Abstract
Maintenance of the mitochondrial genome is essential for proper cellular function. For this purpose, mitochondrial DNA (mtDNA) needs to be faithfully replicated, transcribed, translated, and repaired in the face of constant onslaught from endogenous and environmental agents. While only 13 polypeptides are encoded within mtDNA, the mitochondrial proteome comprises over 1500 proteins that are encoded by nuclear genes and translocated to the mitochondria for the purpose of maintaining mitochondrial function. Regulation of mtDNA and mitochondrial proteins by epigenetic changes and post-translational modifications facilitate crosstalk between the nucleus and the mitochondria and ultimately lead to the maintenance of cellular health and homeostasis. DNA methyl transferases have been identified in the mitochondria implicating that methylation occurs within this organelle; however, the extent to which mtDNA is methylated has been debated for many years. Mechanisms of demethylation within this organelle have also been postulated, but the exact mechanisms and their outcomes is still an active area of research. Mitochondrial dysfunction in the form of altered gene expression and ATP production, resulting from epigenetic changes, can lead to various conditions including aging related neurodegenerative disorders, altered metabolism, changes in circadian rhythm, and cancer. Here, we provide an overview of the epigenetic regulation of mtDNA via methylation, long and short non-coding RNAs, and post-translational modifications of nucleoid proteins (as mitochondria lack histones). We also highlight the influence of xenobiotics such as airborne environmental pollutants, contamination from heavy metals, and therapeutic drugs on mtDNA methylation.
Keywords: mitochondrial epigenetics, mtDNA methylation, non-coding RNAs, mitochondrial post-translational modifications, xenobiotics
1. Introduction
1.1. Mitochondrial DNA (MtDNA)
Mitochondria are key organelles in the cell that carry out many important functions necessary for cell survival. These functions include adenosine triphosphate (ATP) production, metal homeostasis, regulation of cellular metabolism, and cellular respiration (Kaniak-Golik and Skoneczna 2015; Lee and Han 2017). Mitochondria are not only essential, but also unique in that they contain their own DNA within the mitochondrial matrix, separate from the genomic nuclear DNA (Yasukawa and Kang 2018). Mitochondrial DNA (mtDNA) is circular, comprises 16,569 bp, and is double stranded where one strand is purine rich (termed the heavy strand) and the complementary strand is rich in pyrimidines (called the light strand) (Asin-Cayuela and Gustafsson 2007). First sequenced in 1981, mtDNA is inherited maternally and encodes for 13 polypeptides (subunits NADH dehydrogenase (ND1–6) including ND4 and ND4L, of respiratory complex I, cytochrome c oxidase subunit I–III (CO1–3) of respiratory complex IV, subunits adenosine triphosphate 6 and 8 (ATP6 and ATP8) of F1F0 ATPase, and cytochrome B (CYB) of respiratory complex III that are required for the generation of ATP via oxidative phosphorylation (OXPHOS). MtDNA additionally encodes for 22 transfer RNAs (tRNAs) and 2 ribosomal RNAs (rRNAs) (Calvo and Mootha 2010; Yasukawa and Kang 2018). MtDNA also possesses a noncoding control region (NCR) of ~1.1 kb that has elements to regulate replication and two major transcription initiation sites required to generate polycistronic transcripts (Shutt and Gray 2006; Asin-Cayuela and Gustafsson 2007). Unlike nuclear DNA (nDNA), mtDNA is not protected by arrangement into nucleosome structures; instead, it is organized into nucleoprotein complexes known as nucleoids. The primary structural constituent of mitochondrial nucleoids is mitochondrial transcription factor A (TFAM), an abundant protein, which as its name suggests, is involved with mtDNA transcription in addition to packaging mtDNA into nucleoids (Wang and Bogenhagen 2006; Nicholls and Gustafsson 2018). While each mitochondrion contains multiple copies of mtDNA, the stoichiometric amounts of TFAM required for the maintenance of mtDNA has been debated (reviewed in Shokolenko and Alexeyev 2017). However, recent super-resolution microscopy experiments suggest that each nucleoid is approximately 100 nm in diameter and comprises 1 copy of mtDNA packaged by multiple TFAM molecules (Kukat et al. 2011; Kukat et al. 2015).
1.2. Mitochondrial DNA Replication, Transcription, and Repair
The mitochondrial genome is maintained independently of the nuclear genome by proteins that are translocated to the organelle for purposes such as DNA replication, transcription, translation, and repair. Barring the 13 polypeptides encoded by mtDNA, the mitochondrial proteome comprises an estimated 1158 proteins as predicted by Mitocarta 2.0 (Calvo et al. 2016) that are encoded by nuclear genes and imported into the organelle via various import pathways (Becker et al. 2012; Dudek et al. 2013). The regulation of expression of these proteins could be dependent upon epigenetic, post-transcriptional, or post-translational modifications either within the nucleus or the mitochondrion.
The NCR within mtDNA harbors an origin of replication on the heavy strand (OH) as well as a second origin on the light strand (OL), which is located approximately 11 kb away from OH (Fig. 1, reviewed in Young and Copeland 2016; Falkenberg 2018). The exact mechanism of mitochondrial replication is unknown and there are conflicting theories about the process (reviewed in Falkenberg 2018; Yasukawa and Kang 2018). The three main players involved with the process of mtDNA replication are DNA polymerase gamma (Polγ), the mitochondrial helicase, TWINKLE, and the mitochondrial single-stranded DNA binding protein, mtSSB (Lee et al. 2009; reviewed in Graziewicz et al. 2006; Falkenberg 2018; Yasukawa and Kang 2018). The Polγ holoenzyme is a heterotrimer consisting of two identical accessory subunits (Polγ-B) and one catalytic subunit (Polγ-A) where the accessory subunits enhance the processivity and catalytic function of Polγ-A (Gray and Wong 1992; Johnson et al. 2000; Carrodeguas et al. 2002; reviewed in Kaguni 2004). The DNA helicase, TWINKLE, forms a hexameric donut shaped structure and is required at the replication fork where it functions ahead of Polγ and unwinds the DNA template for replication (Korhonen et al. 2004; Milenkovic et al. 2013; Fernández-Millán et al. 2015). Until recently, Polγ was thought to be the sole polymerase within the mitochondrion that was responsible for both mtDNA replication and repair; however, at least 4 other DNA polymerases have been noted to play a role in mtDNA maintenance namely, DNA polymerase beta, PrimPol, polymerase theta, and polymerase zeta (Singh et al. 2015; Wisnovsky et al. 2016; Sykora et al. 2017; Bailey et al. 2019; reviewed in Bailey and Doherty 2017; Kaufman and Van Houten 2017; Krasich and Copeland 2017).
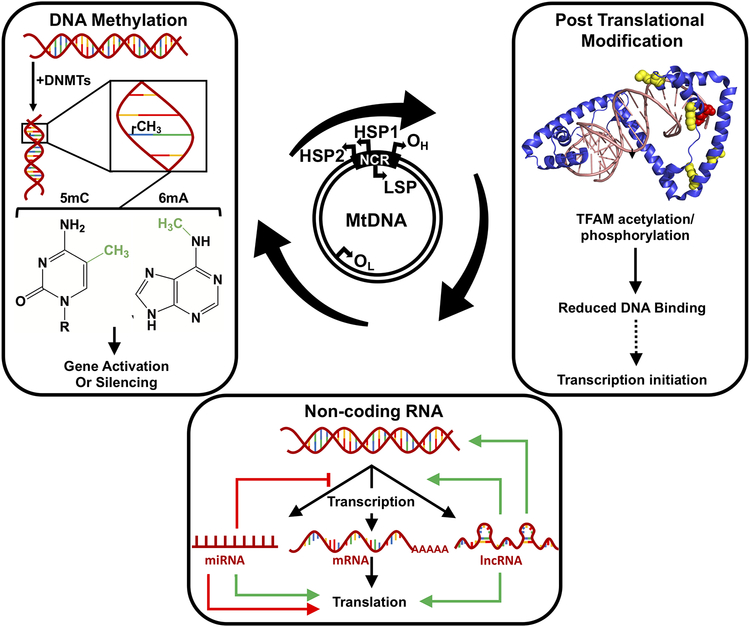
Figure 1:
Overview of the epigenetic regulation of mtDNA. The 16.569 kbp mitochondrial genome is replicated from 2 origins OH and OL found 11 kb apart on the heavy and light strands, respectively. MtDNA encodes for 13 polypeptide sequences, 22 tRNAs, and 2 rRNAs whose transcription is controlled by the heavy strand promoters (HSP1 and HSP2) and the light strand promoter (LSP). Most of these control elements are present within the non-coding region, NCR. The main modifications that regulate gene expression (gene activation or silencing) within the mitochondria include DNA methylation (cytosine or adenine where 5mC is 5-methyl cytosine and 6mA is 6-methyl adenine), non-coding RNAs, and post-translational modifications of nucleoid proteins. In the post-translational modification panel of the figure, the crystal structure of TFAM bound to DNA is indicated (PDB ID:3TQ6, Rubio-Cosials et al. 2011) with the acetylation and phosphorylation sites within the high mobility group 1 domain highlighted by yellow, and red spheres, respectively. The dotted arrow indicates an outcome that is hypothesized for which there is no current evidence. In the non-coding RNA panel, black arrows are representative of cellular processes, green arrows indicate increased regulation, red arrow represents a decrease in regulation, and inhibition is represented with a red line.
MtDNA transcription is still an active area of research as several unknown variables related to initiation, elongation, and termination remain to be elucidated. As genetic information is contained within both strands of mtDNA, the bidirectionality of mtDNA transcription in Xenopus laevis was noted, a decade after the seminal work by Attardi and colleagues (Aloni and Attardi 1971; Storrie and Attardi 1973; Bogenhagen and Yoza 1986). Studies involving in vitro reconstitution of mtDNA transcription minimally requires TFAM, mitochondrial transcription factor B2 (TFB2M), and mitochondrial RNA polymerase (POLRMT) where the precise role of TFAM has been debated. Transcription of mtDNA is initiated at one of three promoter regions, 2 located on the heavy strand (HSP1 and HSP2) and 1 located on the light strand (LSP) where an overwhelming majority of transcripts are produced by transcription from HSP1 (Fig. 1, reviewed in Shokolenko and Alexeyev 2017). Of the mitochondrial transcription components, TFAM appears to be regulated via post-translational modifications, where the precise implications of these modifications on epigenetic regulation of mitochondrial transcription is unknown (Lu et al. 2013; King et al. 2018).
