Abstract
Mass spectrometry imaging (MSI) enables label-free spatial mapping of hundreds of biomolecules in tissue sections. This capability provides valuable information on tissue heterogeneity that is difficult to obtain using population-averaged assays. Despite substantial developments in both instrumentation and methodology, MSI of tissue samples at single-cell resolution remains challenging. Herein, we describe a protocol for robust imaging of tissue sections with a high (better than 10-μm) spatial resolution using nanospray desorption electrospray ionization (nano-DESI) mass spectrometry, an ambient ionization technique that does not require sample pretreatment before analysis. In this protocol, mouse uterine tissue is used as a model system to illustrate both the workflow and data obtained in these experiments. We provide a detailed description of the nano-DESI MSI platform, fabrication of the nano-DESI and shear force probes, shear force microscopy experiments, spectral acquisition, and data processing. A properly trained researcher (e.g., technician, graduate student, or postdoc) can complete all the steps from probe fabrication to data acquisition and processing within a single day. We also describe a new strategy for acquiring both positive- and negative-mode imaging data in the same experiment. This is achieved by alternating between positive and negative data acquisition modes during consecutive line scans. Using our imaging approach, hundreds of high-quality ion images were obtained from a single uterine section. This protocol enables sensitive and quantitative imaging of lipids and metabolites in heterogeneous tissue sections with high spatial resolution, which is critical to understanding biochemical processes occurring in biological tissues.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Introduction
MSI enables label-free spatial mapping of molecules directly from the surface of tissue samples, which makes it a powerful analytical technique for applications in biology, clinical research, and drug discovery1–8. Unlike population-averaged assays such as liquid chromatography–mass spectrometry (LC–MS), MSI provides insights into tissue heterogeneity, thereby addressing one of the central challenges in biology. A major goal in this field is to image biological tissues at single-cell resolution. Although MALDI MSI has been performed with a spatial resolution down to ~1–2 μm (refs.9,10), imaging at a resolution of better than 10 μm using ambient ionization techniques, which do not require any special sample pretreatment, is still in its infancy. The experimental protocol for high-resolution MSI of tissue sections using the ambient nano-DESI technique described herein summarizes key steps and challenges associated with high-resolution nano-DESI MSI experiments.
Development of the protocol
Nano-DESI MSI is an ambient ionization technique based on localized liquid extraction, which enables sensitive and quantitative analysis of molecules from a sample and does not require any special sample pretreatment. This technique has been widely used for imaging of biological samples ranging from animal and human tissues to living microbial colonies on agar plates11–20. In nano-DESI MSI, the spatial resolution is determined by the size of the liquid bridge formed between two glass capillaries, which is controlled by the size of the capillaries, their position, and the flow rate11,16,17. The capillaries constitute a proximity probe (nano-DESI probe), which must be positioned at a controlled distance above the sample surface to prevent the probe from either crashing or losing contact with the sample. In particular, high spatial resolution nano-DESI MSI requires automated and precise control of the distance between the nano-DESI probe and the sample throughout the imaging experiment.
We have developed a strategy for controlling the probe-to-sample distance using a three-point plane mode, in which the vertical position of the sample is automatically adjusted to maintain a constant probe-to-sample distance based on the defined tilt of the flat sample surface12. This approach enables automated imaging of relatively flat tissues (e.g., brain tissue) with a spatial resolution of 20–100 μm. A different approach has been developed for nano-DESI MSI experiments with a spatial resolution of better than 10 μm. In particular, we have demonstrated that the distance between the sample and nano-DESI probe can be precisely controlled to within 1 μm by using shear force microscopy20. Proof-of-concept experiments performed using this approach demonstrated imaging of lung and brain tissue sections with a spatial resolution of better than 10 μm (ref.15). In another study, we used this approach to obtain first insights into the localization of lipids and metabolites in pancreatic islets and surrounding exocrine tissue16. That study demonstrated reproducible and robust high-resolution imaging of tissue samples using nano-DESI MSI.
Two major improvements over the previously published protocol are reported herein21. First, the nano-DESI probe is assembled using two finely pulled capillaries. A primary capillary pulled to an o. d. of ~15 μm continuously delivers the working solvent to the sample. Meanwhile, a pulled nanospray capillary with a tip o.d. of ~25 μm delivers molecules extracted from the sample to a mass spectrometer inlet and ionizes them by electrospray ionization. To avoid clogging of the capillaries, we optimized their fabrication process, which allowed us to use the pulled capillaries continuously for more than a week. This is particularly important for imaging tissue sections with high throughput and spatial resolution. Second, shear force microscopy is combined with nano-DESI MSI to ensure control of the distance between the nano-DESI probe and sample to within 1 μm. This approach is critical to imaging of both flat and rough tissues with high spatial resolution.
Overview of the procedure
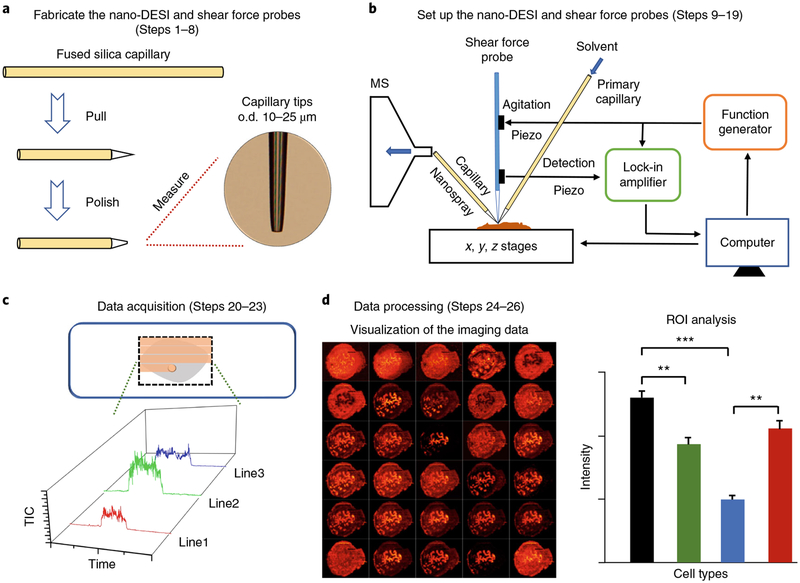
In this protocol, we describe a workflow for robust imaging of tissue sections with a spatial resolution of better than 10 μm using nano-DESI MSI (Fig. 1). Specifically, a high-resolution nano-DESI probe comprises two finely pulled capillaries with o.d. of 15–25 μm, and an additional capillary pulled to an o.d. of 10 μm is used as a shear force probe. The key steps for fabricating the primary capillary and nanospray capillary (Steps 1–8) are illustrated in Fig. 2a,b. The nano-DESI probe is positioned in front of a mass spectrometer inlet and used to extract and ionize endogenous molecules from the sample. The small size of the liquid bridge can be obtained by appropriately adjusting the relative positions of the primary and nanospray capillaries, as well as the position of the nanospray capillary relative to the mass spectrometer inlet (Steps 9–13), which is necessary to achieve a resolution of better than 10 μm.
Fig. 1 |. Overall workflow for high-resolution imaging of biological tissues using nano-DESI MSI.
a, The nano-DESI and shear force probes are fabricated by pulling and polishing the capillary tips to a final size (Steps 1–8). b, Schematic drawing of the nano-DESI MSI source. The nano-DESI probe composed of the primary and nanospray capillaries is positioned in front of the mass spectrometer (MS) inlet, and the shear force probe positioned in close proximity to the nano-DESI probe is used to maintain a constant probe-to-sample distance (Steps 9–19). b adapted with permission from Nguyen, S. N., Liyu, A. V., Chu, R. K., Anderton, C. R. & Laskin, J. Constant-distance mode nanospray desorption electrospray ionization mass spectrometry imaging of biological samples with complex topography. Anal. Chem. 89, 1131–1137, copyright (2016) American Chemical Society. c, Nano-DESI MSI data are acquired by scanning the tissue with the nano-DESI probe in lines shown as orange highlights in the inset (Steps 20–23). d, The imaging data are visualized and processed using MSI QuickView (Steps 24–26). **P < 0.01; ***P < 0.001.
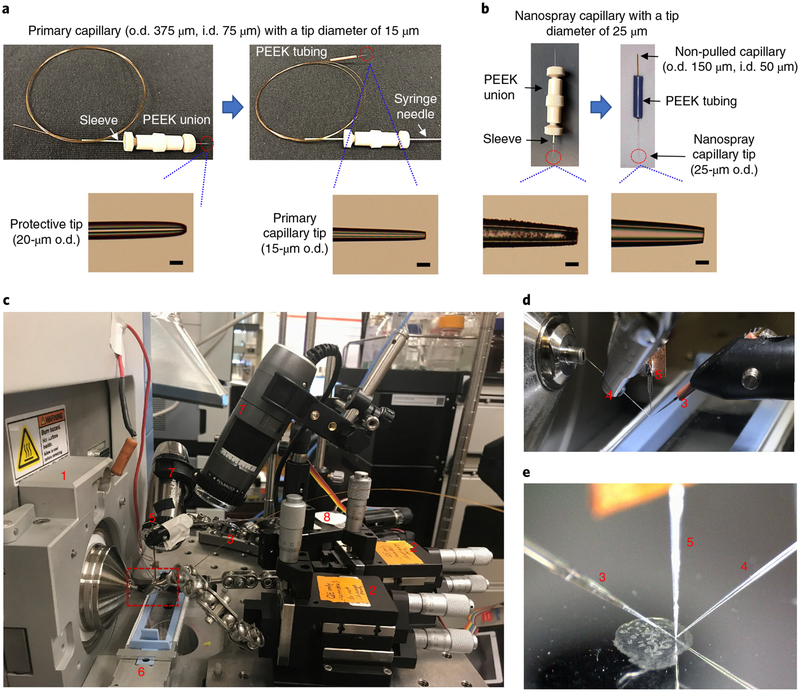
Fig. 2 |. Platform for high-resolution imaging of biological tissues using nano-DESI MSI.
a, Fabrication of the primary capillary. Top panel shows the key stages for fabricating the primary capillary. Bottom panels show photos of the pulled tips. Scale bars, 25 μm. b, Fabrication of the nanospray capillary. Top panel shows the key stages for fabricating the nanospray capillary. Bottom panels show photos of the pulled tips. Scale bars, 25 μm. c, Photo of the imaging platform, highlighting (1), instrument mounting flange; (2), xyz 500 inline micro-positioner; (3), primary capillary; (4), nanospray capillary; (5), shear force probe; (6), sample holder; (7), Dino-Lite microscope; and (8), motorized micro-positioner (for shear force probe). d, Zoomed-in picture of the probes, corresponding to the red dashed box in c. e, A photo of the probes and sample recorded by a side-view microscope during the imaging experiment.
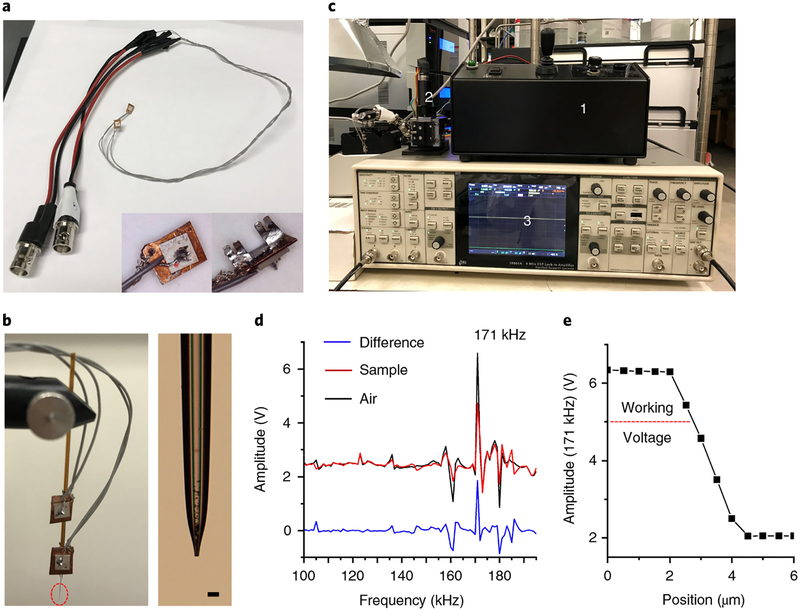
The shear force probe positioned in close proximity to the nano-DESI probe is used to maintain a constant distance between the nano-DESI probe and the sample throughout the imaging experiment (Steps 14–19). The experimental setup of the constant-distance-mode nano-DESI MSI is illustrated in Fig. 2c–e, and the main components of the shear force microscopy are shown in Fig. 3. Shear force microscopy enables measurement of the topography of the sample using a sharp oscillating probe22,23. In our approach, this technique is used to maintain a constant probe-to-sample distance15,16,20. Specifically, two piezoelectric ceramic plates are attached to the shear force probe (Fig. 3a–c). The upper plate (agitation piezo) is connected to a function generator and used to induce the probe oscillation. The lower plate (detection piezo), positioned closer to the sample, is connected to a lock-in amplifier to detect the amplitude of the shear force probe vibration at a selected frequency. Shear force microscopy experiments are performed by monitoring the amplitude of a selected resonant probe oscillation that is strongly affected by shear forces between the sample and the probe. This resonant frequency is determined by acquiring probe excitation spectra away from (in air) and on the sample surface (Fig. 3d). Subsequently, an approach curve is obtained by measuring the signal amplitude at the selected resonant frequency while moving the sample toward the shear force probe, and the working voltage is set to ~80% of its maximum value with a tolerance of ±5% to prevent the probe from crashing or losing contact with the sample (Fig. 3e). In brief, constant-distance-mode imaging is performed by selecting a frequency of an appropriate mode of oscillation and maintaining its amplitude at a constant value, using a computer-controlled closed-feedback loop.
