SUMMARY
Many adult tissues contain resident stem cells, such as the Pax7+ satellite cells within skeletal muscle, that regenerate parenchymal elements following damage. Tissue-resident mesenchymal progenitors (MPs) also participate in regeneration, although their function and fate in this process are unclear. Here, we identify Hypermethylated in cancer 1 (Hic1) as a marker of MPs in skeletal muscle and further show that Hic1 deletion leads to MP hyperplasia. Single cell RNA-seq and ATAC-seq analysis of Hic1+ MPs in skeletal muscle shows multiple subpopulations, which we further show have distinct functions and lineage potential. Hic1+ MPs orchestrate multiple aspects of skeletal muscle regeneration by providing stage-specific immunomodulation, trophic and mechanical support. During muscle regeneration, Hic1+ derivatives directly contribute to several mesenchymal compartments including Col22a1-expressing cells within the myotendinous junction. Collectively, these findings demonstrate that HIC1 regulates MP quiescence, and identifies MP subpopulations with transient and enduring roles in muscle regeneration.
Keywords: Quiescence, tissue regeneration, lineage tracing, mesenchymal progenitors, tendon, skeletal muscle, pericytes, myotendinous junction, scRNA-seq, scATAC-seq
Graphical Abstract

eTOC:
Multiple stem/progenitor populations, including stromal mesenchymal progenitors (MPs) participate in skeletal muscle regeneration. We found Hic1 is a functional marker for MP quiescence, and Hic1+ MPs coordinate multiple facets of the muscle regeneration program and contribute to several mesenchymal lineages, including “myotenocytes”.
INTRODUCTION
Mesenchymal progenitors (MPs) are thought to regulate tissue maintenance and regeneration by providing trophic support to tissue specific stem cells (Ding et al., 2012; Mendez-Ferrer et al., 2010; Sigal et al., 2017; Zepp et al., 2017; Zhao et al., 2017), a concept that has supported their use in cell therapy approaches. A clear example of such a role, stems from studies in adult skeletal muscle, whose efficient regeneration requires the coordinated action of distinct tissue-resident stem/progenitor cell populations (Murphy et al., 2011). Figuring prominently in this scenario are Pax7+ satellite cells, which are essential for post-natal muscle formation and regeneration following damage (Lepper et al., 2011; Sambasivan et al., 2011). In addition to satellite cells, we and others have identified a population of mesenchymal fibro/adipogenic progenitors (FAPs) within the muscle interstitium that also plays an important role in regeneration (Joe et al., 2010; Murphy et al., 2011; Uezumi et al., 2010). Kardon and colleagues used Tcf7l2-based transgenic tools to identify and conditionally ablate this population, showing that a decrease of ~ 45% in these cells was sufficient to dysregulate satellite cell differentiation which negatively impacted muscle regeneration (Murphy et al., 2011). In addition to playing a positive role in regeneration, these progenitors are a main source of the pathogenic extracellular matrix (ECM)-expressing cells underlying muscle fibrosis, which is often observed in aging, chronic disease or in conditions in which regeneration is impaired (Lemos et al., 2015; Uezumi et al., 2011).
Despite the clear evidence of important roles for stromal progenitors in health and disease, our understanding of their contribution to regenerating tissues is limited by the lack of reliable markers to define them. Mesenchymal populations within various tissues have been identified using a variety of markers including Nestin-GFP, Gli1, Grem1, Lepr, PDGFRA, Sca1 (LY6A) and CD34, and combinations thereof (Joe et al., 2010; Kramann et al., 2015; Mendez-Ferrer et al., 2010; Uezumi et al., 2010; Worthley et al., 2015; Zhou et al., 2014). Cre lines based on these markers have provided fundamental insights into their lineage contributions in vivo (Chen et al., 2017; Scott and Underhill, 2016). Mesenchymal cells with potential stem cell activity, or mesenchymal stem cells (MSCs), were originally identified in bone marrow (BM) (Bianco, 2014; Bianco and Robey, 2015). Within this tissue, Lepr expression has been used to identify mesenchymal cells that contribute to the adipogenic and the osteogenic lineages (Zhou et al., 2014), and like LepR, Grem1 expression can also be used to identify BM-MSCs with endogenous osteogenic lineage potential (Worthley et al., 2015). Gli1 appears to mark an MSC-like population in bone and across multiple tissues, where Gli1+ cells generate myofibroblasts in fibrosis (Kramann et al., 2015; Schneider et al., 2017). While these markers allowed a preliminary glimpse of the in vivo role of MPs, it is unclear whether they label specialized, possibly lineage committed subsets of cells, and a marker capable of reliably identifying immature progenitors is currently lacking. As a consequence, the extent of MP heterogeneity and the molecular mechanisms modulating MP fate and function are poorly understood. Here, we identify the gene Hypermethylated in Cancer 1 (Hic1) as a marker of MPs in skeletal muscle. Newly generated transgenic tools based on this marker allow the lineage tracing and molecular characterization of these cells and their progeny, highlighting their role as a signaling hub in coordinating the response to damage, and revealing the extent of their contributions to mature lineages in homeostasis and regeneration. In addition, findings related to Hic1 expression and deletion reveal a fundamental role for this factor in regulating MP quiescence and as a consequence the abundance of tissue resident MPs at homeostasis.
RESULTS
HIC1 marks MPs within skeletal muscle
To identify MP-specific markers we fractionated whole muscle into multiple populations (Figures 1A and S1A) and focused on the analysis of the Lin− (CD31−CD45−Ter119−) LY6A+ population which we previously showed was enriched for MPs (Joe et al., 2010). RNA-seq analysis was applied to these fractions (entire population - popRNA-seq) to identify markers enriched in the Lin−LY6A+ fraction (Figure 1B). As expected, within this fraction there was a clear enrichment of several known MP-related markers including Pdgfra, Thy1 and Gli1 (Figures S1B and Table S1). Interestingly, pericyte markers including Rgs5, Pdgfrb, Mcam, Cspg4 and Kcnj8 (Armulik et al., 2011) were substantially enriched in the Lin−LY6A− fraction as were markers reflective of the tenogenic lineage (Scx, Mkx and Tnmd) (Huang et al., 2015) (Figures S1C and S1D). This fraction also contained a strong satellite signature associated with expression of Pax7 (Dumont and Rudnicki, 2017)(Figures 1B and S1D). Within the Lin−LY6A+ fraction we noted a substantial enrichment in the transcript for Hic1, whose expression also tightly correlated with that of Gli1, a marker expressed in MSC-like cells across many tissues (Figures 1B and 1C) (Kramann et al., 2015). Hic1 encodes for a transcriptional repressor with potential tumor suppressor activity that has been shown to directly regulate genes involved in the cell cycle (Chen et al., 2003; Fleuriel et al., 2009; Van Rechem et al., 2010). It was originally identified based on observations that the HIC1 locus was hypermethylated and transcriptionally silenced in diverse human tumors (Wales et al., 1995). Characterization of Hic1 in the developing mouse embryo showed that Hic1 is predominantly restricted to mesenchyme within various tissues and organs (Grimm et al., 1999; Pospichalova et al., 2011). In previous studies, we have also shown that Hic1 is a downstream target gene of the retinoic acid (RA) signaling pathway (Hassan et al., 2017), and that RA itself is a potent regulator of the mesenchymal phenotype (Dranse et al., 2011; Hoffman et al., 2006; Weston et al., 2002). For these various reasons, the role of Hic1 in MP biology was further explored.
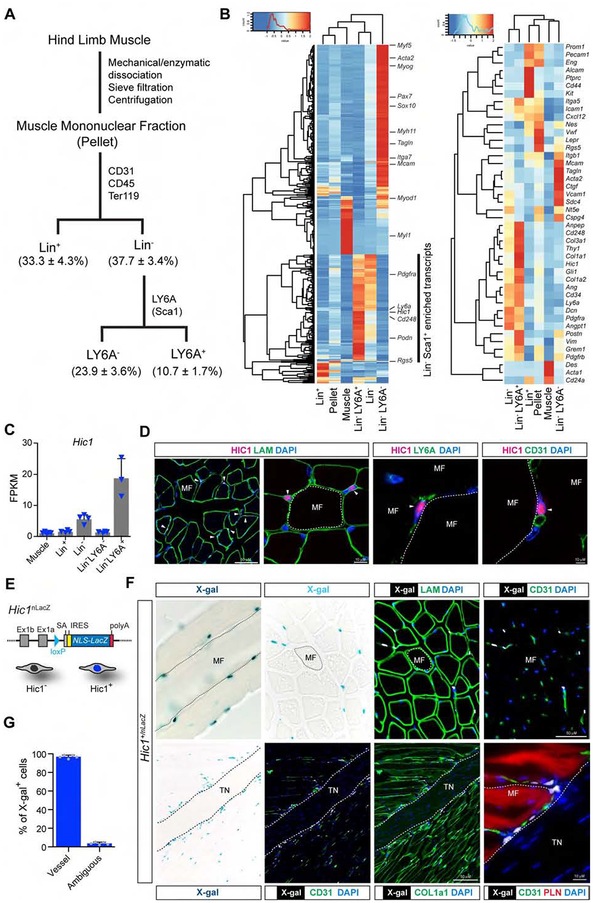
Figure 1:
Identification of Hic1 as an enriched transcript in MPs. A, schematic overview of the strategy used to purify MPs from TA muscle. Numbers in parentheses indicate percent of the total mononuclear fraction from 3 independent isolations (see Figure S1A for markers and sorting gates). B, heat map from RNA-seq analysis of the various fractions indicated in A. Select genes associated with various cell types within muscle are shown in the right panel, with Hic1 showing enrichment in the Lin− LY6A+ fraction. C, Hic1 transcript abundance in the different fractions from the RNA-seq analyses (n = 3–4, data represent the mean ± SD). D, representative image of anti-HIC1 staining of 8 wk adult TA muscle counterstained with Laminin, LY6A or CD31. White arrowheads, HIC1+ cells. E, schematic representation of the Hic1 null allele that expresses nuclear LacZ (see Figure S1E for Hic1 knock-in allele description). F, the distribution of Hic1+ cells was evaluated in representative whole-mount (upper left panel) and in situ X-gal stained sections of TA muscles and the myotendinous junction (bottom series) from Hic1+/nLacZ mice. PLN, phalloidin+ muscle fibre. G, Hic1+ cells were enumerated from stained samples in F (n = 5, data represent the mean ± SD). MF, myofibre; TN, tendon.
To validate the observed enrichment of Hic1 in the MP fraction of the RNA-seq data, immunofluorescence (IF) was carried out on adult tibialis anterior (TA) muscle. HIC1 was only detected within the nuclei of blood vessel associated LY6A+ cells in the muscle interstitium (Figure 1D). Analysis of a unique mouse strain containing a nuclear LacZ knock-in Hic1nLacZ/+ (Figures 1E and S1E), confirmed the presence of Hic1+ cells within the interstitial space (Figure 1F) in association with capillaries (Figures 1F and 1G). Within arterioles and arteries, Hic1+ cells were observed in the adventitia (Figure S1F) and collectively, 96.9 ± 0.8% of the Hic1+ cells exhibited a perivascular location (Figure 1G). Infrequent vessel-associated Hic1+ cells were also detected in the periphery of tendons (Figure 1F) and on vessels within the myotendinous junction (Figure 1F). In summary, Hic1/HIC1 is expressed in muscle resident MPs.
HIC1 regulates MP quiescence
In adult murine skeletal muscle under homeostatic conditions cellular turnover is limited, and the vast majority of satellite stem cells and MPs are in a quiescent state. Following muscle injury-induced activation, these populations enter the cell cycle and generate progeny to support tissue regeneration. To investigate the participation of Hic1+ MPs in muscle regeneration, a notexin (NTX)-induced skeletal muscle injury model was employed to produce acute myofiber degeneration within the TA muscle. As noted above, prior to injury, X-gal+ cells can be observed within the muscle interstitium (Figure 2A). Within the first few days post-injury (p.i.), as previously reported (Lemos et al., 2015), a pronounced increase in mononuclear cells was observed consistent with inflammation (Figures 2A and 2B). Unexpectedly, rather than the typical increase in MPs that would be apparent at these early time points (Lemos et al., 2015), a significant decrease in X-gal+ cells was observed within the injured areas (Figures 2A and 2C). However, at ~ 14 d p.i., the frequency and distribution of X-gal+ cells returned to pre-injury levels (Figures 2A and 2C). This pattern of X-gal+ cells pre and p.i. was also observed in in vitro cultures of Hic1+/nLacZ MPs following scratch-induced wound of a confluent cell monolayer (Figure S2A). Consistent with pre-injury distribution in vivo, > 96% of the X-gal+ cells were found associated with CD31+ blood vessels at 14 d p.i. (Figures S2B and S2C). Thus, Hic1 expression was readily detectable in growth-arrested MPs, decreased in activated MPs, and following appearance of regenerated myofibres or wound closure in vitro, re-emerged in attendant growth arrested MPs. Collectively, this pattern indicates that Hic1 expression is associated with the quiescent MP phenotype (Figure S2D). Consistent with this, Coller et al. (Coller et al., 2006) identified Hic1 as a quiescence program-associated gene in human lung fibroblasts and heterotopic expression of HIC1 has been shown to reduce colony forming activity (Van Rechem et al., 2010; Wales et al., 1995).