Given the proximity of mtDNA to the sites of OXPHOS at the inner membrane of the mitochondria, it is not surprising that mtDNA is more prone to oxidized DNA damage when compared to its nuclear counterpart. Like the nucleus, mitochondria are also equipped with pathways to repair damaged DNA, however, not all components of nuclear DNA repair pathways are found within the mitochondrion. For instance, the base excision repair (BER) pathway appears to be the most abundant pathway within the mitochondria and repairs small, non-bulky lesions such as alkylated and oxidized DNA bases (reviewed in Prakash and Doublié 2015; Saki and Prakash 2017). Only seven of the eleven known DNA glycosylases that initiate BER have been identified in the mitochondria and recently, evidence presented by the Wilson and Bohr groups indicated that the resynthesis step after damage removal can be performed by DNA polymerase beta during repair of mtDNA (Prasad et al. 2017; Sykora et al. 2017; reviewed in Prakash and Doublié 2015). Various proteins involved in homologous recombination and translesion DNA synthesis have also been identified within the mitochondria (Sage et al. 2010; Dmitrieva et al. 2011; García-Gómez et al. 2013; Singh et al. 2015). While nucleotide excision repair, which is involved with the repair of bulky adducts, has not been defined as a bonafide pathway within the mitochondria, the protein Xeroderma pigmentosum group D (XPD) was observed within the organelle and appears to protect mtDNA from oxidative DNA damage (Liu et al. 2015). Similarly, the proteins that initiate mismatch repair in the nucleus have not been detected in the mitochondria, however, low levels of MMR-like activity were observed previously (Mason et al. 2003) in addition to the detection of MLH1, a protein essential for nuclear MMR (Martin et al. 2010).
Despite the presence of DNA repair pathways in the organelle, as mitochondria possess multiple copies of mtDNA, mutated mtDNA molecules exist simultaneously with undamaged mtDNA leading to heteroplasmy, blurring the line between mitochondrial disease and normalcy. Furthermore, crosstalk between the nucleus and mitochondria is essential for mitochondrial health; and because the mitochondrial proteome comprises both nuclear and mitochondrial proteins, mitochondrial dysfunction resulting from mutated nuclear proteins can also contribute to mitochondrial diseases. A growing body of evidence suggests that xenobiotic agents including environmental factors, drugs, and food additives contribute to mitochondrial disease by causing epigenetic modifications to mtDNA. In this review article, we focus our discussion on the epigenetic regulation of mtDNA as well as highlight the impact of xenobiotics on the process.
2. Epigenetic Regulation of Mitochondrial DNA
Epigenetic changes are inheritable changes that could result in altered gene expression without any modification to the original DNA sequence. While both nuclear DNA as well as mtDNA can be regulated via epigenetic mechanisms, the understanding of mtDNA epigenetics has only recently gained recognition. The most common epigenetic mechanisms include non-coding RNAs and covalent modifications on either DNA or proteins. DNA can be epigenetically modified via methylation, while histones and other proteins can be post-translationally modified via acetylation, phosphorylation, methylation, sumoylation, ubiquitination, and PARylation. Epigenetic modifications explain differential gene expression in various cell types and tissues within an organism. These modifications can be impacted by a number of factors such as environmental exposures, which result in alterations to various cellular processes that could eventually lead to disease. Three major epigenetic mechanisms, discussed in this review article, that regulate gene expression within the mitochondrion include DNA methylation, non-coding RNAs, and post-translational modifications of nucleoid-associated proteins (Fig. 1).
2.1. The enigma of mtDNA methylation
DNA methylation is the addition of a methyl group from S-adenosyl-methionine to a DNA base usually cytosine (C) or adenine (A) that is catalyzed by specialized enzymes called DNA methyltransferases (DNMTs) (reviewed in Maresca et al. 2015). While cytosine methylation occurs at position C-5 resulting in 5-methylcytosine (5mC), the exocyclic NH2 group of adenine gets methylated at position 6, which is converted into N6-methyladenine (6mA; Figs. 1 and 2). Cytosine methylation is more predominant in eukaryotes and both 5mC and 6mA in eukaryotes often result in the silencing/activation of gene transcription (Fig. 1). In the nucleus, cytosine methylation predominantly occurs within CpG dinucleotides located on CpG islands, which are repeat sequences of ~500–1500 bp in length. However, in the mitochondria, methylation occurs within CpG dinucleotides as CpG islands are virtually absent owing to the smaller size of mtDNA and its short non-coding control region. Methylation has also been observed within non-CpG (CpA, CpT, and CpC) sites within nuclear DNA in specific cell types such as stem cells, oocytes, neurons, and glial cells (Jang et al. 2017). Adenine methylation is more common in prokaryotes and serves as a mechanism to differentiate between self and non-self DNA in order to destroy the latter. However, 6mA has also been observed in higher eukaryotes including humans (Iyer et al. 2016; Xiao et al. 2018).
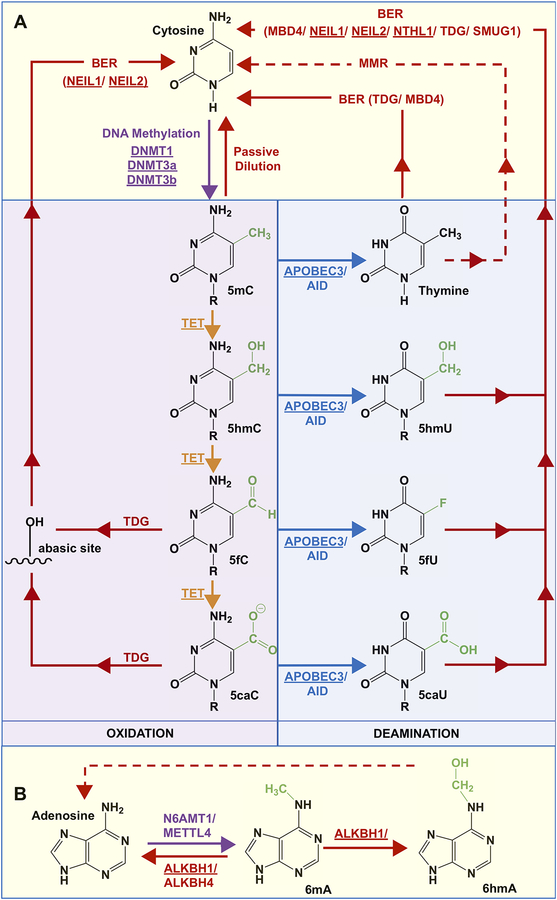
Figure 2:
Potential pathways of (de)methylation of cytosine and adenine. A) DNA methyltransferase (DNMT) enzymes 1, 3a, and 3b convert C to 5mC. Oxidation of 5mC by the TET enzymes results in 5hmC, 5fC, and 5caC; in the nucleus, the monofunctional TDG recognizes and catalyzes the removal of 5fC or 5caC, and the resulting abasic sites are converted to cytosine by NEIL-mediated BER. Other possibilities to restore the original cytosine base include active deamination of 5mC, 5hmC, 5fC and 5caC by AID or APOBEC3 to yield thymine, 5hmU, 5fU, and 5caU, respectively, which are excised by a DNA glycosylase and reverted to cytosine by BER. Given that several glycosylases have overlapping substrate preferences, MBD4, NEIL1, NEIL2, NTHL1, TDG, or SMUG1 can facilitate the repair of the deaminated bases. In the mitochondria given that TDG, SMUG1, and MBD4 are not present, 5mC that is deaminated to T yielding a T-G mispair can be repaired via mismatch repair, but this remains unknown (dotted red line). B) In the nucleus, A can be methylated to 6mA by N6AMT1/METTL4, and this process can be reversed by the ALKBH1/4 demethylases. N6-hydroxymethyladenine (6hmA) can also be formed by the oxidation of 6mA by ALKBH1, which may be converted to A via a mechanism that is yet unknown (dotted red line). Proteins that are underlined have been observed in the mitochondria.
Global or local DNA methylation can be detected using variations of a single or a combination of three main approaches (recently reviewed in Kurdyukov and Bullock 2016). The first approach involves restriction digestion using methylation sensitive and insensitive enzymes followed by sequencing. The second method is affinity purification using a methyl-CpG-binding domain (MBD) or an antibody that specifically recognizes methylated bases (5mC/5hmC). The third approach uses bisulfite sequencing where the DNA is first treated with sodium bisulfite, which converts unmodified C but not 5mC to uracil (U), followed by PCR amplification and sequencing. Currently, this latter technique is considered to be the “gold standard” for DNA methylation; however, a caveat of the technique is the detection of false positives due to the incomplete conversion of unmodified cytosine to uracil (reviewed in Zilberman and Henikoff 2007).
In the nucleus, the three main DNMTs, DNMT1 (methylation maintenance), and DNMT3a and 3b (de novo methylation) are responsible for cytosine methylation (Fig. 2; reviewed in Jin and Robertson 2013; Lyko 2018). These enzymes share a conserved catalytic motif and genetic knockouts of DNMT1 or 3a/3b indicate that these enzymes are essential for viability (Li et al. 1992; Okano et al. 1999). Demethylation of 5mC can be achieved either by passive dilution through multiple rounds of replication or active modification (i.e. oxidation/ deamination) followed by recognition by a DNA glycosylase enzyme that catalyzes the first step of DNA BER (Fig. 2A). Demethylation of 5mC is initiated via oxidation by one of the three ten-eleven translocation (TET1/2/3) dioxygenases or via deamination by activation induced deaminase (AID) or the apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3) (Fig. 2A; reviewed in Bayraktar and Kreutz 2018). The oxidation products of 5mC by TETs form 5-hydroxymethylcytosine (5hmC), which can be further oxidized to form 5-formylcytosine (5fC) and subsequently to 5-carboxylcytosine (5caC; Fig. 2A, reviewed in Wu and Zhang 2017; Bayraktar and Kreutz 2018). 5fC and 5caC can then be recognized and excised by thymidine DNA glycosylase (TDG) forming an abasic (AP) site, which allows for AP endonuclease 1 (APE1)- or endonuclease eight-like (NEIL)-mediated BER to restore the original cytosine base (Fig. 2A, reviewed in Drohat and Coey 2016; Bochtler et al. 2017). In one study, substrate turnover by TDG was reportedly enhanced by the NEIL enzymes (Schomacher et al. 2016). The thymine, formed due to AID or APOBEC3-mediated deamination of 5mC, can be restored to cytosine by either TDG or methyl-CpG-binding domain protein 4 (MBD4)-mediated BER. 5hmC may also be deaminated to 5hmU, which can be recognized by TDG, single-strand selective monofunctional uracil-DNA glycosylase (SMUG1), MBD4 or the NEIL1 DNA glycosylase and repaired via BER (reviewed in Bayraktar and Kreutz 2018). It is not clear whether active demethylation occurs via deamination of 5fC to 5fU and 5caC to 5caU, which are also known substrates of the TDG, SMUG1, endonuclease three-like DNA glycosylase 1 (NTHL1), and NEIL1 glycosylases. Enzymes associated with adenine methylation and demethylation have been recently identified in the nucleus. DNMTs, N-6 adenine-specific DNA methyltransferase 1 (N6AMT1), and methyltransferase like 4 (METTL4) can facilitate adenine methylation, while AlkB homolog 1 (ALKBH1) and ALKBH4 can facilitate 6mA demethylation (Fig. 2B, Xiao et al. 2018; Kweon et al. 2019). The mechanism of adenine (de)methylation remains largely unknown, and the existence of N6-hydroxymethyladenine (6hmA), an oxidation product of 6mA formed by ALKBH1, is currently acknowledged in mammalian cells and tissues (Xiong et al. 2018).
