Abstract
Histone deacetylase (HDAC) isozymes modulate numerous regulatory signals and pathways in biological systems, hence serving as targets for drug design. For example, HDAC6 is the cytosolic tubulin deacetylase and its inhibition compromises microtubule dynamics, leading to cancer cell cycle arrest and apoptosis. The design of inhibitors that selectively target HDAC6 is desirable to avoid side effects resulting from the inhibition of off-target HDACs. High resolution X-ray crystal structures of HDAC6 have accelerated structure-based approaches to drug design targeting HDAC6. Crystal structure analysis reveals that the tubulin deacetylase domain of human HDAC6 (catalytic domain 2, also known as CD2) is very similar to that of HDAC6 CD2 from Danio rerio (zebrafish, designated zCD2). Thus, zCD2 is a valid surrogate of human HDAC6 CD2, the actual drug target; moreover, zCD2 is much more easily prepared and crystallized. A plasmid containing the zCD2 construct for heterologous expression in Escherichia coli is available through Addgene (#122031). In this chapter, we review the preparation, purification, and crystallization of zCD2-inhibitor complexes. These methods enable the rapid acquisition of structural data regarding optimal zinc-binding groups, capping groups, and linkers in the discovery of new and selective HDAC6 inhibitors.
Keywords: zinc enzyme, epigenetics, tubulin, protein crystallography, drug design
1. INTRODUCTION
Reversible protein acetylation is a common post-translational modification that rivals phosphorylation in the regulation of biological function (Kouzarides et al., 2000; Choudhary et al., 2009; Norvell et al., 2010). The acetylation of lysine side chains in histone and non-histone proteins is observed in cellular functions such as transcription (Grunstein, 1997; Drazic et al., 2016), signaling (Parra & Verdin, 2010; Gao & Alumkal, 2010; Lopéz et al., 2016), and metabolism (Zhang et al., 2010; Wang et al., 2010; Choudhary et al., 2014). Acetylome function requires three protein components: a “writer”, i.e., a histone acetyltransferase (HAT), also known as a lysine acetyltransferase (KAT); a “reader”, i.e., a regulatory protein that specifically binds to the acetyllysine moiety, such as a bromodomain-containing protein; and an “eraser”, i.e., a histone deacetylase (HDAC), also known as a lysine deacetylase (KDAC) (Roth et al., 2001; Lombardi et al., 2011; Marmorstein et al., 2014; Verdin et al, 2015; McCullough et al., 2016).
The HDACs are particularly intriguing in that they are targets for cancer chemotherapy as well as the treatment of atherosclerosis and neurodegenerative diseases (Su, et al., 2008; Witt et al., 2009; Villagra et al., 2010; Mottama et al., 2015; Didonna & Opal, 2015; Leucker et al., 2017). The HDAC inhibitors Romidepsin, Vorinostat, Belinostat, and Panobinostat are currently approved for clinical use, and others are in various stages of clinical trials (Dokmanovic et al., 2007; West et al., 2014; Ma et al., 2016; Eckschlager et al., 2017).
Phylogenetic analysis of human HDACs indicates three distinct groupings of the eleven metal-dependent isozymes: the class I isozymes, HDAC1, HDAC2, HDAC3, and HDAC8; the class II isozymes, further subdivided into the class IIa isozymes (HDAC4, HDAC5, HDAC7, HDAC9) and the class IIb isozymes (HDAC6, HDAC10); and the class IV isozyme HDAC11 (Gregoretti et al., 2004). These enzymes can utilize Zn2+, Fe2+, or Co2+ for catalytic activity in vitro, but Zn2+, or possibly Fe2+, is the most likely cofactor utilized in vivo as demonstrated for HDAC8 (Gantt et al., 2006). Interestingly, HDACs are evolutionarily related to the arginases, which require Mn2+ for catalytic activity (Ash et al., 2000; Christianson, 2005; Lombardi et al., 2011; Hai et al., 2017). Parenthetically, the class III HDACs, more commonly known as sirtuins, are NAD+-dependent enzymes that are structurally and mechanistically distinct from the metal-dependent HDACs (Denu, 2005; Yuan & Marmorstein, 2012).
Unique among the metal-dependent deacetylases is HDAC6, which is the only isozyme that contains two catalytic domains, designated CD1 and CD2 (Grozinger et al., 1999; Verdel & Khochbin, 1999; Zhang et al., 2006; Zou et al., 2006). Predominantly localized in the cytoplasm (Bertos et al., 2004), HDAC6 CD2 is the tubulin deacetylase, inhibition of which compromises microtubule dynamics to result in cell cycle arrest and apoptosis (Hubbert et al., 2002; Haggarty et al., 2003). Thus, HDAC6 is a target for the development of isozyme-selective inhibitors (Dallavalle et al., 2012; Szyk et al., 2014; Seidel et al., 2015). The recently determined crystal structures of human CD2 (Hai & Christianson, 2016) and zebrafish CD2 (Hai & Christianson, 2016; Miyake et al., 2016) have accelerated the structure-based design of inhibitors, leading to a new understanding of structure-affinity and structure-selectivity relationships. To date, more than 30 crystal structures of HDAC6 CD2–inhibitor complexes have been reported (Hai & Christianson, 2016; Miyake et al., 2016; Porter et al., 2017, 2018a,b,c; Bhatia et al., 2018; Mackwitz et al., 2018).
In this chapter, we outline the design of a construct of HDAC6 CD2 from Danio rerio (zebrafish) and the preparation of a suitable vector for heterologous expression in Escherichia coli to generate copious amounts of soluble and catalytically-active protein. Henceforth, we refer to this construct as zCD2. The zCD2 construct is a valid surrogate of human HDAC6 CD2 since these orthologues share 59% amino acid sequence identity. Structural comparison of zCD2 and human HDAC6 CD2 reveals essentially identical structures – all active site residues are conserved except for N530 and N645 of zCD2 at the rim of the active site, which correspond to D567 and M682 in the human enzyme (Hai & Christianson, 2016). Furthermore, zCD2 is more readily crystallized in complexes with diverse inhibitors, and crystals of zCD2–inhibitor complexes routinely diffract to high or ultra-high resolution. For example, the highest resolution structure reported to date is that of the complex with trichostatin A determined at 1.05 Å resolution (Porter et al., 2017). Accordingly, this chapter concludes with a summary of our approach to the crystallization of zCD2–inhibitor complexes to enable the study of structure-affinity and structure-selectivity relationships.
2. DESIGN OF THE CONSTRUCT AND EXPRESSION VECTOR
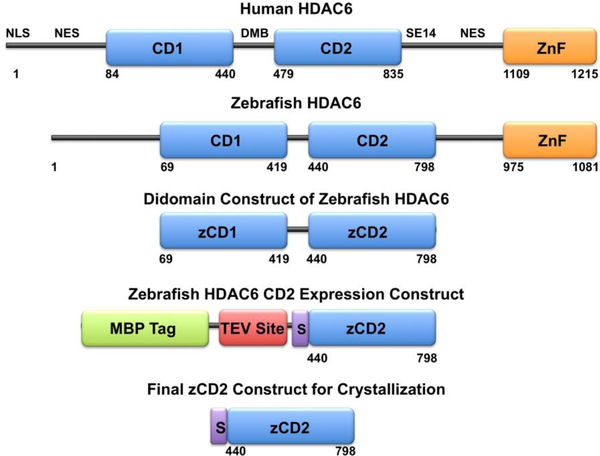
The actual drug target, human HDAC6, shares a close structural and functional relationship with zebrafish HDAC6, as demonstrated in enzymatic assays and X-ray crystal structure determinations of the CD2 domains of the human and zebrafish proteins (Hai & Christianson, 2016). The full-length proteins share similar primary structures, although the zebrafish enzyme is smaller than the human enzyme. Primary structure analysis provides a starting point for the design of a suitable zCD2 construct for crystallization and X-ray crystal structure determination, as first achieved by Hai & Christianson (2016). The zCD2 protein is first prepared as a fusion construct with maltose binding protein, after which the fusion tag is cleaved to generate pure zCD2 (Figure 1).
