Abstract
Primary Sjögren’s syndrome (pSS) is an autoimmune disease in which the underlying cause has yet to be elucidated. The main objective of this study was to determine the T cell receptor (TCR) repertoires of individual infiltrating T helper (Th)-1 and 17 cells of pSS patients using single-cell analysis. Single-cell analysis of ex-vivo infiltrating T cells demonstrated that pSS patients had higher frequencies of activated Th17 cells. Single-cell TCR sequencing revealed that TCRβ variable (TRBV)3–1/joint (J)1–2 (CLFLSMSACVW) and TRBV20–1/J1–1 (SVGS-TAIPP*T) were expressed by activated Th1 and Th17 cells in both cohorts. Uniquely, TCRα variable (TRAV)8–2/J5 (VVSDTVLETAGE) was expressed by Th1 cells present only in patients and complementarity-determining region (CDR)3α-specific motif (LSTD*E) present in both Th1/Th17 cells. The study demonstrates that both activated Th1 and Th17 cells of pSS patients showed restricted clonal diversities of which two CDR3 motifs were present in controls and patients, with another two motifs unique to pSS.
Keywords: Sjögren’s syndrome, T cell receptor, Single-cell analysis, Th1, Th17, IFN-γ, IL-17A
1. Introduction
Sjögren’s Syndrome (SS) is a complex autoimmune disorder characterized by the inflammation of secretory glands, namely the lacrimal and salivary glands leading to keratoconjunctivitis sicca (KCS) and xerostomia i.e. dry eyes and dry mouth [1]. SS manifests in patients as either primary SS (pSS), in which patients suffer from only SS symptoms, or secondary SS which occurs in association with other autoimmune diseases. The disease progression is marked by the infiltration of lymphocytes of which some foci organize into germinal center-like formations in the glands causing periductal aggregates, inflammation, and apoptosis [2–4]. In human patients, focus scores (FS) of ≥1 were found to be strongly correlated with seropositive anti-SSA/SSB, rheumatoid factor, and the SS ocular component, but not with salivary/lacrimal gland hypofunction [5]. The αβ T cell receptor (TCR) is most prominently expressed on infiltrating CD4+ T helper cells (Th) [6–9]. Recent studies have indicated that interferon (IFN)-γ-producing Th1 cells and interleukin (IL)-17A-producing Th17 cells are essential to the development of the autoimmune response and secondary manifestations [10–12].
Patients suffering from SS exhibit high levels of IFN-γ and IFN-responsive factors. The upregulation of the IFN pathway induces the activation of macrophages, natural killer (NK) cells and CD8+ T cells. Additionally, it induces vascular adhesion molecule-1 (VCAM-1), L-selectin, lymphocyte function-associated antigen-1 (LFA-1), and other molecules that can trigger the homing of immune cells to the glands [13]. Earlier studies, which used an animal model of SS, demonstrated that IFN-γ plays a role in early disease development by attracting invasive lymphocytes and hindering gland development; this, in turn, exacerbates glandular dysfunction. In addition to IFN-γ, Th17 cells have been shown to contribute profoundly to the disease pathogenesis, where it induces isotypic antibody switching, recruits neutrophils, and induces proliferation. Recent studies indicate the essential function of Th17 cells in sexual dimorphism in the SS mouse model, specifically the promotion of sialadenitis in females and alteration of plasma cells and germinal center B cell populations [14,15].
Proliferation and activation of the effector T cells are initiated by recognition of a peptide antigen in a specific interaction via the TCR in the context of the major histocompatibility complex (MHC) expressed on antigen presenting cells [16]. TCRs are heterodimeric membrane proteins that comes in two forms, αβ and γδ, the former of which is present in the SGs in 70% of infiltrating T cells [16]. During the immune response, antigen-specific interactions lead to proliferation of only reactive T cells, clonotypic restriction, and loss in diversity of the T cell repertoire [17–19]. A productive immune response would be thus characterized by a rich, robust T cell clonal repertoire and/or a restricted distribution of T cell clones. Diversity of the TCR is generated from the unique pairing of variable (V) and joining (J) gene segments for the α chain or V, diversity (D), and J in the case of the β chain, which generates on the order of 1014 possible combinations [20,21]. Each chain possesses a hypervariable region or complementarity determining region 3 (CDR3), which contains the amino acids (AA) responsible for interaction with the antigen. In this study, using single-cell analysis, we investigated the comprehensive TCR repertoires of individual infiltrating T cells in labial salivary gland (LSG) biopsies by analyzing the TCRαβ transcripts of CD4+ and CD8+ T cells that produce either IFN-γ or IL-17A. Our results indicate that the TCR repertoire of the Th1 and Th17 cells in the LSG of pSS patients exhibited reduced diversity in the CDR3 regions, expressing predominantly TRBV3–1/J1–2 (CFLFLSMSACVW), TRBV20–1/J11 (SVGSTAIPP*T), and TRAV8–2/J5(CVVSDTVLETAGE), the latter of which (along with CDR3α-specific motif ‘LSTD*E’) is unique to pSS patients. These clones indicate an antigen-mediated maturation of effector T cells to a common autoantigen in pSS. These findings allow us to understand the mechanism of autoantigen recognition and brings us closer to understanding the underlying etiology of SS.
2. Material and methods
2.1. Human subjects
Participants underwent extensive serologic and histological evaluations as standard of care as previously described [22]. Following the initial evaluation by a rheumatologist, each patient was referred to the Oral Medicine Clinic at the University of Florida for a review of his or her medical history, an oral examination, an unstimulated whole salivary flow rate, and a LSG biopsy. pSS patients were defined using the 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for pSS [23]. Patients with symptoms indicative of pSS (n = 9) who had a cumulative score of ≥4, in which a score of 3 for focal lymphocytic sialadenitis with a focus score of ≥1 foci/4 mm2 and seropositive for anti-SSA/Ro, and score of 1 for an unstimulated salivary flow rate of ≤0.1 mL/min. Non-pSS controls or sicca controls (SC) (n = 20) were defined as those who complained of dry mouth/eyes, but did not meet criteria for pSS with the cumulative score of < 4 [23]. All participants’ clinical profiles were presented in Table 1. All procedures were reviewed and approved by the University of Florida Institutional Review Board.
Table 1.
Patients’ clinical profiles.
| Patients | Age (years) | Sex | Disease | SFR (mL/ min) |
Focus Score |
ANA | SSA | SSB |
|---|---|---|---|---|---|---|---|---|
| 235 | 40 | F | SC | 0.130 | 0 | − | + | − |
| 150 | 44 | F | SC | 0.150 | 0 | + | − | − |
| 252 | 46 | F | SC | 0.110 | 0 | − | − | − |
| 255 | 47 | F | SC | 0.020 | 0 | − | − | + |
| 257 | 49 | F | SC | 0.048 | 0 | − | − | − |
| 251 | 51 | F | SC | 0.190 | 0 | + | + | − |
| 236 | 52 | F | SC | 0.047 | 0 | − | − | − |
| 264 | 52 | F | SC | 0.380 | 0 | − | − | − |
| 265 | 54 | F | SC | 0.210 | 0 | − | − | − |
| 215 | 55 | F | SC | 0.012 | 0 | − | − | − |
| 229 | 55 | F | SC | 0.020 | 0 | − | − | − |
| 243 | 57 | F | SC | 0.002 | 0 | − | − | − |
| 220 | 57 | F | SC | 0.070 | 0 | − | − | − |
| 259 | 67 | F | SC | 0.152 | 0 | − | − | − |
| 242 | 68 | F | SC | 0.280 | 0 | + | − | − |
| 239 | 73 | F | SC | 0.048 | 0 | − | − | − |
| 261 | 70 | M | SC | 0.080 | 0 | + | − | − |
| 246 | 72 | M | SC | 0.312 | 0 | − | − | − |
| 262 | 75 | M | SC | 0.400 | 0 | − | + | − |
| 248 | 76 | M | SC | 0.002 | 0 | − | − | − |
| 247 | 36 | F | pSS | 0.134 | 1 | + | − | + |
| 241 | 48 | F | pSS | 0.176 | 5 | + | + | − |
| 227 | 50 | F | pSS | 0.030 | 9 | + | + | − |
| 254 | 53 | F | pSS | 0.007 | 3 | + | − | − |
| 226 | 58 | F | pSS | 0.020 | 2 | − | − | − |
| 249 | 59 | F | pSS | 0.054 | 1 | + | + | − |
| 258 | 69 | F | pSS | 0.104 | 1 | + | − | − |
| 260 | 66 | M | pSS | 0.080 | 0 | − | + | − |
| 263 | 61 | F | pSS | 0.068 | 1 | − | − | − |
Summary of patient profiles of the individuals in this study. SC: sicca. control, pSS: primary Sjögren’s Syndrome, SFR: unstimulated saliva flow rate, Focus score is based on a scale of 1–9 where 9 represents severe in, ANA: Antinuclear antibody, SSA: anti-SSA/Ro, SSB: anti-SSB/La.