The mechanism of DNA methylation and demethylation in the mitochondria is still not clearly understood. Over the last decade, mtDNA methylation has been extensively scrutinized as several studies have identified this modification in cell lines and tissue samples of mouse and human origin, in both normal as well as disease conditions, in young vs. aged mice, as well as under various conditions of oxidative stress, nutrition, and environmental exposure (Chestnut et al. 2011; Shock et al. 2011; Dzitoyeva et al. 2012; Bellizzi et al. 2013; Pirola et al. 2013; Wong et al. 2013; Ghosh et al. 2014; Baccarelli and Byun 2015; Saini et al. 2017). Controversial reports observe either the presence or complete absence of mtDNA methylation in the non-coding control region within a short 7S DNA fragment that results in a three-stranded displacement or D-loop structure, as well as within several rRNA, tRNA and protein encoding genes (Hong et al. 2013; Wong et al. 2013; Ghosh et al. 2014; Liu et al. 2016; Mechta et al. 2017; Mposhi et al. 2017; Matsuda et al. 2018; Owa et al. 2018). It is advantageous to measure methylation within the mitochondrial D-loop because this region harbors the HSP and LSP promotor elements and is easily accessible to proteins during mtDNA replication and transcription where methylation can directly impact these processes (van der Wijst et al. 2017). DNA methylation in the mitochondria was first reported almost five decades ago and has been a topic of controversy ever since (reviewed in Maresca et al. 2015; D’Aquila et al. 2017; Mposhi et al. 2017). In earlier studies in the 1970s, the presence of 5mC was reported in mouse and hamster cell lines (Nass 1973), as well as in bovine and rat liver mtDNA (Kirnos and Vaniushin 1976). The mtDNA of mammals, fishes, and birds displayed varying levels of 5mC in a species-dependent manner (Vanyushin and Kirnos 1977; Nobre et al. 1978). Concurrent studies also reported the complete absence of mtDNA methylation in cultured cells and in various organisms including calf, rat, yeast, and Paramecium (Cummings et al. 1974; Groot and Kroon 1979). Years after the first observation, in the 1980s, a study reported that 2–5% of mtDNA at CCGG sites were fully methylated in human fibroblast cells (Shmookler Reis and Goldstein 1983) and a separate study noted methylation in 3–5% of CpG sites in mouse mtDNA (Pollack et al. 1984). In contrast to these observations, in 2004, one study reported the non-existence of mtDNA methylation in cancer cell lines and tissue samples from gastrointestinal cancer patients using bisulfite-PCR–single-stranded DNA conformation polymorphism analysis and posited that mitochondrial methylation is likely a rare event (Maekawa et al. 2004). Antibodies against methylated bases 5mC and 5hmC allowed for immunoprecipitation and ELISA-like methods to detect the presence of these modifications in the mitochondria of various cultured cells and tissues (Chestnut et al. 2011; Shock et al. 2011; Chen et al. 2012; Dzitoyeva et al. 2012). In order to study the functional effect of mtDNA methylation on transcription, either bacterial CpG methyl transferase or Chorella virus GpC DNA methyl transferase were targeted to the mitochondria and while cytosine was methylated at both CpG and GpC sites, a decrease in HSP1 and HSP2 regulated gene expression was observed only in the context of GpC methylation (van der Wijst et al. 2017). In another study, overexpression of DNMT1 resulted in the modified transcription of particular genes where MT-ND6 expression from LSP was downregulated, MT-ATP6 and MT-CO1 expression were unaltered, and expression of MT-ND1 from HSP was upregulated (Shock et al. 2011).
It has been suggested that the levels of mitochondrial methylation observed initially, were overestimated due to inefficient bisulfite conversion of circular supercoiled mtDNA, and it was recommended that mtDNA should be linearized prior to the detection of methylation (Liu et al. 2016). MtDNA methylation levels may also be overestimated due to either nuclear DNA contamination in mtDNA preparations or nuclear mitochondrial sequences (NUMTs), which are regions of the mitochondrial genome that translocated to the nuclear genome over time and thus share a high sequence homology with their mitochondrial counterpart (Woischnik and Moraes 2002). A recent study using bisulfite sequencing after mtDNA linearization and whole genome shotgun sequencing did not detect significant levels of CpG and non-CpG methylation. Experimental conditions such as primer design, incomplete bisulfite conversion, and template purity/topology appear to play a critical role in the precise analysis of mtDNA methylation (Owa et al. 2018).
To date, most studies aimed at identifying methylation in mtDNA have focused efforts on determining methylation at CpG sites, however, non-CpG methylation and adenine methylation within mtDNA has also been observed (Bellizzi et al. 2013; Bianchessi et al. 2016; Blanch et al. 2016; Koh et al. 2018). The presence of 6mA in the human mitochondrial genome was reported in 2018 using 6mA-crosslinking-exonuclease-sequencing. The mitochondrial genome was found to contain >8000 times higher 6mA than the nuclear genome, usually present within a ‘6mAT’ dinucleotide motif. The distribution of 6mA was found to be uniform throughout mtDNA with no bias towards clusters. The functional implications of 6mA were associated with mtDNA melting and the subsequent recruitment of mtSSB to mtDNA (Koh et al. 2018). The endosymbiotic theory of mitochondrial origin combined with the presence of an ~8000-fold increase in 6mA compared to nuclear DNA indicates the possibility that mtDNA methylation occurs predominantly on adenine. The numerous conflicting reports surrounding cytosine methylation further supports this hypothesis.
The earliest evidence of DNMT activity in mitochondria was observed in vertebrates including loach embryos, rat liver, ox, as well as in mouse and hamster cells (Vanyushin et al. 1971; Vanyushin and Kirnos 1976; Vanyushin and Kirnos 1977). The expression of various DNMTs in different cell types is also controversial and appears to be preferential depending upon the cell and tissue type being studied (Saini et al. 2017; reviewed in Maresca et al. 2015). Currently, DNMT1, DNMT3a, DNMT3b, and two TET enzymes (TET1 and TET2) have been identified within the mitochondria using immunofluorescence and western blotting procedures (Chestnut et al. 2011; Shock et al. 2011; Chen et al. 2012; Dzitoyeva et al. 2012; Bellizzi et al. 2013; Wong et al. 2013; reviewed in Maresca et al. 2015). The methyl transferase responsible for adenine methylation in mitochondria has not yet been identified however, the demethylase ALKBH1 was identified in the mitochondria in 2018 where it plays a role in regulating oxidative phosphorylation (Koh et al. 2018). The presence of TET1 and TET2 in the mitochondria indicates that mtDNA demethylation occurs via oxidation-mediated active demethylation (Fig. 2). Whether deamination-mediated demethylation takes place in mitochondria is not clear. APOBEC3 has been recently identified in the mitochondria implicating the possibility of deamination of 5mC in mtDNA (Wakae et al. 2018). The main DNA glycosylases involved with the removal of the oxidation/deamination products of 5mC include TDG as well as two other DNA glycosylases, SMUG1, and MBD4, are absent in the mitochondria (reviewed in Prakash and Doublié 2015). Therefore, the next steps in the repair of oxidation/deamination products of methylated bases, remains an area for further research and raises the possibility that other DNA glycosylases such as the NEIL1, NEIL2, or NTHL1 enzymes that are present in the mitochondria could compensate for the lack of TDG in the mitochondria and initiate the repair process. A recent study suggested that the NEIL1 enzyme can recognize and excise 5caC (Slyvka et al. 2017), a result that has been previously disputed (Fig. 2A, Moréra et al. 2012). A separate study reported that 5hmU and 5fU, the further oxidation/ deamination products of 5mC, are also known substrates for NEIL1 (Fig. 2A, (Zhang et al. 2005). Yet another possible avenue of repair for the thymine base generated opposite guanine as a result of deamination of 5mC, could be mismatch DNA repair-like activity that can recognize and repair G-T mismatches, but this still remains to be elucidated (Fig. 2A, Mason et al. 2003; de Souza-Pinto et al. 2009).
2.2. Impact of xenobiotics on mtDNA methylation
Mitochondrial dysfunction caused by alteration and damage to mtDNA is believed to be one of the major underlying mechanisms in the development of disease. With the acceptance that epigenetic modifications such as methylation play a role in mtDNA regulation, the effects of xenobiotic agents on the levels of mtDNA methylation, in particular, 5mC and 5hmC levels, have also been explored. MtDNA methylation can be affected by exposure to various external factors including air pollutants, metals, cigarette smoke, dietary oils, food supplements, and therapeutic drugs.
Airborne pollution and cigarette smoke can cause various diseases including birth defects, cardiovascular diseases, respiratory diseases, neurological disorders, and cancer (Stingone et al. 2014; Fetterman et al. 2017; Boovarahan and Kurian 2018). Airborne particulate matter can be classified into three categories based on their size: coarse (PM10 <10 μm diameter), fine (PM2.5 <10 μm diameter), and ultrafine or nano, (PM0.2; <0.2 μm). These particulates can methylate the mitochondrial D-loop region and mitochondrial genes leading to an increased risk for disease. For example, the exposure of steel workers to PM10 has been associated with hypermethylation of the MT-TF (phenylalanine tRNA) and MT-RNR1 (12S rRNA) genes in peripheral blood (Byun et al. 2013). Similarly, prenatal exposure to PM2.5 in air pollution has been correlated with increased placental mtDNA methylation in the D-loop and MT-RNR1 (Janssen et al. 2015). In contrast, the exposure of PM2.5, produced from welding, decreases blood mtDNA methylation in the D-loop region (Byun et al. 2016). In another study, exposure to fumes from welding has been linked to hypomethylation in the D-loop and MT-TF and increases blood pressure (Xu et al. 2017). Lastly, smoking during pregnancy has been associated with methylation of the placental D-loop region as well as MT-RNR1 gene impacting health of newborns (Armstrong et al. 2016; Janssen et al. 2017).