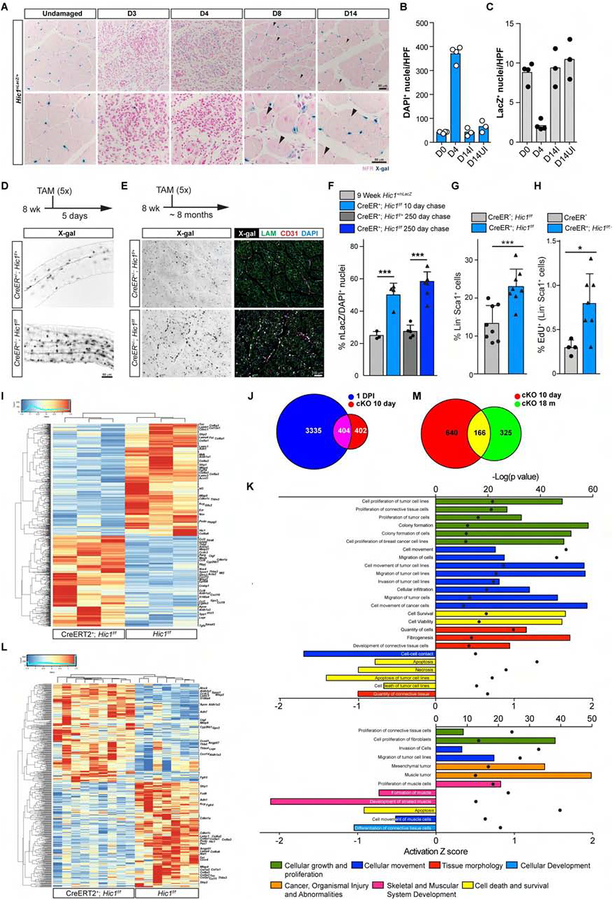
Figure 2:
HIC1 regulates MP quiescence: deletion of Hic1 leads to an increase in MP number and an activated MP-like phenotype. A, chronological series of representative X-gal stained muscle sections following TA muscle injury in Hic1nLacZ/+ mice. Arrowheads, centrally located nuclei in newly regenerated myofibres. B, enumeration of DAPI nuclei per high powered field (hpf) from A. UI, uninjured control; I, injured. C, quantification of X-gal stained Hic1+ cells from A (data represent the mean ± SD, n = 3–4). D, representative whole-mount images of X-gal stained myofibres 5 d post-TAM treatment from 9 wk old male mice. E, representative histological images of X-gal stained cells from Hic1-deleted and non-deleted TA muscles at 8 months post-TAM. F, enumeration of X-gal stained cells from TA muscles (n = 3–5, data represent the mean ± SD, one-way ANOVA Bonferroni post-test, ***p<0.001 relative to 9 wk Hic1+/nLacZ baseline). G, FCM analysis of Lin−LY6A+ MPs in TA muscle from Hic1f/f and Hic1-deleted mice 10 d post-TAM injection (see Figure S2E for sorting gates and markers) (n = 8, data represent the mean ± SD, unpaired t-test ***p<0.001). H, FCM analysis of EdU incorporation in Lin−LY6A+ MPs from wild-type and Hic1-deleted mice (n ≥ 4). Mice received TAM at 2 m of age and 3 consecutive daily doses (0.5 mg/mouse) of EdU starting on day 5 post-TAM and were collected for analysis 24 h after the last EdU injection (n = 4–7, data represent the mean ± SD, unpaired t-test *p<0.05). I, heatmap of differentially expressed genes (Cuffdiff, q< 0.05) from RNA-seq analysis of sorted MPs from 2 m TA muscles, (n = 3). J, pairwise Venn diagram plots of overlapping gene expression between the indicated datasets. The 1 d p.i. dataset was generated using a comparison of 0 d MPs to 1 d p.i. as described in Figure 5A. K, IPA interrogation of transcriptomic data, from data shown in I (top panel) and L (bottom panel). L, heatmap of differentially expressed genes (Cuffdiff, q< 0.05) from RNA-seq analysis of whole 18 m TA muscles (TAM at 2 m) (n = 7–9). M, pairwise Venn diagram plots of overlapping gene expression between samples described in H and K.
To test the role of Hic1 in quiescence, Hic1 was ubiquitously deleted in 9-week old mice (UBC-CreERT2; Hic1f/f or +). Following deletion, the nLacZ transcript reflects Hic1 promoter activity. Skeletal muscle tissues were collected at various times post-tamoxifen (TAM) injection and X-gal stained to identify Hic1+ cells (Figures 2D–2G). Under these conditions, an increase in X-gal+ cells relative to untreated controls was apparent by 5 d following TAM administration (Figure 2D) and remained stable for longer periods of time up to 8 months (Figure 2E). In particular, when whole-mount TA muscles were examined, numerous clusters of X-gal+ cells were observed in the Hic1 cKO samples, whereas the control samples exhibited a more even distribution of X-gal+ cells (Figure 2D). The increase in MP frequency following the conditional deletion of Hic1 was independently confirmed by flow cytometric enumeration of the Lin−LY6A+ fraction (Figures 2G and S2E). Consistent with this increase in X-gal+ cells immediately following TAM treatment, an ~ 2.7-fold increase in EdU (5-ethynyl-2’-deoxyuridine) incorporation indicative of proliferation was detected in the Lin−LY6A+ population in the TA muscles from Hic1 cKO samples in comparison to controls (Figure 2H). The increase in X-gal+ cells observed at 10 d post-TAM extended to animals examined 250 d later with similar abundance (Figure 2F). Thus, deletion of Hic1 leads to a rapid increase in MPs which is maintained over time, resulting in a higher number of Hic1-expressing cells in skeletal muscle at steady state. Analysis of RNA-seq data (Table S2) of sorted Lin−LY6A+ cells at 10 d post-TAM showed Hic1 deletion efficiency of 88.5% and an enrichment for genes associated with MP activation (Figure 2I and below). Comparison of differentially expressed genes in Hic1-deleted cells versus damage-induced activation revealed that > 50% of the differentially-expressed genes in the Hic1-deleted MPs overlapped with that of 1 d p.i.-activated MPs (Figure 2J and see below). Consistent with this activated MP phenotype, pathway analysis identified programs associated with cell survival, proliferation, migration and invasion in Hic1-deleted MPs versus control (Figures 2I and 2K). Thus, deletion of Hic1 leads to a partially activated MP phenotype (Figures 2I–2K, and S2F). Taken together, these findings indicate that HIC1 directly regulates quiescence and through this mechanism, the number of MPs present at homeostasis.
Hic1 deletion affects skeletal muscle regeneration
Conditional deletion of Hic1 leads to a > 2-fold increase in skeletal muscle MPs and an expression signature reflective of MP activation. To assess the impact of this modified MP state on skeletal muscle regeneration, NTX-induced injury was carried out on 3 m and 14 m old Hic1-deleted mice which received TAM treatment at 2 m of age. In both instances, myofiber regeneration was negatively influenced by Hic1 deletion, in that the average myofiber cross-sectional area was significantly reduced by 14.7% and 10.7% in the 3 m and 14 m cKOs, respectively (Figures S2G and S2I). This decrease was associated with an increase in the number of smaller myofibres in cKOs, both at 3 and 14 m (Figures S2G and S2I). Furthermore, picrosirius red staining (PSR) showed no significant differences in extracellular collagen in both control and cKOs TA muscles following injury in 3 m and 14 m mice (Figures S2G and S2H). To assess the impact of Hic1 deletion on muscle homeostasis, RNA-seq was carried out on 18 m old wt and cKO TA muscle samples (TAM at 2 m) (Table S3). Under these conditions Hic1 deletion was 92.4%. Surprisingly, several genes associated with the muscle satellite cell compartment (Marcelle et al., 1994; Yin et al., 2013) including Pax7, Cxcr4, Sdc3, Itga7 and Fgfr4 were significantly down-regulated in the cKO TA muscle in comparison to control samples (Figure 2L and S2J, and Table S3). A reduction in satellite cell number and/or decreased Pax7 expression have both been shown to compromise muscle regeneration (Gunther et al., 2013; Lepper et al., 2011; Sambasivan et al., 2011; von Maltzahn et al., 2013). Further inspection of all RNA-seq samples (cKO 10 d and 18 m) revealed significant down-regulation of multiple ECM-related genes, including collagens including Col5a1, 6a1, 6a2, 6a3, 6a6 in the cKO muscles (Figures 2I, 2L and S2F, S2J), the encoded products of which contribute to the satellite cell niche (Eklund et al., 2001; Hicks et al., 2014; Mohassel et al., 2018; Tierney et al., 2016; Urciuolo et al., 2013; Zou et al., 2014). Collagens 5 and 6 are also abundantly produced by the Lin−LY6A+ populations (Figure S2K). Consistent with these observations, pathway analysis of the 18 m TA muscles also revealed a negative impact on the development and formation of striated muscle in the cKO compared to control samples (Figure 2K). Furthermore, the Hic1-deleted phenotype appears reasonably stable as > 1/3 of the differentially expressed genes, which include several of the aforementioned collagen genes, in the 18 m cKO samples overlap with that of the 10 d sorted cKO samples (Figure 2M). In aggregate, these observations indicate that modulation of the MP compartment as described herein leads to smaller regenerated myofibres indicative of impaired muscle regeneration.
Hic1 marks multiple MP populations in skeletal muscle
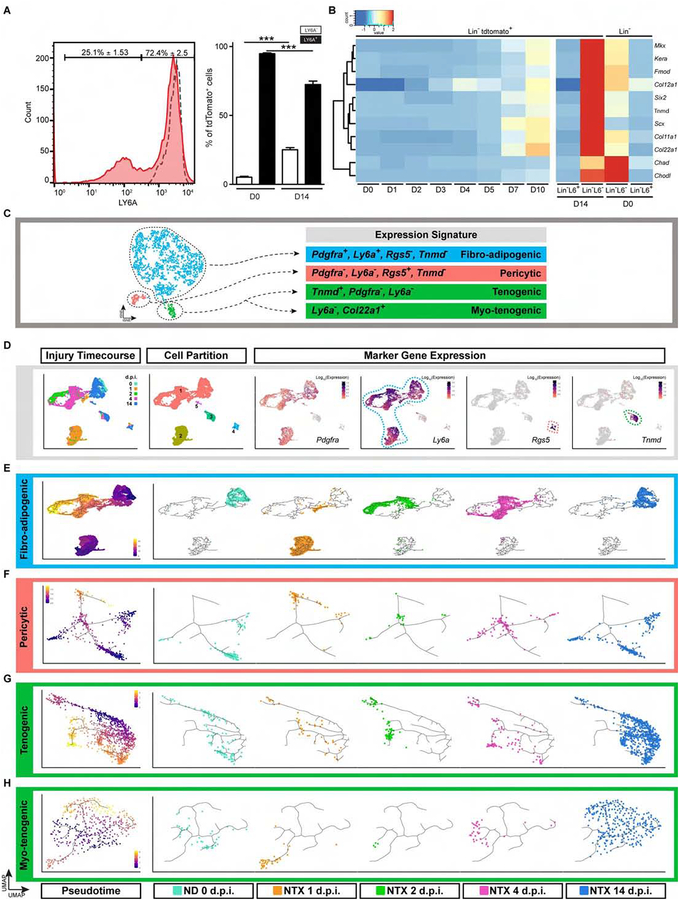
To more effectively mark, isolate and enable fate mapping of Hic1+ cells in muscle regeneration, a CreERT2 knock-in line was generated and bred to a conditional tdTomato reporter line, Rosa26LSL-tdTom (Figures 3A and S3A). Flow cytometry (FCM) analysis of tdTomato+ cells immediately after TAM treatment showed that 94.8 ± 0.7% of this population was Lin−LY6A+ FAPs (Figure 3B), whereas tdTomato+ cells represented 84.7% ± 1.4% of the Lin−LY6A+ fraction. In addition, the transcriptome of tdTomato+ cells highly correlated (R2=.93) with that of the Lin−LY6A+ fraction from Figure 1B (Figures 1B and S3B), suggesting extensive overlap between the two subsets. Indeed, labelled cells were enriched for the expression of MP and stem/progenitor cell markers such as Cd34, Gli1, Pdgfra and Thy1, but lacked markers associated with a myogenic cell fate such as Pax7 (Figure S3C).
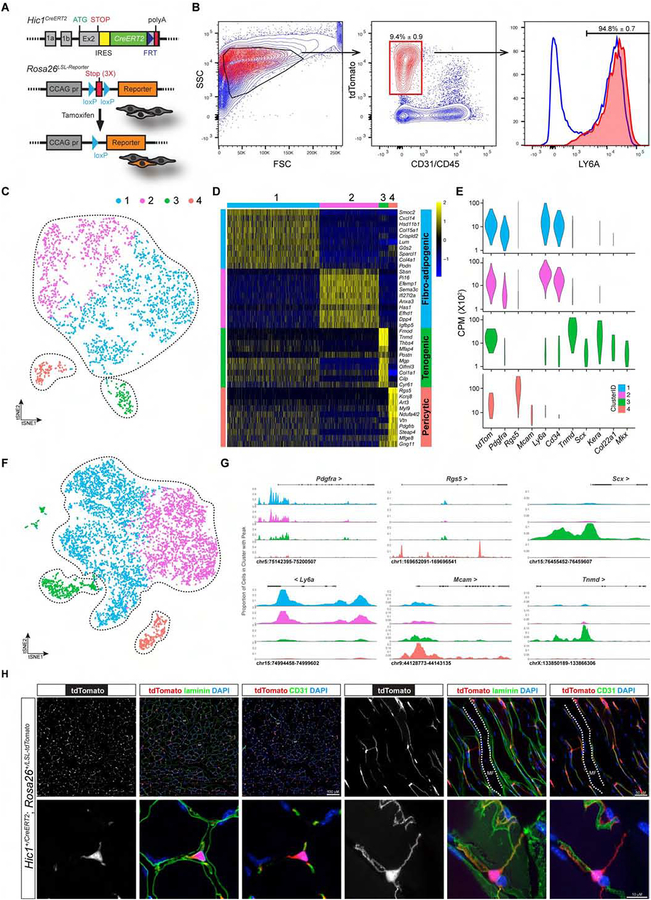
Figure 3:
Generation and use of a Hic1CreERT2 knock-in allele to characterize Hic1+ MPs. A, overview of the Hic1CreERT2 knock-in allele and associated lineage-tracing strategy (see Figure S3A for description of Hic1 knock-in allele). B, FCM analysis of enzymatically dissociated TA muscles from Hic1CreERT2; Rosa26LSL-tdTomato mice post-TAM (n ≥ 4, data reflect the mean ± SD). C, t-SNE plot of scRNA-seq data from enriched tdTomato+ MPs 10 d post-TAM induction. D, heat-map of scRNA-seq data showing enriched genes in the 4 different clusters. E, violin plots showing expression of select lineage-associated genes from the scRNA-seq data. F, tSNE plot of scATAC-seq from enriched tdTomato+ MPs using a similar TAM injection regime as in D. The clusters indicative of the 4 populations were colored using the same convention as in C–E. G, genome browser tracks displaying the promoter sum signal around the indicated gene loci from the 4 defined clusters. H, representative transverse and longitudinal sections of the TA muscle at 5 d post-TAM, counterstained as indicated.