Figure 1.
The primary structure of full-length human (Homo sapiens) HDAC6 reveals domain organization as follows: NLS, nuclear localization signal; NES, nuclear export signal; CD1, catalytic domain 1; DMB, dynein motor binding domain; CD2, catalytic domain 2; SE14, Ser-Glu tetradecapeptide repeat; ZnF, zinc-finger domain. Catalytic domain 2 is the tubulin deacetylase domain. The primary structure of full-length zebrafish (Danio rerio) HDAC6 reveals similar domain organization. Truncation of the zebrafish enzyme to a CD1-CD2 construct, followed by the substitution of CD1 with a maltose binding protein (MBP) fusion tag along with a tobacco etch virus (TEV) protease cleavage site, yields the final expression construct with the additional peptide segment SNAGG (S) at the N-terminus of zCD2. Cleavage with TEV protease yields the final zCD2 construct used for assay and crystallization.
2.1. Equipment
T100™ thermal cycler (Bio-Rad)
EC3000P series 90 programmable electrophoresis power supply (Thermo Fisher Scientific)
Transilluminator FBTIV-88 (Fisher Scientific)
Digital dry bath (USA Scientific)
MiniSpin® plus (Eppendorf)
Isotemp™ waterbath (Fisher Scientific)
Innova® 40 incubator shaker (New Brunswick)
Isotemp™ incubator (Fisher Scientific)
2.2. Materials
D. rerio (zebrafish) HDAC6 gene (residues 60–798, Uniprot F8W4B7). Note that zCD2 corresponds to residues 440–798.
pET28a(+) vector with tobacco etch virus (TEV) protease-cleavable N-terminal His6-maltose binding protein-tag (from Dr. Scott Gradia, University of California, Berkeley; Addgene #29656)
- Oligonucleotide primers for ligation independent PCR cloning of residues 440–798 of zCD2 (Integrated DNA Technologies)
- Forward primer: 5’-TACTTCCAATCCAATGCATCTAGTCCGATTACCGGTCTG-3’ (italics correspond to first seven residues of zCD2, codon-optimized)
- Reverse primer: 5’-TTATCCACTTCCAATGTTATTAACGCAGTGAAGACCAAAACGG-3’ (italics correspond to last seven residues of zCD2, codon-optimized)
- There should be 14–18 base pairs of complementarity with the zCD2 sequence; ensure that the melting temperatures of the two primers are within 5°C.
Deoxynucleotide triphosphate (dNTP) 10 mM mixture, PCR grade (Invitrogen #18–427-013)
PfuUltra II Fusion HS DNA polymerase (Agilent Technologies #600672–51)
0.8 – 1.2% w/v agarose gel (Fisher Scientific #BP1356–100)
Tris-Acetate-EDTA 50x buffer (pH 8.4) (Boston Bio Products #BM-250)
1-kb DNA ladder (New England Biolabs #N3232S)
Gel loading dye (6X) purple with SDS (New England Biolabs #B7024S)
10X CutSmart® buffer (New England Biolabs #B7204S)
- Restriction endonuclease Sspl-HF (20 units/μL, New England Biolabs #R0132S)
- One unit is defined as the amount of enzyme required to digest 1 μg of λ DNA in 1 hour at 37°C in a total reaction volume of 50 μL.
SpinSmart™ PCR Purification and Gel Extraction Column Kit (Thomas Scientific Inc. #1158P45)
10X NEBuffer™ 2.1 (New England Biolabs #B7202S)
Bovine Serum Albumin (Fisher Scientific #BP9706100)
Dithiothreitol (Fisher Scientific #BP172–5)
T4 DNA polymerase (New England Biolabs #M0203S)
dGTP, 100 mM solution (Invitrogen #10–218-014)
dCTP, 100 mM solution (Invitrogen #10–217-016)
HyPure™ Molecular Biology Grade Water (HyClone #SH3053801)
Ethylenediaminetetraacetic acid (EDTA) (ACROS Organics #AC30160100)
SYBR® Safe DNA Gel Stain (Invitrogen #S33102)
NEB® 5α Competent E. coli High Efficiency strain (New England Biolabs #C2987H)
Microbiology Media: Lysogeny Broth (Fisher BioReagents #BP1426–500)
Lysogeny Broth (LB) plates with 50 μg/mL kanamycin (GoldBio #K-120–100)
QIAprep Spin Miniprep Kit (Qiagen #27106)
BL21(DE3) Competent E. coli strain (New England Biolabs #C2527H)
Eppendorf™ 0.2-mL thin-walled PCR tubes (Fisher Scientific #E0030124260)
2.3. Procedure
-
1.
Begin by preparing a 50 μL sample of the zCD2 insert. The insert and all required PCR reagents are contained in a 0.2-mL thin-walled PCR tube. The quantities or final concentrations used are as follows:
Plasmid DNA: 10 ng
Primers: 0.50 μM
dNTP mixture: 0.25 mM
PfuUltra II Fusion HS DNA polymerase: 3.0 units
-
2.
Set the T100™ thermal cycler to the following parameters:
Stage 1: 95°C, 2 min, 1 cycle
-
Stage 2: 95°C, 30 s, 4 cycles
55°C, 30 s (approximately 5°C below primer melting temperature)
72°C, 1.5 min
-
Stage 3: 95°C, 30 s, 28 cycles
55°C, 30 s (approximately 5°C below primer melting temperature)
72°C, 1.5 min
Stage 4: 72°C, 10 min, 1 cycle
Stage 5: 20°C, hold
-
3.
Place the PCR tube in the T100™ thermal cycler and run the program.
-
4.
Use gel electrophoresis to verify that the PCR reaction was successful using the following materials:
0.8–1.2% w/v agarose gel
Tris-Acetate-EDTA 1x buffer
1-kb DNA ladder (Dilute 1 μL ladder with 4 μL H2O and 1 μL gel loading dye)
SYBR® Safe DNA Gel Stain (Add 5 μL into gel for band visualization)
zCD2 insert (Dilute 5 μL insert with 1 μL gel loading dye)
-
5.
Run 5 μL of the plasmid sample on the agarose gel for 40 min at 100 Volts using the EC3000P Series 90 programmable electrophoresis power supply.
Ensure that a band corresponding to the zCD2 insert is present using the Transilluminator FBTIV-88.
Set the remaining 45 μL of the zCD2 insert sample aside until the vector is ready.
-
6.
For vector linearization, prepare a reaction mixture with the following final concentrations or quantities of reagents in a final volume of 100 μL in water:
3 μg of vector DNA
1X CutSmart® buffer
60 units Sspl-HF
H2O to bring to 100 μL
-
7.
Incubate at 37°C for 1 h using the digital dry bath. At this point, a PCR cleanup step should be run using the QIAprep Spin Miniprep Kit (use the protocol included with the kit) for both the PCR insert and vector. Once complete, both the zCD2 insert and vector are ready for T4 polymerase digestion.
-
8.