Metal ions in the environment such as chromium and arsenic that accumulate due to occupational exposure or polluted drinking water, can alter patterns of mtDNA methylation. Hypomethylation in the MT-TF and MT-RMR1 genes was observed following chromium exposure in chrome plating workers (Yang et al. 2016). Likewise, arsenic exposure was associated with hypomethylation of the D-loop and MT-ND6 in an Indian population exposed to high concentration of arsenic present in drinking water (Sanyal et al. 2018). Furthermore, arsenic exposure also resulted in an increase in mtDNA copy number, upregulation in the expression of the nuclear-encoded TFAM gene, and increased expression of the mitochondrial proteins ND4 and ND6 (Sanyal et al. 2018). Another environmental agent used in building materials, electronics, furniture, vehicles, and plastics known as polybrominated diphenyl ethers (PBDEs), are flame retardant substances that can cause neurological impairment. Upon exposure to PBDEs, a decrease in 5mC levels within the MT-CO2 gene was observed and may likely contribute to neuronal toxicity (Byun et al. 2015).
Maternal diet can alter the mtDNA methylation levels of newborns, modulating their mitochondrial OXPHOS capacity, which may have long-term consequences on energy homeostasis. For example, protein deficiency during pregnancy alters mtDNA methylation levels in a sex-specific manner and betaine supplementation has been implicated in hypomethylation in the D-loop region and overexpression of mitochondrial encoded OXPHOS genes in newborn piglets (Jia et al. 2013; Jia et al. 2015). Dietary lipid concentration can also affect mtDNA methylation as observed in large yellow croaker fish, where high lipid diet increases D-loop methylation, but low lipid diet reduces methylation in MT-RMR1 (Liao et al. 2016). Similarly, the consumption of olive oil and perilla oil increases methylation in MT-ND4L and MT-TR (arginine tRNA) in the liver of large yellow croaker. However, methylation of MT-RNR1 was decreased upon intake of perilla oil. The modulation of lipid metabolism in response to high lipid content present in these oils has been suggested to impair mitochondrial function (Liao et al. 2016).
Pharmaceutical agents can cause mitochondrial dysfunction by altering expression of DNMT/TET enzymes and consequently causing changes in the levels of mtDNA methylation. Valproic acid (VPA), an anticonvulsant and mood stabilizer, is often used in the treatment of epilepsy and may cause liver toxicity due to mitochondrial dysfunction (Silva et al. 2008). In order to understand the underlying mechanism of hepatotoxicity, the effects of VPA on mtDNA methylation were tested using mammalian cultured cells. A reduction of 5hmC levels in mtDNA was observed following VPA treatment on mouse fibroblast cells and was associated with decreased mRNA and protein levels of TET1 (Chen et al. 2012). In a separate study using primary human hepatocytes, hypomethylation of 7 mitochondrial genes was observed upon VPA treatment and within the same study, two nuclear genes crucial for DNA methylation, DNMT and MAT (methionine adenosyltransferase), were found to be hypermethylated, resulting in reduced levels of the DNMT enzymes and S-adenosyl methionine (SAM). This may impact mtDNA methylation downstream, which is suggestive of crosstalk between the nucleus and mitochondria after VPA exposure (Wolters et al. 2017). The downstream molecular events after VPA exposure were also explored in human hepatocytes, where upregulation of DNMT caused transient hypermethylation in mtDNA and downregulation of mitochondrial proteins resulting in impaired mitochondrial function. In this study, crosstalk between the nucleus and mitochondria was observed where the mitochondrial MT-CO2 gene initiates a complex cascade of interactions mediated by the nuclear genes FN1, MYC and CPT1A. Most of these genes participate in various mitochondrial functions such as electron transport, apoptosis, mitochondrial import, and fatty acid oxidation, the impairment of which can lead to mitochondrial dysfunction (Wolters et al. 2018).
Epigenetic events are also involved in adaptive responses against chemotherapeutic drugs. A low dose of a chemotherapeutic drug, doxorubicin, downregulates DNMT1 expression in rat cardiac myocytes, resulting in mtDNA hypomethylation, which leads to upregulation of mitochondrial encoded proteins (Ferreira et al. 2019). This low dose of doxorubicin treatment increases cellular resistance against a higher subsequent dose and was believed to be an epigenetic-linked mitochondrial adaptive response facilitating effective anticancer treatment (Ferreira et al. 2019). Another therapeutic drug that elicits mitochondrial dysfunction via changes in epigenetic markers is the antiviral nucleo(t)side analog (NA), that is used to treat patients with chronic Hepatitis B virus infection (CHB). A potential cause of NA-induced mitochondrial pathogenesis may be the epigenetic changes observed in CHB patients, such as hypermethylation of the mitochondrial D-loop region that correlates with overexpression of DNMT1 (Madeddu et al. 2017).
2.2. Mitochondrial Gene Regulation by Non-coding RNAs
Non-coding RNAs (ncRNAs) are RNA molecules that lack the ability to be translated into proteins (reviewed in Mattick and Makunin 2006). The most common ncRNAs are tRNAs, and rRNAs, which are essential for protein synthesis. Other ncRNAs such as long and small ncRNAs regulate gene expression through epigenetic mechanisms and can also act as signaling molecules to regulate essential cellular and biological processes (reviewed in Huang and Zhang 2014; Wei et al. 2017). NcRNAs involved in the epigenetic regulation of mitochondrial gene expression can be encoded by both the nuclear and mitochondrial genomes, however, it is not clear whether the mtDNA encoded ncRNAs are derived from mitochondrial genes integrated into the nuclear genome (i.e. NUMTs) or are actually transcribed inside the mitochondria (reviewed in Vendramin et al. 2017). The levels of ncRNAs can be altered manipulating cellular and mitochondrial functions during various types of cancer making them a potential target for cancer therapy (reviewed in Bienertova-Vasku et al. 2013; Matsui and Corey 2017; Sas-Chen et al. 2017; Olavarria et al. 2018; Zhao et al. 2018).
Long non-coding RNAs (lncRNAs):
LncRNAs encode for transcripts that are >200 bp and have a characteristic structure containing multiple domains for RNA binding, protein binding, and possibly DNA binding (reviewed in Mercer and Mattick 2013). Similar to mRNA, they can also undergo post-transcriptional modifications such as alternative splicing, 5’ end capping, and 3’ polyadenylation (reviewed in Mercer and Mattick 2013). Several lncRNA encoded by the mitochondrial genome have been identified and participate in the regulation of mitochondrial gene expression or act as retrograde signaling molecules to maintain activity of nuclear genes (reviewed in Dong et al. 2017; Zhao et al. 2018). The first 3 mtDNA encoded lncRNA identified were a mitochondrial sense ncRNA (SncmtRNA) and two antisense ncRNAs (ASncmtRNAs-1 and −2), and they contain sense and antisense strands of 16s rRNA gene respectively, connected to a 5’ leader fragment derived from respective complementary strands. These lncRNAs are transported into the nucleus as retrograde signaling molecules and play an important role in mito-nuclear crosstalk. Both sense and antisense lncmtRNAs are overexpressed in normal proliferating cells, however in tumor cells, SncmtRNAs are upregulated whereas ASncmtRNAs are downregulated indicating that SncmtRNAs function in cell cycle progression, while ASncmtRNAs may act as tumor suppressors (Villegas et al. 2007; Burzio et al. 2009; Landerer et al. 2011). The knockdown of ASncmtRNAs induces apoptotic cell death of cancer cells via reduced expression of an apoptosis inhibitor, surivivin, suggesting that ASncmtRNAs are promising targets for cancer therapy (Vidaurre et al. 2014). After the initial discovery, three more mtlncRNAs, ND5, ND6, and CYB were identified, which are encoded by their respective mitochondrial genes. The expression of these lncRNA is regulated by the nuclear encoded mitochondrial RNaseP complex. The exact function of these lncRNAs is not known, however, they form intermolecular duplexes with their functional mRNA counterparts, indicating their role in stabilizing mRNA or regulating their expression (Rackham et al. 2011). In another study, an attempt was made to identify the potential role of lncRNA as biomarkers in heart disease. Transcriptome analysis revealed the abundance of 7 putative lncRNAs encoded by the mitochondrial genome, out of which, two lncRNAs were found to be significantly downregulated in heart patients. One studied of these two lncRNAs, called LIPCAR (Long Intergenic noncoding RNA predicting CARdiac remodeling) (Kumarswamy et al. 2014), is a chimeric lncRNA molecule encoded by the antisense regions of the MT-CYB and MT-CO2 genes (reviewed in Dong et al. 2017). LIPCAR levels were increased in patients with late stage myocardial infarction, suggesting that it may be a putative biomarker for chronic cardiovascular diseases and heart failure mortality (Kumarswamy et al. 2014). Two additional mtlncRNA, MDL1 (Mitochondrial D-Loop 1) and MDL1AS (MDL1 Anti Sense), were identified recently where MDL1 covers the antisense region of the tRNAPro gene and the entire mitochondrial D-loop region, while MDL1AS is the antisense transcript of MDL1. These lncRNAs were proposed to be precursors of transcription initiation RNAs (tiRNA) and are thought to play a role in the regulation of mitochondrial gene expression; however their exact function is yet to be determined (Gao et al. 2018).
Some lncRNA molecules are also encoded by nuclear DNA and transported into the mitochondria where they can interact with mitochondrial proteins and regulate mitochondrial biosynthesis, metabolism, and apoptosis. One such lncRNA is the RNA component of the RNA processing endoribonuclease (RMRP), that is transported by RNA binding proteins HuR, GRSF1, and PNPase in the mitochondria, where it participates in mtDNA replication and transcription. Lowering the RMRP levels led to a reduction in the basal oxygen consumption rate as well as levels of the ATP6, COX1, and CYTB proteins (Wang et al. 2010; Noh et al. 2016).
