Abstract
Background
Somatic embryogenesis (SE) is a process of somatic cells that dedifferentiate to totipotent embryonic stem cells and generate embryos in vitro. Longan SE has been established and wildly used as model system for studying embryogenesis in woody plants, SE-related genes had been characterized. In spite of that, a comprehensive overview of SE at a molecular level is still absent. To understand the molecular mechanisms during longan SE, we examined the transcriptome changes by using Illumina HiSeq from the four distinct developmental stages, including non-embryogenic callus (NEC), embryogenic callus (EC), incomplete compact pro-embryogenic cultures (ICpEC), globular embryos (GE).
Results
RNA-seq of the four samples generated a total of 243.78 million high quality reads, approximately 81.5% of the data were mapped to longan genome. The cDNA libraries of NEC, EC, ICpEC and GE, generated 22,743, 19,745, 21,144, 21,102 expressed transcripts, 1935, 1710, 1816, 1732 novel transcripts, 2645, 366, 505, 588 unique genes, respectively. Comparative transcriptome analysis showed that a total of 10,642, 4180, 5846 and 1785 genes were differentially expressed in the pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, ICpEC_vs_GE, respectively. Among them, plant hormones signalling related genes were significantly enriched, especially the auxin and cytokinin signalling components. The transcripts of flavonoid biosynthesis related genes were mainly expressed in NEC, while fatty acid biosynthesis related genes mainly accumulated in early SE. In addition, the extracelluar protein encoding genes LTP, CHI, GLP, AGP, EP1 were related to longan SE. Combined with the FPKM value of longan nine tissues transcription, 27 SE specific or preferential genes (LEC1, LEC1-like, PDF1.3, GH3.6, AGL80, PIN1, BBM, WOX9, WOX2, ABI3, et al.) and 28 NEC preferential genes (LEA5, CNOT3, DC2.15, PR1–1, NsLTP2, DIR1, PIP1, PIP2.1, TIP2–1, POD-P7 and POD5 et al.) were characterized as molecular markers for longan early SE. qRT-PCR validation of SE-related genes showed a high correlation between RNA-seq and qRT-PCR data.
Conclusion
This study provides new insights into the role of the transcriptome during early SE in longan. Differentially expressed genes reveal that plant hormones signalling, flavonoid and fatty acid biosynthesis, and extracelluar protein related genes were involved in longan early SE. It could serve as a valuable platform resource for further functional studies addressing embryogenesis in woody plants.
Keywords: Dimocarpus longan, Somatic embryogenesis, Illumina HiSeq, Auxin and cytokinin, Molecular marker gene, qRT-PCR
Background
Longan (Dimocarpus longan Lour.), a tropical/subtropical evergreen fruit tree within the Sapindaceae family, native to South China and Southeast Asia, is now widely cultivated in Southeast Asia, South Asia, Australia and Hawaii [1]. Logan embryo development status was close association with the seed size, fruit-set rate, fruit production and quality. Base on the observation of histological and cytological, the change of endogenous hormones and polyamines, proteomics analysis of the isozymes and proteins, molecular biology researches on SE-related genes mRNA differential display, homologous cloning, and expression pattern by qRT-PCR have been used to illuminate the potential regulation mechanism of longan SE [2]. However, elucidating the embryo development mechanism at a molecular level remains a great challenge due to its highly genetic heterozygosity and difficulties in accessibility of early embryos in vivo [3]. Plant SE shares close similarities at almost all development stages to normal zygotic embryogenesis [4, 5], SE has been wildly used as a model system to study the molecular regulation mechanism of early embryogenesis in plants [6]. The longan SE system has been established and extensively used as a model system for investigating embryogenesis in woody plants, which revealed that the concentration of 2,4-D was the key factor in controlling longan high-consistency SE [1, 7, 8].
Over the last few years, the expression profiles of SE related genes and other differentially expressed genes during SE had been extensively excavated by RNA-seq sequencing in various species, including Gossypium hirsutum [9–12], Arabidopsis [13, 14], Maize [15], Norway spruce [16, 17], Coconut plam [18], Brazilian pine [19], Eleutherococcus senticosusk [20], Camphor tree [21], Strawberry [22], Rice [23], Lilium pumilum [24], Mangosteen [25], Papaya [26], and Triticum aestivum [27]. Meanwhile, the comparative proteome analysis during SE also characterized numerous proteins that associated with SE in many plant species, such as Maize [28], Papaya [29], Cacao [30], Sugarcane [31], Musa. spp. [32], and Gossypium hirsutum [33]. The transcriptome and proteome analysis of plant SE revealed several molecular regulation mechanisms of SE, and a large number of potential key factors of embryogenesis. Numerous genes and proteins that playing an important role in somatic embryogenesis have been reported, such as Somatic embryogenesis receptor-like kinase (SERK) [34–36], Leafy Cotyledon [36–38], BABYBOOM [36, 39, 40], WUSCHEL [41, 42], WUSCHEL homeobox 2 [36, 43], AGAMOUS-like 15 [44, 45], and late embryogenesis abundant (LEA) protein [26].
To date, the transcript profiling of longan embryogenic callus (EC) had been illuminated by Lai and Lin [46], which revealed numerous embryogenesis-related and reproductive growth related unigenes in EC. Lin and Lai [47] had identified and profiled the conserved and novel miRNA during longan SE by using Solexa sequencing combined with computational, and qRT-PCR methods, and the potential roles of 20 conserved and 4 novel miRNA in longan SE were described by their tissue or stage-specific expression profiling. Recently, longan draft genome sequences become available [48], which provided the comprehensive genomic information for studying the molecular regulation of SE. Transition from NEC to EC, and from EC to somatic embryo are the key steps of SE. However, the molecular regulation mechanisms during longan SE remain largely unknown. To elucidate the molecular mechanism in the transition from NEC to EC, and during early SE by investigating the expression profiling using Illumina RNA-seq technology, and to identify the molecular marker genes during SE. This RNA-seq of comparative transcriptome analysis will gain new insight into the molecular and developmental mechanisms of longan SE.
Results
RNA-Seq analysis of longan early SE aligned with the Dimocarpus longan draft genome
To provide a comprehensive understanding of longan SE at a transcriptional level, we sequenced the four cDNA libraries constructed from the four in vitro embryo developmental stages (NEC, EC, ICpEC, and GE, Fig. 1). A total of 243,783,126 clean reads (comprising approximately 24.38 G of nucleotides) were obtained after data cleaning and quality checks. After aligned with longan reference genome [48], 48,798,229 (81.62%), 52,623,741 (81.1%), 48,346,067 (81.14%), and 48,871,200 (82.08%) reads in four cDNA libraries were mapped to longan reference genome, respectively. Among these, 44,655,772 (74.69%), 48,333,703 (74.50%), 44,490,292 (74.67%), and 44,924,511 (74.45%) reads were uniquely mapped to one location, respectively. Meanwhile, 34,380,246 (57.51%), 35,386,494 (54.54%), 30,535,088 (51.25%), and 29,214,788 (49.07%) reads in four cDNA libraries were mapped to gene, respectively. A summary of mapping statistics obtained for each sample is given in Table 1.
Fig. 1.
The synchronized cultures during longan SE. NEC: non-embryogenic callus; EC: Friable-embryogenic callus; ICpEC: Incomplete compact pro-embryogenic cultures; GE: Globular embryos. Bars = 50 μm
Table 1.
Statistics of reads generated by transcriptome sequencing of longan SE
| Sample Name | Total Clean reads | Total Reads Map to Genome | Genome Mapping Rate (%) | Total Reads Map to Gene | Gene Mapping Rate (%) | Expressed Transcripts | Expressed Exon | Novel Transcripts | Extend Gene | Alternative Splicing |
|---|---|---|---|---|---|---|---|---|---|---|
| NEC | 59,785,854 | 48,798,229 | 81.62 | 34,380,246 | 57.51 | 22,743 | 110,864 | 1935 | 10,281 | 35,084 |
| EC | 64,876,258 | 52,623,741 | 81.11 | 35,386,494 | 54.54 | 19,745 | 103,200 | 1710 | 9092 | 19,056 |
| ICpEC | 59,580,846 | 48,346,067 | 81.14 | 30,535,088 | 51.25 | 21,144 | 107,592 | 1816 | 9197 | 36,446 |
| GE | 59,540,168 | 48,871,200 | 82.08 | 29,214,788 | 49.07 | 21,102 | 107,971 | 1732 | 9638 | 39,768 |
The transcribed regions/units of four different stages samples were constructed independently, generated 22,743, 19,745, 21,144, and 21,102 expressed transcripts, showed 57.89, 50.26, 53.82, and 53.71% overlapped with longan genome (39,282 genes), respectively. After filtering out short sequences which less than 180 bp and low sequencing depth that lower than two, 1935, 1710, 1816, and 1732 novel transcripts in four samples were detected, respectively. Among these, 1025, 819, 832, and 806 novel genes were identified as coding RNAs, and 910, 891, 984, and 926 novel genes were identified as non-coding RNAs in longan genome.
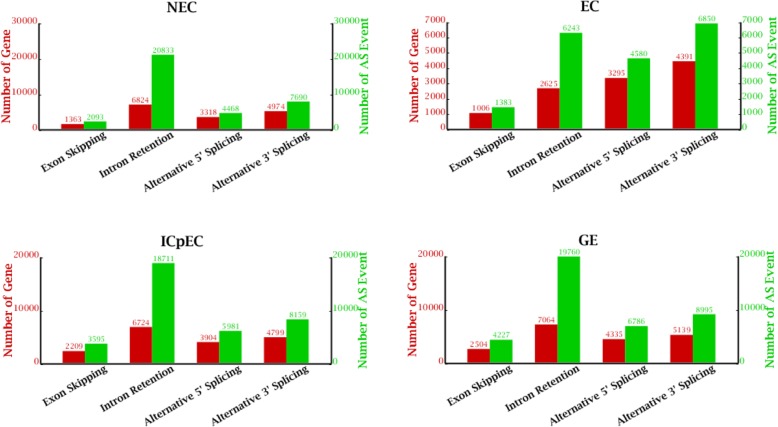
Alternative splicing (AS) events represented in our transcriptome were predicted by TopHat2. We analyzed the exon level of the four samples, 110,864, 103,200, 107,592, and 107,971 expressed exon were detected (Table 1). A total of 130,354 AS events were checked across the four stages, including exon skipping, intron retention, alternative 5′ splicing and alternative 3′ splicing. The largest number of AS events were detected in GE (39,768), followed by ICpEC (36,446), and NEC (35,084), and the smallest in EC (19,056). Exon skipping is the least type in all samples, and intron retention is the most popular type of AS events in NEC, ICpEC and GE (Fig. 2).
Fig. 2.
Alternative splicing events in the four stages of SE
Global analysis of gene expression across the four distinct developmental processes
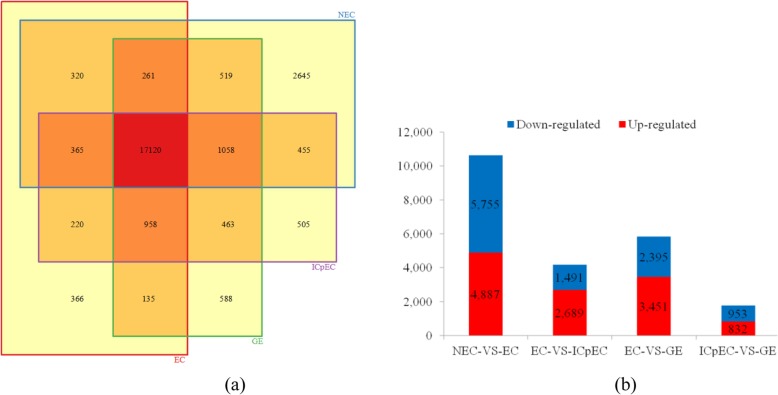
There were 22,743, 19,745, 21,144, and 21,102 expressed genes in NEC, EC, ICpEC, and GE stage. Among these, more than 75.3% of the expressed genes were present in all four developmental stages, while 2645 genes were only expressed in NEC. However, only 366, 505 and 588 genes were unique present in EC, ICpEC, and GE stage, respectively (Fig. 3a), which suggested that distinct spatial transcriptional patterns were present in the four developmental processes. To evaluate the differences of molecular response among four samples, gene expression were normalized to FPKM by RSEM software. After filtering with FPKM> 60, a total of 2961 (11.40%), 3445 (13.26%), 3445 (13.26%), and 3442 (13.25%) genes were highly expressed in NEC, EC, ICpEC, and GE, respectively (Table 2). The Top10 most enriched (FPKM) genes were range from 5476 to 58,812, 2766 to 15,114, 2343 to 10,330, and 2091 to 4004, respectively. The top 20 most expressed genes from the four libraries were shown in Tables 3, SE-related genes such as leafy cotyledon 1 (LEC1), leafy cotyledon 1-like (L1L), Protodermal factor 1 (PDF1), lipid transfer protein (LTP), Heat-Shock protein 90 (HSP90), chitinase (CHI), Indole-3-acetic acid-amido synthetase GH3.6, glutathione S-transferase (GST), root meristem growth factor 3 (RGF3) were highly expressed in EC, ICpEC or GE stage.
Fig. 3.
Statistical analysis of differentially expressed unigenes in NEC and early SE stages. a The venn diagram of expressed genes in four developmental stages. b Statistic of Up/Down regulated genes in pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE
Table 2.