Fig. 3 |. Shear force microscopy.
a, Photo of piezoelectric ceramic plates with cables. The back and front views of the piezo plate are shown in bottom right corner. b, Setup of the shear force probe. Left, the upper plate is used to induce the probe oscillation that connects to the sine out (+) of the lock-in amplifier (i.e., the internal oscillator). The lower plate is used to detect the amplitude of the probe vibration that connects to the A input of the amplifier. Right, the zoomed-in view of the probe tip, corresponding to the red dashed circle in the left panel. Scale bar, 25 μm. c, Photo of the main components for performing shear force microscopy, highlighting (1) a stepping controller for the motorized micro-positioner; (2) a motorized micro-positioner assembled with a shear force probe; (3) a lock-in amplifier. d, Excitation spectra acquired with the probe kept in air (air, black trace) and positioned on the tissue surface (sample, red trace). Blue trace (difference) represents the difference spectrum between the air and the tissue surface. e, An approach curve at 171 kHz is established by plotting the amplitude of the shear force probe vibration as a function of the distance between the sample and the probe.
The imaging data are collected by scanning the sample under the nano-DESI probe in lines, and each line scan is saved as a separate Xcalibur raw file (Steps 20–23). These line scans are subsequently visualized and processed using MSI QuickView (Steps 24–26). MSI QuickView is a custom software for processing and visualizing the data acquired using Xcalibur. A detailed description of the software can be found in our previous report12.
Mouse uterine tissue sections were used in this study to demonstrate the performance of our imaging approach (e.g., spatial resolution, throughput, and coverage). Mouse uterine tissue is an excellent model system that contains several distinct cell types distributed over a small cross-sectional area of 1–2 mm. These include luminal epithelial (LE) and glandular epithelial (GE) cells, stroma (S) cells surrounding the LE and GE, and myometrial (Myo; inner circular and outer longitudinal muscle layers) cells surrounding the stroma24. The molecular signatures of these cells are described in this protocol. The results presented herein indicate that nano-DESI MSI with high spatial resolution is a powerful analytical tool for obtaining high-quality images of hundreds of molecules in biological tissues.
Advantages and limitations
Advantages of the protocol described herein include (i) the ability to image different types of tissue samples with high spatial resolution, (ii) no need for sample pretreatment beyond sectioning of a frozen tissue and attaching the slices to a glass slide, (iii) good coverage of lipids and metabolites, and (iv) accurate determination of the spatial localization of molecules by using normalization to the signal of the internal standard. A resolution of better than 10 μm has been achieved using this protocol and can be further improved (see the ‘Spatial resolution’ section for details). We also have developed a strategy for acquiring both positive- and negative-mode imaging data in the same experiment. This is achieved by alternating between positive and negative data acquisition modes during consecutive line scans. Using this approach, we obtained high-resolution ion images of >500 distinct features in each uterine section (~250 features in each ionization mode), some of which are shown in Fig. 4. This approach improves the throughput of the imaging data acquisition at the expense of a slightly reduced spatial resolution. For the sample shown in Fig. 4, which has a size of 2.5 mm × 1.7 mm, the acquisition time is ~4 h when using a scan rate of 20 μm/s and a line spacing of 15 μm. A total of 116 lines were collected, and ~900 spectra (pixels) were acquired for each line, generating a total of 104,400 pixels with a pixel size of ~2.5 μm (x direction) × 10 μm (y direction).
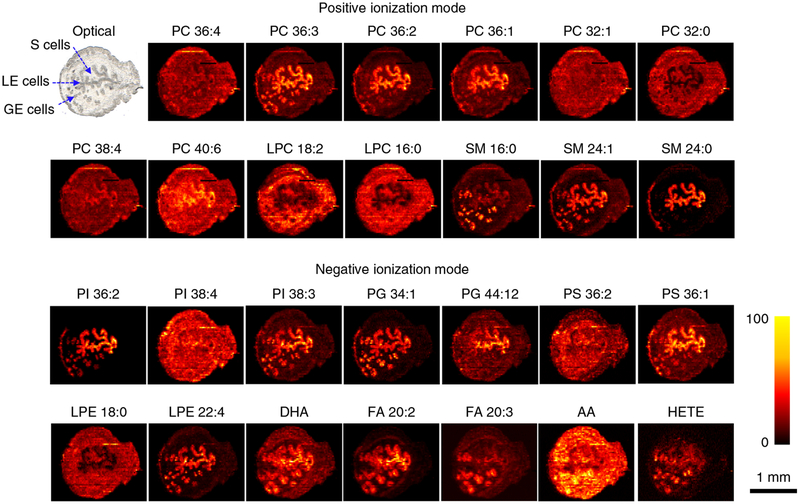
Fig. 4 |. Representative ion images of endogenous molecules acquired in uterine tissues.
Both positive- and negative-mode images are obtained from the same section. This is achieved by alternating between positive and negative data acquisition modes during consecutive line scans (see Steps 20–23 for details). In the positive mode, each image represents the sodium adduct ([M+Na]+) of a unique phospholipid. In negative mode, each image represents the [M−H]− ion of a unique analyte. AA, arachidonic acid (FA 20:4); DHA, docosahexaenoic acid (FA 22:6); HETE, hydroxyeicosatetraenoic acid; LPE, lysophosphatidylethanolamine. The intensity scale changes from black (low) to yellow (high).
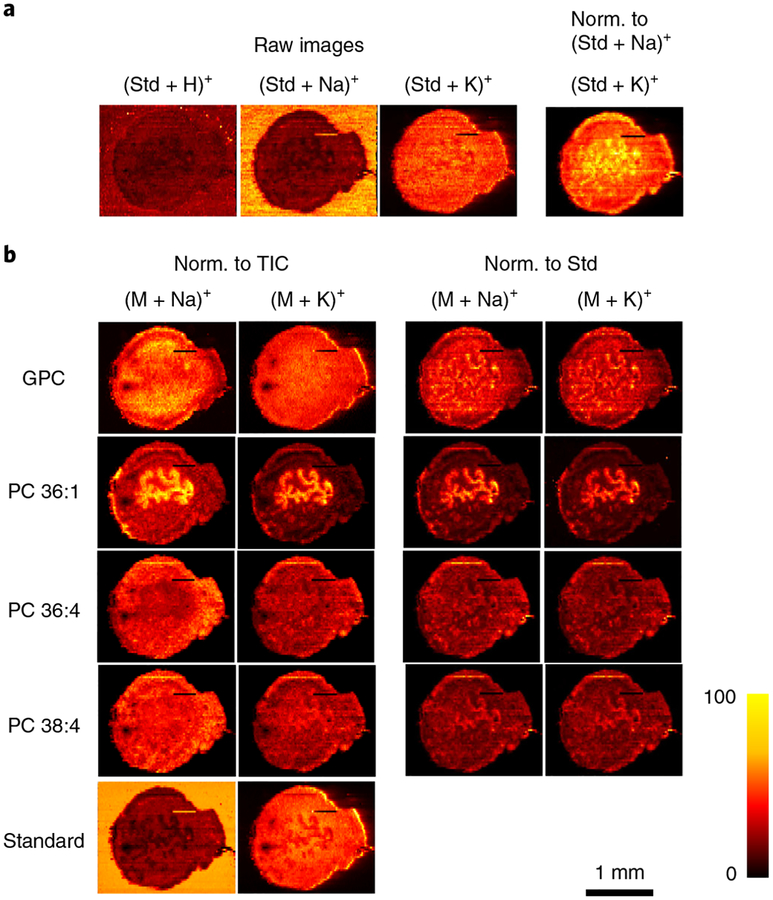
The ability to generate ion images free of matrix effects is a unique advantage of liquid extraction–based ambient ionization techniques such as nano-DESI5. Matrix effects originating from competitive ionization of analytes mixtures extracted from the sample present a major challenge to the accurate determination of chemical gradients in biological tissues5,25,26. Owing to matrix effects, ionization efficiencies of analyte molecules may vary substantially in different regions of a tissue section, which may affect the experimentally measured spatial distributions. Lanekoff et al.27 demonstrated that matrix effects in nano-DESI MSI can be evaluated by adding appropriate internal standards to the working solvent. Furthermore, normalization of the signals of endogenous molecules to the appropriate signal of the standard effectively compensates for matrix effects and enables accurate measurement of concentration gradients in the sample. Matrix effects in nano-DESI MSI of uterine tissues are illustrated in Fig. 5a. In this experiment, lysophosphatidylcholine (LPC) 19:0 was added as an internal standard to the working solvent at a 200 nM concentration. Ion images obtained for different adducts of the internal standard show that although it was supplied at a constant rate to the sample during the imaging experiment, the abundances of LPC 19:0 ([M+Na]+ and [M+H]+) are substantially and non-uniformly suppressed on the tissue sample. The stronger suppression of these signals in LE cells indicates that the ionization of the standard is more strongly suppressed by the molecules extracted from this region of the tissue. By contrast, the abundance of the potassium adduct of LPC 19:0 ([M+K]+) is substantially enhanced on the sample because a higher concentration of K+ is present in the tissue as compared to the solvent. The ratio of [LPC 19:0+K]+ to [LPC 19:0+Na]+ shows that the concentration ratio of alkali ions (K+ and Na+) is not constant throughout the tissue section. Specifically, we observe an increase in the amount of K+ ions relative to Na+ ions in the LE cells and the surrounding tissue. The variation in the abundance of alkali metal salts (e.g., Na+ and K+) may also affect the observed ion images (especially for the positive ionization mode). Therefore, developing an approach that efficiently compensates for matrix effects is critical to obtaining accurate molecular localization in tissue samples. Ion images are often normalized to the total ion current (TIC). However, normalization to the TIC does not compensate for matrix effects (Fig. 5b). By contrast, normalization to the internal standard reveals the actual distribution of endogenous molecules in the tissue provided the standard and extracted analytes experience similar ion suppression during ionization. Figure 5b shows that normalization affects ion images obtained for glycerylphosphorylcholine (GPC), phosphatidylcholine (PC) 36:1, PC 36:4, and PC 38:4. Close correspondence between ion images obtained for [M+Na]+ and [M+K]+ ions of the endogenous molecules when using normalization to the corresponding adduct of the standard (e.g., LPC 19:0) corroborates the assertion that such normalization efficiently compensates for matrix effects and reveals accurate concentration gradients in the sample.
Fig. 5 |. Compensation for matrix effects in nano-DESI MSI by adding internal standards to the solvent.
a, The use of the internal standard (Std; LPC 19:0) indicates the matrix effects originating from competitive ionization of analytes, as well as variations in the abundance of alkali metal salts. Raw images describe the ion images without any normalization (Norm.). b, Ion images of [M+Na]+ and [M+K]+ of endogenous GPC, PC 36:1, PC 36:4, PC 38:4, and standard LPC 19:0 (Standard). Ion images are normalized to the TIC in the left two columns and to the standard LPC 19:0 (Std) in the right two columns. By normalizing to the corresponding LPC 19:0 adducts, exactly the same ion images are obtained for [M+Na]+ and [M+K]+ ions of endogenous lipids, which are free of matrix effects and present their actual distribution in uterine tissue.
