Abstract
Today, the treatment of bacterial infections is a major challenge, due to growing rate of multidrug-resistant bacteria, complication of treatment and increased healthcare costs. Moreover, new treatments for bacterial infections are limited. Oligonucleotide aptamers are single stranded DNAs or RNAs with target-selective high-affinity feature, which considered as nucleic acid-based affinity ligands, replacing monoclonal antibodies. The aptamer-based systems have been found to be talented tools in the treatment of microbial infections, regarding their promising anti-biofilm and antimicrobial activities; they can reduce or inhibit the effects of bacterial toxins, and inhibit pathogen invasion to immune cell, as well as they can be used in drug delivery systems. The focus of this review is on the therapeutic applications of aptamers in infections. In this regard, an introduction of infections and related challenges were presented, first. Then, aptamer definition and selection, with a brief history of aptamers development against various pathogens and toxins were reviewed. Diverse strategies of aptamer application in drug delivery, as well as, the effect of aptamers on the immune system, as the main natural agents of human defense against pathogens, were also discussed. Finally, the future trends in clinical applications of this technology were discussed.
Keywords: Aptamer, SELEX, Single stranded DNA or RNA, Antibiotic resistance, Drug delivery, Biofilm
Introduction
Despite significant advances in the prevention and treatment of infectious diseases, expanding day-to-day, however, infections are still one of the leading causes of death, worldwide [1]. During the past few years, the number of just approved antibacterial drugs with a new mechanism of action becomes to decline [2, 3]. Incorrect use of antibiotics leads to side effects, and more importantly, the development of bacterial drug resistance [4]. Due to the increased incidence of antibiotic-resistant bacterial strains, antibiotic selection for infection control is restricted. Antibiotic-resistant bacteria cause numerous death every year and have a direct impact on the economy [5]. Moreover, biofilm-forming bacteria, which result in chronic infections, have developed increased resistance against antibiotic treatments and host defense systems [6]. Therefore, new strategies for infection control and developing of novel antibiotics with less serious side effects, low toxicity and high efficacy seem necessary [7].
The small single-stranded DNAs or RNAs that bind their specific targets with high affinity and selectivity, are called aptamers and produced by a method named systematic evolution of ligands by exponential enrichment (SELEX) or other modified SELEX strategies [8–10]. More than other diverse analytical and clinical applications in diagnosis and treatment, aptamers have also been evaluated as an antimicrobial agent [11]. Today, aptamers are considered in a range of clinical evaluations for a wide range of human disorders. They are powerful and flexible tools for therapeutic goals [12, 13]. In this review, we focused on the role of aptamers in the treatment of microbial infections.
Aptamer and SELEX
Aptamers; definition and history
Aptamers are small single-stranded oligonucleotides (DNAs or RNAs) with low molecular weight (ranging 5–40 kDa), which form a specific three-dimensional structure. Compared with antibodies, aptamers have minor complexity and immunogenicity, in addition to higher affinity and specificity against variety of targets (proteins, cells or small molecules). As well, they have more flexibility and therefore, can attach to interior non-accessible epitopes that cannot be easily targeted by antibodies. They also, show more stability, in addition to the fact that after denaturation, they can return to their proper primary conformation without loss of activity [14]. Several important attributes of aptamers make them a promising alternative to antibodies, including having thermal and chemical stability, simple chemical synthesis that reduce the cost of production and at the same time, it is possible to made modification through chemical synthesis process to make them more adaptable for different applications [15]. siRNAs, fluorophores, radioisotopes, electrochemical systems as well as various nanoparticles can be coupled with aptamers [15, 16]. In addition, aptamers can block the function of target proteins and as a result, they are promising candidates to develop new therapeutics for infections [15].
Aptamers have applied in diverse valuable fields of life sciences, for example, they have been developed as new drug, diagnostic and a bio-imaging agent, drug delivery agent, analytical reagent, food inspection and hazard detection [17]. In addition, aptamers functionalities are diverse, including their ability to block the interaction of receptor–ligand or activating the function of target receptors, as well as they can also act as promising targeting carriers to selectively deliver therapeutic agents to the target cells [18]. However, several disadvantages have been also reported for aptamers. They are sensitive to enzymatic degradation by nucleases in cells and blood circulation. Correspondingly, there are challenges regarding the topical or systemic administration of these drugs [19]. Low molecular weight and short time of aptamers residence in the circulation emerge significant challenges regards to renal filtration control [20]. Some drawbacks can be overcome by appropriated strategies, for example aptamers can be formulated with a large moieties to increase the molecular weight and reduce the renal filtration [21]. The chemical modifications or adding specific functional groups in order to improve aptamers lifetime and their stability in the body, can occur primarily in the SELEX protocol or through post-SELEX modification [22]. One of the challenge resolving strategy is to conjugate the aptamer with nanomaterials that can decrease various of the challenges ahead [19].
In vitro SELEX method
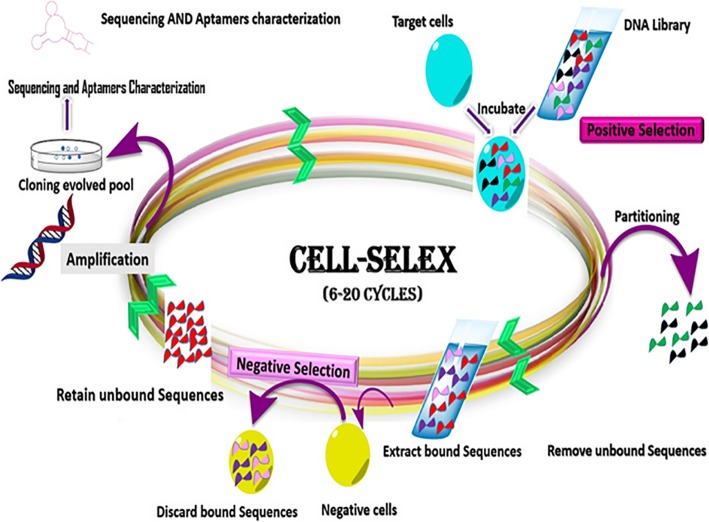
Aptamers are produced through an in vitro procedure, which specifically isolates aptamers for a target of interest and involves repetitive selection-amplification rounds, termed as SELEX [23]. The main advantage of this procedure is to select aptamers, as nucleic acid-based affinity ligands, without any prior knowledge of the target. One of the modified SELEX procedures is the Cell-SELEX, which is used to discover specific aptamers against a whole cell (Fig. 1). In other words, cells are considered as the target in selection process [24]. An oligonucleotide library consists of 1012 to 1016 distinct sequences. The random region of library is usually 40–100 bases long, flanked by constant primer sequences at both ends, facilitating PCR amplification [25]. The separation of the target-bound sequences from unbound ones via an affinity-based partitioning method and the amplification of the bound sequences by PCR are the important phases of SELEX method. Then, amplified sequences are converted into single strand forms and single stranded oligonucleotides enter the next cycle of SELEX, same as the previous round. The selection cycles are repeated several times to achieve high-affinity sequences in an enriched pool. The identity of DNA pool individuals is determined by cloning and then, sequencing [26] or by next generation sequencing techniques [27]. Several negative selection cycles are developed through a SELEX process to reduce the nonspecific bound oligonucleotides, to enhance the specificity of evolved pool. This type of SELEX has been extensively applied to develop aptamers against bacteria or other pathogenic cells. Low diversity of the DNA library sequence, PCR reagent contamination, contamination of the eluted sequences from the target cells, strong non-specific binding, low frequency of target molecules on the cells and the change in a cellular state can affect negatively the enrichment of the aptamer sequences [28].
Fig. 1.
Schematic presentation of DNA aptamer selection using SELEX method
Anti-bacterial DNA Aptamers
Antimicrobial activities of aptamers have been reported in a number of studies. Some of aptamers targeting bacterial infections are listed in Table 1 and several other important examples were discussed in following paragraphs.
Table 1.