For the T4 polymerase digestion, prepare two separate 40 μL reactions with the following final concentrations or quantities:
1X NEBuffer™ 2.1
10 μg Bovine Serum Albumin
5 mM dithiothreitol
3 units of T4 DNA polymerase
2.5 mM dNTP (dGTP for vector and dCTP for insert)
Vector for one reaction and zCD2 insert for the other reaction. Fill remaining 40 μL of each reaction with vector or insert (concentration does not matter).
-
9.
Incubate at room temperature for 30 min.
-
10.
Heat at 75°C for 20 min using the digital dry bath.
-
11.
Spin reactions at top speed (13.4 RPM) for 2 min at 4°C using the MiniSpin® plus.
-
12.
Remove and save the supernatant (the pellet is precipitated T4 polymerase).
-
13.
Ethanol precipitation of the supernatant from the previous step; save the pellet.
-
14.
Redissolve each DNA pellet in 12 μL of ultra-pure water; do not combine pellets.
-
15.
Set up multiple vector:insert ratios (volume:volume) to maximize cloning efficiency, e.g., 1:4, 1:6, 1:8, 1:10, 1:12; use 1:0 as a control.
-
16.
Incubate the vector:insert reactions at room temperature for 30 min.
-
17.
Add 1 μL of 25 mM EDTA solution to each tube.
-
18.
Incubate at room temperature for 15 min. Once complete, the final plasmid is ready to be transformed.
-
19.
Transform E. coli strain NEB® 5α by mixing 1 μL of plasmid with 50 μL of cells.
-
21.
Allow the plasmid to settle in solution for 30 min while being chilled on ice.
-
22.
Heat shock cells for 40 s at 37°C using the Isotemp™ waterbath.
-
23.
Immediately place plasmid back on ice for 10 min.
-
24.
Add 200 μL Lysogeny Broth to plasmid.
-
25.
Shake at 250 RPM and 37°C for 1 h in the Innova® 40 incubator shaker.
-
26.
Plate 200 μL or streak out plasmid onto a 50 μg/mL kanamycin plate.
-
27.
Place plate upside down in Isotemp™ incubator at 37°C for 16 h. Placing the plate upside down ensures that bacteria grow across the media rather than down into the media, and also avoids the accumulation of condensation on the surface of the media.
-
28.
Colonies containing your plasmid should appear. Prepare these colonies for sequencing using the QIAprep Spin Miniprep Kit (use the protocol included with the kit) and submit sample to a local DNA sequencing center with the following 3 primers:
Primer 1: T7 promoter: 5’-TAATACGACTCACTATAGGG-3’. This primer should be available through your DNA sequencing center.
Primer 2: T7 terminator: 5’-GCTAGTTATTGCTCAGCGG-3’. This primer should be available through your DNA sequencing center.
Primer 3: Internal: 5’-GCAGGTATTAACGCCGCCAG-3’. This primer must be ordered. Due to the length of the MBP-zCD2 construct, an internal primer is needed to read the full DNA sequence. Primer 3 is specifically designed to begin reading the DNA sequence between Primer 1 and Primer 2. Specifically, Primer 3 will begin reading nucleotides near the end of the MBP tag and continue to read the DNA sequence corresponding to most of the zCD2 protein. Without Primer 3, there would be a gap in the DNA sequencing. Thus, all 3 primers are required.
-
29.
After DNA sequence verification, transform zCD2 into E. coli strain BL21(DE3) for protein expression using the same procedure outlined in steps 19–27.
2.4. Notes
Protein fusion tags enhance the solubility of recombinantly expressed proteins in E. coli (Costa et al., 2013). Here, we use maltose-binding protein as the solubility-enhancing tag (Figure 1). Additionally, the SNAGG segment is inserted between the C-terminus of maltose binding protein and the N-terminus of zCD2. This results from the ligation independent cloning, and may also facilitate TEV protease access (Hai & Christianson, 2016). Any alteration of the expression system, tag, or construct may lead to alternative results. The zCD2 plasmid is available through Addgene (#122031).
3. Expression and Purification of zCD2
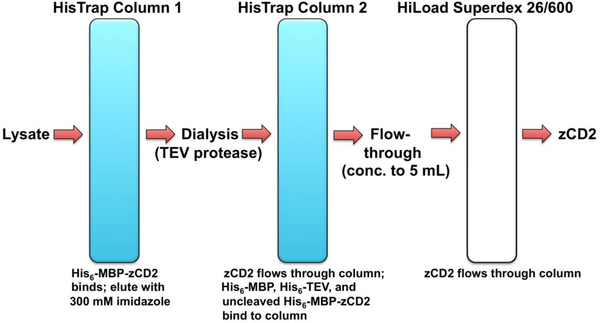
Here, we describe an updated approach for the expression and purification of zCD2. The originally reported purification protocol yielded sufficiently pure protein for crystallization and structure determination of several zCD2-inhibitor complexes (Hai & Christianson, 2016). However, endogenous maltose binding protein contamination after the final purification step was a recurring problem. Accordingly, the purification scheme was revised to maximize zCD2 yield and purity (Figure 2).
Figure 2.
The purification scheme for zCD2 utilizes a HisTrap™ HP column, off-column digestion with 6 mg/ml TEV protease, a second HisTrap™ HP column for further purification, followed by a HiLoad™ Superdex™ 26/600 200 column.
3.1. Equipment
Autoclave (STERIS® Healthcare)
Innova® 40 incubator shaker (New Brunswick)
WPA CO8000 cell density meter (Biochrom)
Sorvall LYNX 6000 centrifuge (Thermo Fisher Scientific)
Revco® ultima PLUS −80° freezer (Thermo Fisher Scientific)
Vortex-Genie® 2 (Scientific Industries)
Sonicator (Qsonica)
5 mL HisTrap™ HP column (GE Healthcare)
ÄKTA Prime Plus FPLC (GE Healthcare)
HiLoad™ Superdex™ 26/600 200 pg column (GE Healthcare)
PowerPac™ basic gel machine (Bio-Rad)
150 mL Superloop (GE Healthcare)
5 mL Superloop (GE Healthcare)
Magnetic Stirrer RT Basic (Thermo Fisher Scientific)
3.2. Materials
2XYT media broth, powder (Research Products International Corp. #50-489-141)
50 μg/mL kanamycin (GoldBio #K-120–100)
D-(+)-Glucose, anhydrous (ACROS Organics #AC410955000)
zCD2 BL21(DE3) Competent E. coli strain (as transformed in section 2.3)
Fisher Bioreagents Microbiology Media: Lysogeny Broth, Miller (Thermo Fisher Scientific #BP1426-500)
Zinc sulfate heptahydrate 99% (ZnSO4) (ACROS Organics #AC424605000)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (GoldBio #I2481C50)
Potassium phosphate dibasic anhydrous powder (K2HPO4) (Fisher Scientific #P288–100)
Sodium chloride (NaCl) (Fisher Chemical #S271–500)
Imidazole (Fisher Chemical #03196–500)
Tris-(2-carboxyethyl)phosphine (TCEP)-HCl (GoldBio #TCEP25)
Glycerol (Fisher Chemical #G33–500)
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Alfa Aesar #AAA1477709)
Potassium chloride (KCl) (Fisher Chemical #P217–500)
Magnesium chloride (MgCl2) (Thermo Fisher Scientific #AB-0359)
Lysozyme from chicken egg white (Sigma-Aldrich #NC0826086)
Deoxyribonuclease 1, bovine pancreas (Alfa Aesar #AAJ61061MC)
cOmplete™ Protease Inhibitor Cocktail Tablets (MiiliporeSigma #NC0962311)
Tobacco etch virus (TEV) protease (R&D Systems #4469TP200)
Regenerated cellulose (RC) dialysis tubing 6–8 KD molecular weight cut-off (Spectra/Por® #132665)
0.22-μM Millex®-GV filter unit (MilliporeSigma #SAMP2GVNB)
Boston Bio Products Inc. MES-SDS Running Buffer (Thermo Fisher Scientific #NC9041372)
HyPure™ Molecular Biology Grade Water (HyClone #SH3053801)
Amicon® ultra-15 Centrifugal Filter Unit 10 kDa molecular weight cutoff (EMD Millipore #UFC901096)
Polar Ware 250B Stainless Steel Griffin Style Beaker 250 mL Capacity (Stoelting #1526Q30EA)
3.3. Procedure
Prepare 12 L of 2XYT media by adding 31 grams of 2XYT media to each of 12 2-L baffled flasks. Each 2 L flask should contain only 1 L of water, so that the flasks do not overflow upon shaking. Autoclave the flasks after they are prepared with water and media.
