Abstract
Two new siderophores, madurastatin D1 and D2, together with (−)-madurastatin C1, the enantiomer of a known compound, were isolated from marine-derived Actinomadura sp. The presence of an unusual 4-imidazolidinone ring in madurastatins D1 and D2 inspired us to sequence the Actinomadura sp. genome and to identify the mad biosynthetic gene cluster, knowledge of which enables us to now propose a biosynthetic pathway. Madurastatin D1 and D2 are moderately active in antimicrobial assays with M. luteus.
Graphical Abstract
Siderophores represent a crucial class of bacterial secondary metabolites that are employed as iron chelators by microorganisms to facilitate the absorption of poorly soluble environmental iron.1 Siderophores feature high binding affinities toward Fe(III) and often contain multiple N, O functionalities1. They can also be employed by pathogenic bacteria to acquire iron, the fundamental limiting nutrient for life, from their host organisms and influence the cellular iron level of the hosts.2
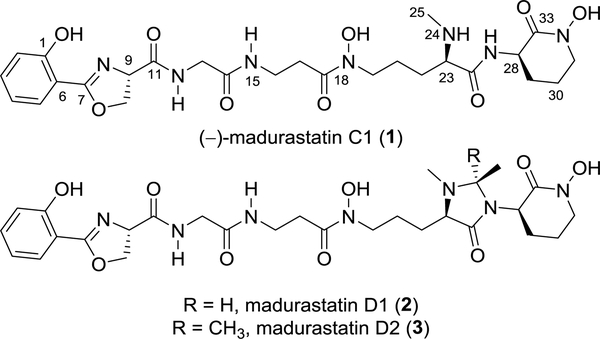
The bacterial genus Actinomadura from the phylum Actinobacteria has been reported to produce a series of phenolate-hydroxamate siderophores,2 including madurastatins,3–5 and maduraferrin.6 The madurastatins were first characterized as aziridine-containing pentapeptide siderophores; however, structural revision on the aziridine ring was suggested by Thorson and Shaaban7 based on their analyses of siderophores with 2-(2-hydroxyphenyl)oxazoline moieties. NMR data of the isolated madurastatin C1 (also designated as MBJ-0034) from bacteria cell culture and synthetic analogs with aziridine- and 2-(2-hydroxyphenyl)oxazoline moieties were compared by Hall in 2017,8 which confirmed the structural revision proposed by Thorson and Shaaban. The absolute configuration of madurastatin C1 (1) (Figure 1) was also established by Hall.8
Figure 1.
Structures of madurastatin C1 (1), D1 (2) and D2 (3).
As part of our goal to discover new natural products from marine invertebrate associated bacteria9–11, our attention was drawn to strain WMMA-1423, a marine Actinomadura sp. cultivated from the sponge Tedania sp., after its crude extract was found to inhibit the growth of both MRSA and Bacillus subtillis on agar plates. We isolated two new madurastatins, madurastatin D1 (2) and D2 (3) (Figure 1), as well as the enantiomer [1, (−)-madurastatin C1] of the known compound madurastatin C1 by bioassay guided isolation using MRSA (ATCC #33591) as the test strain. Though only slightly active against MRSA (vide infra), fractions containing 1–3 were sufficiently active in agar plate assays as to peak our interest. Madurastatin D1 (2) and D2 (3) feature the cyclic 4-imidazolidinone in contrast to the linear α-amino amide in madurastatin C1. Both 2 and 3 exhibited lower MICs against M. luteus than 1, suggesting that the additional heterocycle increases antibacterial activity. Due to the unusual cyclization bridging two nitrogens, whole genome sequencing of strain WMMA-1423 was conducted and a biosynthetic pathway to madurastatins D1 and D2 was proposed.