While FAPs comprise the vast majority of cells labelled with this approach, FCM data indicates that about 5% of tdTomato+ cells are LY6A−, implying additional cell types also express Hic1. To further explore the heterogeneity within Hic1-expressing cells, 2,173 Lin−tdTomato+ cells were profiled using single cell (sc)RNA-seq. Cluster analysis revealed four subsets, a major one comprised of two sub-populations expressing FAP markers (Cd34, Ly6a, Pdgfra) as expected, and two smaller subsets characterized by absent/low Ly6a expression (Figures 3C–3E). The FAP population sub-clustered into 2 populations, and this was in part due to increased expression of genes associated with the ECM in the FAP1 (blue), such as collagens Col4a1, 4a2, Col6a1, 6a2, 6a3 and Col15a1, and numerous other ECM genes including Lum, Sparcl1, Podn, Smoc2, Mgp and Bgn (Figure 3D). In contrast, FAP2 (magenta) was enriched for transcripts involved in multiple biological programs and cell signaling pathways (Sfrp4, Igfbp5, Sema3c, Dpp4, Tgfrb2 and Wnt2) (Figure 3D). The latter 2 Ly6alo populations were also low for other FAP markers (Cd34 and Pdgfra) but were each enriched for markers reflective of tenogenic cells (Scx, Mkx and Tnmd) or pericytes (Rgs5, Kcnj8, Mcam and Pdgfrb) (Figures 3D–3E). To complement these analyses and further characterize the nature of the tdTomato+ populations, chromatin accessibility was investigated using scATAC-seq (Assay for Transposase-Accessible Chromatin). Similar to that observed with scRNA-seq, a tenogenic and pericytic population along with 2 FAP sub-populations were identified from the analysis of 7,273 nuclei (Figure 3F). Expectedly, transcripts indicative of the individual clusters were also associated with regions of open chromatin (Figure 3G). Interestingly, lineage-defining genes such as Scx and Rgs5 within the minor clusters were restricted to their corresponding clusters, indicating that these populations may exhibit restricted or unique potential in comparison to the FAP clusters (Figure 3G). Collectively, these findings indicate that Hic1 is expressed in at least 3 phenotypically distinct mesenchymal cell populations within skeletal muscle.
In situ analysis of TdTomato+ cells revealed a basement membrane shared with CD31+ capillaries (Figures 3H and S3D), and further enabled the visualization of a surprisingly intricate network of delicate dendritic processes decorating host and neighboring vessels. Similar to that observed with analysis of Hic1nLacZ/+ cell distribution (Figures 1F and 1G), tdTomato+ cells were restricted to the adventitia in arteries (Figure S3E). Within the tdTomato+ population, numerous Pdgfra+ (PdgfraH2b-EGFP) cells can also be visualized reflective of FAPs (Figure S3D). Under homeostatic conditions these cells appeared quiescent, as shown by the negligible levels (0.17 ± 0.06%, n = 3) of EdU incorporation. Furthermore, consistent with the transcriptomic features of quiescent stem and progenitor cells (Cheung and Rando, 2013), genes associated with cell cycle entry and progression (i.e. Birc5, Ccna2, Ccnb1, Ccne2 and Mki67), while not transcribed in all 4 clusters (Figures 5H–5I and S5D), contained robust peaks indicative of open chromatin domains (Figure S3G). Consistent with these properties, sorted tdTomato+ cells exhibited robust colony forming activity with a frequency of 1 in 17 ± 5.7, a frequency congruent with that reported by other groups for skeletal muscle (Joe et al., 2010) and LepR+ cells in the bone marrow (Zhou et al., 2014). Collectively, these findings demonstrate that Hic1 is expressed by multiple quiescent mesenchymal populations.
Figure 5:
The activated MP phenotype displays stage-specific activities indicative of a coordinated response to injury. A, RNA-seq analysis of tdTomato+ enriched MPs at various time points p.i. Select gene profiles of identified cellular programs are shown. Cxcl5, cytokines; Mki67, cell cycle; Postn, provisional matrix; Lamc1, basement membrane. B, heatmap from popRNA-seq of differentially-expressed cytokine genes after injury. C, representative image of anti-CXCL5 staining in control and NTX-injured TA muscles from Hic1;tdTomato reporter mice. Graph inset, quantification of CXCL5+tdTomato+ cells p.i. (n ≥ 3, data reflect the mean ± SD). D, scRNA-seq t-SNE clustering of cells at the indicated time points p.i. E, heat-map of scRNA-seq hierarchically clustered genes. The vertical colored lines reflect the programs indicated in A. T, tenogenic cluster; P, pericytic cluster. F, t-SNE plots of scRNA-seq data for Pdgfra and Cxcl5. The pericytic and tendogenic lineages are indicated by salmon and green dashed lines, respectively. G, analysis of cytokine gene chromatin accessibility and gene expression using scATAC-seq and scRNA-seq, respectively. Genome browser tracks displaying the promoter sum signal around the indicated gene loci from the 4 defined clusters from scATAC-seq (D0) are shown along with the corresponding tSNE plot of the scRNA-seq expression data. H, heatmap from popRNA-seq highlighting genes associated with cell proliferation. I, t-SNE plots of clusters expressing Mki67 at different times p.i. J, analysis of EdU+, tdTomato+ cells at 0 and 4 d p.i. including representative images of stained sections (see Figure S5C for sorting gates and markers) (n ≥ 3, data reflect the mean ± SD, unpaired t-test ***p<0.001). K, multiple genes associated with provisional ECM production and turnover are coordinately expressed starting at D1 through to D14 (see Figure S5E for additional genes). L, distribution of Postn and Lox transcripts in t-SNE derived clusters of single cells (scRNA-seq). M, genome browser tracks showing the promoter sum signal around the Lox and Postn gene loci within the 4 different clusters (scATAC-seq) from tdTomato+ cells at D0. N, representative images of IF staining of 0, 4 and 14 d p.i. TA muscles from Hic1; tdTomato reporter mice with anti-POSTN.
Hic1+ cells are highly dynamic and do not contribute to parenchymal lineages following injury
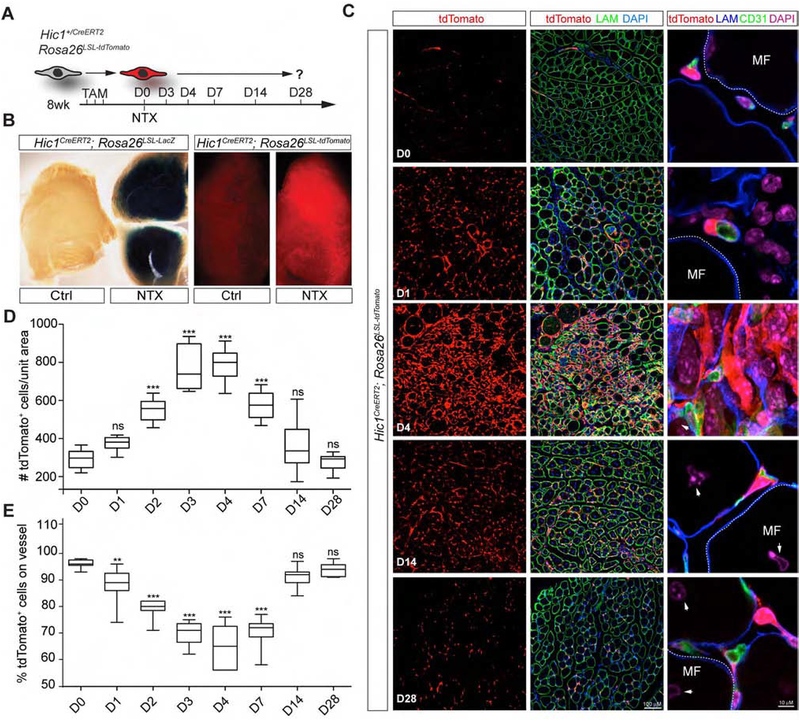
The availability of a reporter system labelling multiple mesenchymal lineages allows the morphological and molecular characterization of their response to damage. To this end, reporter expression was visualized at different time points following muscle damage (Figure 4A). In whole-mount preparations of the TA muscle 3 d p.i., a substantial increase in reporter gene expression was observed using either conditional tdTomato or LacZ (Rosa26LSL-LacZ) reporters (Figure 4B). Collection at various time points p.i. revealed dynamic changes within the traced population (Figures 4C and S4A–4B). A significant increase in cell number compared to uninjured controls was observed starting 2 d after damage (Figure 4D). This correlated with an ~ 9-fold increase in labelled cells that were no longer associated with vessels (Figures 4D–4E and S4A–S4B). The number of tdTomato+ cells peaked at 3–4 d p.i. with a total increase of ~2.7-fold over baseline. At this stage, tdTomato+ cells exhibited a markedly modified morphology, forming an extensive interlaced network of processes that circumscribed the regenerating myofibres (Figure S4B). By 7 d p.i., tdTomato+ cell number began to recede, and by 14 d p.i., it had returned to baseline (Figure 4D and 4E). At 28 d p.i., tdTomato+ cell morphology and distribution were indistinguishable from uninjured controls (Figures 4C–D and S4B). As shown in Figures 4C and S4C, no appreciable tdTomato+ myofibres were detected, indicating that the mesenchymal lineages traced in the Hic1CreERT2; tdTomato conditional reporter mice do not contribute to this lineage. Consistent with these findings, scRNA-seq and scATAC-seq revealed negligible transcript expression and an absence of open chromatin within the loci for muscle program-specific factors such as Pax7, MyoD1 and Myf5 (Figure S4D). In aggregate, our findings show that the Hic1 expressing cells are highly dynamic following damage.
Figure 4:
Marked transformation of the quiescent MP phenotype following injury-induced activation. A, experimental plan for lineage tracing of MPs using the Hic1CreERT2; reporter mice and NTX induced muscle injury. B, two different Cre-dependent LoxP-stop-LoxP (LSL) reporter lines were used to follow MP activation after injury. Representative stained, whole mount and imaged TA muscles are shown in non-damaged and 3 d p.i. C, immunodetection of indicated markers within representative TA muscle sections at various time points after NTX-induced injury (see Figure S4B for additional time points). D and E, quantification of tdTomato+ abundance (D) and distribution (E) (n = 3). One-way ANOVA Bonferroni post-test **p<0.01, ***p<0.001, relative to baseline. ns, not significant.
Hic1+ cells coordinate multiple aspects of skeletal muscle regeneration
To better understand the function of Hic1+ MPs during muscle regeneration, RNA-seq and scRNA-seq was used to characterize the nature of their transcriptome 1, 2, 3, 4, 5, 7, and 10 d p.i. (Table S4). Bioinformatic analyses revealed a dynamic temporal succession of transcriptional programs indicative of multiple diverse functions such as cytokine production, cell proliferation, trophic factor production, extracellular matrix production and remodeling, and basement membrane synthesis (Figure 5A). Within the first 24 h p.i. numerous cytokine genes were upregulated, including those associated with neutrophil recruitment (Cxcl1 and 5), and adhesion, recruitment, expansion and maturation of monocytes (Cxcl2, Cxcl14, Csf1 and Ccl7) (Figure 5B). Interestingly, most cytokine production was transient, peaked at 24 h p.i., and quickly diminished thereafter (Figures 5A–B). At this time point, 29 ± 3.0% of the tdTomato+ cells were positive for the neutrophil chemotactic cytokine CXCL5 (Figure 5C). ScRNA-seq analysis of 4,820 cells at 1 d p.i. revealed that this and most other cytokine transcripts were abundant in the FAP sub-clusters, and nearly absent in the pericytic and tenogenic clusters (Figures 5D–F and S5A–B). While transcripts for many of these cytokine genes were either low or undetectable prior to injury, robust peaks indicative of open chromatin were identified in some of the uninjured 0 d clusters indicating that these genes are poised for transcription (Figure 5G).
On the second day following injury, numerous genes associated with cell proliferation (Ccnd1, Ccnd2, Ccnd3, Cdk4 and Mki67) were coordinately upregulated (Figure 5H). These transcripts peaked on 3 and 4 d p.i., consistent with the increased cell number (Figure 4D) and EdU incorporation (Figures 5J and S5C), as well as with the peak of DNA synthesis previously reported for FAPs by Joe et al. (Joe et al., 2010). At 2 d p.i., cells expressing proliferation-related genes clustered separately and represented ~ 15% of the profiled population (377 cells out of 2,556 cells) (Figure 5I). Transcript signatures representative of all the three Hic1-expressing lineages identified in undamaged tissue were detected within cell cycle regulator-expressing clusters, indicating that they all participated in cell proliferation (Figure S5D). Transcripts associated with the cell cycle were greatly reduced in all clusters by 4 d p.i., with negligible detection at 14 d p.i. (Figures 5I and S5D).