A summary of aptamers selected against bacterial cells or toxins using SELEX technique, since 2015 till today
| Target type | Aptamer Name | Target | Aptamer Type | Sequence of the Aptamer (5′ to 3′) | Kd (nM) | Ref |
|---|---|---|---|---|---|---|
| Bacteria | Lyd-3 | S. pneumoniae | DNA |
TGACGAGCCCAAGTTACCTGCCCCCGAACCATACCACACG ATGCCCCGTACCCCAGCCACCAGAATCTCCGCTGCCTACA |
661.8 ± 111.3 | [29] |
| Apt3 | S. choleraesuis | DNA | GGCAGGACAACAGCGTGTAGTATCAGCTTACGGTG | 41 ± 2 | [11] | |
| BB16-11f | B. brevea | DNA | CTCCCAGGCCGTTGGGGCGTTGCCTGCGT |
18.66 ± 1.41 |
[30] | |
| CCFM641–5 | B. bifidumb | DNA | TGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGC | 10.69 ± 0.89 | [18] | |
| SAL 26 | S.typhimurium | DNA | TAGCTCACTCATTAGGCACATTTGTGGCACCAAATTTGAATTAATCAAGACAGTGTGGTGCATAGTTAAGCCAGCC | 123 ± 23 | [31] | |
| Bacterial toxin | CT916 | cholera toxin | DNA | GGCAAAAAGGATTGCCCAGGTCTGCTGTCTAGCCGGATTC | 48.5 ± 0.5 | [32] |
| S3 | SEA | DNA | CCCGCCTCTGAGCATTATTAATGTTATACCTTACGGCTGG | 36.93 ± 7.29 | [33] | |
| ML12 | LF | DNA | CGAGGGAGACGCGAACCTTCTCGCCTTGGG | 11.0 ± 2.7 | [34] | |
| C10 | enterotoxin C1 | DNA |
AGCAGCACAGAGGTCAGATGTATACTTCTAAAATTTGTTTGT ATCTACGATGTTCTTCGTCCTATGCGTGCTACCGTGAA |
65.14 ± 11.64 | [35] |
aBifidobacterium. Breve (B. breve)
bBifidobacterium. Bifidum (B. bifidum)
The multidrug-resistant (MDR) strains of bacteria such as Salmonella have developed due to inappropriate use of antibiotics. Accordingly, novel anti-microbial agents are required to fight these resistant strains. In a report, the effect of aptamer on colony formation of S. typhimurium and S. enteritidis was studied that indicated 30 min incubation of specific aptamer with bacteria could result in a significant growth inhibition. The antimicrobial effect of this aptamer might be the result of bacterial cell wall depolarization [36].
Mycobacterium tuberculosis (MTB), the bacterium that causes tuberculosis, is another important human pathogen that developed various mechanisms to multiply and survive resistantly in the lungs by confusing the immune system [37]. The polyphosphate kinase (PPK) gene in MTB regulates the intracellular metabolism of inorganic polyphosphate (polyP), which plays a main role in bacterial persistence. The inhibition of PPK activity resulted in the interference of polyP-dependent processes. Aptamer G9 is a therapeutic agent developed against MTB and at the concentration of 1 μM could completely inhibit PPK2 protein of bacteria. Through this process, it exhibited promising antimicrobial activity against MTB [38].
Aptamer-based therapeutic applications
Aptamers in the inhibition of biofilm formation
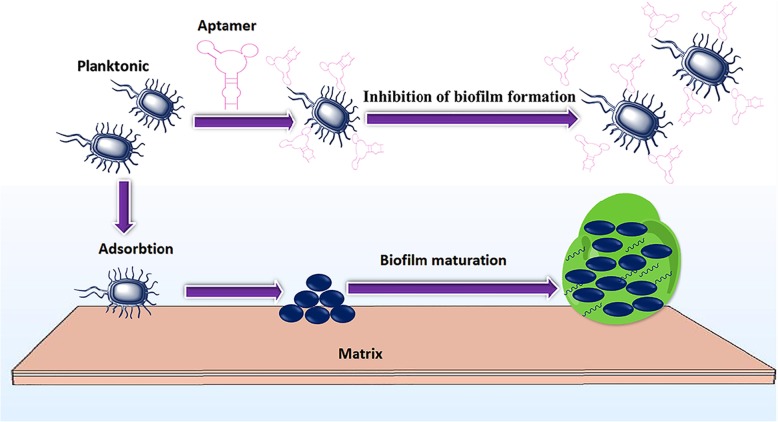
Biofilms are persistent communities of microbes that embedded in a matrix of exopolysaccharide and can adhere to surfaces. Biofilms are important in human infections that are hard to treat and cannot be easily eradicated with current antibiotics [39]. Chronic infections by biofilm-forming strains are associated with the accumulation of bacteria, which have a much more complex antibiotic resistance profile, due to the failure of antibiotics in penetrating the polysaccharide layer, as well as, biofilm resistaant against the human immune system [6]. Treatment of biofilm infections is currently a serious problem and antibiotic therapy alone is often inadequate for complete treatment. Biofilms are shown in more than 65% of microbial infections [40]. Motility and initial attachment are believed to be the major features for biofilm formation. It was found that the binding of specific aptamer to the flagella of Salmonella choleraesuis could result in the restriction of bacterial rotational frequency, due to the increment of the electrostatic repulsion of cells and surfaces and therefore, preventing the formation of mature biofilms (Fig. 2). In addition, it was shown that in the presence of aptamer, cells were more easily attacked by antibiotics. In this study, it was indicated that the pretreatment of biofilm forming S. choleraesuis with aptamer at the concentration of 1.1 μM, could substantially decrease the amount of ampicillin needed for the similar inhibition, as well as, prevented the development of antibiotic resistant strains [41]. It could be concluded that using aptamer pretreatment could eliminate the need for highly potent antibiotics in high dose.
Fig. 2.
schematic presentation of aptamer effect as an anti-biofilm agent. Aptamer has led to an early stage suppression of biofilm formation. In the absence of aptamer, flagella-mediated motility results in the formation of mature biofilms
Streptococcus pneumoniae is another fundamental pathogen that is involved in the majority of bacterial pneumonias and is the major cause of meningitis, septicemia, otitis media and sinusitis. All wild-type strains are able to form biofilms, in which state they are more invasive for lungs and brain [29, 42]. Among the known DNA aptamers against S. pneumoniae (Lyd-1, Lyd-2 and Lyd-3), Lyd-3 aptamer was reported as an effective preventative therapy with anti-biofilm activity, and in combination therapy with appropriate antibiotics, could prevent bacterial colonization. Lyd-3 aptamer at the concentration of 1 μM could reduce the biofilm formation from 100 to 35.8% [29]. The results of this study suggested that specific aptamers, as novel anti-biofilm agents can help to combat the increase rate of antibiotic resistance. Therefore, aptamers can offer a tremendous opportunity for effective treatment of chronic infection, and also, serve a better eradication of biofilm formation bacteria. In general, using aptamers with anti-biofilm activity can lessen the dose and duration of antibiotic therapy in chronic and resistant infections, thus, reduce the risk of abuse and misuse of antibiotic administration [43].
Aptamers for the inhibition of microbial toxins
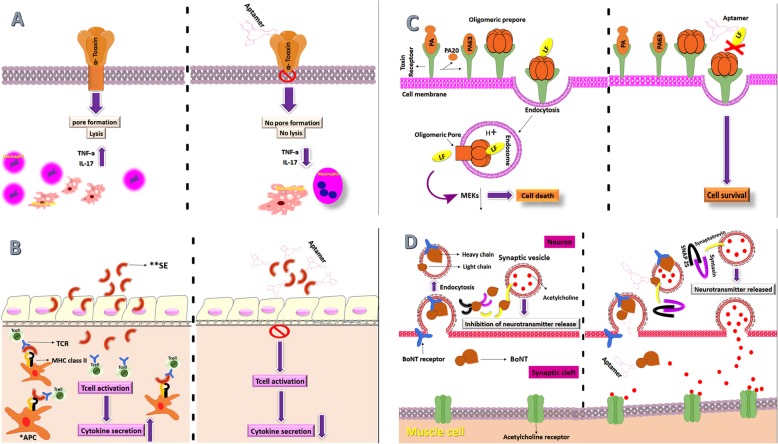
Toxins are powerful bioactive molecules that are produced by various bacterial pathogens and play a key role in the pathogenesis of bacteria. They are essential virulence factors allowing the organism to harm the host, through various physio-pathological pathways. Some toxins may target macrophages and neutrophils, resulting in the reduction of intrinsic immune responses and as a result, provide a suitable environment for active bacterial proliferation [44]. Bacterial toxins may degrade signaling pathways and disrupt the structure of host cells and lead to the persistence of infection [45]. Aptamers have been demonstrated as powerful inhibitory agents against bacterial toxins (Fig. 3).