Gene expression levels given in FPKM during longan SE
| FPKM Interval | NEC | EC | ICpEC | GE |
|---|---|---|---|---|
| ≤0.1 | 3900(15.01%) | 6910(26.60%) | 5364(20.65%) | 5391(20.75%) |
| 0.11–1 | 3587(13.81%) | 3075(11.84%) | 3241(12.48%) | 3384(13.03%) |
| 1.01–3 | 2706(10.42%) | 1957(7.53%) | 2439(9.39%) | 2362(9.09%) |
| 3.01–15 | 6440(24.79%) | 4774(18.38%) | 5413(20.84%) | 5208(20.05%) |
| 15.01–60 | 6384(24.57%) | 5817(22.39%) | 6076(23.39%) | 6191(23.83%) |
| 60.01–100 | 1278(4.92%) | 1431(5.51%) | 1601(6.16%) | 1573(6.06%) |
| ≥100 | 1683(6.48%) | 2014(7.75%) | 1844(7.10%) | 1869(7.19%) |
Table 3.
The top 20 most expressed genes in NEC, EC, ICpEC, GE library
| NO. | Gene_id | FPKM_NEC | Description |
|---|---|---|---|
| 1 | Dlo_008315.1 | 58,812.43 | repetitive proline-rich cell wall protein 2 |
| 2 | Dlo_019949.1 | 36,215.45 | Late embryogenesis abundant protein Lea5 |
| 3 | Dlo_008311.1 | 11,187.75 | unknow protein |
| 4 | Dlo_028175.1 | 10,885 | unknow protein |
| 5 | Dlo_011615.1 | 10,317.69 | extensin-2-like |
| 6 | Dlo_030517.1 | 8931.79 | chitinase CHI |
| 7 | Dlo_004649.1 | 7800.33 | metallothionein |
| 8 | Dlo_024177.1 | 6055.72 | chitinase |
| 9 | Dlo_017033.1 | 5645.16 | pathogenesis-related protein 1 |
| 10 | Dlo_008997.3 | 5476.22 | unknow protein |
| 11 | Dlo_009172.1 | 5469.19 | osmotin-like protein I |
| 12 | Dlo_021620.1 | 4483.38 | peroxidase 4 |
| 13 | Dlo_003142.1 | 4116.17 | unknow protein |
| 14 | Dlo_030075.1 | 3732.29 | Wound-induced protein WIN1 precursor |
| 15 | Dlo_030519.1 | 3587.55 | chitinase CHI |
| 16 | Dlo_022694.1 | 3546.1 | 14 KDa proline-rich protein DC2.15-like |
| 17 | Dlo_030516.1 | 3288.7 | chitinase |
| 18 | Dlo_030074.1 | 3170.89 | PR-4 protein |
| 19 | Dlo_011076.1 | 2572.89 | ubiquitin C |
| 20 | Dlo_011004.1 | 2367.71 | non-specific lipid-transfer protein 2-like |
| NO. | Gene_id | FPKM_EC | Description |
| 1 | Dlo_030812.1 | 15,114.88 | Protodermal factor 1.3 PDF1.3 |
| 2 | Dlo_013012.1 | 7392.37 | lipid transfer protein |
| 3 | Dlo_030517.1 | 5117.6 | chitinase CHI |
| 4 | Dlo_025725.1 | 4537.79 | Pollen-specific protein C13 precursor |
| 5 | Dlo_020986.1 | 4136.45 | Indole-3-acetic acid-amido synthetase GH3.6 |
| 6 | Dlo_011615.1 | 4031.23 | extensin-2-like |
| 7 | Dlo_021620.1 | 3793.82 | peroxidase 4 |
| 8 | Dlo_032035.1 | 2962.58 | histone H4-like |
| 9 | Dlo_031913.1 | 2959.02 | lipid binding protein |
| 10 | Dlo_003789.1 | 2766.68 | EXORDIUM-like 2 EXL2 |
| 11 | Dlo_005176.1 | 2363.28 | omega-6 fatty acid desaturase |
| 12 | Dlo_026048.1 | 2219.4 | root meristem growth factor 3 RGF3 |
| 13 | Dlo_033433.1 | 2202.74 | unknow |
| 14 | Dlo_019526.1 | 2145.11 | unknow |
| 15 | Dlo_017203.1 | 2144.67 | Hsp90 |
| 16 | Dlo_026351.1 | 2131.12 | peptidase |
| 17 | Dlo_017092.1 | 2118.54 | transcription factor leafy cotyledon1 |
| 18 | Dlo_020190.1 | 2035.05 | cysteine protease |
| 19 | Dlo_007905.1 | 2000.4 | small ubiquitin-related modifier 2-like |
| 20 | Dlo_012332.1 | 1967.95 | 26S proteasome complex subunit DSS1 |
| NO. | Gene_id | FPKM_ICpEC | Description |
| 1 | Dlo_030812.1 | 10,330.16 | protodermal factor 1.3 |
| 2 | Dlo_026048.1 | 6620.49 | root meristem growth factor 3 RGF3 |
| 3 | Dlo_031913.1 | 4694.68 | lipid binding protein |
| 4 | Dlo_013012.1 | 4301.99 | lipid transfer protein |
| 5 | Dlo_008315.1 | 4140.34 | proline-rich cell wall protein 2-like PRP2 |
| 6 | Dlo_032146.1 | 3401.87 | NADH dehydrogenase (ubiquinone)1 beta subcomplex 7 |
| 7 | Dlo_028379.1 | 2815.75 | dehydrin 1 |
| 8 | Dlo_025725.1 | 2772.74 | Pollen-specific protein C13 precursor |
| 9 | Dlo_020986.1 | 2439.09 | Indole-3-acetic acid-amido synthetase GH3.6 |
| 10 | Dlo_021620.1 | 2343.85 | peroxidase 4 |
| 11 | Dlo_019476.1 | 2300.16 | unknow |
| 12 | Dlo_028328.1 | 2106.48 | high mobility group box 1 |
| 13 | Dlo_017203.1 | 1958.55 | Hsp90 |
| 14 | Dlo_019638.1 | 1958.53 | elongation factor 1-alpha |
| 15 | Dlo_030608.1 | 1823.94 | unknow |
| 16 | dlo_034323.1 | 1818.75 | histone H1 |
| 17 | Dlo_011615.1 | 1789.67 | extensin-2-like |
| 18 | Dlo_010406.1 | 1773.3 | transcription factor BTF3 |
| 19 | Dlo_017539.1 | 1684.81 | histone H2B.1-like |
| 20 | Dlo_030517.1 | 1598.6 | chitinase CHI |
| NO. | Gene_id | FPKM_GE | Description |
| 1 | Dlo_013012.1 | 4004.61 | lipid transfer protein |
| 2 | Dlo_026048.1 | 3762.46 | root meristem growth factor 3 RGF3 |
| 3 | Dlo_021620.1 | 3304.97 | peroxidase 4 |
| 4 | Dlo_031913.1 | 2957.57 | lipid binding protein |
| 5 | Dlo_032146.1 | 2784.3 | NADH dehydrogenase (ubiquinone)1 beta subcomplex 7 |
| 6 | Dlo_028379.1 | 2676 | dehydrin 1 |
| 7 | Dlo_014867.1 | 2300.44 | argonaute 4 |
| 8 | Dlo_008315.1 | 2218.56 | proline-rich cell wall protein 2-like PRP2 |
| 9 | Dlo_030608.1 | 2100.52 | unknow |
| 10 | Dlo_012964.1 | 2091.04 | extensin, proline-rich protein |
| 11 | Dlo_032870.1 | 2072.74 | glutathione S-transferase parC-like |
| 12 | Dlo_025725.1 | 2018.61 | Pollen-specific protein C13 precursor |
| 13 | Dlo_019476.1 | 1997.45 | unknow |
| 14 | Dlo_015927.1 | 1907.93 | unknow |
| 15 | Dlo_030812.1 | 1899.07 | protodermal factor 1.3 |
| 16 | Dlo_028328.1 | 1848.42 | high mobility group box 1 |
| 17 | Dlo_018634.1 | 1743.25 | 60S ribosomal protein L27Ae |
| 18 | Dlo_019638.1 | 1709.96 | elongation factor 1-alpha |
| 19 | Dlo_017203.1 | 1696.73 | Hsp90 |
| 20 | Dlo_020821.1 | 1665.7 | leafy cotyledon1-like |
To reveal the potential key genetic factors involved in early SE, we filtered the significantly differentially expressed genes (DEGs) with |log2fold change| ≥ 1 and FDR < 0.001 between these four pairwise comparisons as follow: NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE. Among these four comparisons (Fig. 3b), a total of 10,642, 4180, 5846 and 1785 DEGs were identified, respectively. Compared with NEC, EC had 4887 up-regulated and 5755 down-regulated genes. Compared with EC, ICpEC had 2689 up-regulated and 1491 down-regulated genes, GE had 3451 up-regulated and 2395 down-regulated genes. Compared with ICpEC, GE had 832 up-regulated and 953 down-regulated genes. DEGs analysis revealed that longan transcriptome undergoes significantly dynamic changes during SE, particularly during the transition period from NEC to EC. Therefore, the longan SE transcriptome datasets given here may serve as a valuable molecular resource for future studies.
Functional classification of DEGs base on GO and KEGG
To evaluate the potential functions of the DEGs, we used GO terms assignment to classify the functions of DEGs in pairwise comparisons under three GO main categories: biological process, cellular component and molecular function (Additional file 1: Figure S1). In all pairwise comparisons, the term with the largest proportion in “biological process” was ‘metabolic process’, followed by ‘cellular process’, ‘single-organism process’, ‘respond to stimulus’ and ‘localization’, the term with the largest proportion in “cellular component” were ‘cell’ and ‘cell part’, followed by ‘organelle’ and ‘membrane’, the term with the largest proportion in “molecular function” was ‘catalytic activity’, followed by ‘binding’, ‘transporter activity’, ‘molecular transducer activity’ and ‘nucleic acid binding transcription factor activity’ .
To investigate the biological pathways of the DEGs, we used the KEGG database to classify the DEGs function with emphasis on biological pathways (Additional file 2: Figure S2). According to KEGG annotation, 6516 DEGs (NEC_vs_EC) were assigned to 128 pathways, 2514 DEGs (EC_vs_ICpEC) were assigned to 126 pathways, 3555 DEGs (EC_vs_GE) were assigned to 126 pathways, 1062 DEGs (ICpEC_vs_GE) were assigned to 111 pathways. The annotated changes in all comparisons were mainly enriched in ‘metabolic pathway’ (21.38, 22.43, 23.12 and 25.52%, respectively), ‘biosynthesis of secondary metabolites’ (11.97, 11.46, 11.70 and 14.52%, respectively), ‘plant-pathogen interaction’ (8.01, 8.23, 7.59 and 6.40%, respectively) and ‘plant hormone signal transduction’ (5.22, 5.41, 5.40 and 8.38%, respectively) pathway. Furthermore, dozens of genes involved in ‘flavonoid biosynthesis’, ‘phenylpropanoid biosynthesis’, ‘zeatin biosynthesis’, ‘fatty acid biosynthesis’ and ‘biosynthesis of unsaturated fatty acids’.
Differential expression analysis of plant hormones signaling pathway related genes during longan SE
Based on the KEGG and other annotation, plant hormone signal transduction, zeatin biosynthesis and tryptophan metabolism were the representative pathways in our study. A large number of genes invovled in auxin (97 DEGs) and cytokinin (94 DEGs) biosynthesis and signal transduction pathway were differentially expressed when compared EC with NEC (Additional file 3: Figure S3) and early SE. For example, the expression level of PIN1, IAA (IAA6, IAA6-like, IAA9, IAA11, IAA14, IAA16, IAA29, IAA31 and IAA33), ARFs (ARF1, ARF1-like, ARF2, ARF2-like, ARF5, ARF10, ARF16, ARF17, ARF18, ARF18–1 and ARF24), GH3 (GH3.6, GH3.1, GH3.17), and three SAUR, genes involved in auxin signal transduction, were significantly up-regulated from NEC to EC, most of them remained highly expression in EC, ICpEC and GE stages. Nevertheless, AUX1, TIR1, IAA (IAA1, IAA4, IAA13, IAA26, IAA26-like, IAA27), ARFs (ARF4, ARF4-like, ARF10-like), GH3.9 and GH3.17-like, and 12 SAUR were mainly expressed in NEC stage and down-regulated in EC. From EC to ICpEC and GE stages, AUX1 (Dlo_024286.1, Dlo_031956.2), IAA (IAA4, IAA14, IAA26-like, IAA27, IAA13), ARFs (ARF4, ARF4-like, ARF10-like), two SAUR showed noteworthy up-regulated expression (Fig. 4a). In IAA biosynthesis, except PAI, Trp synthesis key genes ASA, IGS, TSA, TSB, were up-regulated in EC and remained high expression during early SE. CYP83B1, one ST5a, five YUCCAs, three CYP71A13 and NIT showed NEC-specific expression pattern. Three YUCCAs, three AAO1, one NIT, CYP71A13 and three ST5a were up-regulated in EC and remained high during early SE, and YUCCA_Dlo_013505.1 kept up-regulated during early SE (Fig. 4b).
Fig. 4.