Shortly following the beginning of MP expansion, transcripts related to the production of ECM proteins became notable (Figure 5K). Numerous transcripts reflective of the production of provisional ECM, known to be transiently deposited during successful muscle regeneration (Serrano and Munoz-Canoves, 2010) were present, with Periostin (Postn) being highly abundant (Figure 5L) both at the transcript and protein levels (Figures 5L and 5N). Similarly, Lox which is involved in collagen synthesis was also abundantly induced with similar kinetics to Postn (Figures 5K and 5L). At the single cell level, the transcripts for Postn and Lox were highly expressed in the FAP population which exhibited a myofibroblast-like signature (Acta2, Col1a1, Col1a2, etc.) (Figures 5K and S5E). Interestingly, this population was also enriched for expression of Adam12, a gene associated with fibrogenic cells in skin and muscle (Dulauroy et al., 2012) (Figure S5E). Postn transcripts were detected within all three Hic1-labelled lineages to varying extents, however, Rgs5+ pericytic cells showed the least amount. Congruent with these findings, peaks indicative of open chromatin were evident in these gene loci within the 4 cell clusters, albeit the Rgs5+ cluster exhibited markedly smaller peaks in comparison to the other clusters (Figure 5M). ECM production was in decline by 7 d p.i., a time at which numerous proteases associated with matrix remodeling were induced, such that by 10 d p.i. much of the provisional matrix had been removed and regenerated myofibres began to emerge (Figure 5N). By 14 d p.i., extracellular POSTN was no longer detectable (Figure 5N). Thus, Hic1+ derivatives transiently acquire a myofibroblast-like phenotype and secrete an ECM-rich scaffold, providing a favorable microenvironment to promote myofibre regeneration.
Interestingly, several genes (Col8a2, Col14a1, Col15a1, Fbln1, Fbln5, Hspg2, Lama2, Lama4, Lamc1, Lamb2 and Nid2) associated with the basal lamina including the collagen genes down-regulated in the cKO samples (Col5 and Col6 genes) were expressed with clearly distinct kinetics relative to cytokine and cell cycle transcripts (Figures 5A–5B and 5H, and S5F–S5H). These transcripts were abundant in quiescent MPs, quickly declined following activation (Figure S5F–S5H) and slowly increased during ECM remodeling and returned to baseline levels as late as 14 d p.i. Thus, changes in expression of basement membrane-associated genes in Hic1+ FAPs corresponded with the disruption and restoration of the laminin sheath that envelopes myofibres and MPs (Figures 4C and S4B).
Hic1+ MP progeny contribute to multiple mesenchymal lineages in regenerated skeletal muscle
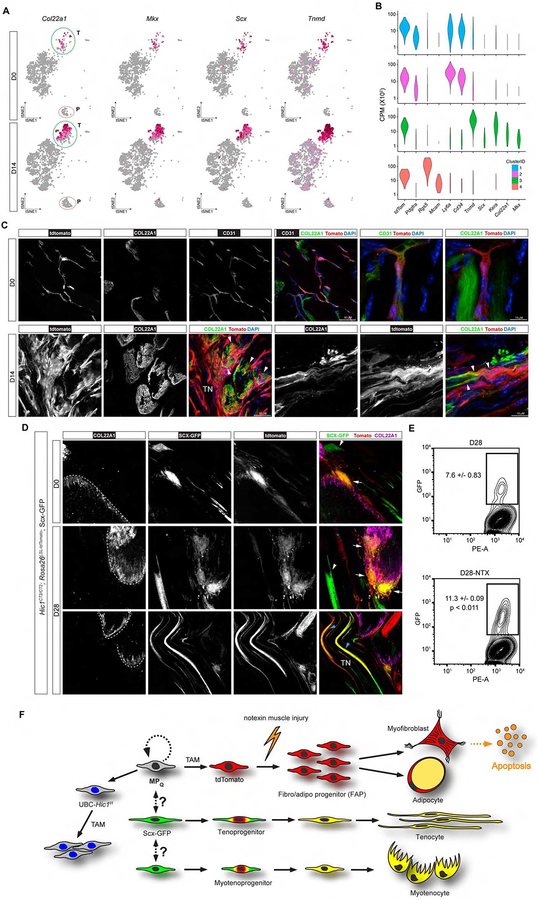
To test if the role of Hic1+ MPs extended beyond transient FAP modulation of the regenerative microenvironment, p.i. tissue samples were examined for the presence of differentiated progeny. In successfully regenerated muscle, overall MP frequency and distribution was indistinguishable from the uninjured control (Figures 4 and S4B). However, FCM analysis at 14 d p.i. revealed a substantive increase in the LY6A− subset of tdTomato+ cells (25.1 ± 1.53% versus 5.2 ± 0.4% in uninjured muscle) (Figure 6A). PopRNA-seq analyses at this time point also revealed an increase in the expression of genes associated with pericytes (Rgs5, Mcam, Notch3, Kcnj8, etc.) (Figure S7A) as well as with tenocytes (Tnmd, Mkx, Scx, etc.) (Figure 6B). Interestingly Col22a1, a gene expressed within the myotendinous junction (Charvet et al., 2013; Felsenthal and Zelzer, 2017; Koch et al., 2004; Subramanian and Schilling, 2015), was also substantially increased. Similar to the FCM analyses, within the scRNA-seq clusters, the LY6A− pericytic and tenogenic sub-populations both increase from 5.5% at 0 d to 7.2% at 14 d p.i. and 5.6% at 0 d to 22.1% at 14 d p.i., respectively. To study the fate of the individual sub-clusters in pseudotime, the dataset was input to Monocle3 and this generated multiple trajectories reflective of the originally defined clusters observed at 0 d (Figures 6C–D and S6A). The tenogenic population was further subdivided manually based on Col22a1 expression (Figure 6C and S6C). Genes were filtered based on expression level and variability to order the cells in pseudotime (Figure 6E–6H). Pseudotime analysis of the FAPs after injury revealed a highly dynamic transcriptional program that transitioned through multiple states with the 14 d p.i. population returning to a state resembling 0 d (Figures 6E and S6B). In fact, the 1 d p.i. FAP population underwent such a marked transcriptional change that they formed their own distinct trajectory (Figure 6D–6E and S6B). Similarly, the other 3 populations exhibit pronounced transcriptional changes indicative of modified biological states (Figures 6F–6H and S6D–F). Congruent with the FAP pseudotime trajectory, some cells returned to a pre-injury state (i.e. overlap between 14 d p.i. and 0 d), whereas others occupied a position distinct from 0 d (Figures 6F–6H and S6D–F). Collectively, these analyses revealed that following injury, all MPs produce progeny with modified transcriptional signatures. In FAPs, this program returned to baseline, whereas the pericytic and two tenogenic populations generated progeny that adopted new and enduring transcriptional programs.
Figure 6:
Hic1-tracked MPs directly contribute to multiple mesenchymal cell types within the regenerated muscle. A, FCM analysis of LY6A expression in tdTomato+ cells 14 d p.i. shows a significant increase in LY6A+tdTomato+ cells. The dashed line represents the undamaged sample at 0 d (n ≥ 3, data reflect the mean ± SD, one-way ANOVA Bonferroni post-test, *** p<0.001). B, popRNA-seq analysis of tdTomato+ enriched fraction from D14 TA muscle p.i. shows enrichment of transcripts associated with tendon (Scx, Mkx, Tnmd and Kera) and the MTJ (Col22a1). C, tSNE overview of Hic1;tdTomato sub-populations and cluster-marker genes. D, UMAP (left panel) with cells colored by library ID indicative of the time of collection (see Figure S6A for UMAP colored by cluster). Cell partitions identified through Monocle3 and the representative markers (Pdgfra, Ly6a, Rgs5 and Tnmd) used to identify the cell types. E, UMAP plot (left panel) of pseudotime trajectories for the FAP subpopulation. The right panels contain plots of the individual time points (library ID). F-H, replicate of what is shown in D for the pericytic (F), tenogenic (G) and myotenogenic (H) populations.
To experimentally test the cell fates suggested by pseudotime analyses, the presence of the aforementioned 3 stable populations was examined in p.i. samples. Consistent with the popRNA-seq, the scRNA-seq profiling revealed the presence of a bona fide tenogenic signature (Mkx, Scx and Tnmd) within the tdTomato+ Ly6a− subpopulation that also contained appreciable Col22a1 transcript (Figures 7A and 7B). Within skeletal muscle, Col22a1 is restricted to the myotendinous junction (MTJ), and is thought to be expressed and produced by the myofibres (Subramanian and Schilling, 2015). Knockdown of Col22a1 in zebrafish leads to weak and dysfunctional MTJs that are susceptible to strain-induced muscle detachment from the tendon (Charvet et al., 2013). However, the origin of Col22a1 within the MTJ has not been well defined. Interestingly, following damage tdTomato+ cells were found embedded in a COL22a1-rich ECM at the distal ends of myofibres, which they enveloped with processes that also inserted into the tendon (Figure 7C). In contrast, participation of tdTomato+ cells to these basket-like anatomical structures was not observed in undamaged tissue, in which the traced cells associated with MTJs were restricted to CD31+ blood vessels, with no appreciable contribution to the junction itself or to tenocytes (Figures 1F and 7C). As noted above, the Col22a1-expressing cells appeared to share properties with the tenogenic lineage, and this was further evaluated by the introduction of an Scx-GFP allele into the Hic1; tdTomato reporter background. Within the muscle mononuclear fractions, Col22a1 was also enriched > 10 fold in the Lin−LY6A− fraction versus the whole muscle sample (Table S1). In the un-injured state, spindle-shaped GFP+tdTomato+ cells were detected in close proximity to the MTJ (Figure 7D). At 28 d p.i., GFP+, tdTomato+ cells were identified within the COL22a1-rich MTJ (Figure 7D). However, as observed in Figure 7C, these cells now acquired a distinct morphology consisting of finger-like projections that interdigitated with processes emanating from the distal end of the myofibre (Figure 7D). Following injury, tdTomato+, GFP+ elongated tenocytes could also be found within the tendon (Figure 7D). As indicated in the scRNA-seq analyses, this tdTomato+GFP+ population significantly expanded following injury (Figure 7E). In aggregate, our data strongly suggests that tenogenic cells within the MTJ are derived from Hic1-expressing progenitors and represent modified tenocytes.
Figure 7:
Hic1-marked MPs directly contribute to regeneration of the myotendinous junction. A, t-SNE plots from scRNAseq-analyses of Hic1;tdTomato sorted MPs. Tendon-expressed genes are shown from 2,173 and 3,527 profiled cells from D0 and D14, respectively. B, violin plots of gene expression for the clusters shown in Figure 3E. C, representative images of anti-COL22a1 stained sections from uninjured and 14 d p.i. TA muscles (white arrowheads, tdTomato+ cells embedded in COL22a1 rich matrix). D, visualization of GFP (Scx-GFP) and tdTomato expression in representative images from within MTJ regions that were identified by anti-COL22A1 staining. White arrowheads indicate Scx-GFP single positive cells and white arrows denote GFP+tdTomato+ cells. Blue arrowheads indicate GFP+tdTomato+ tenocytes. E, FCM analysis of GFP+tdTomato+ cells prior to injury and at 28 d p.i. (n ≥ 3, data reflect the mean ± SD, unpaired t-test). F, overarching schematic of Hic1+ MP participation in muscle regeneration.
In addition to the tenogenic cluster, another prominent Hic1-expressing subpopulation in undamaged tissue was defined as pericyte-like based on the expression of Rgs5, Acta2, Mcam, Notch3, Kcnj8 and Tagln (Figures S7A and S7B). Fourteen days p.i. this subpopulation increased modestly, from 5.5% to 7.3% of the total tdTomato+ scRNA-seq profiled cells. Consistent with this, infrequent ACTA2+, TAGLN+ or MCAM+ pericytes positive for tdTomato were evident at 14 d p.i. (Figure S7C).
Within regenerated muscle, tdTomato+ cells contributed with variable extents to other mesenchymal cell types. Rare interstitial tdTomato+ perilipin+ adipocytes were observed; consistent with the rarity of adipocyte generation during successful regeneration in mice, they were relatively infrequent and we did not detect a prominent adipogenic signature in the popRNA-seq nor scRNA-seq analyses (Figures 5D–5E and S7D–S7E). This may also reflect the inherent challenges in isolating, sorting and capturing adipocytes for RNA-seq analyses. Interestingly, scATAC-seq analyses of the 4 clusters revealed peaks indicative of open chromatin at gene loci (Cebpa, Cebpb and Cebpd) associated with the adipogenic lineage (Figure S7F). Cebpb and Cebpd are linked with adipogenic lineage potential, whereas Cebpa is up-regulated following adipocyte commitment and differentiation (Cristancho and Lazar, 2011). To further assess adipogenic lineage within the Hic1+ clusters, an enrichment strategy was implemented based on robust detection and discrimination of FAPs (tdTomato+LY6A+) and tenogenic progenitors (TPs) (tdTomato+LY6A−Scx-GFP+) (Figure S7G). The ability of these cells to differentiate into adipocytes was assessed following culture in adipogenic media. While FAPs exhibited appreciably more Cebpa transcript than TPs at 0 day (Figure S7H), both lines were able to generate adipocytes in vitro (Figures S7I–J) consistent with the epigenetic analyses (Figure S7F). Tenogenic potential was assessed using a similar strategy with culture in tenogenic differentiation media. Scx was abundantly expressed in the TPs at 0 d of culture in comparison to FAPs (Figure S7H). Culture in fibrogenic media (growth media supplemented with TGFβ1) or tenogenic media led to robust induction of Scx in tenogenic progenitors, with a muted response being apparent in the FAP cultures (Figure S7L). Interestingly, the marked induction of Scx observed in the TPs in the fibrogenic media was not associated with a differentiated tenogenic phenotype, as Tnmd was markedly reduced under these conditions even in comparison to the adipogenic media culture conditions. Consistent with these observations, tendon-like elongated tdTomato+ GFP+ structures were only identified in the TP cultures with tenogenic media (Figure S7L). In aggregate, lineage potential as assessed by in vitro culture and differentiation of FAP and TPs is congruent with their in vivo lineage potential as defined by chromatin accessibility.