Small non-coding RNAs:
Most common small non-coding RNAs studied within the mitochondria are microRNAs (miRNAs). MiRNAs are usually transcribed in the nucleus from the non-coding DNA regions as primary miRNA transcripts (pri-miRNA), which is then processed by Drosha/DGC8 complex to form precursor miRNA (pre-miRNA). The pre-miRNAs are then transported into the cytoplasm by exportin 5 and further processed by DICER to form mature miRNAs in the cytoplasm. The mature miRNAs then associate with argonaute 2 (AGO2) and localize to their respective target mRNA sites (reviewed in Macfarlane and Murphy 2010). They bind to the 3’- untranslated region (UTR) of the target mRNA via sequence complementarity and recruit the RNA-induced silencing complex (RISC) that either represses translation or promotes the degradation of the target mRNA resulting in decreased protein expression (reviewed in Macfarlane and Murphy 2010). In some cases, additional mechanisms of function are also proposed where miRNA can activate transcription or upregulate protein expression (discussed in Ramchandran and Chaluvally-Raghavan 2017; Vaschetto 2018). The miRNAs identified in the mitochondria are collectively termed mito-microRNAs (mitomiRs) (Bandiera et al. 2011). MitomiRs are short single stranded RNA molecules ~17–25 bp nucleotides in length that are encoded by the nuclear genome and translocated into the mitochondria, however, a small subset can also be encoded by the mitochondrial genome (Sripada et al. 2012; Ro et al. 2013). The exact mechanism of nuclear encoded mitomiRs transport is largely unknown, and some studies suggest that PNPase or AGO2 may be involved with the transport process (Wang et al. 2010; Bandiera et al. 2011; Zhang et al. 2014; Shepherd et al. 2017). MitomiRs regulate expression of proteins encoded by nuclear as well as mtDNA and for the purposes of this review, we will discuss only the latter (reviewed in Bandiera et al. 2013; Srinivasan and Das 2015). MitomiRs can enhance or repress protein expression at both transcriptional and translational levels resulting in the modulation of mitochondrial metabolic activity and cellular homeostasis. For instance, specific base pairing between miR-2392 and mtDNA in an AGO2-dependent manner inhibits mtDNA transcription partially, resulting in the downregulation of OXPHOS and the concomitant upregulation of glycolysis. This metabolic reprogramming is believed to trigger chemo-resistance during cisplatin chemotherapy in tongue squamous cell carcinoma (Fan et al. 2019). At the translational level, mitomiRs downregulate most mtDNA encoded proteins. For example, miR-181c downregulates MT-CO1 by targeting the 3’ end of its mRNA resulting in the remodeling of complex IV and leading to cardiac dysfunction (Das et al. 2012; Das et al. 2014). Similarly, miR-378 overexpression in type I diabetes downregulates the mitochondrial encoded F0 component MT-ATP6 (Jagannathan et al. 2015). Another mitomiR, miR-214, has been implicated in the pathogenesis of chronic kidney disease (CKD) by disrupting OXPHOS due to downregulation of MT-ND4L and MT-ND6 and is suggested to be a potential therapeutic target and biomarker for CKD (Bai et al. 2019).
Some mitomiRs can also upregulate the expression of mitochondrial genes; for instance, the upregulation of MT-CO1 and MT-ND1 was observed by the translocation of miR-1 and miR-1a-3p to the mitochondria (Zhang et al. 2014; He et al. 2019). Another case of enhanced translation by a mitomiR was observed in spontaneous hypertensive rats, where, upon reduced levels of the CYTB protein, miR-21 is overexpressed and translocated inside the mitochondria to counteract the downregulated CYTB protein level. Exogenous expression of miR-21 also increased the CYTB protein level without changing the mRNA levels. This was believed to be a compensatory mechanism against high blood pressure and could be used in the treatment of hypertension-associated diseases (Li et al. 2016).
2.3. Post-translational modifications of mitochondrial nucleoid proteins
In the nucleus, epigenetic changes in the form of post-translational modifications of histone proteins alter gene transcription and maintain cell-type-specific expression patterns. These modifications include lysine methylation, phosphorylation, acetylation, ubiquitylation, sumoylation, and PARylation (reviewed in Bannister and Kouzarides 2011). In the mitochondria where histone proteins are absent, epigenetic changes to nucleoid proteins play an important role in the regulation of mtDNA gene expression. It is estimated that approximately 63% of proteins that are localized within the mitochondrion contain lysine acetylation sites, and in one study conducted in 2011, 216 phospho-peptides were identified from mitochondrial preparations (Zhao et al. 2011). This latter number is likely higher given the increasing number of proteins that have been identified within the mitochondrion since the study was conducted. Mitochondrial proteins can be acetylated both enzymatically and non-enzymatically. Non-enzymatic acetylation is favored due to a high concentration of acetyl CoA and alkaline pH in the mitochondria. The enzymatic acetylation inside mitochondria has been recently acknowledged as four acetyl transferases ACAT1, MOF, GCN5L1, and PCAF have been identified in the mitochondria and are responsible for regulation of acetylation levels of mitochondrial proteins (Fan et al. 2014; Chatterjee et al. 2016; Wang et al. 2017; Savoia et al. 2019). Deacetylation is predominantly performed by the NAD+-dependent sirtuin family of enzymes, specifically SIRT3, while two-additional mitochondrial sirtuin enzymes, SIRT4 and SIRT5, remove longer acyl groups including succinyl, malonyl, and lipoyl groups (reviewed in He et al. 2012). A number of kinases and protein phosphatases have also been identified within the mitochondria and regulate protein phosphorylation and dephosphorylation (reviewed in Lim et al. 2016).
TFAM, the main structural constituent of mitochondrial nucleoids is one example of a mitochondrial protein that gets post-translationally modified via acetylation, O-linked glycosylation, and phosphorylation (Suarez et al. 2008; Lu et al. 2013; King et al. 2018). Two serine residues (S55 and S56) within the high-mobility group 1 (HMG1) domain are primary sites for phosphorylation, while four lysine residues (K62, K76, K111, and K118) also within HMG1 appear to be sites for acetylation (Fig. 1). Phosphorylation and acetylation both regulate aspects of mtDNA dynamics by altering the binding affinity of TFAM to DNA where reduced binding of TFAM to DNA causes increased diffusion of the protein on DNA and decreased mtDNA compaction. The extent to which mtDNA is compacted, ultimately influences mtDNA replication and transcription. Acetylation and phosphorylation sites have also been identified in other mitochondrial nucleoid associated proteins including mtSSB, and DNA Polγ; however, the exact role of these posttranslational modifications in regulating mtDNA metabolism have not been elucidated thus far (Matsuoka et al. 2007; Choudhary et al. 2009; Zhou et al. 2013).
3. Concluding Remarks
Mitochondria are perhaps one of the most intriguing organelles within a cell. Not only do they contain their own DNA that is maternally inherited, but their endosymbiotic origin makes them evolutionarily unique (reviewed in Gray 2012). The double-membraned organelle, much like its prokaryotic ancestor, is capable of supporting processes such as replication, protein production to generate ATP, and possesses the machinery to drive its own division. While only a small fraction of proteins found within the organelle are encoded by mtDNA, a majority of the mitochondrial proteome is encoded within the nucleus making crosstalk between the two organelles necessary and inevitable. Furthermore, depending upon the cell type, many mitochondria exist within a single cell and each mitochondrion has multiple copies of mtDNA leading to heteroplasmy where wild-type mtDNA molecules coexist with those that may have acquired mutations. Aspects of mito-nuclear crosstalk that signal dysfunction and the extent to which heteroplasmy influences mitochondrial function and disease, still remain unclear. From the standpoint of epigenetic modifications, studies investigating the role of mtDNA methylation on gene expression and disease outcomes, should take into account mtDNA copy number as a potential confounding variable (reviewed in Byun and Baccarelli 2014; Stimpfel et al. 2018).
The technical inadequacy of current methods used to detect methylation within the mitochondria has sparked a long-standing debate in the field as to the existence of methylation within the organelle. However, it is now generally accepted that gene expression within the mitochondria is regulated by epigenetic mechanisms, where several facets of the process remain to be elucidated presenting an active and exciting area of research. For instance, the complex interplay between the TET and APOBEC3 enzymes leading to restoration of cytosine via BER is far from clear and requires further scrutiny. Furthermore, the specific DNA glycosylase required to initiate BER in the mitochondria in the absence of TDG, SMUG1, and MBD4 that partake in nuclear restoration of methylated bases, also needs to be elucidated.
Whether mtDNA methylation leads to a particular disease condition or occurs as a consequence of disease, is a perplexing correlation for many researchers and is an area that is still in its infancy. Several diseases that have been associated with differential mtDNA methylation include aging related neurodegenerative disorders (Alzheimer’s diseases, Parkinson’s etc.), cancer, obesity, diabetes, and cardiovascular disease (reviewed in Mposhi et al. 2017). The treatment of such diseases may benefit from pharmaceutical drugs that can specifically target the drivers of mitochondrial epigenetic modifications.
Acknowledgements
The authors would like to thank Dr. Robert W. Sobol for providing constructive feedback and critically evaluating this manuscript.
Funding
NS and AP are supported by grants from the National Institutes of Health (NIEHS grant 5R00ES024417-04 to AP and 1R01ES030084 - 01 to AP). NS and AP were also supported by funds provided by the University of South Alabama Mitchell Cancer Institute.