Fig. 3.
Schematic presentation of aptamer function against different microbial toxins. A) After binding and oligomerization, in the way to create a pore, toxin heptamer inserts into the target cell and leading to cell lysis. In the presence of aptamer, after binding and oligomerization, pore formation does not occur, which inhibit cell lysis. B) *APC: Antigen presenting cell and **SE: Staphylococcal Enterotoxins. SE bound to MHC class II and TCR is expressed on CD4+ T cells. MHC class II, SEs and TCR interactions may result in hyper activation of the T cells, leading to the excessive proliferation of T cells and the uncontrolled burst of numerous pro inflammatory cytokines and chemokines. Aptamer inhibits T-cell activation, Therefore, the production of cytokines does not occur. C) PA (protective antigen) binds to the Anthrax Toxin Receptor that then interacts with LF to form the lethal toxin. Translocation of LF through the PA heptamer channel into the host cell cytosol results in cell death. Aptamers block the active site of LF and lead to cell survival. D) BONT (botulinum) is internalized into endosomes. In cytosol, proteolysis by the light chain cleavages SNARE proteins (synaptobrevin, SNAP25 and syntaxin) in the neurons and prevents release of the acetylcholine. Aptamers were bounded to light chain of toxin and caused a strong inhibition of endopeptidase activity
Staphylococcus aureus has been the most important human pathogen among the staphylococci with virulent toxin, by far. Although this organism is frequently a part of the normal human microflora, it can cause important opportunistic infections under the suitable conditions. S. aureus may cause a wide range of infections and quickly becomes resistant against many antibiotics [46]. One of the key virulence factors of S. aureus is alpha toxin, which can form heptameric pores in target cell membranes and cause tissue damage [46, 47]. Overexpression of TNF-α shows a major role in the destruction of epithelial cells and cell death, which caused by S. aureus alpha-toxin [48]. Four aptamers, AT-27, AT-33, AT-36 and AT-49 were developed, which specifically prevented the alpha toxin-induced cytolysis and activation of TNF-α and IL-17. The viability of cells in the presence of toxin after treatment with these four aptamers increased from 50 to 60% to 85–90% [47]. Moreover, enterotoxin B (SEB) from S. aureus can cause staphylococcal toxic shock syndrome (TSS), which is rarely observed with other enterotoxins types. SEB, one of the virulence agents of S. aureus, acts as superantigen, and stimulates T cells to produce a storm of cytokines, which manifested to clinical complications and food poisoning [49]. A11 is a reported aptamer that prevented SEB-mediated expression of inflammatory cytokines genes, which mediating septic shock. A11 could significantly inhibit SEB activity at the onset of the toxicity cascade, prior to pathological activation of T cells [50].
Another example is S3 aptamer, which could bind to Staphylococcal Enterotoxin A (SEA), as the main component of S. aureus pathogenesis. It effectively neutralized SEA, as well as, significantly decreased cytokine secretion. The blockage of the toxic cascade, caused by SEA, may be an effective approach for the treatment of SEA-related illnesses [33].
Bacillus anthracis is another important pathogen of the genus Bacillus that causes the emergence of anthrax. The virulence of B. anthracis depends on the production of anthrax toxin. The anthrax toxins consist of three components. One of them is protective antigen (PA), and the active PA heptamer can bind one or more molecules of edema factor (EF), lethal factor (LF) or both that facilitates the pass of this complex into the cell. EF with PA forms edema toxin. A main virulence factor of B. anthracis is the LF with zinc metalloprotease activity and is capable of cleaving a kinase enzyme, known as mitogen-activated protein kinase (MEK) family. Blocking the interaction between LF and MEK1 leads to the reduction of LF toxicity. An ssDNA aptamer (ML12) was reported that could bind strongly to and was an impressive inhibitor of LF. ML12 by blocking the active site of LF, prevented the protease activity and served as a promising drug candidate for neutralization of LF toxicity [34].
Another important toxin related syndrome is Botulism, caused by Clostridium botulinum. Botulinum neurotoxins (BoNT) are one of the most lethal toxins for humans, which also considered in biological weapons. Types A, B and E are the main causes of human illness and type A is the strongest one among other serotypes. This toxin is a 150 kDa protein, which is cleaved into 100 kDa and 50 kDa proteins linked by a disulfide bond; light chains (LCs) with zinc-endopeptidase activity and a nontoxic subunit (heavy chain). The carboxyl-terminal of the botulinum heavy chain binds to the target nerve cells and stimulates endocytosis of the toxin molecule. The LCs inactivates the proteins that regulate the release of acetylcholine, and results in the blocking of neurotransmission at peripheral cholinergic synapses. Three aptamers for BoNT were reported that were bounded to LC with inhibition constants in the low nM range and could cause a strong inhibition of endopeptidase activity [51].
Aptamers as intracellular delivery vehicles
Conventional therapy with antibiotics had faced serious restrictions, including low intracellular drug concentrations, loss of the drug through efflux pumps or enzymatic degradation and development of multi-drug resistance (MDR) strains. Drug delivery systems have introduced numerous profits, including improving treatment by increasing the efficacy and duration of drug activity, increasing patient satisfaction by reducing the required dose of drug, adaptation with convenient routes of administration, delivering to the target site to reduce the unwanted effects, and the reduction in cost of treatment that are generally due to their good stability, expanded loading capacity and improved physicochemical features [52].
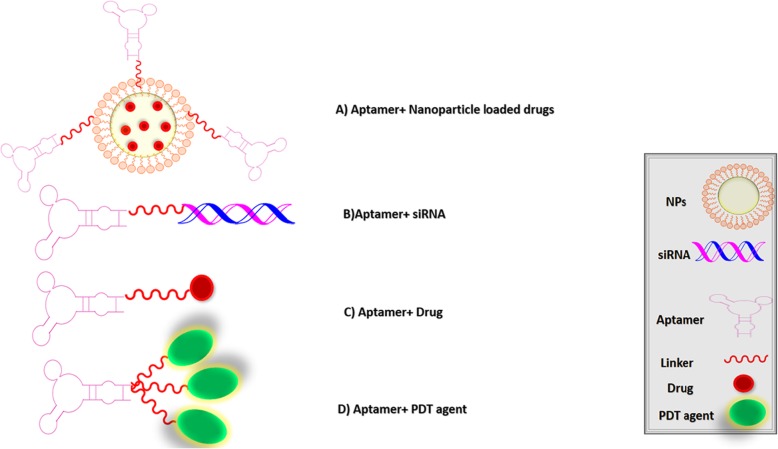
Aptamers can bind specifically to cell surface receptor molecules. Therefore, aptamer-based delivery systems can improve the cellular entrance of therapeutic agents. Aptamers can be designed for a wide range of applications through coupling with nanoparticles, drugs or other nucleic acids (nano carrier/ drug carrier) and play a major role in cell internalization (Fig. 4) [53].
Fig. 4.
Schematic presentation of the therapeutic applications of aptamers for microbial infections
Aptamer-nanoparticle (NPs) conjugates
NPs are small particles with the size of 1–100 nm, which exhibited the potential of antimicrobial activities. Thus, they are appropriately suited to fight microbial infections [54]. Antibiotics encapsulated in NPs have a large capacity to be replaced with antibiotics in free form. There are numerous benefits in nano-encapsulation strategy such as very-small size, a big surface-zone-to-mass ratio, high loading capacity and high reactivity, protection of antibiotics against physical inactivation, improvement in antibiotics pharmacokinetics, facilitation of the antibiotic release in infection area and reducing the required dose of drug [55, 56]. The assembly of aptamers- nanoparticles can result in an increased targeting and more efficient therapeutics [57]. Most NPs possess acceptable biocompatibility, and they can defend nucleic acids from the destruction caused by nuclease digestion. In this regard, aptamer conjugated NPs are promising tools in drug delivery [58].