After damage, numerous non-vessel associated tdTomato+ cells were observed within the muscle endo, ecto and perimysium, suggesting that the progeny of Hic1-expressing cells also contributes to the connective tissue sheath components of the regenerated muscle (Figure S7M).
In summary, Hic1-expressing cells contribute to multiple stromal elements following muscle damage, including pericytes, tenocytes, adipocytes and specialized MTJ cells.
DISCUSSION
MPs are emerging as central players in tissue homeostasis and regeneration. Herein, we have defined a solitary MP marker, Hic1, that has enabled interrogation of this population in muscle regeneration. Specifically, Hic1 marks quiescent MPs and consistent with this, deletion of Hic1 leads to MP expansion. Within muscle, Hic1 identifies 3 populations with distinct functions and fates in tissue homeostasis and regeneration (Figure 7F). The major FAP sub-population plays a prominent but transient role in muscle regeneration, whereas TPs generate stable derivatives critical for muscle function. Interestingly, a recent study which profiled the mononuclear cellular composition of skeletal muscle also identified 3 distinguishable mesenchymal cell types, referred to as FAP, tenocyte and smooth muscle mesenchymal cells (SMMC; Itga7+, Vcam1−) (Giordani et al., 2019). These correspond to the Hic1+ FAP, tenogenic and pericytic populations, respectively, indicating that Hic1 identifies all the mesenchymal cell constituents within skeletal muscle.
Hic1 and MP Quiescence
Quiescence is a key defining property of many stem and progenitor cells, but not of their post-mitotic derivatives, and is typically defined by the ability of a growth-arrested cell to enter the cell cycle. Quiescent MPs can be activated to enter the cell cycle by a number of signals, including but not limited to inflammation and damage, and expand several-fold to produce a microenvironment and progeny to support regeneration and restoration of tissue function. Following regeneration, quiescent MPs “re-appear” in preparation for tissue renewal or the next regenerative event. In this regard, quiescence is vital to MP function; however, with the possible exception of the hedgehog pathway, the mechanisms regulating this fundamental property are poorly defined. Within muscle MPs, hierarchical clustering revealed an almost identical pattern of expression for Hic1 with that of the downstream hedgehog effector Gli1. In the lung, hedgehog signaling has been shown to regulate quiescence of lung mesenchyme, and deletion of Smo leads to an inappropriate but limited expansion of this compartment (Peng et al., 2015). Similarly, conditional deletion of Hic1 leads to an immediate expansion of MPs within the TA muscle. Furthermore, loss of Hic1 leads to induction of a transcriptional program that overlaps appreciably with multiple components of the MP activation transcriptome following injury, but does not completely mimic MP activation as this likely relies, in part, on signals present within injured muscle. In this regard, the restriction of Hic1 to quiescent MPs reflects its function and the periodic need of MPs to enter the cell cycle.
MPs in muscle homeostasis and regeneration
Muscle interstitial stromal cells are emerging as critical cellular constituents important in skeletal muscle maintenance, renewal and regeneration. A variety of strategies and Cre drivers have been used to ablate this population which leads to reduced numbers and/or aberrant function of muscle satellite cells, culminating in deficient muscle regeneration (Murphy et al., 2011; Roberts et al., 2013; Wosczyna et al., 2019). Specific ablation of FAPs using a PdgfraCreERT2 allele in combination with a Cre dependent diptheria toxin fragment A led to a sustained and substantial reduction in FAPs that contributed to muscle atrophy under homeostasis and impaired muscle regeneration following injury (Wosczyna et al., 2019). Herein, conditional deletion of Hic1 in MPs was associated with an activated MP phenotype that also compromised muscle regeneration. Collectively, these studies support a fundamental role for MPs in regulating multiple aspects of muscle physiology in adult mice. This is likely accomplished through both direct and indirect mechanisms, including the production of trophic factors and ECM components. Transcriptomic analyses of Hic1-deleted MPs following short or long-term deletion showed significant down-regulation of multiple genes encoding ECM proteins. For instance, mutations in Col6a1, Col6a2 and Col6a3 are associated with myopathies in humans and deletion of Col6a1 in mice caused impaired satellite cell function and muscle regeneration (Mohassel et al., 2018; Urciuolo et al., 2013). Col5a1, Col5a2 and Col5a3 were all down-regulated, and COL5a1 has recently been reported to be a fundamental component of the satellite cell niche (Baghdadi et al., 2018). In addition to these genes, numerous other ECM constituents were also down-regulated following Hic1 deletion including collagens (Col4a1, Col8a1, Col8a2, Col12a1 and Col15a1) and core basement membrane components (Hspg2, Lama4, Lamc1, Nid1 and Vtn), many of which have been associated with muscle deficits (Eklund et al., 2001; Hicks et al., 2014; Labelle-Dumais et al., 2019; Mohassel et al., 2018; Xu et al., 2010; Yurchenco and Patton, 2009; Zou et al., 2014). Furthermore, these genes are all expressed by MPs (Table S1), with FAPs representing the most prominent source especially for Collagens 5a1–5a3 and 6a1–6a6 (Figure S2K and Table S1). It should be noted, that many of these genes are also expressed by muscle satellite cells to varying extents and the contribution of MP-derived ECM components to the satellite cell niche remains to be clarified. In aggregate, these observations suggest that MPs impact muscle homeostasis, renewal and regeneration in part through their direct regulation of ECM composition.
Hic1 identifies tenogenic and myotenogenic progenitors
Identification of TPs in situ has proved somewhat elusive. Hic1 identifies a sub-population of blood vessel-associated Scx-GFP positive cells within the peritenon. Following a skeletal muscle hypercontraction injury, the Hic1+ tenogenic population expanded, and fate mapping revealed a contribution to mature tenocytes. Unexpectedly, within this tenogenic compartment a specialized tenocyte subset was also identified that expressed the Col22a1 transcript, indicative of the MTJ (Koch et al., 2004). Following injury, lineage traced ScxGFP+tdTomato+ cells were visualized within the MTJ where they adopted a distinct phenotype reminiscent of previously characterized MTJ-associated cells (Figure 7F) (Subramanian et al., 2018). The scRNA-seq analyses demonstrated that this population exhibits a unique but overlapping transcriptional program with that of tenogenic progenitors in addition to an exclusive but stable morphology. Thus, we suggest adoption of the term myotenocyte for this population which constitutes the interface between myofibres and tendons. The ability to label and manipulate this population provides an experimental foundation for interrogation of the myotenocyte life cycle in health and disease.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. T. Michael Underhill (tunderhi@brc.ubc.ca).
Mouse lines generated in this study are available upon request by contacting the lead contact.
EXPERIMENTAL MODEL DETAILS
Mice
The Hic1f and Hic1CreERT2 mice were generated using standard gene targeting methodology. For the Hic1f mice a targeting vector was constructed that contained multiple components, including LoxP sites flanking the major coding exon 2 of Hic1, a PGK-neo cassette flanked by FRT sites distal to exon 2 and upstream of the 3’ LoxP site. An IRES-NLS-LacZ cassette distal to the 3’ LoxP site (downstream of exon 2) was added to enable efficient identification of Hic1 deleted cells along with cells in which the Hic1 promoter is active. This targeting construct was introduced into 129Sv/Pas ES cells and properly targeted ES cells were identified through a combination of PCR and southern analysis. Four targeted ES cell clones were used for subsequent injection into ~ 60 C57BL/6 blastocysts which led to 10 males with chimerism ranging from 8–100%. These male chimeras were bred with female Flippase-expressing mice for deletion of the PGK-neo cassette, and PGK-neo deleted mice were subsequently back-crossed to C57BL/6 mice for > 10 generations. The Hic1CreERT2 mice were generated using similar methodology and involved introduction of an IRES-CreERT2 cassette into the 3’ untranslated region of the Hic1 gene. The CreERT2 gene was removed from plasmid pCAG-CreERT2 (Addgene #14797) and introduced into a targeting vector based on C57Bl/6 DNA sequences along with a FRT flanked PGKneo cassette. This vector was electroporated into C57Bl/6 ES cells and successfully targeted clones were injected into C57Bl/6 blastocysts. Male chimeras were subsequently bred to Flippase-expressing mice to delete the PGK-neo cassette. These mouse lines were both generated under contract by GenOway Inc. and were created in and/or interbred into a pure C57BL/6 background (> 10 backcrosses). Other mouse lines used include: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Jax stock number 007914; herein referred to as RosaLSL-tdTomato), B6.Cg-Tg(UBC-cre/ERT2)1Ejb/2J (Jax stock number 008085; herein referred to as UBC-CreERT2), B6.129S4-Pdgfratm11(EGFP)Sor/J (Jax stock number 007669), C57BL/6J (Jax stock number 000664) and ScxGFP(Pryce et al., 2007). All mice were maintained in (or as noted backcrossed to) a C57BL/6 background.
For lineage tracing experiments, Hic1CreERT2 mice were interbred with RosaLSL-tdTomato mice (Ai14 line, JAX stock 007914) to generate Hic1CreERT2/CreERT2 or +; Rosa26LSL-tdTomato/+. To induce CRE-ERT2 nuclear translocation, 8–9-week old mice were injected intraperitoneally with 100 mg/kg of Tamoxifen in 100 μL sunflower oil daily for 5 consecutive days. Muscle injury studies were initiated in these animals following a > 14 d washout period.
For experiments involving conditional deletion of Hic1, Hic1f/f mice were interbred with UBC-CreERT2 mice to generate Wt or UBC-CreERT2; Hic1f/f mice. TAM was administered to mice > 8 wks of age as described above.
Mice were housed under standard conditions (12 hr light/dark cycle) and provided food and water ad libitum. For all experiments, litter mates of the same sex were randomly assigned to experimental groups. Animals were maintained and experimental protocols were conducted in accordance with approved and ethical treatment standards of the Animal Care Committee at the University of British Columbia.
MP primary cell culture
Primary cultures of FAPs and TPs were established from dissociated hind limb muscle of Hic1CreERT2; Rosa26LSL-tdTomato; Scx-GFP mice. Mice were IP injected with TAM at 8 weeks. After a 10-day washout period, whole limb muscle was dissected and dissociated as described below for isolation of the tdTomato+ MP population. The single cell suspension was fractionated by FACS. To isolate MPs, Lin−tdTomato+ cells were selected. This MP population was further fractionated into a LY6A positive (FAP) and a negative population from which the Scx-GFP+ (tenogenic) sub-population was obtained. Tenogenic MPs were collected and used to seed a single 96 well plate and maintained in MesenCult expansion media (StemCell Technologies) and incubated at 37° C in a 5% CO2 and 5% O2 environment. Enriched FAPs were seeded into a single well of a 24-well plate. MP sub-populations were expanded sufficiently to produce a T75 flask of confluent cells. To assess lineage potential, cells were subsequently plated for in a 24 well format at P4 under various differentiation conditions. Cells were allowed to reach confluence before collection at 0 d or addition of differentiation media. To promote myofibroblast differentiation, cells were cultured in MesenCult expansion media containing 5 ng/mL TGFβ1. For adipogenic assays, MesenCult adipogenic media was employed. For tenocyte formation, cells were maintained in MesenCult tenogenic differentiation media. Media was changed every three days until D21 when cells were lysed for RNA (Qiagen RNeasy). Replicate wells were fixed with 2% PFA for visualization of endogenous fluorescent reporter protein or fixed and stained with oil red o, counterstained with hematoxylin and images collected using an Axiovert S100 (Zeiss) inverted microscope with a QImaging Retiga 1300 camera.
METHOD DETAILS
Muscle injury and evaluation
Muscle injury of the tibialis anterior muscles was induced by intramuscular injection of 0.2 μg of notexin snake venom extract (Latoxan) in 20 μL PBS (10 μg/mL) (Joe et al., 2010). Myofibre cross-sectional area measurements were carried out on Picrosirius red stained TA muscle sections using ImageJ. Individual myofibres were defined by Picrosirius red stained muscle basal lamina. The extent of interstitial collagen was quantified as the percent of Picrosirius red stained area per field.
Generation of anti-HIC1 antibody
A rabbit polyclonal antibody to HIC1 was generated based on the carboxyl 25 amino acids of HIC1. This region exhibits 96% amino acid identity between mouse and human. A peptide (C25; AEVLSQGAHLAAGPDSRTIDRF) was conjugated to keyhole limpet hemocyanin and used to immunize 2 rabbits in complete Freud’s adjuvant. Collected serum was subsequently affinity purified on a C25 peptide-containing column.