The fabrication and alignment of the nano-DESI and shear force probes are currently the most difficult but critical steps in setting up the high-resolution imaging experiment. Finely pulled glass capillaries are fragile and must be handled with care. Alignment of the primary and nanospray capillaries relative to each other is fairly straightforward. However, because the solvent flow in the probe is assisted by the vacuum of the instrument, alignment of the probe relative to the mass spectrometer inlet to obtain a stable flow and ion signal is still tedious. Future developments will address this limitation to ensure that the alignment and stability of the nano-DESI probe do not depend on the vacuum of a mass spectrometer. For example, a pneumatically assisted nano-DESI approach described by Duncan et al.28 may be implemented to propel the solvent through the nanospray capillary independent of the instrument vacuum.
A major limitation of the current data acquisition software used in our laboratory is that a rectangular area, which includes the entire tissue section (Figs. 4 and 5) is analyzed in each experiment and 1/4–1/3 of the acquisition time is spent scanning the surrounding glass rather than the sample. Commercial MSI instruments enable imaging of an area of an arbitrary shape determined by the user. We are modifying our data acquisition software to enable an automated adjustment of the start position and end position of each line on the basis of the sample shape.
Applications of the protocol
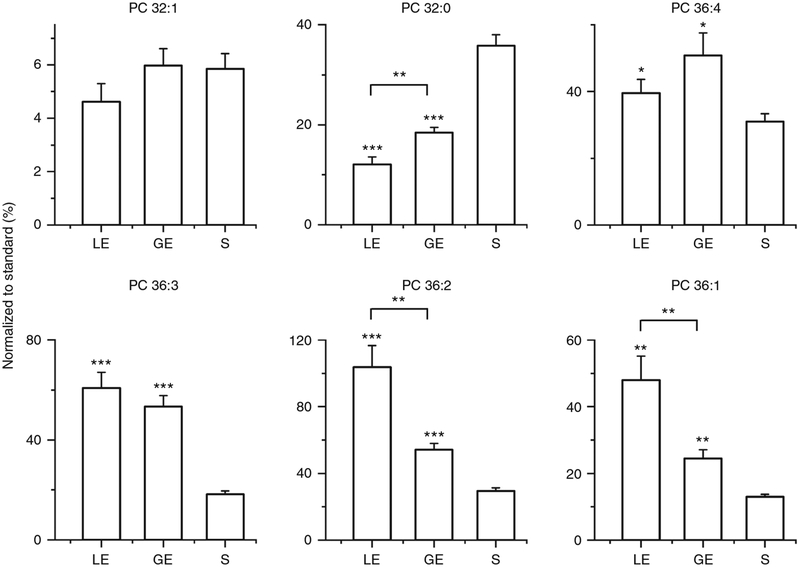
The protocol presented herein is generally applicable to imaging any type of thin tissue section with high spatial resolution. Previously, we used a similar protocol to determine the spatial distribution of PC, phosphatidylethanolamine (PE), sphingomyelin (SM), phosphatidylinositol (PI), and phosphatidylserine (PS), fatty acids (FAs) and other metabolites in individual pancreatic islets, which are difficult to analyze using traditional mass spectrometry approaches because of their small size (~100 μm) and molecular heterogeneity16. Several lipids (e.g., PC 36:2, PC 38:4, and SM 16:0) were found to be substantially enhanced in islets but not in exocrine tissue, suggesting that these lipids may play a role in islet biology.
In this study, high-quality ion images of lipids and metabolites were obtained for uterine tissue sections (Fig. 4). We observed distinct patterns of phospholipid localization to the heterogeneous cell types (LE, GE, and stroma) of the uterus. In particular, remarkably different localizations were observed for several lipids with the same length of acyl chain but different numbers of double bonds. For example, we observed a slight enhancement of PC 36:4 and substantial enhancement of PC 36:3 in both LE and GE cells, whereas PC 36:2 and PC 36:1 are substantially enhanced only in LE cells. These results illustrate how lipid composition can change across cell types, thereby giving insight into the membrane architecture and lipid synthesis and signaling pathways of diverse cells with unique biological functions.
Alternative methods
Commercially available MSI systems used for tissue imaging are dominated by secondary ion mass spectrometry (SIMS), MALDI, and DESI. Among these popular techniques, SIMS is characterized by the highest spatial resolution (better than 1 μm) but suffers from extensive fragmentation of analyte molecules29,30. Recent developments in cluster beam sources have enabled SIMS imaging of intact lipids, including labile cardiolipins, with a spatial resolution of better than 10 μm (refs.31,32). Soft ionization of analyte molecules is advantageous for their identification. MALDI is the most widely used soft ionization technique in MSI experiments. Although most MALDI MSI studies have been performed with a moderate spatial resolution (~50–150 μm), several groups have achieved a spatial resolution of better than 10 μm, which enables imaging of most animal tissues with single-cell resolution9,10,33. For example, Spengler et al.9 have developed an atmospheric pressure (AP) MALDI MSI platform that can achieve a spatial resolution of ~1.4 μm. Zavalin et al.33 successfully integrated a transmission geometry MALDI source with time-of-flight mass spectrometry that enables imaging of tissue proteins with a resolution of 2.5 μm.
Ambient ionization techniques that do not require special sample pretreatment before analysis have also been used for imaging of tissue samples with high spatial resolution5,6,15,34. DESI is the most popular ambient ionization technique that has been used to image different types of tissues, including brain, bladder, prostate, spinal cord, and kidney4–6. The spatial resolution used in DESI MSI experiments is typically 150–200 μm. However, a spatial resolution of 35 μm has been obtained by carefully optimizing the experimental conditions35. We have developed a constant-distance-mode nano-DESI MSI technique for imaging tissue samples at a resolution of better than 10 μm (refs.15,16), which is described in detail in this protocol. Yang et al.34 have reported a spatial resolution of 8.5 μm using single-probe mass spectrometry. MSI of hippocampal tissue with a resolution of 2.9–5.1 μm has been achieved using an AP nanoparticle and plasma-assisted laser desorption ionization (AP-nanoPALDI) source36.
Although a direct comparison between the results of high-resolution MSI obtained using different ionization techniques has not been reported, in our experience they provide similar coverage of endogenous analytes. Typically, mass spectra of phospholipids observed using different MSI techniques applied to similar tissue sections are quite similar. However, it is reasonable to assume that there is a certain degree of complementarity between laser-based and liquid extraction–based ionization approaches. For example, the relatively energetic extraction of molecules from tissue sections using MALDI and other laser-based ionization techniques may be advantageous for the detection of strongly bound molecules that may be difficult to observe using liquid extraction–based techniques. Preliminary results from our laboratory indicate that some molecules not observed in MALDI MSI can be detected using nano-DESI. More detailed comparative studies are necessary to draw conclusions regarding analyte coverage provided by these techniques.
Commercially available MSI sources suitable for imaging with high spatial resolution are expensive but provide the advantage of easy operation and facile image visualization capabilities. By contrast, custom-designed systems are relatively inexpensive (the estimated cost of a high-resolution nano-DESI source is <$25,000) and can be readily adapted to different mass spectrometers, but are often more difficult to operate. Efficient compensation for matrix effects by using an extraction solvent containing internal standards is a unique advantage of liquid extraction–based techniques such as DESI and nano-DESI that is important to both relative and absolute quantification of the imaging data.
Experimental design
Spatial resolution
Spatial resolution refers to the minimum distance required to distinguish between two adjacent features and is a key parameter describing the performance of MSI techniques. The spatial resolution of MALDI MSI is often defined to be equal to the size of the laser spot1. Meanwhile, a different definition of the spatial resolution is typically used in DESI and nano-DESI MSI experiments, in which data are acquired as a collection of line scans rather than individual spots. In particular, the upper limit of the spatial resolution in these experiments is estimated from the distance across which the sharpest distinct features in the image show a change in the relative signal intensity from 20 to 80% (refs.11,37).
Several factors affect the spatial resolution of nano-DESI MSI, including the size of the liquid bridge formed between the sample and the nano-DESI probe, the scan rate at which the sample is moved under the probe, and the acquisition rate of the mass spectrometer. The size of the liquid bridge is a key factor that determines the size of the sampled area at each point during the imaging experiment. A smaller liquid bridge can be obtained by (i) using finely pulled primary and nanospray capillaries; (ii) balancing the solvent flow between the primary and nanospray capillaries; and (iii) adjusting the relative position and angle of the primary capillary and the nanospray capillary. In a successful high-resolution nano-DESI MSI experiment, a small and stable liquid bridge must be maintained throughout the entire experiment, which lasts from ~30 min to many hours. Herein, we provide a detailed description of how to achieve a small and stable liquid bridge. The spatial resolution is also limited by the scan rate at which the sample is moved under the nano-DESI probe and the acquisition rate of the mass spectrometer, which determine the smallest size of each pixel (i.e., the best spatial resolution that can be achieved in an imaging experiment). If the size of the liquid bridge is smaller than the pixel size determined by these two parameters, the spatial resolution can be improved by reducing the scan rate and increasing the acquisition rate. For example, the acquisition rate of a Q-Exactive HF-x (QE) mass spectrometer (typically, ~130 ms per spectrum) is six times faster than that of an LTQ Orbitrap mass spectrometer (typically, ~800 ms per spectrum). For a scan rate of 20 μm/s, the pixel sizes are ~2.6 and ~16 μm on a QE and LTQ Orbitrap instruments, respectively. It follows that, using the same acquisition time, it is possible to obtain a six times higher spatial resolution on a QE instrument. In this study, imaging experiments were performed on a QE instrument using a scan rate of 20 μm/s.
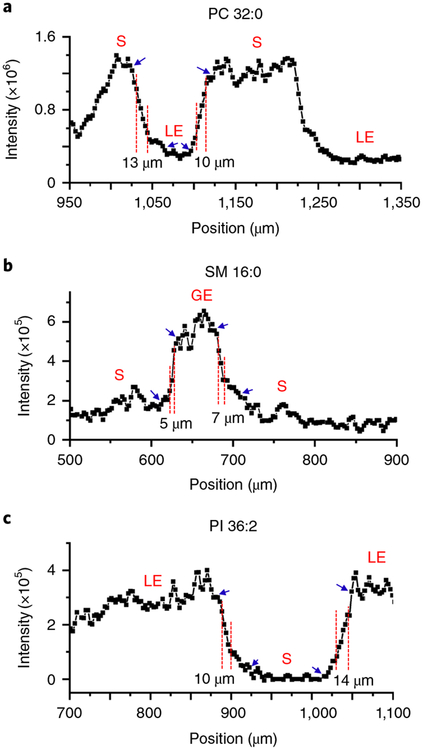
The spatial resolution is estimated as the distance over which the signal intensity increases from 20 to 80% of the maximum value, as described earlier. Accurate measurement of the spatial resolution requires the presence of steep chemical gradients in the sample. In mouse uterine tissue sections, relatively steep chemical gradients were observed for PC 32:0 and PI 36:2 on the boundary between LE and stroma cells. Furthermore, a substantial chemical gradient for SM 16:0 was found on the boundary between GE and stroma cells (Fig. 6). On the basis of these chemical gradients, the estimated spatial resolution of our current experimental setup ranges from 5 to 14 μm, with an average of 9.8 ± 3.4 μm (n = 6). This value, which corresponds to an upper limit estimate of the spatial resolution, is consistent with our previous reports15,16.
Fig. 6 |. Representative line profiles of PC 32:0, SM 16:0, and PI 36:2 acquired on uterine tissue.
a, Chemical gradients for PC 32:0 are observed on the boundaries between S and LE cells. b, Chemical gradients for SM 16:0 are observed on the boundaries between S and GE cells. c, Chemical gradients for PI 36:2 are observed on the boundaries between S and LE cells. The spatial resolution of our approach ranges from 5 to 14 μm, estimated as the distance over which the signal intensity increases from 20 to 80% of the maximum value (labeled with red dashed lines). Blue arrows present the maximum (100%) and minimum (0%) values.
Sample preparation
Optimal cutting temperature compound (OCT) is a common embedding material used for tissue sectioning for histological staining. However, OCT is not recommended for MSI because it strongly suppresses mass spectrometry signals of endogenous analyte molecules38. To solve this problem, an alternative protocol for preparing thin tissue sections has been developed for MSI, in which carboxymethylcellulose (CMC) is used as an embedding compound instead of OCT39. Alternatively, we have used a method for sectioning freshly frozen tissues, in which very small amounts of OCT and/or water are used to help mount the tissue in the cryostat. In this method, it is important to ensure that the blade does not come into contact with OCT. We usually use the CMC-based protocol to section fragile tissues (e.g., mouse pancreas or lungs) and the second method for other tissues (e.g., mouse uterus).