2.2. Single-cell microengraving process
LSG biopsies were obtained and washed 5X in RPMI complete medium. Single lymphocyte isolation of LSG biopsies were isolated in RPMI medium supplemented with 1 mg/mL collagenase and 1 mg/mL DNase as previously described [14]. Lymphocytes were cultured in RPMI with 3 Units/μL IL-2, anti-CD3/CD28 beads (Thermo Scientific), 500 ng/mL Ionomycin sulfate and 50 ng/mL PMA (Sigma-Aldrich) for 4 h at 37 °C. Cells were then stained using anti-CD8-APC, anti-CD3-FTIC, anti-CD4-PE (Biolegend), and Calcein violet 405 nm Live/Dead (Life Technologies). Following surface staining, cells were suspended in 300 μL complete medium were deposited onto the arrays of nanowells and imaged as previously described [22]. The nanowells were hybridized with the capture slide coated with anti-IL-17A (eBioscience) and anti-IFN-γ (MABTECH). After 2 h, the capture slide was coated with anti-IL-17A-Dylight 488 and anti-IFN-γ-Dylight 594. The slide was imaged using the Genepix 4400A scanner (Molecular Devices). Single-cell data analysis was performed as described previously [22,24].
2.3. Sequencing of TCRs expressed by single T cells using nested RT-PCR
Individual T cells secreting IFN-γ or IL-17A were isolated using a micromanipulator, were stored in PBS with 2% BSA and 200 U/mL Rnasin RNAse inhibitor (Promega), and frozen at −20 °C. These were then used individually for reverse transcription (RT) reaction per manufacturer’s protocol (iScript cDNA Sythesis Kit, Biorad) supplemented with 0.1% Triton X100 detergent to facilitate lysis. Following RT, PCR was performed using a protocol and primers described previously [25]. In brief, a 1:50 dilution of the cDNA produced from single-cell RT reaction was used as template for PCR using Primestar Taq (Clontech). Successful reactions were determined by gel electrophoresis and band size confirmation. PCR products were purified as instructed by manufacturer (QIAquick PCR Purification Kit, Qiagen) and sequenced using Sanger sequencing (Eton Bio).
2.4. Diversity indices
TCR repertoire diversity was determined using Shannon’s Entropy (SE) as well as Simpson’s Diversity Index (SD). Both indices account for the population’s richness and uniformity or homogeneity of the population, which are two important components of repertoire diversity. The richness of a population calculated by SE [26] is defined by its total number of species (for example, CDR3 AA sequences), whereas homogeneity calculated by the SD [27] which measures the distribution of the species (for example, CDR3 amino-acid sequences) in the population. SE depicts the diversity of a population by measuring uniformity; higher values are more uniform and thus normally distributed. In contrast, SD approximates diversity and indicates overlap, where a lower value indicates greater diversity and 0 indicates no clonal overlap.
2.5. Bioinformatic analysis of TRA and TRB hypervariable region and motif analyses
T receptor gene and allele identification was determined inputting the nucleotide sequence into the IgBLAST (TR sequence analysis) tool (http://www.ncbi.nlm.nih.gov/igblast/). Additionally, nucleotide sequences were translated with ExPASy (web.expasy.org/translate) Frame alignment was determined by AA sequence generated by IgBLAST. Most CDR3 sequences were also generated by IgBLAST; the remaining sequences were determined by IMGT criteria (starting at TGY (cysteine codon) followed by a variable sequence of approximately 10–14 AAs and ending at TTG (tryptophan codon). Motif analysis was performed on the CDR3 AA sequences with MEME Suite (www.memesuite.org), where statistical significance was provided based on the input data group.
2.6. Statistical analysis
Data was analyzed using unpaired two-tailed Mann-Whitney test (GraphPad Prism) to determine the statistical significance. In all cases, p values ≤ 0.05 were considered significant. TCR Clonal diversity was determined with Shannon’s entropy as well as Simpson’s Diversity Index.
3. Results
3.1. Increased frequency of active IL-17A-producing Th17 cells in the LSG of pSS patients using single-cell analysis
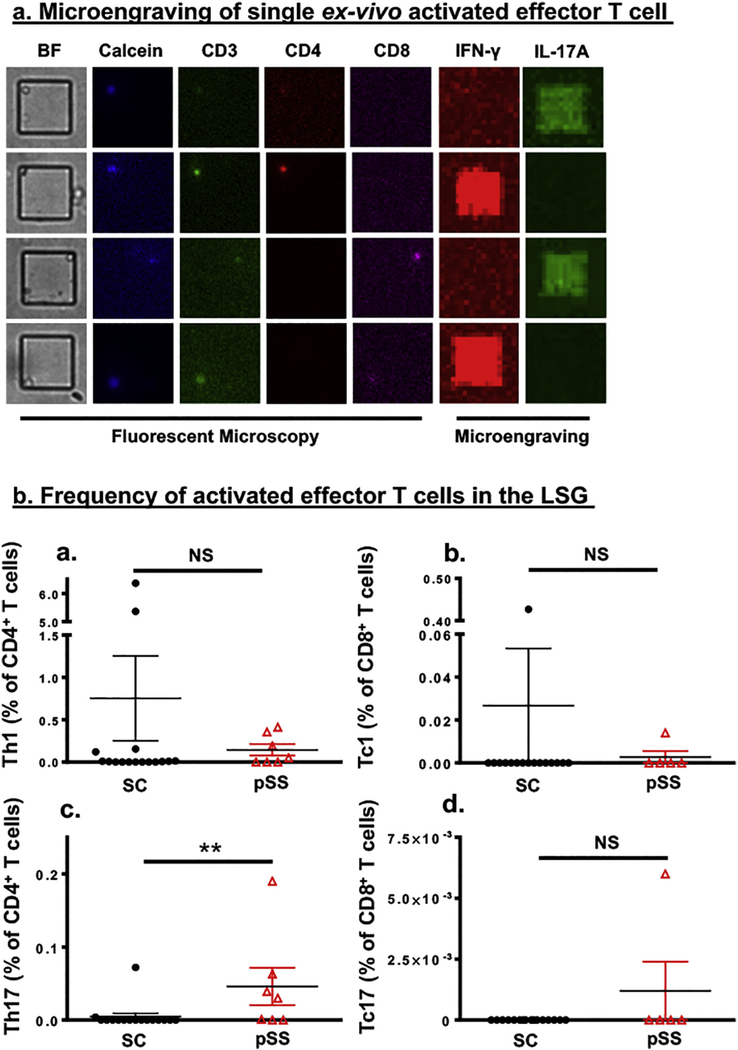
Glandular infiltrating effector T cells that produce either IFN-γ or IL-17A have been implicated in the etiology and the clinical manifestations of SS [28–32]. Current techniques, including immunostaining and flow cytometry, have identified a significant presence of these cell populations in the labial salivary glands (LSGs) of SS patients. However, due to the small size of the LSG biopsies, the complete profiling of the effector T cell populations ex-vivo is limited. As a result, in this study, single-cell analysis was utilized to identify and examine live ex-vivo effector T cells in LSG biopsies. Single-cell suspensions from LSGs were isolated from pSS patients and sicca controls (SC). Specific subsets of activated effector T cells were identified and microengraved for active secretion of IL-17A and IFN-γ with the following makers: CD3+CD4+IFN-γ+ (Th1), CD3+CD4+IL-17A+ (Th17), CD3+CD8+IFN-γ+ (Tc1), and CD3+CD8+ IL-17A+ (Tc17) (Figure (Fig. 1a). As presented in Fig. 1b and Supplementary Table S1, control subjects appear to exhibit a higher, but statistically insignificant, frequency of Th1 (0.753% vs 0.143%) and Tc1 (0.027% vs 0.003%) cells than pSS patients, whereas pSS patients had a significant increase of Th17 cells over SC subjects. Examining total cell counts yielded similar result (Supplementary Fig. S1). The data indicated that ex vivo examination of live LSG cells using single-cell analysis reveals marked expansion of activated Th17 cells in pSS patients.
