aiken C. Maturation-induced cloaking of neutralization epitopes on HIV-1 particles. PLoS Pathog. 2011;7(9):e1002234. 10.1371/journal.ppat.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rabi SA, Laird GM, Durand CM, et al. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest. 2013;123(9):3848–3860. 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wyma DJ, Kotov A, Aiken C. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J Virol. 2000; 74(20):9381–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jones DR, Suzuki K, Piller SC. A 100-amino acid truncation in the cytoplasmic tail of glycoprotein 41 in the reference HIV type 1 strain RF. AIDS Res Hum Retroviruses. 2002;18(7):513–517. 10.1089/088922202317406664. [DOI] [PubMed] [Google Scholar]
- 173.Shimizu H, Hasebe F, Tsuchie H, Morikawa S, Ushijima H, Kitamura T. Analysis of a human immunodeficiency virus type 1 isolate carrying a truncated transmembrane glycoprotein. Virology. 1992;189(2):534–546. [DOI] [PubMed] [Google Scholar]
- 174.Kodama T, Wooley DP, Naidu YM, et al. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63(11):4709–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63(10):4395–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jouvenet N, Neil SJD, Zhadina M, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83(4):1837–1844. 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yondola MA, Fernandes F, Belicha-Villanueva A, et al. Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J Virol. 2011;85(6):2480–2491. 10.1128/JVI.02188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Neil SJD, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 179.Van Damme N, Goff D, Katsura C, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Perez-Caballero D, Zang T, Ebrahimi A, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139(3):499–511. 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Veillette M, Désormeaux A, Medjahed H, et al. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol. 2014;88(5):2633–2644. 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Arias JF, Heyer LN, von Bredow B, et al. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A. 2014;111(17):6425–6430. 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pham TN, Lukhele S, Hajjar F, Routy J-P, Cohen EA. HIV Nef and Vpu protect HIV-infected CD4 + T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology. 2014;11(1):15 10.1186/1742-4690-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe. 2012;12(5):633–644. 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. Stimulation of NF-κB activity by the HIV restriction factor BST2. J Virol. 2013;87(4):2046–2057. 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Früh K. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6(5):e1000913. 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang F, Wilson SJ, Landford WC, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6(1):54–67. 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J Virol. 1996;70(2):820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J Virol. 1996;70(12):8285–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bour SP, Aberham C, Perrin C, Strebel K. Lack of effect of cytoplasmic tail truncations on human immunodeficiency virus type 2 ROD env particle release activity. J Virol. 1999;73(1):778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bour S, Akari H, Miyagi E, Strebel K. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology. 2003;309(1):85–98. [DOI] [PubMed] [Google Scholar]
- 192.Hauser H, Lopez LA, Yang SJ, et al. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology. 2010;7:51 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Le Tortorec A, Neil SJD. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol. 2009;83(22):11966–11978. 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Serra-Moreno R, Jia B, Breed M, Alvarez X, Evans DT. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe. 2011;9(1):46–57. 10.1016/j.chom.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Morrison JH, Guevara RB, Marcano AC, et al. Feline immunodeficiency virus envelope glycoproteins antagonize tetherin through a distinctive mechanism that requires virion incorporation. J Virol. 2014;88(6):3255–3272. 10.1128/JVI.03814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Celestino M, Calistri A, Del Vecchio C, et al. Feline tetherin is characterized by a short N-terminal region and is counteracted by the feline immunodeficiency virus envelope glycoprotein. J Virol. 2012;86(12):6688–6700. http://dx.doiorg/10.1128/JVI.07037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yin X, Hu Z, Gu Q, et al. Equine tetherin blocks retrovirus release and its activity is antagonized by equine infectious anemia virus envelope protein. J Virol. 2014;88(2):1259–1270. 10.1128/JVI.03148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]