Supplement the media in each 2-L flask with 50 μg/mL kanamycin and 0.02 M glucose after flasks cool down to room temperature.
Overnight cultures of zCD2 should be run for 16 h at 250 RPM and 37°C prior to the growth. To do this, pick 1 single colony from the plate of zCD2 E. coli BL21(DE3) cells and place it into 100 mL of Lysogeny Broth supplemented with 50 μg/mL kanamycin (to prevent bacterial contamination).
The next day, transfer 5 mL of zCD2 culture into each 2-L flask.
Grow at 37°C and 250 RPM in the Innova® 40 incubator shaker (RPM = 250) until the OD600 reaches approximately 0.75. This can be checked using the WPA CO8000 cell density meter and should be checked every hour.
Turn the shaker temperature down to 18°C for 20 min. This should correspond to an inducing OD600 of approximately 1.0.
Supplement each flask with 200 μM ZnSO4 and 100 μM IPTG.
Grow for 18 h at 18°C and 250 RPM.
- On the third day, spin down cells using the Sorvall LYNX 6000 centrifuge at 5,000 RPM (9,778g) and freeze cell pellets in the Revco® ultima PLUS −80° freezer. The growth phase is now complete.
- An average 6-L growth yields approximately 40 g of cell pellet.
- The cell pellet can be stored in the −80° freezer.
-
For zCD2 purification, make the following buffers fresh on the day of purification using the final concentrations and quantities listed below:
-
Buffer A (1.0 L):
50 mM K2HPO4 (pH 8.0)
300 mM NaCl
30 mM imidazole
1 mM TCEP
5% glycerol
H2O to bring to 1.0 L
-
Buffer B (1.0 L):
50 mM K2HPO4 (pH 8.0)
300 mM NaCl
300 mM imidazole
1 mM TCEP
5% glycerol
H2O to bring to 1.0 L
-
Buffer C (1.0 L):
50 mM HEPES (pH 7.5)
100 mM KCl
1 mM TCEP
5% glycerol
H2O to bring to 1.0 L
-
Buffer D (150 μL):
50 mM K2HPO4 (pH 8.0)
300 mM NaCl
10 mM MgCl2
1 mM TCEP
5% glycerol
0.1 mg/mL lysozyme
50 μg/mL DNAse
2 protease inhibitor tablets
H2O to bring to 150 μL
-
Buffers are similar to those used in the originally reported purification protocol (Yang & Christianson, 2016) with minor alterations:
30 mM of background imidazole is added to Buffer A to prevent nonspecific binding to the column.
The originally-reported Tris elution buffer is no longer used. HEPES is now used as the buffer instead of Tris.
-
Retrieve the frozen zCD2 cell pellet from the −80°C freezer and resuspend in 100 mL of Buffer D in a stainless steel beaker. For optimal mixing, Buffer D should be added 10 mL at a time and vortexed until the entire zCD2 pellet is mixed homogenously.
-
Break open the cells using a sonicator set with the following parameters:
-
Qsonica Sonicator:
Amplitude = 30%
Process time = 8.0 min
Pulse on time = 1.0 s
Pulse off time = 2.0 s
It is important to prevent overheating and killing the cells. To prevent this, use ice at all times throughout this step. Mix the cells every 2 min to ensure complete lysis. The sonicator tip should be placed near the bottom of the cell suspension.
-
Once sonication is complete, centrifuge the cells for 1 h at 15,000 RPM (38,625g) using the Sorvall LYNX 6000 centrifuge. Inspect the cell pellet carefully. If there is white material present, this indicates the presence of insoluble protein. Ideally, a clean beige-colored pellet will be observed after centrifugation (Figure 3).
Pour clear lysate into a 150 mL superloop.
-
Prior to injecting the lysate onto the column, the column must first be equilibrated with buffer.
Attach a 5 mL HisTrap™ HP column to the ÄKTA Prime Plus FPLC.
Run 5 column volumes of Buffer B over the HisTrap™ HP column.
Run 5 column volumes of Ultra-pure water over the HisTrap™ HP column.
Run 5 column volumes of Buffer A over the HisTrap™ HP column. The column is now equilibrated.
Inject the zCD2 lysate over the HisTrap™ HP column at 1 mL/minute collecting 10-mL fractions while lysate is being applied.
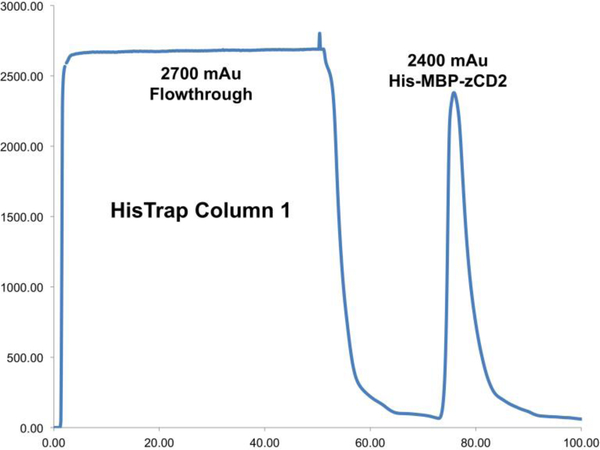
After the chromatogram returns to baseline, run a 20% buffer B wash (60 mM imidazole) on the load setting while collecting 5-mL fractions. This will remove contaminants, since His6-tagged proteins typically elute at approximately 200 mM imidazole.
Run a 100% Buffer B wash (300 mM imidazole) while collecting 5-mL fractions. This is the peak corresponding to the maltose binding protein-zCD2 fusion protein (Figure 4).
-
Collect the 100% Buffer B wash fractions and add 6 mg/mL of TEV protease.
Protein must be kept on ice.
Avoid over-concentrating (dark yellow color) or concentrating for extended periods of time to avoid protein precipitation.
Load standard RC dialysis tubing (6–8 kD molecular weight cut-off) with the mixture prepared in the previous step and gently stir overnight at 60 RPM using a magnetic stirrer. If white precipitant is visible the next morning, then some zCD2 precipitated out of solution.