Fig. 1.
Microengraving shows greater infiltration by activated Th17 cell in the labial salivary glands of pSS patients. a) Microengraving of single ex-vivo activated effector T cell. Representative fluorescent microscopy coupled with microengraving of secreted cytokines from isolated individual T cell. Fluorescent antibody staining was performed with anti-CD3-FITC (green) anti-CD4-PE (red), anti-CD8-APC (Magenta), and Calcein violet-405 (blue), a marker of viable cells. Secreted cytokines were captured during microengraving and detected with anti-IFN-γ (red) and anti-IL-17A (green). b) Quantification of activated effector T cells isolated from the LSG of SC subjects () and pSS patients () expressing (a) CD3+CD4+IFN-γ+(Th1), (b) CD3+CD8+IFN-γ+(Tc1), (c) CD3+CD4+IL-17+ (Th17), and (d) CD3+CD8+IL-17+ (Tc17). The frequency in percentage was determined by using the percentage (multiplied by 100) of the total number of Th1, Th17, Tc1, and Tc17 cells from wells with single live cells among the total number of wells with single CD4+ or CD8+ cells. Statistics were performed using an unpaired two-tailed Mann-Whitney test. Significance was determined as **p < 0.01, and NS: not significant.
3.2. Loss of TCR repertoire diversity on activated Th1 and Th17 cells is associated with Sjögren’s syndrome
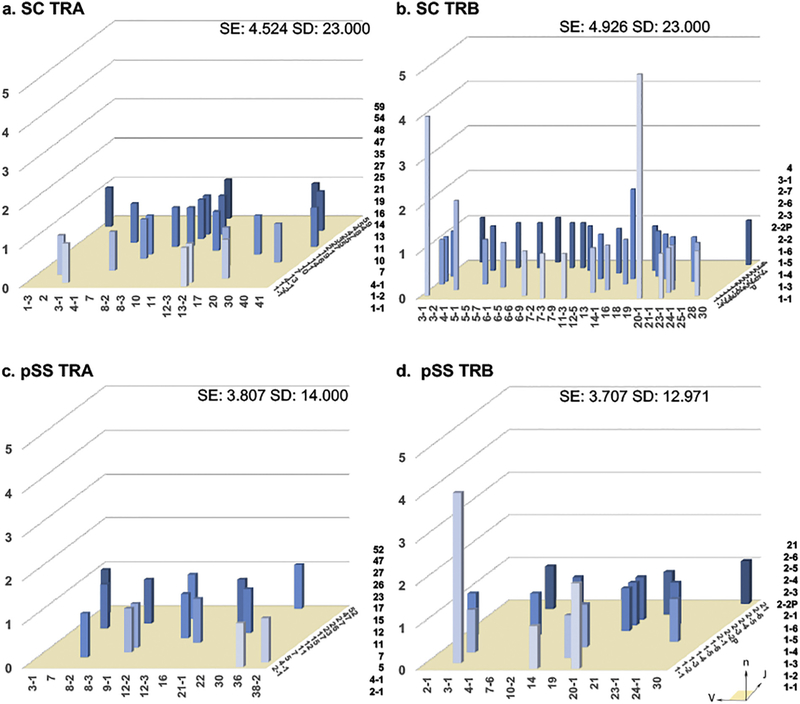
To explore the TCR repertoires of effector T cells of pSS patients, ex vivo Th1 and Th17 cells were examined for TCR gene rearrangements. After microengraving, nested PCR was performed with primers that target the CDR3 hypervariable regions to examine the TCRs of individual cells. Sequences were aligned to the IMGT database via the IgBLAST tool to determine the V/J (and D) genes; the diversity of whose combinations were calculated for each group with SE and SD. The diversity reflects the progression of the autoimmune response where a lower diversity indicates clonal expansion with positive selection for antigen-experienced effector T cells. V/J combinations are shown in Fig. 2 as a representation of the total repertoire of infiltrating effector T cells from SC and pSS patients. SC subjects had slightly higher SE values than pSS patients for both TRA (4.524 vs 3.807, respectively, Fig. 2a and c) and TRB (4.926 vs 3.707, respectively, Fig. 2b and d) TCR repertoires. Likewise, SC subjects had a greater SD for TRA (23.000 vs 14.000, respectively, Fig. 2a and c) and TRB (23.000 vs 12.971, Fig. 2b and d). While SC and pSS repertoires exhibited similar gene usage for TRA repertoires, pSS patients showed a restriction in TRBJ gene usage, specifically TRBV (14 and 13 TRBJ alleles for SC and pSS, respectively as opposed to 27 and 12 TRVB alleles, respectively). A single high frequency pairing TRBV3–1/J1–2 was present in both SC and pSS; TRBV20–1/J1–1 was present at significantly elevated levels only in SC repertoires. There are no unique pairings found at elevated levels exclusively in pSS patients. This reveals that pSS has reduced evenness between the frequencies of specific CDR3 AA sequences, thus clonally restricted, while SC subjects have greater richness, or total number, of CDR3 AA sequences.
Fig. 2.
Loss of normal distribution in V/J pairing is associated with pSS patients. Total LSG T-lymphocyte three dimensional histograms of TCR gene V/J combinations for SC TRA (a) and SC TRB (b) genes, as well as pSS TRA (c) and TRB (d) genes. The x-axis represents the analyzed Vα or Vβ gene segment, the z-axis is the analyzed Jα or Jβ gene segments, the y-axis indicates the total reads of specific V/J combination. V and J genes were designated by the disambiguated IMGT nomenclature for each gene subgroup. TCR repertoire was quantified by entropy (SE = Shannon’s Entropy) and diversity (SD = Simpson’s Diversity Index).