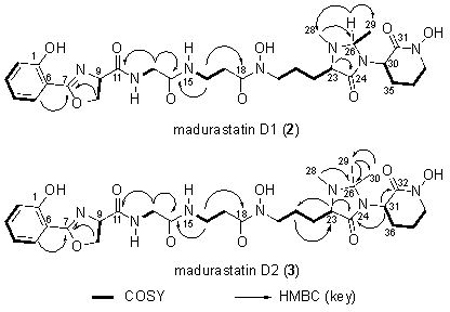
HRMS data suggested the molecular formula of C28H39N7O9 (m/z = 618.2876, [M+H]+) for madurastatin D1 (2). Since Fe has a unique isotope distribution (54Fe:56Fe:57Fe ≈ 6:92:2), the observation of Fe(III) adduct ions [M-2H+Fe]+ (m/z = 671.1991, Figure S14) was a clear indicator of compound 2’s ability of binding iron and further confirmation with regard to the assigned molecular ion. Compared to the known madurastatin C1, 2 had one additional degree of unsaturation. By comparing of 1H and 13C NMR data (Table 1, Figure S3−S7) between the reported madurastatin C18 and 2, The tertiary carbon C-26 (74.7 ppm) and the methyl group carbon C-29 (19.7 ppm) were observed only in 2, which have HSQC correlation to H-26 (4.01 ppm) and H-29 (1.21 ppm), respectively. H-26 showed COSY correlations to H-29, and it also showed HMBC correlations to C-28 (37.9 ppm, N-Me group attached to N-25) and to C-29, which suggested the connection between N-25 and C-26. In light of these data, the structure of 2 was assigned as containing the 1, 2-dimethyl-4-imidazolidinone cyclic structure.
Table 1.
1H and 13C NMR data for 2 and 3 (600 MHz for 1H, 125 MHz for 13C, d6-DMSO)
 | ||||
|---|---|---|---|---|
| Posi tion |
2 | 3 | ||
| δC, type | δH, (J in Hz) | δC, type | δH, (J in Hz) | |
| 1 | 159.1 | 159.1 | ||
| 2 | 116.7, CH | 7.01, d (8.4) | 116.7, CH | 6.98, d (7.6) |
| 3 | 134.1, CH | 7.47, ddd (8.4, 7.3, 1.6) | 134.1, CH | 7.45, t (7.6) |
| 4 | 119.1, CH | 6.95, t (7.5) | 119.0, CH | 6.91, t (7.6) |
| 5 | 128.1, CH | 7.64, dd (7.6, 1.4) | 128.1, CH | 7.63, d (7.4) |
| 6 | 110.0 | 110.0 | ||
| 7 | 165.9 | 165.9 | ||
| 9 | 67.5, CH | 5.01, dd (10.4, 7.7) | 67.4, CH | 5.00, dd (10.2. 8.0) |
| 10 | 69.5, CH2 | 4.65, dd (10.3, 8.5) 4.51, t (8.0) |
69.4, CH2 | 4.63, dd (10.2, 8.1) 4.51, t (8.3) |
| 11 | 170.2 | 170.2 | ||
| 12 | 8.61, t (5.9) | 8.56, s | ||
| 13 | 42.2, CH2 | 3.73, dd (16.6, 6.0) 3.67, dd (16.6, 6.0) |
42.2, CH2 | 3.75, d (16.0) 3.66, d (16.0) |
| 14 | 168.4 | 168.4 | ||
| 15 | 8.01, t (5.4) | 7.97, s | ||
| 16 | 34.7, CH2 | 3.24, dd (6.6, 6.0) | 34.8, CH2 | 3.25, dd (7.0, 5.6) |
| 17 | 31.9, CH2 | 2.56–2.48, m | 31.7, CH2 | 2.50–2.55, m |
| 18 | 170.8 | 170.6 | ||
| 20 | 47.4, CH2 | 3.51–3.42, m | 51.1, CH2 | 3.41–3.53, m |
| 21 | 21.6, CH2 | 1.70–1.43, m | 20.9, CH2 | 1.36–1.46, m |
| 22 | 26.6, CH2 | 1.53–1.43, m 1.98–1.82, m |
25.0, CH2 | 1.85–1.94, m |
| 23 | 64.8, CH | 2.88, m | 62.0, CH | 2.93, t (5.0) |
| 24 | 171.6 | 169.9 | ||
| 26 | 74.7, CH | 4.01, q (5.0) | 77.9 | |
| 28 | 37.9, CH3 | 2.29, s | 32.5, CH3 | 2.24, s |
| 29 | 19.7, CH3 | 1.21, d (5.2) | 25.7, CH3 | 1.29, s |
| 30 | 51.8, CH | 4.32–4.24, m | 19.8, CH3 | 1.11, s |
| 31 | 162.4 | 51.7, CH | 3.91, dd (6.0, 5.4) | |
| 32 | 163.3 | |||
| 33 | 51.1, CH2 | 3.51–3.57, m 3.41–3.46, m |
||
| 34 | 21.6, CH2 | 1.87–1.79, m | 47.5, CH2 | 3.43–3.47, m |
| 35 | 21.3, CH2 | 2.03–1.95, m | 21.3, CH2 |
1.67–1.73, m 1.84–1.93, m |
| 36 | 25.5, CH2 | 2.34–2.42, m 1.67–1.73, m |
||