Fluorescence-activated cell sorting methodology
MPs were released from various muscles of the hind limb using a previously described protocol with modifications (Joe et al., 2010). Freshly dissected TA muscles were sequentially digested in a volume of 250μL per TA containing 500 U/mL Collagenase II (Sigma C-6685–5G) for 30 mins followed by a cocktail of 1.5 U/mL Collagenase D (Roche 11 088 882 001) and 2.4 U/mL Dispase II (Roche 04 942 078 001) for 60 mins. Digested material was subsequently triturated by pipetting and passed through a 40 m cell strainer. The resultant cell suspension was washed with FACS buffer (PBS containing 2 mM EDTA and 2% FBS) and centrifuged at 500 × g for 5 mins. To enrich for MPs from this whole muscle mononuclear suspension, cells were selected for the expression of various cell surface markers including CD45, CD31, Ter119 or LY6A. The lin− fraction was considered to be negative for CD45−, Ter119− and CD31−. This was accomplished by incubation with a cocktail containing anti-Ter119–488 (Ablab 48-0031-01, 1:200), anti-CD45-PerCP (BD Pharmingen 557235, 1:400) and anti-CD31-FITC (eBioscience 11-0311-85, 1:400). The Lin− population was further sub-fractionated based on LY6A expression by the addition of anti-Ly-6A/E (Sca-1)-PE-Cy7 (eBioscience 25-5981-82, 1:3000) to the above antibody cocktail. This mixture containing cells and antibodies was subsequently incubated on ice in the dark for 30 mins. Afterwards, 15 mls of FACS buffer was added to dilute the antibodies prior to centrifugation at 500 × g for 5 mins. The pellet was resuspended in FACS buffer containing Hoechst 33342 (Sigma B2261) to a final concentration of 4 M. Stained cells were sorted using a BD Influx, and Hoechst and forward/side scatter parameters were used to identify viable single cells for all FACS enrichments. Sorted cells were collected into sort media [DMEM, 20% FBS, L-glutamine (2 mM), penicillin (100 units/ml) and streptomycin (100 μg/ml)] in cooled siliconized microcentrifuge tubes (Fisher Scientific; 02-681-320). For isolation of RNA, cells were microfuged at 500 × g for 5 mins and the isolated cell pellet was lysed in RNAzol (Sigma R4533).
To enrich for tdTomato-labelled Hic1+ MPs a similar FACS strategy was employed with the following antibody cocktail, anti-Ter119–647 (Ablab 67-0031-01, 1:200), anti-CD31-APC (BD Biosciences 551262, 1:400), anti-CD45–647 (Ablab 67-0047-01, 1:400). These antibodies were included to aid enrichment of tdTomato+ cells and reduce confounding cell contaminants. Furthermore, to attenuate the signal associated with tdTomato reporter expression in transgenic MPs and improve sorting selectivity, the PECy7 configuration was used to gate the tdTomato+ population.
To enrich for WT and Hic1 cKO MPs the above described approach was used with some modifications. The staining cocktail was altered to further enhance MP enrichment and included: anti-Ter119–647 (Ablab 67-0031-01, 1:200), anti-CD31-APC (BD Biosciences 551262, 1:400), anti-CD117-APC (eBioscience 17-1172-82, 1:500), anti-CD11b-647 (Ablab 67-0055-01, 1:500), anti-F4/80–647 (Ablab 67-0035-05, 1:500), anti-CD45–647 (Ablab 67-0047-01, 1:400), anti-LY6A/E (Sca-1)-PE-Cy7 (eBioscience 25-5981-82, 1:3000).
Cell cycle analysis
To enable detection of proliferating cells, standard methodology involving analysis of the incorporation of the nucleoside triphosphate analogue 5-ethynyl-2’-deoxyuridine (EdU) was used. For these purposes, animals were injected intraperitoneally with 0.5 mg of EdU (ThermoFisher E10415) in 250 μL PBS daily during the 3 d pulse period after Hic1 deletion or following muscle injury. EdU+ cells were subsequently detected 24 h after the last EdU injection by FCM and/or in histological samples using the Click-iT Plus EdU Pacific Blue Flow Cytometry Assay kit (ThermoFisher C10636) and the Click-iT Plus EdU Alexa Fluor 647 Imaging kit (ThermoFisher C10640), respectively, both as per manufacturer instructions. For analysis of EdU incorporation in MPs from the Hic1 cKO and wild-type samples, cells were counter-stained as per the enrichment strategy for Lin−LY6A+ MPs.
Whole-mount LacZ staining
LacZ activity in some tissue samples was detected using established whole-mount X-gal staining (Scott and Underhill, 2016). Briefly, to enhance penetration of the fixative in whole tissues, dissected tissues were cut into ~ 4 mm3 pieces and fixed for 18–24 h at 4°C in LacZ fixative (100 mM MgCl2, 0.2 % glutaraldehyde, 5 mM ethylene glycol tetra-acetic acid in PBS). Samples were subsequently washed 3 times for 30 mins each in PBS then permeabilized in a solution containing 2 mM MgCl2, 0.01% deoxycholate, 0.02% NP40, 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide overnight at 37°C. This was followed by an overnight incubation in LacZ staining solution (2 mM MgCl2, 0.01% Deoxycholate, 0.02% NP40, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) at 37°C. Samples were post fixed in 4% PFA overnight at 4°C and visualized and/or paraffin embedded for histological analysis. Six μm sections were prepared and counterstained with nuclear fast red for analysis.
Immunofluorescence and/or in situ X-gal staining
For in situ X-gal staining on histological sections, tissues were prepared in the following manner. Mice were terminally anesthetized by intraperitoneal injection of Avertin (400 mg/kg) and fixed by intracardial perfusion of cold PBS followed by cold LacZ fixative as described above (Scott and Underhill, 2016), before dissection and immersion of tissue samples in LacZ fixative for 3 h on ice.
To enable detection of native tdTomato expression in processed tissue samples, mice were terminally anesthetized by intraperitoneal injection of Avertin and fixed by intracardial perfusion of 10 mM EDTA in PBS followed by 4% paraformaldehyde. Tissues were immersed in 4% paraformaldehyde for 48 h at 4°C.
For cryosectioning the above collected and fixed samples were washed with PBS then incubated through a cryoprotective series of sucrose solutions of increasing concentration from 10–50% for ≥ 3 h each before embedding into OCT compound (Tissue Tek 4583). Tissues were immersed into OCT in disposable plastic cryomolds (Polysciences 18646A) and frozen in an isopentane bath cooled by liquid nitrogen. Cryosections were cut (Leica CM3050S) at a thickness of 5–30 m and mounted onto Superfrost Plus slides (VWR 48311–703). This material was used for either LacZ and/or IF staining.
For detection of LacZ on sections, in situ LacZ staining with X-gal was carried out. Slides were thawed at rt, washed 3 × 10 mins in PBS and then incubated overnight at 37°C in a humidified chamber with the aforementioned X-gal staining solution. Slides were counter-stained with nuclear fast red and mounted with Aqua Polymount. For additional IF staining of LacZ stained samples, freshly stained slides were subsequently washed with PBS for 3 × 5 mins and IF staining was carried out as described below.
For IF staining, slides were thawed at room temperature, washed 3 × 10 mins in PBS and incubated for 1 h in PBS containing 10 mg/mL sodium borohydride (Sigma 213462) to quench autofluorescence. Following this treatment, slides were washed with PBS and incubated in block solution containing 2.5% BSA (Sigma A7030) and 2.5% Goat serum (Gemini 100–190) for 90 mins at room temperature prior to incubation in primary antibody (listed below) overnight at 4°C. Alexa Fluor conjugated secondary antibodies were typically diluted 1:500 and applied to the slides for 45 mins. After each antibody incubation, 3 × 5 min PBS washes were performed and sections were counterstained with DAPI (600 nM) and mounted with Aqua Polymount (Polysciences 18606).
For detection of HIC1 with anti-HIC1 (C25; 1:2000 dilution) a modified tyramide staining strategy was used. Cryosectioned material was stained using TSA Kit #16 (ThermoFisher T20926) as per manufacturer’s instructions from step 2.4 to step 4.4 with the following modifications. Either anti-LY6A (Abcam51317, 1:200) or anti-CD31 (BD 550274, 1:50) was included in the antibody cocktail in step 3.3 and Alexa Fluor 555 goat anti-rat IgG (ThermoFisher A21434, 1:500) in step 3.6. Furthermore step 4.2 was performed overnight at 4°C. To highlight the basement membrane using an antibody of the same species without cross reactivity, separate consecutive antibody incubations were performed instead. Between steps 4.3 and 4.4 the sections were incubated with rabbit anti-laminin (Ab11575) for 90 mins, followed by 3 × 5 mins PBS washes. Laminin staining was subsequently detected without amplification by incubation with goat anti-rabbit 488 (ThermoFisher A11034) for 45 mins, and the slides were counterstained with DAPI (ThermoFisher D3571) and mounted with Aqua Polymount.
Image acquisition and quantification
Widefield epi-fluorescence microscopy images were collected on an Olympus BX63 compound microscope. Confocal images were collected using either a Nikon Eclipse Ti inverted microscope equipped with a C2Si confocal system or Nikon Ti-E inverted microscope with an A1R HD25 confocal scanning head and acquired in Nikon Elements software. A Zeiss LSM 800 with AiryScan and ZEN blue software was used to acquire images in Figure 7D. Brightness and contrast of images within panels were adjusted to comparable levels in Fiji software (ImageJ version 2.0.0-rc-68/1.52e).
For enumeration and analysis of the distribution of tdTomato+ cells in time course experiments, a total of 15 images were analyzed per time point (5 images/mouse; 3 mice/time point). TdTomato+ cell bodies defined by DAPI nuclear stain were enumerated and apposition to vessels assigned per cell. The distribution of a low percentage of cells (1–2%) was considered ambiguous and they were not included in the enumeration. Enumeration and distribution of X-gal positive nuclei in samples from Hic1nLacZ/+ mice was carried out in a similar manner. For Figure S2A and S2B, proximity to vessels was quantified using Nikon NIS Elements general analysis MinDistanceTo function.
The enumeration of nLacZ-expressing cells in skeletal muscle following Hic1 deletion relied on DAPI-stained nuclei density. The number of nLacZ+ nuclei were normalized to the total number of DAPI+ nuclei within the counted region. For these purposes, a minimum of 3 regions were randomly selected from images of each sample (n = 4–5) for analysis.
For images of X-gal counterstained with IF, single monochrome images for each fluorescence channel were acquired. Color and monochrome images of identical X-gal stained regions were collected under bright-field. The bright-field monochrome image was subsequently inverted and then merged with the IF images using Fiji software (ImageJ version 2.0.0-rc-68/1.52e) to generate composite images.
To evaluate CXCL5 expression in “activated” MPs, the percentage of doubly positive tdTomato+ CXCL5+ cells in injured TA muscles was counted. Cells counts were carried out on a minimum of two 1 mm2 images cropped from stitched images of one whole muscle section for all samples. Injured areas at this time point were selected by increased mononuclear infiltrate.
Quantitative gene expression analysis
To investigate the expression of genes associated with differentiation, 250 ng of total RNA was converted to cDNA as per the standard protocol using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ThermoFisher 4368813). Taqman-based quantitative PCR was used to measure transcript abundance using TaqMan Fast Advanced Master Mix (Applied Biosystems, ThermoFisher 4444557) with PrimeTime Predesigned qPCR Assay (Integrated DNA Technologies) primer probe sets for Tnmd (Mm.PT.58.13530921), Acta2 (Mm.PT.58.16320644), Cebpa (N007678.1.pt.Cebpa), Scx (Mm.PT.58.31750069) and Tbp (Mm.PT.39a.22214839). Quantitative PCR was carried out on a ViiA7 thermocycler for 55 cycles. Run data was analyzed using QuantStudio version 1.3 (Applied Biosystems). Further calculations including normalization to Tbp were performed using Excel (Microsoft). Graphs were generated using Prism 5 (Graphpad) and Illustrator CC (Adobe).
Scratch wound healing assay
To generate Hic1+/nLacZ MPs for culture, whole hindlimb muscle from 9 week Hic1+/nLacZ mice were dissociated and 300K cells were FACS enriched as described above for the Hic1 KO cell isolation. Cells were expanded by sub-culturing twice a week in sort media (described above) supplemented with 20 ng/ml FGF2 (R&D Systems, Cat. 133-FB). At passage 4, 20K were seeded per well into three 96 well ImageLock plates (Essen Biosciences) pre-coated with 2% gelatin (Sigma; G-1890 made up in MilliQ water). Each experiment involved the use of 3 plates. After the cells were allowed to growth arrest for 24 h, plate one was collected as the D0 pre-injury condition, fixed with 0.2% glutaraldehyde (Sigma) for 15 min and stained with X-gal for 24 h at 37°C, using the solutions described above for slide staining. Stained cells were post fixed with formalin for 24 h at room temperature and imaged on an Axiovert S100 (Zeiss) microscope with a QImaging Retiga 1300 camera. In the remaining plates, wounds were created in each well with a 96-well WoundMaker Tool according to the provided protocol (Essen Bioscience; Cat. 4493). For collection of an intermediate time, plates were fixed and stained at 1 d p.i. at which time the wound was partially closed. To collect a “healed” sample, the scratched plate was cultured for an additional 2 d for a total culture time of 3 d, at which point is was fixed, stained with X-gal and imaged as described above.
Gene expression profiling – popRNA-seq
Total RNA was isolated using RNAzol (Sigma R4533) as per manufacturer’s instructions with the following modifications: StepV.2.-linear polyacrylamide (GenElute, Sigma 56575) was added as a carrier to the RNA solution prior to the addition of cold isopropanol. Samples were incubated overnight at −20° C to support precipitation of total RNA. Precipitated RNA was centrifuged at 21,000 × g for 30 mins at 4° C. SUPERase-IN RNase inhibitor (ThermoFisher AM2696) was added to 1:20 to the resuspended RNA solution. Sample integrity was tested on an Agilent Bioanalyzer 2100 RNA 6000 Nano kit (5067–1511). RNA samples with a RNA Integrity Number > 8 were used to prepare libraries following the standard protocol for the TruSeq Stranded mRNA library kit (Illumina) on the Illumina Neoprep automated microfluidic library prep instrument. Paired end sequencing was performed on the Illumina NextSeq 500 using the High Output 150 cycle Kit.