Antimicrobial peptides (AMPs) are considered as a suitable drug candidate to treat MDR bacteria, but they are unstable in vivo and have a short half-life with low penetration into mammalian cells [59]. HPA3PHis, as an antimicrobial peptide, loaded onto a gold nanoparticle conjugated with Vibrio vulnificus specific DNA aptamer. The resulted composite promoted bacterial degradation by destroying the integrity of the V. vulnificus membrane, thus, caused bacterial cell death. In this study, following intravenous injection, AuNP-AptHis-HPA3PHis induced 0% mortality in infected mice, compared to the HPA3PHis alone (100% mortality). AuNP-AptHis exhibited bactericidal activity in concentrations more than 5 nM. AuNP-AptHis-HPA3PHis had several advantages; 1) The AuNP-AptHis enabled the efficient intracellular delivery of HPA3PHis to the host; 2) It increased the stability of HPA3PHis by protecting it from proteolysis; 3) It helped avoiding short-term reiterative administrations and reducing overall cost of therapy; 4) This system was simple and did not show any evident host toxicity [60]. On similar note, Yeom, J.-H et al. reported a C-terminally hexa-histidine-tagged A3-APO (A3-APOHis) AMP, which were loaded onto AuNPs conjugated with His-tag DNA aptamer (AuNP-AptHis) in order to successfully and effectively deliver the antimicrobial peptide into Salmonella-infected HeLa cells. Anti- His-tag aptamer improved the attachment of His-tag-AMP to NPs. The system was designed to improve AMP stability and its intracellular delivery and had potential impact in the elimination of intracellular Salmonella typhimurium infection by disrupting the bacterial cell. In vivo evaluation of this system in mice also indicated promising results [59].
The effect of aptamer-conjugated nanoparticles on the biofilm formation has been also evaluated. In a report, single-walled carbon nanotubes (SWNTs) with significant antimicrobial activity were utilized as a nanomaterial to detect Pseudomonas aeruginosa as well as it could increase the toxic effect of antimicrobial agents on bacterial biofilms. A specific aptamer (PA-ap1) against P. aeruginosa with the binding affinity in nM range had been developed. The conjugation of PA-ap1 to SWNTs could produce an effective concentration of the particles around bacterial cells, resulting in the disruption of bacteria accumulation. Compared with PA-ap1 and SWNTs alone, PA-ap1-conjugated SWNTs, caused a 36-fold higher inhibition of the biofilm formation. So, PA-ap1 increased the efficiency of anti-biofilm activity of SWNT [61].
Another effective aptamer- nanoparticle complex against biofilm was also reported. Graphene oxide (GO), is an extensively used chemically modified graphene derivative, which has anti-microbial activity. A research group was studied the conjugation of four different aptamer sequences (ST-1, ST-2, ST-3, ST-4) with GO in order to apply as an anti-biofilm agent in S. typhimurium model. ST-1 aptamer -conjugated graphene oxide (ST-1-GO) provides a powerful platform to impede biofilm formation through a targeted manner, showing the synergistic effect of aptamer and GO against biofilm formation. By using ST-1-GO, effective anti-biofilm concentration of GO was decreased from 125 mg/L to 100 mg/L, while the anti-biofilm effect was enhanced from 66.4 to 71.4%. Actually, GO entry into the biofilm was facilitated by aptamer ST-3 and they could reduce the cellular membrane potential to inhibit bacterial growth at the same time. The results support that ST-1 aptamer can be a therapeutic candidate for drug delivery [62].
Aptamer-siRNA/miRNA conjugates
Short interfering RNA (siRNA) are 21–23 base pairs (bp) in length and as an exogenous agent can experimentally manipulate gene expression. siRNAs, compared to similar therapeutic agents, exhibited minimal toxicity and high stability. siRNAs had demonstrated to have a promising modulatory effect on virulence, drug resistance and pathogenesis [63]. The use of siRNA–aptamer conjugates for drug delivery can result in the increase of favorable therapeutic efficacy and safety [24].
During a viral challenge, infected cells encoding miRNAs is capable of influencing the host cell RNAi system and induce transcriptional gene silencing (TGS) at the viral genome, thus forming latency in viral infection. In addition, the presentation of exogenous siRNA, miRNA and shRNA into infected cells that target integrated viral promoters can significantly repress viral transcription through TGS [64]. In HIV-1-infected cells, gp120 aptamer -LTR-362 siRNA conjugates induced TGS with a ~ 10-fold suppression of viral p24 levels and also, reduced CD4+ T cell depletion. Actually, gp120 aptamer functionally delivered LTR-362 siRNA and induced TGS of HIV-1. Generally, receptor-directed aptamers can deliver small RNAs that functionally modulate HIV-1 infection as well as viral transcription [65].
Aptamer- drug conjugates
According to the presence, as well as easily add functional groups to the terminals of oligonucleotide sequences, drug molecules can be easily bound to aptamers [66]. For example, ampicillin, by itself, is ineffective against biofilm formation in S. choleraesuis infections. Aptamer - ampicillin conjugate could produce synergistic results against biofilms. Aptamer as the antibiotic carrier, helped ampicillin to penetrate into the biofilm and kill the bacteria or reduce biofilms’ tolerance to drugs [11].
Tetracycline-loaded hydrogels were considered for controlled release by using DNA oligonucleotide as aptamer similar agent, that increased tetracycline loading and sustained its release. The study was designed based on the physicochemical features of nucleic acid sequences and possible interaction between oligonucleotide and tetracycline. This oligonucleotide could rise the partition coefficient of tetracycline between hydrogels and drug loading solutions and also, it could slow the release of tetracycline from hydrogels to increase bacterial inhibition [67].
Aptamer conjugates for photodynamic therapy (PDT)
PDT is a non-invasive technique includes three items: visible light, a photosensitizer (PS) and oxygen and has approved as an antimicrobial therapy in the treatment of infections [68, 69]. The studies discussed on the applications of aptamer-conjugated to a PS indicated that this complex could increase the selectivity of PDT [70]. Photothermal therapy (PTT) belongs to the group of phototherapy that produces reactive oxygen species, thereby inducing oxidative stress. They are slightly invasive by low toxicity for the nearby healthy tissue, in which photon energy is converted into heat [71, 72].
Methicillin resistant S. aureus (MRSA) is a famous pathogen in producing serious infections that resistant to many of the common antibiotics. In order to increase the MRSA treatment efficiency, an aptamer-functionalized gold nanorods (Apt@Au NRs) was introduced as a targeted PTT. It specifically bound to MRSA cells, resulting in recognition and inactivation of MRSA cells through hyper-thermia. The immobilization of aptamer on the surface of AuNRs led to the increase in targeted MRSA cell- binding affinity [73]. Light strongly is absorbed into the gold nanoparticles and causes the quick conversion of photon energy into heat [71].
Aptamer as an inhibitor of immune cells’ invasion
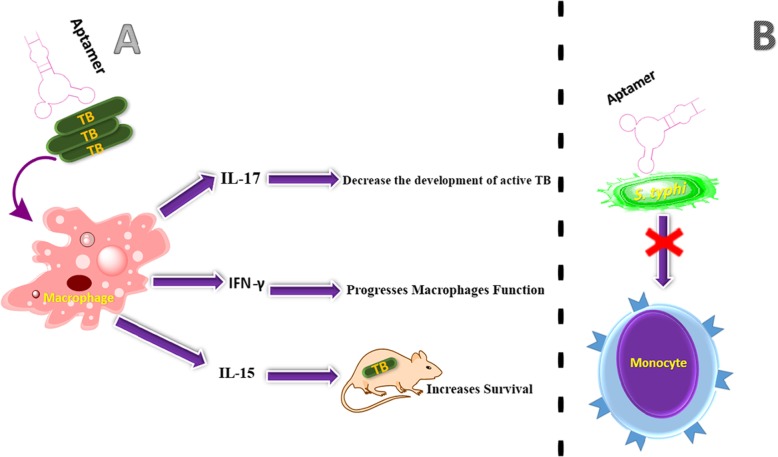
Bacterial specific aptamers can be a promising agent in the prevention of pathogen invasion to immune cell (Fig. 5). Some of the examples were illustrated in following section.
Fig. 5.
Schematic presentation of aptamer effect as an inhibitor of immune cell invasion
A) The aptamer binding to MTB leads to the inhibition of virulent MTB invasion to macrophages.
B) The aptamer with S.typhi leads to the entrance inhibition of bacteria into human monocytic cells.