-
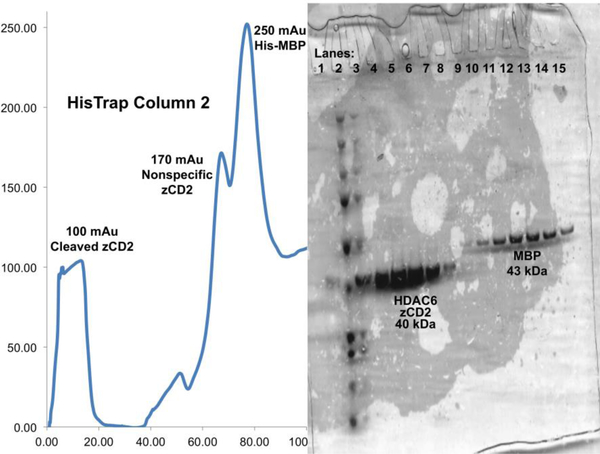
Apply the mixture from the previous step to a HisTrap™ HP column and repeat steps 16–17. Due to nonspecific binding of zCD2 to the second HisTrap™ HP column, do not run a 100% Buffer B wash. Instead, elute with 200 mL on a 0–100% Buffer B gradient at 1 mL/min to ensure clear separation of nonspecific zCD2 and maltose binding protein (Figure 5).
The uncleaved zCD2 should bind to the column; however, zCD2 can bind nonspecifically as well.
Caution: maltose binding protein (42.5 kDa) has a similar molecular weight to zCD2 (40.2 kDa). SDS-PAGE should always be run prior to the next step to ensure that the correct protein is being isolated.
-
Collect and concentrate all zCD2-containing fractions to a final volume of 5 mL using a 0.22-μM Millex®-GV filter unit.
A 15 mL centrifugal filter unit with a molecular weight cut off of 10,000 kDa should be used.
Concentrate at 4,000 x g in 10 minute cycles to ensure zCD2 does not precipitate.
Keep protein on ice and concentrate at 4 °C.
Inject 5 mL of zCD2 into a 5 mL superloop attached to the ÄKTA Prime Plus FPLC.
Attach a HiLoad™ Superdex™ 26/600 200 pg column to the ÄKTA Prime Plus FPLC.
Equilibrate the Superdex™ column with 360 mL Buffer C at 1 mL/min.
Inject 5 mL of zCD2 onto the Superdex™ column at 1 mL/min.
Collect 5-mL fractions at 1 mL/min.
Run 360 mL Buffer C at 1 mL/min.
Run SDS-PAGE on desired fractions in MES-SDS Running Buffer using a PowerPac™ basic gel machine, running at 200 V for 35 min.
-
Concentrate protein to 10 mg/mL using the same procedure outlined above in section 3.22, with one exception: do not filter zCD2 using the 0.22-μM Millex®-GV filter unit when concentrated to 10 mg/mL to avoid losing protein inside the filter. Instead, zCD2 will be filtered prior to crystallization trials, as discussed in section 5.3.
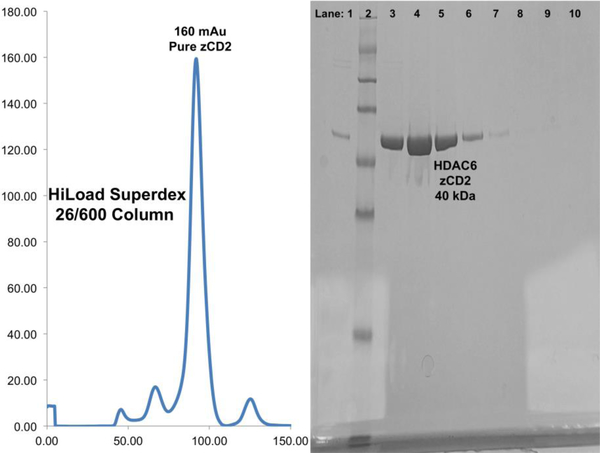
Dilute zCD2 51 fold and use dilution factor = 51, molar extinction coefficient = 38390, and molecular weight = 40234.53 to calculate the concentration.
Protein is typically better than 90% pure (Figure 6). If it is not, then the protein can be run over the HisTrap™ HP column and the HiLoad™ Superdex™ 26/600 200 pg column a second time (steps 15–30).
Figure 3.
The cell pellet should be beige in color with no white streaks.
Figure 4.
Cell lysate should max out the UV detector at approximately 3,000 mAu with a prominent protein peak following elution. This is the maltose binding protein-zCD2 fusion protein. A representative HisTrap™ HP column chromatogram is shown.
Figure 5.
The zCD2 protein should no longer have a His6 tag and therefore should no longer bind to the HisTrap™ HP column. However, some zCD2 may bind nonspecifically, which is easily discernible by SDS-PAGE. A representative HisTrap™ HP column chromatogram from the second purification step and gel are shown.
Figure 6.
The zCD2 protein should elute from the sizing column at approximately 150–250 mAu without maltose binding protein contamination. A representative sizing chromatogram and gel are shown.
3.4. Notes
On average, 500 μL of zCD2 at a concentration of approximately 15–20 mg/mL is obtained from a 6-L preparation.
4. Activity Assay with a Fluorogenic Substrate
The zCD2 sample prepared as described in section 3 is suitable for use in a variety of biochemical and biophysical assays, e.g., isothermal titration calorimetry measurements of inhibitor binding (Porter et al., 2018c) and measurements of catalytic activity (Hai & Christianson, 2016). A discontinuous liquid chromatography-mass spectrometry assay has been used to measure zCD2 activity with non-fluorogenic substrates (Hai & Christianson, 2016). Below, we outline a common method for measuring zCD2 activity using a fluorogenic substrate.
4.1. Equipment
UV-VIS spectrophotometer (Agilent Technologies)
Infinite® M1000Pro plate reader (Tecan)
4.2. Materials
1.0 μM zCD2 in Buffer C (see section 3.3.10)
Tris-HCl 1.0 M solution (pH 8.0) (Thermo Fisher Scientific #AAJ22638AE)
Sodium chloride (NaCl) (Fisher Chemical #S271–500)
Potassium chloride (KCl) (Fisher Chemical #P217–500)
Magnesium chloride (MgCl2) (Thermo Fisher Scientific #AB-0359)
Trichostatin A (Tocris Bioscience #14–061)
Bovine pancreatic trypsin (Millipore-Sigma #65–022-5MU)
Dimethyl Sulfoxide (DMSO), HPLC grade (Fisher Chemical #D159–4)
HyPure™ Molecular Biology Grade Water (HyClone #SH3053801)
- 300 μM Ac-RHKAcKAc-AMC fluorogenic substrate in assay buffer (KAc = acetyllysine, AMC = aminomethylcoumarin). Two possible commercial sources of this assay substrate are as follows:
- GenScript Biotech Corp.: $398.57 for 25.0–29.0 mg; average unit cost = $14.84/mg
- Enzo Life Sciences, Inc.: $201.00 for ~0.40 mg; average unit cost = ~$500/mg
Nunc™ 384-well black optical bottom plate (Fisher Scientific #12–566-2)
Fluor de lys deacetylated standard (Enzo Life Sciences #BML-KI142–0030)
4.3. Procedure
- Make 50 mL of fresh assay buffer using the following final concentrations:
- 50 mM Tris-HCl (pH 8.0)
- 137 mM NaCl
- 2.7 mM KCl
- 1.0 mM MgCl2
- H2O to bring up to 50 mL
- Make 50 mL of fresh developer solution using the following final concentrations:
- 10 μM trichostatin A (any tight-binding HDAC inhibitor will suffice)
- 1 μM trypsin
- DMSO to bring up to 50 mL
- Ensure protein concentration is correct (1.0 μM) by rechecking prior to use by using UV-VIS spectroscopy as discussed in section 3.3.30.
- Frozen aliquots of protein do not always have the same concentration due to concentrated areas prior to freezing. Do not assume the concentration of zCD2 prior to freezing will be the same as thawed aliquots of zCD2.
- Dilute the protein to 1.0 μM using assay buffer (use dilution factor = 51, molar extinction coefficient = 38390, and molecular weight = 40234.53 to calculate the concentration).