Heatmap of the differentially expressed genes in auxin and cytokinin signaling pathway during longan SE. a Auxin signal transduction; b Cytokinin signal transduction; c IAA biosynthesis; d Zeatin biosynthesis. The heatmap was clustered by pearson method of Mev4.90 software. Heatmap indicate the gene expression level by Log2[FPKM+ 1] with a rainbow color scale, each row represents a single gene, the IDs and names of selected DEGs are indicated to the right of the histograms, and each column represents a sample
As showed in Fig. 4c, TRIT1, a gene involved in cis-zeatin synthesis was up-regulated from NEC to GE. CisZOG family involved in Cis-zeatin O-glycosylation were highly expressed in NEC, and significantly down-regulated from NEC to EC. During early SE, five CisZOG were up-regulated from EC to ICpEC, four CisZOG were down-regulated from ICpEC to GE. In trans-zeatin biosynthesis, six IPT1,4, five CYP735A, four CKX, three UGT76C were noteworthy down-regulated from NEC to EC; two IPT1,4, four CYP735A, one CKX, three UGT76C were up-regulated in EC. During early SE, IPT1,4 family, five CYP735A, two CKX, four UGT76C were up-regulated during early SE with minimal FPKM. Among the cytokinin signal pathway, two A_ARR, 10 B_ARR, 15 CRE1 were mainly expressed in NEC, and down-regulated in EC. One A_ARR, five B_ARR, seven CRE1 were up-regulated in EC. 13 CRE1, seven B_ARR and all A_ARR showed up-regulated expression during early SE, two B_ARR and five CRE1 were down-regulated during early SE (Fig. 4d).
In addition, numerous genes involved in abscisic acid, gibberellin, ethylene, salicylic acid, jasmonic acid and brassinosteroid signal transduction pathway were differentially expressed during longan SE (Additional file 4: Figure S4; Additional file 5: Table S1 a-h). Such an observation suggested an essential role of hormones and their complicated crosstalk during early SE. Therefore, the plant hormones signaling pathway may be the key regulator during longan early SE.
Flavonoids and fatty acid biosynthesis related genes were differential expressed during longan SE
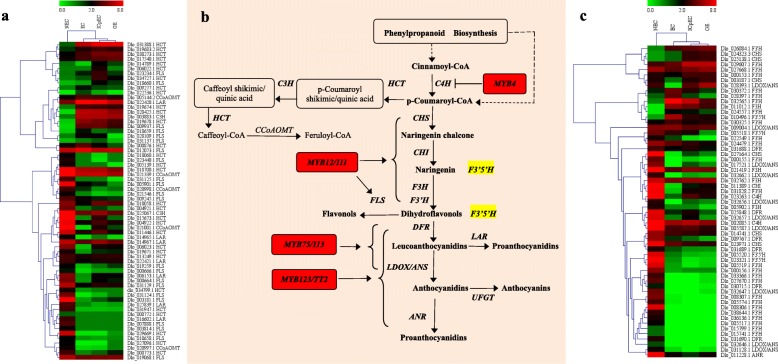
Flavonoid biosynthesis and fatty acid biosynthesis were the representative KEGG pathways, a total of 125 significant DEGs were assigned to ‘flavonoid biosynthesis’ across the early SE processes (Fig. 5). In the transition from NEC to EC, the flavonoid biosynthesis key genes, C4H, CHS, CHI, F3H, F3’5’H, DFR, LDOX/ANS, ANR, LAR, CCoAOMT were mainly expressed in NEC, while drastic down-regulated from NEC to EC and remained very low expression level in ICpEC and GE stages, except that F3H_Dlo_011012.1, F3’5’H_ Dlo_010496.1, LAR_ Dlo_022420.1, CCoAOMT_ Dlo_005144.2 were up-regulated in EC, but down-regulated during early SE. Besides, most of the FLS and F3’H family were mainly expressed in NEC, significantly down-regulated in EC and kept low FPKM during early SE, especially, 15 F3’H and 9 FLS belonged to NEC-specific genes. Only four FLS and six F3’H were up-regulated from NEC to EC and then down-regulated or kept low expression level during early SE (Additional file 6: Table S2).
Fig. 5.
Simplified diagram of flavonoid biosynthetic pathway. a Cluster analysis of expression profiles of HCT, C3H, CCoAOMT, FLS and LAR. b Simplified diagram of flavonoid biosynthetic pathway. c Cluster analysis of expression profiles of C4H, CHS, CHI, F3H, F3’H, F3’5’H, DFR, LDOX/ANS and ANR. The heatmaps was clustered by pearson method of Mev4.90 software. Heatmaps indicate the gene expression levels by Log2 [FPKM+ 1] with a rainbow color scale, each row represents a single gene, and each column represents a sample. The IDs and names of selected DEGs are indicated to the right of the histograms
Several R2R3-MYB transcription factors are involve in the regulation of flavonoid biosynthesis in Arabidopsis [49–51]. For example, AtMYB11, − 12, − 111 regulated flavonol biosynthesis by up-regulated CHS, CHI, F3H, F3’H and FLS [49, 52]. AtMYB75, − 90, − 113, − 114 controlled anthocyanin biosynthesis in vegetative [53]. AtMYB123 controlled the biosynthesis of proanthocyanidins in the seed coat [54]. MtMYB5, − 14 played the key role in seed coat polymer biosynthesis [55]. AtMYB4 negative controlled sinapate ester biosynthesis through down-regulated C4H in a UV-dependent manner [56]. In our study, 11 R2R3-MYB transcripts were differentially expressed. During longan SE, MYB12 and MYB111 were barely expressed in NEC, significant up-regulated from NEC to EC and remained high during early SE. MYB75, MYB113, MYB4 and MYB123 were significant down-regulated in EC, and kept relative low expression during early SE.
The fatty acid composition rapidly changed during SE in Daucus carota [57], and Gossypium hirsutum [33]. In our study, a total of 35 fatty acid biosynthesis related genes were differently expressed during SE (Additional file 7: Table S3). From NEC to EC, except ACCase (Dlo_000360.1), three FabG, two FabZ, SAD (Dlo_031652.1), most of the ACCase, FabD, FabF, FabG, FabZ, FabI, FatB and SAD were significantly up-regulated in EC. During early SE, most of the DEGs remained high expression, part of them with slightly up/down-regulated expression. For example, ACCase (Dlo_023270.1) and SAD (Dlo_019646.1) were up-regulated from NEC to EC, and highly expressed during early SE. Our results indicated that flavonoids were mainly expressed in NEC, while fatty acid were mainly accumulated in early SE stages, especially in EC.
Extracellular protein encoding genes effect on the transition from NEC to EC
It had been reported that extracellular protein germins and germin-like (GLPs), Arabinogalactan proteins (AGPs), chitinases (CHIs), lipid transfer proteins (LTPs) and glycoprotein were critical to SE, and can be served as protein marker during early SE [58]. In our study, 16 CHIs were differentially expressed, and most of them were preferential expressed in NEC, and remarkable down-regulated in EC, only seven CHIs were up-regulated during early SE with low FPKM. Among the 14 identified LTPs, only LTP (Dlo_013012.1, Dlo_013014.1) were highly and specific expressed in early SE, most of them were mainly expressed in NEC and down-regulated from NEC to EC. Meanwhile, 12 GLPs and two secreted glycoprotein genes (EP1-like) were mainly expressed in NEC and kept very low FPKM during early SE. Except AGP10 was first up-regulated in EC and down-regulated during early SE, most of the AGPs were down-regulated in EC, and kept relative low expression level during early SE (Additional file 8: Table S4). The results indicated that most of the extracellular protein encoding genes were mainly expressed in NEC, they were predicted to involve in the transition from NEC to EC.
Characterization of molecular markers for longan SE
Several genes have been reported to molecular marker of SE, such as somatic embryogenesis receptor-like kinase (SERK), leafy cotyledon1 (LEC1), BABYBOOM (BBM), wuschel (WUS), WUS-homeobox (WOX). In order to characterize the full-scale of molecular markers for early SE, the comparative analysis of FPKM in nine tissues of longan [48], including root, stem, leaf, flower, flower bud, young fruit, pericarp, pulp and seed [48] were employed to select the molecular marker genes during SE. For our purposes here, it is crucial to identify the reliable molecular marker genes for distinguishing NEC stage from EC, ICpEC and GE stages. In our study, several embryogenesis-labeled genes that had been reported previously were differentially expressed in each stage (Additional file 9: Table S5). However, some of them showed down-regulated or slightly up-regulated in EC, and kept low expression level from NEC to GE, such as late embryogenesis abundant protein (LEA14A, LEAD34, LEA76), SERK1, SERK3, WUS, WOX5, WOX3, AIL6, AGL15, CLV1, EMB8, suggesting that they were unseemly markers for longan SE.
In our study, a total of 55 genes were identified as representative molecular markers, which were closely related to SE, can be classified as two main categories: NEC markers and SE molecular markers by their specific expression profiles in all test-samples (Table 4). The SE marker genes were barely or undetected in NEC, highly expressed during early SE, they also can be divided into SE-specific and SE-expressed genes. The SE-specific genes were highly transcribed only in somatic embryos, including LEC1, LEC2, WOX9, WOX2, Agamous-like 80 (AGL80), PIN-FORMED1 (PIN1), BBM, PLETHORA2 (PLT2), mannan endo 1,4-beta-mannosidase7 (MAN7), Glycine-rich protein 5 (GRP-5), GRF-interacting factor 2 (GIF2), root meristem growth factor 3 (RGF3), 60S ribosomal protein L17e (RPL17e), zeta-carotene desaturase (ZDS), 3-ketoacyl-CoA synthase (KCS), CYP78A5, CYP87A3 and three unknown genes (DlU1, DlU2, DlU3) (Table 4). These SE-specific genes might play a key role in longan SE. The SE-expressed genes were similar to SE-specific genes, except that these genes also highly expressed in one or some tested tissues included in this study, including LEC1-like (L1L), ABA-insensitive protein 3 (ABI3), FUSCA3 (FUS3), Indole-3-acetic acid-amido synthetase (GH3.6), Protodermal factor 1.3 (PDF1.3), Lipid transfer protein (LTP, Dlo_013012.1) and Lipid binding protein (LBP). For instance, L1L, FUS3 and ABI3 showed very strong transcription level not only in somatic embryos but also in seed. GH3.6 was highly expressed in flower, PDF1.3 and LBP showed high expression level in pulp, LTP also highly transcribed in pulp, flower bud, flower and stem, suggesting their multifunctional on SE and other development processes (Table 4).
Table 4.