RNA-seq Bioinformatic Analyses
Illumina sequencing output generated bcl files were de-multiplexed by bcl2fastq2. De-multiplexed read sequences were then aligned to the Mouse Genome mm10 reference sequence using TopHat splice junction mapper with Bowtie 2 (http://ccb.jhu.edu/software/tophat/index.shtml) or STAR (https://www.ncbi.nlm.nih.gov/pubmed/23104886) aligners. Assembly and differential expression was estimated using Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/). For some datasets, enriched pathways or processes were identified using Ingenuity Pathway Analysis (IPA). For these purposes, IPA was performed on gene lists that had been filtered for a p-value < 0.05 using Cufflinks. Hierarchical clustering of popRNA-seq data and heatmap generation were performed with VisRseq (Younesy et al., 2015). Venn diagrams were generated using the VennDiagram package for R.
Single Cell RNA-seq
Single cell suspensions were generated as described for pop-RNA-seq with the following downstream modifications. Target cells were enriched by FACS sorting into 0.22 m vacuum filtered SC collection media (DMEM containing 5% FBS) with propidium iodide (1 μg/ml). Viable target cells were subsequently further purified and debris reduced by sorting a second time and collected into 10 L of SC collection media. Cells were counted and quality control was determined by hemocytometer. If > 98% of visible objects were verified to be single tdTomato+ cells the suspension was input into a Chromium Controller (10X Genomics), captured and libraried with the Chromium single cell 3’ reagent kit v2 (10X Genomics). cDNA libraries were sequenced on a Nextseq 500 (Illumina) to a minimum depth of 50,000 reads per cell. A transgenic reference genome was generated by the concatenation of the sequences for tdTomato to the mm10 reference genome and subsequent use of the cellranger mkref pipeline. Demultiplexing, alignment to the modified mm10 reference genome, principal component analysis, clustering, non-linear reduction (tSNE) and differential expression was performed using the cellranger count pipeline (10X Genomics). Aggregated library datasets were created using the cellranger aggr pipeline. Graphical output was generated using the cellrangerRkit R package (10x Genomics) and the output from the geneBCmatrix (cellrangerRkit) was formatted using an in-house developed R function to generate violin plots and plotted using the grammar of graphics 2 R package. A parallel set of principal component and downstream analyses were performed using the Seurat R package (Satija lab, NYU)(Satija et al., 2015). The Seurat dataset was subsetted based on lineage markers (Figure 6) to generate individual Monocle cell datasets (Monocle3 R package (Cao et al., 2019; Qiu et al., 2017a; Qiu et al., 2017b; Trapnell et al., 2014)). Pseudotime trajectories were subsequently generated using reverse-graph embedding (DDRtree) to analyze cell fate of distinct MP subsets.
Single cell ATAC seq
The input for scATAC-seq requires cell nuclei. To isolate single nuclei, ~ 100,000 fresh FACS purified Hic1; tdTomato reporter+ cells were collected as per scRNA-seq sorting conditions and lysed according to the described protocol (10x Genomics, https://support.10xgenomics.com/single-cell-atac) for 5 min. Nuclei were quantified using a Countess II FL automated cell counter (ThermoFisher) and 10,000 nuclei were targeted for transposition and capture using a 10x Chromium controller. ScATAC-seq libraries were prepared according to the Chromium Single Cell ATAC Reagent Kits User Guide (10x Genomics; CG000168 Rev B). Single cell libraries were sequenced on a Nextseq500 (Illumina) using 2 × 75PE kit to produce 2 × 50 reads to a sequencing saturation of > 80%. The cellranger-atac single cell ATAC pipeline version 1.1.0 was used to generate fastq files from the sequencer output bcl files and further perform read filtering and alignment, detection of accessible chromatin peaks, dimensionality reduction, cell clustering and differential accessibility analyses. Quality metrics for the scATAC-seq such as insert size distribution, enrichment around the transcriptional start site and a tSNE heatmap of fragments per cell are shown in Figure S6G. Plots were generated using Loupe Cell Browser (10X genomics, v. 3.1.0).
QUANTIFICATION AND STATISTICAL ANALYSES
All data are represented as mean ± SD, the sample number and statistical analyses are indicated in the figure legends. Sample size determination was based on anticipated variability and effect size that was observed in the investigator’s lab for similar experiments. For quantification of myofibre cross-sectional area, picosirius red staining between wt and Hic1 cKO mice (Figure S2G–I) and the X-gal+ cell counts (Figure 2F) the individual performing the counts was blinded to sample identity. Analysis of differential gene expression between wt and Hic1 cKO samples involved the use of CuffDiff and a q value of < 0.05 was considered significant. For pairwise comparisons, unpaired two-tailed t tests were used to calculate P values, p < 0.05. For comparison of >2 means, one-way ANOVA with Boneferroni’s correction was employed, p < 0.05. Both statistical methods were carried out using Prism (GraphPad Software v. 5).
DATA AND CODE AVAILABILITY
All sequence data described in this paper have been deposited in GEO: GSE110038. Software used to analyze the data is either freely or commercially available.
Supplementary Material
Table S1. RNA-seq of skeletal muscle-derived mononuclear fractions (related to Figure 1)
Table S2. RNA-seq of control and Hic1 10 d cKO MPs (related to Figure 2)
Table S3. RNA-seq of 18 m control and Hic1 cKO whole skeletal muscle (related to Figure 2)
Table S4. RNA-seq of sorted MPs from skeletal muscle at various time points following injury (related to Figure 5)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Ly-6A/E (Sca-1)-PE-Cy7 | eBioscience | Cat# 25-5981-82, RRID:AB_469669 |
| anti-Ter119-Alexa488 | UBC Ablab | Cat# 48-0031-01, clone ter119 |
| anti-CD31-PerCP | eBioscience | Cat# 46-0311-82, RRID:AB_1834429 |
| anti-CD45-PerCP | BD Pharmingen | Cat# 557235, RRID:AB_396609 |
| anti-CD31-FITC | eBioscience | Cat# 11-0311-85, RRID:AB_465013 |
| anti-CD45.2-FITC | eBioscience | Cat# 11-0454-85, RRID:AB_465062 |
| anti-Ter119-Alexa647 | UBC Ablab | Cat# 67-0031-01, clone ter119 |
| anti-CD31-APC | BD Biosciences | Cat# 551262, RRID:AB_398497 |
| anti-CD117-APC | eBioscience | Cat# 17-1172-82, RRID:AB_469433 |
| anti-CD11b-Alexa647 | UBC Ablab | Cat# 67-0055-01, clone M1/70 |
| anti-F4/80-Alexa647 | UBC Ablab | Cat# 67-0035-05, clone BM8 |
| anti-CD45-Alexa647 | UBC Ablab | Cat# 67-0047-01, clone 13/2 |
| anti-Laminin | Abcam | Cat# ab11575, RRID:AB_298179 |
| anti-CD31 | BD | Cat# 550274, RRID:AB_393571 |
| anti-LY6A (Sca1) | Abcam | Cat# ab51317, RRID:AB_1640946 |
| anti-CXCL5 | Abcam | Cat# ab198505 |
| anti-Perilipin | Cell Signaling | Cat# 9349S, RRID:AB_10829911 |
| anti-Periostin | Abcam | Cat# ab14041, RRID:AB_2299859 |
| anti-Tagln | Abcam | Cat# ab14106, RRID:AB_443021 |
| anti-αSMA | Abcam | Cat# ab5694, RRID:AB_2223021 |
| anti-Collagen 1a1 | Abcam | Cat# ab34710, RRID:AB_731684 |
| anti-Collagen 22a1 | generously provided by Ronen Schweitzer | N/A |
| anti-Collagen 22a1 | Abcam | Cat# ab121846, RRID:AB_11128704 |
| anti-CD146/MCAM | Abcam | Cat# ab75769, RRID:AB_2143375 |
| Alexa Fluor 488 goat anti-mouse IgG (H+L) | ThermoFisher | Cat# A11029, RRID:AB_2534088) |
| Alexa Fluor 555 goat anti-rat IgG (H+L) | ThermoFisher | Cat# A21434, RRID:AB_2535855 |
| Alexa Fluor 647 goat anti-rat IgG (H+L) | ThermoFisher | Cat# A21247, RRID:AB_141778 |
| Alexa Fluor 488 donkey anti-rat IgG (H+L) | ThermoFisher | Cat# A11055, RRID:AB_2534102 |
| Alexa Fluor 488 goat anti-rabbit IgG (H+L) | ThermoFisher | Cat# A11034, RRID:AB_2576217 |
| Alexa Fluor 594 goat anti-rabbit IgG (H+L) | ThermoFisher | Cat# A11037, RRID:AB_2534095 |
| Alexa Fluor 647 goat anti-rabbit IgG (H+L) | ThermoFisher | Cat# A21245, RRID:AB_2535813 |
| Alexa Fluor 405 donkey anti goat IgG | Abcam | Cat# ab175665, RRID:AB_2636888 |
| anti-HIC1 C25 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sunflower oil | Sigma | S5007 |
| Tamoxifen | Sigma | T5648 |
| 2,2,2 tribromoethanol | Sigma | T48402 |
| tert amyl alcohol | Sigma | 152463 |
| Collagenase II | Sigma | C-6685–5G |
| Collagenase D | Roche | 11 088 882 001 |
| Dispase II | Roche | 04 942 078 001 |
| Hoescht 33342 | Sigma | B2261 |
| Propidium iodide | ThermoFisher | P1304MP |
| 5-ethynyl-2’-deoxyuridine (EdU) | ThermoFisher | E10415 |
| Notexin | Latoxan | L8104-NOTEXIN |
| DAPI | ThermoFisher | D3571 |
| Oil Red O | Sigma | O0625 |
| DMEM | Gibco | 11965–092 |
| FBS | Gibco | 1455501 |
| Pen Strep | Gibco | 151140–112 |
| L-glutamine | Gibco | 25030 |
| FGF2 | R&D Systems | 133-FB |
| RNAzol | Sigma | R4533y |
| Superase | ThermoFisher | AM2696 |
| Linear polyacrylamide | GenElute, Sigma | 56575 |
| Formaldehyde | Avantor Performance Materials | 2106–01 |
| Pparaformaldehyde | PolySciences Inc | 00380–1 |
| Glutaraldehyde | Sigma | G5882 |
| AquaPolymount | PolySciences Inc | 18606 |
| Gelatin | Sigma | G-1890 |
| OCT compound | Sakura Finetek | 4583 |
| TGFB1 | R&D Systems | 240-B |
| Goat serum | Gemini | 100–190 |
| Bovine serum Albumin | Sigma | A7030 |
| Sodium borohydride | Sigma | 213462 |
| Magnesium chloride | Sigma | M2393 |
| NP40 | Bio Basic Inc | NDB0385 |
| Potassium ferricyanide | Sigma | P8131 |
| Potassium ferrocyanide | Sigma | P3289 |
| Ethylene glycol tetra-acetic acid | Sigma | E3889 |
| Deoxycholate | Sigma | D6750 |
| 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside | Invitrogen | 15520–018 |
| Tris-HCl (pH 7.4) | Millipore-Sigma | T2194 |
| Sodium chloride | Millipore-Sigma | 59222C |
| Magnesium chloride | Millipore-Sigma | M1028 |
| Nonidet P40 substitute | Millipore-Sigma | 74385 |
| MACS BSA stock solution | Miltenyl Biotec | 130-091-376 |
| Tween-20 | Sigma | P9416 |
| Digitonin | ThermoFisher | BN2006 |
| Nuclei buffer | 10x Genomics | 2000153 |
| Critical Commercial Assays | ||
| AlexaFluor 647 Tyramide SuperBoost Kit | ThermoFisher | T20926 |
| High Capacity cDNA Reverse Transcription Kit | Applied Biosystems, ThermoFisher | 4368813 |
| TaqMan Fast Advanced Master Mix | Applied Biosystems, ThermoFisher | 4444557 |
| EdU flow | ThermoFisher | C10636 |
| EdU imaging | ThermoFisher | C10640 |
| Rneasy | Qiagen | 74106 |
| MesenCult expansion kit (mouse) | StemCell Technologies | 05513 |
| MesenCult adipogenic Differentiation Kit (Mouse) | StemCell Technologies | 05507 |
| MesenCult tenogenic differentiation media | StemCell Technologies | N/A |
| Woundmaker scratch assay module | Essen | 4493 |
| Bioanalyzer 2100 RNA 6000 Nano kit | Agilent | 5067–1511 |
| TruSeq Stranded mRNA library kit | Illumina | RS-122–2103 |
| Chromium Single Cell A Chip Kit, 48 rxns | 10x Genomics | 120236 |
| Chromium Single Cell 3’ Library & Gel Bead Kit v2, 16rxns | 10x Genomics | 120237 |
| Chromium i7 Multiplex Kit 96 rxns | 10x Genomics | 120262 |
| Chromium Chip E Single Cell ATAC Kit | 10x Genomics | 1000086 |
| Chromium i7 Multiplex Kit N, Set A 96 rxns | 10x Genomics | 1000084 |
| Chromium Single Cell ATAC Library & Gel Bead Kit | 10x Genomics | 1000111 |
| NSQ 500/550 Hi Output KT v2.5 (150 CYS) | Illumina | 20024907 |
| NSQ 500 hi- Output KT v2 (150 CYS) | Illumina | FC-404–2002 |
| NSQ 500/550 Hi Output KT v2.5 (75 CYS) | Illumina | 20024906 |
| NSQ 500 hi- Output KT v2 (75 CYS) | Illumina | FC-404–2005 |
| Deposited Data | ||
| Gene Expression Omnibus (GEO:GSE110038) | this paper | GSE110038 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | Jax stock number 000664 | |
| B6.Cg-Tg(UBC-cre/ERT2)1Ejb/2J | Jax stock number 008085 | |
| Hic1f/f | this paper | N/A |
| Hic1+/− | this paper | N/A |
| Hic1CreERT2 | this paper | N/A |
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jax stock number 007914 | |
| B6.129S4-Pdgfratm11(EGFP)Sor/J | Jax stock number 007669 | |
| ScxGFP | Pryce et al., 2007 | |
| Oligonucleotides | ||
| PrimeTime Predesigned qPCR Assay Tnmd | Integrated DNA Technologies | Mm.PT.58.13530921 |
| PrimeTime Predesigned qPCR Assay Acta2 | Integrated DNA Technologies | Mm.PT.58.16320644 |
| PrimeTime Predesigned qPCR Assay Cebpa | Integrated DNA Technologies | N007678.1.pt.Cebpa |
| PrimeTime Predesigned qPCR Assay Scx | Integrated DNA Technologies | Mm.PT.58.31750069 |
| PrimeTime Predesigned qPCR Assay Tbp | Integrated DNA Technologies | Mm.PT.39a.22214839 |
| Software and Algorithms | ||
| NIS elements | Nikon | N/A |
| Cellsense | Olympus | N/A |
| OpenLab | Meyer instruments | N/A |
| Prism | GraphPad | N/A |
| QuantStudio | Applied Biosystems | N/A |
| Fiji | Schindelin, J et al 2012; PMID:22743772 | https://imagej.net/Fiji/Downloads |
| Illustrator Creative Cloud | Adobe | N/A |
| FlowJo | BD | N/A |
| Basespace | Illumina | N/A |
| Bowtie 2 | Langmead, B et al 2012; PMID:22388286 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR | Dobin, A et al. 2013; PMID:23104886 | https://github.com/alexdobin/STAR |
| Cufflinks | Trapnell, C et al. 2010; PMID:20436464 | http://cole-trapnell-lab.github.io/cufflinks/getting_started/ |
| Cuffdiff | Trapnell, C et al. 2013; PMID:23222703 | http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/ |
| Ingenuity Pathway Analysis | Qiagen bioinformatics | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Excel | Microsoft | N/A |
| Cellranger 2.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Cellranger-atac-1.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R project for Statistical Computing | The R Project for Statistical Computing | https://www.r-project.org/ |
| CellrangerRkit | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat for R | The Comprehensive R Archive Network | https://cran.r-project.org/web/packages/Seurat/index.html |
| Monocle3 for R | Bioconductor | https://cole-trapnell-lab.github.io/monocle3/ |
| VisR | Younesy et al., 2015; PMID:26328469 | https://visrsoftware.github.io/ |
| VennDiagram | The Comprehensive R Archive Network | https://cran.r-project.org/web/packages/VennDiagram/index.html |
| ggplot2 | The Comprehensive R Archive Network | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Loupe Cell Browser | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Other | ||
Highlights:
Hic1 marks multiple quiescent mesenchymal progenitor (MP) subsets within skeletal muscle.