Inhibits the invasion to macrophage
Some of infectious pathogens invade the host immune cells, which made the treatment complex. For example, during the past 200 years, there are almost 1 billion deaths caused by MTB (an intracellular bacterium), which survives and multiply within macrophages. Most of the macrophages are killed by the bacilli that makes it harder to control infection in MTB-HIV coinfection and in the cases of appearance of multiple drug resistant strains [74, 75]. After the activation of macrophages, MTB begins to multiply within cells, then, other macrophages have developed an enhanced ability to kill the bacilli infected cells. Since, invasion to the macrophages is a major step of the MTB infection process, specific aptamers against virulent markers of MTB can prevent its invasion to the immune cell. NK2 aptamer as MTB specific DNA aptamer, with a Kd in the nM range, considerably inhibited virulent MTB invasion to macrophage, in vitro, and virulent MTB that was affected by aptamer could stimulate IFN-γ, IL-15 and IL-17 secretion from macrophage [76]. IFN-γ led to enhance macrophages function against MTB. IL-15 and IL-17 increased survival of MTB - infected mice and decreased the development of active MTB, respectively [77–79].
Inhibits the invasion to monocytic cell
Salmonella enterica serovar typhi (S. typhi) produces a feverish disease called typhoid fever. IVB pili is expressed by S. typhi, which is the main character in the development of epidemics of typhoid fever. Monocytes and macrophages are critical effector molecules that play a crucial role in defense against Salmonella bacterial infections. Several aptamers were recently developed that specifically bind to type IVB pili and can interfere with bacterial pathogenesis, such as inhibit the bacterial aggregation or inhibit the entry of bacteria into human monocytic cells. In this study, RNA aptamers generated based on SELEX procedure, which one of them, S-PS8.4, exhibited the binding affinity in the nM range for S. typhi type IVB pili [80].
Application of Aptamers in the treatment of viral infections
Viral infections are a threat to human health. Effective and early treatment can improve the prognosis of the disease. Due to repeated virus mutations and viruses escaping the immune system, many viral drugs are ineffective. In addition, many antiviral drugs can cause severe side effects or may interfere with other drug agents and lead to low efficacy. As a targeted treatment, aptamers can be applied against a variety of viruses. They can be effective in antiviral therapy through several methods [81]. They can suppress viral nucleic acid replication by inhibiting nucleocapsid assembly that reduces extracellular DNA. Apt.No.28 against hepatitis B virus (HBV) exhibited this feature [82]. Aptamers can impede virus binding to host cells. As an example, the influenza virus hemagglutinin binds to the sialic acid receptor of the host cell, playing a fundamental role in initiating of viral infections. A DNA aptamer, A22, obstructed influenza virus infection by blocking the virus attachment to host cells [83]. Also, T14 and F34 aptamers prevent rabies virus from entering the host cells by blocking the interaction between virus and host cell receptors [84]. Moreover, in the penetration process of herpes simplex virus, the gD protein is a major determinant of herpes simplex virus-1 (HSV) entry. Two aptamers were selected, which interfered the interaction of the gD protein and the HSV-1 target cell receptor [85].
In fact, various enzymes are important in the virus replication cycle. Enzymes are attractive targets that can be targeted by antiviral aptamers [81]. RNA aptamers have been selected which could inhibit the enzyme activity of HIV tat and reverse transcriptase [86], hepatitis C virus (HCV) NS3 protease/helicase [87], NS5B RNA-dependent RNA polymerase [88], SARS (severe acute respiratory syndrome) coronavirus (SCV) NTPase/ Helicase [nsP10] [89]. Some aptamers developed as antiviral therapeutic agents are summarized in Table 2.
Table 2.
A summary of several aptamers developed for the treatment of viral infections by SELEX method
| Organism | Type of aptamer | Target | Aptamer therapeutic effect | Ref |
|---|---|---|---|---|
| HBV | DNA | Core protein |
-Prevent the assembly of the nucleocapsid -function by suppressing HBV replication |
[82] |
| HCV | DNA | Envelope Protein | -Inhibition of virus binding to host cells | [90] |
| Influenza A virus | DNA | Hemagglutinin protein |
-Blocking the binding of virus to target cell receptors -prevention of the virus invasion into the host cells |
[83] |
| Ebola Virus | RNA | VP35 | -Disrupt the eVP35− NP interaction | [91] |
| HIV-1 | RNA | gag protein | -Inhibit HIV-1 replication | [92] |
| HSV-1 | RNA | gD protein | -Blocking the gD functions | [85] |
| Vaccinia virus | DNA | Hemagglutinin protein |
-Recognize proteins expressed on the surface of VV- infected cells |
[93] |
| Influenza B Virus | RNA | Hemagglutinin protein | -Inhibited HA-mediated membrane fusion | [94] |
| SCV | RNA | nsP10 | -Inhibit double-stranded DNA unwinding activity of the helicase | [89] |
| Rabies virus | DNA |
Blocking the interaction between rabies virus and permissive host cell receptors |
-Reduced viral replication in an in vitro infection model | [84] |
VP35 viral protein 35, NP nucleoprotein, nsP10 NTPase/Helicase
Moreover, aptamers may be applicable in drug development. One example in this regard is RNA aptamers developed against aminoglycoside by in vitro selection to discover the interactions between aminoglycoside and natural RNAs, including HIV RNAs, which may have similar mechanism in their interactions. The in vitro selected aminoglycoside aptamers seem to have valuable applications in the discovery of the pathogenesis and treatment of HIV infection, including molecular mechanism of interaction with aminoglycoside antibiotics, and screening of efficient anti-HIV drug candidates [95].
Application of Aptamers in the treatment of parasitic infections
Parasitic infections having a serious impact on the health, worldwide [96]. Many of the drugs used today are not very effective and some of them can cause severe complications, so trying to find novel agents to control parasitic infections has great attention [97]. There are various strategies on the development of parasite-specific aptamers. They can prevent the interaction between the parasite and the host. Trypanosoma cruzi is a parasite that invade host cell. Ulrich, et al. identified an aptamer that could block receptor-ligand interactions between T. cruzi and epithelial monkey kidney LLC-MK2 host cells and thus, inhibit cell invasion [98]. Aptamers can inhibit the function of proteins [83]. Erythrocyte membrane protein 1 (EMP1) is a key factor in the pathogenicity of the Plasmodium falciparum. This protein involved in parasite adhesion to erythrocytes. Specific RNA aptamers are able to detect this protein on the surface of infected erythrocytes. So the aptamers act as an anti-rosetting drug [99]. Other aptamers that target protein are AptLiH2A#1 and AptLiH2A#2 that distinguished the LiH2A protein. These ssDNA aptamers bind to Leishmania infantum H2A with high affinity and does not identify other Leishmania proteins [100]. Another example for the inhibition of the function of proteins are poly (A)-binding protein (PABP) DNA aptamers (ApPABP#3, ApPABP#7 and ApPABP#11). They were able to bind PABP that is an essential protein for leishmania infantum survival. ApPABP#11, degrades the binding of PABP to poly(A), this feature may be applied in regulating the function of PABP in vivo [101]. EhCFIm25 is critical for parasite virulence and survival, EhCFIm25 silencing could reduce parasite mobility and induced cell death. RNA aptamers (C4 and C5) containing the GUUG motif were isolated, which inhibited Entamoeba histolytica’s growth by blocking of EhCFIm25 [102]. Several aptamers developed against parasitic infections are summarized in Table 3.
Table 3.