The expression profile of candidate markers during SE and in nine tissues of longan
| Gene_id | Gene name | FPKM value | Putative protein function/homology | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NEC | EC | ICpEC | GE | Root | Stem | Leaf | Flower | Flower bud | Young fruit | Pericarp | Pulp | Seed | |||
| Dlo_017092.1 | LEC1 | 0 | 2118.54 | 1262.61 | 727.62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.52 | leafy cotyledon 1 transcription factor |
| Dlo_020821.1 | L1L | 0.21 | 511.04 | 393.39 | 1665.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 209.01 | leafy cotyledon 1-like protein |
| Dlo_022316.1 | WOX9 | 1.34 | 1602.12 | 1424.69 | 665.41 | 0.3 | 0 | 0 | 0 | 0.52 | 1.5 | 0 | 5.98 | 0.67 | WUSCHEL-related homeobox 9 |
| Dlo_032045.1 | WOX2 | 0.48 | 141.61 | 83.91 | 59.36 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 | WUSCHEL-related homeobox 2 |
| Dlo_011527.1 | BBM | 6.28 | 367.26 | 484.12 | 511.04 | 9.86 | 0.28 | 0.1 | 0.12 | 0.19 | 0.34 | 0.14 | 0 | 0.67 | AP2/ERF, BABYBOOM |
| Dlo_004646.1 | PLT2 | 3.94 | 491.37 | 610.1 | 759.4 | 0.41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | AP2/ERF, PLT2 |
| Dlo_012160.1 | ABI3 | 0 | 82.94 | 177.78 | 374.51 | 0 | 0 | 0 | 0.13 | 13.28 | 0 | 0 | 0 | 119.83 | ABA-insensitive protein 3 |
| Dlo_022372.1 | LEC2 | 0 | 45.35 | 55.63 | 51.22 | 0.11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.4 | leafy cotyledon 2 transcription factor |
| Dlo_024008.1 | FUS3 | 0 | 5.75 | 37.93 | 260.7 | 0 | 0 | 0 | 0 | 0.35 | 0 | 0 | 0 | 60.65 | FUSCA3 |
| Dlo_017585.1 | AGL80 | 0 | 829.45 | 590.14 | 278.12 | 0.62 | 0.58 | 0.82 | 0.49 | 0 | 0.29 | 0.84 | 0.58 | 1.56 | Agamous-like MADS-box protein 80 |
| Dlo_020986.1 | GH3.6 | 0.06 | 4136.45 | 2439.09 | 1630.99 | 0 | 1.2 | 0.06 | 843.22 | 0.32 | 0 | 0.25 | 0.2 | 0.14 | IAA-amido synthetase GH3.6 |
| Dlo_020694.1 | PIN1 | 3.23 | 216.9 | 96.91 | 100.29 | 0 | 0 | 0 | 0.07 | 0 | 0 | 0 | 0 | 3.04 | Auxin efflux carrier component 1 |
| Dlo_030812.1 | PDF1.3 | 15.81 | 15,114.88 | 10,330.16 | 1899.07 | 0.68 | 0 | 0.16 | 2.4 | 60.18 | 76.88 | 21.19 | 680.91 | 16.67 | Protodermal factor 1.3 |
| Dlo_027182.2 | GRP-5 | 0 | 1699.47 | 1094.49 | 874.66 | 0 | 0.35 | 0 | 0 | 0.32 | 0 | 0 | 0 | 8.03 | glycine-rich protein 5 |
| Dlo_032565.1 | CYP78A5 | 0.97 | 320.4 | 102.08 | 12.72 | 31.26 | 0 | 1.85 | 0.09 | 6.93 | 1.78 | 0.4 | 0.47 | 4.39 | Cytochrome P450 family 78A5 |
| Dlo_017331.1 | CYP87A3 | 0 | 274.33 | 130.99 | 25.01 | 0 | 0.11 | 0 | 5.98 | 0.63 | 0.22 | 0.22 | 4.42 | 0.18 | Cytochrome P450 family 87A3 |
| Dlo_026048.1 | RGF3 | 0 | 2219.4 | 6620.49 | 3762.46 | 0 | 0 | 0 | 3.27 | 1.96 | 0 | 0 | 0 | 0 | root meristem growth factor 3 |
| Dlo_026819.1 | GIF2 | 0 | 726.78 | 454.72 | 339.9 | 0 | 0.94 | 0 | 1.08 | 6.39 | 0.45 | 0.3 | 1.79 | 12.93 | GRF1-interacting factor 2, GIF2 |
| Dlo_013012.1 | LTP | 6.56 | 7392.37 | 4301.99 | 4004.61 | 3.92 | 1182.18 | 5.16 | 1241.54 | 1954.47 | 354.17 | 62.65 | 4225.76 | 67.83 | Lipid transfer protein |
| Dlo_031913.1 | LBP | 3.92 | 2959.02 | 4694.68 | 2957.57 | 1.19 | 25.49 | 12.3 | 28.54 | 27.78 | 32.46 | 4.93 | 428.03 | 5.16 | Lipid binding protein |
| Dlo_025851.1 | MAM7 | 3.43 | 1112.42 | 436.33 | 119.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | mannan endo-1,4-beta-mannosidase 7 |
| Dlo_029005.1 | ZDS | 0 | 1819.85 | 1213.68 | 932.22 | 0.15 | 0.15 | 0.11 | 0.76 | 0.81 | 20.69 | 0.42 | 14.54 | 2.2 | zeta-carotene desaturase |
| Dlo_023272.1 | KCS | 0 | 511.05 | 371.46 | 205.05 | 0 | 0 | 0.08 | 0 | 0.1 | 1.36 | 0 | 1.49 | 0.09 | 3-ketoacyl-CoA synthase (fatty acid) |
| Dlo_032570.1 | RPL17 | 0 | 1081.06 | 1123.65 | 990.78 | 48.99 | 40.71 | 0 | 0 | 3.48 | 3.17 | 0 | 0 | 0 | 60S ribosomal protein L17 |
| Dlo_033433.1 | DlU1 | 3.52 | 2202.74 | 1163.64 | 830.54 | 14.46 | 1.61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Unknown 1 |
| Dlo_026772.1 | DlU2 | 0 | 50.68 | 397.92 | 1516.17 | 0.97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.24 | Unknown 2 |
| Dlo_028569.1 | DlU3 | 0 | 558.23 | 418.54 | 121.81 | 0.78 | 7.07 | 0 | 3.14 | 4.43 | 3.26 | 3.67 | 0 | 0 | Unknown 3 |
| Dlo_019949.1 | LEA5 | 36,215.45 | 27.67 | 145.54 | 256.16 | 3165.31 | 6822.69 | 861.14 | 7685.55 | 11,712.56 | 3205.39 | 24,022.25 | 1205.56 | 3737.57 | Late embryogenesis abundant protein 5 |
| Dlo_008311.1 | CNOT3 | 11,187.75 | 12.96 | 27.95 | 44.39 | 306.61 | 11,545.36 | 933.45 | 1611.48 | 393.14 | 1560.72 | 2650.38 | 7249.71 | 1296.04 | CCR4-NOT transcription complex 3 |
| Dlo_028175.1 | DlU4 | 10,885 | 0.31 | 2.94 | 3.3 | 5220.16 | 576.03 | 3.61 | 217.29 | 35.02 | 4434.9 | 1942.11 | 5575.02 | 396.85 | Unknown 4 |
| Dlo_017033.1 | PR1–1 | 5645.16 | 40.36 | 26.32 | 10.4 | 2601.85 | 122.2 | 1.45 | 86.79 | 22.85 | 0.96 | 361.66 | 577.38 | 17.99 | Pathogenesis-related protein 1–1 |
| Dlo_022694.1 | DC2.15 | 3546.1 | 0.29 | 0 | 0 | 965.82 | 92.92 | 9.21 | 930.75 | 46.71 | 788.54 | 400.61 | 1323.15 | 301.29 | 14 kDa proline-rich protein DC2.15 |
| Dlo_030516.1 | CHI | 3288.7 | 12.16 | 54.25 | 8.31 | 2.21 | 1.5 | 0.7 | 0.62 | 0.45 | 0 | 0.46 | 1.65 | 0.19 | Chitinase |
| Dlo_030074.1 | PR-4 | 3170.89 | 0.31 | 2.48 | 1.62 | 517.32 | 105.29 | 0.59 | 90.21 | 85.99 | 0.79 | 10.68 | 4.68 | 25.33 | PR-4 protein |
| Dlo_028350.1 | CAT | 2411.98 | 15.42 | 41.81 | 55.28 | 4254.18 | 1219.17 | 398.73 | 293.83 | 464.27 | 651.57 | 3884.71 | 264.3 | 319.86 | Catalase |
| Dlo_011004.1 | NsLTP2 | 2367.71 | 0 | 0 | 0 | 1410.22 | 836.82 | 1.46 | 171.89 | 732.06 | 69.9 | 148.79 | 31.47 | 45.31 | Non-specific lipid-transfer protein 2 |
| Dlo_032927.1 | PIP2.1 | 2191.16 | 8.96 | 7.95 | 4.38 | 2253.88 | 162.85 | 81.62 | 45.29 | 21.99 | 140.46 | 842.74 | 1733.29 | 217.06 | Aquaporin PIP2.1 |
| Dlo_011314.1 | POD P7 | 2007.14 | 3.73 | 4.04 | 2.59 | 67.72 | 32.71 | 1.25 | 33.7 | 10.84 | 1.82 | 24.34 | 4.33 | 1.23 | Peroxidase P7-like |
| Dlo_020889.1 | DlU5 | 1534.05 | 0.27 | 18.34 | 62.55 | 1224.15 | 79.36 | 8.05 | 83.74 | 14.28 | 2100.25 | 1136.71 | 29,080.32 | 892.17 | Unknown 5 |
| Dlo_009170.1 | OSM1 | 1459.85 | 8.09 | 7.15 | 4.64 | 1450.79 | 3031.81 | 58.61 | 3028.02 | 3208.04 | 144.45 | 3184.56 | 2337.21 | 430.64 | Osmotin-like protein 1 |
| Dlo_006330.1 | EXLB1 | 1145.83 | 1.14 | 4.4 | 3.19 | 303.02 | 2258.39 | 7.29 | 32.98 | 200.08 | 12.5 | 543.39 | 0.26 | 111.72 | Expansin-like B1 |
| Dlo_003365.1 | DIR1 | 1049.5 | 0 | 0 | 0 | 614.07 | 12.2 | 5.3 | 35.6 | 37.19 | 284.21 | 88.68 | 0.7 | 24.18 | Lipid-transfer protein DIR1 |
| Dlo_024175.1 | CHI | 983.75 | 0.14 | 0 | 1.5 | 3965.12 | 6.77 | 5.9 | 46.39 | 47.66 | 1.53 | 10.77 | 5.23 | 5.17 | class I chitinase |
| Dlo_011438.1 | PIP1 | 812.41 | 0.56 | 1.69 | 2.71 | 1138.83 | 1183.76 | 441.46 | 491.94 | 345.78 | 1651.58 | 806.49 | 239.44 | 651.72 | Aquaporin PIP1 |
| Dlo_014725.1 | PME1 | 753.54 | 0 | 1.19 | 0.57 | 501.17 | 0.62 | 0.96 | 0.53 | 0 | 16.39 | 1.39 | 21.11 | 8.61 | Pectinesterase precursor |
| Dlo_027164.1 | CHS | 610.74 | 0.1 | 1.76 | 0.91 | 1988.95 | 12.84 | 204.09 | 222.61 | 19.12 | 40.45 | 5.95 | 0.96 | 122.71 | chalcone synthase |
| Dlo_023321.1 | TLP1 | 573.07 | 0.8 | 0.14 | 0.26 | 39.55 | 75.91 | 0.5 | 162.54 | 45.36 | 2.27 | 2.42 | 0.79 | 0.99 | thaumatin-like protein |
| Dlo_000377.1 | POD5 | 566.61 | 1.32 | 1.3 | 1.41 | 79.76 | 728.88 | 601.84 | 3319.92 | 905.76 | 380.12 | 225.83 | 1440.49 | 117.02 | Peroxidase 5 |
| Dlo_004275.1 | TIP2–1 | 546.09 | 0 | 0 | 0 | 55.57 | 139.05 | 71.7 | 147.25 | 132.03 | 102.82 | 84.76 | 0.26 | 45.97 | aquaporin MIP family TIP2–1 |
| Dlo_017039.1 | PR1-like | 462.02 | 7.87 | 3.65 | 2.14 | 52.13 | 26.21 | 0 | 121.87 | 14.14 | 137.08 | 3.17 | 0.42 | 0 | Pathogenesis-related protein 1 |
| Dlo_017003.1 | GAST-like | 459.7 | 0.24 | 5.45 | 5.45 | 28.63 | 219.33 | 1.56 | 1181.5 | 449.48 | 0.89 | 3.56 | 67.29 | 4.4 | Gibberellic Acid Stimulated Transcript-like |
| Dlo_012593.1 | DlU6 | 433.86 | 0 | 0 | 0 | 0 | 0 | 0.64 | 0 | 0.73 | 1.88 | 0.74 | 0 | 0 | Unknown 6 |
| Dlo_005429.1 | ERF114 | 352.05 | 0 | 2.51 | 1.98 | 9.51 | 137.66 | 32.36 | 98.04 | 49.11 | 1.79 | 2.92 | 0.53 | 3.31 | ethylene-responsive transcription factor |
| Dlo_032871.1 | GST | 293.99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.07 | 0 | 0 | glutathione S-transferase |
| Dlo_014570.1 | GLP3 | 259.25 | 0.18 | 4.45 | 4.26 | 107.7 | 0.72 | 4.66 | 2.57 | 1.99 | 0 | 0 | 0 | 0.23 | germin-like protein 3 |
On the contrary, 28 representative NEC marker genes were highly and preferentially expressed in NEC, barely or undetected in EC, ICpEC and GE, including LEA5, CCR4-NOT transcription complex subunit 3 (CNOT3), pathogenesis-related protein (PR1–1, PR1-like, PR4), 14 kDa proline-rich protein DC2.15 (DC2.15), chitinases (CHI: Dlo_030517.1, Dlo_024175.1), catalase (CAT), Lipid transfer proteins (NsLTP2, DIR1), aquaporins (PIP1, PIP2.1, TIP2–1), peroxidases (POD-P7, POD5), osmotin-like protein 1 (OSM1), expansin-like B1 (EXLB1), Pectinesterase precursor (PME1), chalcone synthase (CHS), thaumatin-like protein (TLP1), Gibberellic Acid Stimulated Transcript-like (GAST1), ethylene-responsive transcription factor 114 (ERF114), glutathione S-transferase (GST, Dlo_032871.1), germin-like protein 3 (GLP3), and three unknown genes (DlU4, DlU5, DlU6) (Table 4). The NEC-specific marker genes maybe the key inhibitor of the transition from NEC to EC, while the SE markers may function on SE development.
qRT-PCR verification of selected molecular markers
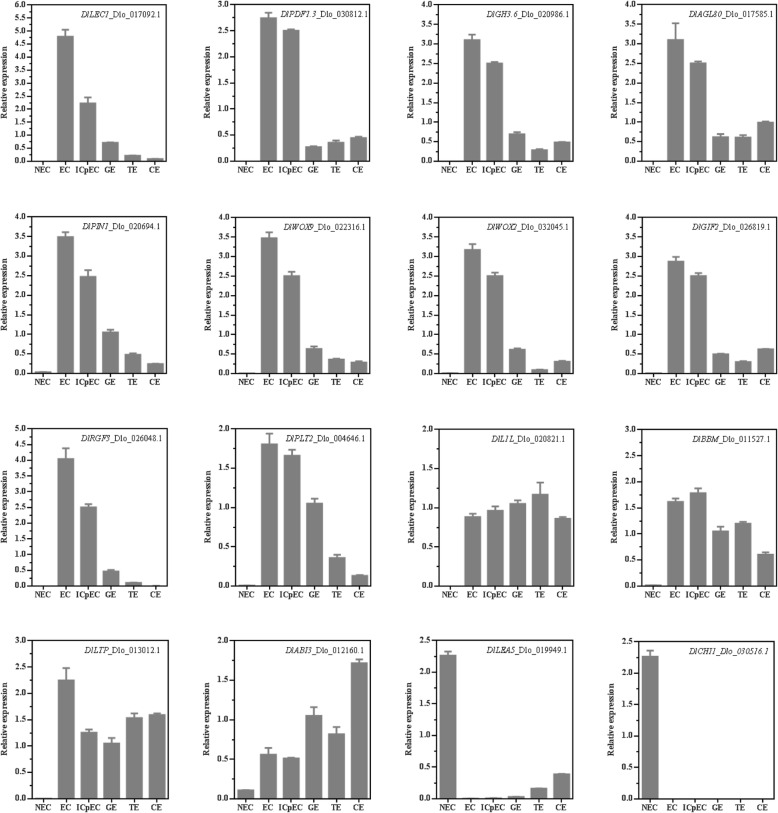
To experimentally confirm that the molecular markers were indeed expressed and played a key role during longan SE, 16 molecular markers, including 8 transcription factors DlLEC1_Dlo_017092.1, DlL1L_Dlo_020821.1, DlABI3_Dlo_012160.1, DlWOX9_ Dlo_022316.1, DlWOX2_Dlo_032045.1, DlAGL80_Dlo_017585.1, DlBBM_Dlo_011527.1 and DlPLT2_Dlo_004646.1, auxin metabolism gene DlGH3.6_ Dlo_020986.1, auxin polar transport gene DlPIN1_Dlo_020694.1, 3 meristem growth regulation genes DlPDF1.3_ Dlo_030812.1, DlRGF3_Dlo_026048.1, DlGIF2_Dlo_026819.1, 2 extracellular protein encoding genes DlLTP_Dlo_013012.1 and DlCHI_Dlo_030516.1, a late embryogenesis abundant protein gene DlLEA5_Dlo_019949.1, were selected for qRT-PCR identification in the synchronized cultures at distinct developmental stages during longan SE, including NEC, EC, ICpEC, GE, torpedo-shaped embryos (TE) and cotyledonary embryos (CE).