Additional factors that must be considered during sample preparation include the thickness of the section, the number of sections, and sample delivery and storage. The thickness of the tissue section is not critical to nano-DESI MSI. Although the typical thickness of tissue sections is 10 μm, the procedure described here is applicable to both thinner and thicker sections, ranging in thickness from a few microns to hundreds of microns. For thicker sections, the shear force feedback must be adjusted to ensure that the response of the shear force probe is fast enough to allow the probe to maintain constant distance during imaging of a rough surface.
We recommend preparing 10–15 consecutive sections for each sample type. Some sections can be used for the optimization of the instrument parameters and working solvent. Other sections can be used for nano-DESI MSI and staining. Frozen tissue sections can be shipped on dry ice, and the received sections should be stored at −80 °C before the imaging experiments.
Solvent selection
The solvent composition has a direct influence on lipid and metabolite coverage in nano-DESI MSI. A 9:1 methanol–water mixture is the commonly used solvent, which provides comprehensive lipid coverage in most tissue samples. As an example, we have identified a total of 284 lipids and metabolites in mouse lung tissues using nano-DESI MSI, which is comparable to LC–tandem MS lipidomics (263 lipids in total)40. However, the 9:1 methanol–water mixture does not efficiently extract triacylglycerols or sterols from tissue samples. This problem can be addressed by optimizing the solvent composition. For example, triacylglycerols should be detectable when using a more hydrophobic solvent as compared with the 9:1 methanol–water mixture. It has been demonstrated that the addition of silver ions to the solvent significantly enhances the ionization of prostaglandins41. Furthermore, reactive analysis can be combined with nano-DESI MSI to improve ionization of molecules that are difficult to ionize using ESI42. Minor modifications of the protocol would be required for high-resolution imaging of proteins and peptides in tissue sections. Specifically, before the imaging experiments, the samples must be rinsed with organic solvents to remove lipids and the working solvent optimized for protein extraction/ionization43,44. This washing step enables the detection of proteins and peptides by removing lipids, the most abundant components in cells. However, we do not recommend adding the washing step when this protocol is used for imaging of lipids and metabolites. It is likely that even gentle rinsing methods will wash away or displace some lipids and metabolites, which is detrimental to determining their original spatial gradients using imaging.
Quantitative nano-DESI MSI
As discussed earlier, efficient compensation for matrix effects is achieved in nano-DESI MSI experiments by adding internal standards to the working solvent. Normalization of the signals to the appropriate internal standard provides accurate concentration gradients of endogenous molecules in tissue samples. Therefore, selection of internal standards is critically important to quantitative nano-DESI MSI. Isotopically labeled molecules are excellent reference compounds for these experiments. For example, in this study, arachidonic acid-d8 (AA-d8) was used as an internal standard for quantification of AA and other FAs. However, isotopically labeled compounds are available for only a few molecules observed in MSI experiments. In our studies, we often use phospholipids with odd acyl chains as internal standards. Specifically, LPC 19:0 and PS (17:0/17:0) are used as internal standards for endogenous PC and PS species, respectively. Other commercially available lipids can be used as internal standards as long as they do not interfere with the endogenous compounds. For absolute quantification, the summed normalized abundance of an individual endogenous compound is compared to the related signal obtained for a solution of a homogenized tissue section spiked with the same internal standard as described in detail in our previous papers13,14.
Materials
Biological tissues
Uterine tissues from 2-month-old female mice were snap-frozen, and frozen sections with a thickness of 10 μm were made with a cryostat. The mice were maintained on a C57BL6 mixed background and housed in the vivarium at the Cincinnati Children’s Hospital Medical Center according to NIH and institutional guidelines for laboratory animals. Mice were housed in negative-airflow polycarbonate cages with corn cob beddings. They were provided with double-distilled autoclaved water ad libitum and rodent diet (LabDiet 5010) ! CAUTION All studies involving animals must be reviewed and approved by the relevant institutional animal care and use committees and must conform to local and national laws. The protocol for our study was approved by the Cincinnati Children’s Hospital Research Foundation Institutional Animal Care and Use Committee.
Reagents
Methanol (Optima LC-MS grade; Fisher Chemical, cat. no. A456–4) ! CAUTION Methanol is flammable and hazardous; it should be handled in a fume hood.
Ethanol (200 proof; Decon Laboratories, cat. no. DSP-MD.43)
Ultrapure water obtained from Milli-Q Direct 8 Water Purification System (Millipore Sigma, cat. no. ZR0Q008WW)
Pierce LTQ Velos ESI positive ion calibration solution (Thermo Fisher Scientific, cat. no. 88324)
-
Pierce LTQ ESI negative ion calibration solution (Thermo Fisher Scientific, cat. no. 88323)
Standards ▲ CRITICAL All three standards listed below should be appropriately stored according to the manufacturer’s instructions.
LPC 19:0 (Avanti Polar Lipids, cat. no. 855776P)
PI (16:0/16:0) (Avanti Polar Lipids, cat. no. 850141)
AA-d8 (Sigma-Aldrich, cat. no. 735000–5MG)
Equipment
Flexible fused-silica capillary (i.d. 50 μm, o.d. 150 μm; Polymicro Technologies, cat. no. TSP050150)
Flexible fused-silica capillary (i.d. 75 μm, o.d. 375 μm; Polymicro Technologies, cat. no. TSP075375)
Flexible fused-silica capillary (i.d. 200 μm, o.d. 790 μm; Polymicro Technologies, cat. no. 106815–0067)
Union assembly, MicroTight polyetheretherketone (PEEK; Idex Health & Science, cat. no. P-720)
MicroTight sleeve, green (0.025 inch o.d. × 0.0155 inch i.d.; Idex Health & Science, cat. no. F-185)
MicroTight sleeve, yellow (0.025 inch o.d. × 0.007 inch i.d.; Idex Health & Science, cat. no. F-181)
PEEK tubing (1/16 inch o.d. × 0.020 inch i.d.; Idex Health & Science, cat. no. 1532L)
PEEK tubing (1/16 inch o.d. × 0.010 inch i.d.; Idex Health & Science, cat. no. 1531BL)
Plastic tubing cutter (Idex Health & Science, cat. no. A-327)
Cleaving stone (1 inch × 1 inch; Polymicro Technologies, cat. no. 1068680064)
Laser-based micropipette puller (Sutter Instrument, cat. no. P-2000/F)
Kimwipes (Kimberly-Clark Professional, cat. no. 34155)
Legato 180 syringe pump (KD Scientific, cat. no. 788180) ▲ CRITICAL This model of syringe pump provides the smooth, consistent solvent flow necessary for maintaining a stable liquid bridge.
Syringe (2.5 mL; model no. 1002 LTN SYR, cemented needle, 22 gauge, 2 inch, point style 3; Hamilton, cat. no. 81416)
Microforge–grinding center with 20× wide-field (WF) eyepiece and 4×, 10×/0.25, 20×/4.8 mm, and 40×/3.3 mm objective lenses (Microdata Instrument, model no. MFG-5) ▲ CRITICAL This device is used to prepare and examine the primary and nanospray capillaries, along with the shear force probe. It should be operated according to the manufacturer’s instructions.
Microscope with a magnification range of 700×–900× (Dino-Lite Digital Microscope, cat. no. AM4515T8)
Digital microscope (Dino-Lite 5MP Edge; Dino-Lite, cat. no. AM7915MZTL)
Digital microscope (Dino-Lite Edge; Dino-Lite, cat. no. AM4515ZTL)
Camera holder (Dino-Lite, cat. no. MSHD-P1 or MSHD-M1)
Miniature optical mounting posts (50.8 mm; Newport, cat. no. M-TSP-2)
Miniature optical mounting posts (76.2 mm; Newport, cat. no. M-TSP-3)
Adjustable-angle post for 0.312-inch posts (Newport, model no. MCA-2)
Right-angle post clamp for 0.312-inch posts (Newport, model no. MCA-1)
Miniature optical post holder (Newport, model no. MSPH-1.5)
Inline micro-positioners with micrometers (Quater Research & Development, model no. XYZ 500 MIM (inline))
Magnetic base for XYZ 500 (Quater Research & Development, cat. no. 20232)
Articulating arm with an alligator clip (Siskiyou, model no. MXB-3i, cat. no. 20200000E)
Inline micro-positioners for XYZ 500 with motorized option (Quater Research & Development, model no. XYZ 500 MIMT (inline))
Microstepping controller (Quater Research & Development, cat. no. 20411) ▲ CRITICAL Read the manufacturer’s manual carefully before operating the controller for the first time.
Dual-phase lock-in amplifier with rack mount kit (4 MHz; Stanford Research Systems, model no. SR865A)
Daflon ultra-flexible microminiature PTFE hook-up wire (Daburn, cat. no. 2452/30)
Piezo ceramic plate (3 × 3 × 0.55 mm; STEMiNC, cat. no. SMPL3W3T05410)
Micropipette metal holder (Digi-Key Electronics, cat. no. 952-1476-1-ND)
WireJewelry dead soft copper sheet metal (sheet size, 6 × 6 inch, 0.01 inch (30 gauge) thick, Amazon)
Silver conductive epoxy adhesive (MG Chemicals, cat. no. 8331–14g)
Pomona BNC (Bayonet Neil–Concelman) female plug to flying leads (Newark Element14, cat. no. 94F1506)
Instrument mounting flange (Prosolia)
Motorized stage (50-mm travel, RS-232 with accessory kit; Zaber, model no. T-LSM050A-KT03)
Motorized stage (25-mm travel, RS-232; Zaber, model no. T-LSM025A)
Motorized linear stage (25-mm travel, RS-232 with motor extension cable; Zaber, model no. LSM025A-T4-MC04)
A-series stepper motor controller (Zaber, model no. A-MCA-K07U)
Multifunction I/O device (National Instruments, model no. USB-6009)
Olympus optical microscope (Olympus, model no. DP71)
Array slide holders, blue, pack of 20 (Camlab, cat. no. 1158383)
Disposable scintillation vials (20 mL; Dan Yang Hua Fang Paper, cat. no. XB15288–65)
Digital vortex mixer (VWR)
Low-power pathology slide scanner (Meyer Instruments, model no. PathScan Enabler IV)
Freezer (set to −80 °C; 13 cubic feet, upright, 120 V/60 Hz; VWR, cat. no. 10160–724)
Electrostatic discharge–safe stainless-steel tweezers set (Delcast; 8 pieces; Amazon, cat. no. TZ-8X)
Digital multimeter with true RMS (Fluke, model no. 115)
Instant adhesive glue (0.07 oz; 3M Scotch-Weld; 3M, cat. no. CA4)
Fisherbrand general-purpose label tape (Thermo Fisher Scientific, cat. no. 15937)
Candle
Crocodile clip
Nitrile powder-free gloves
General purpose masking tape
E-Projects 10-MΩ resistor (Amazon, cat. no. 100EP51210M0)
Gold Seal Plain Microscope Slides (ThermoFisher Scientific, cat. no. 3057–02)
Software
DinoCapture 2.0 (https://www.dinolite.us/downloads/)
MSI QuickView (available from the authors upon request)
Custom LabVIEW program (available from the authors upon request)
Reagent setup
Solvent preparation
Prepare 5 mL of a 9:1 (vol/vol) methanol–water mixture in a 20-mL scintillation vial. Add 5 μL of 200 μM LPC 19:0 (an internal standard for positive-mode experiments), 5 μL of 200 μM PI (16:0/16:0) (an internal standard for negative-mode experiments), and 5 μL of 300 μM AA-d8 (an internal standard for negative–mode experiments) to the mixture. Vortex the solution vigorously. The final concentrations of LPC 19:0, PI (16:0/16:0), and AA-d8 are 200, 200 and 300 nM, respectively. The solvent can be stored for a week at room temperature (22–26 °C) or for several months at −20 °C. ▲ CRITICAL The methanol–water mixture should not be prepared in a plastic tube in order to avoid contamination from plastic materials. Always use a glass vial to prepare and store the mixture. ▲ CRITICAL To avoid potential contamination, the mixture should be prepared and handled only while wearing nitrile powder-free gloves.
Equipment setup
Laser-based micropipette puller
We use a P-2000 micropipette puller to fabricate the primary capillaries and shear force probes. The device is a microprocessor-controlled CO2 laser-based puller. The use of a CO2 laser enables fabrication of capillary tips with different geometries (such as shapes and sizes) using fused-silica (quartz) glass capillaries. The pulling parameters can be found in Table 1. These parameters can be adjusted to obtain different tip sizes. ▲ CRITICAL Read the manufacturer’s manual carefully before operating the puller for the first time. ▲ CRITICAL The retro mirror should be regularly checked and cleaned with ethanol and a Kimwipe.
Table 1 |.