A molecular formula of C29H41N7O9 (m/z = 632.3025, [M+H]+) was suggested by HRMS data for madurastatin D2 (3). Fe(III) adduct ions [M+Fe+Na-3H]+ (m/z = 707.1941) were also observed (Figure S15). The NMR data (Table 1, Figures S8–S12) of 3 was compared to 2 and, since these two compounds have the same number of degrees of unsaturation, a similar 4-imidazolidinone cyclic structure was also proposed for 3 despite a few differences. The major difference was that the tertiary C-26 carbon in 2 was replaced by a tetrasubstituted C-26 (77.9 ppm) in 3. Two methyl groups (C-29 and C-30) showed HMBC correlations to C-26 and to each other as well, suggesting shared connectivity to the C-26. These two methyl groups also showed different 13C chemical shifts (25.7 ppm for C-29, 19.8 ppm for C-30) and 1H chemical shifts (1.29 for H-29, 1.11 for H-30), which suggested both of them are connected to a carbon that belongs to a cyclic system. Thus, the structure of 3 was proposed with a similar 1, 2, 2-trimethyl-4-imidazolidinone cyclic structure. Interestingly, the difference in methylation status at C-26 was found to induce slight, but noticeable, differences in the 13C shifts for the N-hydroxy lactams of compounds 2 and 3 as well as a ~ 5 ppm difference in the 13C shifts observed at C-28; these are likely attributable to conformational restrictions encountered by 3 but not 2.
The siderophoric properties of compounds 1–3 were assessed on the basis of the chrome azurol S (CAS) assay (Figure S16).9 Deferoxamine mesylate was also analyzed and served as a positive Fe(III) binding control. The colorimetric CAS assay revealed that 1 and 2 bind iron with efficiencies comparable to deferoxamine mesylate whereas compound 3, bearing the geminal-dimethyl moiety also bound iron but with noticeably impaired affinity relative to 1 and 2. This trend in iron binding supports the notion that C-26 dimethylation imparts conformational limitations to 3 that are lacking in compounds 1 and 2.
The absolute configuration of madurastatin C1 was determined by Hall and coworkers8 using the Marfey’s method (Figure S17).12 The 13C chemical shifts (Figure S2) of 1 were identical to those reported for madurastatin C1 although the optical rotation of 1 was found to be inverted from that of madurastatin C1. The C-9 configuration of madurastatin C1 was shown to be 9R by Marfey’s method.8,12 Thus, we determined that 1 is the enantiomer of madurastatin C1 and has stereocenter assignments as 9S, 23R, 28R. Notably, enantiomeric natural product pairings are well known and have been rigorously reviewed.13 Since 2 and 3 were produced by the same organism, both compounds were proposed to have the same configuration as found in 1 [9S, 23R, 30R (31R for 3)]. Further efforts to determine the configuration of C-26 in 2 employed molecular modeling and density functional theory (DFT) NMR calculations of the two models (23R, 26R, 30R- and 23R, 26S, 30R-). Spartan 14 (v.1.1.7 Wavefunction Inc. 2014) was used to identify the conformer Boltzmann distribution for each diastereoisomer using molecular mechanics. Gaussian 09 was then used to optimize the local geometry of the low energy conformers based on DFT energy calculation (B3LYP/6–31G(d,p)), and the NMR chemical shifts of different stereoisomers were calculated using GIAO method (Table S1). NMR chemical shifts were referenced to TMS and benzene using the multistandard method.14 The DP4 probability method15 was used to compare the calculated 13C NMR chemical shifts of the two models with the experimental 13C chemical shifts and yielded a 100% probability for the R compared to the S configuration (Table S2).