Base on the qRT-PCR results, all selected genes were expressed at varying levels at different development stages (Fig. 6). The selected molecular markers DlLEC1, DlPDF1.3, DlGH3.6, DlPIN1, DlWOX9, DlWOX2, DlGIF2, DlRGF3, DlPLT2 and DlAGL80 were barely or undetected in NEC, while they mainly expressed during early SE, they all highest expressed in EC and then down-regulated during SE, showed relative low expression in TE and CE, indicated that those molecular markers played an important role in EC induction and maintainance. Meanwhile, DlL1L, DlBBM, DlABI3 and DlLTP were highly expressed or up-regulated during SE processes, and minimally or undiscovered expressed in NEC, suggested that those marker genes may positive regulated the longan SE development. In addition, the transcription level of DlLEA5 and DlCHI were highly and specific expressed in NCE, they may the inhibitor of the transition from NEC to EC. qRT-PCR validation of SE-related genes also showed a high correlation between RNA-seq and qRT-PCR data (Additional file 10: Table S6).
Fig. 6.
qRT-PCR verification of the selected molecular markers during longan SE. Non-embryogenesis callus (NEC), friable-embryogenesis callus (EC), incomplete compact pro-embryogenic cultures (ICpEC), globular embryos (GE), torpedo-shaped embryos (TE) and cotyledonary embryos (CE). DlFSD, DlEF1a, and Dlelf4a are used as reference genes. Data are means±SD (n = 3)
Discussion
Auxin and cytokinin play an important role in longan SE
It is well know that auxin and cytokinin (CTK) were key factors of plant cell division, differentiation, and SE induction [59]. Meanwhile, the level of endogenous IAA and CTK were influenced by the application of exogenous auxin and CTK [3, 10, 60–62]. Auxin was consider as a central regulator in SE, probably due to the establishment of auxin gradients during SE induction [9]. So far, the exogenous application of auxin during SE has been well documented [9, 10, 27, 60]. Among the auxin, 2,4-dichlorophenoxyacetic acid (2,4-D) was most effective and widely used for induction of SE in several plants [63–65]. The level of endogenous IAA was correlated with pro-embryogenic mass formation and high-frequency SE competency [66]. Previous study had also proved that dynamic change of endogenous IAA was among the first signals leading to the induction of SE [67].
Over the past 20 years, longan SE has been established and widely used as model system for embryogenesis in woody plants, high concentration of 2,4-D in MS medium was require for inducing EC from immature zygotic embryo, while it suppressed the further development of SE, moreover, 2,4-D and KT were the key factors in long term maintenance of longan EC [1, 7, 8]. Subsequent studies revealed that controlling the doses of 2,4-D could synchronized regulated the developmental processes of longan SE, withdrawal of 2,4-D from the medium triggered further embryo development [46, 68–70]. The level of endogenous IAA and CTK in early SE stages (EC, ICpEC and GE) were much higher than NEC stage, IAA level reached the peak in GE and then significantly decreased at later stages. In addition, the level of IAA higher than CTK at the same stage during early SE. The results indicated that high level of endogenous IAA and lower level of CTK were essential for early SE [3]. However, the molecular mechanism responsible for the endogenous IAA and CTK level changing during SE, and potential crosstalk with each other or other factors remains poorly-understood.
The increase of IAA during longan early SE might be due to the increased biosynthesis and transition of endogenous auxin precursor [9]. The tryptophan (Trp) dependent IAA biosynthesis was an important pathway in higher plants, exogenous applied the doses of Trp and IAA had similar enhancement during rice SE [71]. In our study, the expression level of ASA, IGS, TSA, TSB, the key genes in Trp synthesis, were drastic up-regulated in EC and remained high in early SE, only PAI showed NEC specific with low FPKM, suggested that the level of Trp during early SE was higher than NEC, high IAA level might be due to high level of auxin precursor during early SE. YUCCAs family encoding key enzymes in IAA biosynthesis, were required for SE induction in Arabidopsis [72], and three YUCCAs and AAO1, one NIT, CYP71A13 and three ST5a, showed up-regulated expression from NEC to EC, two YUCCAs, AAO1, ST5a were down-regulated during early SE, while YUCCA_Dlo_013505.1 kept up-regulated expression during early SE. Other IAA synthesis genes were mainly expressed in NEC with minimal FPKM. The increase of IAA level may due to these differentially expressed genes during early SE. However, more evidences is needed to prove the relationship between these DEGs and increased IAA level.
During SE induction of C. canephora, the balance of free IAA and IAA conjugates was essential for embryogenic potential [73], the conjugation of auxin was synthesized by GH3 family [74], we found that GH3 family genes were minimal expressed in NEC, most of them dramatic up-regulated in EC and down-regulated during early SE, indicated that the conjugated IAA also played an important role in longan SE. Previous studies had revealed that auxin transports was complex and highly regulated for embryogenic development [75]. TIR1 mediated Aux/IAA proteins degradation and auxin-regulated transcription in the present of auxin [76], while TIR1 genes were down-regulated in EC and remained low during longan early SE. AUX1, which mediated influx of IAA into cells, were mainly expressed in NEC, and down-regulated during early SE. PIN1 played a fundamental role in maintaining the embryonic auxin gradients [77], were up-regulated in EC and kept high in ICpEC and GE in our study.
Auxin transcriptionally activated Aux/IAA, GH3 and SAUR family, the Aux/IAA family had 29 members in Arabidopsis, but not all members were induce by auxin [78]. SAUR was the most abundant family of early auxin-inducible genes, but only few members had been functional characterized, OsSAUR39 was reported to negatively regulate auxin biosynthesis and transport [79]. ARF showed strongly disturbance during zygotic embryo development [80, 81], and ARF5 seemed to be importance for SE [82]. Further transcript analysis during SE revealed that the components of auxin signaling: Aux/IAA, ARF, SAUR and other auxin-responsive genes were wildly modulated during SE [9, 13]. In our study, 11 ARF significantly up-regulated in EC and remained high during early SE, three ARF first down-regulated in EC and then up-regulated during SE. IAA family showed the similar expression pattern during SE, while most of SAUR were mainly expressed in NEC.
Other than auxin being a main inducer of SE, exogenously supplied CTK to induce SE was well established in a lot of species [83–85]. Large numbers of transcripts involved in zeatin biosynthesis and signal transduction were differentially expressed during cotton SE [10]. Meanwhile, endogenous CTK level were higher in SE than in NEC [3]. From NEC to EC, a total of 40 DEGs implicated in cytokinin signal transduction, including 22 CRE1 (seven up-regulated and 15 down-regulated), 15 B-ARR (five up-regulated and 10 down-regulated), 3 A-ARR (one up-regulated and two down-regulated). During early SE, a total of 32 DEGs involved in cytokinin signaling pathway, most of them were up-regulated during early SE. In zeatin synthesis pathway, TRIT1 was up-regulated from NEC to ICpEC, most of CisZOG, IPT1,4, CYP735A, CKX, UGT76C were down-regulated in EC and remained low during early SE. However, IAA and zeatin biosynthesis and signal transduction related genes showed complex and integrated regulation during SE, further study of these genes is required in longan SE.
SE-related molecular marker genes play a key role during longan SE
The molecular marker genes for longan SE belong to several distinct functional categories, they can be used to mark the embryogenic potential of plant cells and study various biochemical and physiological processes of plant embryogenesis and development. A number of transcription factors (TFs) had been reported as key factors in SE induction. In Brassica napus, LEC1, LEC2, FUS3, ABI3, WOX9, WOX2, BBM, genes belonged to TFs, were identified as molecular markers for early microspore embryogenesis [86]. In our study, 10 molecular markers were TFs (DlLEC1, DlL1L, DlLEC2, DlABI3, DlFUS3, DlWOX9, DlWOX2, DlAGL80, DlBBM, DlPLT2), their functions on embryogenesis had been well characterized in various plants. Ectopic expression of LEC1 was sufficient to trigger embryogenic potential and to induce somatic embryo from Arabidopsis leaf surface [87]. ZmLEC1 was used as a reliable marker for early SE in maize as its expression pattern during SE was similar to that of AtLEC1 during zygotic embryogenesis [88]. Mutational analyses in Arabidopsis showed that LEC genes were essential for induction of SE [37]. Ectopic expression of a carrot C-LEC1 which was driven by AtLEC1 promoter, rescued the defects of lec1–1 mutant [89]. Moreover, ectopic-expressed AtLEC1 in tobacco induced the start of embryogenic transition [90]. The LEC1-like (L1L) was most closely related to LEC1 and required for normal embryo development, ectopic-expressed L1L in Arabidopsis can complement LEC1 functions [91]. Meanwhile, L1L expression was mainly accumulated in the early stage SE of Theobroma cacao [92], Vitis vinifera [93], and Helianthus annuus [94].
LEC2, ABI3, FUS3 were B3 domain-containing transcription factors, ectopically expressed AtABI3 do not induced SE but endowed the embryo with traits to seedling [95]. BBM and PLT2 were clustered to AP2/ERF transcription factor family, their functions on embryogenesis and root meristem were overlap [96–98]. Over-expression of BBM triggered spontaneous somatic embryo formation in Arabidopsis thaliana and Brassica napus, BBM was server as a marker for embryogenesis cells in Brassica napus [96]. Recently study show that BBM and PLT2 induced SE in a quantitatively and context dependent manner by LEC1-ABI3-FUS3-LEC2 (LAFL) network, and LAFL/AGL15 were required for BBM mediated embryogenesis [40]. In this assay, DlLEC1, DlLEC2, DlFUS3 were early SE-specific genes, DlL1L, DlBBM, DlABI3, DlPLT2 were highly expressed during the SE processes, they can be used as remarkable markers for longan early SE.. To date, AGL15 was the only MADS-BOX member which preferentially expressed in developing embryos and promote the initiation of SE [44, 45], and AGL80 was essential for the central cell and endosperm development [99]. However, DlAGL15 was considered as poor marker. Firstly, we suggested another MADS-BOX gene DlAGL80, a SE-specific gene as a new marker for longan early SE.
WUS was a critical regulator for stem cell fate in the shoot apical meristem [100]. Over-expression of AtWUS initiated the acquisition of embryogenic competence in Gossypium hirsutum [41, 42]. WUS was suggested as a useful gene marker for SE initiation [101]. Meanwhile, WOX genes marked cell fate during early embryogenesis in Arabidopsis [102], WOX2 was used as potential marker during early SE [103]. STIMPY/WOX9 played an important role in promoting cell proliferation and preventing precocious differentiation in emerging seedlings [104]. WOX2 and WOX9 were highly expressed at the early stage of SE in Picea abies, they may function together on conifer embryo patterning [105]. In addition, DlWUS was isolated from embryogenic callus and expressed in all the stage of SE, which consistent with our transcriptome date suggested that DlWUS genes were poor markers during longan SE. qRT-PCR verification demonstrated that DlWOX2 and DlWOX9 were specific expressed in early SE and down-regulated during SE, they might played an importance role in longan early SE.
PIN1 was involved in auxin polar transport and cellular differentiation during embryogenesis [106, 107]. Antisense expression of PIN1 disrupted the formation of somatic embryos and reduced the expression of SE-related genes, indicated that PIN1 was essential for SE induction [108]. GH3.6 was contribute to maintain auxin homeostasis by converting excess IAA to IAA-amino acid conjugates, over-expression of GH3.6 significantly enhanced the accumulation of IAA-Asp [109]. DlPIN1 and DlGH3.6 were specific expressed in early SE and down-regulated during SE.
In our study, CYP78A5 and CYP87A3 were most abundant in EC and follow by ICpEC stage. In B. napus, CYP78A5 was identified as an early marker for microspore-derived embryos development [86]. PDF1.3 was closely related to Arabidopsis Protodermal factor 1, a gene exclusively expressed in L1 layer of vegetative, inflorescence, floral meristems and specific-expressed in protodermal cell during embryogenesis which related to cell fate determination [110]. In addition, AtGRP-5 was associated with somatic embryo formation in Arabidopsis and eggplant [111]. RGF3 and GIF2 were key genes of cell proliferation, showed SE-specific expression pattern during early SE. RGF3 belonged to root meristem growth factors family that played the redundant role in maintaining the post-embryonic root stem cell niche and by positive regulating cell proliferation [112]. GIF2 was required for cell proliferation and lateral organs grow [113]. Those SE-related genes DlPIN1, DlGH3.6, DlPDF1.3, DlGRP-1, DlRGF3 and DlGIF2 can be use to mark the early stage of longan early SE. Furthermore, DlLBP, DlKCS, DlZDS and DlRPL17, DlMAN7 and DlU1, DlU2, DlU3 were specific accumulated in early SE, despite that no functions on SE have been published yet for them, suggested that they might be the key genes for longan SE.