Initiate reaction by combining 25 μL of 1.0 μM zCD2 in assay buffer with 25 μL of 300 μM Ac-RHKAcKAc-AMC fluorogenic substrate in assay buffer in a Nunc® 384-well black optical bottom plate. Note: the substrate is light sensitive so keep covered with aluminum foil or place inside a drawer while reaction occurs.
After 30 min, add 50 μL developer solution to quench the reaction.
- Analyze activity using an Infinite® M1000Pro plate reader with the following parameters:
- λex = 360 nm
- λem = 455 nm
- Avoid bubbles for accurate readings.
Compare signal to a standard curve prepared using the Fluor de lys deacetylated standard.
4.4. Notes
Assays should be run in triplicate. Statistical tests, such as confidence intervals, standard error, R-squared values, etc., can be used to assess data quality. If air bubbles are created in the reaction mixtures in the wells of the Nunc® 384-well black optical bottom plate, fluorescence emission readings will be incorrect. It is best to run each trial with an inhibitor control, protein control, and buffer control.
5. Crystallization of zCD2
To date, more than 30 crystal structures of zCD2 have been deposited in the Protein Data Bank (PDB). The highest resolution structure has been determined at 1.05 Å resolution (PDB accession code 5WGI) and the lowest resolution structure has been determined at 2.88 Å resolution (PDB accession code 5G0J). On average, the resolution expected for crystalline zCD2 complexes is approximately 1.90 Å. The zCD2 protein crystallizes predominantly in orthorhombic space group P212121 (26%) and monoclinic space group P21 (23%); orthorhombic space group P21212 (16%) is also observed. Seven additional space groups have been encountered less often. A summary of zCD2 space group parameters is found in Table 1 and photographs of typical zCD2 crystals are found in Figure 7.
Table 1.
Summary of zCD2 crystal parameters
| PDB Code |
Resolution (Å) |
Space Group |
a (Å) | b (Å) | c (Å) | α (°) | β (°) | γ (°) |
|---|---|---|---|---|---|---|---|---|
| 5EEM | 2.00 | P1 | 48.5 | 55.4 | 74.2 | 73.6 | 89.8 | 82.6 |
| 5EF7 | 1.90 | P1 | 48.7 | 56.6 | 74.8 | 106 | 90.1 | 97.1 |
| 5WGI | 1.05 | P21 | 48.4 | 69.7 | 50.2 | 90.0 | 110.4 | 90.0 |
| 5EEI | 1.32 | P21 | 54.8 | 83.6 | 86.8 | 90.0 | 98.0 | 90.0 |
| 6DVM | 1.47 | P21 | 78.4 | 95.2 | 98.0 | 90.0 | 98.9 | 90.0 |
| 5G0H | 1.60 | P21 | 50.0 | 48.2 | 73.8 | 90.0 | 103.2 | 90.0 |
| 5EFN | 1.80 | P21 | 55.0 | 83.9 | 86.9 | 90.0 | 98.1 | 90.0 |
| 6DVL | 2.10 | P21 | 55.0 | 82.6 | 88.9 | 90.0 | 98.5 | 90.0 |
| 6DVN | 2.20 | P21 | 78.6 | 95.5 | 98.2 | 90.0 | 98.6 | 90.0 |
| 5WGK | 1.82 | P22121 | 51.7 | 84.0 | 94.6 | 90.0 | 90.0 | 90.0 |
| 6CGP | 2.50 | P22121 | 45.3 | 65.0 | 140.5 | 90.0 | 90.0 | 90.0 |
| 5EEK | 1.59 | P21212 | 83.9 | 94.4 | 51.7 | 90.0 | 90.0 | 90.0 |
| 5EFJ | 1.73 | P21212 | 83.5 | 94.1 | 51.5 | 90.0 | 90.0 | 90.0 |
| 5EFK | 1.82 | P21212 | 83.3 | 94.7 | 51.6 | 90.0 | 90.0 | 90.0 |
| 5EFB | 2.54 | P21212 | 83.4 | 94.4 | 51.6 | 90.0 | 90.0 | 90.0 |
| 5EF8 | 2.60 | P21212 | 65.2 | 140.4 | 91.9 | 90.0 | 90.0 | 90.0 |
| 6CSP | 1.24 | P212121 | 74.7 | 91.8 | 96.5 | 90.0 | 90.0 | 90.0 |
| 6CSR | 1.62 | P212121 | 74.9 | 92.0 | 96.4 | 90.0 | 90.0 | 90.0 |
| 6CSS | 1.70 | P212121 | 74.8 | 92.0 | 96.6 | 90.0 | 90.0 | 90.0 |
| 5WGL | 1.70 | P212121 | 87.2 | 88.0 | 119.0 | 90.0 | 90.0 | 90.0 |
| 5EEN | 1.86 | P212121 | 76.0 | 95.5 | 96.3 | 90.0 | 90.0 | 90.0 |
| 6CSQ | 2.03 | P212121 | 74.6 | 92.0 | 96.6 | 90.0 | 90.0 | 90.0 |
| 5EFH | 2.16 | P212121 | 74.7 | 92.4 | 96.2 | 90.0 | 90.0 | 90.0 |
| 5EFG | 2.25 | P212121 | 75.1 | 91.8 | 96.2 | 90.0 | 90.0 | 90.0 |
| 6MR5 | 1.85 | C2 | 159.0 | 46.5 | 96.4 | 90.0 | 119.2 | 90.0 |
| 5G0I | 1.99 | C21 | 162.6 | 52.8 | 186.4 | 90.0 | 108.9 | 90.0 |
| 5WGM | 1.75 | C2221 | 97.8 | 174.3 | 149.1 | 90.0 | 90.0 | 90.0 |
| 6CW8 | 1.90 | C2221 | 48.3 | 128.4 | 260.4 | 90.0 | 90.0 | 900. |
| 5W5K | 2.70 | C2221 | 101.3 | 170.9 | 151.4 | 90.0 | 90.0 | 120 |
| 5G0J | 2.88 | P3221 | 187.2 | 187.2 | 102.7 | 90.0 | 90.0 | 120 |
| 6DVO | 1.98 | P65 | 97.2 | 97.2 | 78.9 | 90.0 | 90.0 | 120 |
Figure 7.
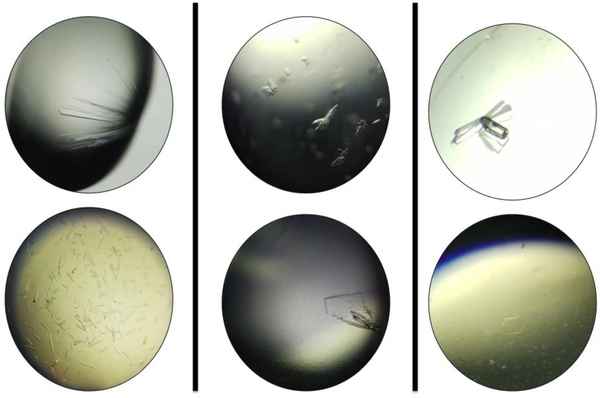
Examples of crystals of zCD2-inhibitor complexes. (A) Long thin rods are most common. These have yielded crystal structures. (B) Stacked plate-like clusters and smaller crystals are observed less often, but have nonetheless yielded crystal structures. (C) Prisms and hexagons are also observed in crystallization trials, but to date these have always been salt crystals.