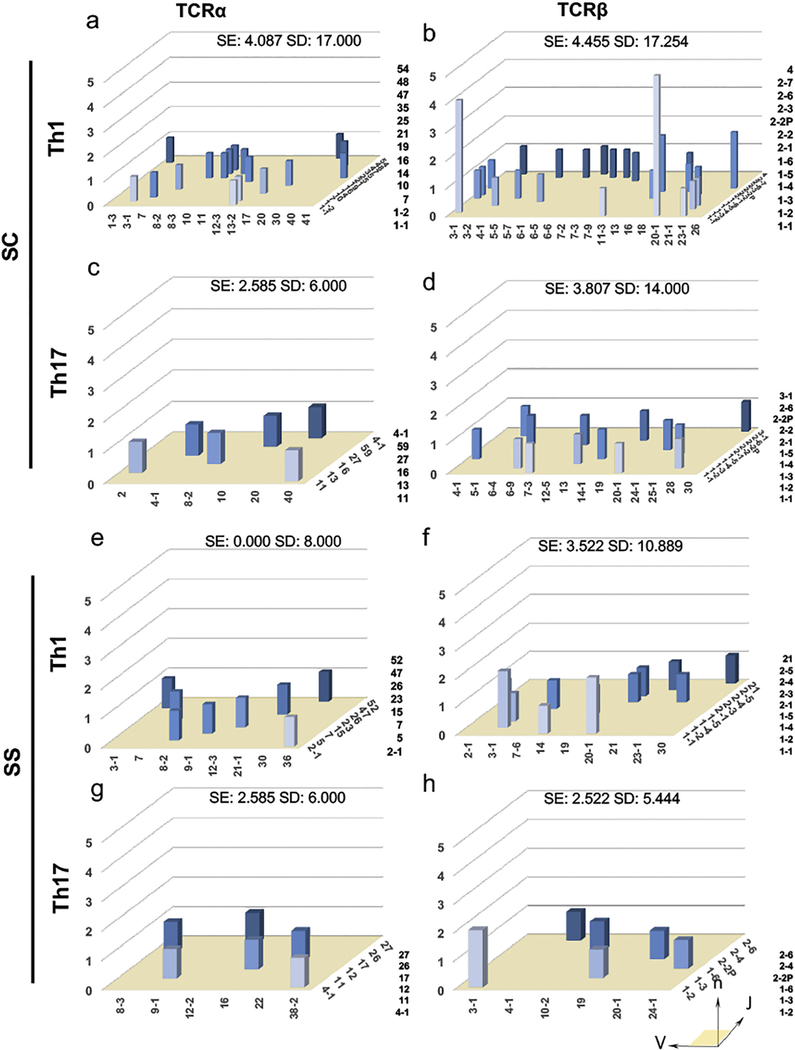
3.3. Distribution V/J pairing contributed by activated Th1 and Th17 reveals restricted Th1 TCRα and Th17 TCRβ repertoires
The V/J pairings of Th1 and Th17 cells were examined for the overall sample richness. The repertoire evenness which measures the relative abundance of the receptor sequences in a repertoire using SE values. As presented in Fig. 3. SE values were greater for Th1 TCRα/β and Th17 TCRβ of SC subjects in comparison to pSS patients (SC Th1 TCRα: 4.087, TCRβ: 4.455, Th17 TCRα: 2.585, TCRβ: 3.807 compared to pSS repertoires Th1 TCRα: 0.000, TCRβ: 3.522, Th17 TCRα: 2.585, TCRβ: 2.522). Furthermore, Th1 TCRα was severely depressed in pSS patients and SE values of Th17 TCRα were similar in both groups which suggests that the extent to which a few of the Th17 TCRα sequences present in SC and pSS patients is comparable (Fig. 3). Using SD to measure diversity, the data illustrated that the overall diversity of the SC repertoires in comparison to the pSS repertoires (Fig. 3, Th1 TCRα: 17.00 vs 8.000; Th17 TCRβ: 14.00 vs 5.444; Th1 TCRβ: 17.254 vs 10.889, SC to pSS, respectively). However, Th17 TCRα repertoires of both SC and pSS patients were equally rich, with a relatively low diversity (Fig. 3, SD of 6.000).
Fig. 3.
Th1 and Th17 cells from the pSS patients show reduced diversity in V/J pairing. Three dimensional histograms of TCR gene V/J combinations derived from SC Th1 cells (a)TRA and (b) TRB genes, Th17 cells (c) TRA and (d) TRB genes, pSS Th1 cells (e) TRA and (f) TRB genes, Th17 cells (g) TRA and (h) TRB genes. The x-axis represents the analyzed Vα or Vβ gene segment, the z-axis is the analyzed Jα or Jβ gene segments, the y-axis indicates the total reads of specific V/J combination. V and J genes were designated by the unambiguous IMGT nomenclature for each gene subgroup.
The TRBV3–1/J1–2 pairing is present only in SC Th1 cells, but in Th1 and Th17 subsets of pSS. Meanwhile, the TRBV20–1/J1–1 pairing was present in Th1 and Th17 of SC subjects, and Th17 cells of pSS patients. These data suggest an enhanced richness and evenness of SC TCRα and TCRβ repertoires of Th1 cells when compared to pSS patients. These findings are consistent with autoimmune patients experiencing positive selection for antigen-experience T cells. Notably, both SC and pSS patients experienced low diversity and uniformity of the Th17 TCRα repertoire, indicating both groups were going through this positive selection event.
3.4. High frequency of CDR3 sequences were found in activated Th1 and Th17 cells of both SC and SS patients
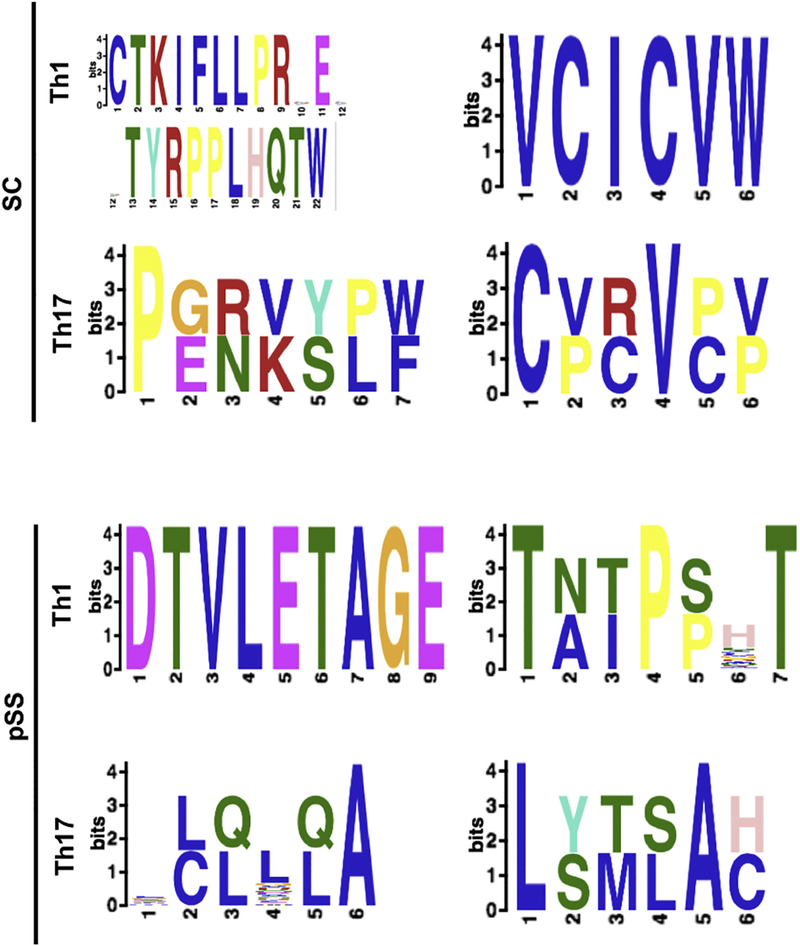
Convergent mechanisms of epitope maturation will lead to common CDR3 AA sequences. Non-template N nucleotide addition yields a higher diversity upon the myriad of sequences derived from V/J recombination. To understand the repertoire diversity in hypervariable regions, we examined the AA sequences in the CDR3 regions of pSS and SC populations. SC subjects had no identical CDR3α clones (Table S2 and Table S3). Only 23% of these sequences had premature stop codons in the CDR3α and 26% of CDR3β chain repertoires in the Th1 cells. Where no premature stop codons existed in the CDR3α of Th17 cells, 43% of the CDR3β chain repertoires of Th17 cells in SC subjects contained stop codons. 21% of Th1 CDR3β repertoires and 25% of CDR3α from Th17 cells of pSS repertoires contained premature stop codons. In pSS Th17 cells, 33% of the CDR3α repertoires contained a premature stop codon compared to 14% of the CDR3β repertoires. There were no overlapping CDR3α sequences in Th17 cells (Table S4 and Table S5). Of TCR clones expressed by Th1 and Th17 cells with productive gene arrangements, four conserved clones were identified, in which one clone was shared between SC and pSS patients, one was specific only to SC subjects, and two were unique to pSS patients. The most common CDR3 motif was derived from the TRBV3–1/J1–2 combination, which yielded “CFLFLSMSACVW’, and was present in all CDR3β repertoires of pSS and SC patients, except SC Th17 cells. The next most common motif was derived from TRBV20–1/J1–1; ‘SVGSTAIPP*T’ was present in both activated Th1 and Th17 CDR3β repertoires of SC subjects, but only in activated Th1CDR3β repertoires of pSS patients. The two pSS-unique motifs identified, ‘VVSDTVLETAGE’ from TRAV8–2/J5 and ‘LSTD*E’ from varying CDR3α chains. The latter was present only in the CDR3α chain of activated Th1 cells of pSS patients and accounted for 25% of the CDR3α repertoire. The two common CDR3β motifs, “CFLFLSMSACVW” and ‘SVGSTAIPP*T’, imply that there is a similar antigen in the glands of both SC and pSS patients, however the presence of the two unique motifs, ‘VVSDTVLETAGE’ and ‘LSTD*E’, in the glands of pSS patients is evidence of a unique autoantigen to SS.