Pulling parameters for the fabrication of the primary capillary and shear force probe
| Program | Line | Heat (°C) | Fil | Vel | Del (ms) | Pull | Notes |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 550 | 4 | 50 | 135 | 50 | Primary capillary with an ~15-μm opening and a long taper |
| 2 | 650 | 4 | 50 | 130 | 30 | ||
| 3 | 550 | 4 | 50 | 130 | 30 | ||
| 2 | 1 | 550 | 4 | 60 | 130 | 50 | Primary capillary with an ~15-μm opening and a short taper |
| 2 | 650 | 4 | 60 | 130 | 30 | ||
| 3 | 550 | 4 | 60 | 130 | 30 | ||
| 3 | 1 | 650 | 4 | 70 | 130 | 60 | Shear force probe with an ~5-μm opening |
| 2 | 650 | 4 | 60 | 130 | 60 | ||
| 3 | 650 | 4 | 70 | 130 | 70 |
Del, delay; Fil, filament; Vel, velocity.
Mounting a Dino-Lite camera onto the MFG-5 microforge
The MFG-5 microforge is used to observe and polish pulled capillary tips. Select the 20× WF eyepiece and the 10×/0.25 objective lens. The 200× magnification allows us to examine capillary tips with an o. d. of ≥1 μm. However, the microforge cannot be used to record and measure the size of the tip. To solve this problem, a Dino-Lite microscope with a 700×–900× magnification should be installed on the microforge. Specifically, we mount a 0.312-inch optical post on the arm located on the left side of the grinder position adjustment and connect it to another optical post, using a right-angle post clamp. The microscope is mounted onto the stand using a Dino-Lite metal holder. The working distance of the microscope is only 6 mm; it must be positioned very close to the capillary tip. Adjust both the microscope and the tip to obtain a good focus on the tip. Connect the microscope to a computer’s USB port. Install DinoCapture 2.0 for operating the microscope. The modified device is very convenient for examining, polishing, and measuring capillary tips. Attach a 1.0-μm grade grinding disk to the top of the grinding center. Adjust the grinding speed to between 5 and 7. Turn it on with the foot switch and polish the tip while examining it under the microscope. ▲ CRITICAL The Dino-Lite camera should be properly calibrated by following the manufacturer’s manual. ▲ CRITICAL To keep the precision grinder and microscopes clean, the microforge should be covered with a plastic cover while it is not in use.
Nano-DESI imaging platform
Our imaging platform comprises an instrument mounting flange, micro-positioners, xyz motorized stages, a custom-made sample holder, Dino-Lite digital microscopes, and a shear force microscope (Fig. 2c). Attach the mounting flange (1, Fig. 2c) to a mass spectrometer (such as a QE instrument). The spray voltage is applied to the primary capillary through a high-voltage cable (the red cable in Fig. 2c). Note that a 10-MΩ resistor has been integrated into the high-voltage cable to avoid potential electric shock induced by a high spray voltage. A custom-made 20 cm × 20 cm stainless-steel plate mounted onto the flange is used to support the micro-positioners and microscopes. Three micro-positioners need to be assembled as follows: (i) attach a magnetic base to the micro-positioner to hold it tightly to the stainless-steel plate, and (ii) install an articulating arm with an alligator clip into the micro-positioner. The length of the arm should be adjusted according to your needs. Specifically, two XYZ 500 inline micro-positioners (2, Fig. 2c) are used to position the primary (3, Fig. 2c) and nanospray capillaries (4, Fig. 2c), respectively. Another motorized XYZ 500 micro-positioner (8, Fig. 2c) is used to position the shear force probe (5, Fig. 2c). The motorized xyz stage is used to position the sample relative to the nano-DESI probe. It is composed of a T-series stage with 50-mm travel (x), a T-series stage with 25-mm travel (y), and a T4 stage with 25-mm travel (z). A stepper motor controller (A-MCA) is connected to the z stage, which enables faster and more accurate control of the movement of the stage. Mount the z stage onto the flange using M3 screws, which can be found in the package of the Zaber stage. The y stage is attached to the z stage by vertical mounting, and the x stage is mounted on the top of the y stage using M3 screws. The x and y stages share the same power supply and serial port. The xy stages and z stage are connected to the computer’s USB port with a T-USBDC cable, which can be found in the packages of the Zaber stages. Mount a sample holder (6, Fig. 2c) onto the x stage. A microscope glass slide containing tissue sections will be mounted onto the sample holder. The sample positioning xyz stage is controlled using a custom LabVIEW program developed at Pacific Northwest National Laboratory. Dino-Lite microscopes (7, Fig. 2c) are used to monitor the nano-DESI probe during imaging experiments. To build a stand for the microscope, mount a miniature optical post holder onto the stainless-steel plate using an M6 screw, insert a 76.2- or 50.8-mm optical post, connect to another optical post (76.2 or 50.8 mm) using a right-angle post clamp, and mount the third optical post onto the second one, using an adjustable-angle post clamp. The size of the stand can be adjusted according to your needs. Attach the microscope to the stand, using a Dino-Lite camera holder. The Dino-Lite microscopes are connected to the computer’s USB port. The DinoCapture 2.0 software should be installed in order to operate the microscope. The shear force microscope will be discussed in the next section. ! CAUTION The ion transfer tube and the surrounding surface are very hot. Avoid touching them with your hands.
Shear force microscope
The shear force microscope includes a finely pulled capillary, two piezoelectric ceramic plates with cables, a motorized micro-positioner with a stepping controller, and a 4-MHz dual-phase lock-in amplifier (Fig. 3a–c). The shear force probe is produced by pulling a fused-silica capillary (200-μm i.d., 790-μm o.d.) to an ~10-μm o.d. using program 3 of the P-2000 micropipette puller (Table 1). To mount the piezo ceramic plate (Fig. 3a), cut a small (~4.5 mm × 6-mm) piece of 0.01–inch-thick copper plate and solder the copper plate, a 0.02-inch-o.d. wire and a metal holder together at 400 °C. Solder another wire to the positive side of a 3 mm × 3-mm piezo plate (labeled with a red spot in the inset of Fig. 3a), using a temperature of 250–270 °C for 2 s. Glue the piezo plate and copper plate together using silver conductive epoxy adhesive. Connect the wires to a BNC female plug (connect the negative side and copper plate to the shield of the BNC, and the positive side to the center). Prepare another piezoelectric plate following the same steps. The lock-in amplifier is a key component that enables precise measurement of the small signals generated by the vibrating shear force probe. A lock-in internal oscillator is included in the amplifier and can be used to generate the reference signal in a frequency range from 1 mHz to 4 MHz, which allows the use of the amplifier for different types and sizes of piezo plates and shear force capillaries. Typical settings for the device are summarized in Table 2. ▲ CRITICAL The lock-in amplifier should be properly operated by following the manufacturer’s instructions. ▲ CRITICAL Improper soldering (such as use of a temperature of ≥300 °C or a long heating time) could damage the conductive surface and insulator of the piezo plate. ▲ CRITICAL Only a small amount of the conductive adhesive should be used to glue the piezo and copper plate together to prevent its spread over the insulator, which could cause a short of the piezo plate.
Table 2 |.
Parameters for the lock-in amplifier
| Signal path settings | Reference settings | ||
|---|---|---|---|
| Sensitivity | 0–10 mV (adjustable based on the detected signals) | Source | Internal |
| Amplitude | 500 mV | ||
| Time constant | 10 ms | Sine out | + |
| Input range | 300 mV | Output settings | |
| Voltage | A/AC/float | CH1 output | X without expand |
| Filter | 24 dB | CH2 output | Y without expand |
| Input | Voltage | ||
A, single-ended voltage connection; AC, alternating current coupling.
Mass spectrometer
The imaging platform is compatible with multiple Thermo Fisher Scientific mass spectrometers, including the LTQ XL, LTQ/Orbitrap, and QE (Thermo Electron) instruments. The nano-DESI MSI experiments on pancreatic islets were performed on an LTQ/Orbitrap mass spectrometer. A detailed description of instrument parameters can be found in our previous paper16. Experiments with mouse uterine tissue sections were performed on a QE mass spectrometer. By alternating between positive and negative data acquisition modes during consecutive line scans, both positive- and negative-mode mass spectra were acquired in the same experiment. A high voltage of +3.2 kV and a radiofrequency (RF) funnel voltage of +100 V were applied in positive mode, whereas a voltage of −3.2 kV and an RF funnel voltage of −100 V were used in negative mode. For both modes, mass spectra were acquired in the range of m/z 150−2,000, with a mass resolution of 60,000 at m/z 200; automatic gain control (AGC) was set to 1 × 106, and the maximum injection time was 200 ms; the heated capillary was held at 250 °C. ▲ CRITICAL To ensure high mass accuracy, the instrument should be frequently calibrated using the ion calibration solutions.
Procedure
! CAUTION The high-resolution nano-DESI probe and shear force probe are very sharp. They should be fabricated and handled only while wearing nitrile powder-free gloves, a lab coat, and safety glasses.
Fabrication of the high-resolution primary capillary ● Timing 1 h
▲ CRITICAL The primary capillary is made from a fused-silica capillary (o.d. 375 μm; i.d. 75 μm). Both sides of the primary capillary should be pulled to produce tips with an o.d. of 15–20 μm. One side of the pulled capillary delivers the working solvent. Pulling the other side of the capillary helps prevent its clogging. The procedure is illustrated in Fig. 2a. First, one side of the primary capillary is pulled and inserted into the syringe needle. Next, the other side of the capillary is pulled.
-
1
Pull the primary capillary to make a tip with o.d. of 15–20 μm. Turn on the P-2000 puller and select program 2 (Table 2). Cut 60–70 cm of a fused-silica capillary (o.d. 375 μm; i.d. 75 μm), and wipe it with ethanol, using a Kimwipe. To make an ~2-cm transparent zone, use a candle to burn the capillary coating and carefully remove the burned coating with ethanol and a Kimwipe. The transparent zone is ~3 cm away from one end of the capillary. Place the capillary in a V-groove of the puller bar, adjust the position of the capillary to ensure that only the transparent zone of the capillary is in the laser path, and tighten the clamping knobs. Close the lid, and press the ‘pull’ key on the keypad. The capillary should be separated in seconds. Remove the pulled capillary from the puller bar. Examine and measure the tip, using an Olympus DP71 microscope or the modified MFG-5 microforge (‘Equipment setup’). The tip should have an o.d. of 15–20 μm with a short taper (Fig. 2a).
▲ CRITICAL STEP Using the right technique for pulling a capillary with the expected shape is critical. We recommend that readers watch the tutorial videos for the P-2000 puller that can be found on the Sutter website (https://www.sutter.com/2.html). The videos are very helpful for mastering the pulling skills.
▲ CRITICAL STEP As the tip is inserted into the syringe needle, it should have the same or a little larger i.d. as compared to the other side of the primary capillary, which ensures the primary capillary will not become clogged for a long time. A short taper of this tip sustains higher flow under the same pressure.
? TROUBLESHOOTING
-
2
Connect the pulled capillary to the syringe. Insert a green MicroTight sleeve (0.025-inch o.d.; 0.0155-inch i.d.) into a MicroTight PEEK union. Insert the unpulled side of the capillary through the union (screw without sleeve < union body < screw with sleeve). Make sure that the pulled side will be connected to the syringe with ~0.5 cm of the capillary tip extending out of the union (Fig. 2a). Fill a syringe with 1.5–2 mL of working solvent, slowly slide the tip into the syringe needle, and carefully rotate the union to secure the connection.
▲ CRITICAL STEP Do not break the tip when sliding it into the needle; otherwise, the primary capillary will become clogged quickly. Insert the tip into the syringe needle under a microscope.
-
3
Check that the pulled tip is open by pushing the solvent through the syringe by hand. You should observe the solvent flowing out of the capillary within 1 min. Insert the capillary into a ~1.5-cm-long PEEK tubing (1/16-inch o.d.; 0.020-inch i.d.), and tape them with general purpose masking tape.
▲ CRITICAL STEP Insert the capillary into the PEEK tubing before pulling; otherwise, it is easy to break the pulled capillary when inserting it into the PEEK tubing.