The most common problem encountered in the cocrystallization of zCD2 with inhibitors is ensuring that the desired inhibitor is bound in the active site of the crystalline complex. The zCD2-acetate complex (PDB accession code 5EFG) crystallizes in orthorhombic space group P212121 with unit cell dimensions a = 75 Å, b = 92 Å, and c = 96 Å; oftentimes, but not always, acetate is observed bound to the active site Zn2+ ion instead of the inhibitor included in the cocrystallization experiment. This can occur when acetate is included in the crystallization conditions, but it also can occur when acetate is not contained in the protein solution or crystallization buffer. Presumably, acetate can be carried through the protein purification procedure in such instances. It is also notable that the same orthorhombic unit cell dimensions are found for successfully prepared zCD2–inhibitor complexes (see, for example, structures with PDB accession codes 6CSP, 6CSQ, 6CSR, 6CSS, and 5EFH (Table 1)). Thus, zCD2 crystals with these unit cell dimensions must undergo complete diffraction data collection and electron density map calculation before it can be determined whether or not the inhibitor is bound in the enzyme active site.
5.1. Equipment
Mosquito® crystallization robot (TTP Labtech)
4°C cold room
S8APO KL 300 LED Microscope (Leica Microsystems)
5.2. Materials
zCD2 protein (10 mg/mL) (as purified in section 3.3)
HDAC inhibitors (40 mM) solubilized in DMSO
Buffer C (section 3.3.10)
EMD Millipore™ Ultrafree™ Centrifugal Device with Durapore™ Membrane (MilliporeSigma #UFC30GV00)
- Precipitant solutions
- PEG/Ion 1 Screen™ (Hampton Research #HR2–126)
- PEG/Ion 2 Screen™ (Hampton Research #HR2–098)
- PEG RX™ 1 (Hampton Research #HR2–082)
- PEG RX™ 2 (Hampton Research #HR2–084)
Ethylene glycol, 100% solution (Hampton Research #HR2–621)
Eppendorf™ 0.2-mL thin-walled PCR tubes (Fisher Scientific #E0030124260)
5.3. Procedure
Fresh protein is best for crystallization trials; however, several structures have been obtained from protein stocks frozen at −80°C and thawed to room temperature. In fact, we have successfully crystallized protein samples that have been through two freeze-thaw cycles, so zCD2 appears to be relatively robust. Thaw the desired amount of protein required for each tray. Keep protein on ice.
Keep inhibitor solutions off of ice (otherwise the DMSO will freeze). It is important for the inhibitors to be solubilized in DMSO. Avoid inhibitors prepared as trifluoroacetic acid salts, because these have yielded structures of zCD2-acetate complexes instead of complexes with the desired inhibitor.
Once thawed, add zCD2 to a microcentrifuge tube with a final concentration of 10 mg/mL in buffer C. This protein concentration is generally most successful in yielding crystals. If too much precipitation results, then the protein concentration can be lowered, e.g., to 5 mg/mL.
- Add the desired inhibitor solution such that the final concentration of inhibitor is 2 mM. Note that some inhibitors are not particularly soluble, in which case the inhibitor will precipitate from solution to yield a cloudy mixture (Figure 8). If this occurs, there are three options:
- Reduce the final inhibitor concentration to 1 mM or its maximum solubility.
- Gently heat the solution to ensure that more inhibitor becomes solubilized (but do not overheat; maximum temperature = 37°C).
- Spin down the solution to obtain a maximally saturated solution.
Filter each enzyme–inhibitor stock solution using a centrifugal filter unit.
Prepare the desired 96-well plate for crystallization screening. Note that polyethylene glycol (PEG)-based screens tend to yield more crystals than other crystallization precipitants.
- Set up crystallization trays using the Mosquito® crystallization robot using the following guidelines:
- Sitting-drop vapor diffusion method
- 1:1 ratio (volume:volume) of protein–inhibitor solution:precipitant solution
- Store trays at 4°C in a cold room
Long, thin rod-shaped crystals appear within 2–3 days (see examples in Figure 7).
Ethylene glycol (15–25%) is a satisfactory cryoprotectant to facilitate crystal looping and flash-cooling for storage prior to X-ray diffraction data collection.
Figure 8.
Protein-inhibitor solutions should be clear (a clear solution indicates a fully soluble inhibitor). If the solution is white or cloudy, the inhibitor is not fully soluble. An insoluble inhibitor can result in a lack of crystal formation or crystallization of the unliganded enzyme.
5.4. Notes
The cold room temperature of 4°C is recommended since the majority of zCD2 crystals form at this temperature. However, a zCD2–inhibitor complex might preferentially crystallize at room temperature, so if no crystals appear in an initial screen at 4°C, proceed with a crystallization screen at a higher temperature. Hydroxamate-bearing inhibitors appear to cocrystallize more readily with zCD2 than inhibitors bearing alternative zinc-binding groups. Hydroxamate-zinc binding geometry is either bidentate (through the C=O and N–O– groups) or monodentate (through the N–O– group). While a third monodentate binding interaction is possible through the ionized hydroxamate NH group, as observed in complexes with the zinc enzyme carbonic anhydrase (Scolnick et al., 1997), this binding mode has not been observed to date in any HDAC–hydroxamate complex.
6. Summary
The preparation, assay, and crystallization of zCD2 is important due to its structural similarity to human HDAC6 CD2, a validated drug target for cancer chemotherapy and other diseases. Since the original report of the preparation and crystal structure determination of zCD2 and its inhibitor complexes (Hai & Christianson, 2016), the protein purification protocol has been optimized as described herein. This protocol has ensured success in the cocrystallization of zCD2 with a varied array of inhibitors. These structures provide important clues with regard to the structural basis of inhibitor affinity and selectivity for zCD2 and human HDAC6 CD2.
Acknowledgements
We thank Drs. Yang Hai, Nicholas Porter, and Stephen Shinsky for helpful discussions and comments on the manuscript. We are especially grateful to Dr. Yang Hai for his preparation of the original zCD2 construct and crystal structure determination (Hai & Christianson, 2016). This research was supported by NIH grant GM49758.