3.5. Motif analysis reveals conserved amino acids in the hypervariable CDR3 regions of the pSS
MEME analysis was employed to determine the likelihood of AA identity in each position of a motif, specifically the pairings above (TRBV3–1/J1–2, TRBV20–1/J1–1, and TRAV8–2/J5) with the aforementioned motifs to determine any underlying mechanisms for CDR3 AA selection (Table 2). Both “CFLFLSMSACVW” and ‘SVGSTAIPP*T’ motifs were conserved and shared by SC and pSS patients. The ‘LSTD*E’was identified in only pSS patients and expressed by both Th1 and Th17 cells. Whereas, ‘VVSDTVLETAGE’ motif was only identified in Th1 of pSS patients. The MEME analysis of this motif showed the DTVLE core section, which provides two strongly hydrophobic AAs (valine and leucine) flanked by two charged hydrophilic AAs.
Table 2.
MEME Analysis of high frequency hypervariable regions from SC and pSS patient’s reveal common and unique motifs.
| CDR3 amino acid motif | E-value | Cell Type | V-J |
|---|---|---|---|
 |
1.4e−037 | Various | Vβ3–1/Jβ1–2 |
 |
1.1e−021 | Th1 (Th17)* | Vβ20–1/Jβ1–1 |
 |
1.5e−001 | pSS Th1** | Vα30/Jα52 |
 |
4.3e+002 | pSS ** | Vα-Jα Various |
Amino acid motif analysis was performed on the high frequency CDR3 sequences from both SC and pSS patients. Bit height corresponds to the likelihood of the amino acid in each position. Blue – hydrophobic, uncharged amino acids, Red – positively charge hydrophilic amino acids, Green – Neutral hydrophilic amino acids, Magenta – negatively charged hydrophilic amino acids, Orange-glycine, and Yellow—proline. E-value indicates the model confidence for the amino acid in that position. Cell type indicates the phenotype associated with the motif, V/J indicates recombination gene segment pairings.
Indicates a bias towards presence in Th1 cells.
Present in only pSS patients.
To discern any discriminating selective differences between activated Th1 and Th17 cells in controls and pSS patients, GLAM2 algorithm was employed to examine the motifs of the full repertoires of activated Th1 and Th17 cells (Fig. 4). While SC CDR3α motifs do not contain a shared motif neither between Th1 and Th17, nor with any pSS cells, pSS Th17 and Th1 cells shared a profile. The internal portion of the Th1 motif “VLETA” aligned directly with “LQLQA” motif, where both neutral sequences consist of a hydrophobic, non-polar AA is followed by another of that kind or a polar AA (glutamic acid), a variable AA, a polar AA, then a conserved hydrophobic alanine. SC Th1 CDR3β 6-AA motif “VCICVW” is a completely conserved hydrophobic, non-polar AA sequence. Both SC and pSS Th17 CDR3β 6-AA motifs have a conserved starting hydrophobic, non-polar AA (cysteine and lysine, respectively), with another in the fourth or fifth position (valine or alanine, respectively). For SC Th17 CDR3β, the second, fifth, and sixth positions are either an aliphatic AA like the SC Th1 CDR3β sequence, or a cyclic AA (proline), but in any case hydrophobic and non-polar. The third position however, can also contain a basic, hydrophilic AA (arginine). Th17 pSS CDR3β, at positions two, three, four, and six, vary widely between polar and non-polar AAs. Similarly, though the pSS Th1 CDR3β starts and ends with a conserved polar AA (threonine), positions two, three, and five fluctuate between polar and non-polar AAs, flanking a conserved, non-polar cyclic residue (proline). Also of note, is that several motifs contain premature stop codons: SC Th1 CDR3α, pSS Th17 CDR3α, and pSS Th1 CDR3β. Half of the sequences from both SC and pSS patients were six AAs (with no bias to cell or chain type). The short motifs imply little commonality in sequences from these repertoires. The ‘DTVLETAGE’ sequence at nine AAs is fairly long, but biased due to the heavy contribution from the ‘CVVSDTVLETAGE’ motif identified in Table 2, which may indicate an antigenic response. The highly conserved, longest common motif was the CDR3α from activated Th1 cells in SC patients, indicating a specific antigen response. Interestingly, one motif, ‘VVSDTVLETAGE’, is unique to only pSS and is characterized by hydrophobic valine-leucine pairing flanked by the polar aspartic and glutamic acids. The results indicate in addition to restricted repertoires, there are conserved AAs in the CDR3 motifs which suggest common antigen or antigen recognition mechanism in SC subjects and SS patients.
Fig. 4.
TCR motif analyses of Th1 and Th17 cells show conserved amino acids in CDR3 regions. GLAM2 motif analyses were performed on TCR repertoires of Th1 and Th17 in pSS and controls. Bit height corresponds to AA identity likelihood. Blue – hydrophobic, neutral amino acids (AAs), Red – positively charge hydrophilic AAs, Green – Neutral hydrophilic AAs, Magenta – negatively charged hydrophilic AAs, Orange-glycine, Teal-Tyrosine, Pink-Histidine (positively charged moderately hydrophobic), and Yellow—Proline.