A summary of several aptamers developed for the treatment of parasitic infections by SELEX method
| Organism | Type of aptamer | Target | Aptamer therapeutic effect | Ref |
|---|---|---|---|---|
| E. histolytica | RNA | Polyadenylation factor EhCFIm25 | Inhibited parasite proliferation and led to cell death | [102] |
| L. infantum | DNA | rLiPABP | Affecting the physiological role of PABP | [101] |
| P. falciparum | RNA | EMP1 | Disruption and sequestration of parasites Reduced rosette formation | [99] |
| T. cruzi | RNA | Cell adhesion | Inhibit cell invasion | [98] |
| T. brucei | RNA | VSG proteins | Directing antibodies to the surface of the parasite | [103] |
rLiPABP recombinant 6xHIS-LiPABP protein, EMP1 erythrocyte membrane protein 1, VSG variant surface glycoproteins
Conclusions and perspectives
Despite significant advances in the treatment of infectious diseases over the past half century, there are still many problems associated with the development of effective therapies and overcoming the side effects of antibiotics and pathogen drug resistance. Aptamers could provide a strong tool for the expansion of new therapeutic factors with the ability to block the function of pathogen microorganisms, also, they are simple and cheap therapeutic agents with less side effects compared to traditional antibiotics. Different therapeutic approaches of aptamers application have been reported in the field of microbial infections. For example, aptamer could inhibit biofilm formation or inhibit the functionality of microbial toxins, therefore, they could be used as new therapeutic tools to combat chronic infections. In addition, aptamers could be conjugated with nanoparticles, siRNA/miRNA, therapeutic drugs, and PDT agents to formulate a new therapeutic composite, which specifically kill bacterial cells. Application of aptamer technology in the treatment of microbial infections is still in its primal phase, and various challenges need to be overcome. This review provides recently developed aptamer-based technologies for microbial infections that illustrated this promising technology as an actual alternative to the traditional approaches in infection therapy. Aptamers are applicable tools in the detection and defense against unknown pathogens (bacterial, fungal, and viral agents) and can become a valuable anti-infective therapeutic tool for clinicians in recent future.
Acknowledgements
NA
Authors’ contributions
Sh. A., M. P., R. R., M. T. and A. B. contributed in data gathering. Sh. A. wrote the main text and drew the Figs. M. T. edited the final version of the text. All authors read and confirmed the final file before submission.
Funding
NA
Availability of data and materials
NA
Ethics approval and consent to participate
NA
Consent for publication
NA
Competing interests
There is no conflict of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maryam Tabarzad, Email: m_tabarzad@sbmu.ac.ir.
Abbas Bahador, Email: ab.bahador@gmail.com.
References
- 1.Soares NC, Bou G, Blackburn JM. Proteomics of microbial human pathogens. Front Microbiol. 2016;7:1742. doi: 10.3389/fmicb.2016.01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Maisch T. Antimicrobial photodynamic treatment—a helpful approach in immunocompromised patients. Photodiagn Photodyn Ther. 2011;8(2):178. doi: 10.1016/j.pdpdt.2011.03.179. [DOI] [Google Scholar]
- 4.Komerik N, MacRobert AJ. Photodynamic therapy as an alternative antimicrobial modality for oral infections. J Environ Pathol Toxicol Oncol. 2006;25(1–2):487–504. doi: 10.1615/jenvironpatholtoxicoloncol.v25.i1-2.310. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Webster TJ. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J Orthop Res®. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Brooks BD, Brooks AE. Therapeutic strategies to combat antibiotic resistance. Adv Drug Deliv Rev. 2014;78:14–27. doi: 10.1016/j.addr.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 8.Tabarzad M, Jafari M. Trends in the design and development of specific Aptamers against peptides and proteins. Protein J. 2016;35(2):81–99. doi: 10.1007/s10930-016-9653-2. [DOI] [PubMed] [Google Scholar]
- 9.Tabarzad M, Kazemi B, Vahidi H, Aboofazeli R, Shahhosseini S, Nafissi-Varcheh N. Challenges to design and develop of DNA aptamers for protein targets. I. Optimization of asymmetric PCR for generation of a single stranded DNA library. Iran J Pharm Res. 2014;13(SUPPL):133–141. [PMC free article] [PubMed] [Google Scholar]
- 10.Vahidi H, Nafissi-Varcheh N, Kazemi B, Aboofazeli R, Shahhosseini S, Tabarzad M. Challenges to design and develop of DNA aptamers for protein targets. II. Development of the aptameric affinity ligands specific to human plasma coagulation factor viii using SEC-SELEX. Iran J Pharm Res. 2017;16(2):734–741. [PMC free article] [PubMed] [Google Scholar]
- 11.Lijuan C, Xing Y, Minxi W, Wenkai L, Le D. Development of an aptamer-ampicillin conjugate for treating biofilms. Biochem Biophys Res Commun. 2017;483(2):847–854. doi: 10.1016/j.bbrc.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Urmann K, Arshavsky-Graham S, Walter J-G, Scheper T, Segal E. Whole-cell detection of live lactobacillus acidophilus on aptamer-decorated porous silicon biosensors. Anal. 2016;141(18):5432–5440. doi: 10.1039/C6AN00810K. [DOI] [PubMed] [Google Scholar]
- 13.Marton S, Cleto F, Krieger MA, Cardoso J. Isolation of an aptamer that binds specifically to E. coli. PLoS One. 2016;11(4):e0153637. doi: 10.1371/journal.pone.0153637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groff K, Brown J, Clippinger AJ. Modern affinity reagents: recombinant antibodies and aptamers. Biotechnol Adv. 2015;33(8):1787–1798. doi: 10.1016/j.biotechadv.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Kadioglu O, Malczyk AH, Greten HJ, Efferth T. Aptamers as a novel tool for diagnostics and therapy. Investig New Drugs. 2015;33(2):513–520. doi: 10.1007/s10637-015-0213-y. [DOI] [PubMed] [Google Scholar]
- 16.Mokhtarzadeh A, Tabarzad M, Ranjbari J, de la Guardia M, Hejazi M, Ramezani M. Aptamers as smart ligands for nano-carriers targeting. TrAC Trends Anal Chem. 2016;82:316–327. doi: 10.1016/j.trac.2016.06.018. [DOI] [Google Scholar]
- 17.Song K-M, Lee S, Ban C. Aptamers and their biological applications. Sens. 2012;12(1):612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16(3):181. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Yigit MV, Mazumdar D, Lu Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Deliv Rev. 2010;62(6):592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wandtke T, Woźniak J, Kopiński P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7(2):751–780. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C, et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. 2004;21(12):2234–2246. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 22.Röthlisberger Pascal, Hollenstein Marcel. Aptamer chemistry. Advanced Drug Delivery Reviews. 2018;134:3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Ohuchi S. Cell-SELEX technology. BioResearch Open Access. 2012;1(6):265–272. doi: 10.1089/biores.2012.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo K-T, Ziemer G, Paul A, Wendel H. CELL-SELEX: novel perspectives of aptamer-based therapeutics. Int J Mol Sci. 2008;9(4):668–678. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorobyeva M, Davydova A, Vorobjev P, Pyshnyi D, Venyaminova A. Key aspects of nucleic acid library design for in vitro selection. Int J Mol Sci. 2018;19(2):470. doi: 10.3390/ijms19020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Blank M. Next-generation analysis of deep sequencing data: bringing light into the black box of SELEX experiments. Nucleic Acid Aptamers: Springer; 2016. pp. 85–95. [DOI] [PubMed] [Google Scholar]
- 28.Özalp VC, Bilecen K, Kavruk M, Avni ÖH. Antimicrobial aptamers for detection and inhibition of microbial pathogen growth. Future Microbiol. 2013;8(3):387–401. doi: 10.2217/fmb.12.149. [DOI] [PubMed] [Google Scholar]
- 29.Bayraç AT, Donmez SI. Selection of DNA aptamers to Streptococcus pneumonia and fabrication of graphene oxide based fluorescent assay. Anal Biochem. 2018;556:91–98. doi: 10.1016/j.ab.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Hu L, Wang L, Lu W, Zhai Q, Fan D, Liu X, et al. Selection, identification and application of DNA aptamers for the detection of Bifidobacterium breve. RSC Adv. 2017;7(19):11672–11679. doi: 10.1039/C6RA27672E. [DOI] [Google Scholar]
- 31.Lavu PSR, Mondal B, Ramlal S, Murali HS, Batra HV. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica serovar typhimurium. ACS Comb Sci. 2016;18(6):292–301. doi: 10.1021/acscombsci.5b00123. [DOI] [PubMed] [Google Scholar]