Abbreviations:
- 5caC
5-carboxylcytosine
- 5fC
5-formylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- 6hmA
N6-hydroxymethyladenine
- 6mA
N6-methyladenine
- A
adenine
- AGO2
argonaute 2
- AID
activation induced deaminase
- ALKBH
AlkB homolog
- APE1
AP endonuclease 1
- APOBEC3
apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3
- ASncmtRNA
mitochondrial antisense ncRNA
- ATP
adenosine triphosphate
- BER
base excision repair
- C
cytosine
- CHB
chronic Hepatitis B virus infection
- CKD
chronic kidney disease
- CO/CoX
cytochrome c oxidase
- CYB/CYTB
cytochrome B
- DNMT
DNA methyltransferase
- G
guanine
- GRSF1
G-Rich RNA Sequence Binding Factor 1
- HSP
heavy strand promoter
- HuR
human antigen R
- LIPCAR
long Intergenic noncoding RNA predicting CARdiac remodeling
- LncRNA
long non-coding RNA
- LSP
light strand promoter
- MBD4
methyl-CpG-binding domain protein 4
- MDL1
mitochondrial D-Loop 1
- MDL1AS
mitochondrial D-Loop 1 anti Sense
- METTL4
methyltransferase like 4
- miRNA
microRNA
- mitomiR
mitochondrial microRNA
- MLH1
MutL homolog 1
- MMR
mismatch repair
- mtDNA
mitochondrial DNA
- mtSSB
mitochondrial single-stranded DNA binding protein
- N6AMT1
N-6 adenine-specific DNA methyltransferase 1
- NA
nucleo(t)side analog
- NADH
nicotinamide adenine dinucleotide reduced
- NCR
noncoding control region
- ncRNAs
non-coding RNAs
- ND
NADH dehydrogenase
- nDNA
nuclear DNA
- NEIL
endonuclease eight-like protein
- NTHL1
endonuclease three-like DNA glycosylase 1
- NUMTs
nuclear mitochondrial sequences
- OH
origin of replication on the heavy strand
- OL
origin of replication on the light strand
- OXPHOS
oxidative phosphorylation
- PBDEs
polybrominated diphenyl ethers
- PM
particulate matter
- PNPase
Polynucleotide phosphorylase
- POLRMT
mitochondrial RNA polymerase
- Polγ
DNA polymerase gamma
- pre-miRNA
precursor microRNA
- pri-miRNA
primary microRNA
- RISC
RNA-induced silencing complex
- RMRP
RNA processing endoribonuclease
- RNaseP
ribonuclease P
- RNaseH2
ribonuclease H2
- rRNAs
ribosomal RNAs
- SMUG1
single-strand selective monofunctional uracil-DNA glycosylase
- SncmtRNA
mitochondrial sense non coding RNA
- TDG
thymidine DNA glycosylase
- TET
ten-eleven translocation enzyme
- TFAM
transcription factor A
- TFB2M
mitochondrial transcription factor B2
- tiRNA
transcription initiation RNAs
- tRNAs
transfer RNAs
- U
uracil
- UTR
untranslated region
- VPA
Valproic acid
- XPD
Xeroderma pigmentosum group D
Footnotes
Conflict of interest
None
REFERENCES CITED
- Aloni Y, Attardi G. 1971. Expression of the mitochondria genome in HeLa cells. IV. Titration of mitochondrial genes for 16 s, 12 s and 4 s RNA. J Mol Biol 55(2):271–276. [DOI] [PubMed] [Google Scholar]
- Armstrong DA, Green BB, Blair BA, Guerin DJ, Litzky JF, Chavan NR, Pearson KJ, Marsit CJ. 2016. Maternal smoking during pregnancy is associated with mitochondrial DNA methylation. Environ Epigenet 2(3):dvw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asin-Cayuela J, Gustafsson CM. 2007. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci 32(3):111–117. [DOI] [PubMed] [Google Scholar]
- Baccarelli AA, Byun HM. 2015. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clin Epigenetics 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Chen H, Ding D, Song R, Lin J, Zhang Y, Guo Y, Chen S, Ding G, Jia Z, Huang S, He JC, Yang L, Zhang A. 2019. MicroRNA-214 promotes chronic kidney disease by disrupting mitochondrial oxidative phosphorylation. Kidney Int 95(6):1389–1404. [DOI] [PubMed] [Google Scholar]
- Bailey LJ, Bianchi J, Doherty AJ. 2019. PrimPol is required for the maintenance of efficient nuclear and mitochondrial DNA replication in human cells. Nucleic Acids Res 47(8):4026–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey LJ, Doherty AJ. 2017. Mitochondrial DNA replication: a PrimPol perspective. Biochem Soc Trans 45(2):513–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandiera S, Matégot R, Girard M, Demongeot J, Henrion-Caude A. 2013. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med 64:12–19. [DOI] [PubMed] [Google Scholar]
- Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chrétien D, Munnich A, Lyonnet S, Henrion-Caude A. 2011. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6(6):e20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar G, Kreutz MR. 2018. The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders. Front Mol Neurosci 11:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Böttinger L, Pfanner N. 2012. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci 37(3):85–91. [DOI] [PubMed] [Google Scholar]
- Bellizzi D, D’Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G. 2013. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res 20(6):537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchessi V, Vinci MC, Nigro P, Rizzi V, Farina F, Capogrossi MC, Pompilio G, Gualdi V, Lauri A. 2016. Methylation profiling by bisulfite sequencing analysis of the mtDNA Non-Coding Region in replicative and senescent Endothelial Cells. Mitochondrion 27:40–47. [DOI] [PubMed] [Google Scholar]
- Bienertova-Vasku J, Sana J, Slaby O. 2013. The role of microRNAs in mitochondria in cancer. Cancer Lett 336(1):1–7. [DOI] [PubMed] [Google Scholar]
- Blanch M, Mosquera JL, Ansoleaga B, Ferrer I, Barrachina M. 2016. Altered Mitochondrial DNA Methylation Pattern in Alzheimer Disease-Related Pathology and in Parkinson Disease. Am J Pathol 186(2):385–397. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Kolano A, Xu GL. 2017. DNA demethylation pathways: Additional players and regulators. Bioessays 39(1):1–13. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Yoza BK. 1986. Accurate in vitro transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol Cell Biol 6(7):2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boovarahan SR, Kurian GA. 2018. Mitochondrial dysfunction: a key player in the pathogenesis of cardiovascular diseases linked to air pollution. Rev Environ Health 33(2):111–122. [DOI] [PubMed] [Google Scholar]
- Burzio VA, Villota C, Villegas J, Landerer E, Boccardo E, Villa LL, Martínez R, Lopez C, Gaete F, Toro V, Rodriguez X, Burzio LO. 2009. Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc Natl Acad Sci U S A 106(23):9430–9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Baccarelli AA. 2014. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum Genet 133(3):247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Benachour N, Zalko D, Frisardi MC, Colicino E, Takser L, Baccarelli AA. 2015. Epigenetic effects of low perinatal doses of flame retardant BDE-47 on mitochondrial and nuclear genes in rat offspring. Toxicology 328:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Colicino E, Trevisi L, Fan T, Christiani DC, Baccarelli AA. 2016. Effects of Air Pollution and Blood Mitochondrial DNA Methylation on Markers of Heart Rate Variability. J Am Heart Assoc 5(4):e003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Panni T, Motta V, Hou L, Nordio F, Apostoli P, Bertazzi PA, Baccarelli AA. 2013. Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol 10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. 2016. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44(D1):D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK. 2010. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 11:25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrodeguas JA, Pinz KG, Bogenhagen DF. 2002. DNA binding properties of human pol gammaB. J Biol Chem 277(51):50008–50014. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Seyfferth J, Lucci J, Gilsbach R, Preissl S, Böttinger L, Mårtensson CU, Panhale A, Stehle T, Kretz O, Sahyoun AH, Avilov S, Eimer S, Hein L, Pfanner N, Becker T, Akhtar A. 2016. MOF Acetyl Transferase Regulates Transcription and Respiration in Mitochondria. Cell 167(3):722–738.e723. [DOI] [PubMed] [Google Scholar]
- Chen H, Dzitoyeva S, Manev H. 2012. Effect of valproic acid on mitochondrial epigenetics. Eur J Pharmacol 690(1–3):51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. 2011. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci 31(46):16619–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325(5942):834–840. [DOI] [PubMed] [Google Scholar]
- Cummings DJ, Tait A, Goddard JM. 1974. Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta 374(1):1–11. [DOI] [PubMed] [Google Scholar]
- D’Aquila P, Montesanto A, Guarasci F, Passarino G, Bellizzi D. 2017. Mitochondrial genome and epigenome: two sides of the same coin. Front Biosci (Landmark Ed) 22:888–908. [DOI] [PubMed] [Google Scholar]
- Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C. 2014. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One 9(5):e96820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C. 2012. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 110(12):1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. 2009. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 8(6):704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva NI, Malide D, Burg MB. 2011. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am J Physiol Regul Integr Comp Physiol 301(3):R632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yoshitomi T, Hu JF, Cui J. 2017. Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics Chromatin 10(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drohat AC, Coey CT. 2016. Role of Base Excision “Repair” Enzymes in Erasing Epigenetic Marks from DNA. Chem Rev 116(20):12711–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Rehling P, van der Laan M. 2013. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta 1833(2):274–285. [DOI] [PubMed] [Google Scholar]
- Dzitoyeva S, Chen H, Manev H. 2012. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging 33(12):2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg M 2018. Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays Biochem 62(3):287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Alečković M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, Chen J. 2014. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell 53(4):534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Tian T, Chen W, Lv X, Lei X, Zhang H, Sun S, Cai L, Pan G, He L, Ou Z, Lin X, Wang X, Perez MF, Tu Z, Ferrone S, Tannous BA, Li J. 2019. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res 79(6):1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Millán P, Lázaro M, Cansız-Arda Ş, Gerhold JM, Rajala N, Schmitz CA, Silva-Espiña C, Gil D, Bernadó P, Valle M, Spelbrink JN, Solà M. 2015. The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res 43(8):4284–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LL, Cunha-Oliveira T, Veloso CD, Costa CF, Wallace KB, Oliveira PJ. 2019. Single nanomolar doxorubicin exposure triggers compensatory mitochondrial responses in H9c2 cardiomyoblasts. Food Chem Toxicol 124:450–461. [DOI] [PubMed] [Google Scholar]
- Fetterman JL, Sammy MJ, Ballinger SW. 2017. Mitochondrial toxicity of tobacco smoke and air pollution. Toxicology 391:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Tian X, Chang H, Sun Y, Wu Z, Cheng Z, Dong P, Zhao Q, Ruan J, Bu W. 2018. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion 38:41–47. [DOI] [PubMed] [Google Scholar]
- García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, Blanco L. 2013. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell 52(4):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sengupta S, Scaria V. 2014. Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion 18:58–62. [DOI] [PubMed] [Google Scholar]