Conditional deletion of Hic1 leads to MP hyperplasia and an activated MP phenotype.
Hic1+ MPs generate transit amplifying progeny post-injury that support regeneration.
Following injury, select Hic1+ progeny persist and regenerate the myotendinous junction.
ACKNOWLEDGEMENTS
This work was supported by the following grants: Canadian Institutes of Health (CIHR) PJT-149026 and PJT-148816 (TMU), Terry Fox New Frontiers Program Project Grant 1021 (TMU), National Institutes of Health grant CA231652 (TMU) and CIHR MOP 97856 (FMVR). MA was supported by a UBC graduate scholarship. We would like to thank Dr. Arthur Sampaio (Stem Cell Technologies) for providing the tenogenic differentiation media. We would also like to thank the following for technical assistance: Heejung Choi, Germain Ho, Amanda Yang, Lin Yi, Ingrid Barta, Petra Schreiner, Daniel Tausan, and the following facilities, BRC-seq, ubcFLOW cytometry, AbLab, BRC genotyping and BRC transgenic unit. We also thank Dr. M. Koch for providing the anti-COL22a1 antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors have no competing interests to declare.
REFERENCES
- Armulik A, Genove G, and Betsholtz C (2011). Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21, 193–215. [DOI] [PubMed] [Google Scholar]
- Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, and Mourikis P (2018). Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P (2014). “Mesenchymal” stem cells. Annu Rev Cell Dev Biol 30, 677–704. [DOI] [PubMed] [Google Scholar]
- Bianco P, and Robey PG (2015). Skeletal stem cells. Development 142, 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet B, Guiraud A, Malbouyres M, Zwolanek D, Guillon E, Bretaud S, Monnot C, Schulze J, Bader HL, Allard B, et al. (2013). Knockdown of col22a1 gene in zebrafish induces a muscular dystrophy by disruption of the myotendinous junction. Development 140, 4602–4613. [DOI] [PubMed] [Google Scholar]
- Chen KG, Johnson KR, and Robey PG (2017). Mouse Genetic Analysis of Bone Marrow Stem Cell Niches: Technological Pitfalls, Challenges, and Translational Considerations. Stem Cell Reports 9, 1343–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, Watkins DN, Herman JG, Mankowski JL, and Baylin SB (2003). Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet 33, 197–202. [DOI] [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Sang L, and Roberts JM (2006). A new description of cellular quiescence. PLoS Biol 4, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, and Lazar MA (2011). Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, and Morrison SJ (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranse HJ, Sampaio AV, Petkovich M, and Underhill TM (2011). Genetic deletion of Cyp26b1 negatively impacts limb skeletogenesis by inhibiting chondrogenesis. J Cell Sci 124, 2723–2734. [DOI] [PubMed] [Google Scholar]
- Dulauroy S, Di Carlo SE, Langa F, Eberl G, and Peduto L (2012). Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat Med 18, 1262–1270. [DOI] [PubMed] [Google Scholar]
- Dumont NA, and Rudnicki MA (2017). Characterizing Satellite Cells and Myogenic Progenitors During Skeletal Muscle Regeneration. Methods Mol Biol 1560, 179–188. [DOI] [PubMed] [Google Scholar]
- Eklund L, Piuhola J, Komulainen J, Sormunen R, Ongvarrasopone C, Fassler R, Muona A, Ilves M, Ruskoaho H, Takala TE, and Pihlajaniemi T (2001). Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci U S A 98, 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenthal N, and Zelzer E (2017). Mechanical regulation of musculoskeletal system development. Development 144, 4271–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, and Leprince D (2009). HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. The international journal of biochemistry & cell biology 41, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, Wan R, Corneau A, Tajbakhsh S, Cheung TH, and Le Grand F (2019). High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol Cell 74, 609–621 e606. [DOI] [PubMed] [Google Scholar]
- Grimm C, Sporle R, Schmid TE, Adler ID, Adamski J, Schughart K, and Graw J (1999). Isolation and embryonic expression of the novel mouse gene Hic1, the homologue of HIC1, a candidate gene for the Miller-Dieker syndrome. Hum Mol Genet 8, 697–710. [DOI] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, and Braun T (2013). Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell stem cell 13, 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HM, Kolendowski B, Isovic M, Bose K, Dranse HJ, Sampaio AV, Underhill TM, and Torchia J (2017). Regulation of Active DNA Demethylation through RAR-Mediated Recruitment of a TET/TDG Complex. Cell Rep 19, 1685–1697. [DOI] [PubMed] [Google Scholar]
- Hicks D, Farsani GT, Laval S, Collins J, Sarkozy A, Martoni E, Shah A, Zou Y, Koch M, Bonnemann CG, et al. (2014). Mutations in the collagen XII gene define a new form of extracellular matrix-related myopathy. Hum Mol Genet 23, 2353–2363. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Garcha K, Karamboulas K, Cowan MF, Drysdale LM, Horton WA, and Underhill TM (2006). BMP action in skeletogenesis involves attenuation of retinoid signaling. J Cell Biol 174, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH, Lu HH, and Schweitzer R (2015). Molecular regulation of tendon cell fate during development. J Orthop Res 33, 800–812. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, and Rossi FM (2010). Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology 12, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Schulze J, Hansen U, Ashwodt T, Keene DR, Brunken WJ, Burgeson RE, Bruckner P, and Bruckner-Tuderman L (2004). A novel marker of tissue junctions, collagen XXII. J Biol Chem 279, 22514–22521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, and Humphreys BD (2015). Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell stem cell 16, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle-Dumais C, Schuitema V, Hayashi G, Hoff K, Gong W, Dao DQ, Ullian EM, Oishi P, Margeta M, and Gould DB (2019). COL4A1 Mutations Cause Neuromuscular Disease with Tissue-Specific Mechanistic Heterogeneity. Am J Hum Genet 104, 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos DR, Babaeijandaghi F, Low M, Chang CK, Lee ST, Fiore D, Zhang RH, Natarajan A, Nedospasov SA, and Rossi FM (2015). Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med 21, 786–794. [DOI] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, and Fan CM (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelle C, Eichmann A, Halevy O, Breant C, and Le Douarin NM (1994). Distinct developmental expression of a new avian fibroblast growth factor receptor. Development 120, 683–694. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, and Frenette PS (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohassel P, Foley AR, and Bonnemann CG (2018). Extracellular matrix-driven congenital muscular dystrophies. Matrix Biol 71–72, 188–204. [DOI] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, and Kardon G (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, and Morrisey EE (2015). Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospichalova V, Tureckova J, Fafilek B, Vojtechova M, Krausova M, Lukas J, Sloncova E, Takacova S, Divoky V, Leprince D, et al. (2011). Generation of two modified mouse alleles of the Hic1 tumor suppressor gene. Genesis 49, 142–151. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, and Schweitzer R (2007). Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn 236, 1677–1682. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017a). Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017b). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EW, Deonarine A, Jones JO, Denton AE, Feig C, Lyons SK, Espeli M, Kraman M, McKenna B, Wells RJ, et al. (2013). Depletion of stromal cells expressing fibroblast activation protein-alpha from skeletal muscle and bone marrow results in cachexia and anemia. J Exp Med 210, 1137–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, and Galy A (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RK, Mullally A, Dugourd A, Peisker F, Hoogenboezem R, Van Strien PMH, Bindels EM, Heckl D, Busche G, Fleck D, et al. (2017). Gli1+ Mesenchymal Stromal Cells Are a Key Driver of Bone Marrow Fibrosis and an Important Cellular Therapeutic Target. Cell stem cell 20, 785–800 e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RW, and Underhill TM (2016). Methods and Strategies for Lineage Tracing of Mesenchymal Progenitor Cells. Methods Mol Biol 1416, 171–203. [DOI] [PubMed] [Google Scholar]
- Serrano AL, and Munoz-Canoves P (2010). Regulation and dysregulation of fibrosis in skeletal muscle. Exp Cell Res 316, 3050–3058. [DOI] [PubMed] [Google Scholar]
- Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, Nusse R, Amieva MR, and Meyer TF (2017). Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature 548, 451–455. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kanzaki LF, Galloway JL, and Schilling TF (2018). Mechanical force regulates tendon extracellular matrix organization and tenocyte morphogenesis through TGFbeta signaling. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, and Schilling TF (2015). Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. Development 142, 4191–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney MT, Gromova A, Sesillo FB, Sala D, Spenle C, Orend G, and Sacco A (2016). Autonomous Extracellular Matrix Remodeling Controls a Progressive Adaptation in Muscle Stem Cell Regenerative Capacity during Development. Cell Rep 14, 1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, and Tsuchida K (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature cell biology 12, 143–152. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, Yamaguchi M, Ogawa R, Matev MM, Miyagoe-Suzuki Y, et al. (2011). Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 124, 3654–3664. [DOI] [PubMed] [Google Scholar]
- Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, et al. (2013). Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat Commun 4, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guerardel C, and Leprince D (2010). Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol 30, 4045–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, and Rudnicki MA (2013). Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A 110, 16474–16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, and Baylin SB (1995). p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med 1, 570–577. [DOI] [PubMed] [Google Scholar]
- Weston AD, Chandraratna RA, Torchia J, and Underhill TM (2002). Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J Cell Biol 158, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y, et al. (2015). Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell 160, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosczyna MN, Konishi CT, Perez Carbajal EE, Wang TT, Walsh RA, Gan Q, Wagner MW, and Rando TA (2019). Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep 27, 2029–2035 e2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Ichikawa N, Kosaki K, Yamada Y, Sasaki T, Sakai LY, Kurosawa H, Hattori N, and Arikawa-Hirasawa E (2010). Perlecan deficiency causes muscle hypertrophy, a decrease in myostatin expression, and changes in muscle fiber composition. Matrix Biol 29, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, and Rudnicki MA (2013). Satellite cells and the muscle stem cell niche. Physiol Rev 93, 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younesy H, Moller T, Lorincz MC, Karimi MM, and Jones SJ (2015). VisRseq: R-based visual framework for analysis of sequencing data. BMC Bioinformatics 16 Suppl 11, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD, and Patton BL (2009). Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des 15, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, and Morrisey EE (2017). Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Cai S, Shin K, Lim A, Kalisky T, Lu WJ, Clarke MF, and Beachy PA (2017). Stromal Gli2 activity coordinates a niche signaling program for mammary epithelial stem cells. Science 356. [DOI] [PubMed] [Google Scholar]
- Zhou BO, Yue R, Murphy MM, Peyer JG, and Morrison SJ (2014). Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell stem cell 15, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Zwolanek D, Izu Y, Gandhy S, Schreiber G, Brockmann K, Devoto M, Tian Z, Hu Y, Veit G, et al. (2014). Recessive and dominant mutations in COL12A1 cause a novel EDS/myopathy overlap syndrome in humans and mice. Hum Mol Genet 23, 2339–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RNA-seq of skeletal muscle-derived mononuclear fractions (related to Figure 1)
Table S2. RNA-seq of control and Hic1 10 d cKO MPs (related to Figure 2)
Table S3. RNA-seq of 18 m control and Hic1 cKO whole skeletal muscle (related to Figure 2)
Table S4. RNA-seq of sorted MPs from skeletal muscle at various time points following injury (related to Figure 5)
Data Availability Statement
All sequence data described in this paper have been deposited in GEO: GSE110038. Software used to analyze the data is either freely or commercially available.