SERK played a key role in the acquisition of embryogenic competence in plant cells, DcSERK was identified as a suitable marker for SE as it only abundant in embryogenic cultures and ceased after the globular stage, but not in any other tissues [34]. In Dactylis glomerata, SERK showed the similar expression pattern with DcSERK and used as a convenient marker for cells competent to form embryos in monocots [114]. AtSERK1 was highly expressed during Arabidopsis embryogenic cell formation and early embryogenesis, suggested that AtSERK1 was sufficient to mark embryogenic competence in culture [115]. However, ZmSERKs were detected in non-embryogenic callus [116], and the identification of SERK genes in rice [117], and wheat [118], suggested that their functions were not limit to embryogenesis. In our study, DlSERKs were expressed not only in SE stages, but also in non-embryogenic callus and other tissues.
LEA5 belonged to the fifth group of late embryogenesis abundant proteins gene, were abundant in late embryogenesis of mature seed, and involved in the abiotic stresses responses [119]. CNOT3, might be a new CCR4-NOT complex gene in plant, which had proved in regulation of cell division in HeLa cell, while its functions on plant was poorly-understood. 14 kDa proline-rich protein DC2.15 was connected with the initiation of embryogenesis by the removal of auxin [120]. Aquaporins were the major channels of water transport pass through biological membranes, and involved in cell expansion, organ movement and elongation [121]. It is widely acceptable that the extracellular proteins (such as GLPs, LTPs, CHIs) were required for plant differentiation and morphogenesis, they were used as protein markers for SE [58]. Our study revealed that a total of 28 transcripts were specific and extreme-highly expressed in NEC, while barely or undetected during early SE stages, for example, the LEA5, CNOT3, DC2.15, PIP1;2, PIP2;1, GLP3, NsLPT, CAT, POD, GST, et al., these genes might play an important role in the transition from NEC to EC. However, some of these markers belonged to the certain gene family with distinct expression patterns during SE, further study of these genes function on longan SE is required.
Conclusions
In summary, our study generated a high resolution transcriptome datasets for longan SE. A comparative analysis of global gene expression patterns during early SE stages provided subsets of DEGs that regulated SE in longan. Our study revealed the expression profiles of genes involved in plant hormones such as auxin and cytokinin signaling pathway, flavonoid and fatty acid biosynthesis pathway, extracellular protein, as well as the representative molecular marker genes, indicating their possible roles in longan SE. This transcriptomic data provides new insights into future functional studies, as a means of studying the molecular mechanisms in SE.
Materials and methods
Plant material and RNA extraction
The synchronized cultures at different developmental stages, including non-embryogenic callus (NEC), friable-embryogenic callus (EC), incomplete compact pro-embryogenic cultures (ICpEC), globular embryos (GE), torpedo-shaped embryos (TE) and cotyledonary embryos (CE) of D. longan ‘Honghezi’ were obtained following previously methods [1, 7, 8, 68]. To obtained.the synchronized cultures of NEC, EC, ICpEC, GE and TE, embryogenic calli was transferred to MS basal medium (2% sucrose and 6 g/L agar, pH 5.8) supplemented with 0.2% activated carbon and 1.0 mg/L, 0.5 mg/L, 0.1 mg/L, 0.05 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), respectively. Embryogenic calli was transferred to MS basal medium (5% sucrose and 6 g/L agar, pH 5.8) to obtained the synchronized cultures of CE. The synchronized cultures of different stage were cultured in three biological replicates, each replicates consisting of 10 culture bottles, and were confirmed by the histological observations as shown schematically in Fig. 1. We collected the NEC, EC, ICpEC and GE samples from 5 bottles in each of the three replicates, and stored at − 80 °C for RNA extraction.
Total RNA was extracted separately from NEC, EC, ICpEC and GE in the three biological replicates using Trizol Reagent (Invitrogen, USA), then DNase I was used to digest any genomic DNA. Extracted RNAs were quantified by Agilent 2100 bioanalyzer (Agilent Technologies, USA) and evaluated the integrality by denaturing agarose gel electrophoresis and ethidium bromide staining. RNA samples with A260/A280 ratios between 1.9~2.1, 28S/18S ratios ≥1.0, and integrity numbers (RINs) more than 8.5 were selected to construct cDNA libraries. The RNA of the three biological replicates were mixed in equal amounts and used for cDNA library construction.
Library construction and RNA sequencing
After purification with oligo (dT)25-attached magnetic beads, the mRNA was interrupted into short fragments by divalent cations under elevated temperature. Then, these cleaved RNA fragments were used to synthesize first-strand cDNA using a random hexamer primer and the SuperScript III (Invitrogen, USA) reverse transcriptase. The second-strand cDNA was subsequently synthesized using random primers and end repaired, then adaptors were ligated by T4 DNA ligase after adenylation at the 3′-end. Eventually, suitable adaptor-ligated fragments were selected as templates for PCR amplification to generate the final cDNA library. The four resulting cDNA libraries were quantified by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and qRT-PCR (ABI StepOnePlus Real-Time PCR System, USA), and then RNA Sequencing (RNA-seq) was carried out with an Illumina HiSeq™ 2000 system at The Beijing Genomics Institute (BGI, Shenzhen, China). The entire set of raw reads was submitted to NCBI Sequence Read Archive under the accession number: PRJNA565345.
RNA-Seq reads mapping and differential expression
The raw reads were cleaned by removing adapter reads, reads containing poly-N larger than 10%, and low quality reads (QPhred < 20). Cleaned reads were then aligned to the longan reference genome using Bowtie software (http://bowtie-bio.sourceforge.net/index.shtml) and TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml), read count for each gene was then obtained after mapping. Gene expression levels for each sample were estimated by RSEM (RNA-Seq by Expectation Maximization) software [122]. The expression levels of matched genes in each cDNA library were derived and normalized to FPKM (Fragments Per Kilobase of exon per Million fragments mapped) [123]. The differentially expression analysis of the pairwise comparison of RNA-Seq libraries were confirmed by using the Poisson Distribution analysis method [124], and the False Discovery Rate (FDR) was used to determine the P value threshold. The unique reads with the absolute value of log2 (Fold_Change) ≥ 1 and the FDR < 0.001 were used as the thresholds to define as differentially expressed genes (DEGs) in the pairwise comparisons (NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, ICpEC_vs_GE).
Expression annotation and functional analysis of DEGs
Gene function was annotated based on the databases of Blast Nr (NCBI non-redundant protein sequences), GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes database). The GO and KEGG functional enrichment analysis of DEGs were performed to identify which DEGs were significantly enriched in GO terms or KEGG pathways. GO terms with corrected P-value ≤0.05 were considered as significantly enriched terms. The KEGG enrichment was determined by Rich factor, Q-value, and the number of enriched genes in this pathway. Q-value ≤0.05 was defined as those with genes that showed significant differential expression.
Quantitative real-time PCR analysis
For qRT-PCR validation, 500 ng total RNA extracted from each stage of longan SE (NEC, EC, ICpEC, GE, TE and CE) were transcribed into cDNA with random primers and Oligo dT primer using the SYBR Ex Script™ kit (Takara, China), Sixteen unique transcripts with potential roles in longan SE were chosen and their specific primers were designed using DNAMAN 7.0. qRT-PCR was performed on the LightCycler 480 instrument (Roche Applied Science, Switzerland) in a total volume of 20 μL in each well containing 10 μL of 2× SYBR Premix Ex Taq™; 0.8 μL of each specific primer (100 nM); 1.0 μL of cDNA template (in a 1:10 dilution); and 7.4 μL of ddH2O. The PCR conditions were: denaturation for 60 s at 95 °C, and then 40 cycles of 10 s at 95 °C and 20 s between 58 °C and 61 °C in function of the Tm of the primers. Primer annealing specificity was examined and verified by melting curve analysis. Four-point standard curves of a fivefold dilution series (1:5 to 1:625) from pooled cDNA were used to calculate PCR efficiency. The reactions were performed in 96-well PCR plates and each experiment consisted of three biological replicates. The expressive abundance of the sixteen selected genes were calculated relative to the expression of reference genes DlFSD, DlEF-1a, and Dlelf4a. Data were further processed in MS Excel. Gene names, primer sequences, product sizes, and annealing temperatures are given in Additional file 11: Table S7.
Supplementary information
Additional file 1: Figure S1. Gene Ontology functional classification for the pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE.
Additional file 2: Figure S2. Statistic of KEGG pathway enrichment for the pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE. (DOC 230 kb)
Additional file 3: Figure S3. Plant hormone signal transduction pathway in the comparison of NEC_vs_EC. Red frame represents a transcript with increased levels of expression and green frame represents transcripts with decreased levels of expression. The image was obtained from http://www.genome.jp/kegg/.
Additional file 4: Figure S4. Heatmap of the differentially expressed genes in plant hormone signal transduction during longan SE. (a). Abscisic acid signal transduction; (b). Gibberellin signal transduction; (c). Ethylene signal transduction; (d). Salicylic acid signal transduction; (e). Brassinolide signal transduction; (f). Jasmonic acid signal transduction. Heatmaps indicate the gene expression levels by Log2[FPKM + 1] with a rainbow color scale. The IDs and names of selected DEGs are indicated to the right of the histograms.
Additional file 5: Table S1. Differentially expressed genes involved in plant hormones signaling pathway during longan SE. S1-a: Auxin signaling pathway; S1-b: Cytokinin signaling pathway; S1-c: Abscisic acid signal transduction; S1-d: Gibberellin signal transduction; S1-e: Ethylene signal transduction; S1-f: Salicylic acid signal transduction; S1-g: Jasmonic acid signal transduction; S1-h: Brassinosteroid signal transduction. (XLS 151 kb)
Additional file 6: Table S2. Differentially expressed genes involved in flavonoid biosynthesis. (XLS 50 kb)
Additional file 7: Table S3. Differentially expressed genes involved in fatty acid biosynthesis during longan SE. (XLS 26 kb)
Additional file 8: Table S4. The distinct expression pattern of extracellular protein encoding genes during longan SE.
Additional file 9: Table S5. The expression pattern of molecular marker genes during longan SE.
Additional file 10: Table S6. Comparing differential expression genes from RNA-seq and qRT-PCR during longan SE.
Additional file 11: Table S7. Primers used for real-time quantitative PCR.
Abbreviations
- ABI3
Abscisic acid insensitive 3
- AGL80
Agamous-like 80
- ARF
Auxin response factor
- BBM
Babyboom
- CE
Cotyledonary embryos
- CHI
Chitinase
- DEGs
Differentially expressed genes
- Dl
Dimocarpus longan
- EC
Embryogenic callus
- FDR
False discovery rate
- FPKM
Fragments Per Kilo-base of exon per Million fragments mapped
- FSD
Fe-SOD
- FUS3
FUSCA3
- GE
Globular embryos
- GH3.6
Indole-3-acetic acid-amido synthetase
- GLP
Germine-like protein
- GO
Gene Ontology
- GST
Glutathione S-transferase
- ICpEC
Incomplete compact pro-embryogenic cultures
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LEA5
Late embryogenesis abundant protein 5
- LEC
Leafy cotyledon
- LTP
Lipid transfer protein
- NEC
Non-embryogenic callus
- PDF1.3
Protodermal factor 1
- PIN1
PIN-FORMED1
- qRT-PCR
Quantitative real-time PCR
- RNA-seq
RNA sequencing
- SE
Somatic embryogenesis
- TE
Torpedo-shaped embryos
- WOX
WUSCHEL-related homeobox
Authors’ contributions
ZXL and YLL conceived the research, participated in its design and coordination, and helped to draft the manuscript. YKC participated in its design, carried out the experimental work and wrote the manuscript. XPX and XHX helped to draft the manuscript. XPX, ZXL, and ZHZ prepared the materials. YLL, ZXL, YKC, XHX analyzed the data and revised the paper. All authors read and approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was funded by the National Natural Science Foundation of China (31572088, 31672127), the Natural Science Funds for Distinguished Young Scholars in Fujian Province (2015 J06004), the Research Funds for the program for New Century Excellent Talents in Fujian Province University (20151104).
Availability of data and materials
The Illumina sequence data from this study have been submitted to the NCBI sequence read archive under the accession number [PRJNA565345]. All the supporting data are included in Additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declared that they have no competing financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yukun Chen, Email: cyk68@163.com.
Xiaoping Xu, Email: byxxp310107@163.com.
Zhuanxia Liu, Email: Liuzx0701@163.com.
Zihao Zhang, Email: zhangzihao863@163.com.
Xu XuHan, Email: xxuhan@163.com.
Yuling Lin, Email: buliang84@163.com.
Zhongxion Lai, Email: laizx01@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6393-7.