- 32.Frohnmeyer E, Frisch F, Falke S, Betzel C, Fischer M. Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA. J Biotechnol. 2018;269:35–42. doi: 10.1016/j.jbiotec.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Wu D, Chen Z, Zhang X, Yang X, Yang CJ, et al. Inhibition of the superantigenic activities of staphylococcal enterotoxin a by an aptamer antagonist. Toxicon. 2016;119:21–27. doi: 10.1016/j.toxicon.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Lahousse M, Park H-C, Lee S-C, Ha N-R, Jung I-P, Schlesinger SR, et al. Inhibition of anthrax lethal factor by ssDNA aptamers. Arch Biochem Biophys. 2018;646:16–23. doi: 10.1016/j.abb.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Chen X, Duan N, Wu S, Wang Z, Wei X, et al. Selection and characterization of DNA aptamers against Staphylococcus aureus enterotoxin C1. Food Chem. 2015;166:623–629. doi: 10.1016/j.foodchem.2014.06.039. [DOI] [PubMed] [Google Scholar]
- 36.Kolovskaya OS, Savitskaya AG, Zamay TN, Reshetneva IT, Zamay GS, Erkaev EN, et al. Development of bacteriostatic DNA aptamers for salmonella. J Med Chem. 2013;56(4):1564–1572. doi: 10.1021/jm301856j. [DOI] [PubMed] [Google Scholar]
- 37.Chuang Y-M, Belchis DA, Karakousis PC. The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. MBio. 2013;4(3):e00039–e00013. doi: 10.1128/mBio.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shum KT, Lui ELH, Wong SCK, Yeung P, Sam L, Wang Y, et al. Aptamer-mediated inhibition of mycobacterium tuberculosis polyphosphate kinase 2. Biochem. 2011;50(15):3261–3271. doi: 10.1021/bi2001455. [DOI] [PubMed] [Google Scholar]
- 39.Hall CW, Mah T-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 40.Percival S. Importance of biofilm formation in surgical infection. Br J Surg. 2017;104(2):e85–e94. doi: 10.1002/bjs.10433. [DOI] [PubMed] [Google Scholar]
- 41.Ning Y, Cheng L, Ling M, Feng X, Chen L, Wu M, et al. Efficient suppression of biofilm formation by a nucleic acid aptamer. Pathog Dis. 2015;73(6):ftv034. doi: 10.1093/femspd/ftv034. [DOI] [PubMed] [Google Scholar]
- 42.Henriques-Normark B, Tuomanen EI. The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med. 2013;3(7):a010215. doi: 10.1101/cshperspect.a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Mao B, Wu M, Liang J, Deng L. Influence of aptamer-targeted antibiofilm agents for treatment of Pseudomonas aeruginosa biofilms. Antonie Van Leeuwenhoek. 2018;111(2):199–208. doi: 10.1007/s10482-017-0941-4. [DOI] [PubMed] [Google Scholar]
- 44.do Vale A, Cabanes D, Sousa S. Bacterial toxins as pathogen weapons against phagocytes. Front Microbiol. 2016;7:42. doi: 10.3389/fmicb.2016.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson J, Schurr M, LeBlanc C, Ramamurthy R, Buchanan K, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad Med J. 2002;78(918):216–224. doi: 10.1136/pmj.78.918.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thay B, Wai SN, Oscarsson J. Staphylococcus aureus α-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One. 2013;8(1):e54661. doi: 10.1371/journal.pone.0054661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vivekananda J, Salgado C, Millenbaugh NJ. DNA aptamers as a novel approach to neutralize Staphylococcus aureus α-toxin. Biochem Biophys Res Commun. 2014;444(3):433–438. doi: 10.1016/j.bbrc.2014.01.076. [DOI] [PubMed] [Google Scholar]
- 48.Liang X, Ji Y. Involvement of alpha5beta1-integrin and TNF-alpha in Staphylococcus aureus alpha-toxin-induced death of epithelial cells. Cell Microbiol. 2007;9(7):1809–1821. doi: 10.1111/j.1462-5822.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- 49.Ortega E, Abriouel H, Lucas R, Gálvez A. Multiple roles of Staphylococcus aureus enterotoxins: pathogenicity, superantigenic activity, and correlation to antibiotic resistance. Toxins. 2010;2(8):2117–2131. doi: 10.3390/toxins2082117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Gan L, Jiang L, Zhang X, Yang X, Chen M, et al. Neutralization of Staphylococcal enterotoxin B by aptamer antagonist. Antimicrob Agents Chemother. 2015:59(4):2072-2077. [DOI] [PMC free article] [PubMed]
- 51.Chang T-W, Blank M, Janardhanan P, Singh BR, Mello C, Blind M, et al. In vitro selection of RNA aptamers that inhibit the activity of type a botulinum neurotoxin. Biochem Biophys Res Commun. 2010;396(4):854–860. doi: 10.1016/j.bbrc.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes GA. Chapter 2: Nanostructure-Mediated Drug Delivery. Nanomedicine in Cancer: Jenny Stanford Publishing; 2017. p. 47–72.
- 53.Gopinath SC, Lakshmipriya T, Chen Y, Arshad MM, Kerishnan JP, Ruslinda A, et al. Cell-targeting aptamers act as intracellular delivery vehicles. Appl Microbiol Biotechnol. 2016;100(16):6955–6969. doi: 10.1007/s00253-016-7686-2. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang F, Liu B, Lu J, Li F, Li D, Liang C, et al. Progress and challenges in developing aptamer-functionalized targeted drug delivery systems. Int J Mol Sci. 2015;16(10):23784–23822. doi: 10.3390/ijms161023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kavruk M, Celikbicak O, Ozalp V, Borsa B, Hernandez F, Bayramoglu G, et al. Antibiotic loaded nanocapsules functionalized with aptamer gates for targeted destruction of pathogens. Chem Commun. 2015;51(40):8492–8495. doi: 10.1039/C5CC01869B. [DOI] [PubMed] [Google Scholar]
- 57.Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ, Langer R, Farokhzad OC. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26(8):442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Wu Y, Phillips JA, Liu H, Yang R, Tan W. Carbon nanotubes protect DNA strands during cellular delivery. ACS Nano. 2008;2(10):2023–2028. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeom J-H, Lee B, Kim D, Lee J-k, Kim S, Bae J, et al. Gold nanoparticle-DNA aptamer conjugate-assisted delivery of antimicrobial peptide effectively eliminates intracellular Salmonella enterica serovar Typhimurium. Biomaterials. 2016;104:43–51. doi: 10.1016/j.biomaterials.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 60.Lee B, Park J, Ryu M, Kim S, Joo M, Yeom J-H, et al. Antimicrobial peptide-loaded gold nanoparticle-DNA aptamer conjugates as highly effective antibacterial therapeutics against Vibrio vulnificus. Sci Rep. 2017;7(1):13572. doi: 10.1038/s41598-017-14127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y, Wang X, Duan N, Xia Y, Wang Z, Che Z, et al. Selection and characterization, application of a DNA aptamer targeted to Streptococcus pyogenes in cooked chicken. Anal Biochem. 2018;551:37–42. doi: 10.1016/j.ab.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 62.Mao B, Cheng L, Wang S, Zhou J, Deng L. Combat biofilm by bacteriostatic aptamer-functionalized graphene oxide. Biotechnol Appl Biochem. 2018;65(3):355–361. doi: 10.1002/bab.1631. [DOI] [PubMed] [Google Scholar]
- 63.Gong F-Y, Zhang D-Y, Zhang J-G, Wang L-L, Zhan W-L, Qi J-Y, et al. SiRNA-mediated gene silencing of MexB from the MexA-MexB-OprM efflux pump in Pseudomonas aeruginosa. BMB Rep. 2014;47(4):203. doi: 10.5483/BMBRep.2014.47.4.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sampey GC, Guendel I, Das R, Jaworski E, Klase Z, Narayanan A, et al. Transcriptional gene silencing (TGS) via the RNAi machinery in HIV-1 infections. Biol. 2012;1(2):339–369. doi: 10.3390/biology1020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Lazar D, Li H, Xia X, Satheesan S, Charlins P, et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics. 2018;8(6):1575. doi: 10.7150/thno.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Catuogno S, Esposito C, de Franciscis V. Aptamer-mediated targeted delivery of therapeutics: an update. Pharmaceuticals. 2016;9(4):69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Soontornworajit B, Zhang Z, Chen N, Wang Y. Enhanced loading and controlled release of antibiotics using nucleic acids as an antibiotic-binding effector in hydrogels. Biomacromolecules. 2012;13(7):2202–2210. doi: 10.1021/bm3006227. [DOI] [PubMed] [Google Scholar]
- 68.Huang L, Dai T, Hamblin MR. Antimicrobial photodynamic inactivation and photodynamic therapy for infections. Photodynamic Therapy: Springer; 2010. pp. 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robertson CA, Evans DH, Abrahamse H. Photodynamic therapy (PDT): a short review on cellular mechanisms and cancer research applications for PDT. J Photochem Photobiol B Biol. 2009;96(1):1–8. doi: 10.1016/j.jphotobiol.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Mallikaratchy P, Tang Z, Tan W. Cell specific aptamer–photosensitizer conjugates as a molecular tool in photodynamic therapy. ChemMedChem. 2008;3(3):425–428. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang Xiaohua, El-Sayed Mostafa A. Plasmonic photo-thermal therapy (PPTT) Alexandria Journal of Medicine. 2011;47(1):1–9. doi: 10.1016/j.ajme.2011.01.001. [DOI] [Google Scholar]
- 72.Scheer A, Kirsch M, Ferenz KB. Perfluorocarbons in photodynamic and photothermal therapy. J Nanosci Nanomed. 2017;1(1):21–27.