Previous reports have shown that madurastatin C1 inhibits the growth of M. luteus.4 Therefore, compounds 1–3 were tested for antibacterial activity against M. luteus. Notably, compounds 2 and 3 both inhibited M. luteus with MICs of 25.3 and 25.8 μM, respectively; the MIC of compound 1, devoid of the imidazolidinone, was 108.1 μM. Thus, the 4-imidazolidinone clearly benefits the antibacterial activity of these agents although efforts to elucidate the specific mechanisms at play are beyond the scope of this report. Noteworthy is that, although MRSA proved important during bioassay-guided fractionation, none of the new madurastatins inhibited MRSA with MICs less than 100 μM.
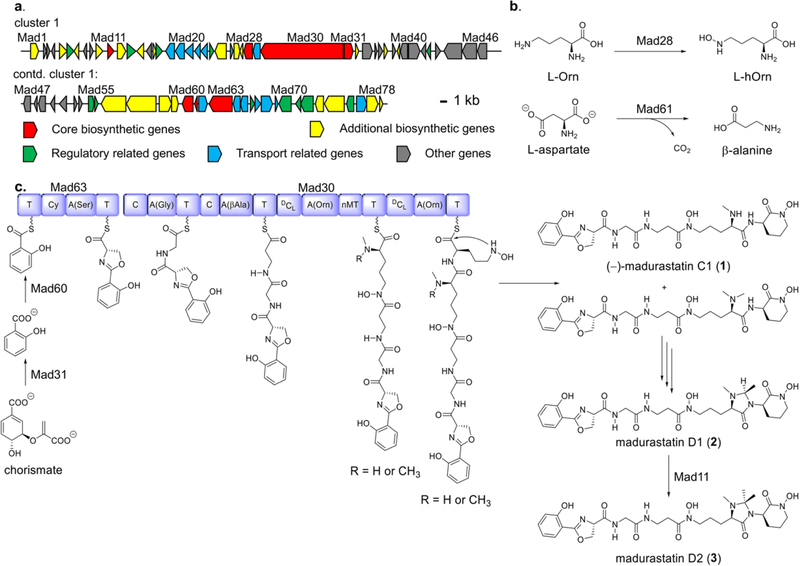
To identify the biosynthetic cluster for madurastatin D1 (1) biosynthesis, the whole genome of Actinomadura sp. WMMA-1423 was sequenced (GenBank accession number CP041244) and analyzed using antiSMASH (version 5.0.0 rc1).16–17 Two gene clusters (57.9 kb and 46.7 kb, respectively) were identified as the nonribosomal peptide synthetases (NRPSs), transport and regulatory genes for (−)-madurastatin C1 (1), madurastatin D1 (2) and madurastatin D2 (3) production. Although antiSMASH identified these as separate clusters, they were found to be co-localized in the genome (12.1 kb apart from each other) and thus, represent one cluster. The architecture and annotation of the madurastatin (mad) biosynthetic gene cluster shown in Figure 2 and Table S3. The mad cluster consists of two NRPS genes (Figure 2a, red, mad30, mad63), two chain initiation genes (red, mad31, mad60), two amino acid tailoring genes (red, mad28, mad61), one S-adenosyl-methionine (SAM)-dependent methyltransferase gene (red, mad11) as well as additional biosynthetic (yellow), transport (teal), regulation-related (green) and other genes (grey); full annotations are provided in Table S3). Most of the proteins encoded in the mad cluster are homologous to those involved in cahuitamycin,18 albachelin,19 and amychelin20 biosynthetic pathways. The genome analysis has allowed us to put forth a hypothesis with regard to the possible biosynthesis of madurastatins, where a SAM-dependent mechanism is involved in the biosynthesis of madurastatin D2 (Figure 2c, Table S3). Further biosynthetic studies to more completely understand the assembly of 1–3, with special emphasis on the unique imidazolidinone of 2 and 3, are clearly warranted.