? TROUBLESHOOTING
-
4
Pull the primary capillary to make a second tip with an o.d. of ~15 μm. Place the capillary into the puller and pull the capillary with program 1 shown in Table 1. Examine and measure the tip using an Olympus DP71 microscope or the modified MFG-5 microforge. If the size of the tip is >20 μm, you should pull the capillary again. If the size of the tip is <15 μm, use the MFG-5 microforge to polish the tip. Hold the capillary securely in the pipette holder of the microforge, turn on the LED lamps, and adjust the focusing knob to focus on the capillary. Adjust the grinder manipulators until you see the grinder in the lower portion of the view. Turn on the grinder by pressing the foot switch and move the capillary down onto the grinder. Grind the capillary for 2 s and switch off the grinder. Move the capillary up and measure it using the Dino-Lite microscope (‘Equipment setup’). Repeat these steps until a good size (~15 μm) is obtained. It is very important to slowly push the solvent through the capillary with a syringe to flush it during grinding. The solvent flow helps prevent dust particles produced during grinding from entering the capillary tip. Remove the general purpose tape and move the PEEK tubing all the way through until ~1 cm of the capillary extends out. Use a small amount of instant adhesive glue to fix the tubing to the capillary. A photograph of the primary capillary is shown in Fig. 2a.
▲ CRITICAL STEP It may be difficult to securely hold a 375-μm o.d. capillary using the pipette holder of the microforge. In this case, glue the PEEK tubing to the capillary before polishing the capillary.
▲ CRITICAL STEP If the capillary becomes clogged during polishing, you could try to heat the tip over a flame to remove the clog.
▲ CRITICAL STEP Use only a small drop of instant adhesive to glue the PEEK tubing to the capillary; make sure the glue does not contaminate the capillary tip.
? TROUBLESHOOTING
Fabrication of the high-resolution nanospray capillary ● Timing 30 min
▲ CRITICAL The nanospray capillary is made from a fused-silica capillary (o.d. 150 μm; i.d. 50 μm). The nanospray capillary tip is pulled by hand, and the size of the pulled tip is not reproducible, so it is often necessary to polish the pulled tip to the final size. It may be difficult to remove dust particles that accumulate inside the capillary tip during grinding. Our strategy is to prepare a capillary with two pulled tips, connect it to a syringe, and flush the dust by pushing a solvent through the capillary. Finally, the capillary is cut to a length of 1.5–1.8 cm.
-
5
Prepare a 5- to 6-cm-long capillary with two pulled tips. Cut a 60- to 70-cm-long fused-silica capillary (o.d. 150 μm; i.d. 50 μm), wipe the capillary with ethanol using a Kimwipe, and pull it manually by holding the capillary with one hand over a candle flame for 2–3 s and pulling the other end of the capillary with tweezers to separate it. The length of each pulled capillary is ~5–6 cm. Observe and measure the o.d. of the pulled tips using an Olympus DP71 microscope or the MFG-5 microforge. If the size of the pulled tip is ~25 μm, skip Step 6 and perform Step 7. If the tip has an o. d. of <25 μm, grind it to ~25 μm using the microforge.
! CAUTION Keep your hands away from the flame to avoid burns when pulling.
-
6
(Optional) Clean the pulled and polished capillary. To remove any glass particles inside the capillary that were caused by grinding (Fig. 2b), connect it to a syringe filled with ethanol and flush. Specifically, cut a yellow MicroTight sleeve (0.025 inch o.d.; 0.007 inch i.d.) into two equal parts, insert one-half of the sleeve into a MicroTight PEEK union, insert the non-ground side of the capillary through the union (screw with sleeve < union body < screw without sleeve) until ~0.5-cm of the capillary tip extends out of the union, and tighten the screws. Make sure the non-ground end of the capillary is connected to the syringe (Fig. 2b). Slide the non-ground end all the way into the needle of the syringe filled with ethanol. Flush the line for 5–10 s. You should observe the solvent flowing out of the capillary. Loosen the screws, hold the ground side of the capillary, and pull it out of the union. Do not touch the capillary tip. To remove the outside glass particles, put a droplet of ethanol on the edge of a Kimwipe and gently touch the capillary tip with the wet region of the Kimwipe. Make sure that the paper is not folded. A single-layer paper will not break the tip. Examine the capillary tip under a microscope. A clean, flat capillary tip should be obtained (Fig. 2b).
-
7
Prepare the nanospray capillary with a final length of 1.5–1.8 cm and an o.d. of 25 μm. Insert the non-ground end of the capillary into a ~0.5-cm-long PEEK tubing (1/16 inch o.d.; 0.010 inch i.d.), pull the capillary through the tubing until ~0.5-cm of capillary tip (ground end) extends out, and glue the PEEK tubing to the capillary, using a small amount of instant adhesive. Wait for 2–3 min to let the glue dry. Cut the capillary to a final length of 1.5–1.8 cm. Make sure the capillary has a flat end. To save time, prepare three or four nanospray capillaries in parallel. An example of the nanospray capillary is shown in Fig. 2b.
▲ CRITICAL STEP This step should be performed in an area without a strong air flow, because a steady flame will make the pulling much easier.
▲ CRITICAL STEP Both sides of the capillary should be pulled. Considering that the flushing process takes only 5–10 s, the size of the non-ground end connected to a syringe is not critical and could be in a range of 5–40 μm. However, using a smaller tip (5–15 μm) helps to avoid clogging.
Fabrication of the shear force probe ● Timing 20 min
-
8
Make the shear force probe from a fused-silica capillary (o.d. 790 μm; i.d. 200 μm). Cut 60 cm of a fused-silica capillary (o.d. 790 μm; i.d. 200 μm) and wipe it with ethanol, using a Kimwipe. Place the capillary into the P-2000 puller and use program 3 shown in Table 1 to pull it. Cut the pulled capillary to a final length of ~5 cm. Note that two 5-cm-long pulled capillaries can be prepared from each pulling. Examine and measure the capillary tips using the MFG-5 microforge. Grind them to a final o.d. of ~10 μm. Carefully remove the burned coating that often remains at the edges, using a Kimwipe with ethanol. To clean the capillary tip, put a droplet of ethanol on the edge of a Kimwipe and gently touch the capillary tip with the wet region. Observe the tip again, using the microforge, to make sure it is clean and not broken.
? TROUBLESHOOTING
■ PAUSE POINT The fabricated nano-DESI and shear force capillaries can be stored for future use. It is recommended to make multiple capillaries at once, including multiple primary capillaries, nanospray capillaries, and shear force probes. In the case of breakage, a broken capillary can be quickly replaced with a spare capillary. To avoid possible degradation of standards, the primary capillary should be prepared no more than a week before the imaging experiments.
Setup of the nano-DESI probe ● Timing 1–2 h
-
9
Attach the nano-DESI imaging platform to a mass spectrometer (such as a QE instrument).
-
10
Mount and position the nanospray capillary. Mount the nanospray capillary onto an XYZ 500 inline micro-positioner (2, Fig. 2c) and place it in front of the mass spectrometer entrance. First, adjust the Dino-Lite microscopes to focus on the mass spectrometer entrance and move the capillary close to the center of the entrance, using the micro-positioner. Next, adjust the microscopes to focus on the pulled tip of the capillary. To clearly see a tip with an o.d. of 15–25 μm, the microscopes should be positioned as close to the tip as possible to make sure the microscopes will not block the movement of the stage during the experiment. Typical magnifications for the side-view and top-view microscopes are 60–80× and 30–40×, respectively.
-
11
Mount and position the primary capillary. Mount the primary capillary onto a second XYZ 500 inline micro-positioner (2, Fig. 2c) and place the micro-positioner on the platform next to the first one. Place the syringe connected to the primary capillary into a syringe pump and slide the motor until it touches the plunger. Click the ‘fast forward’ button on the keypad of the pump softly and release it quickly. Repeat the steps until the solvent flows out of the capillary. Turn on the syringe pump and set the flow rate to 0.5 μL/min. Use the micro-positioner to move the primary capillary to bring it into contact with the nanospray capillary. Adjust the relative position of the capillaries and the position of the nanospray capillary relative to the mass spectrometer inlet in order to obtain a stable solvent flow through the nanospray capillary. Specifically, the primary capillary needs to touch the nanospray capillary and should be positioned a few microns below the nanospray capillary. Moving the nanospray capillary closer to the mass spectrometer inlet increases the solvent flow through the nanospray capillary, and moving it further from the inlet decreases the flow. The angle between the two capillaries has only a small effect on the spatial resolution. The recommended angles of the primary capillary and nanospray capillary range from 90° to 120°. When any of these angles is used, a spatial resolution of better than 10 μm can be achieved by carefully adjusting the relative position of the capillaries, as well as the position of the nanospray capillary relative to the mass spectrometer inlet. An angle of ~100° was used for imaging the uterine tissue sections described in this study (Fig. 2d,e).
▲ CRITICAL STEP Do not disconnect the primary capillary from the syringe unless the solvent needs to be replaced. Reconnecting the primary capillary to the syringe must be performed with care to avoid damaging the capillary tip, which may result in clogging of the primary capillary.
▲ CRITICAL STEP Do not press the ‘fast forward’ button of the syringe pump too hard, because the primary capillary may become disconnected from the syringe when subjected to high flow rates. Check the settings of the pump carefully before starting the pump, including syringe type, size, and flow rate.
-
12
Attach the high-voltage cable to the syringe needle using a crocodile clip, and turn on the mass spectrometer to check whether a stable MS signal has been achieved. The MS signal should be stable as long as the solvent flow is stable. A high spray voltage can increase the solvent flow through the nanospray capillary. Move the nanospray capillary further from the entrance to achieve a stable flow, if necessary, after applying the high voltage.
! CAUTION A high spray voltage of 3–4 kV is applied here. Make sure to put the mass spectrometer into standby mode or change the voltage to 0 V before touching the crocodile clip of the high-voltage cable.
? TROUBLESHOOTING
-
13
Land the nano-DESI probe on a surface (glass or sample) to check the stability of the liquid bridge. Mount a microscope glass slide containing thin tissue sections (such as mice uterine tissue) onto a sample holder (6, Fig. 2c) and move the sample toward the nano-DESI probe until the surface is ~20–30 μm under the probe. The position of the sample should be monitored using the Dino-Lite microscopes. Move the sample up to the probe step by step, with a step size of 2 μm, until a liquid bridge is observed. Refine the relative position of the capillaries and the position of the nanospray capillary relative to the mass spectrometer inlet in order to obtain a small, stable liquid bridge. In general, lowering the position of the primary capillary relative to the nanospray capillary increases the size of the liquid bridge, and bringing it up decreases the size of the liquid bridge. The position of the nanospray capillary relative to the mass spectrometer inlet is also critically important to achieving a small liquid bridge due to the vacuum of the mass spectrometer. For example, moving the nanospray capillary closer to the mass spectrometer inlet decreases the size of the liquid bridge by increasing the solvent flow due to vacuum suction. The positioning of the two capillaries is iterative and based on the stability of the signal and the size of the liquid bridge. After a few iterations, a stable small liquid bridge can be obtained. Record the vertical position of the sample holder once a small stable liquid bridge has formed. Retract the sample with a step size of 2 μm until the droplet is separated, and record the distance between the liquid bridge formation and separation. Repeat the landing process several times. The z position and the distance should be reproducible. Typically, the distance between the points of liquid bridge formation and separation is ~4–8 μm. If the distance is <4 μm or >8 μm, either loss of contact with the sample or formation of a large liquid droplet may occur during the imaging experiment.
▲ CRITICAL STEP To avoid damage to the nano-DESI probe, the sample should be moved toward the probe with caution. We found that performing the final approach between the sample and the nano-DESI probe in 2-μm steps helps to avoid damaging the probe. The movement of the sample should be monitored with the microscopes.
? TROUBLESHOOTING
Assembly and positioning of the shear force probe ● Timing 1 h
-
14
Mount two piezo ceramic plates (Fig. 3a) onto the shear force capillary. Insert the pulled shear force capillary through the holder connected to the first piezo plate and squeeze the holder with tweezers to securely hold the capillary. Position the plate 2–2.5 cm above the capillary tip. Insert the capillary through the holder connected to the second piezo plate. Position the second piezo plate 0.5–1 cm above the capillary tip. The distance between the plates should be between 0.5 and 1 cm. Mount the shear force probe to a motorized XYZ 500 inline micro-positioner (Fig. 3b,c). Connect the upper plate (agitation piezo) to the sine out (+) of the lock-in amplifier using a BNC cable. Connect the lower plate (detection piezo) to the A input of the amplifier using a BNC cable.
▲ CRITICAL STEP The connection between the copper plate and wire may be fragile. Use a multimeter to check all the connections after mounting. Specifically, verify that both the negative sides of the piezo plate and copper plate are connected to the shield of the BNC and the positive side of the piezo plate is connected to the core of the BNC. The core should be insulated from the shield.