4. Discussion
Sjögren’s syndrome is a complex autoimmune immune disease with multi-factorial etiologies. The temporal influx of effector T cells with potent inflammatory cytokines, like IFN-γ and IL-17A contributes to various pertinent patho-immunological aspects of the disease. In the present study, using functional single-cell analysis, we were able to identify and characterize different subsets of activated effector T cells in the LSG of patients. Additionally, we were able to examine the TCR repertoires of individual effector T cells, identified by their signature secreted cytokines, concomitantly. The results indicate that activated Th17 cells are highly prevalent in the glands of pSS patients and that patients exhibit restricted Th1 and Th17 TCR repertoires. There are two shared public clonal TCR motifs between controls and pSS patients (CFLFLSMSACVW and SVGSTAIPP*T); there are two unique TCR motifs in pSS (CVVSDTVLETAGE and LSTD*E).
Earlier studies using gene expression and immunostaining demonstrated that Th1 and IFN-γ were elevated in the LSG, which positively correlated with high infiltration scores and longer disease duration in patients [33–35]. Additionally, SS patients who developed lymphoma exhibited even higher Ifn-γ transcripts compared to both (non-lymphoma) SS and healthy controls [36]. A recent study suggested that IFN-γ was inversely related to patient–reported fatigue [37]. In contrast, animal models of SS did not show temporal upregulation of IFN-γ or Th1 cells, however Ifn-γ ablation impeded the disease progression [14,15]. As presented, controls and pSS patients showed no significant differences of ex vivo Th1 or Tc1 cells in the LSG using single-cell analysis. This result could be attributed to the direct quantification of IFN-γ secretion from a single live T cell, instead of the entire gland or artifacts of immunostaining. Interestingly, significant increases in local Th17 but not Tc17 cells were observed. IL-17A is involved in the proliferation, maturation and recruitment of neutrophils during the initial insult to the glands. Furthermore, IL-17A can function as a B cell helper by inducing a strong proliferative response in B cells and triggering antibody production with class switching and germinal center formation [38,39]. Hypergammaglobulinemia, circulating autoantibodies, and ectopic germinal center (GC) are some of the unique hallmarks during the clinical stage of SS [40–42]. Th2 and follicular helper T (Tfh) cells are positively correlated with increased lymphocytic foci and GCs [43]. Immunostaining of LSG of patients at this stage indicated that the majority of infiltrating cells were CD20+ B cells, with very few CD3+ cells [28]. This study did not examine the effector Th2/Tfh cells, but it could explain the low numbers of other effector T cells in the LSG. Other contributing factors to the low frequencies of Th1, Th17, Tc1, and Tc17 cells are possibly related to the concept that only activated ex-vivo T cells that were actively secreting IFN-γ and IL-17A were examined, and not inactive or transitional T cells. At this particular stage of the disease, their roles are dispensable to maintain the manifestations. One potential question which has not well understood or investigated in the LSG is differentiation of recirculating memory T cells present in the vascular system of the LSG and resident memory T cells in the parenchyma in human patients. A recent study by our group used animal models to suggest that the majority of the effector T cells are resident or gland-infiltrating T cells and not from circulation [44]. Nevertheless, in this study, the influx of activated Th17 cells marks a unique phenotype of SS compared to other autoimmune diseases.
The TCVα repertoire of infiltrating T cells is restricted with limited heterogeneity, specifically Vα17.1, Vα2, and Vα11.1 were found predominantly in the LSG of pSS [45]. The most commonly conserved motifs of the CDR3 region contained GGPKT and VD*G motifs, or ST*TLRNEQ, in the case of TRBV2 and TRBV13. There was, however, little evidence of TCR gene distal skewing or any distinct grouping of V/J genes in our study. A recent study [46] examined the TCR repertoires of individual memory T cells in the LSGs of pSS showed a number of similar TCRs identified in our study, specifically TRAV8–2, 12–3, 12–2, 16, and TRBV30, 20–1, 19, 7–6, 14, 20–1, 3–1, and 24–1. Interestingly, both studies showed the abundance and expansion of TRBV3–1 and TRBV20–1. There are also differences in the TCR sequences identified in both studies. These variations could be attributed by the cell populations examined, i.e. general memory T cells versus subsets of ex-vivo T cells and comparing TCRs between LSGs of pSS and peripheral blood of healthy controls versus comparing TCRs of LSGs between pSS and non-pSS cohorts. Our study has found the two most prevalent CDR3 clonotypes observed (CFLFLSMSACVW and SVGSTAIPP*T) were present in Th17 and Th1 cells in both pSS and SC patients. This implies that both Th1 and Th17 cells may have a common selective pressure for a common autoantigen in the glands, while only pSS selected for the third motif (CVVSDTVLETAGE) present, outlining a unique autoantigen. This unique autoantigen could facilitate the immune cascade which results in SS. More importantly, the unique motif was only expressed by Th1 cells. Ample evidence indicates that early exposure to viruses contribute significantly to the pathogenesis of the disease [47–50]. This study identified a unique CDR3 motif expressed by Th1 cells of pSS patients that produce IFN-γ, a potent anti-viral cytokine. It is likely that there was an adaptive rearrangement or co-evolution of CDR3 genes by Th1 cells in response to viral exposure. The relationship between TCR diversity and SS remains complex. Further work is needed to explore this interesting evolutionary process.
The interaction of CDR3 with the SS-specific autoantigens has yet to be elucidated. However, hydrophobic AAs in the N regions play a part in binding autoantigens [51,52]; hypothetically by promotion of additional interaction with the MHC-peptide complex resulting in a stronger binding [53]. In this manner the findings are consistent, in that hydrophobic or partially hydrophobic AAs could recognize or interact intimately with SS-specific autoantigens. Interestingly, two of the clonotypes identified (CFLFLSMSACVW and SVGSTAIPP*T) present a pattern of serine or threonine (uncharged polar AAs) and then two or more prolines flanked by hydrophobic AAs. The second of these clones has a frame shift leading to a premature stop codon, but still maintains a conserved pattern. The appearance of proline in the CDR3 region is not thoroughly documented. Ciudad et al. have shown that prolines were associated with cleavage sights during autoantigen processing and loading of HLA-DR complexes [54]. Furthermore, Kurien et al. have determined that prolidase (an enzyme that is responsible for the proper processing of collagen proline dipeptide) deficiency is associated with elevated anti-nuclear antibodies, anti-Ro, and anti-dsDNA in systemic lupus erythematosus [55]. Perhaps improper processing of proline dipeptides in SS patients prevents the negative selection of TCRs with the dipeptide allowing autoreactive T cells to persist and activate upon interaction with antigen presenting cells.
5. Conclusions
In summary, pSS patients develop a greater frequency of activated glandular Th17 cells and have a limited repertoire diversity of TCRα/β in activated Th1 cells. Three CDR3 motifs were identified of which one is unique to pSS. The limited and clonal expanded repertoires suggest a restricted antigenic exposure. Further work is need to determine the evolution and function of these TCR-specific glandular Th1 and Th17 cells.
Supplementary Material
Acknowledgements
Conceived and designed the experiments: CQN., SS, CMS, IB., and AV.
Funding
This study was supported financially in part by PHS grants DE023433, DE018958, AI122182 (CQN) from the National Institutes of Health (NIH).
Abbreviations:
- SS
Sjögren’s Syndrome
- Th
T helper
- Tc
T cytotoxic
- TCR
T cell receptor
- CDR3
complementarity determining region 3
Footnotes
Competing financial interests
The authors have no competing financial interests regarding the subjects in this study.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2018.04.009.