References
- 1.Lai Z, Chen C, Zeng L, Chen Z. Somatic embryogenesis in longan [Dimocarpus longan Lour.] In: Jain SM, Gupta PK, Newton RJ, editors. Somatic Embryogenesis in Woody Plants: Volume 6. Dordrecht: Springer Netherlands; 2000. pp. 415–431. [Google Scholar]
- 2.Lai ZX, He Y, Chen YT, Cai YQ, Lai CC, Lin YL, Lin XL, Fang ZZ. Molecular Biology and Proteomics during Somatic Embryogenesis in Dimocarpus longan Lour. Leuven: 2010-05-13 2010: International Society for Horticultural Science (ISHS); 2010. pp. 95–102. [Google Scholar]
- 3.Lai Z, Chen C. Changes of endogenous phytohormones in the process of somatic embryogenesis in longan (Dimocarpus longan Lour.) Chin J Trop Crops. 2002;23(2):41–47. [Google Scholar]
- 4.Dodeman VL, Ducreux G, Kreis M. Zygotic embryogenesis versus somatic embryogenesis. J Exp Bot. 1997;48(8):1493–1509. doi: 10.1093/jxb/48.8.1493. [DOI] [Google Scholar]
- 5.Willemsen V, Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annu Rep Genet. 2004;38:587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman JL. Somatic embryogenesis: a model for early development in higher plants. Plant Cell. 1993;5(10):1411–1423. doi: 10.2307/3869792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai Z, Pan LZ, Chen ZG. Establishment and maintenance of longan embryogenic cell lines. J Fujian Agric Fort Univ. 1997;26(2):160–167. [Google Scholar]
- 8.Lai ZX, Chen ZG. Somatic embryogenesis of high frequency from longan embryogenic calli. J Fujian Agric Fort Univ. 1997;26(3):271–276. [Google Scholar]
- 9.Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012;12:110. doi: 10.1186/1471-2229-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z, Zhang C, Zhang X, Liu C, Wu Z, Yang Z, Zhou K, Yang X, Li F. Transcriptome profiling reveals Auxin and Cytokinin regulating somatic embryogenesis in different sister lines of cotton cultivar CCRI24. J Integr Plant Biol. 2013;55(7):631–642. doi: 10.1111/jipb.12073. [DOI] [PubMed] [Google Scholar]
- 11.Jin F, Hu L, Yuan D, Xu J, Gao W, He L, Yang X, Zhang X. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol J. 2014;12(2):161–173. doi: 10.1111/pbi.12123. [DOI] [PubMed] [Google Scholar]
- 12.Cao A, Zheng Y, Yu Y, Wang X, Shao D, Sun J, Cui B. Comparative Transcriptome analysis of SE initial dedifferentiation in cotton of different SE capability. Sci Rep. 2017;7(1):8583. doi: 10.1038/s41598-017-08763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gliwicka M, Nowak K, Balazadeh S, Mueller-Roeber B, Gaj MD. Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS One. 2013;8(7):e69261. doi: 10.1371/journal.pone.0069261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickramasuriya AM, Dunwell JM. Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genomics. 2015;16:301. doi: 10.1186/s12864-015-1504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvo SAGD, Hirsch CN, Buell CR, Kaeppler SM, Kaeppler HF. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS One. 2014;9(10):e111407. doi: 10.1371/journal.pone.0111407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakovlev IA, Lee Y, Rotter B, Olsen JE, Skrøppa T, Johnsen Ø, Fossdal CG. Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet Genomes. 2014;10(2):355–366. doi: 10.1007/s11295-013-0691-z. [DOI] [Google Scholar]
- 17.Yakovlev IA, Carneros E, Lee Y, Olsen JE, Fossdal CG. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta. 2016;243(5):1237–1249. doi: 10.1007/s00425-016-2484-8. [DOI] [PubMed] [Google Scholar]
- 18.Rajesh MK, Fayas TP, Naganeeswaran S, Rachana KE, Bhavyashree U, Sajini KK, Karun A. De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing. Protoplasma. 2016;253(3):913–928. doi: 10.1007/s00709-015-0856-8. [DOI] [PubMed] [Google Scholar]
- 19.Elbl P, Lira BS, Andrade SCS, Jo L, Dos Santos ALW, Coutinho LL, Floh EIS, Rossi M. Comparative transcriptome analysis of early somatic embryo formation and seed development in Brazilian pine, Araucaria angustifolia (Bertol.) Kuntze. Plant Cell Tiss Org Cult. 2015;120(3):903–915. doi: 10.1007/s11240-014-0523-3. [DOI] [Google Scholar]
- 20.Tao L, Zhao Y, Wu Y, Wang Q, Yuan H, Zhao L, Guo W, You X. Transcriptome profiling and digital gene expression by deep sequencing in early somatic embryogenesis of endangered medicinal Eleutherococcus senticosus maxim. Gene. 2016;578(1):17–24. doi: 10.1016/j.gene.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 21.Shi X, Zhang C, Liu Q, Zhang Z, Zheng B, Bao M. De novo comparative transcriptome analysis provides new insights into sucrose induced somatic embryogenesis in camphor tree (Cinnamomum camphora L.) BMC Genomics. 2016;17:26. doi: 10.1186/s12864-015-2357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao LM, Zhang J, Hou Y, Yao Y, Ji QL. RNA-Seq screening of differentially expressed genes during somatic embryogenesis in Fragaria×ananassa Duch 'Benihopp'. J Hortic Sci Biotech. 2015;90:671–681. doi: 10.1080/14620316.2015.11668730. [DOI] [Google Scholar]
- 23.Indoliya Y, Tiwari P, Chauhan AS, Goel R, Shri M, Bag SK, Chakrabarty D. Decoding regulatory landscape of somatic embryogenesis reveals differential regulatory networks between japonica and indica rice subspecies. Sci Rep. 2016;6:23050. doi: 10.1038/srep23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Xue B, Gai M, Song S, Jia N, Sun H. Small RNA and Transcriptome sequencing reveal a potential miRNA-mediated interaction network that functions during somatic embryogenesis in Lilium pumilum DC. Fisch Front Plant Sci. 2017;8:566. doi: 10.3389/fpls.2017.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdavi-Darvari F, Noor NM. New insight into early somatic embryogenesis of Mangosteen (Garcinia mangostana) through de novo and comparative Transcriptome analyses. Trop Plant Biol. 2017;10(1):30–44. doi: 10.1007/s12042-016-9182-3. [DOI] [Google Scholar]
- 26.Jamaluddin ND, Mohd Noor N, Goh H. Genome-wide transcriptome profiling of Carica papaya L. embryogenic callus. Physiol Mol Biol Plants. 2017;23(2):357–368. doi: 10.1007/s12298-017-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu Z, Chen J, Sun J, Dong Z, Yang X, Wang Y, Xu H, Zhang X, Chen F, Cui D. De novo assembly and comparative analysis of the transcriptome of embryogenic callus formation in bread wheat (Triticum aestivum L.) BMC Plant Biol. 2017;17(1):244. doi: 10.1186/s12870-017-1204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varhaníková M, Uvackova L, Skultety L, Pretova A, Obert B, Hajduch M. Comparative quantitative proteomic analysis of embryogenic and non-embryogenic calli in maize suggests the role of oxylipins in plant totipotency. J Proteomics. 2014;104:57–65. doi: 10.1016/j.jprot.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Vale EDM, Heringer AS, Barroso T, Ferreira ATDS, Da Costa MN, Perales JEA, Santa-Catarina C, Silveira V. Comparative proteomic analysis of somatic embryo maturation in Carica papaya L. Proteome Sci. 2014;12:37. doi: 10.1186/1477-5956-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandra Pila Quinga L, Heringer AS, Pacheco De Freitas Fraga H, Do Nascimento Vieira L, Silveira V, Steinmacher DA, Guerra MP. Insights into the conversion potential of Theobroma cacao L. somatic embryos using quantitative proteomic analysis. Sci Hortic. 2018;229:65–76. doi: 10.1016/j.scienta.2017.10.005. [DOI] [Google Scholar]
- 31.Heringer AS, Reis RS, Passamani LZ, de Souza-Filho GA, Santa-Catarina C, Silveira V. Comparative proteomics analysis of the effect of combined red and blue lights on sugarcane somatic embryogenesis. Acta Physiol Plant. 2017;39(2):52. doi: 10.1007/s11738-017-2349-1. [DOI] [Google Scholar]
- 32.Kumaravel M, Uma S, Backiyarani S, Saraswathi MS, Vaganan MM, Muthusamy M, Sajith KP. Differential proteome analysis during early somatic embryogenesis in Musa spp. AAA cv. Grand Naine. Plant Cell Rep. 2017;36(1):163–178. doi: 10.1007/s00299-016-2067-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Cheng W, Tian W, Li Y, Liu F, Xue F, Zhu Q, Sun Y, Sun J. iTRAQ-based comparative proteomic analysis provides insights into somatic embryogenesis in Gossypium hirsutum L. Plant Mol Biol. 2018;96(1):89–102. doi: 10.1007/s11103-017-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt ED, Guzzo F, Toonen MA, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124(10):2049. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- 35.Savona M, Mattioli R, Nigro S, Falasca G, Della Rovere F, Costantino P, De Vries S, Ruffoni B, Trovato M, Altamura MM. Two SERK genes are markers of pluripotency in Cyclamen persicum mill. J Exp Bot. 2012;63(1):471–488. doi: 10.1093/jxb/err295. [DOI] [PubMed] [Google Scholar]
- 36.Rupps A, Raschke J, Rümmler M, Linke B, Zoglauer K. Identification of putative homologs of Larix decidua to BABYBOOM (BBM), LEAFY COTYLEDON1 (LEC1), WUSCHEL-related HOMEOBOX2 (WOX2) and SOMATIC EMBRYOGENESIS RECEPTOR-like KINASE (SERK) during somatic embryogenesis. Planta. 2016;243(2):473–488. doi: 10.1007/s00425-015-2409-y. [DOI] [PubMed] [Google Scholar]
- 37.Gaj MD, Zhang S, Harada JJ, Lemaux PG. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta. 2005;222(6):977–988. doi: 10.1007/s00425-005-0041-y. [DOI] [PubMed] [Google Scholar]
- 38.Nic-Can GI, López-Torres A, Barredo-Pool F, Wrobel K, Loyola-Vargas VM, Rojas-Herrera R, De-la-Peña C. New insights into somatic embryogenesis: leafy cotyledon1, baby boom1 and WUSCHEL-related homeobox4 are epigenetically regulated in Coffea canephora. PLoS One. 2013;8(8):e72160. doi: 10.1371/journal.pone.0072160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Florez SL, Erwin RL, Maximova SN, Guiltinan MJ, Curtis WR. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 2015;15(1):121. doi: 10.1186/s12870-015-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muiño JM, Angenent GC, Boutilier K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017;175:848–857. doi: 10.1104/pp.17.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouchabké-Coussa O, Obellianne M, Linderme D, Montes E, Maia-Grondard A, Vilaine F, Pannetier C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013;32(5):675–686. doi: 10.1007/s00299-013-1402-9. [DOI] [PubMed] [Google Scholar]
- 42.Zheng W, Zhang X, Yang Z, Wu J, Li F, Duan L, Liu C, Lu L, Zhang C, Li F. AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS One. 2014;9(1):e87502. doi: 10.1371/journal.pone.0087502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tvorogova VE, Lebedeva MA, Lutova LA. Expression of WOX and PIN genes during somatic and zygotic embryogenesis in Medicago truncatula. Russ J Genet. 2015;51(12):1189–1198. doi: 10.1134/S1022795415120121. [DOI] [PubMed] [Google Scholar]
- 44.Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol. 2008;146(4):1663–1672. doi: 10.1104/pp.108.115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Q, Zheng Y, Perry SE. AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 2013;161(4):2113–2127. doi: 10.1104/pp.113.216275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai Z, Lin Y. Analysis of the global transcriptome of longan (Dimocarpus longan Lour.) embryogenic callus using Illumina paired-end sequencing. BMC Genomics. 2013;14(1):561. doi: 10.1186/1471-2164-14-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Y, Lai Z. Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.) Plos One. 2013;8(4):e60337. doi: 10.1371/journal.pone.0060337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Min J, Lai R, Wu Z, Chen Y, Yu L, Cheng C, Jin Y, Tian Q, Liu Q, et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience. 2017;6(5):1–14. doi: 10.1093/gigascience/gix023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Czemmel S, Heppel SC, Bogs J. R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma. 2012;249(2):109–118. doi: 10.1007/s00709-012-0380-z. [DOI] [PubMed] [Google Scholar]
- 51.Escaray FJ, Passeri V, Perea-García A, Antonelli CJ, Damiani F, Ruiz OA, Paolocci F. The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of Lotus species. Planta. 2017;246(2):243–261. doi: 10.1007/s00425-017-2696-6. [DOI] [PubMed] [Google Scholar]
- 52.Wang F, Kong W, Wong G, Fu L, Peng R, Li Z, Yao Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol Genet Genomics. 2016;291(4):1545–1559. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez Antonio, Zhao Mingzhe, Leavitt John M., Lloyd Alan M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 54.Lepiniec L, Debeaujon I, Routaboul J, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 55.Liu C, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 2014;165(4):1424–1439. doi: 10.1104/pp.114.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19(22):6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren GS, Fowler MW. Changing fatty acid composition during somatic embryogenesis in cultures of Daucus carota. Planta. 1979;144(5):451–454. doi: 10.1007/BF00380121. [DOI] [PubMed] [Google Scholar]
- 58.Tchorbadjieva MI. Protein Markers for Somatic Embryogenesis. In: Mujib A, Šamaj J, editors. Somatic Embryogenesis. Plant Cell Monographs. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. pp. 215–233. [Google Scholar]
- 59.Nic-Can GI, Loyola-Vargas VM. The Role of the Auxins During Somatic Embryogenesis. In: Loyola-Vargas VM, Ochoa-Alejo N, editors. Somatic Embryogenesis: Fundamental Aspects and Applications. Cham: Springer International Publishing; 2016. pp. 171–182. [Google Scholar]
- 60.Vondráková Z, Eliášová K, Fischerová L, Vágner M. The role of auxins in somatic embryogenesis of Abies alba. Cent Eur J Biol. 2011;6(4):587–596. [Google Scholar]
- 61.Michalczuk L, Ribnicky DM, Cooke TJ, Cohen JD. Regulation of Indole-3-acetic acid biosynthetic pathways in carrot cell cultures. Plant Physiol. 1992;100(3):1346–1353. doi: 10.1104/pp.100.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribnicky DM, Ilic N, Cohen JD, Cooke TJ. The effects of exogenous Auxins on endogenous Indole-3-acetic acid metabolism (the implications for carrot somatic embryogenesis) Plant Physiol. 1996;112(2):549–558. doi: 10.1104/pp.112.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sofiari E, Raemakers CJJM, Kanju E, Danso K, van Lammeren AM, Jacobsen E, Visser RGF. Comparison of NAA and 2,4-D induced somatic embryogenesis in cassava. Plant Cell Tiss Org Cult. 1997;50(1):45–56. doi: 10.1023/A:1005844414258. [DOI] [Google Scholar]