- Gray H, Wong TW. 1992. Purification and identification of subunit structure of the human mitochondrial DNA polymerase. J Biol Chem 267(9):5835–5841. [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol 4(9):a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. 2006. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev 106(2):383–405. [DOI] [PubMed] [Google Scholar]
- Groot GS, Kroon AM. 1979. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in -CCGG- sequences. Biochim Biophys Acta 564(2):355–357. [DOI] [PubMed] [Google Scholar]
- He R, Ding C, Yin P, He L, Xu Q, Wu Z, Shi Y, Su L. 2019. MiR-1a-3p mitigates isoproterenol-induced heart failure by enhancing the expression of mitochondrial ND1 and COX1. Exp Cell Res 378(1):87–97. [DOI] [PubMed] [Google Scholar]
- He W, Newman JC, Wang MZ, Ho L, Verdin E. 2012. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 23(9):467–476. [DOI] [PubMed] [Google Scholar]
- Hong EE, Okitsu CY, Smith AD, Hsieh CL. 2013. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol 33(14):2683–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Zhang R. 2014. Regulatory non-coding RNAs: revolutionizing the RNA world. Mol Biol Rep 41(6):3915–3923. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Aravind L. 2016. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38(1):27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE, Martinez I, Hollander JM. 2015. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ Cardiovasc Genet 8(6):785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shin WJ, Lee JE, Do JT. 2017. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes (Basel) 8(6):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. 2015. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: An ENVIRONAGE birth cohort study. Epigenetics 10(6):536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Gyselaers W, Byun HM, Roels HA, Cuypers A, Baccarelli AA, Nawrot TS. 2017. Placental mitochondrial DNA and CYP1A1 gene methylation as molecular signatures for tobacco smoke exposure in pregnant women and the relevance for birth weight. J Transl Med 15(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Li R, Cong R, Yang X, Sun Q, Parvizi N, Zhao R. 2013. Maternal low-protein diet affects epigenetic regulation of hepatic mitochondrial DNA transcription in a sex-specific manner in newborn piglets associated with GR binding to its promoter. PLoS One 8(5):e63855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Song H, Gao G, Cai D, Yang X, Zhao R. 2015. Maternal Betaine Supplementation during Gestation Enhances Expression of mtDNA-Encoded Genes through D-Loop DNA Hypomethylation in the Skeletal Muscle of Newborn Piglets. J Agric Food Chem 63(46):10152–10160. [DOI] [PubMed] [Google Scholar]
- Jin B, Robertson KD. 2013. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol 754:3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Tsai Y, Graves SW, Johnson KA. 2000. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry 39(7):1702–1708. [DOI] [PubMed] [Google Scholar]
- Kaguni LS. 2004. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem 73:293–320. [DOI] [PubMed] [Google Scholar]
- Kaniak-Golik A, Skoneczna A. 2015. Mitochondria-nucleus network for genome stability. Free Radic Biol Med 82:73–104. [DOI] [PubMed] [Google Scholar]
- Kaufman BA, Van Houten B. 2017. POLB: A new role of DNA polymerase beta in mitochondrial base excision repair. DNA Repair (Amst) 60:A1–A5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA, Hashemi Shabestari M, Taris KH, Pandey AK, Venkatesh S, Thilagavathi J, Singh K, Krishna Koppisetti R, Temiakov D, Roos WH, Suzuki CK, Wuite GJL. 2018. Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res 46(7):3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnos MD, Vaniushin BF. 1976. [Isolation of mitochondrial DNA, purified of nuclear DNA, from animal tissues (degree of methylation and level of pyrimidine nucleotide clustering--criteria of purity)]. Biokhimiia 41(1):68–74. [PubMed] [Google Scholar]
- Koh CWQ, Goh YT, Toh JDW, Neo SP, Ng SB, Gunaratne J, Gao YG, Quake SR, Burkholder WF, Goh WSS. 2018. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res 46(22):11659–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. 2004. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J 23(12):2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasich R, Copeland WC. 2017. DNA polymerases in the mitochondria: A critical review of the evidence. Front Biosci (Landmark Ed) 22:692–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, Kühlbrandt W, Larsson NG. 2015. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A 112(36):11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson NG, Jakobs S. 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A 108(33):13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. 2014. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 114(10):1569–1575. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Bullock M. 2016. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel) 5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon SM, Chen Y, Moon E, Kvederaviciutė K, Klimasauskas S, Feldman DE. 2019. An Adversarial DNA N6 -Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol Cell 74:1–10 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landerer E, Villegas J, Burzio VA, Oliveira L, Villota C, Lopez C, Restovic F, Martinez R, Castillo O, Burzio LO. 2011. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol (Dordr) 34(4):297–305. [DOI] [PubMed] [Google Scholar]
- Lee SR, Han J. 2017. Mitochondrial Nucleoid: Shield and Switch of the Mitochondrial Genome. Oxid Med Cell Longev 2017:8060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kennedy WD, Yin YW. 2009. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell 139(2):312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69(6):915–926. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang X, Wang F, Zhou L, Yin Z, Fan J, Nie X, Wang P, Fu XD, Chen C, Wang DW. 2016. MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation 134(10):734–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao K, Yan J, Mai K, Ai Q. 2016. Dietary lipid concentration affects liver mitochondrial DNA copy number, gene expression and DNA methylation in large yellow croaker (Larimichthys crocea). Comp Biochem Physiol B Biochem Mol Biol 193:25–32. [DOI] [PubMed] [Google Scholar]
- Lim S, Smith KR, Lim ST, Tian R, Lu J, Tan M. 2016. Regulation of mitochondrial functions by protein phosphorylation and dephosphorylation. Cell Biosci 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Du Q, Chen L, Fu G, Li S, Fu L, Zhang X, Ma C, Bin C. 2016. CpG methylation patterns of human mitochondrial DNA. Sci Rep 6:23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fang H, Chi Z, Wu Z, Wei D, Mo D, Niu K, Balajee AS, Hei TK, Nie L, Zhao Y. 2015. XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage. Nucleic Acids Res 43(11):5476–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49(1):121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 19(2):81–92. [DOI] [PubMed] [Google Scholar]
- Macfarlane LA, Murphy PR. 2010. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 11(7):537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeddu G, Ortu S, Garrucciu G, Maida I, Melis M, Muredda AA, Mura MS, Babudieri S. 2017. DNMT1 modulation in chronic hepatitis B patients and hypothetic influence on mitochondrial DNA methylation status during long-term nucleo(t)side analogs therapy. J Med Virol 89(7):1208–1214. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Taniguchi T, Higashi H, Sugimura H, Sugano K, Kanno T. 2004. Methylation of mitochondrial DNA is not a useful marker for cancer detection. Clin Chem 50(8):1480–1481. [DOI] [PubMed] [Google Scholar]
- Maresca A, Zaffagnini M, Caporali L, Carelli V, Zanna C. 2015. DNA methyltransferase 1 mutations and mitochondrial pathology: is mtDNA methylated? Front Genet 6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. 2010. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell 17(3):235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PA, Matheson EC, Hall AG, Lightowlers RN. 2003. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res 31(3):1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Yasukawa T, Sakaguchi Y, Ichiyanagi K, Unoki M, Gotoh K, Fukuda K, Sasaki H, Suzuki T, Kang D. 2018. Accurate estimation of 5-methylcytosine in mammalian mitochondrial DNA. Sci Rep 8(1):5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Corey DR. 2017. Non-coding RNAs as drug targets. Nat Rev Drug Discov 16(3):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316(5828):1160–1166. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. 2006. Non-coding RNA. Hum Mol Genet 15 Spec No 1:R17–29. [DOI] [PubMed] [Google Scholar]
- Mechta M, Ingerslev LR, Fabre O, Picard M, Barrès R. 2017. Evidence Suggesting Absence of Mitochondrial DNA Methylation. Front Genet 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20(3):300–307. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Matic S, Kühl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. 2013. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet 22(10):1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moréra S, Grin I, Vigouroux A, Couvé S, Henriot V, Saparbaev M, Ishchenko AA. 2012. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res 40(19):9917–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mposhi A, Van der Wijst MG, Faber KN, Rots MG. 2017. Regulation of mitochondrial gene expression, the epigenetic enigma. Front Biosci (Landmark Ed) 22:1099–1113. [DOI] [PubMed] [Google Scholar]
- Nass MM. 1973. Differential methylation of mitochondrial and nuclear DNA in cultured mouse, hamster and virus-transformed hamster cells. In vivo and in vitro methylation. J Mol Biol 80(1):155–175. [DOI] [PubMed] [Google Scholar]
- Nicholls TJ, Gustafsson CM. 2018. Separating and Segregating the Human Mitochondrial Genome. Trends Biochem Sci 43(11):869–881. [DOI] [PubMed] [Google Scholar]
- Nobre F, Vichi FL, Nogueira JL, Salgado W, Evora PR. 1978. [The heart rate in chronic chagas’ heart disease. 2. The sinus node rate in Chagas patients with heart failure]. Arq Bras Cardiol 31(3):181–184. [PubMed] [Google Scholar]
- Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, Kim J, Curtis J, Moad CA, Wohler CM, Indig FE, de Paula W, Dudekula DB, De S, Piao Y, Yang X, Martindale JL, de Cabo R, Gorospe M. 2016. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev 30(10):1224–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99(3):247–257. [DOI] [PubMed] [Google Scholar]
- Olavarria JV, Burzio VA, Borgna V, Lobos-Gonzalez L, Araya M, Guevara F, Villota C, Burzio LO. 2018. Long Noncoding Mitochondrial RNAs (LncmtRNAs) as Targets for Cancer Therapy. Mitochondrial DNA-New Insights: IntechOpen. p doi: 10.5772/intechopen.75453 [DOI] [Google Scholar]
- Owa C, Poulin M, Yan L, Shioda T. 2018. Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: Sources and avoidance of false-positive detection. PLoS One 13(2):e0192722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. 2013. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 62(9):1356–1363. [DOI] [PubMed] [Google Scholar]