Figure 2.
(a) Organization of mad cluster. (b) Proposed functions of Mad28 and Mad61 as building block producers, and (c), Proposed biosynthetic pathway to 1–3. Domain annotations: A, adenylation; C, condensation; Cy, cyclization; DCL, epimerization/condensation; T, thiolation; nMT, N-methylation.
Mad28, Mad61 are likely ornithine N-monooxygenase and aspartate 1-decarboxylase enzymes, responsible for generating the l-N-OH-Orn (l-hOrn) and β-alanine moieties, respectively. Mad31 is homologous to salicylate synthase AmcL in amychelin biosynthesis (59% identity) and Mad60 is homologous to the salicylate-AMP ligase CahJ (67% identity) in cahuitamycin biosynthesis. Together, Mad31 and Mad60 likely catalyze the formation of a Mad63-salicylate conjugate starting from chorismate. Mad63 contains one set of C, A, T domains and very likely installs the oxazoline ring since it shows 58% identity to CahA; CahA contains a cyclization domain that creates the oxazoline ring in cahuitamycins. We envision that the growing chain is further extended by Mad30, which contains four sets of C, A, T domains and one N-methylation (nMT) domain; antiSMASH analysis revealed that two of the C domains function as DCL domains (Fig. 2c). The A domains in Mad30 are proposed to activate Gly, β-Ala and l-hOrn substrates. We envision that epimerization of each l-hOrn, accounting ultimately for the R stereocenters found in 1–3, likely results from the dual epimerase/condensation activities of the DCL domains.21
The nMT domain located between the third and fourth A domains is envisioned to catalyze both mono- and dimethylation of the α-amine group of the initially added d-hOrn. Both mono- and dimethylated intermediates would undergo further condensation to add on the final hOrn unit. Intramolecular nucleophilic substitution is then proposed to install the N-hydroxy lactam (with liberation from Mad30) to afford (−)-madurastatin C1 (1) and the dimethylated variant of 1; both are possible intermediates en route to madurastatin D1 (2) although we favor the dimethylated species based on its complete absence during the course of compound isolations. We envision that N-Me oxidation may play a central role in imidazolidinone assembly, from the dimethylated version of 1, although it is not yet clear which gene/s in the mad cluster would code for this chemistry (Figure 2c). In proceeding from 2 to 3, we invoke the SAM-dependent methyltransferase Mad11; SAM-dependent methylation at both sp2 and sp3 carbon adjacent to heteroatoms are well known.22–25 At present, Mad11 appears to be the most likely candidate for converting monomethylated 2 into dimethylated madurastatin D2 (3).
In summary, we report the isolation and structural elucidation of madurastatin D1 (2) and D2 (3), two new madurastatin siderophores that showed in vitro activity against M. luteus. To the best of our knowledge, only five other madurastatin analogs3–5 have been isolated from Actinomadura sp., and none of them contain the 4-imidazolidinone cyclic moiety. The biosynthetic origins for this unique structure are currently unclear although we hypothesize that a combination of methyltransferase and oxidase chemistries may prove central. While scientifically intriguing, further studies will be required to test this biosynthetic hypothesis.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from the University of Wisconsin-Madison School of Pharmacy and the Graduate School at the University of Wisconsin. This work was also funded by the NIH Grant U19AI109673 and U19AI142720. We would like to thank the Analytical Instrumentation Center at the School of Pharmacy, University of Wisconsin-Madison for the facilities to acquire spectroscopic data. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supporting Information (PDF)
The authors declare no conflict of interest.