-
15
Connect the motorized micro-positioner to a stepping controller. Place the micro-positioner on the platform next to the micro-positioner holding the primary capillary (2, Fig. 3c). Move the sample away from the nano-DESI probe and adjust the articulating arm to position the shear force probe close to the nano-DESI probe. Use the stepping controller to finely position the shear force probe in close proximity to the nano-DESI probe. The stepping controller allows precise control of the movement of the micro-positioner down to 0.1 μm/s in any direction (x, y, or z). The distance between the nano-DESI and shear force probe should be within 10–20 μm. The tip of the shear force capillary should be positioned 50 μm or more below the tip of the nano-DESI probe to ensure the nano-DESI probe does not touch the sample surface when performing frequency scanning on the sample surface (Step 16).
-
16
Perform frequency scanning experiments. Turn on the lock-in amplifier and make sure that the settings are as shown in Table 2. Excitation spectra showing the frequency response of the probe are collected in the frequency range of 50–200 kHz with the probe resonating in air and directly in contact with the sample surface. Typical results are shown in Fig. 3d. The excitation spectra are acquired at least two times to check for reproducibility.
▲ CRITICAL STEP To obtain the frequency response of the shear force probe on the surface, it must be carefully brought into contact with the sample by moving the sample toward the probe in small steps.
? TROUBLESHOOTING
-
17
Obtain an approach curve by measuring the signal amplitude at the optimal frequency while moving the sample toward the shear force probe. For controlling the sample-to-probe distance, select the frequency with the maximum signal difference between the sample surface and air (171 kHz in Fig. 3d). In our setup, the optimal frequency is usually found ~170 kHz. Set the optimal frequency as the reference frequency of the amplifier. Position the sample ~30 μm below the shear force probe. Use the controller to move the probe toward the sample surface. When you observe an amplitude change on the screen of the amplifier, indicating that the probe has started sampling the surface, continue moving the shear force probe down by another 2–4 μm until you observe a stable signal. Set this position as the end position for the approach curve acquisition and then move the probe up (away from the sample surface) by 10 μm and set the current position as the start position for the approach curve acquisition. Move the probe, with a step of 0.3 μm, from the start position to the end position and measure the amplitude of the signal recorded by the lock-in amplifier at each step.
▲ CRITICAL STEP To protect the probe and the sample, avoid touching the sample surface with the nano-DESI probe when measuring the approach curve.
? TROUBLESHOOTING
-
18
Set the working voltage and slope of the shear force probe. The working voltage, which determines the distance between the shear force probe and the sample, is selected by examining the approach curve. For example, for the approach curve shown in Fig. 3e, a working voltage of 5.0 V, with a tolerance of ±0.2 V, is selected to ensure that the shear force probe is positioned 0.5 ± 0.2 μm above the sample surface. The shear force probe should be positioned between 0.5 and 1.5 μm above the surface. If it is positioned out of this range, the shear force probe will either crash into the sample or lose contact with the sample during the imaging experiment. The working slope determines the speed at which the system responds to the height variations of the sample. The slope corresponding to the line range of the approach curve can be used as a reference value for setting the working slope. For a tissue section with a thickness of ≤10 μm, the working slope is set to a value much lower than the slope of the approach curve (typically, 0.5–1.0 μm/V). Thicker sections require a higher responding speed (i.e., higher working slope) to prevent the shear force probe from crashing into the edge of the tissue sample. This value needs to be optimized for thicker sections before the imaging experiments.
-
19
Align the nano-DESI probe and the shear force probe. Use the stepping controller to move the shear force probe up to a position above the nano-DESI probe and land the nano-DESI probe on the glass surface. A small liquid bridge should be observed. Use the stepping controller to move the shear force probe down to the surface until you observe an amplitude change on the screen of the amplifier. Typically, the distance between the shear force probe and the nano-DESI probe should be between 10 and 20 μm, and the shear force probe should be positioned ~2–4 μm below the nano-DESI probe to ensure that the nano-DESI probe does not have direct contact with the sample surface. Acquire several line scans on the glass surface to check the stability of the liquid bridge. The distance between the nano-DESI probe and surface can be adjusted during the line scan by refining the position of the shear force probe. Specifically, the sample surface can be brought closer to the nano-DESI probe by moving the shear force probe up. Conversely, the nano-DESI probe can be moved farther away from the sample surface by moving the shear force probe down.
Data acquisition ● Timing variable (~4 h for a uterine section with a size of 2 mm × 1.5 mm)
-
20
Define the imaging region, scan rate, and line spacing (x and y directions). The imaging region should include either part of or the entire tissue section to be analyzed. The start position of the line scans should be set as close to the tissue edge as possible; the end position should be set at least 0.2 mm beyond the tissue edge to ensure the analyte molecules extracted from the edge have enough time to be transferred to the mass spectrometer. Typical scan rate and line spacing used in high-resolution nano-DESI MSI experiments are 5–40 μm/s and 10–30 μm, respectively. A scan rate of 20 μm/s and line spacing of 15 μm were used for imaging the sample shown in Fig. 4. We note that the average size of the liquid bridge (i.e., spatial resolution) is ~10 μm in the x and y directions, and ~4 pixels or spectra (pixel size: 2.5 μm (x) × 10 μm (y)) can be acquired from each sampling spot. It is very likely that there will be oversampling along the x axis. However, it is difficult to provide a conclusive statement regarding the oversampling because the actual size of the liquid bridge is difficult to establish. A line spacing of 15 μm was used in the imaging experiments to avoid oversampling in the y direction.
-
21
Define the vertical start position (zstart) and the move-out position (zout). zstart is the position from which the sample will be moved toward the shear force probe at the beginning of each line. zout is the vertical position of the sample to which it is moved at the end of each line. Typical zstart and zout positions are ~20 and 200 μm below the nano-DESI probe, respectively. Keeping zout farther away from the sample helps prevent crashing of the probe into the sample when the probe moves from line to line, whereas setting zstart closer to the sample helps reduce the time required for landing the probe on the sample. The speed of the z stage is set to 50 μm/s. After each line scan, the sample holder moves down to zout at 50 μm/s and then to the next xy start position at 1 mm/s. At the start of each line scan, the sample holder is first moved from zout to zstart at 50 μm/s and then moved up to the shear force probe. The approach of the sample to the shear force probe is controlled by the LabVIEW program using the working voltage selected in Step 18 at a frequency selected in Step 17 as a feedback.
-
22
Set up a run-sequence using Xcalibur. Open the software and create a method for each ionization mode (positive and negative). The methods include the optimized parameters and can be used in many experiments. Make sure that the run time of both methods is the same and is slightly shorter than the time of the line scan calculated by the LabVIEW program. You can choose to put the instrument into standby mode automatically after the sequence is completed. Create a run sequence and load the method. Each line scan corresponds to one sample in the sequence. In the sequence created for imaging of the sample shown in Fig. 4, a positive-mode method was used for the odd lines, and a negative-mode method was used for the even lines, which enabled acquisition of both positive- and negative-mode data in the same imaging experiment.
-
23
Start the acquisition. Ensure that there is sufficient solvent in the syringe for the entire acquisition. Click the ‘run sequence’ button in Xcalibur, and when the instrument shows ‘waiting for a contact closure’, start the line scan over the sample, using the LabVIEW program. When the probe lands on the surface, the contact closure signal is generated by a multifunction I/O device (USB-6009), which starts the data acquisition. We recommend using the microscopes to regularly monitor a few line scans during the imaging experiment. Typically, 8–10 small samples (such as pancreatic islets; surface area: 0.5 mm × 0.5 mm) or 2–3 larger samples (such as uterine samples; surface area: 2 mm × 2 mm) can be imaged in 1 d. Redo Step 13 (check the stability of the liquid bridge), Step 17 (obtain approach curve), and Step 19 (align the nano-DESI and shear force probe) before the imaging experiments each day.
? TROUBLESHOOTING
Data analysis ● Timing variable (6 h–1 d for a uterine section with a size of 2 mm × 1.5 mm)
-
24
Assign MS peaks. To generate a mass list of peak assignments, open several raw files using Xcalibur and select a representative line scan. Average mass spectra should be collected from both the tissue and solvent. View the spectrum list, select all peaks, and export the exact mass with a certain threshold (typically 0.5% in our experiments). Align the exact mass lists from the tissue and solvent, remove isotopic peaks and peaks originating from the solvent. Usually, solvent peaks are strongly suppressed on the tissue. This allows us to eliminate solvent peaks on the basis of the ratio of intensities in the spectra of the solvent and tissue. An intensity ratio (tissue versus solvent) of >3 is typically used to classify peaks originating from the tissue. The selected peaks observed on the tissue are tentatively assigned using the Lipid Maps (http://www.lipidmaps.org/tools/ms/lm_mass_form.php) and Metlin (https://metlin.scripps.edu/landing_page.php?pgcontent=advanced_search) databases on the basis of accurate m/z values (those with an error of <5 p.p.m.), and the assignments are confirmed using MS/MS data acquired over the tissue using data-dependent acquisition after each imaging experiment.
-
25
Visualize the raw files using MSI QuickView. Briefly, load the raw files into the software, define the aspect ratio of the sampled area, load the mass list to be visualized, generate ion images for each mass, and save the ion images. The positive mode and negative data acquired from uterine tissues should be processed and visualized separately. The ion images can be normalized to either the TIC or the signal of the standard (LPC 19:0 for positive mode; AA-d8 for FAs in negative mode; and PI (16:0/16:0) for phospholipids in negative mode).
-
26
Perform region of interest (ROI) analyses. ROI analyses can be performed on the imaging data using MSI QuickView. Select an ROI and export average mass spectra. We recommend dividing the region into three sub-regions of similar size to obtain an average value and standard deviation. Compile the exported data into an Excel file and normalize the intensities of the endogenous molecules to the intensity of the corresponding standard. The normalized intensities are proportional to the actual concentrations free of matrix effects. Furthermore, statistics analysis (e.g., two-tailed Student’s t-test) can be performed on those data.
Troubleshooting
Troubleshooting advice can be found in Table 3.
Table 3 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1,4,8 | The size and shape of the pulled tip are not reproducible | The puller has not warmed up | Turn the puller on and let it warm up for at least 15 min before pulling the tips |
| Dirty retro mirror | Clean the mirror with Kimwipes and ethanol | ||
| The capillary is not correctly positioned | Carefully position the capillary in the puller to ensure it is correctly placed in the V-groove of the puller bar | ||
| 3 | The solvent is not flowing through the capillary when pushed with a syringe | One of the connections is leaking The pulled tip is closed | Check the connections Disconnect the tip from the syringe, and examine it under microscope. If the tip is closed, repeat Steps 1–3 |
| 12,13 | MS signals are unstable | Unstable flow of the solvent through the nanospray capillary The solvent flow delivered by the primary capillary is too low |
Balance the solvent flow between the primary and nanospray capillary by adjusting the position of the nanospray capillary relative to the mass spectrometer inlet. If air bubbles are observed inside the nanospray capillary, move the capillary further away from the mass spectrometer inlet |
| Examine the primary capillary tip under a microscope. If the tip is clogged, pull the capillary to make a new tip. If the new tip becomes clogged again quickly, most likely the other side of the capillary is broken. Repeat Steps 1–4 to make a new primary capillary | |||
| 16,17 | A sudden and marked change occurs in the amplitude of the shear force probe vibration at a selected frequency | The shear force capillary may be broken One of the wire connections may be broken |
Examine the shear force capillary under a microscope. If the tip is broken, replace it with a new probe Check the connections using a multimeter, and reconnect the broken wire |
| 23 | The nano-DESI probe crashes into the sample or loses contact with the sample during data acquisition | The vertical position of the nano-DESI probe relative to the shear force probe needs to be readjusted | If the nano-DESI probe crashes into the sample, which means the nano-DESI probe is positioned too close to the sample, move the shear force probe down closer to the sample using a stepping controller. This will increase the distance between the sample and the nano-DESI probe |
| If the nano-DESI probe loses contact with the sample, move the shear force probe up further away from the sample, using a stepping controller, until the liquid bridge is formed between the nano-DESI probe and the sample |
Timing
Steps 1–7, fabrication of the primary and nanospray capillaries: ~1.5 h
Step 8, fabrication of the shear force probe: 20 min
Steps 9–13, setup of the nano-DESI probe: 1–2 h
Steps 14–19, assembly and positioning of the shear force probe: 1 h
Steps 20–23, data acquisition: variable, depending on the number of samples and the size of each sample (~4 h for the sample shown in Fig. 4)
Steps 24–26, data analysis: variable, depending on the size of the collected imaging data (6 h–1 d for the sample in Fig. 4)
Anticipated results
Using this protocol, we were able to determine the localization of lipids and metabolites in brain15, lung15, pancreatic islets16, and uterus with a resolution of better than 10 μm. Typically, hundreds of lipids and metabolites are detected in a single sample, including PC, PE, SM, PI, PS, phosphatidylglycerol (PG), FAs, and other metabolites. PC and SM species are often detected in positive ionization mode, whereas PI, PS, PG, and FAs are observed in negative ionization mode. For both the pancreatic islets and uterine tissue sections, we found that PC species (e.g., PC 34:1, PC 36:2, PC 38:4, and their analogs) are the most abundant compounds in positive mode, whereas the most abundant species observed in negative mode include FAs (e.g., linoleic acid, arachidonic acid, docosahexaenoic acid, and their analogs) and PI species (e.g., PI 38:4 and its analogs).