- 73.Ocsoy I, Yusufbeyoglu S, Yılmaz V, McLamore ES, Ildız N, Ülgen A. DNA aptamer functionalized gold nanostructures for molecular recognition and photothermal inactivation of methicillin-resistant Staphylococcus aureus. Colloids Surf B: Biointerfaces. 2017;159:16–22. doi: 10.1016/j.colsurfb.2017.07.056. [DOI] [PubMed] [Google Scholar]
- 74.Gagneux S. Ecology and evolution of mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(4):202. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 75.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2(5):352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 76.Chen F, Zhang X, Zhou J, Liu S, Liu J. Aptamer inhibits mycobacterium tuberculosis (H37Rv) invasion of macrophage. Mol Biol Rep. 2012;39(3):2157–2162. doi: 10.1007/s11033-011-0963-3. [DOI] [PubMed] [Google Scholar]
- 77.Khan TA, Mazhar H, Saleha S, Tipu HN, Muhammad N, Abbas MN. Interferon-gamma improves macrophages function against M. tuberculosis in multidrug-resistant tuberculosis patients. Chemother Res Pract. 2016:7295390. [DOI] [PMC free article] [PubMed]
- 78.Maeurer MJ, Trinder P, Hommel G, Walter W, Freitag K, Atkins D, et al. Interleukin-7 or interleukin-15 enhances survival of mycobacterium tuberculosis-infected mice. Infect Immun. 2000;68(5):2962–2970. doi: 10.1128/IAI.68.5.2962-2970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Zhang M, Liao M, Graner MW, Wu C, Yang Q, et al. Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T cells. Am J Respir Crit Care Med. 2010;181(7):734–742. doi: 10.1164/rccm.200909-1463OC. [DOI] [PubMed] [Google Scholar]
- 80.Pan Q, Zhang X-L, Wu H-Y, He P-W, Wang F, Zhang M-S, et al. Aptamers that preferentially bind type IVB pili and inhibit human monocytic-cell invasion by Salmonella enterica serovar typhi. Antimicrob Agents Chemother. 2005;49(10):4052–4060. doi: 10.1128/AAC.49.10.4052-4060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou X, Wu J, Gu J, Shen L, Mao L. Application of Aptamers in virus detection and antiviral therapy. Front Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Z, Zhang J, Pei X, Zhang Q, Lu B, Zhang X, et al. An aptamer targets HBV core protein and suppresses HBV replication in HepG2. 2.15 cells. Int J Mol Med. 2014;34(5):1423–1429. doi: 10.3892/ijmm.2014.1908. [DOI] [PubMed] [Google Scholar]
- 83.Jeon SH, Kayhan B, Ben-Yedidia T, Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J Biol Chem. 2004;279(46):48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 84.Liang H-R, Hu G-Q, Zhang T, Yang Y-J, Zhao L-L, Qi YL, et al. Isolation of ssDNA aptamers that inhibit rabies virus. Int Immunopharmacol. 2012;14(3):341–347. doi: 10.1016/j.intimp.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 85.Gopinath SC, Hayashi K, Kumar PK. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J Virol. 2012;86(12):6732–6744. doi: 10.1128/JVI.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamamoto R, Katahira M, Nishikawa S, Baba T, Taira K, Kumar PK. A novel RNA motif that binds efficiently and specifically to the tat protein of HIV and inhibits the trans-activation by tat of transcription in vitro and in vivo. Genes Cells. 2000;5(5):371–388. doi: 10.1046/j.1365-2443.2000.00330.x. [DOI] [PubMed] [Google Scholar]
- 87.Fukuda K, Vishnuvardhan D, Sekiya S, Hwang J, Kakiuchi N, Taira K, et al. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur J Biochem. 2000;267(12):3685–3694. doi: 10.1046/j.1432-1327.2000.01400.x. [DOI] [PubMed] [Google Scholar]
- 88.Kanamori H, Yuhashi K, Uchiyama Y, Kodama T, Ohnishi S. In vitro selection of RNA aptamers that bind the RNA-dependent RNA polymerase of hepatitis C virus: a possible role of GC-rich RNA motifs in NS5B binding. Virol. 2009;388(1):91–102. doi: 10.1016/j.virol.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 89.Jang KJ, Lee N-R, Yeo W-S, Jeong Y-J, Kim D-E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase. Biochem Biophys Res Commun. 2008;366(3):738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang D, Meng X, Yu Q, Xu L, Long Y, Liu B, et al. Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein. Antimicrob Agents Chemother. 2013;57(10):4937–4944. doi: 10.1128/AAC.00897-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Binning JM, Wang T, Luthra P, Shabman RS, Borek DM, Liu G, et al. Development of RNA aptamers targeting Ebola virus VP35. Biochem. 2013;52(47):8406–8419. doi: 10.1021/bi400704d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A, Prasad VR. RNA aptamers directed to human immunodeficiency virus type 1 gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J Virol. 2011;85(1):305–314. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parekh P, Tang Z, Turner PC, Moyer RW, Tan W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal Chem. 2010;82(20):8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gopinath SC, Sakamaki Y, Kawasaki K, Kumar PK. An efficient RNA aptamer against human influenza B virus hemagglutinin. J Biochem. 2006;139(5):837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 95.Auwardt SL. Use of RNA aptamers for enhanced drug therapy and imaging. Graduate Theses and Dissertations, Iowa State University; 2014. 14074. 10.31274/etd-180810-3480.
- 96.Davydova A, Vorobjeva M, Pyshnyi D, Altman S, Vlassov V, Venyaminova A. Aptamers against pathogenic microorganisms. Crit Rev Microbiol. 2016;42(6):847–865. doi: 10.3109/1040841X.2015.1070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ospina-Villa JD, López-Camarillo C, Castañón-Sánchez CA, Soto-Sánchez J, Ramírez-Moreno E, Marchat LA. Advances on aptamers against protozoan parasites. Genes. 2018;9(12):584. doi: 10.3390/genes9120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ulrich H, Magdesian MH, Alves MJM, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem. 2002;277(23):20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 99.Barfod A, Persson T, Lindh J. In vitro selection of RNA aptamers against a conserved region of the plasmodium falciparum erythrocyte membrane protein 1. Parasitol Res. 2009;105(6):1557–1566. doi: 10.1007/s00436-009-1583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martín ME, García-Hernández M, García-Recio EM, Gómez-Chacón GF, Sánchez-López M, González VM. DNA aptamers selectively target Leishmania infantum H2A protein. PLoS One. 2013;8(10):e78886. doi: 10.1371/journal.pone.0078886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guerra-Pérez N, Ramos E, García-Hernández M, Pinto C, Soto M, Martín ME, et al. Molecular and functional characterization of ssDNA aptamers that specifically bind leishmania infantum pabp. PLoS One. 2015;10(10):e0140048. doi: 10.1371/journal.pone.0140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ospina-Villa JD, Dufour A, Weber C, Ramirez-Moreno E, Zamorano-Carrillo A, Guillen N, et al. Targeting the polyadenylation factor EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica. Sci Rep. 2018;8(1):5720. doi: 10.1038/s41598-018-23997-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lorger M, Engstler M, Homann M, Göringer HU. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot Cell. 2003;2(1):84–94. doi: 10.1128/EC.2.1.84-94.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NA