REFERENCES
- (1).Hider RC; Kong X Nat. Prod. Rep. 2010, 27, 637–657. [DOI] [PubMed] [Google Scholar]
- (2).Miethke M; Marahiel MA Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Harada K; Tomita K; Fujii K; Masuda K; Mikami Y; Yazawa K; Komaki HJ Antibiot. 2004, 57, 125–135. [DOI] [PubMed] [Google Scholar]
- (4).Mazzei E; Iorio M; Maffioli SI; Sosio M; Donadio SJ Antibiot. 2012, 65, 267–269. [DOI] [PubMed] [Google Scholar]
- (5).Kawahara T; Itoh M; Izumikawa M; Sakata N; Tsuchida T; Shin-ya KJ Antibiot. 2014, 67, 577–580. [DOI] [PubMed] [Google Scholar]
- (6).Keller-Schierlein W; Hagmann L; Zähner H; Huhn W Helv. Chim. Acta 1988, 71, 1528–1540. [Google Scholar]
- (7).Shaaban KA; Saunders MA; Zhang Y; Tran T; Elshahawi SI; Ponomareva LV; Wang X; Zhang J; Copley GC; Sunkara M; Kharel MK; Morris AJ; Hower JC; Tremblay MS; Prendergast MA; Thorson JS J. Nat. Prod. 2017, 80, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tyler AR; Mosaei H; Morton S; Waddell PG; Wills C; McFarlane W; Gray J; Goodfellow M; Errington J; Allenby N; Zenkin N; Hall MJ Nat. Prod. 2017, 80, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhang F; Barns K; Hoffmann FM; Braun DR; Andes DR; Bugni TS J. Nat. Prod. 2017, 80, 2551–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wyche TP; Standiford M; Hou Y; Braun D; Johnson DA; Johnson JA; Bugni TS Mar. Drugs 2013, 11, 5089–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wyche TP; Piotrowski JS; Hou Y; Braun D; Deshpande R; Mcllwain S; Ong IM; Myers CL; Guzei IA; Westler WM; Andes DR; Bugni TS Angew. Chem. Int. Ed. 2014, 53, 11583–11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Marfey P Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar]
- (13).Finefield JM; Sherman DH; Kreitman M; Williams RM Angew. Chem. Int. Ed. 2012, 51, 4802–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sarotti AM; Pellegrinet SC J. Org. Chem. 2009, 74, 7254–7260. [DOI] [PubMed] [Google Scholar]
- (15).Smith SG; Goodman JM J. Am. Chem. Soc. 2010, 132, 12946–12959. [DOI] [PubMed] [Google Scholar]
- (16).Blin K; Shaw S; Steinke K; Villebro R; Ziemert N; Lee SY; Medema MH; Weber T Nucleic Acids Res. 2019, 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Blin K; Wolf T; Chevrette MG; Lu X; Schwalen CJ; Kautsar SA; Duran HGS; de los Santos ELC; Kim HU; Nave M; Dickschat JS; Mitchell DA; Shelest E; Breitling R; Takano E; Lee SY; Weber T; Medema MH Nucleic Acids Research, 2017, 45, W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Park SR; Tripathi A; Wu J; Schultz PJ; Yim I; McQuade TJ; Yu F; Arevang C–J; Mensah AY; Tamayo-Castillo G; Xi C; Sherman DH Nat. Commun. 2016, 7:10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kodani S; Komaki H; Suzuki M; Hemmi H; Ohnishi-Kameyama M Biometals, 2015, 28, 381–389. [DOI] [PubMed] [Google Scholar]
- (20).Seyedsayamdost MR; Traxler MF; Zheng S–L; Kolter R; Clardy J J. Am. Chem. Soc. 2011, 133, 11434–11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rausch C; Hoof I; Weber T; Wohlleben W; Huson DH BMC Evol. Biol. 2007, 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Grove TL; Benner JS; Radle MI; Ahlum JH; Landgraf BJ; Krebs C; Booker SJ Science 2011, 332, 604–607. [DOI] [PubMed] [Google Scholar]
- (23).Buckel W; Thauer RK Angew. Chem. Int. Ed. 2011, 50, 10492–10494. [DOI] [PubMed] [Google Scholar]
- (24).Kim HJ; McCarty RM; Ogasawara Y; Liu Y–N; Mansoorabadi SO; LeVieux J Liu H–W J. Am. Chem. Soc. 2013, 135, 8093–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Woodyer RD; Li G; Zhao H; van der Donk WA Chem. Commun. 2007, 28, 359–361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.