High-quality ion images of lipids and metabolites have been obtained using the protocol described herein. By alternating between positive- and negative-mode acquisition, we were able to obtain high-resolution ion images of >500 distinct features for each uterine section (~250 features in each ionization mode). Major classes of compounds observed in these experiments include phospholipids (e.g., PC, SM, PI, and PS species), FAs, and other metabolites. Figure 4 shows representative ion images acquired for mouse uterine tissue sections. Our data reveal substantially different localization of phospholipids to uterine stroma, LE, GE, and Myo cells. For example, PC 36:2, PC 36:1, SM 24:0, and PG 44:12 are substantially enhanced in LE cells; SM 16:0 is enhanced in GE cells; PC 36:3, PI 36:2, PI 38:3, PG 34:1, SM 24:1, and LPE 22:4 are enhanced in both LE and GE cells. Meanwhile, PC 32:0, LPC 16:0, and LPE 18:0 are markedly suppressed in both LE and GE cells, and enhanced in stroma cells. Remarkably different distributions are observed for both phospholipids and FAs differing by only one double bond, such as PC 32:1 versus PC 32:0, PC 36:4 versus PC 36:3, PC 36:3 versus PC 36:2, PI 38:4 versus PI 38:3, arachidonic acid (FA 20:4) versus FA 20:3, and FA 20:3 versus FA 20:2. These results demonstrate that minor variations in the FA chain length and saturation may have a pronounced effect on the localization of both phospholipids and FAs in biological tissues.
Ion images allow us to accurately determinate the relationships between phospholipids and FAs, and between different phospholipids in the same types of uterine cells. For example, depending on the lipid class, arachidonate-containing phospholipids were found to have different localization patterns. Specifically, both PC 36:4 (16:0/20:4) and PC 38:4 (18:0/20:4) are slightly enhanced in LE cells, whereas PI 38:4 (18:0/20:4) is suppressed in LE cells (Fig. 4), which is consistent with previous study45. Furthermore, arachidonic acid (FA 20:4) released by cytosolic phospholipase A2α (cPLA2α) from those phospholipids46 has a similar distribution to that of PI 38:4. The observed suppression of FA 20:4 in LE cells is associated with a substantial enhancement of the hydroxyeicosatetraenoic acid (HETE) in the same area, which is a metabolite of arachidonic acid produced by arachidonate-5-lipoxygenase47. It has been demonstrated that FA 20:4 and its metabolites (e.g., prostaglandin and 5-HETE) play a critical role in the process of embryo development24,48,49. In addition, we found that in comparison to phospholipids showing substantial enhancement in LE and GE cells (e.g., PC 36:2 and PI 38:3), FAs show distinctly different spatial patterns with a broadened shape around both LE and GE cells (Fig. 4). This interesting observation may be related to epithelial–stromal crosstalk and merits further investigation.
ROI analyses provide additional insights into the localization of lipids and metabolites to different types of cells in the tissue. Figure 7 shows the relative abundances of representative phospholipids in uterine LE, GE, and stroma cells. We observe that the GE cells have a 52% higher level (t-test, P < 0.001) of PC 32:0 compared to the LE cells. The levels of PC 36:4 in the LE and GE cells are 28 and 64% higher, respectively, than that of the stroma cells (P < 0.05). Our data indicate that small changes in the abundance of phospholipids can be determined using ROI analysis. The protocol presented herein can be readily extended to other types of biological systems and conditions. For example, MSI with a spatial resolution of 100–150 μm showed remarkable alterations in phospholipid distributions in distinct cellular areas of the uterus (e.g., antimesometrial side and mesometrial side) during days 4–8 of pregnancy that were associated with abnormal mouse embryo implantation19,45,50. High-resolution nano-DESI MSI can be used to examine molecular signatures of embryo implantation with single-cell resolution. This protocol also can be used for imaging of drugs and their metabolites in dosed animal tissues13.
Fig. 7 |. Quantitative analysis of endogenous PC species in different types of uterine cells.
y axis presents the normalized intensities of PC species to LPC 19:0. t-test analysis was performed for LE versus S cells and GE versus S cells, and the resulting P values are shown above the corresponding bars; *P < 0.05, **P < 0.01, ***P < 0.001. For PC 32:0, PC36:2 and PC 36:1, t-test analysis was also performed for LE versus GE cells; the resulting P values are displayed using square brackets.
In summary, our imaging approach enables sensitive and quantitative imaging of lipids and metabolites in different types of tissue sections with high throughput and spatial resolution. This protocol will provide important insights into biochemical processes occurring in biological tissues of interest for drug discovery, molecular biology, and clinical research.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under award UG3HL145593 (HuBMAP Program, J.L. and K.E.B.-J.), the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, grant R21 HD084788 to K.E.B.-J.), and NIH grants (DA006668 and HD068524 to S.K.D.).
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0237-4.
Peer review information Nature Protocols thanks Pierre Chaurand and Livia Schiavinato Eberlin for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The data presented in this study are available from the corresponding author upon reasonable request.
Code availability
The LabVIEW and MSI QuickView programs are available from the corresponding author upon reasonable request.
References
- 1.Norris JL & Caprioli RM Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev 113, 2309–2342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaysse P-M, Heeren R, Porta T & Balluff B Mass spectrometry imaging for clinical research–latest developments, applications, and current limitations. Analyst 142, 2690–2712 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Chaurand P Imaging mass spectrometry of thin tissue sections: a decade of collective efforts. J. Proteom 75, 4883–4892 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Wu C, Dill AL, Eberlin LS, Cooks RG & Ifa DR Mass spectrometry imaging under ambient conditions. Mass Spectrom. Rev 32, 218–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laskin J & Lanekoff I Ambient mass spectrometry imaging using direct liquid extraction techniques. Anal. Chem 88, 52–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feider CL, Krieger A, DeHoog RJ & Eberlin LS Ambient ionization mass spectrometry: recent developments and applications. Anal. Chem 91, 4266–4290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubakhin SS, Jurchen JC, Monroe EB & Sweedler JV Imaging mass spectrometry: fundamentals and applications to drug discovery. Drug Discov. Today 10, 823–837 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Greer T, Sturm R & Li L Mass spectrometry imaging for drugs and metabolites. J. Proteom 74, 2617–2631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kompauer M, Heiles S & Spengler B Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 14, 90–96 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zavalin A et al. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J. Mass Spectrom 47, 1473–1481 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Laskin J, Heath BS, Roach PJ, Cazares L & Semmes OJ Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 84, 141–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanekoff I et al. Automated platform for high-resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 84, 8351–8356 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Lanekoff I et al. Imaging nicotine in rat brain tissue by use of nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 85, 882–889 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Lanekoff I, Thomas M & Laskin J Shotgun approach for quantitative imaging of phospholipids using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 86, 1872–1880 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Nguyen SN et al. Towards high-resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry coupled to shear force microscopy. J. Am. Soc. Mass. Spectrom 29, 316–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin R et al. High spatial resolution imaging of mouse pancreatic islets using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 90, 6548–6555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergman H-M, Lundin E, Andersson M & Lanekoff I Quantitative mass spectrometry imaging of small-molecule neurotransmitters in rat brain tissue sections using nanospray desorption electrospray ionization. Analyst 141, 3686–3695 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Duncan KD & Lanekoff I Oversampling to improve spatial resolution for liquid extraction mass spectrometry imaging. Anal. Chem 90, 2451–2455 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Lanekoff I et al. Trp53 deficient mice predisposed to preterm birth display region-specific lipid alterations at the embryo implantation site. Sci. Rep 6, 33023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen SN, Liyu AV, Chu RK, Anderton CR & Laskin J Constant-distance mode nanospray desorption electrospray ionization mass spectrometry imaging of biological samples with complex topography. Anal. Chem 89, 1131–1137 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Lanekoff I & Laskin J Imaging of lipids and metabolites using nanospray desorption electrospray ionization mass spectrometry. Methods Mol. Biol 1203, 99–106 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Ballesteros Katemann B, Schulte A & Schuhmann W Constant-distance mode scanning electrochemical microscopy (SECM)—part I: adaptation of a non-optical shear-force-based positioning mode for SECM tips. Chem. Eur. J 9, 2025–2033 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Ndobo-Epoy JP, Lesniewska E & Guicquero JP Shear force microscopy with a nanoscale resolution. Ultramicroscopy 103, 229–236 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Wang H & Dey SK Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet 7, 185–199 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Heeren RM, Smith DF, Stauber J, Kukrer-Kaletas B & MacAleese L Imaging mass spectrometry: hype or hope? J. Am. Soc. Mass Spectrom 20, 1006–1014 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hankin JA & Murphy RC Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal. Chem 82, 8476–8484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanekoff I, Stevens SL, Stenzel-Poore MP & Laskin J Matrix effects in biological mass spectrometry imaging: identification and compensation. Analyst 139, 3528–3532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan KD, Bergman H-M & Lanekoff I A pneumatically assisted nanospray desorption electrospray ionization source for increased solvent versatility and enhanced metabolite detection from tissue. Analyst 142, 3424–3431 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bodzon-Kulakowska A & Suder P Imaging mass spectrometry: instrumentation, applications, and combination with other visualization techniques. Mass Spectrom. Rev 35, 147–169 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Gamble LJ & Anderton CR Secondary ion mass spectrometry imaging of tissues, cells, and microbial systems. Micros Today 24, 24–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian H et al. Gas cluster ion beam time-of-flight secondary ion mass spectrometry high-resolution imaging of cardiolipin speciation in the brain: identification of molecular losses after traumatic injury. Anal. Chem 89, 4611–4619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian H et al. Secondary-ion mass spectrometry images cardiolipins and phosphatidylethanolamines at the subcellular level. Angew. Chem 131, 3188–3193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zavalin A, Yang J, Hayden K, Vestal M & Caprioli RM Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal. Bioanal. Chem 407, 2337–2342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao W, Pan N & Yang Z High resolution tissue imaging using the single-probe mass spectrometry under ambient conditions. J. Am. Soc. Mass Spectrom 26, 986–993 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Campbell DI, Ferreira CR, Eberlin LS & Cooks RG Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization. Anal. Bioanal. Chem 404, 389–398 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Kim JY et al. Atmospheric pressure mass spectrometric imaging of live hippocampal tissue slices with subcellular spatial resolution. Nat. Commun 8, 2113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luxembourg SL, Mize TH, McDonnell LA & Heeren RM High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal. Chem 76, 5339–5344 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Schwartz SA, Reyzer ML & Caprioli RM Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom 38, 699–708 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Stoeckli M, Staab D & Schweitzer A Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int. J. Mass Spectrom 260, 195–202 (2007). [Google Scholar]
- 40.Nguyen SN et al. Lipid coverage in nanospray desorption electrospray ionization mass spectrometry imaging of mouse lung tissues. Anal. Chem 91, 11629–11635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan KD et al. Quantitative mass spectrometry imaging of prostaglandins as silver ion adducts with nanospray desorption electrospray ionization. Anal. Chem 90, 7246–7252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C, Ifa DR, Manicke NE & Cooks RG Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal. Chem 81, 7618–7624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garza KY et al. Desorption electrospray ionization mass spectrometry imaging of proteins directly from biological tissue sections. Anal. Chem 90, 7785–7789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu CC, Chou PT & Zare RN Imaging of proteins in tissue samples using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem 87, 11171–11175 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Burnum KE et al. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res 50, 2290–2298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song H et al. Cytosolic phospholipase A2alpha is crucial for ‘on-time’ embryo implantation that directs subsequent development. Development 129, 2879–2889 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Brash AR, Boeglin WE & Chang MS Discovery of a second 15S-lipoxygenase in humans. Proc. Natl. Acad. Sci. USA 94, 6148–6152 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim H et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91, 197–208 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Walsh SW Evidence for 5-hydroxyeicosatetraenoic acid (5-HETE) and leukotriene C4(LTC4) in the onset of labor. Ann. NY Acad. Sci 622, 341–354 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Lanekoff I et al. Three-dimensional imaging of lipids and metabolites in tissues by nanospray desorption electrospray ionization mass spectrometry. Anal. Bioanal. Chem 407, 2063–2071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]