References
- Ash DE, Cox JD, Christianson DW (2000). Arginase: a binuclear manganese metalloenzyme. Metal Ions in Biological Systems. 37, 407–428. [PubMed] [Google Scholar]
- Bertos NR, Gilquin B, Chan GK, Yen TJ, Khochbin S, Yang XJ (2004). Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. Journal of Biological Chemistry. 279, 48246–54. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Krieger V, Groll M, Osko JD, Reβing N, Ahlert H, Borkhardt A, Kurtz T, Christianson DW, Hauer J, Hansen FK (2018). Discovery of the First-in-class dual histone deacetylase-proteasome inhibitor. Journal of Medicinal Chemistry. 61, 10299–10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325, 834–40. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nature Reviews Molecular CellBiololgy. 15, 536–50. [DOI] [PubMed] [Google Scholar]
- Christianson DW (2005). Arginase: structure, mechanism, and physiological role in male and female sexual arousal. Accounts of Chemical Research. 38, 191–201. [DOI] [PubMed] [Google Scholar]
- Costa SJ, Almeida A, Castro A, Domingues L, Besir H. (2013). The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: A comparison with the traditional gene fusion technology. Applied Microbiology and Biotechnology. 97, 6779–91. [DOI] [PubMed] [Google Scholar]
- Dallavalle S, Pisano C, Zunino F. (2012). Development and therapeutic impact of HDAC6-selective inhibitors. Biochemical Pharmacology. 84, 756–65. [DOI] [PubMed] [Google Scholar]
- Denu JM (2005). The Sir 2 family of protein deacetylases. Current Opinion in Chemical Biology. 9, 431–40. [DOI] [PubMed] [Google Scholar]
- Didonna A, Opal P. (2015). The promise and perils of HDAC inhibitors in neurodegeneration. Annals of Clinical and Translational Neurology. 2, 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA (2007). Histone deacetylase inhibitors: overview and perspectives. Molecular Cancer Research. 5, 981–9. [DOI] [PubMed] [Google Scholar]
- Drazic A, Myklebust LM, Ree R, Arnesen T. (2016). Biochimica et Biophysica Acta. The world of protein acetylation. 1864, 1372–401. [DOI] [PubMed] [Google Scholar]
- Eckschlager T, Plch J, Stiborova M, Hrabeta J. (2017). Histone deacetylase inhibitors as anticancer drugs. International Journal of Molecular Sciences. 18, 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt SL, Gattis SG, Fierke CA (2006). Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 45, 6170–8. [DOI] [PubMed] [Google Scholar]
- Gao L, and Alumkal J. (2010). Epigenetic regulation of androgen receptor signaling in prostate cancer. Epigenetics. 5, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV (2004). Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of Molecular Biology. 338, 17–31. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL (1999). Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proceedings of the National Academy of Sciences of the United States of America. 96, 4868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997). Histone acetylation in chromatin structure and transcription. Nature. 389, 349–52. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL (2003). Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proceedings of the National Academy of Sciences of the UnitedStates of America. 100, 4389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai Y, and Christianson DW (2016). Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nature Chemical Biology. 12, 741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai Y, Shinsky SA, Porter NJ, Christianson DW (2017). Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nature Communications. 8, 15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002). HDAC6 is a microtubule-associated deacetylase. Nature. 417,455–8. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000). Acetylation: a regulatory modification to rival phosphorylation? EMBO Journal. 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucker TM, Nomura Y, Kim JH, Bhatta A, Wang V, Wecker A, Jandu S, Santhanam L, Berkowitz D, Romer L, Pandey D. (2017). Cystathionine γ-lyase protects vascular endothelium: a role for inhibition of histone deacetylase 6. American Journal of Physiology-Heart and Circulatory Physiology. 312, H711–H720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi PM, Cole KA, Dowling DP, Christianson DW (2011). Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Current Opinion in Structural Biology. 21, 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopéz JE, Sullivan ED, and Fierke CA (2016). Metal-dependent deacetylases: cancer and epigenetic regulators. ACS Chemical Biology. 11, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Luo Y, Wang Y, Liao C, Ye WC, Jiang S. (2016). Selective histone deacetylase inhibitors with anticancer activity. Current Topics in Medicinal Chemistry. 16, 415–26. [DOI] [PubMed] [Google Scholar]
- Mackwitz MKW, Hamacher A, Osko JD, Held J, Schöler A, Christianson DW, Kassack MU, Hansen FK (2018). Multicomponent synthesis and binding mode of imidazo[1,2-a]pyridine-capped selective HDAC6 inhibitors. Organic Letters. 20, 3255–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, and Zhou MM (2014). Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor Perspectives in Biology. 6, a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough CE, and Marmorstein R. (2016). Molecular basis for histone acetyltransferase regulation by binding partners, associated domains, and autoacetylation. ACS Chemical Biology. 11, 632–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, Melancon BJ, Helquist P, Gut H, Matthias P. (2016). Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nature Chemical Biology. 12, 748–54. [DOI] [PubMed] [Google Scholar]
- Mottama M, Zheng S, Huang TL, Wang G. (2015). Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 20, 3898–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, and McMahon SB (2010). Cell biology: Rise of the rival. Science. 327, 964–5. [DOI] [PubMed] [Google Scholar]
- Parra M, and Verdin E. (2010). Regulatory signal transduction pathways for class IIa histone deacetylases. Current Opinions in Pharmacology. 10, 454–460. [DOI] [PubMed] [Google Scholar]
- Porter NJ, Mahendran A, Breslow R, Christianson DW (2017). Unusual zinc binding mode of HDAC6-selective hydroxamate inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 114, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NJ, Osko JD, Diedrich D, Kurz T, Hooker JM, Hansen FK, Christianson DW (2018a). Histone deacetylase 6-selective inhibitors and the influence of capping groups on hydroxamate-zinc denticity. Journal of Medicinal Chemistry. 61, 8054–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NJ, Shen S, Barinka C, Kozikowski AP, Christianson DW (2018b). Molecular basis for the selective inhibition of histone deacetylase 6 by a mercaptoacetamide inhibitor. ACS Medicinal Chemistry Letters. 9, 1301–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NJ, Wagner FF, and Christianson DW (2018c). Entropy as a driver of selectivity for inhibitor binding to histone deacetylase 6. Biochemistry. 57, 3916–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Denu JM, and Allis CD (2001). Histone acetyltransferases. Annual Review of Biochemistry. 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Scolnick LR, Clements AM, Liao J, Crenshaw L, Hellberg M, May J, Dean TR, Christianson DW (1997). Novel binding mode of hydroxamate inhibitors to human carbonic anhydrase II. Journal of the American Chemical Society. 119, 850–851. [Google Scholar]
- Seidel C, Schnekenburger M, Dicato M, Diederich M. (2015). Histone deacetylase 6 in health and disease. Epigenomics. 7, 103–18. [DOI] [PubMed] [Google Scholar]
- Su H, Altucci L, and You Q. (2008). Competitive or noncompetitive, that’s the question: research toward histone deacetylase inhibitors. Molecular Cancer Therapeutics. 7,1007–12. [DOI] [PubMed] [Google Scholar]
- Szyk A, Deaconesu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, Roll-Mecak A. (2014). Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 157, 1405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, and Khochbin S. (1999). Identification of a new family of higher eukaryotic histone deacetylases Coordinate expression of differentiation-dependent chromatin modifiers. Journal of Biological Chemistry. 274, 2440–2445. [DOI] [PubMed] [Google Scholar]
- Verdin E, and Ott M. (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nature Reviews Molecular Cell Biology. 16, 258–64. [DOI] [PubMed] [Google Scholar]
- Villagra A, Sotomayor EM, and Seto E. (2010). Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 29, 157–73. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 327, 1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, and Johnstone RW (2014). New and emerging HDAC inhibitors for cancer treatment. Journal of Clinical Investigation. 124, 30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Deubzer HF, Milde T, Oehme I. (2009). HDAC family: What are the cancer relevant targets? Cancer Letters. 277, 8–21. [DOI] [PubMed] [Google Scholar]
- Yuan H, and Marmorstein R. (2012). Structural basis for sirtuin activity and inhibition. Journal of Biological Chemistry. 287, 42428–42435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gilquin B, Khochbin S, Matthias P. (2006). Two catalytic domains are required for protein deacetylation. Journal of Biological Chemistry. 281, 201–4. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xiang S, Jo HY, Wang L, Williams KA, Liu W, Hu C, Tong D, Haakenson J, Wang C, Zhang S, Pavlovicz RE, Jones A, Schmidt KH, Tang J, Dong H, Shan B, Fang B, Radhakrishnan R, Glazer PM, Matthias P, Koomen J, Seto E, Bepler G, Nicosia SV, Chen J, Li C, Gu L, Li GM, Bai W, Wang H, Zhang X. (2014). HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levelts of MutSα. Molecular Cell. 55, 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL (2010). Regulation of cellular metabolism by protein lysine acetylation. Science. 327, 1000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Wu Y, Navre M, Sang BC (2006). Characterization of the two catalytic domains in histone deacetylase 6. Biochemical and Biophysical Research Communications. 341, 45–50 [DOI] [PubMed] [Google Scholar]