References
- [1].Fox RI, Sjögren’s syndrome, Lancet 366 (2005) 321–331. [DOI] [PubMed] [Google Scholar]
- [2].Chisholm DM, Mason DK, Labial salivary gland biopsy in Sjögren’s disease, J. Clin. Pathol. 21 (1968) 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Manganelli P, Fietta P, Apoptosis and Sjögren syndrome, Semin. Arthritis Rheum. 33 (2003) 49–65. [DOI] [PubMed] [Google Scholar]
- [4].Mavragani CP, Moutsopoulos HM, Sjögren syndrome, CMAJ 186 (2014) E579–E586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Daniels TE, Cox D, Shiboski CH, Schiodt M, Wu A, Lanfranchi H, Umehara H, Zhao Y, Challacombe S, Lam MY, De Souza Y, Schiodt J, Holm H, Bisio PA, Gandolfo MS, Sawaki T, Li M, Zhang W, Varghese-Jacob B, Ibsen P, Keszler A, Kurose N, Nojima T, Odell E, Criswell LA, Jordan R, Greenspan JS, Associations between salivary gland histopathologic diagnoses and phenotypic features of Sjogren’s syndrome among 1,726 registry participants, Arthritis Rheum. 63 (2011) 2021–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Costa S, Schutz S, Cornec D, Uguen A, Quintin-Roué I, Lesourd A, Berthelot JM, Hachulla E, Hatron P-Y, Goeb V, Vittecoq O, Pers JO,Marcorelles P, Saraux A, Devauchelle-Pensec V, B-cell and T-cell quantification in minor salivary glands in primary Sjögren’s syndrome: development and validation of a pixel-based digital procedure, Arthritis Res. & Ther. 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Corsiero E, Sutcliffe N, Pitzalis C, Bombardieri M, Accumulation of self-reactive Naïve and memory B cell reveals sequential defects in B cell tolerance checkpoints in Sjögren’s syndrome, PLoS One 9 (2014) e114575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kapsogeorgou EK, Christodoulou MI, Panagiotakos DB, Paikos S, Tassidou A, Tzioufas AG, Moutsopoulos HM, Minor salivary gland inflammatory lesions in Sjögren syndrome: do they evolve? J. Rheumatol. 40 (2013) 1566–1571. [DOI] [PubMed] [Google Scholar]
- [9].Manoussakis MN, Boiu S, Korkolopoulou P, Kapsogeorgou EK, Kavantzas N, Ziakas P, Patsouris E, Moutsopoulos HM, Rates of infiltration by macrophages and dendritic cells and expression of interleukin-18 and interleukin-12 in the chronic inflammatory lesions of Sjögren’s syndrome: correlation with certain features of immune hyperactivity and factors associated with high risk of lymphoma development, Arthritis Rheum. 56 (2007) 3977–3988. [DOI] [PubMed] [Google Scholar]
- [10].Kyriakidis NC, Kapsogeorgou EK, Tzioufas AG, A comprehensive review of autoantibodies in primary Sjögren’s syndrome: clinical phenotypes and regulatory mechanisms, J. Autoimmun. 51 (2014) 67–74. [DOI] [PubMed] [Google Scholar]
- [11].Nguyen CQ, Hu MH, Li Y, Stewart C, Peck AB, Salivary gland tissue expression of Interleukin-23 and Interleukin-17 in Sjögren’s syndrome, Arthritis Rheum. 58 (2008) 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nardi N, Brito-Zerón P, Ramos-Casals M, Aguiló S, Cervera R, Ingelmo M, Font, Circulating auto-antibodies against nuclear and non-nuclear antigens in primary Sjögren’s syndrome, Clin. Rheumatol. 25 (2005) 341–346. [DOI] [PubMed] [Google Scholar]
- [13].Nguyen C, Peck AB, The interferon-signature of Sjögren’s syndrome: how unique biomarkers can identify underlying inflammatory and immunopathological mechanisms of specific diseases, Immunother. and Vacc. 4 (2013) 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Voigt A, Esfandiary L, Nguyen CQ, Sexual dimorphism in an animal model of Sjogren’s syndrome: a potential role for Th17 cells, Biol. Open 4 (2015) 1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Voigt A, Esfandiary L, Wanchoo A, Glenton P, Donate A, Craft WF, Craft SL, Nguyen CQ, Sexual dimorphic function of IL-17 in salivary gland dysfunction of the C57BL/6.NOD-Aec1Aec2 model of Sjogren’s syndrome, Sci. Rep. 6 (2016) 38717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zumla A, Mathur M, Stewart J, Wilkinson L, Isenberg D, T cell receptor expression in Sjögren’s syndrome, Ann. Rheum. Dis. 50 (1991) 691–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Elhanati Y, Murugan A, Callan CG Jr, Mora T, Walczak AM, Quantifying selection in immune receptor repertoires, Proc. Natl. Acad. Sci. U.S.A 111 (2014) 9875–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Eltahla AA, Rizzetto S, Pirozyan MR, Betz-Stablein BD, Venturi V, Kedzierska K, Lloyd AR, Bull RA, Luciani F, Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells, Immunol. Cell Biol 94 (2016) 604–611. [DOI] [PubMed] [Google Scholar]
- [19].Kern J, Drutel R, Leanhart S, Bogacz M, Pacholczyk R, Reduction of T cell receptor diversity in NOD mice prevents development of type 1 diabetes but not Sjögren’s syndrome, PLoS One 9 (2014) e112467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Murray JS, An old twist in HLA-A: CDR3α hook up at an R65-joint, Front. Immunol. 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton, Wilson IA, An αβ T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex, Science 274 (1996) 209–219. [DOI] [PubMed] [Google Scholar]
- [22].Esfandiary L, Gupta N, Voigt A, Wanchoo A, Chan EK, Sukumaran S, Nguyen CQ, Single-cell antibody nanowells: a novel technology in detecting anti-SSA/Ro60- and anti-SSB/La autoantibody-producing cells in peripheral blood of rheumatic disease patients, Arthritis Res. & Ther. 18 (2016) 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, Rasmussen A, Scofield H, Vitali C, Bowman SJ, Mariette X, International G. Sjogren’s Syndrome Criteria Working, American College of Rheumatology/European league against rheumatism classification criteria for primary Sjogren’s syndrome: a consensus and data-driven methodology involving three international patient cohorts, Ann. Rheum. Dis. 76 (2017) (2016) 9–16. [DOI] [PubMed] [Google Scholar]
- [24].Nguyen CQ, Ogunniyi AO, Karabiyik A, Love JC, Single-cell analysis reveals isotype-specific autoreactive B cell repertoires in Sjögren’s syndrome, PLoS One 8 (2013) e58127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yousef S, Planas R, Chakroun K, Hoffmeister-Ullerich S, Binder TM, Eiermann TH, Martin R, Sospedra M, TCR bias and HLA cross-restriction are strategies of human brain-infiltrating JC virus-specific CD4+ T cells during viral infection, J. Immunol. 189 (2012) 3618–3630. [DOI] [PubMed] [Google Scholar]
- [26].Shannon CE, The mathematical theory of communication. 1963, MD Comput 14 (1997) 306–317. [PubMed] [Google Scholar]
- [27].Simpson EH, Measurement of diversity, Nature 163 (1949) 1. [Google Scholar]
- [28].Nguyen CQ, Hu MH, Li Y, Stewart C, Peck AB, Salivary gland tissue expression of interleukin-23 and interleukin-17 in Sjogren’s syndrome: findings in humans and mice, Arthritis Rheum. 58 (2008) 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sakai A, Sugawara Y, Kuroishi T, Sasano T, Sugawara S, Identification of IL-18 and Th17 cells in salivary glands of patients with Sjogren’s syndrome, and amplification of IL-17-mediated secretion of inflammatory cytokines from salivary gland cells by IL-18, J. Immunol. 181 (2008) 2898–2906. [DOI] [PubMed] [Google Scholar]