- Pollack Y, Kasir J, Shemer R, Metzger S, Szyf M. 1984. Methylation pattern of mouse mitochondrial DNA. Nucleic Acids Res 12(12):4811–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Doublié S. 2015. Base Excision Repair in the Mitochondria. J Cell Biochem 116(8):1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Çağlayan M, Dai DP, Nadalutti CA, Zhao ML, Gassman NR, Janoshazi AK, Stefanick DF, Horton JK, Krasich R, Longley MJ, Copeland WC, Griffith JD, Wilson SH. 2017. DNA polymerase β: A missing link of the base excision repair machinery in mammalian mitochondria. DNA Repair (Amst) 60:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. 2011. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17(12):2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandran R, Chaluvally-Raghavan P. 2017. miRNA-Mediated RNA Activation in Mammalian Cells. Adv Exp Med Biol 983:81–89. [DOI] [PubMed] [Google Scholar]
- Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW, Zheng H, Lin YM, Moro L, Hsieh JT, Yan W. 2013. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res 23(6):759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cosials A, Sidow JF, Jiménez-Menéndez N, Fernández-Millán P, Montoya J, Jacobs HT, Coll M, Bernadó P, Solà M. 2011. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol 18(11):1281–1289. [DOI] [PubMed] [Google Scholar]
- Sage JM, Gildemeister OS, Knight KL. 2010. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem 285(25):18984–18990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini SK, Mangalhara KC, Prakasam G, Bamezai RNK. 2017. DNA Methyltransferase1 (DNMT1) Isoform3 methylates mitochondrial genome and modulates its biology. Sci Rep 7(1):1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saki M, Prakash A. 2017. DNA damage related crosstalk between the nucleus and mitochondria. Free Radic Biol Med 107:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal T, Bhattacharjee P, Bhattacharjee S. 2018. Hypomethylation of mitochondrial D-loop and ND6 with increased mitochondrial DNA copy number in the arsenic-exposed population. Toxicology 408:54–61. [DOI] [PubMed] [Google Scholar]
- Sas-Chen A, Srivastava S, Yarden Y. 2017. The short and the long: non-coding RNAs and growth factors in cancer progression. Biochem Soc Trans 45(1):51–64. [DOI] [PubMed] [Google Scholar]
- Savoia M, Cencioni C, Mori M, Atlante S, Zaccagnini G, Devanna P, Di Marcotullio L, Botta B, Martelli F, Zeiher AM, Pontecorvi A, Farsetti A, Spallotta F, Gaetano C. 2019. P300/CBP-associated factor regulates transcription and function of isocitrate dehydrogenase 2 during muscle differentiation. FASEB J 33(3):4107–4123. [DOI] [PubMed] [Google Scholar]
- Schomacher L, Han D, Musheev MU, Arab K, Kienhöfer S, von Seggern A, Niehrs C. 2016. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol 23(2):116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd DL, Hathaway QA, Pinti MV, Nichols CE, Durr AJ, Sreekumar S, Hughes KM, Stine SM, Martinez I, Hollander JM. 2017. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J Mol Cell Cardiol 110:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis RJ, Goldstein S. 1983. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J Biol Chem 258(15):9078–9085. [PubMed] [Google Scholar]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. 2011. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A 108(9):3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko IN, Alexeyev MF. 2017. Mitochondrial transcription in mammalian cells. Front Biosci (Landmark Ed) 22:835–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Gray MW. 2006. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet 22(2):90–95. [DOI] [PubMed] [Google Scholar]
- Silva MF, Aires CC, Luis PB, Ruiter JP, IJlst L, Duran M, Wanders RJ, Tavares de Almeida I. 2008. Valproic acid metabolism and its effects on mitochondrial fatty acid oxidation: a review. J Inherit Metab Dis 31(2):205–216. [DOI] [PubMed] [Google Scholar]
- Singh B, Li X, Owens KM, Vanniarajan A, Liang P, Singh KK. 2015. Human REV3 DNA Polymerase Zeta Localizes to Mitochondria and Protects the Mitochondrial Genome. PLoS One 10(10):e0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyvka A, Mierzejewska K, Bochtler M. 2017. Nei-like 1 (NEIL1) excises 5-carboxylcytosine directly and stimulates TDG-mediated 5-formyl and 5-carboxylcytosine excision. Sci Rep 7(1):9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan H, Das S. 2015. Mitochondrial miRNA (MitomiR): a new player in cardiovascular health. Can J Physiol Pharmacol 93(10):855–861. [DOI] [PubMed] [Google Scholar]
- Sripada L, Tomar D, Prajapati P, Singh R, Singh AK. 2012. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7(9):e44873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpfel M, Jancar N, Virant-Klun I. 2018. New Challenge: Mitochondrial Epigenetics? Stem Cell Rev 14(1):13–26. [DOI] [PubMed] [Google Scholar]
- Stingone JA, Luben TJ, Daniels JL, Fuentes M, Richardson DB, Aylsworth AS, Herring AH, Anderka M, Botto L, Correa A, Gilboa SM, Langlois PH, Mosley B, Shaw GM, Siffel C, Olshan AF, Study NBDP. 2014. Maternal exposure to criteria air pollutants and congenital heart defects in offspring: results from the national birth defects prevention study. Environ Health Perspect 122(8):863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B, Attardi G. 1973. Expression of the mitochondrial genome in HeLa cells. IX. Effect of inhibition of mitochondrial protein synthesis on mitochondrial formation. J Cell Biol 56(3):819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez J, Hu Y, Makino A, Fricovsky E, Wang H, Dillmann WH. 2008. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am J Physiol Cell Physiol 295(6):C1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Kanno S, Akbari M, Kulikowicz T, Baptiste BA, Leandro GS, Lu H, Tian J, May A, Becker KA, Croteau DL, Wilson DM, Sobol RW, Yasui A, Bohr VA. 2017. DNA Polymerase Beta Participates in Mitochondrial DNA Repair. Mol Cell Biol 37(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst MG, van Tilburg AY, Ruiters MH, Rots MG. 2017. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci Rep 7(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin BF, Kirnos MD. 1976. Structure of animal mitochondrial DNA: nucleotide composition, pyrimidine clusters, and methylation character. Mol Biol (Mosk) 10(4):715–724. [PubMed] [Google Scholar]
- Vanyushin BF, Kirnos MD. 1977. Structure of animal mitochondrial DNA (base composition, pyrimidine clusters, character of methylation). Biochim Biophys Acta 475(2):323–336. [DOI] [PubMed] [Google Scholar]
- Vanyushin BF, Kiryanov GI, Kudryashova IB, Belozersky AN. 1971. DNA-methylase in loach embryos (Misgurnus fossilis). FEBS Lett 15(4):313–316. [DOI] [PubMed] [Google Scholar]
- Vaschetto LM. 2018. miRNA activation is an endogenous gene expression pathway. RNA Biol 15(6):826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendramin R, Marine JC, Leucci E. 2017. Non-coding RNAs: the dark side of nuclear-mitochondrial communication. EMBO J 36(9):1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre S, Fitzpatrick C, Burzio VA, Briones M, Villota C, Villegas J, Echenique J, Oliveira-Cruz L, Araya M, Borgna V, Socías T, Lopez C, Avila R, Burzio LO. 2014. Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy. J Biol Chem 289(39):27182–27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas J, Burzio V, Villota C, Landerer E, Martinez R, Santander M, Pinto R, Vera MI, Boccardo E, Villa LL, Burzio LO. 2007. Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res 35(21):7336–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakae K, Nishiyama T, Kondo S, Izuka T, Que L, Chen C, Kase K, Kitamura K, Mohiuddin M, Wang Z, Ahasan MM, Nakamura M, Fujiwara H, Yoshizaki T, Hosomochi K, Tajima A, Nakahara T, Kiyono T, Muramatsu M. 2018. Keratinocyte differentiation induces APOBEC3A, 3B, and mitochondrial DNA hypermutation. Sci Rep 8(1):9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, Koehler CM, Teitell MA. 2010. PNPASE regulates RNA import into mitochondria. Cell 142(3):456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Scott I, Zhu L, Wu K, Han K, Chen Y, Gucek M, Sack MN. 2017. GCN5L1 modulates cross-talk between mitochondria and cell signaling to regulate FoxO1 stability and gluconeogenesis. Nat Commun 8(1):523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen DF. 2006. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem 281(35):25791–25802. [DOI] [PubMed] [Google Scholar]
- Wei JW, Huang K, Yang C, Kang CS. 2017. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep 37(1):3–9. [DOI] [PubMed] [Google Scholar]
- Wisnovsky S, Jean SR, Kelley SO. 2016. Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nat Chem Biol 12(7):567–573. [DOI] [PubMed] [Google Scholar]
- Woischnik M, Moraes CT. 2002. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res 12(6):885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters JEJ, van Breda SGJ, Caiment F, Claessen SM, de Kok TMCM, Kleinjans JCS. 2017. Nuclear and Mitochondrial DNA Methylation Patterns Induced by Valproic Acid in Human Hepatocytes. Chem Res Toxicol 30(10):1847–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters JEJ, van Breda SGJ, Grossmann J, Fortes C, Caiment F, Kleinjans JCS. 2018. Integrated ‘omics analysis reveals new drug-induced mitochondrial perturbations in human hepatocytes. Toxicol Lett 289:1–13. [DOI] [PubMed] [Google Scholar]
- Wong M, Gertz B, Chestnut BA, Martin LJ. 2013. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci 7:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Y. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18(9):517–534. [DOI] [PubMed] [Google Scholar]
- Xiao CL, Zhu S, He M, Chen, Zhang Q, Chen Y, Yu G, Liu J, Xie SQ, Luo F, Liang Z, Wang DP, Bo XC, Gu XF, Wang K, Yan GR. 2018. N6 -Methyladenine DNA Modification in the Human Genome. Mol Cell 71(2):306–318.e307. [DOI] [PubMed] [Google Scholar]
- Xiong J, Ye TT, Ma CJ, Cheng QY, Yuan BF, Feng YQ. 2018. N6-Hydroxymethyladenine: a hydroxylation derivative of N6-methyladenine in genomic DNA of mammals. Nucleic Acids Res 47(3):1268–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li H, Hedmer M, Hossain MB, Tinnerberg H, Broberg K, Albin M. 2017. Occupational exposure to particles and mitochondrial DNA - relevance for blood pressure. Environ Health 16(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xia B, Yang X, Ding H, Wu D, Zhang H, Jiang G, Liu J, Zhuang Z. 2016. Mitochondrial DNA hypomethylation in chrome plating workers. Toxicol Lett 243:1–6. [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Kang D. 2018. An overview of mammalian mitochondrial DNA replication mechanisms. J Biochem 164(3):183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MJ, Copeland WC. 2016. Human mitochondrial DNA replication machinery and disease. Curr Opin Genet Dev 38:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. 2005. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 4(1):71–79. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD. 2014. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158(3):607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, León IR, Bak S, Mogensen M, Wrzesinski K, Højlund K, Jensen ON. 2011. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics 10.1074/mcp.M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sun L, Wang RR, Hu JF, Cui J. 2018. The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: New players in an old arena. Crit Rev Oncol Hematol 131:76–82. [DOI] [PubMed] [Google Scholar]
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. 2013. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res 12(1):260–271. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. 2007. Genome-wide analysis of DNA methylation patterns. Development 134(22):3959–3965. [DOI] [PubMed] [Google Scholar]