- 64.Raghavan V. Role of 2,4-dichlorophenoxyacetic acid (2,4-D) in somatic embryogenesis on cultured zygotic embryos of Arabidopsis: cell expansion, cell cycling, and morphogenesis during continuous exposure of embryos to 2,4-D. Am J Bot. 2004;91(11):1743–1756. doi: 10.3732/ajb.91.11.1743. [DOI] [PubMed] [Google Scholar]
- 65.Krishnan SRS, Siril EA. Auxin and nutritional stress coupled somatic embryogenesis in Oldenlandia umbellata L. Physiol Mol Biol Plants. 2017;23(2):471–475. doi: 10.1007/s12298-017-0425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou X, Zheng R, Liu G, Xu Y, Zhou Y, Laux T, Zhen Y, Harding SA, Shi J, Chen J. Desiccation treatment and endogenous IAA levels are key factors influencing high frequency somatic embryogenesis in Cunninghamia lanceolata (lamb.) hook. Front. Plant Sci. 2017;8:2054. doi: 10.3389/fpls.2017.02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas C, Bronner R, Molinier J, Prinsen E, van Onckelen H, Hahne G. Immuno-cytochemical localization of indole-3-acetic acid during induction of somatic embryogenesis in cultured sunflower embryos. Planta. 2002;215(4):577–583. doi: 10.1007/s00425-002-0791-8. [DOI] [PubMed] [Google Scholar]
- 68.Chen CL, Lai ZX. Synchronization regulation of embryogenesis of embryogenic calli and their histological observations in longan (Dimocarpus longan Lour.) J Fujian Agri Fort Univ. 2002;31(2):192–194. [Google Scholar]
- 69.Wang F. Synchronization control of somatic embryogenesis from Embryogenic Calli and Extraction of DNA and RNA in Longan (Dimocarpus longan Lour.) Chin J Trop Crops. 2003;24(3):31–35. [Google Scholar]
- 70.Fang ZZ, Zhang YL, Lai CC, Lai ZX. Developmental regulation of Ran 3'untranslated region during somatic embryogenesis in Dimocarpus longan Lour. Sci Hortic. 2014;176:297–302. doi: 10.1016/j.scienta.2014.07.023. [DOI] [Google Scholar]
- 71.Siriwardana S, Nabors MW. Tryptophan enhancement of somatic embryogenesis in rice. Plant Physiol. 1983;73(1):142–146. doi: 10.1104/pp.73.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bai B, Su YH, Yuan J, Zhang XS. Induction of somatic embryos in Arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Mol Plant. 2013;6(4):1247–1260. doi: 10.1093/mp/sss154. [DOI] [PubMed] [Google Scholar]
- 73.Ayil-Gutiérrez B, Galaz-Ávalos R, Peña-Cabrera E, Loyola-Vargas V. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal Behav. 2013;8(11):e26998. doi: 10.4161/psb.26998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J Biol Chem. 2010;285(39):29780–29786. doi: 10.1074/jbc.M110.146431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 76.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 77.Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell. 2005;17(9):2517–2526. doi: 10.1105/tpc.105.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17(12):3282. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kant S, Bi Y, Zhu T, Rothstein SJ. SAUR39, a small Auxin-up RNA gene, acts as a negative regulator of Auxin synthesis and transport in Rice. Plant Physiol. 2009;151(2):691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rademacher Eike H., Möller Barbara, Lokerse Annemarie S., Llavata-Peris Cristina I., van den Berg Willy, Weijers Dolf. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. The Plant Journal. 2011;68(4):597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 81.Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G, et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell. 2012;22(1):211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 82.Wójcikowska B, Gaj MD. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017;36(6):843–858. doi: 10.1007/s00299-017-2114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viktor Nørgaard J, Krogstrup P. Cytokinin induced somatic embryogenesis from immature embryos of Abies nordmanniana Lk. Plant Cell Rep. 1991;9(9):509–513. doi: 10.1007/BF00232107. [DOI] [PubMed] [Google Scholar]
- 84.Sagare AP, Lee YL, Lin TC, Chen CC, Tsay HS. Cytokinin-induced somatic embryogenesis and plant regeneration in Corydalis yanhusuo (Fumariaceae)-a medicinal plant. Plant Sci. 2000;160(1):139–147. doi: 10.1016/S0168-9452(00)00377-0. [DOI] [PubMed] [Google Scholar]
- 85.Ernst D, Oesterhelt D. Effect of exogenous cytokinins on growth and somatic embryogenesis in anise cells (Pimpinella anisum L.) Planta. 1984;161(3):246–248. doi: 10.1007/BF00982920. [DOI] [PubMed] [Google Scholar]
- 86.Malik MR, Wang F, Dirpaul JM, Zhou N, Polowick PL, Ferrie AMR, Krochko JE. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol. 2007;144(1):134–154. doi: 10.1104/pp.106.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93(7):1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 88.Zhang S, Wong L, Meng L, Lemaux PG. Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize ( Zea mays L.) Planta. 2002;215(2):191–194. doi: 10.1007/s00425-002-0735-3. [DOI] [PubMed] [Google Scholar]
- 89.Yazawa K, Takahata K, Kamada H. Isolation of the gene encoding carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol Biochem. 2004;42(3):215–223. doi: 10.1016/j.plaphy.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 90.Guo F, Liu C, Xia H, Bi Y, Zhao C, Zhao S, Hou L, Li F, Wang X. Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS One. 2013;8(8):e71714. doi: 10.1371/journal.pone.0071714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15(1):5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alemanno L, Devic M, Niemenak N, Sanier C, Guilleminot J, Rio M, Verdeil J, Montoro P. Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta. 2008;227(4):853–866. doi: 10.1007/s00425-007-0662-4. [DOI] [PubMed] [Google Scholar]
- 93.Schellenbaum P, Jacques A, Maillot P, Bertsch C, Mazet F, Farine S, Walter B. Characterization of VvSERK1, VvSERK2, VvSERK3 and VvL1L genes and their expression during somatic embryogenesis of grapevine (Vitis vinifera L.) Plant Cell Rep. 2008;27(12):1799–1809. doi: 10.1007/s00299-008-0588-8. [DOI] [PubMed] [Google Scholar]
- 94.Chiappetta A, Fambrini M, Petrarulo M, Rapparini F, Michelotti V, Bruno L, Greco M, Baraldi R, Salvini M, Pugliesi C, et al. Ectopic expression of LEAFY COTYLEDON1-LIKE gene and localized auxin accumulation mark embryogenic competence in epiphyllous plants of Helianthus annuus x H. tuberosus. Ann Bot. 2009;103(5):735–747. doi: 10.1093/aob/mcn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol. 2000;220(2):412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- 96.Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu C, van Lammeren AAM, Miki BLA, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14(8):1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119(1):109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 98.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449(7165):1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 99.Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN. AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell. 2006;18(8):1862–1872. doi: 10.1105/tpc.106.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 101.Mahdavi-Darvari F, Noor NM, Ismanizan I. Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell Tiss Org Cult. 2015;120(2):407–422. doi: 10.1007/s11240-014-0615-0. [DOI] [Google Scholar]
- 102.Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131(3):657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- 103.Malabadi RB, Nataraja K, Kumar SV, Mulgund GS. Evidence of WUSCHEL (WOX2) gene expression during induction of somatic embryogenesis from apical shoot buds of mature trees of Pinus roxburghii. Res Plant Biol. 2011;1(4):7–12. [Google Scholar]
- 104.Wu X, Dabi T, Weigel D. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol. 2005;15(5):436–440. doi: 10.1016/j.cub.2004.12.079. [DOI] [PubMed] [Google Scholar]
- 105.Palovaara J, Hallberg H, Stasolla C, Hakman I. Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies. New Phytol. 2010;188(1):122–135. doi: 10.1111/j.1469-8137.2010.03336.x. [DOI] [PubMed] [Google Scholar]
- 106.Forestan C, Meda S, Varotto S. ZmPIN1-mediated Auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2009;152(3):1373–1390. doi: 10.1104/pp.109.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Forestan C, Varotto S. The role of PIN Auxin efflux carriers in polar Auxin transport and accumulation and their effect on shaping maize development. Mol Plant. 2012;5(4):787–798. doi: 10.1093/mp/ssr103. [DOI] [PubMed] [Google Scholar]
- 108.Su YH, Zhao XY, Liu YB, Zhang CL, O'Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009;59(3):448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abe M, Takahashi T, Komeda Y. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 1999;40(6):571–580. doi: 10.1093/oxfordjournals.pcp.a029579. [DOI] [PubMed] [Google Scholar]
- 111.Magioli C, Maria Barrôco R, Rocha CAB, Dolores De Santiago-Fernandes L, Mansur E, Engler G, Margis-Pinheiro M, Sachetto-Martins G. Somatic embryo formation in Arabidopsis and eggplant is associated with expression of a glycine-rich protein gene (Atgrp-5) Plant Sci. 2001;161(3):559–567. doi: 10.1016/S0168-9452(01)00443-5. [DOI] [Google Scholar]
- 112.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329(5995):1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 113.Lee BH, Ko J, Lee S, Lee Y, Pak J, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009;151(2):655–668. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Somleva MN, Schmidt EDL, de Vries SC. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep. 2000;19(7):718–726. doi: 10.1007/s002999900169. [DOI] [PubMed] [Google Scholar]
- 115.Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001;127(3):803–816. doi: 10.1104/pp.010324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang S, Liu X, Lin Y, Xie G, Fu F, Liu H, Wang J, Gao S, Lan H, Rong T. Characterization of a ZmSERK gene and its relationship to somatic embryogenesis in a maize culture. Plant Cell Tiss Org Cult. 2011;105(1):29–37. doi: 10.1007/s11240-010-9834-1. [DOI] [Google Scholar]
- 117.Ito Y, Takaya K, Kurata N. Expression of SERK family receptor-like protein kinase genes in rice. BBA-BIOENERGETICS. 2005;1730(3):253–258. doi: 10.1016/j.bbaexp.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 118.Singla B, Khurana JP, Khurana P. Characterization of three somatic embryogenesis receptor kinase genes from wheat, Triticum aestivum. Plant Cell Rep. 2008;27(5):833–843. doi: 10.1007/s00299-008-0505-1. [DOI] [PubMed] [Google Scholar]
- 119.Liu Y, Xie L, Liang X, Zhang S. CpLEA5, the late embryogenesis abundant protein gene from Chimonanthus praecox, possesses low temperature and osmotic resistances in prokaryote and eukaryotes. Int J Mol Sci. 2015;16(11):26978–26990. doi: 10.3390/ijms161126006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aleith F, Richter G. Gene expression during induction of somatic embryogenesis in carrot cell suspensions. Planta. 1991;183(1):17–24. doi: 10.1007/BF00197562. [DOI] [PubMed] [Google Scholar]
- 121.Ma N, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008;148(2):894. doi: 10.1104/pp.108.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Gene Ontology functional classification for the pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE.
Additional file 2: Figure S2. Statistic of KEGG pathway enrichment for the pairwise comparisons of NEC_vs_EC, EC_vs_ICpEC, EC_vs_GE, and ICpEC_vs_GE. (DOC 230 kb)
Additional file 3: Figure S3. Plant hormone signal transduction pathway in the comparison of NEC_vs_EC. Red frame represents a transcript with increased levels of expression and green frame represents transcripts with decreased levels of expression. The image was obtained from http://www.genome.jp/kegg/.
Additional file 4: Figure S4. Heatmap of the differentially expressed genes in plant hormone signal transduction during longan SE. (a). Abscisic acid signal transduction; (b). Gibberellin signal transduction; (c). Ethylene signal transduction; (d). Salicylic acid signal transduction; (e). Brassinolide signal transduction; (f). Jasmonic acid signal transduction. Heatmaps indicate the gene expression levels by Log2[FPKM + 1] with a rainbow color scale. The IDs and names of selected DEGs are indicated to the right of the histograms.
Additional file 5: Table S1. Differentially expressed genes involved in plant hormones signaling pathway during longan SE. S1-a: Auxin signaling pathway; S1-b: Cytokinin signaling pathway; S1-c: Abscisic acid signal transduction; S1-d: Gibberellin signal transduction; S1-e: Ethylene signal transduction; S1-f: Salicylic acid signal transduction; S1-g: Jasmonic acid signal transduction; S1-h: Brassinosteroid signal transduction. (XLS 151 kb)
Additional file 6: Table S2. Differentially expressed genes involved in flavonoid biosynthesis. (XLS 50 kb)
Additional file 7: Table S3. Differentially expressed genes involved in fatty acid biosynthesis during longan SE. (XLS 26 kb)
Additional file 8: Table S4. The distinct expression pattern of extracellular protein encoding genes during longan SE.
Additional file 9: Table S5. The expression pattern of molecular marker genes during longan SE.
Additional file 10: Table S6. Comparing differential expression genes from RNA-seq and qRT-PCR during longan SE.
Additional file 11: Table S7. Primers used for real-time quantitative PCR.
Data Availability Statement
The Illumina sequence data from this study have been submitted to the NCBI sequence read archive under the accession number [PRJNA565345]. All the supporting data are included in Additional files.