- [30].Katsifis GE, Rekka S, Moutsopoulos NM, Pillemer S, Wahl SM, Systemic and local interleukin-17 and linked cytokines associated with Sjogren’s syndrome immunopathogenesis, Am. J. Pathol. 175 (2009) 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hall JC, Casciola-Rosen L, Berger AE, Kapsogeorgou EK, Cheadle C, Tzioufas AG, Baer AN, Rosen A, Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases, Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 17609–17614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, Jacques S, Ba N, Ittah M, Lepajolec C, Labetoulle M, Ardizzone M, Sibilia J, Fournier G. Chiocchia, Mariette X, Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren’s syndrome, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mitsias DI, Tzioufas AG, Veiopoulou C, Zintzaras E, Tassios IK, Kogopoulou O, Moutsopoulos HM, Thyphronitis G, The Th1/Th2 cytokine balance changes with the progress of the immunopathological lesion of Sjogren’s syndrome, Clin. Exp. Immunol. 128 (2002) 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].van Woerkom JM, Kruize AA, Wenting-van Wijk MJ, Knol E, Bihari IC, Jacobs JW, Bijlsma JW, Lafeber FP, van Roon JA, Salivary gland and peripheral blood T helper 1 and 2 cell activity in Sjogren’s syndrome compared with non-Sjogren’s sicca syndrome, Ann. Rheum. Dis. 64 (2005) 1474–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kolkowski EC, Reth P, Pelusa F, Bosch J, Pujol-Borrell R, Coll J, Jaraquemada, Th1 predominance and perforin expression in minor salivary glands from patients with primary Sjogren’s syndrome, J. Autoimmun. 13 (1999) 155–162. [DOI] [PubMed] [Google Scholar]
- [36].Nezos A, Gravani F, Tassidou A, Kapsogeorgou EK, Voulgarelis M, Koutsilieris M, Crow MK, Mavragani CP, Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis, J. Autoimmun. 63 (2015) 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Howard Tripp N, Tarn J, Natasari A, Gillespie C, Mitchell S, Hackett KL, Bowman SJ, Price E, Pease CT, Emery P, Lanyon P, Hunter J, Gupta M, Bombardieri M, Sutcliffe N, Pitzalis C, McLaren J, Cooper A, Regan M, Giles I, Isenberg DA, Saravanan V, Coady D, Dasgupta B, McHugh N, Young-Min S, Moots R, Gendi N, Akil M, Griffiths B, Lendrem DW, Ng WF, Fatigue in primary Sjogren’s syndrome is associated with lower levels of proinflammatory cytokines, RMD Open 2 (2016) e000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mitsdoerffer M, Lee Y, Jager A, Kim HJ, Korn T, Kolls JK, Cantor H, Bettelli E, Kuchroo VK, Proinflammatory T helper type 17 cells are effective B-cell helpers, Proc. Natl. Acad. Sci. U. S. A. 107 (2010) 14292–14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pisitkun P, Ha HL, Wang H, Claudio E, Tivy CC, Zhou H, Mayadas TN, Illei GG, Siebenlist U, Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis, Immunity 37 (2012) 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Voulgarelis M, Skopouli FN, Clinical, immunologic, and molecular factors predicting lymphoma development in Sjogren’s syndrome patients, Clin. Rev. Allergy Immunol 32 (2007) 265–274. [DOI] [PubMed] [Google Scholar]
- [41].Fox RI, Kang HI, Pathogenesis of Sjogren’s syndrome, Rheum. Dis. Clin. N. Am. 18 (1992) 517–538. [PubMed] [Google Scholar]
- [42].Gordon TP, Bolstad AI, Rischmueller M, Jonsson R, Waterman SA, Autoantibodies in primary Sjogren’s syndrome: new insights into mechanisms of autoantibody diversification and disease pathogenesis, Autoimmunity 34 (2001) 123–132. [DOI] [PubMed] [Google Scholar]
- [43].Maehara T, Moriyama M, Hayashida JN, Tanaka A, Shinozaki S, Kubo Y, Matsumura K, Nakamura S, Selective localization of T helper subsets in labial salivary glands from primary Sjogren’s syndrome patients, Clin. Exp. Immunol. 169 (2012) 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wanchoo A, Voigt A, Sukumaran S, Stewart CM, Bhattacharya I, Nguyen CQ, Single-cell analysis reveals sexually dimorphic repertoires of interferon-gamma and IL-17A producing T cells in salivary glands of Sjogren’s syndrome mice, Sci. Rep. 7 (2017) 12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sumida T, Kita Y, Yonaha F, Maeda T, Iwamoto I, Yoshida S, T cell receptor V alpha repertoire of infiltrating T cells in labial salivary glands from patients with Sjogren’s syndrome, J. Rheumatol. 21 (1994) 1655–1661. [PubMed] [Google Scholar]
- [46].Joachims ML, Leehan KM, Lawrence C, Pelikan RC, Moore JS, Pan Z, Rasmussen A, Radfar L, Lewis DM, Grundahl KM, Kelly JA, Wiley GB, Shugay M, Chudakov DM, Lessard CJ, Stone DU, Scofield RH, Montgomery CG, Sivils KL, Thompson LF, Farris AD, Single-cell analysis of glandular T cell receptors in Sjogren’s syndrome, JCI insight 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, Liang Zhou J, Kirou KA, Seshan SV, Moutsopoulos HM, Crow MK, Expression of long interspersed nuclear element 1 Retroelements and induction of type I interferon in patients with systemic autoimmune disease, Arthritis Rheum. 68 (2016) 2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Weller ML, Gardener MR, Bogus ZC, Smith MA, Astorri E, Michael DG, Michael DA, Zheng C, Burbelo PD, Lai Z, Wilson PA, Swaim W, Handelman B, Afione SA, Bombardieri M, Chiorini JA, Hepatitis Delta virus detected in salivary glands of Sjogren’s syndrome patients and recapitulates a Sjogren’s syndrome-like phenotype in vivo, Pathog Immun 1 (2016) 12–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nakamura H, Takahashi Y, Yamamoto-Fukuda T, Horai Y, Nakashima Y, Arima K, Nakamura T, Koji T, Kawakami A, Direct infection of primary salivary gland epithelial cells by human T lymphotropic virus type I in patients with Sjogren’s syndrome, Arthritis Rheum. 67 (2015) 1096–1106. [DOI] [PubMed] [Google Scholar]
- [50].Voigt A, Nguyen CQ, Human T-Lymphotrophic virus type-I: a unique association with myelopathy in Sjogren’s syndrome, Clin. Microbiol 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stadinski BD, Shekhar K, Gómez-Touriño I, Jung J, Sasaki K, Sewell AK, Peakman M, Chakraborty AK, Huseby ES, Hydrophobic CDR3 residues promote the development of self-reactive T cells, Nat. Immunol. 17 (2016) 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, Greenberg PD, Klavinskis LS, Blattman JN, Anderson KS, TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes, Proc. Natl. Acad. Sci. U. S. A. 112 (2015) E1754–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, Greenberg PD, Klavinskis LS, Blattman JN, Anderson KS, TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes, Proc. Natl. Acad. Sci. U. S. A. 112 (2015) E1754–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ciudad MT, Sorvillo N, van Alphen FP, Catalan D, Meijer AB, Voorberg J, Jaraquemada D, Analysis of the HLA-DR peptidome from human dendritic cells reveals high affinity repertoires and nonconventional pathways of peptide generation, J. Leukoc. Biol. 101 (2017) 15–27. [DOI] [PubMed] [Google Scholar]
- [55].Kurien BT, D’Sousa A, Bruner BF, Gross T, James JA, Targoff IN, Maier-Moore JS, Harley IT, Wang H, Scofield RH, Prolidase deficiency breaks tolerance to lupus-associated antigens, Int. J. Rheum. Dis 16 (2013) 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.